Abstract
Background
The intracellular concentration of heavy-metal cations, such as copper, nickel, and zinc is pivotal for the mycobacterial response to the hostile environment inside macrophages. To date, copper transport mediated by P-type ATPases across the mycobacterial plasma membrane has not been sufficiently explored.
Results
In this work, the ATPase activity of the putative Mycobacterium tuberculosis P1B-type ATPase CtpB was associated with copper (I) transport from mycobacterial cells. Although CtpB heterologously expressed in M. smegmatis induced tolerance to toxic concentrations of Cu2+ and a metal preference for Cu+, the disruption of ctpB in M. tuberculosis cells did not promote impaired cell growth or heavy-metal accumulation in whole mutant cells in cultures under high doses of copper. In addition, the Cu+ ATPase activity of CtpB embedded in the plasma membrane showed features of high affinity/slow turnover ATPases, with enzymatic parameters KM 0.19 ± 0.04 µM and Vmax 2.29 ± 0.10 nmol/mg min. In contrast, the ctpB gene transcription was activated in cells under culture conditions that mimicked the hostile intraphagosomal environment, such as hypoxia, nitrosative and oxidative stress, but not under high doses of copper.
Conclusions
The overall results suggest that M. tuberculosis CtpB is associated with Cu+ transport from mycobacterial cells possibly playing a role different from copper detoxification.
Keywords: Mycobacterium tuberculosis, P-type ATPase, CtpB, Plasma membrane, Copper transport
Background
Tuberculosis (TB) is an infectious disease caused by the acid-fast bacillus Mycobacterium tuberculosis, which in turn is the origin of one of the biggest public health problems worldwide [1]. According to the World Health Organization (WHO), there were 10.4 million new cases and 1.3 million deaths by TB in 2017 [1]. Currently, vaccination with the Bacillus Calmette-Guerin (BCG) and chemotherapy are the common strategies for TB control. The tuberculous infection begins when mycobacteria is phagocytosed by macrophages and dendritic cells in the pulmonary alveoli [2]. M. tuberculosis persists inside macrophages, facing adverse conditions such as an acidic pH, reactive oxygen species (ROS) and nitrogen species (RNS), antimicrobial peptides, acid hydrolases, low micronutrients content, and non-physiological concentrations of heavy-metal ions [3, 4].
The concentrations of Cu2+, Fe2+, and Zn2+ inside phagosomes infected with M. tuberculosis increase between 1 and 24 h post-infection [5], until antibacterial levels of Cu+ and Zn2+ are reached [4]. In particular, high Cu+ concentrations generate a ROS response via oxidative stress, protein denaturation (by metal interaction with thiol groups), inactivation of enzymes by substitution of other metal cofactors, and membrane destabilization [6–8]. The intracellular concentration of heavy-metal cations is pivotal for the mycobacterial response to the oxidative burst within macrophages [6, 7]. Indeed, M. tuberculosis has two superoxide dismutases, SodA and SodC, that use Mn2+/Fe2+ and Cu2+ as cofactors, respectively [9, 10]. Other enzymes involved in the mycobacterial response to oxidative stress, such as catalase-peroxidase (KatG), alkyl hydroperoxide reductase (AhpC), and thioredoxins (TrxA and TrxB) use Fe2+ as a cofactor [11]. Therefore, the ion homeostasis of heavy-metal cations is essential for mycobacterial viability. The normal levels of reactive oxygen species (ROS) and nitrogen species (RNS) in macrophages are able to produce DNA and lipid damage, disrupting the activity of proteins containing Fe-S clusters, heavy-metals cations, hemes, thiols, sulfhydryl, or tyrosyl groups in phagocytosed pathogens [12, 13]. The damage mediated by ROS and RNS leads to a general inhibition of the intracellular processes associated with proton-dependent active transport, oxygen utilization, and oxidative phosphorylation [14]. In addition, bacterial pathogens to face the intraphagosomal environment produce enzymes that interfere with the synthesis of reactive species, allow the direct degradation of intermediary products, or participate in the DNA repair [13].
P-type ATPases may play an important role in mycobacterial ion homeostasis, since these usually transport cations against the concentration gradient [15]. M. tuberculosis has many P-type ATPases annotated as possible transporters of heavy-metal cations, suggesting that these proteins could play a relevant role in M. tuberculosis survival [16]. Some mycobacterial P1B-type ATPases have been associated with bacterial detoxification of heavy metals cations, such as Cu+, Zn2+, Co2+, and Ni2+. Specifically, CtpG, CtpV, CtpC, and CtpA are activated in human macrophages infected with M. tuberculosis [17]. In addition, some of these are also involved in M. tuberculosis virulence in animal models [18]. Indeed, CtpC and CtpV respond to an increase in the intraphagosomal concentrations of Cu+ and Zn2+ during the infection process [19].
M. tuberculosis possesses three putative copper transporting P-type ATPases: CtpA, CtpB, and CtpV [16]. The functional differences among these are not well understood. The M. tuberculosis ctpA gene is activated during the tuberculous infection of human cells [20]. On the other hand, the ctpV gene belongs to the cso “copper-sensitive operon”, which includes csoR, rv0968, ctpV, and rv0970 [21] under the control of the Cu+-responding transcriptional regulator CsoR [22]. Interestingly, csoR is part of the regulon of PhoP, one global regulator of M. tuberculosis virulence [23]. Therefore, it has been suggested that P1B-type ATPases are key for controlling the intracellular content of heavy metals and delivering cofactors to extracytoplasmic enzymes [24, 25].
Interestingly, the M. tuberculosis ctpB gene is overexpressed under in vitro stress conditions, such as hypoxia that mimics the hostile intraphagosomal environment [26], suggesting that CtpB could be involved in tuberculous infection progress. However, the enzymatic features of CtpB and its possible role in heavy-metal cations transport across the mycobacterial plasma membrane are unknown. In this work, different functional assays were performed to evaluate the ion specificity of CtpB and its transcriptional response against stress conditions. Our findings suggest that M. tuberculosis CtpB is associated with Cu+ pumping from mycobacterial cells possibly in response to stress conditions. Furthermore, an alternative role of CtpB different from copper detoxification is discussed.
Results
CtpB has typical P-type ATPase copper transporting domains
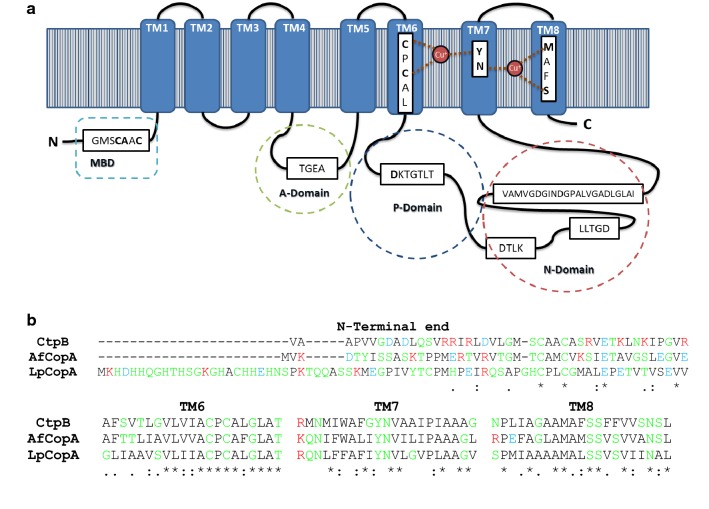
The ctpB gene encodes for a putative protein of 752 amino acids (77.5 kDa) [27]. In silico predictions show that CtpB belongs to the P-type ATPase family and is annotated as an acid anhydride hydrolase (EC 3.6.3.-) that catalyzes the transmembrane movement of substances. The amino acid sequence of CtpB contains eight TM segments arranged in a Type I topology [28], common to P1B-type ATPases (Fig. 1). The three well-known cytoplasmic domains (A: actuator, P: phosphorylation, and N: nucleotide binding) are observed in loops within TM4-5 and TM6-7 segments (Fig. 1).
Fig. 1.
Topology, functional motifs, and domains of M. tuberculosis CtpB. a The predicted topology of CtpB shows eight TM helices (1 to 8), locating the N- and C-terminal ends within the cytoplasmic portion. The cytoplasmic domains are represented by stitched lines and the functional motifs of P1B-type ATPase are shown. The amino acids responsible for cation coordination within TM segments 6, 7 and 8 are highlighted in bold. b The TM helices were predicted using TOPCONS. The multiple alignment of the N-terminal end, TM6, TM7, and TM8 of CtpB and other bacterial characterized Cu+-ATPases (CopA from Archaeoglobus fulgidus (WP_010877980.1) and Legionella pneumophila (YP_095057.1)) was constructed using Clustal Omega. Some of the amino acids involved in the coordination of Cu+ are located within TM6, 7 and 8. The residues are classified as nonpolar (black), polar (green), acid (blue), and basic (red)
Analysis of functional motifs obtained by the InterProScan server [29] also showed that CtpB is a putative copper/heavy-metal P-type ATPase transporter. The characteristic functional motifs reported by Thever and Saier [28] were identified in CtpB, with subtle differences. Specifically, CtpB showed the following functional motifs: TGEA associated with phosphatase activity; CPCAL located in the TM6 segment, providing two Cys necessary to coordinate the cation [30]; DKTGTLT that contains the phosphorylation site and is located in the largest cytoplasmic loop between TM4 and TM5 segments; DTLK and LLTGD motifs that catalyze the transfer of the phosphate group; and finally the motif known as hinge (VAMVGDGINDGPALVGADLGLAI) that provides the necessary flexibility for the conformational changes during the catalytic cycle of the enzyme [16].
Usually, the amino acids responsible for recognizing the transported cation form a binding pocket within TM6, 7 and 8, and display the adequate geometry, size, and charge for cation binding in P1B-type ATPases [15]. An alignment between TM6-8 segments of CtpB and the N-terminal end of the well-known Cu+-ATPases CopA of A. fulgidus and L. pneumophila showed that all the aligned sequences displayed the three characteristic motifs of Cu+-ATPases (CPC, YN, and MXXS) [31], containing six amino acids responsible for the coordination of Cu+ in CtpB within TM6, 7, and 8, respectively (Fig. 1b).
Some P1B-type ATPases have cytoplasmic metal binding domains (MBD) at the N-terminal end [32]. In this case, CtpB also showed the MBD characteristic motif GMSC(S/A)AC of P-type ATPases, where Cys could be responsible for Cu+ coordination. On the other hand, CtpB displayed all the residues responsible for metal coordination present in the Cu+-ATPase of L. pneumophila LpCopA: Cys382, Cys384, and Tyr688 (binding site I), together with Asn689, Met717, and Ser721 (binding site II) [33] (Fig. 1b). Therefore, since Cu+ coordination is normally associated with only three coordination bonds in heavy-metal P-type ATPases [30], the presence of six residues possibly associated with metal coordination suggest that CtpB could be able to coordinate and transport at least two cations during the catalytic cycle.
Mycobacterium smegmatis cells expressing M. tuberculosis CtpB tolerate toxic concentrations of copper
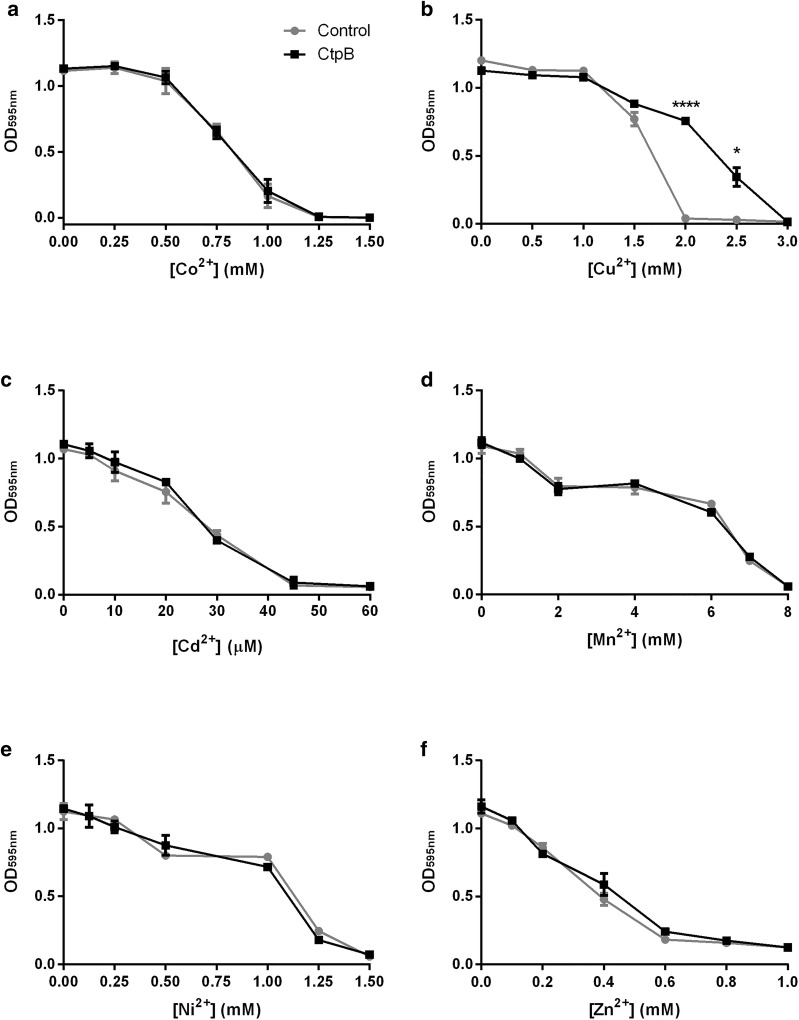
We assessed the tolerance of mycobacterial cells expressing the recombinant protein in cultures supplemented with toxic concentrations of different heavy-metal cations. Since M. smegmatis is an adequate non-pathogenic model for the heterologous expression of M. tuberculosis proteins [34], M. tuberculosis CtpB was cloned and expressed in the plasma membrane of M. smegmatis mc2155. Nevertheless, the overexpressed M. tuberculosis CtpB protein in mycobacterial cells could not be detected from crude protein extracts by SDS-PAGE. Only low amounts of the CtpB protein (approximately 78 kDa) were detected in the cell membrane fraction of induced recombinant cells (M. smegmatis transformed with pALT10) compared to control cells transformed with the expression vector pMV261 (Additional file 1). As shown in Fig. 2, M. smegmatis cells expressing CtpB did not tolerate high doses of Co2+, Cd2+, Mn2+, Ni2+, and Zn2+. In contrast, recombinant mycobacterial cells tolerated high concentrations of Cu2+ ranging from 2.0 to 2.5 mM, which are usually toxic in mycobacteria. Specifically, the recombinant cells tolerated approximately twofold more Cu2+ concentration than control cells (M. smegmatis transformed with pMV261), suggesting that CtpB may be involved in copper pumping from mycobacterial cells.
Fig. 2.
Tolerance of M. smegmatis cells expressing CtpB in presence of high doses of heavy-metal cations. Cultures in LB supplemented with increasing concentrations of cations: a Co2+, b Cd2+, c Cu2+, d Mn2+, e Ni2+, and f Zn2+ were incubated during 72 h at 37 °C and the OD595 was measured. The absorbance measured for mycobacterial cells expressing CtpB (transformed with pALT10) and control cells (transformed with pMV261) is shown in gray and black lines, respectively. The data correspond to the mean OD595 ± SEM from three independent experiments. Significant differences correspond to values of *P ≤ 0.05 and **** P ≤ 0.0001
CtpB shows high affinity/low turnover for Cu+ in the mycobacterial plasma membrane
The ATPase activity stimulated by different heavy-metal cations was measured in the plasma membrane of M. smegmatis mc2155 cells expressing M. tuberculosis CtpB. Since there is a contribution to the ATPase activity from other cell-membrane ATPases, parallel enzymatic reactions using membranes from M. smegmatis mc2155 transformed with the empty vector pMV261 were used as control. Therefore, the enzymatic activity mediated by CtpB was calculated by subtracting the ATPase activity of the control membranes from the ATPase activity of the recombinant membranes [35–37]. Using this experimental approach, it was observed that Cu+ was the only cation that significantly stimulated the ATPase activity of CtpB embedded in the mycobacterial plasma membrane, compared with other divalent heavy metal cations (Fig. 3). No statistically significant differences were found when the enzymatic reactions were supplemented with DTT and without Cu+ compared to the same enzymatic reaction without DTT. Specifically, the Cu+ ATPase activity obtained for CtpB recombinant membranes was 12.31 ± 0.14 nmol/mg-min.
Fig. 3.
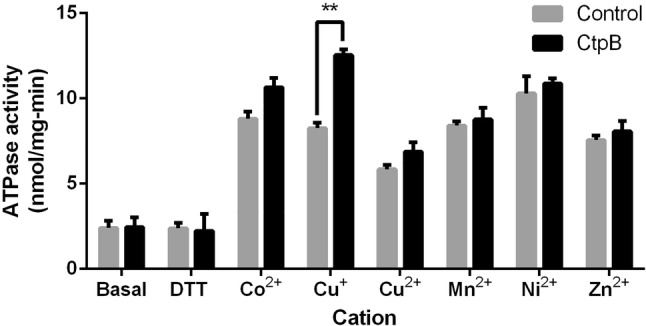
ATPase activity of CtpB in the presence of heavy metal cations. The ATPase activity was measured using plasma membranes isolated from M. smegmatis mc2155 expressing CtpB (transformed with pALT10, black bars) and control cells (M. smegmatis transformed with pMV261, gray bars). The values of ATPase activity were measured separately by supplementing the enzymatic reactions with Co2+, Cu+, Cu2+, Mn2+, Ni2+, and Zn2+ (at 10 μM final concentration). The data correspond to the mean ATPases activity ± SEM derived from measurements of technical triplicates. Significant differences are values of **P ≤ 0.01
On the other hand, Fig. 4 shows that 37 °C is the optimal temperature among the tested temperatures for the enzymatic reaction of CtpB. This optimal temperature is similar to the average temperature of the human body, which in turn is the natural host of M. tuberculosis. Similarly, the observed optimal pH (7.4) is close to neutrality, as well as the observed for other mycobacterial P-type ATPases [35–37]. Finally, CtpB displayed a concentration-dependent Cu+ ATPase activity when the enzymatic reaction was supplemented with Cys at concentrations below 0.5 mM. In conclusion, the optimal conditions for further estimation of CtpB kinetic parameters are 0.5 mM cysteine, pH 7.4, and 37 °C.
Fig. 4.
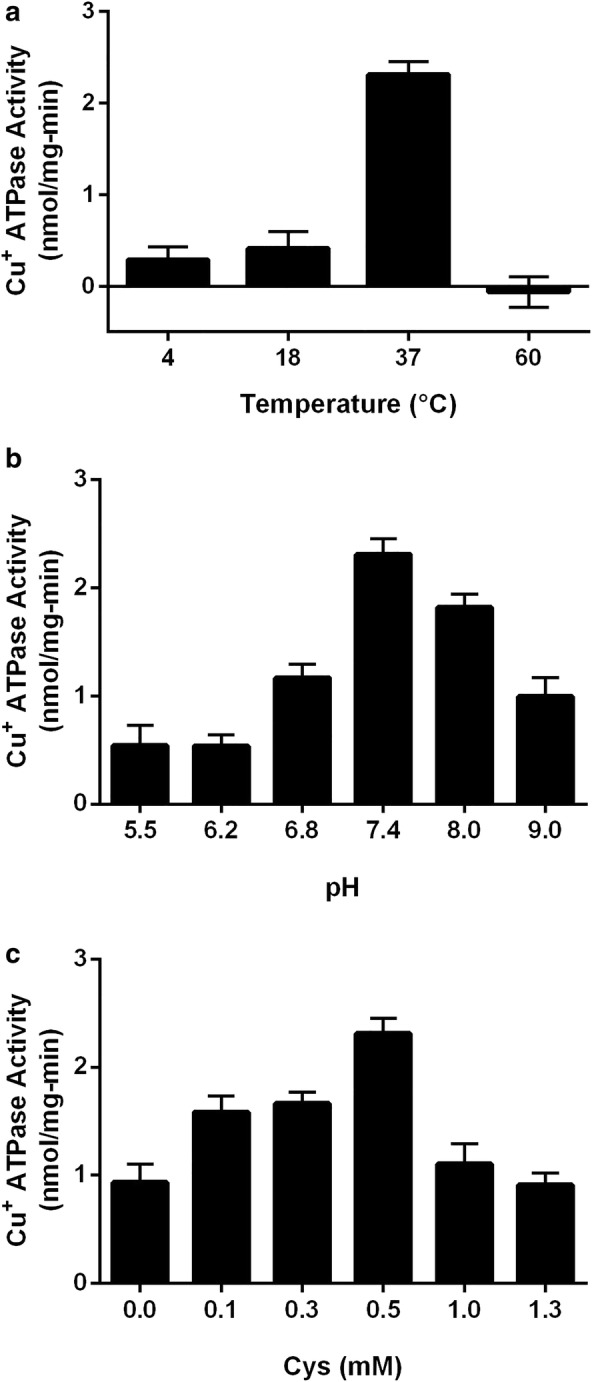
Optimal conditions for Cu+ ATPase activity mediated by CtpB. The Cu+ ATPase activity was measured using plasma membranes isolated from M. smegmatis mc2155 expressing CtpB and membranes isolated from M. smegmatis transformed with the expression vector pMV261. The enzymatic parameters evaluated were: a temperature, b pH, and c cysteine concentration. The data correspond to the mean ATPases activity ± SEM derived from measurements of technical triplicate
The optimal conditions observed for the ATPase activity were used to estimate the kinetic parameters of CtpB. When the concentrations of Mg2+ and ATP are constant, CtpB catalyzes ATP hydrolysis in a Cu+ concentration-dependent manner, following a Michaelis–Menten kinetics (Fig. 5 and Additional file 2). CtpB embedded in the plasma membrane reached the maximal activity in vitro when reactions were supplemented with 5 μM Cu+. In addition, CtpB displayed a Vmax of 2.29 ± 0.10 nmol/mg-min and KM of 0.19 ± 0.04 µM Cu+.
Fig. 5.
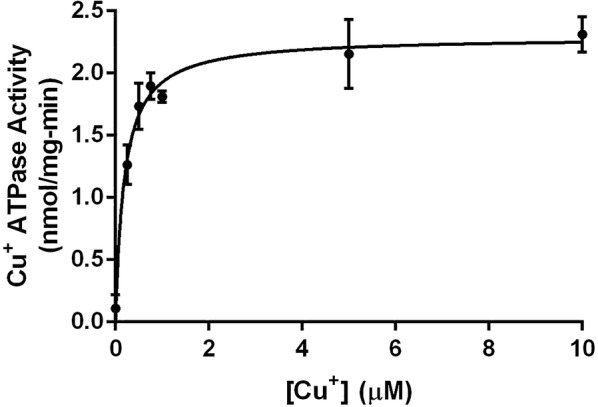
Kinetic parameters of CtpB. The Cu+ ATPase activity of CtpB follows a Michaelis–Menten kinetics. Cu+ was added at different concentrations (0.25 to 10.0 μM) and the released phosphate from the hydrolysis of ATP was quantified in the enzymatic reactions. The kinetic parameters for CtpB embedded in the plasma membrane (KM 0.19 ± 0.04 μM and Vmax 2.29 ± 0.10 nmol/mg min) were calculated using the Prism v6.0 for Windows GraphPad Software, La Jolla California USA, http://www.graphpad.com. The plotted data correspond to the average activity ± SEM from three independent experiments
ctpB disruption does not alter the growth rate and copper accumulation of M. tuberculosis cells
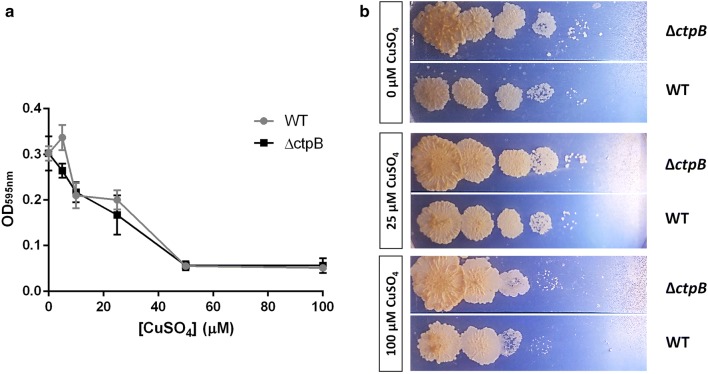
ctpB disruption obtained by recombineering in M. tuberculosis cells (ΔctpB) was corroborated by PCR (Additional file 3). Initially, we conducted assays of mycobacterial tolerance to toxic concentrations of divalent metals to evaluate if CtpB is involved in copper detoxification of mycobacterial cells, which would result in the susceptibility of M. tuberculosis ∆ctpB cells to toxic doses of copper. Notably, the growth rate of ∆ctpB cells cultured in broth or solid medium under high doses of Cu2+ did not show statistically significant differences compared with wild type cells cultured under the same experimental conditions (Fig. 6). The possible role of CtpB in copper detoxification was also evaluated by comparing copper accumulation in wild-type and mutant cells. Similarly, the experiments of atomic absorption showed that there were no significant differences in copper accumulation between M. tuberculosis wild-type and ∆ctpB cells cultured under high doses of copper (Fig. 7). Therefore, if CtpB were responsible for copper detoxification, gene disruption should reduce cation efflux and copper should accumulate inside the ∆ctpB cells. Conversely, both strains showed similar copper accumulation, corroborating that CtpB is not directly associated with copper detoxification in mycobacterial cells. Therefore, the assays of mycobacterial tolerance to high doses of cations and the metal accumulation assays strongly suggest that CtpB is not directly associated with copper detoxification, whereas CtpB could play an alternative role in the copper transport system mediated by P-type ATPases across the mycobacterial plasma membrane.
Fig. 6.
Effect of Cu2+ in the growth rate of ctpB-disrupted M. tuberculosis cells. a The growth rate of M. tuberculosis ΔctpB (black) and M. tuberculosis H37Ra wild-type (gray) cells in modified Sauton medium supplemented with high doses of Cu2+ was measured at OD595. The data are shown as the mean ± SEM from three independent experiments. b Serial dilutions (tenfold) of M. tuberculosis cells (wild type and ΔctpB cells) previously grown until mid-log phase were seeded and grown on 7H11-OADC plates supplemented with 0.25 and 100 μM CuSO4
Fig. 7.
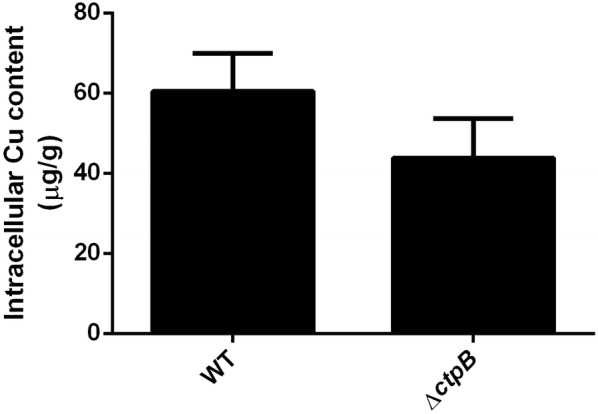
Copper accumulation in whole M. tuberculosis ΔctpB cells. The content of intracellular copper in M. tuberculosis wild-type and ΔctpB cells previously treated with 50 μM of CuSO4 was assessed by AAS-GF. The values of metal accumulation are shown as μg of Cu/g of dry pellet. The data are shown as the mean ± SEM from three independent experiments
CtpB is associated with the response to stress conditions in M. tuberculosis cells
In order to gain further insight into the possible role of CtpB during mycobacterial copper transport, a transcriptional analysis of ctpB was assessed in vitro under experimental conditions mimicking the intraphagosomal environment: the presence of high doses of heavy-metal cations, hypoxia, and redox stress (Fig. 8). We observed that ctpB transcription was activated 4.53 ± 1.12 and 2.94 ± 0.52-fold by redox stressors KCN and sodium nitroprusside, respectively. Similarly, hypoxia strongly induced ctpB transcription (13.02 ± 3.69-fold) compared with cells cultured under complete aeration. Conversely, ctpB transcription was not significantly induced by the presence of toxic doses of heavy-metal cations.
Fig. 8.
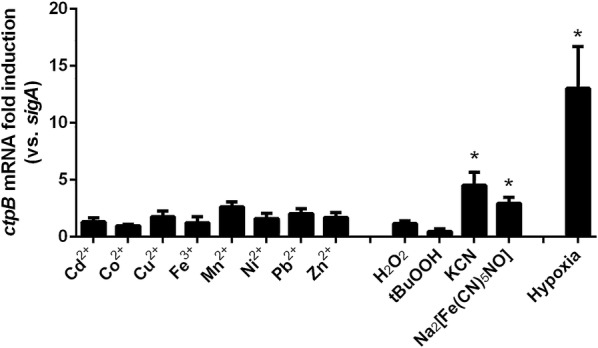
Transcriptional analysis of CtpB under stress conditions. Relative transcription of ctpB from cultures separately supplemented with different concentrations of heavy-metal cations (Cd2+, Co2+, Cu2+, Fe2+, Mn2+, Ni2+, Pb2+, and Zn2+), hypoxia, and 1 mM of different redox stress agents (H2O2, tert butyl hydroperoxide, KCN, and sodium nitroprusside). sigA was used as the normalizer gene. The data were plotted as the average values ± SEM from three independent experiments. The differences (*P < 0.05) were obtained by comparing the values in the presence and absence of the stress conditions
Discussion
Bacterial tolerance against toxic concentrations of metal cations mediated by P-type ATPases has been previously reported in mycobacteria. For example, recombinant M. smegmatis cells overexpressing the putative M. tuberculosis cadmium transporter CtpG tolerate high doses of Cd2+ compared with non-recombinant mycobacterial cells [36]. Similarly, M. smegmatis homologously expressing the putative alkali/alkaline earth cation P-type ATPase transporter Pma1 tolerates toxic concentrations of Na+, K+, and Na+/K+ ions [37]. Regarding P-type ATPases associated with copper transport, we previously reported that mycobacterial cells expressing the putative M. tuberculosis CtpA, a P-type ATPase copper transporter, grow approximately twofold more than non-recombinant cells in the presence of toxic copper concentrations (from 1 to 4 mM), suggesting a possible role of CtpA in copper detoxification in mycobacteria [35]. Therefore, CtpB is likely associated to Cu+ pumping through the mycobacterial plasma membrane, based on the typical P-type ATPase Cu+ transporting domains shown by the bioinformatics analysis of CtpB. In this sense, the fact that M. smegmatis cells expressing CtpB tolerate toxic levels of Cu2+, and that the ATPase activity mediated by this transporter is stimulated by Cu+ but not Cu2+, means that the toxic divalent cation (Cu2+) enters the mycobacteria, where it is reduced to the monovalent form (Cu+) by the reducing environment of the mycobacterial cytoplasm [38]. Thus, Cu+ can be extruded to the external environment by CtpB, like previously observed for M. tuberculosis CtpA [35]. In addition, it is known that Cys chelates copper, which is comparable with the function of Cu+-chaperones that deliver the cation to the ATPases [39]. This suggests that CtpB could prefer the binding of chelated Cu+, which is common in the mycobacterial intracellular environment, where the thiol group of proteins binds Cu+. As a result, the presence of intracellular free Cu+ becomes practically undetectable [40]. On the other hand, the Vmax (2.29 ± 0.10 nmol/mg-min) and KM (0.19 ± 0.04 µM Cu+) of CtpB are lower than those observed for other P1B-type ATPases, such as E. coli CopA (Vmax of = 27.3 ± 4.5 nmol/mg-min) [41] or ATP7A (KM= 0.6 ± 0.04 µM) [42], respectively. This means that CtpB is a less-efficient copper transporter with more affinity and a lower turnover of Cu+ compared to these plasma membrane transporters. However, we cannot rule out that the kinetic parameters of CtpB were measured using plasma membrane fractions, in which other proteins could affect the specific activity of the enzyme and the observed values for Vmax.
It was previously reported that M. tuberculosis CtpV mutant cells do not tolerate toxic doses of copper [6], indicating that CtpV could be the P-type ATPase directly associated with copper detoxification in M. tuberculosis. This finding suggests that the other putative copper P-type ATPase transporters encoded in the M. tuberculosis genome, such as CtpA and CtpB, could play alternative roles during copper transport across the mycobacterial plasma membrane. This is the case of Cu+-ATPases in P. aeruginosa and Sinorhizobium meliloti. Similarly, these bacterial species contain more than two genes coding for Cu+-ATPases in the genome, as with the M. tuberculosis genome. When Cu+-ATPases are encoded by at least two genes in bacteria, one of the encoded enzymes performs an alternative function to metal detoxification, such as metalation of periplasmic and membrane proteins [24]. Indeed, P. aeruginosa contains two paralogous Cu+-ATPases: CopA1 (Vmax 63.1 ± 1.5 nmol/mg min and KM 152.6 ± 7.9 μM Cu+), which mediates copper detoxification, and CopA2 (Vmax 6.7 ± 0.4 nmol/mg min and KM 20.7 ± 3.7 μM Cu+) that is involved in cytochrome c oxidase metalation [43]. Therefore, if the kinetic parameters obtained for CtpB are closer to CopA2 than CopA1, it could be presumed that CtpB may play a protein metalation role, like CopA2. In agreement, knock-out of genes coding for Cu+-ATPases responsible for protein metalation in P. aeruginosa and S. meliloti does not result in a copper-sensitive phenotype [43, 44]. This finding is similar to our observations for M. tuberculosis ∆ctpB cells and enforces the possibility that CtpB is associated with enzyme metalation. Furthermore, it agrees with our results of copper accumulation and mycobacterial tolerance to high doses of cation displayed by M. tuberculosis ∆ctpB mutant cells.
On the other hand, hypoxia and redox stress seem to be correlated in mycobacterial cells. Specifically, it is known that sodium nitroprusside produces nitric oxide (NO), which is able to induce the expression of genes related to hypoxia and affect the aerobic respiration of mycobacteria [45]. NO and CN− inhibit the CtaD subunit of the cytochrome c oxidase, which belongs to the chain of electron transport in mycobacteria. When cytochrome c oxidase is inhibited, mycobacteria respond using the cytochrome bd quinol oxidase, which has a greater affinity for oxygen compared with the cytochrome c oxidase [46]. Therefore, the transcriptional behavior of ctpB described above suggests that the oxidative stressors NO and CN− activate ctpB transcription, possibly facilitating the switching from aerobic respiration to hypoxia. Unlike hypoxia and redox stress, the transcriptional analysis also showed that ctpB was not significantly induced by Cu2+. This finding reinforces the idea that CtpB is associated with a transport function different from copper detoxification. It is possible that CtpB translocates Cu+ from the cytoplasm to the well-known cuproenzymes in the periplasm, such as, superoxide dismutase (SodC), cytochrome oxidase, and multicopper oxidase (MmcO), which use copper as cofactor [10, 24, 47]. In this case, CtpB would be performing an enzyme metalation function, which in turn is involved in the response to hypoxia and redox stress, as suggested by the transcriptional analysis of ctpB in M. tuberculosis cells, along with the growth rate and copper accumulation of M. tuberculosis ∆ctpB cells described above.
Conclusions
Our data indicate that CtpB is a copper (I) transporting P-type ATPase of M. tuberculosis, which induces mycobacterial tolerance to toxic concentrations of Cu2+. However, CtpB shows a metal preference for Cu+ in a metal concentration-dependent manner suggesting that Cu2+ enters the mycobacteria, where it is reduced to Cu+. On the other hand, the Cu+ ATPase activity of CtpB in the plasma membrane shows features of high affinity/slow turnover ATPases similar to Cu+ transporting ATPases involved in protein metalation. Finally, since the ctpB gene is transcriptionally activated under culture conditions that mimicked the stress conditions similar to the intraphagosomal environment, but not under high doses of copper, the overall results suggest that M. tuberculosis CtpB is associated with copper (I) pumping from mycobacterial cells possibly playing a role different from copper detoxification.
Materials and methods
Bioinformatics analysis
The amino acid sequence of CtpB was obtained from Turberculist [27]. The protein topology was predicted with TOPCONS [48], and functional motifs were located as shown by Novoa-Aponte [16]. Clustal OMEGA [49] was used to align the amino acid sequences of M. tuberculosis CtpB with well-characterized bacterial Cu+-ATPases, including CopA from Archaeoglobus fulgidus (WP_010877980.1) and Legionella pneumophila (YP_095057.1). The transmembrane (TM) positions were predicted with TOPCONS server [48].
Bacterial strains and growth conditions
M. tuberculosis H37Ra was grown in Middlebrook 7H9 or 7H11 supplemented with 10% OADC (50 μg/mL oleic acid, 0.5% Bovine albumin Fraction V, 0.2% dextrose, and 0.004% catalase), or Sauton medium supplemented with 0.2% glucose, 30 μM Fe2(SO4)3, and 0.05% Tween-80. M. smegmatis mc2155 cells were grown in LB or Sauton medium supplemented as described above. Mycobacteria were grown until an OD595nm = 0.4 for electroporation experiments or OD600nm = 0.6 for plasma membrane extraction. Escherichia coli TOP10 and HB101 host cells were cultured in LB and used for plasmid constructions. When needed, 7H9 or 7H11 were supplemented with 10% OADC, 0.05% Tween 80, 20 μg/mL kanamycin (Km), and 50 μg/mL hygromycin (Hyg). Likewise, LB was supplemented with 50 μg/mL Km, 100 μg/mL Hyg, 100 μg/mL ampicillin (Amp), 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and 80 μg/mL 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal).
Cloning and expression of M. tuberculosis CtpB
The ctpB gene was amplified by PCR from the genomic DNA of M. tuberculosis H37Rv using the pair of primers pMV-ctpB-dir (5′-TTTTGAATTCGTGGCGGCTCCAGTTGTGG-3′)/pMV-ctpB-rev (5′-TTTTAAGCTTCCTGGATCTGACCCCGACC-3′). The resulting amplimer was cloned in the pGEM-T easy vector (Promega, WI, USA) to obtain the pALT9 recombinant plasmid. The ctpB gen was subcloned in the shuttle vector pMV261 [50] using the restriction sites HindIII–EcoRI, generating the pALT10 plasmid. The integrity of pALT10 was confirmed by automated sequencing. pALT10 and pMV261 were separately transformed in M. smegmatis mc2155 cells by electroporation and, finally, expression of the recombinant protein was induced by incubation of mycobacterial cells at 45 °C for 3 h [37]. The induction of CtpB expression was assessed in cells used to isolate plasma membrane fractions for the ATPase activity assays, including the optimal enzymatic conditions and kinetic parameters of CtpB.
Construction of a M. tuberculosis ctpB mutant
A M. tuberculosis H37Ra ctpB mutant was obtained by recombineering [51]. For the gene mutation, an allelic exchange substrate (AES) was constructed by flanking a HygR cassette with genomic fragments from the upstream and downstream genomic regions of the ctpB gene. Specifically, a 500 bp genomic fragment downstream of ctpB was amplified by PCR using the pair of primers Down-ctpB-dir (5′-TTTTAAGCTTATCGCCGGTGCCGCCATGG-3′)/pMV-ctpB-rev (5′-TTTTCTCGAGTCTGCCGTGGTTGTCGAGC-3′) and cloned in the pGEM-T easy vector to obtain the pALT13 plasmid. This downstream ctpB amplimer was then subcloned into the HindIII and XhoI sites of the pYUB584 vector to obtain the pALT15 plasmid. The upstream region of ctpB was amplified using the pair of primers Up-ctpB-dir (5′-TTTTCCTAGGGCGGGTGTACTGGGTGTCG-3′)/Up-ctpB-rev (5′-TTTTTCTAGATCGACGCGGCTGGCACAGG-3′) and cloned into the AvrII and XbaI sites of pALT15, obtaining the pALT16 plasmid. Finally, the ctpB AES (2987 pb fragment) was isolated from pALT16 by double digestion with XhoI and AvrII. A recombineering strain was separately obtained by electroporation of M. tuberculosis H37Ra cells with the pJV53 vector [51]. The recombineering strain was grown in 7H9-OADC-Km-Tween 80 at 37 °C until an OD595nm = 0.6, then supplemented with 1.5% glycine and 0.2% acetamide, and incubated at 37 °C, 80 rpm during 18 h. Subsequently, 200 ng of ctpB AES were electroporated in the resulting mycobacterial cells. Finally, mutant cells (ΔctpB) were selected by hygromycin resistance on 7H11-OADC-Km-Hyg plates.
Mycobacterial tolerance to different concentration of divalent cations
The tolerance of mycobacterial cells to divalent cations was assessed under high doses of the ions that could be potentially transported by CtpB [35]. Briefly, 200 µL of mycobacterial cultures of M. smegmatis mc2155 separately transformed with pALT10 or pMV261 (control cells) were cultured in LB supplemented with 0.05% Tween-80, 25 mg/mL Km, and different concentrations of heavy metal salts, with incubation at 37 °C and 80 rpm for 72 h. Bacterial growth was measured at OD595nm using the iMark ™ Microplate Absorbance Reader plate reader (Bio-Rad, USA), until an OD595nm = 0.05. The tested concentrations of salts were CdCl2 (5 to 60 μM), CoCl2 (0.25 to 1.5 mM), CuSO4 (0.5 to 3 mM), MnSO4 (1 to 8 mM), NiSO4 (0.125 to1.5 mM), and ZnSO4 (0.1 to 1 mM). Each concentration of cation and strain were tested by triplicate.
The copper sensitivity of the mutant strain (M. tuberculosis H37Ra ∆ctpB) was assessed by growing the mutant cells under different CuSO4 concentrations (5 to 100 μM) using the wild type strain as control cells. Mutant cells were grown in 3 mL of Sauton (initial OD595nm = 0.05) supplemented with 0.2% glucose, 30 μM Fe2(SO4)3, 0.05% Tween-80, and different concentrations of CuSO4. Cultures were incubated at 37 °C and 80 rpm for 3 weeks and the bacterial growth was estimated by measuring the OD595nm. The absorbance values were corrected by subtracting the absorbance of the negative growth control (samples without bacteria). Each copper concentration was tested in three independent experiments. An assay of mycobacterial tolerance to cations was also carried out on solid 7H11-OADC medium supplemented with CuSO4 (0.25 and 100 μM). Briefly, mycobacterial cultures were grown until exponential phase (OD595nm = 0.5) and serially diluted in modified Sauton medium. Subsequently, 5 μL of each dilution were spotted on the agar plates. Plates were incubated at 37 ℃ for 3 weeks. This assay was done in three independent experiments.
ATPase activity of the plasma membrane
Plasma membranes were isolated from cultures (1 L, OD595nm = 0.6) of M. smegmatis mc2155 cells separately transformed with pALT10 or pMV261, as previously described by León-Torres [35], with modifications. Briefly, mycobacterial cells expressing CtpB were collected by centrifugation and washed twice with washing buffer (10 mM MOPS, 250 mM sucrose, pH 7.4). The obtained pellets were then resuspended in 2 mL of lysis buffer (10 mM MOPS–KOH, 0.3 mM EDTA, 1 mM PMSF, and 1 mg/mL lysozyme, pH 7.4) and incubated at room temperature for 30 min. Cells were lysed with a Mini Beadbeater-16 (Biospec, USA) by 4 pulses of 2 min each. The membrane fraction was then obtained by differential centrifugation at 4 ℃: first at 25,000g during 30 min, followed by centrifugation of the obtained supernatant at 10,000g during 90 min. The pellet, that corresponds to the plasma membrane fraction, was resuspended in resuspension buffer (10 mM MOPS–KOH, 250 mM sucrose, and 1 mM PMSF, pH 7.4). The membrane protein was quantified by the Bradford-Zor-Selinger method [52] and subsequently used for the enzymatic assays.
The ATPase activity stimulated by heavy-metal cations was estimated using plasma membranes isolated from M. smegmatis mc2155 cells transformed with pALT10 or pMV261 (control cells) [35]. The enzymatic reactions (50 μL) were performed in the reaction buffer (40 mM MOPS–TRIS, 3 mM MgCl2, 5 mM NaN3, 0.25 mM Na2MoO4, and 0.5 mM Cys, pH 7.4) using 20 μg of plasma membrane protein, and supplemented with 10 μM (final concentration) of the tested heavy metal cation (CoCl2, CuSO4 supplemented with 2.5 mM DTT, CuSO4, MnSO4, NiSO4, and ZnSO4). The enzymatic reactions were initiated by the addition of 3 mM Na2ATP, followed by incubation at 37 °C for 30 min. The ATPase activity was stopped by adding 100 μL of stop solution (3% ascorbic acid, 0.5% ammonium molybdate, and 3% SDS in 1.0 M HCl) and 150 μL of 3.5% bismuth citrate and 3.5% sodium citrate in 2.0 M HCl. Subsequently, the released inorganic phosphate (Pi) was quantified [53]. The difference between the total ATPase activity and the activity obtained from samples supplemented with no cations (basal activity) was considered as the ATPase activity stimulated by the tested cations. Enzymatic reactions supplemented with DTT and without Cu+ were also performed as controls of possible DTT interference in the enzymatic reaction. The ATPase activity of samples was performed by triplicate.
Kinetic parameters of CtpB
The ATPase activity associated with CtpB was calculated as the difference between the activity of the membrane from recombinant cells (transformed with pALT10) and the activity of membrane isolated from the cells transformed with the empty vector pMV261 (control). Since we noticed that values of the enzymatic activity could vary depending on the plasma membrane isolate and it was lost over time, the same plasma membrane preparation was used to estimate the optimal enzymatic conditions and the kinetic parameters. To obtain the optimal conditions for CtpB activity, the enzymatic reactions were incubated at 37 ℃ during 30 min with the following conditions: 20 μg of plasma membrane protein, 40 mM MOPS–TRIS (pH 7.4), 3 mM MgCl2, 10 μM CuSO4, 5 mM NaN3, 0.25 mM Na2MoO4, 0.5 mM Cys, 2.5 mM DTT, and 3 mM Na2ATP. To establish the optimum pH for the enzymatic reaction, samples were tested from pH 5.5 to 9.0, using MES–KOH for pH 5.5 and 6.2, MOPS–KOH for pH 6.8 and 7.4, and Tris–HCl for pH 8 and 9. Similarly, the optimum temperature was tested between 4 and 60 °C, and the optimal concentration of Cys was evaluated by testing concentrations from 0.1 to 10 mM. After that, enzymatic kinetics were performed under the optimal reaction conditions and varying the CuSO4 concentration from 0.25 to 10 μM. The values of KM and Vmax were calculated using the nonlinear least squares regression for Michaelis–Menten enzyme kinetics using Prism 6 version 6.0 h for Windows 10, GraphPad Software, La Jolla California USA, http://www.graphpad.com.
Copper accumulation assay
Assays of copper accumulation in mycobacteria were performed using the M. tuberculosis H37Ra ∆ctpB and wild-type strains, following the procedure reported by Raimunda [54], with some modifications. Briefly, 100 mL of mycobacterial culture was grown in 7H9-OADC-Tween 80 at 37 °C until an OD595 = 0.5. The cultured cells were then collected by centrifugation, washed thrice with one volume of modified Sauton medium, resuspended in 30 mL of the same medium, and aliquoted in samples of 5 mL. Two samples were processed in parallel, one of them was supplemented with 50 μM CuSO4 and the other without a cation. Samples were then incubated at 37 °C and 80 rpm for 3 h. The cells were collected by centrifugation and washed 4 times with one volume of 150 mM NaCl. The pellet was then dried at 37 °C for 24 h and weighed. The cells were mineralized with 500 μL of HNO3 (ultrapure quality, Merck) at 80 °C for 1 h and at room temperature overnight. Subsequently, 20 μL of 30% H2O2 were added to the samples. Finally, the copper content in the samples was determined by graphite furnace atomic absorption spectroscopy (GF-AAS) by High-Resolution Continuum Source Electrothermal Atomic Absorption Spectrometry in a contrAA® 700 D (Analytikjena, Germany) equipment, following the manufacturer’s instructions. Separately, seven copper standards (4, 10, 20, 40, 60, 80, and 100 µg/kg) were prepared from a copper stock for GF-AAS of 1000 mg/kg (Acros Organics) in 0.5% HNO3. Samples were mixed with 5 μL of modifier (0.1% Pb(NO3)2 and 0.05% Mg(NO3)2) and then quantified for copper content at 324.754 nm using pyrolysis and atomization temperatures of 1100 and 1500 °C, respectively. Metal accumulation is reported as μg of accumulated Cu+/g dry cell pellet (μg Cu+/g); the value obtained from the control sample (without cation addition) was subtracted. The assay was performed in three independent experiments. Statistical significance was determined using the Holm-Sidek method (alpha 5.000%), using Prism 6 version 6.0 h for Windows 10, GraphPad Software, La Jolla California USA, http://www.graphpad.com.
Transcriptional analysis of ctpB by qPCR
The transcriptional behavior of ctpB was analyzed from cells cultured under the presence of toxic concentrations of heavy-metal cations, ROS, RNS, and hypoxia. M. tuberculosis H37Ra cells grown until exponential phase (OD595 = 0.6) were washed and resuspended in Sauton medium. The cells were then intoxicated supplementing the cultures separately with CdCl2 (10, 25, and 40 μM), CoCl2 (0.45, 2, and 3.5 mM), CuSO4 (20, 50, and 500 μM), Fe2(SO4)3 (0.2, 0.41, 0.45, and 2 mM), MnSO4 (0.2, 0.4, 1.0, and 15 mM), NiSO4 (0.6, 1, and 2.5 mM), Pb(NO3)2 (0.5 mM) or ZnSO4 (40, 200, and 500 μM). In the case of redox stressors, cells were exposed to 1 mM of H2O2, tert-butyl-hydroperoxide (tBuOOH), KCN, or sodium nitroprusside (Na2[Fe(CN)5NO]). Then, the cells were incubated for 3 h at 37 °C and 80 rpm, harvested and used for RNA isolation. Similarly, the cultures were also subjected to a non-replicative persistence 2 (NRP-2) or anaerobic stage, as described by Wayne and Hayes [55], and compared with standard cultures. In this case, a parallel culture supplemented with methylene-blue (1.5 µg/mL) was used as an indicator of oxygen depletion.
RNA from M. tuberculosis cultures was isolated using the TRIzol method (Invitrogen, USA) [56]. Total RNA was dissolved in DEPC-treated H2O and stored at − 70 °C. cDNA from all samples was prepared using 2 µg of RNA and the RevertAid First Strand cDNA Synthesis kit (Thermo Scientific, USA). ctpB transcription was quantified by qRT-PCR using the specific primers (5′-ACATCCATGGAAACGCTGAT-3′/5′-CAAGAACGAAGACGGTCACA-3′) and normalized to the mean value of sigA gene expression (5′-CCTACGCTACGTGGTGGATT-3′/5′-TGGATTTCCAGCACCTTCTCC-3′). Fluorescence was quantified using the iQ™ SYBR®Green Supermix kit (Bio-Rad, USA) on the CFX-96 thermocycler (Bio-Rad, USA). To validate the qRT-PCR assays and determine their efficiency, serial dilutions of genomic DNA of M. tuberculosis H37Ra were tested. Quantification was performed four times for each condition, and results were analyzed using a calibration curve in which the regression and efficiency values were close to 1 and 2, respectively. In all cases, the relative quantification was analyzed using the Pfaffl method [57]. Reactions were performed in triplicate from two independent experiments, and differences between the experimental data were considered significant at P < 0.05.
Supplementary information
Additional file 1. M. tuberculosis CtpB expression in M. smegmatis plasma membrane. SDS-PAGE analysis of CtpB in the mycobacterial plasma membrane. 12% SDS-PAGE analysis was performed with the membrane fraction of the different cells: pALT10, M. smegmatis expressing CtpB; pMV261, M. smegmatis transformed with the expression vector pMV261 (control without CtpB). The arrow shows a membrane protein of approximately 78.7 kDa corresponding to the molecular weight of the CtpB recombinant protein.
Additional file 2. Enzymatic reaction progress of CtpB. We monitored the Cu+ ATPase reaction progress by measuring the released Pi (nmol) at different enzymatic reaction times. The assay was performed using 10.0 μM Cu+ and enzymatic reactions times between 0 and 60 min. The plotted data correspond to the average released Pi (nmol) ± SEM from three independent experiments.
Additional file 3. Disruption of ctpB in M. tuberculosis H37Ra cells. a) Schematic representation of the M. tuberculosis H37Ra ctpB genomic region, indicating the primers used to verify the mutant genotype (negative control). b) Schematic representation of the M. tuberculosis H37Ra genome, where ctpB is interrupted by a Hyg cassette (positive control); the primers used for PCR amplification are shown. c) PCR of the genomic DNA isolated from three possible M. tuberculosis H37Ra ΔctpB colonies using different primer combinations. Genomic DNA from the wild-type strain (WT) and pALT16 plasmid (AES) were used as controls.
Acknowledgements
This work was supported by the División de Investigación Bogotá (DIB)-Universidad Nacional de Colombia, grants FP44842-108-2016. AL was a Fellow of the Colciencias program “Doctorados Nacionales”, Colombia.
Abbreviations
- AES
Allelic exchange substrate
- GF-AAS
Graphite furnace atomic absorption spectroscopy
- OD
Optical density
- qRT-PCR
Reverse transcription quantitative PCR
- RNS
Reactive nitrogen species
- ROS
Reactive oxygen species
- TB
Tuberculosis
- TM
Transmembrane
- WT
Wild type
Authors’ contributions
AL and CS conceived this study, drafted the manuscript and revised the manuscript. AL, EA, and EC performed the experiments. AL, CS, and EC contributed to data interpretation and preparation of the manuscript. All authors read and approved the final manuscript.
Funding
We are thankful to the División de Investigación Bogotá (DIB)-Universidad Nacional de Colombia and Colciencias for their financial support of this work.
Availability of data and materials
All the data analyzed during this study are included in this article.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40659-020-00274-7.
References
- 1.WHO . Global tuberculosis report 2018. Geneva: World Health Organization; 2018. [Google Scholar]
- 2.Cambier CJ, Falkow S, Ramakrishnan L. Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell. 2014;159(7):1497–1509. doi: 10.1016/j.cell.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 3.Soldati T, Neyrolles O. Mycobacteria and the intraphagosomal environment: take it with a pinch of salt(s)! Traffic. 2012;13(8):1042–1052. doi: 10.1111/j.1600-0854.2012.01358.x. [DOI] [PubMed] [Google Scholar]
- 4.Botella H, Stadthagen G, Lugo-Villarino G, de Chastellier C, Neyrolles O. Metallobiology of host–pathogen interactions: an intoxicating new insight. Trends Microbiol. 2012;20(3):106–112. doi: 10.1016/j.tim.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Wagner D, Maser J, Lai B, Cai Z, Barry CE, Zu Bentrup KH, et al. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell’s endosomal system. J Immunol. 2005;174(3):1491–1500. doi: 10.4049/jimmunol.174.3.1491. [DOI] [PubMed] [Google Scholar]
- 6.Ward SK, Abomoelak B, Hoye EA, Steinberg H, Talaat AM. CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol Microbiol. 2010;77(5):1096–1110. doi: 10.1111/j.1365-2958.2010.07273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neyrolles O, Wolschendorf F, Mitra A, Niederweis M. Mycobacteria, metals, and the macrophage. Immunol Rev. 2015;264(1):249–263. doi: 10.1111/imr.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rowland JL, Niederweis M. Resistance mechanisms of Mycobacterium tuberculosis against phagosomal copper overload. Tuberculosis (Edinb). 2012;92(3):202–210. doi: 10.1016/j.tube.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harth G, Horwitz MA. Export of recombinant Mycobacterium tuberculosis superoxide dismutase is dependent upon both information in the protein and mycobacterial export machinery. A model for studying export of leaderless proteins by pathogenic mycobacteria. J Biol Chem. 1999;274(7):4281–4292. doi: 10.1074/jbc.274.7.4281. [DOI] [PubMed] [Google Scholar]
- 10.Spagnolo L, Toro I, D’Orazio M, O’Neill P, Pedersen JZ, Carugo O, et al. Unique features of the sodC-encoded superoxide dismutase from Mycobacterium tuberculosis, a fully functional copper-containing enzyme lacking zinc in the active site. J Biol Chem. 2004;279(32):33447–33455. doi: 10.1074/jbc.M404699200. [DOI] [PubMed] [Google Scholar]
- 11.Agranoff D, Krishna S. Metal ion transport and regulation in Mycobacterium tuberculosis. Front Biosci. 2004;9:2996–3006. doi: 10.2741/1454. [DOI] [PubMed] [Google Scholar]
- 12.Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci U S A. 2000;97(16):8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shiloh MU, Nathan CF. Reactive nitrogen intermediates and the pathogenesis of Salmonella and mycobacteria. Curr Opin Microbiol. 2000;3(1):35–42. doi: 10.1016/S1369-5274(99)00048-X. [DOI] [PubMed] [Google Scholar]
- 14.Fang FC. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol. 2004;2(10):820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 15.Palmgren MG, Nissen P. P-type ATPases. Annu Rev Biophys. 2011;40:243–266. doi: 10.1146/annurev.biophys.093008.131331. [DOI] [PubMed] [Google Scholar]
- 16.Novoa-Aponte L, Leon-Torres A, Patino-Ruiz M, Cuesta-Bernal J, Salazar LM, Landsman D, et al. In silico identification and characterization of the ion transport specificity for P-type ATPases in the Mycobacterium tuberculosis complex. BMC Struct Biol. 2012;12:25. doi: 10.1186/1472-6807-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novoa-Aponte L, Soto Ospina CY. Mycobacterium tuberculosis P-type ATPases: possible targets for drug or vaccine development. Biomed Res Int. 2014;2014:296986. doi: 10.1155/2014/296986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100(22):12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forrellad MA, Klepp LI, Gioffre A, Sabio y Garcia J, Morbidoni HR, de la Paz Santangelo M, et al. Virulence factors of the Mycobacterium tuberculosis complex. Virulence. 2013;4(1):3–66. doi: 10.4161/viru.22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar M, Khan FG, Sharma S, Kumar R, Faujdar J, Sharma R, et al. Identification of Mycobacterium tuberculosis genes preferentially expressed during human infection. Microb Pathog. 2011;50(1):31–38. doi: 10.1016/j.micpath.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Ward SK, Hoye EA, Talaat AM. The global responses of Mycobacterium tuberculosis to physiological levels of copper. J Bacteriol. 2008;190(8):2939–2946. doi: 10.1128/JB.01847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu T, Ramesh A, Ma Z, Ward SK, Zhang LM, George GN, et al. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat Chem Biol. 2007;3(1):60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalo-Asensio J, Mostowy S, Harders-Westerveen J, Huygen K, Hernandez-Pando R, Thole J, et al. PhoP: a missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS ONE. 2008;3(10):e3496. doi: 10.1371/journal.pone.0003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Argüello JM, Patel SJ, Quintana J. Bacterial Cu(+)-ATPases: models for molecular structure–function studies. Metallomics. 2016;8(9):906–914. doi: 10.1039/C6MT00089D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padilla-Benavides T, Long JE, Raimunda D, Sassetti CM, Argüello JM. A novel P(1B)-type Mn2+ -transporting ATPase is required for secreted protein metallation in mycobacteria. J Biol Chem. 2013;288(16):11334–11347. doi: 10.1074/jbc.M112.448175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho SH, Goodlett D, Franzblau S. ICAT-based comparative proteomic analysis of non-replicating persistent Mycobacterium tuberculosis. Tuberculosis (Edinb). 2006;86(6):445–460. doi: 10.1016/j.tube.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Kapopoulou A, Lew JM, Cole ST. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb). 2011;91(1):8–13. doi: 10.1016/j.tube.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Thever MD, Saier MH., Jr Bioinformatic characterization of p-type ATPases encoded within the fully sequenced genomes of 26 eukaryotes. J Membr Biol. 2009;229(3):115–130. doi: 10.1007/s00232-009-9176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev. 2009;109(10):4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Argüello JM. Identification of ion selectivity determinants in heavy metal transport P1B-type ATPases. J Membr Biol. 2003;195:93–108. doi: 10.1007/s00232-003-2048-2. [DOI] [PubMed] [Google Scholar]
- 32.Drees SL, Beyer DF, Lenders-Lomscher C, Lubben M. Distinct functions of serial metal-binding domains in the Escherichia coli P1 B -ATPase CopA. Mol Microbiol. 2015;97(3):423–438. doi: 10.1111/mmi.13038. [DOI] [PubMed] [Google Scholar]
- 33.Andersson M, Mattle D, Sitsel O, Klymchuk T, Nielsen AM, Moller LB, et al. Copper-transporting P-type ATPases use a unique ion-release pathway. Nat Struct Mol Biol. 2014;21(1):43–48. doi: 10.1038/nsmb.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andreu N, Soto CY, Roca I, Martin C, Gibert I. Mycobacterium smegmatis displays the Mycobacterium tuberculosis virulence-related neutral red character when expressing the Rv0577 gene. FEMS Microbiol Lett. 2004;231(2):283–289. doi: 10.1016/S0378-1097(04)00008-4. [DOI] [PubMed] [Google Scholar]
- 35.León-Torres A, Novoa-Aponte L, Soto CY. CtpA, a putative Mycobacterium tuberculosis P-type ATPase, is stimulated by copper (I) in the mycobacterial plasma membrane. Biometals. 2015;28(4):713–724. doi: 10.1007/s10534-015-9860-x. [DOI] [PubMed] [Google Scholar]
- 36.Lopez M, Quitian LV, Calderon MN, Soto CY. The P-type ATPase CtpG preferentially transports Cd(2 +) across the Mycobacterium tuberculosis plasma membrane. Arch Microbiol. 2017;200(3):483–492. doi: 10.1007/s00203-017-1465-z. [DOI] [PubMed] [Google Scholar]
- 37.Ayala-Torres C, Novoa-Aponte L, Soto CY. Pma1 is an alkali/alkaline earth metal cation ATPase that preferentially transports Na(+) and K(+) across the Mycobacterium smegmatis plasma membrane. Microbiol Res. 2015;176:1–6. doi: 10.1016/j.micres.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 38.Lewinson O, Lee AT, Rees DC. A P-type ATPase importer that discriminates between essential and toxic transition metals. Proc Natl Acad Sci USA. 2009;106(12):4677–4682. doi: 10.1073/pnas.0900666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y, Mandal AK, Bredeston LM, González-Flecha FL, Argüello JM. Activation of Archaeoglobus fulgidus Cu + -ATPase CopA by cysteine. BBA. 2007;1768(3):495–501. doi: 10.1016/j.bbamem.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 40.Nies DH, Herzberg M. A fresh view of the cell biology of copper in enterobacteria. Mol Microbiol. 2013;87(3):447–454. doi: 10.1111/mmi.12123. [DOI] [PubMed] [Google Scholar]
- 41.Padilla-Benavides T, George Thompson AM, McEvoy MM, Argüello JM. Mechanism of ATPase-mediated Cu+ export and delivery to periplasmic chaperones: the interaction of Escherichia coli CopA and CusF. J Biol Chem. 2014;289(30):20492–20501. doi: 10.1074/jbc.M114.577668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hung YH, Layton MJ, Voskoboinik I, Mercer JF, Camakaris J. Purification and membrane reconstitution of catalytically active Menkes copper-transporting P-type ATPase (MNK; ATP7A) Biochem J. 2007;401(2):569–579. doi: 10.1042/BJ20060924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.González-Guerrero M, Raimunda D, Cheng X, Argüello JM. Distinct functional roles of homologous Cu(+) efflux ATPases in Pseudomonas aeruginosa. Mol Mirobiol. 2010;78(5):1246–1258. doi: 10.1111/j.1365-2958.2010.07402.x. [DOI] [PubMed] [Google Scholar]
- 44.Patel SJ, Padilla-Benavides T, Collins JM, Argüello J. Functional diversity of five homologous Cu+ -ATPases present in Sinorhizobium meliloti. Microbiology. 2014;160:1237–1251. doi: 10.1099/mic.0.079137-0. [DOI] [PubMed] [Google Scholar]
- 45.Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198(5):705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boshoff HI, Barry CE., 3rd Tuberculosis—metabolism and respiration in the absence of growth. Nat Rev Microbiol. 2005;3(1):70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- 47.Quintanar L, Stoj C, Taylor AB, Hart PJ, Kosman DJ, Solomon EI. Shall we dance? how a multicopper oxidase chooses its electron transfer partner. Acc Chem Res. 2007;40(6):445–452. doi: 10.1021/ar600051a. [DOI] [PubMed] [Google Scholar]
- 48.Tsirigos KD, Peters C, Shu N, Kall L, Elofsson A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015;43(W1):W401–W407. doi: 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, et al. New use of BCG for recombinant vaccines. Nature. 1991;351(6326):456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 51.van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nat Methods. 2007;4(2):147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 52.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 53.Cariani L, Thomas L, Brito J, del Castillo JR. Bismuth citrate in the quantification of inorganic phosphate and its utility in the determination of membrane-bound phosphatases. Anal Biochem. 2004;324(1):79–83. doi: 10.1016/j.ab.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 54.Raimunda D, González-Guerrero M, Leeber BW, 3rd, Argüello JM. The transport mechanism of bacterial Cu(+)-ATPases: distinct efflux rates adapted to different function. Biometals. 2011;24(3):467–475. doi: 10.1007/s10534-010-9404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64(6):2062–2069. doi: 10.1128/IAI.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rustad TR, Roberts DM, Liao RP, Sherman DR. Isolation of mycobacterial RNA. In: Parish T, Brown AC, editors. Mycobacteria protocols. 2. Totowa: Humana Press; 2009. pp. 13–22. [DOI] [PubMed] [Google Scholar]
- 57.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26(6):509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. M. tuberculosis CtpB expression in M. smegmatis plasma membrane. SDS-PAGE analysis of CtpB in the mycobacterial plasma membrane. 12% SDS-PAGE analysis was performed with the membrane fraction of the different cells: pALT10, M. smegmatis expressing CtpB; pMV261, M. smegmatis transformed with the expression vector pMV261 (control without CtpB). The arrow shows a membrane protein of approximately 78.7 kDa corresponding to the molecular weight of the CtpB recombinant protein.
Additional file 2. Enzymatic reaction progress of CtpB. We monitored the Cu+ ATPase reaction progress by measuring the released Pi (nmol) at different enzymatic reaction times. The assay was performed using 10.0 μM Cu+ and enzymatic reactions times between 0 and 60 min. The plotted data correspond to the average released Pi (nmol) ± SEM from three independent experiments.
Additional file 3. Disruption of ctpB in M. tuberculosis H37Ra cells. a) Schematic representation of the M. tuberculosis H37Ra ctpB genomic region, indicating the primers used to verify the mutant genotype (negative control). b) Schematic representation of the M. tuberculosis H37Ra genome, where ctpB is interrupted by a Hyg cassette (positive control); the primers used for PCR amplification are shown. c) PCR of the genomic DNA isolated from three possible M. tuberculosis H37Ra ΔctpB colonies using different primer combinations. Genomic DNA from the wild-type strain (WT) and pALT16 plasmid (AES) were used as controls.
Data Availability Statement
All the data analyzed during this study are included in this article.