Abstract
Our previous finding that the BET inhibitor (BETi) JQ1 increases levels of the DNA damage marker γH2AX suggested that JQ1 might enhance the sensitivity of tumor cells to PARP inhibitors (PARPi), which are selectively toxic to cells that harbor relatively high levels of DNA damage. To address this hypothesis, we evaluated the effect of a BETi (JQ1 or I-BET762) combined with a PARPi (olaparib or veliparib) in KKU-055 and KKU-100 cholangiocarcinoma (CCA) cell lines and of JQ1 with olaparib in a xenograft model of CCA.
Each combination was more effective than any of the four drugs as single agents. Combination indices ranged from 0.1 to 0.8 at the ED50 for all combinations, indicating synergy and demonstrating that synergy was not limited to a specific combination. Mechanistically, downregulation of BETi molecular targets BRD2 or BRD4 by shRNA sensitized CCA cells to BETi as single agents as well as to the combination of a BETi + a PARPi.
Our data indicate that combinations of a BETi with a PARPi merit further evaluation as a promising strategy for CCA.
Keywords: cholangiocarcinoma, BET inhibitors, PARP inhibitors, c-Myc, RNAi, combination indices
1. Introduction
Cholangiocarcinoma (CCA) is a rare, aggressive neoplasm arising from the epithelial layer of the biliary tract [1], and is the second most common primary hepatic malignancy after hepatocellular carcinoma [2]. CCA is usually diagnosed at late disease stage and present with symptoms including jaundice, fatigue and abdominal pain [2]. Patients who receive the current standard of care, resection followed by gemcitabine with or without cisplatin, see the greatest survival benefit [1]. However, 90% of patients are ineligible for resection [3]. The 5-year survival for patients with CCA is ~30% [2], with relapse occurring 2-3 years following resection [2] and development of chemoresistance [3-5]. This study focuses on identifying a novel combination of agents with synergistic cytotoxicity in vitro and anti-tumor activity in vivo in CCA.
BET inhibitors (BETi) such as JQ1 and I-BET762 function as acetylated lysine (K-Ac) mimetics that bind to the K-Ac binding pocket of BET protein family members (BRD2, BRD3, BRD4 and BRDT) to competitively inhibit the association of BET proteins with K-Ac residues of chromatin-associated histones, thereby inhibiting recruitment of transcriptional complexes to genomic loci that mediate expression of multiple proteins. Proteins whose expression is BET-dependent reportedly vary among tumor types [6-9].
We recently reported the efficacy of the BETi JQ1, with two patient-derived xenograft (PDX) models of CCA [10]. We observed that 50 mg/kg JQ1 administered daily to mice bearing CCA2 tumors suppressed tumor growth (P<0.001). We also observed a concomitant decrease in expression of c-Myc and its transcriptional target Chk1. Further, we made the novel observation that JQ1 increased levels of the DNA damage marker γH2AX and induced apoptosis as reflected by increases in cleaved caspase-3 and cleaved PARP. Because PARP inhibitors (PARPi) are known to be selectively toxic to cells deficient in DNA double strand break repair [11-16] and with elevated levels of DNA damage, we hypothesized that BETi + PARPi would exert synergistic cytotoxicity. The current study evaluates the potency of the BETi (JQ1 or I-BET762) with the PARPi (olaparib or veliparib) in CCA cell lines and efficacy of JQ1 + olaparib in a xenograft model of CCA. This study also determines the effect of level of expression of BETi targets BRD2 or BRD4 on the potency of BETi ± PARPi in CCA cells.
2. Materials and Methods
2.1. Ethics statement
Animal protocols were approved by the University of Alabama at Birmingham Animal Care and Use Committee.
2.2. Cell Culture and Compounds
KKU-055 (JCRB1551) and KKU-100 (JCRB1568) cholangiocarcinoma cell lines were purchased from the Japanese Cancer Research Resources Bank (JCRB) (National Institute of Biomedical Innovation, Japan). Cells were cultured in DMEM (Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA, USA) and 2 mM L-glutamine (Fisher Scientific). Both CCA cell lines were tested for mycoplasma using MycoAlert™ PLUS Mycoplasma Detection Kit (Lonza, Walkersville, MD, USA) and were negative. JQ1 (HY-13030, MedChem Express, Monmouth Junction, NJ, USA), I-BET762 (HY-13032, MedChem Express), olaparib (HY-10162, MedChem Express) and veliparib (ABT-888, Enzo Life Sciences, Farmingdale, NY, USA) were prepared as solutions in DMSO. Final concentrations of DMSO in in vitro experiments were <0.3%.
2.3. In Vitro Cell Viability Assay
Cell viability assays were carried out as described previously [17, 18]. Briefly, cells were seeded in 96-well plates and allowed to adhere for 24 hours. Serial dilutions of BET inhibitors (JQ1 or I-BET762) and/or PARPi (olaparib or veliparib) were added to the culture medium for 96 hours. AlamarBlue (Fisher Scientific) was added in accordance with manufacturer instructions. Fluorescence was read on a PerkinElmer Victor X5 microplate reader at 560nm excitation and 590nm emission wavelengths. IC50 values were calculated using GraphPad Prism 7.0 (San Diego, CA, USA). Combination indices (CI) were calculated using CompuSyn 1.0 software with values <1.0 indicating synergism [19]. Three independent experiments were performed with quadruplicated wells.
2.4. Clonogenic Assay
Survival Fraction (Fig.1):
Cells were plated in a confluency between 50 to 150 cells and 100 to 1,500 cells into 6-well plates for KKU-055 and KKU-100, respectively, and allowed to adhere overnight. Cells were exposed to drug for 72 hours, washed with PBS, given fresh non-drug containing media and allowed to grow for an additional 14 days (total of 18 days in culture). Cells were then formalin fixed and stained with 0.025% crystal violet. Colonies of greater than 50 cells were counted. Control (DMSO) plating efficiency (PE) was calculated using: PE = {(# of colonies formed)/(# of cells plated) × 100}. Percent clonogenic survival, the number of colonies that formed after treatment, was calculated using the equation: Percent clonogenic survival = {(# of colonies formed )/(# of cells plated × PE) × 100} [20].
Colony formation (Fig.3):
Two thousand KKU-055 or KKU-100 cells were plated per well in 24-well plates and allowed to adhere overnight. Cells were exposed to DMSO (<0.3%) or various concentrations of JQ1 (0.1 μM, 1 μM or 10 μM), olaparib (0.1 μM, 1 μM or 10 μM), or JQ1 + olaparib (1:1) for 72 hours, washed with PBS, and grown in drug-free media for an additional 14 days. Cells were then formalin fixed and stained with 0.025% crystal violet. Plates were imaged using an Epson scanner. Three independent experiments were performed.
2.5. qRT-PCR Assay
Trizol-chloroform extraction was used to isolate total RNA. qRT-PCR was performed as previously described [17, 21]. Three independent experiments were performed. Primers used are listed in Table S1.
2.6. Immunoblotting Analysis
Cell lysates were prepared in NP-40 (Fisher Scientific) or RIPA buffer (MilliporeSigma, St. Louis, MO, USA) containing protease inhibitors (Fisher Scientific). Primary antibodies used were: c-Myc (5606S, Cell Signaling, 1:1,000), GAPDH (2118S, Cell Signaling, 1:1,000), vinculin (v4505, MilliporeSigma, 1:10,000), Chk1 (A300-298AT, Bethyl, 1:5,000), BRD2 (5848, Cell Signaling, 1:1,000), BRD4 (13440, Cell Signaling, 1:1,000), cleaved PARP (5625, Cell Signaling, 1:1,000) and γH2AX (9718S, Cell Signaling, 1:1,000). Secondary antibodies used were: HRP goat anti-rabbit IgG (6721, Abcam, 1:50,000) and HRP anti-mouse IgG (7076, Cell Signaling, 1:5,000). Immunoblots were quantitated using ImageStudio Lite 5.2. Data were first normalized to respective loading controls and then to DMSO control.
2.7. Cell Cycle Analysis
KKU-055 cells were exposed to JQ1 (30μM), olaparib (5μM), veliparib (10μM), JQ1 (30μM) + olaparib (5μM) or JQ1 (30μM) + veliparib (10μM) for 48 hours. Cells were harvested, centrifuged and added drop-wise into ice-cold 70% ethanol while vortexing. The cells were incubated at 4°C overnight. The next day, cells were centrifuged, and the precipitate was incubated with propidium iodide-Triton X-100 resuspension buffer in PBS (0.1% Triton X-100, 200 μg/ml RNAase A, and 20 μg/ml propidium iodide) for a minimum of an hour prior to running flow cytometry [22]. Flow cytometry was carried out at the UAB flow cytometry core using a FACSCalibur (BD Biosciences, San Jose, CA, USA) flow cytometry machine. Twenty-thousand cells were analyzed using FlowJo™ (v10.6.1, BD Biosciences) and the Dean-Jett-Fox univariate model.
2.8. Chromatin Immunoprecipitation (ChIP)
ChIP was performed as previously described [17]. Briefly, digested cellular chromatin was immunoprecipitated with a ChIP grade anti-BRD4 or anti-BRD2 antibody (Cell Signaling) or normal rabbit IgG (Cell Signaling) as a negative control. DNA coprecipitated with BRD4 or BRD2 was quantitated using qRT-PCR with primers that bind to a locus in the promoter region of the MYC gene. Data were analyzed relative to the percent input (2%). Two independent experiments were performed. Primers used are listed in Table S1.
2.9. Generation of Stable shRNA-Transfected Cell Lines
Stable shRNA gene knockdown was performed as previously described [17]. KKU-055 cells were plated at low confluency (10%) in 6-well plates and transfected with MISSION shRNA targeted for BRD2, BRD4 (MilliporeSigma) or the control shRNA for GFP (Addgene, Watertown, MA, USA) using PEI (Polysciences Inc., Warrington, PA, USA) for 8 hours. Cells were then washed with PBS and grown in fresh media for 72 hours. Transfected cell populations were selected using puromycin (7.5 μg/ml) (BML-GR312, Enzo Life). The sequences of shRNA oligonucleotides are listed in Table S1.
2.10. In Vivo Drug Efficacy Studies
CB17−/− female SCID mice (4-week-old) were purchased from Taconic Farms (Newton, MA, USA) and housed in the AAALAC accredited vivarium at University of Alabama at Birmingham Research Support Building. KKU-055 cells (5 × 106) in 100 μL PBS were injected into each flank via subcutaneous injection. Mice bearing bilateral tumors were randomized into four groups of 5 mice/group when tumors reached ~200 mm3 [17, 18, 21]. Tumor numbers were VC = 6, JQ1 = 8, olaparib = 7 and JQ1 + olaparib = 9. Intraperitoneal injections of JQ1, olaparib or the combination was given daily for 21 days. Drug solutions were prepared in DMSO and diluted 1:10 into 10% β-cyclodextrin (MilliporeSigma). 10% β-cyclodextrin dissolved in sterile water was utilized for JQ1 and sterile PBS was used for olaparib [17]. Olaparib was administered 30 minutes prior to JQ1 [17]. Tumor volumes were measured three times a week using digital calipers, and tumor volume calculated using the formula v= (π/6) × d3. Results were normalized to Day 0 of drug treatment, and data are expressed as normalized tumor volume. Average mouse body weight (g) was assessed three times a week using a scale throughout the study. Data are presented as mean ± S.E.M. Statistics were done using two-way analysis of variance (ANOVA) using GraphPad Prism 7.
2.11. Statistical Analysis
All statistical analyses were performed using GraphPad Prism 7 software (SanDiego, CA, USA) [17, 21]. Statistical significance was calculated using one-way or two-way-ANOVA. P ≤ 0.05 was considered significant.
3. Results
3.1. JQ1 inhibits CCA cell viability and clonogenic potential.
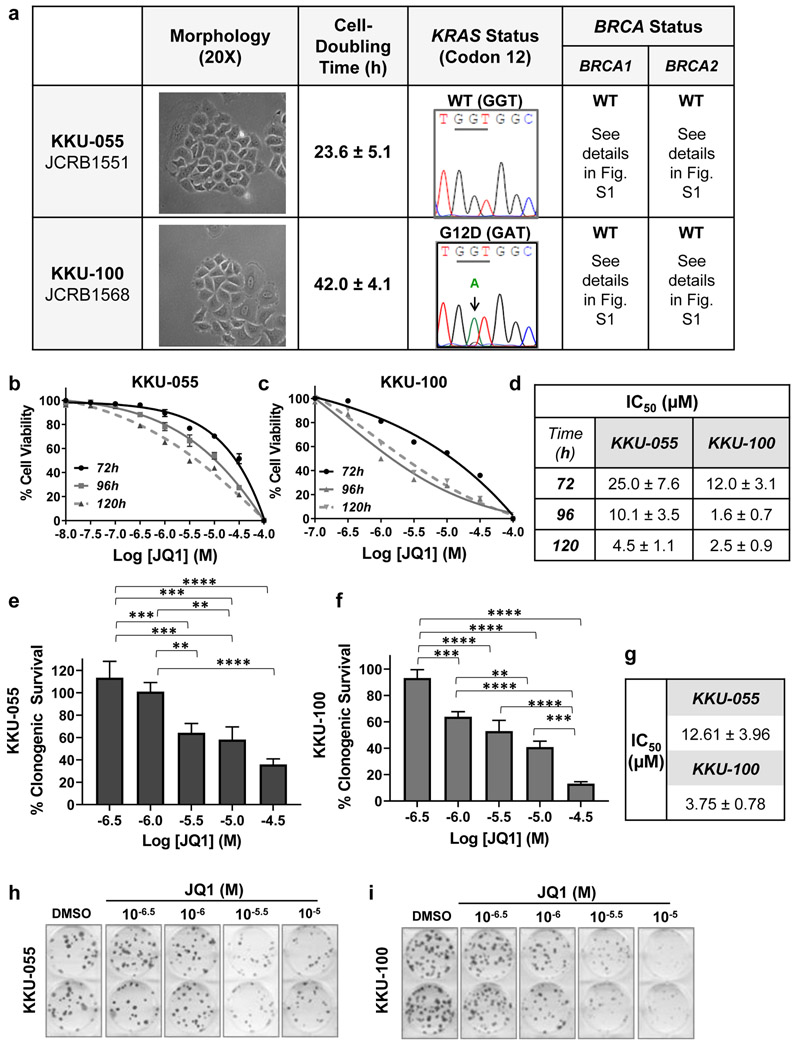
Human cholangiocarcinoma (CCA) cell lines KKU-055 and KKU-100 represent the CCA subtypes intrahepatic and hilar, respectively [23]. Both cell lines have epithelial-like morphology and proliferate in vitro in discrete patches. Cell doubling time was ~24 hours and ~42 hours for KKU-055 and KKU-100 cells, respectively. KKU-100 cells express mutant KRASG12D, while KKU-055 cells express wild type KRAS (Fig.1a). Neither KKU-055 nor KKU-100 cells harbored mutations in six previously characterized hot spots for BRCA1 mutations or in four hotspots for BRCA2 mutations (Fig.1a, Fig.S1) [24]. Because the overall goal of this study was to use CCA models to evaluate the efficacy of BET inhibitors as single agents and in combination, we compared the potency of the BET inhibitors JQ1 and I-BET762 in cell viability and clonogenic assays in vitro assessed efficacy in vivo, and determined the effect of these agents on c-Myc expression and function, using CCA cell lines and a xenograft model.
To assess the potency of JQ1 in vitro, we exposed CCA cell lines to a range of JQ1 concentrations for 72 to 120 hours and assessed cell viability using alamarBlue assays (Figs.1b, 1c). JQ1 decreased cell viability in a dose-dependent manner, and IC50s depended on duration of exposure (Fig. 1d). To also assess the effect of JQ1 on clonogenic potential, we exposed cells to JQ1 for 72 hours, and then propagated them in drug-free medium for an additional 14 days, as detailed in Materials and Methods (Figs. 1e, 1f). Under these conditions, JQ1 decreased the clonogenic potential at micromolar concentrations (P<0.0001) (Fig. 1g). Representative images are shown in Figs.1h and 1i.
Figure 1. JQ1 decreases the viability and the clonogenic potential of KKU-055 and KKU-100 CCA cells in vitro.
(a) Morphology, cell-doubling time, KRAS codon 12 status and BRCA1/2 mutational status of KKU-055 and KKU-100 CCA cell lines. KKU-055 (b) or KKU-100 (c) cells were exposed to the indicated concentrations of JQ1 for 72, 96, or 120 hours. Cell viability was assessed by alamarBlue assays and data are presented as mean ± S.E.M. A minimum of three independent experiments were performed. (d) A table with JQ1 concentration required to inhibit cell viability by 50% (IC50) from Fig.1b and Fig.1c. (e - i) JQ1 inhibited the growth of colony in clonogenic assays. Between 20 and 150 KKU-055 cells (e) or 100 and 1,500 KKU-100 cells (f) were plated, exposed to various concentrations of JQ1 or DMSO for 72 hours, washed with PBS and fresh media added. Cells were incubated in drug-free media for another 14 days. Colonies of >50 cells were counted and quantitated using DMSO as 100%. Three independent experiments were performed. IC50 values were calculated and shown in (g). Representative images of clonogenic assays for KKU-055 (plating cell number = 50) (h) and KKU-100 (plating cell number = 200) (i) cell lines are shown.
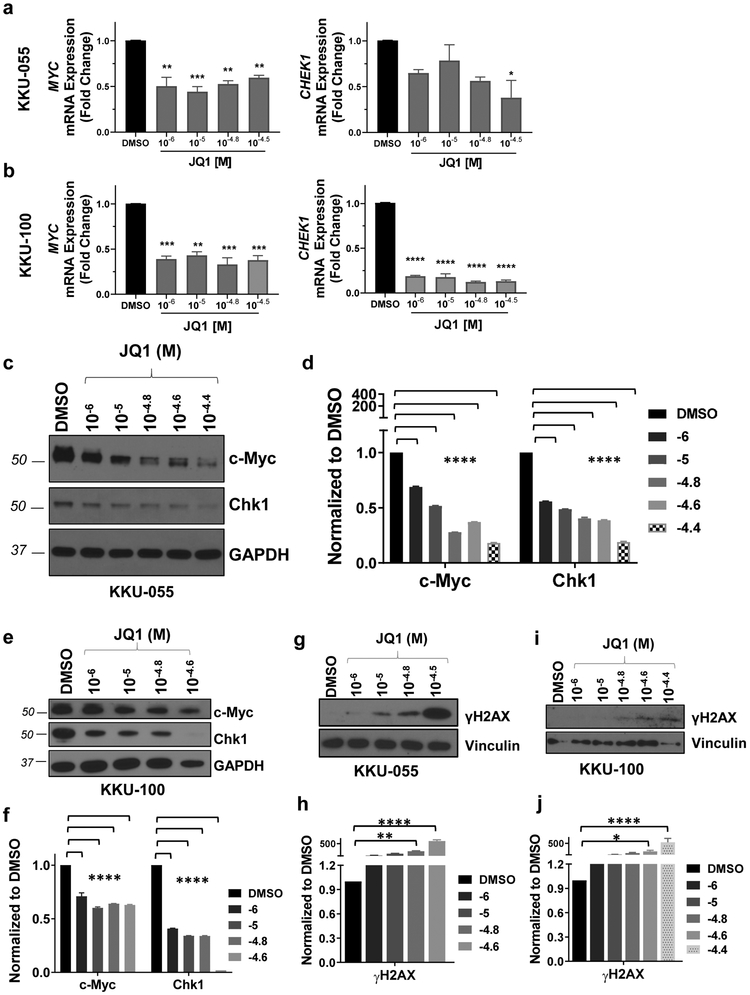
3.2. JQ1 decreases expression of c-Myc and its downstream target Chk1.
Published studies suggest that c-Myc expression depends, at least in part, on BET protein function [25]. Therefore, we evaluated the effect of JQ1 on c-Myc expression. Further, because Chk1 expression was downregulated by JQ1 in an in vivo model of CCA [10] and is regulated by c-Myc [26, 27], we assessed the effect of JQ1 on Chk1 expression, as a measure of c-Myc function.
qRT-PCR assays showed that exposure to 10−6 - 10−4.5 M JQ1 for 72 hours decreased expression of MYC mRNA by >50% (P<0.01) in both cell lines, compared to DMSO controls (Figs.2a, 2b). JQ1 decreased expression of the c-Myc transcriptional target CHEK1 mRNA up to ~60% (P<0.05) and by >80% (P<0.001) in KKU-055 and KKU-100 cells, respectively (Figs. 2a, 2b). We corroborated qRT-PCR results by performing immunoblots (IB) to assess expression of c-Myc and Chk1 protein. JQ1 decreased c-Myc and Chk1 expression in a dose dependent manner, with >50% inhibition of both proteins in both cell lines following exposure to 10−6 - 10−4.4 M JQ1 for 72 hours (Figs. 2c-2f). As we previously reported that JQ1 inhibited the expression of BRCA2 [10], we evaluated the effect of JQ1 on the expression of BRCA2 in KKU-055 and KKU-100 cell lines in vitro. We found that JQ1 (1-32μM for 72 hours) inhibited the expression of BRCA2 mRNA in KKU-100 cells >90% (P<0.0001), and JQ1 (32μM for 72 hours) inhibited the expression of BRCA2 mRNA in KKU-055 cells by ~50% (P<0.01) (Fig. S2).
Figure 2. JQ1 decreases the expression of c-Myc and its transcriptional target Chk1 in both KKU-055 and KKU-100 CCA cell lines.
qRT-PCR shows that JQ1 inhibited the mRNA expression of MYC and CHEK1 in KKU-055 (a) and KKU-100 (b) cell lines. (c) & (e) JQ1 inhibited expression of c-Myc and Chk1 protein in KKU-055 (c) and KKU-100 (e) cell lines. (d) & (f) Immunoblot data in (c) and (e) were quantitated as percent DMSO using ImageStudio Lite (LI-COR Biosciences) and are reported as bar graphs mean ± S.E.M. (g-j) JQ1 increased the level of γH2AX, a marker of DNA damage, in KKU-055 (g) and KKU-100 (i) cell lines. (h) & (j) Immunoblot data in (g) and (i) were quantitated as described above and are reported as bar graphs (mean ± S.E.M.). Analysis was done by one-way ANOVA (*P<0.05, **P<0.01, ***P<0.001, ****p<0.0001).
Based on our previous finding that JQ1 also increases levels of the DNA damage marker γH2AX in pancreatic cancer models and a PDX model of CCA [10, 17], we next assessed the effect of JQ1 on levels of this DNA damage marker, using assay conditions similar to those under which we observed decreases in c-Myc and Chk1. We exposed CCA cells to 10−6 - 10−4.4 M JQ1 for 48-72h and determined levels of expression of γH2AX by immunoblot. JQ1 increased the levels of γH2AX in a dose-dependent manner (Figs. 2g-2j). In light of reports in the literature documenting that DNA repair deficiency or relatively high levels of DNA damage sensitize tumor cells to PARP inhibitors [17, 28-31], we next assessed the potency of BET inhibitors in combination with PARP inhibitors.
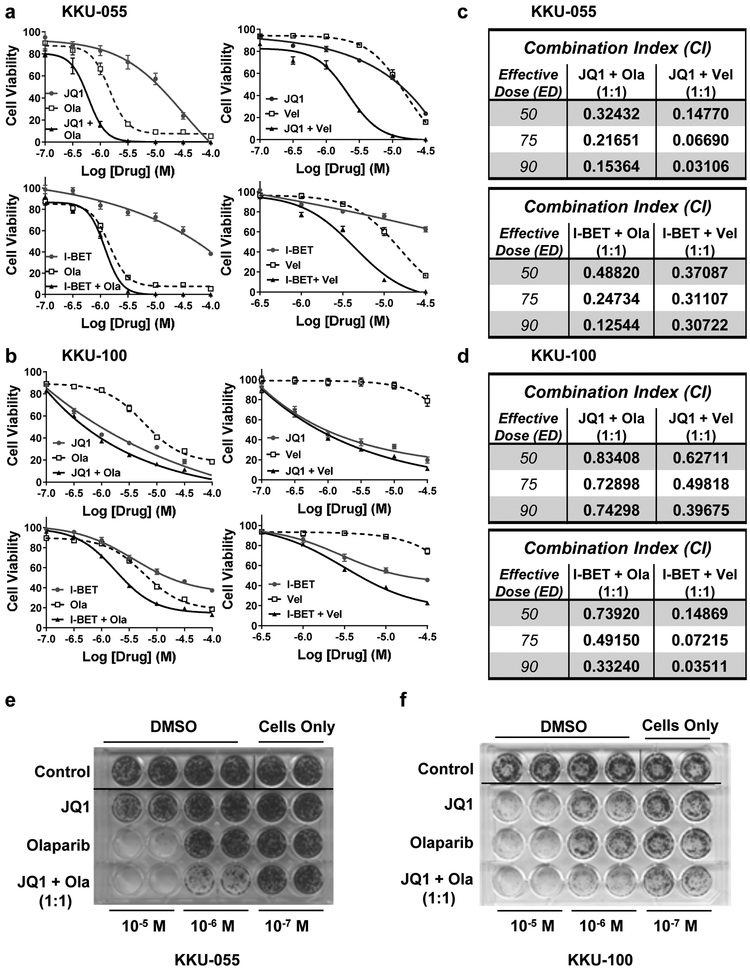
3.3. BETi + PARPi exerts synergistic cytotoxicity in CCA cell lines.
We assessed the potency of the BET inhibitors JQ1 or I-BET762 in combination with PARP inhibitors olaparib or veliparib in alamarBlue cell viability assays. We exposed KKU-055 and KKU-100 cells to a range of BETi + PARPi (10−7 M to 10−4 M) as single agents or the combination at a 1:1 ratio for 96 hours (Figs. 3a, 3b). JQ1 or olaparib as a single agent was more potent than I-BET762 or veliparib in both CCA cell lines. We also calculated combination indices (CI) using CompuSyn (1.0) software, based on Chou and Talalay methodology (Figs.3c, 3d). Combination indices (CI) ranged from 0.03106 to 0.48820 for KKU-055 cells (Fig.3c) and from 0.03511 to 0.83408 for KKU-100 cells (Fig.3d). All indices indicate synergy for all four combinations evaluated. Using clonogenic assays, we observed that the combination of JQ1 + olaparib was more effective than either drug alone in both CCA cell lines (Figs. 3e, 3f). Further, we evaluated the impact of JQ1 ± olaparib or veliparib on levels of protein markers for DNA damage (γH2AX) and apoptosis (cleaved PARP). As shown in Fig.S3, the combinations increased the levels of cleaved PARP >100-fold compared to DMSO controls. We also performed cell cycle analysis using JQ1 ± olaparib or veliparib (Fig.S4). Results agree with data in the literature demonstrating that JQ1 arrests cells in G1; olaparib arrests cells in G2; and veliparib has little, if any, effect on cell cycle distribution [32, 33]. Interestingly, the combination of JQ1 + olaparib arrests cells in G2- similar to olaparib as a single agent and in contrast to JQ1 as a single agent. JQ1 + veliparib had little effect on cell cycle distribution.
Figure 3. A combination of BET and PARP inhibitors are synergistic in KKU-055 and KKU-100 CCA cell lines.
KKU-055 (a) and KKU-100 (b) CCA cell lines were exposed to 1:1 concentration ratio of BET inhibitors (JQ1 or I-BET762) ± PARP inhibitors (olaparib or veliparib) for 96 hours. Cell viability was assessed by alamarBlue as described in Materials and Methods. Data are presented as mean ± S.E.M, in a minimum of three independent experiments with quadruplicated wells. Combination indices (CI) were calculated for (a) and (b) using CompuSyn and presented at ED50, ED75 and ED90 in (c) for KKU-055 and (d) for KKU-100 cell lines. Clonogenic assays for KKU-055 (e) and KKU-100 (f) cell lines were done as described in Materials and Methods, and representative images are shown. Three independent experiments were performed.
Together, the data demonstrate that combinations of a BETi with a PARPi are synergistic in KKU-055 and KKU-100 CCA cells, and that observed synergy was seen with multiple combinations of these classes of agents.
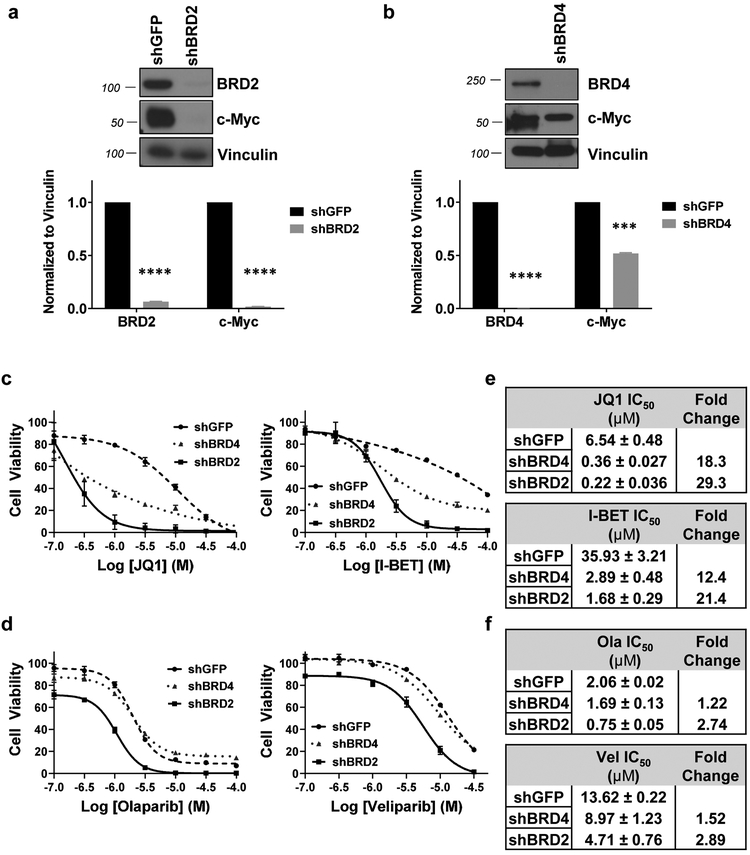
3.4. c-Myc expression was BET-dependent.
Studies in the literature report that BRD4 and BRD2 contribute to regulation of c-Myc expression in multiple cancer types including lung cancer cells, acute leukemia cells, and multiple myeloma cells [25, 34, 35]. We hypothesized that down-regulation of either of these BET proteins would decrease c-Myc expression. To downregulate BRD2 or BRD4 expression, we transfected CCA cells with a shRNA plasmid targeting BRD2 (shBRD2), BRD4 (shBRD4) or GFP (shGFP, negative control). When BRD2 expression was decreased by >95%, we observed a concomitant >95% decrease in c-Myc expression (Fig. 4a). When BRD4 was decreased by >98%, c-Myc expression is also down-regulated by ~50% (Fig. 4b). Our data indicate that c-Myc expression is BRD2- or BRD4-dependent in CCA tumor cells, a novel finding in CCA cells.
Figure 4. Decreased expression of BRD2 (shBRD2) or BRD4 (shBRD4) increased the sensitivity of KKU-055 cells to BET inhibitors (JQ1 or I-BET762) or PARP inhibitors (olaparib or veliparib).
Expression of c-Myc was BRD2 (a) or BRD4 (b) dependent. Quantitation of immunoblots (IB) were done and presented as bar graphs below each IB blots. We used shRNA (see details in Materials and Methods and Table S1 for sequences) to decrease expression of BRD2 (shBRD2) or BRD4 (shBRD4) in KKU-055 cells. (c) Decreased expression of BRD2 or BRD4 increased the sensitivity of KKU-055 cells to JQ1 (left panel) or I-BET762 (right panel). IC50 values were calculated using GraphPad Prism 7 and shown in (e). (d) Decreased expression of BRD2 or BRD4 increased the sensitivity of KKU-055 cells to olaparib (left panel) or veliparib (right panel). IC50 values were calculated using GraphPad Prism 7 and shown in (f). A minimum of three independent experiments were performed and IC50 values presented as mean ± S.E.M.
3.5. Decreased expression of BRD2 or BRD4 increased the sensitivity of CCA cells to BET inhibitors.
To assess whether contrasting levels of expression of JQ1 targets BRD2 or BRD4 affected the sensitivity of CCA cells to JQ1, we exposed shBRD2 and shBRD4 transfectants to a range of JQ1 concentrations (10−7 M to 10−4 M) for 96 hours and compared the viability of shBRD2 and shBRD4 transfectants to control shGFP transfectants (Fig. 4c). We observed that shRNA-transfected KKU-055 cells with lower levels of BRD2 or BRD4 were ~18- to 29-fold more sensitive to JQ1 than shGFP control transfectants (Figs. 4c, 4e). Similar results were observed with the BET inhibitor I-BET762 (Figs. 4c, 4e). BET inhibitors target BET family members BRD2, BRD3, BRD4 and BRDT, with each protein likely contributing to the expression of a different subset of gene products. When one protein is downregulated, the others still comprise molecular targets for BET inhibitors. We interpret the data to indicate that cells with low level BET protein expression are more sensitive to BET inhibitors than cells with high level BET expression. We also observed that shBRD2 transfectants showed less than 3-fold increase in sensitivity to olaparib and veliparib, and shBRD4 transfectants less than 1.5-fold increase in sensitivity to these PARPi (Figs. 4d, 4f). When combined with PARPi, neither shBRD2 nor shBRD4 transfectants reflect the synergistic cytotoxicity we observed with BETi + PARPi in parental CCA cells. Another question we asked was whether BRD2 and BRD4 can complement each other to induce c-Myc expression. We performed chromatin immunoprecipitation (ChIP) assays to examine whether downregulation of one BET protein would alter the binding between the other BET protein and the promoter region of MYC. As shown in Supplemental Fig.S5, we observed no compensatory binding of BRD2 to the MYC promoter when BRD4 is downregulated and no compensatory binding of BRD4 to the MYC promoter when BRD2 is downregulated.
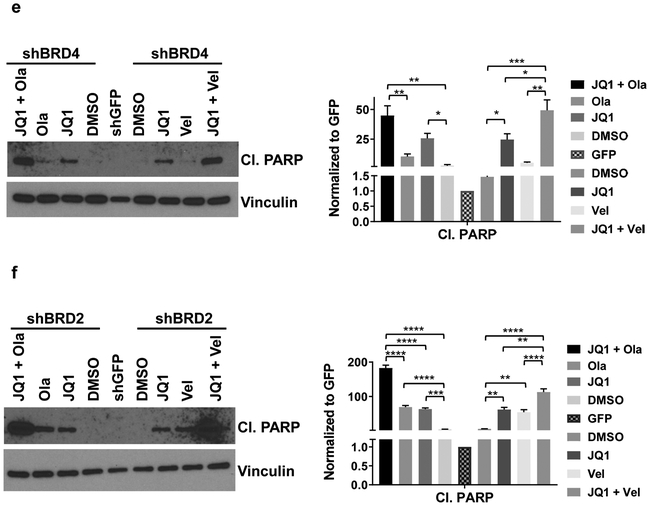
3.6. shBRD2 and shBRD4 transfectants were more sensitive to BETi + PARPi combinations than control transfectants.
Results with JQ1 in shBRD2 and in shBRD4 transfectants predicted that a combination of BETi+ PARPi might be more effective than either drug alone. Data assessing the effect of JQ1 + olaparib, JQ1 + veliparib, I-BET762 + olaparib and I-BET762 + veliparib on cell viability in shBRD2 (Fig. 5a) and shBRD4 (Fig. 5b) were consistent with this hypothesis. We calculated CI values and shown in Figs. 5c. 5d. CI values indicated synergy for all combinations (Figs. 5c, 5d). We also evaluated the impact of JQ1 ± PARPi (olaparib or veliparib) on levels of cleaved PARP, a marker for apoptosis, in shBRD4 and shBRD2 cells (Figs. 5e, 5f). The data show that in both shBRD4 and shBRD2 transfected cells, JQ1 increased the levels of the apoptosis marker cleaved PARP. Further, JQ1 + olaparib or veliparib increased levels of this apoptotic marker more than JQ1 as a single agent. We concluded that CCA cells expressing relatively low levels of BRD2 or BRD4 were more sensitive to the effects of a BETi + PARPi than cells expressing higher levels of these BET proteins, as reflected by induction of increased levels of cleaved PARP.
Figure 5. shBRD2 and shBRD4 transfectants were more sensitive to combinations of BETi + PARPi than shGFP (control) transfectants.
Simultaneous exposure of BET inhibitors (JQ1 or I-BET762) + PARP inhibitors (olaparib or veliparib) induced synergistic cytotoxicity in BRD2 downregulated (shBRD2) (a) or BRD4 downregulated (shBRD4) (b) KKU-055 cells. Cells were exposed to the indicated concentrations of BETi ± PARPi (1:1 ratio) for 96 hours, alamarBlue solution was added, and fluorescence read. Data were normalized to DMSO controls at each time point, with control values = 100%. Each point represents the mean of quadruplicated wells from a minimum of three independent assays. Data are presented as mean ± S.E.M. Combination indices (CI) were calculated for (a) and (b) using CompuSyn and presented at ED50, ED75 and ED90 in (c) for shBRD2 and (d) for shBRD4 KKU-055 cells. (e) shBRD4 or (f) shBRD2 transfectants were exposed to the IC50 values (listed in Figs.4e and 4f) of JQ1, olaparib, veliparib, JQ1 + olaparib or JQ1 + veliparib for 48 hours. Cell lysates were harvested and immunoblots performed. Quantitation and statistics are shown on the right side of the panel. The values presented as mean ± S.E.M analyzed using Prism (one-way ANOVA). *P<0.05, **P<0.01. ***P<0.001, ****p<0.0001.
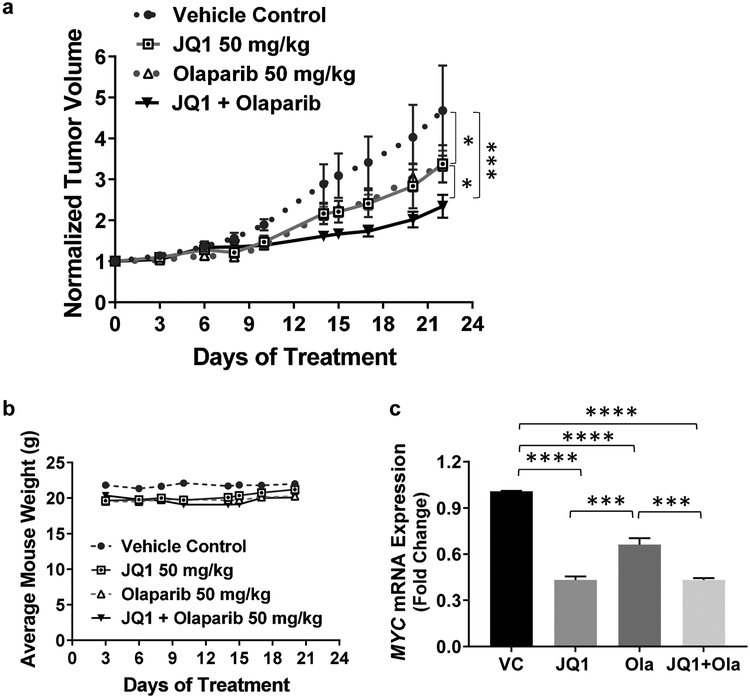
3.7. JQ1 + olaparib suppresses tumor growth in an in vivo KKU-055 CCA model.
Data above indicate that, of the combinations evaluated, KKU-055-derived tumors would be predicted to be sensitive to the combination of JQ1 + olaparib. We addressed this hypothesis directly by injecting 5 million KKU-055 cells, suspended in PBS, into the flanks of SCID mice (Taconic Farms), and allowed tumors of ~200mm3 volume to develop. Tumor-bearing mice were injected intraperitoneally with vehicle control (VC), 50 mg/kg JQ1, 50 mg/kg olaparib or the combination once a day for 21 days. We observed significant differences between each treatment group and the VC group (P<0.001). Importantly, we also observed that JQ1 + olaparib was more effective than JQ1 or olaparib as a single agent (P≤0.05) (Fig.6a), with the nontoxic regimen used for the study (Fig.6b). We also verified that, consistent with in vitro data, JQ1 and the combination of JQ1 + olaparib decreased MYC expression compared to VC (P<0.0001) (Fig. 6c). Interestingly, olaparib reduced the expression of MYC mRNA compared to vehicle control. This finding is consistent with previous reports [36, 37]. Potential explanations for this observation include: 1) PARP-1 could bind directly to the promoter region of MYC to regulate MYC expression; or 2) PARP-1 could interact with E2F-1, and this interaction increases promoter activity of E2F-1 and expression of E2F-1 responsive genes such as MYC.
Figure 6. JQ1 + olaparib suppresses tumor growth in an in vivo KKU-055 CCA model.
(a) Tumor-bearing mice were treated with JQ1 (50 mg/kg daily, i.p.), olaparib (50 mg/kg daily, i.p.), or JQ1 + olaparib, or vehicle (VC daily) for 21 days. Tumor volumes were measured three times per week. Tumor volumes (mm3) were normalized to tumor volumes on Day 0 for each tumor. P values were calculated by two-way ANOVA followed by Tukey post test. *P≤0.05, ***P<0.001. (b) Average body weight per mouse per each treatment group during the treatment period was within 13% for all treatment groups. Average mouse weight was calculated by weighing each cage of mice and dividing by the number of mice per cage. (c) qRT-PCR shows that JQ1 as well as JQ1 + olaparib inhibited the mRNA expression of MYC in an in vivo KKU-055 CCA model. A minimum of three biological experiments were performed and values presented as mean ± S.E.M. ***P<0.001, ****P<0.0001.
In vitro and in vivo data indicate that the combination of BETi + PARPi merits further investigation as potentially effective treatment for patients with CCA tumors.
4. Discussion
This study assesses the anti-proliferative effects of BET bromodomain inhibitors (JQ1 or I-BET762) in combination with PARP inhibitors (veliparib or olaparib) in CCA models in vitro and the efficacy of JQ1 + olaparib in a CCA xenograft model. Each combination of a BETi + PARPi induced synergistic cytotoxicity (CI<1) in both KKU-055 and KKU-100 CCA cell line models. Further, the combination of JQ1 + olaparib had a greater effect than either drug alone in the in vivo model. Mechanistically, JQ1 reduced expression of c-Myc and Chk1 in dose and time dependent manners in vitro. The data demonstrate that BRD2 and BRD4 contribute to the regulation of c-Myc expression in CCA cells, a novel finding in CCA tumor models, and that JQ1 + olaparib have efficacy in preclinical models of this tumor type.
The likely mechanism by which JQ1 decreases c-Myc expression has been postulated to involve competitive inhibition of the K-Ac binding function of the BET protein BRD4 [38-40]. It follows that inhibition of Chk1 expression would be due to inhibition of c-Myc expression, since CHEK1 is a c-Myc transcriptional target. Relevant to these findings, data in the Cancer Genome Atlas (TCGA) suggest that a relatively high c-Myc expression (z-score threshold = ± 2) in CCA tumors correlates with shorter overall survival in CCA (P<0.001) [41], suggesting c-Myc as a potential therapeutic target. Our data demonstrate that JQ1 decreased c-Myc expression, and we propose that BETi may have utility for treatment of this tumor type.
Of particular interest was our finding that shRNA transfectants expressing relatively low levels of BRD2 or BRD4 were relatively sensitive to JQ1 or I-BET762. These data suggest that tumor cells with lower BRD2 or BRD4 expression may be more sensitive to BET inhibitors than cells expressing relatively high levels of these BETi molecular targets. Notably, JQ1 and I-BET762 inhibit all four BET protein family members (BRD2, BRD3, BRD4, BRDT) [38], with varying binding affinities [7]. We postulate that the efficacy of JQ1 depends on simultaneous inhibition of BRD2 and BRD4 in CCA tumors. We further observed that shBRD2 model shows a <3-fold increase in sensitivity to olaparib and veliparib, while shBRD4 model shows <1.5-fold increase in sensitivity to these PARPi. However, when exposed to PARPi neither shBRD2 nor shBRD4 transfectants recapitulate the synergistic cytotoxicity like phenotype observed in parental KKU-055 when exposed to the combinations of BETi with PARPi. Generation of a model with decreased expression of all BET targets, BRD2, BRD3 and BRD4, simultaneously may address our current hypothesis that the observed potency of BETi and synergy between BETi and PARPi depends upon inhibition of multiple members of the BET family as opposed to a single member.
Our data demonstrate that JQ1 decreased c-Myc and increased the levels of DNA damage and apoptosis, and that BETi JQ1 sensitized CCA tumor cells to PARPi olaparib. Further, in addition to inhibiting expression of c-Myc and Chk1, BETi also decreases levels of multiple genes including Ku80, RAD51, BRCA1, WEE1, CDC25B or TOPBP1, that contribute to the DNA damage repair and response [10, 17, 21, 42]. The resulting increase in the levels of DNA damage and apoptosis would be anticipated to sensitize tumor cells to PARPi. Our data indicate that BETi in combination with PARPi may be a promising strategy for CCA.
Supplementary Material
Highlights.
BETi + PARPi exerts synergistic cytotoxicity in cholangiocarcinoma in vitro.
JQ1 + olaparib inhibits growth of cholangiocarcinoma tumors in a preclinical model.
shRNA-mediated decrease in BETi molecular targets BRD2 or BRD4 increases the sensitivity of cholangiocarcinoma cells to BETi ± PARPi.
Acknowledgement
This work was supported by the National Institutes of Health (National Cancer Institute) grants R21CA205501 and R01CA208272 (K.J.Y). We thank Dr. Banchob Spira (Khon Kaen University) for establishment of both KKU-055 and KKU-100 CCA cell lines.
Grant Support: This work was supported by the National Institutes of Health (National Cancer Institute) grants R21CA205501 and R01CA208272 (K.J.Y).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: The authors declare no potential conflict of interest.
References
- [1].Doherty B, Nambudiri VE, Palmer WC, Update on the Diagnosis and Treatment of Cholangiocarcinoma, Current Gastroenterology Reports, 19 (2017) 2. [DOI] [PubMed] [Google Scholar]
- [2].Blechacz B, Cholangiocarcinoma: Current Knowledge and New Developments, Gut and Liver, 11 (2017) 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ramírez-Merino N, Aix SP, Cortés-Funes H, Chemotherapy for cholangiocarcinoma: an update, World J Gastrointest Oncol, 5 (2013) 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cadamuro M, Brivio S, Spirli C, Joplin RE, Strazzabosco M, Fabris L, Autocrine and Paracrine Mechanisms Promoting Chemoresistance in Cholangiocarcinoma, International Journal of Molecular Sciences, 18 (2017) 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Maroni L, Pierantonelli I, Banales JM, Benedetti A, Marzioni M, The significance of genetics for cholangiocarcinoma development, Annals of Translational Medicine, 1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Donati B, Lorenzini E, Ciarrocchi A, BRD4 and Cancer: going beyond transcriptional regulation, Mol Cancer, 17 (2018) 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Selective inhibition of BET bromodomains, Nature, 468 (2010) 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dawson MA, Gudgin EJ, Horton SJ, Giotopoulos G, Meduri E, Robson S, Cannizzaro E, Osaki H, Wiese M, Putwain S, Fong CY, Grove C, Craig J, Dittmann A, Lugo D, Jeffrey P, Drewes G, Lee K, Bullinger L, Prinjha RK, Kouzarides T, Vassiliou GS, Huntly BJ, Recurrent mutations, including NPM1c, activate a BRD4-dependent core transcriptional program in acute myeloid leukemia, Leukemia, 28 (2014) 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shi J, Vakoc CR, The mechanisms behind the therapeutic activity of BET bromodomain inhibition, Molecular Cell, 54 (2014) 728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Garcia PL, Miller AL, Gamblin TL, Council LN, Christein JD, Arnoletti JP, Heslin MJ, Reddy S, Richardson JH, Cui X, van Waardenburg R, Bradner JE, Yang ES, Yoon KJ, JQ1 Induces DNA Damage and Apoptosis, and Inhibits Tumor Growth in a Patient-Derived Xenograft Model of Cholangiocarcinoma, Mol Cancer Ther, 17 (2018) 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Patel AG, Sarkaria JN, Kaufmann SH, Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells, Proc Natl Acad Sci U S A, 108 (2011) 3406–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Min A, Im SA, Yoon YK, Song SH, Nam HJ, Hur HS, Kim HP, Lee KH, Han SW, Oh DY, Kim TY, O'Connor MJ, Kim WH, Bang YJ, RAD51C-deficient cancer cells are highly sensitive to the PARP inhibitor olaparib, Mol Cancer Ther, 12 (2013) 865–877. [DOI] [PubMed] [Google Scholar]
- [13].Basu B, Yap TA, Molife LR, de Bono JS, Targeting the DNA damage response in oncology: past, present and future perspectives, Current opinion in oncology, 24 (2012) 316–324. [DOI] [PubMed] [Google Scholar]
- [14].Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A, Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy, Nature, 434 (2005) 917–921. [DOI] [PubMed] [Google Scholar]
- [15].McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, Giavara S, O'Connor MJ, Tutt AN, Zdzienicka MZ, Smith GC, Ashworth A, Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition, Cancer Res, 66 (2006) 8109–8115. [DOI] [PubMed] [Google Scholar]
- [16].Patel AG, Flatten KS, Schneider PA, Dai NT, McDonald JS, Poirier GG, Kaufmann SH, Enhanced killing of cancer cells by poly(ADP-ribose) polymerase inhibitors and topoisomerase I inhibitors reflects poisoning of both enzymes, The Journal of biological chemistry, 287 (2012) 4198–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Miller AL, Fehling SC, Garcia PL, Gamblin TL, Council LN, van Waardenburg R, Yang ES, Bradner JE, Yoon KJ, The BET inhibitor JQ1 attenuates double-strand break repair and sensitizes models of pancreatic ductal adenocarcinoma to PARP inhibitors, EBioMedicine, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kreitzburg KM, Fehling SC, Landen CN, Gamblin TL, Vance RB, Arend RC, Katre AA, Oliver PG, van Waardenburg R, Alvarez RD, Yoon KJ, FTY720 enhances the anti-tumor activity of carboplatin and tamoxifen in a patient-derived xenograft model of ovarian cancer, Cancer Lett, 436 (2018) 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chou T-C, Drug combination studies and their synergy quantification using the Chou-Talalay method, Cancer Research, 70 (2010) 440–446. [DOI] [PubMed] [Google Scholar]
- [20].Franken NA, Rodermond HM, Stap J, Haveman J, Van Bree C, Clonogenic assay of cells in vitro, Nature Protocols, 1 (2006) 2315–2319. [DOI] [PubMed] [Google Scholar]
- [21].Garcia P, Miller A, Kreitzburg K, Council L, Gamblin T, Christein J, Heslin M, Arnoletti J, Richardson J, Chen D, The BET bromodomain inhibitor JQ1 suppresses growth of pancreatic ductal adenocarcinoma in patient-derived xenograft models, Oncogene, 35 (2016) 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Riccardi C, Nicoletti I, Analysis of apoptosis by propidium iodide staining and flow cytometry, Nature Protocols, 1 (2006) 1458–1461. [DOI] [PubMed] [Google Scholar]
- [23].Tepsiri N, Chaturat L, Sripa B, Namwat W, Wongkham S, Bhudhisawasdi V, Tassaneeyakul W, Drug sensitivity and drug resistance profiles of human intrahepatic cholangiocarcinoma cell lines, World journal of gastroenterology, 11 (2005) 2748–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, Fish P, Harsha B, Hathaway C, Jupe SC, Kok CY, Noble K, Ponting L, Ramshaw CC, Rye CE, Speedy HE, Stefancsik R, Thompson SL, Wang S, Ward S, Campbell PJ, Forbes SA, COSMIC: the Catalogue Of Somatic Mutations In Cancer, Nucleic Acids Res, 47 (2019) D941–d947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Da Costa D, Agathanggelou A, Perry T, Weston V, Petermann E, Zlatanou A, Oldreive C, Wei W, Stewart G, Longman J, BET inhibition as a single or combined therapeutic approach in primary paediatric B-precursor acute lymphoblastic leukaemia, Blood Cancer Journal, 3 (2013) e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hoglund A, Nilsson LM, Muralidharan SV, Hasvold LA, Merta P, Rudelius M, Nikolova V, Keller U, Nilsson JA, Therapeutic implications for the induced levels of Chk1 in Myc-expressing cancer cells, Clinical cancer research : an official journal of the American Association for Cancer Research, 17 (2011) 7067–7079. [DOI] [PubMed] [Google Scholar]
- [27].Wang WJ, Wu SP, Liu JB, Shi YS, Huang X, Zhang QB, Yao KT, MYC regulation of CHK1 and CHK2 promotes radioresistance in a stem cell-like population of nasopharyngeal carcinoma cells, Cancer Res, 73 (2013) 1219–1231. [DOI] [PubMed] [Google Scholar]
- [28].Dedes KJ, Wilkerson PM, Wetterskog D, Weigelt B, Ashworth A, Reis-Filho JS, Synthetic lethality of PARP inhibition in cancers lacking BRCA1 and BRCA2 mutations, Cell Cycle, 10 (2011) 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Underhill C, Toulmonde M, Bonnefoi H, A review of PARP inhibitors: from bench to bedside, Annals of Oncology, 22 (2011) 268–279. [DOI] [PubMed] [Google Scholar]
- [30].Donawho CK, Luo Y, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, Cox BF, DeWeese TL, Dillehay LE, ABT-888, an orally active poly (ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models, Clinical Cancer Research, 13 (2007) 2728–2737. [DOI] [PubMed] [Google Scholar]
- [31].Yin Y, Shen Q, Zhang P, Tao R, Chang W, Li R, Xie G, Liu W, Zhang L, Kapoor P, Chk1 inhibition potentiates the therapeutic efficacy of PARP inhibitor BMN673 in gastric cancer, American Journal of Cancer Research, 7 (2017) 473. [PMC free article] [PubMed] [Google Scholar]
- [32].Jelinic P, Levine DA, New insights into pARP inhibitors' effect on cell cycle and homology-directed DNA damage repair, Mol Cancer Ther, 13 (2014) 1645–1654. [DOI] [PubMed] [Google Scholar]
- [33].Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, Janiszewska M, Huh SJ, Liang Y, Ryan J, Doherty E, Mohammed H, Guo H, Stover DG, Ekram MB, Brown J, D'Santos C, Krop IE, Dillon D, McKeown M, Ott C, Qi J, Ni M, Rao PK, Duarte M, Wu SY, Chiang CM, Anders L, Young RA, Winer E, Letai A, Barry WT, Carroll JS, Long H, Brown M, Liu XS, Meyer CA, Bradner JE, Polyak K, Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer, Nature, 529 (2016) 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Pinz S, Unser S, Rascle A, Signal transducer and activator of transcription STAT5 is recruited to c-Myc super-enhancer, BMC molecular biology, 17 (2016) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Handoko L, Kaczkowski B, Hon CC, Lizio M, Wakamori M, Matsuda T, Ito T, Jeyamohan P, Sato Y, Sakamoto K, Yokoyama S, Kimura H, Minoda A, Umehara T, JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states, Epigenetics, (2018) 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mostocotto C, Carbone M, Battistelli C, Ciotti A, Amati P, Maione R, Poly(ADP-ribosyl)ation is required to modulate chromatin changes at c-MYC promoter during emergence from quiescence, PLoS One, 9 (2014) e102575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Simbulan-Rosenthal CM, Rosenthal DS, Luo R, Samara R, Espinoza LA, Hassa PO, Hottiger MO, Smulson ME, PARP-1 binds E2F-1 independently of its DNA binding and catalytic domains, and acts as a novel coactivator of E2F-1-mediated transcription during reentry of quiescent cells into S phase, Oncogene, 22 (2003) 8460–8471. [DOI] [PubMed] [Google Scholar]
- [38].Anders L, Guenther MG, Qi J, Fan ZP, Marineau JJ, Rahl PB, Lovén J, Sigova AA, Smith WB, Lee TI, Genome-wide localization of small molecules, Nature Biotechnology, 32 (2014) 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, BET bromodomain inhibition as a therapeutic strategy to target c-Myc, Cell, 146 (2011) 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Enomoto K, Zhu X, Park S, Zhao L, Zhu YJ, Willingham MC, Qi J, Copland JA, Meltzer P, Cheng S.-y., Targeting Myc as a therapeutic intervention for anaplastic thyroid cancer, The Journal of Clinical Endocrinology and Metabolism, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cerami J Gao U Dogrusoz BE Gross SO Sumer BA Aksoy A Jacobsen CJ Byrne ML Heuer E. Larsson, The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data, AACR, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Karakashev S, Zhu H, Yokoyama Y, Zhao B, Fatkhutdinov N, Kossenkov AV, Wilson AJ, Simpkins F, Speicher D, Khabele D, Bitler BG, Zhang R, BET Bromodomain Inhibition Synergizes with PARP Inhibitor in Epithelial Ovarian Cancer, Cell Reports, 21 (2017) 3398–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.