Abstract
Cell-based metabolomics was used in a proof-of-concept fashion to investigate the biological effects of contaminants as they traveled from a wastewater treatment plant (WWTP) discharge to a drinking water treatment plant (DWTP) intake in a surface-water usage cycle. Zebrafish liver (ZFL) cells were exposed to water samples collected along a surface-water flowpath, where a WWTP was located ∼14.5 km upstream of a DWTP. The sampling sites included: 1) upstream of the WWTP, 2) the WWTP effluent discharging point, 3) a proximal location downstream of the WWTP outfall, 4) a distal location downstream of the WWTP outfall, 5) the drinking water intake, and 6) the treated drinking water collected prior to discharge to the distribution system. After a 48-h laboratory exposure, the hydrophilic and lipophilic metabolites in ZFL cell extracts were analyzed by proton nuclear magnetic resonance (1H NMR) spectroscopy and gas chromatography-mass spectrometry (GC-MS), respectively. Multivariate statistical analysis revealed distinct changes in metabolite profiles in response to WWTP effluent exposure. These effects on the hydrophilic metabolome gradually diminished downstream of the WWTP, becoming non-significant at the drinking water intake (comparable to upstream of the WWTP, p = 0.98). However, effects on the lipophilic metabolome increased significantly as the river flowed from the distal location downstream of the WWTP to the drinking water intake (p < 0.001), suggesting a source of bioactive compounds in this watershed other than the WWTP. ZFL cells exposed to treated drinking water did not exhibit significant changes in either the hydrophilic (p = 0.15) or lipophilic metabolome (p = 0.83) compared to the upstream site, suggesting that constituents in the WWTP effluent were efficiently removed by the drinking water treatment process. Impacts on ZFL cells from the WWTP effluent included disrupted energy metabolism, a global decrease in amino acids, and altered lipid metabolism pathways. Overall, this study demonstrated the utility of cell-based metabolomics as an effective tool for assessing the biological effects of complex pollutant mixtures, particularly when used as a complement to conventional chemical monitoring.
Keywords: Metabolomics, Cell culture, Wastewater treatment plant effluent, Potable reuse, Water quality
1. Introduction
The past several decades have witnessed a rapidly increasing demand for water resourcesdue to global economic development and an ever-increasing population. In many densely populated or arid regions, the lack of sufficient potable water has led to the use of treated wastewater (e.g., discharges from wastewater treatment plants (WWTPs)) for many non-potable applications, such as agricultural and landscape irrigations and groundwater recharge (USDA, 2013; USEPA, 2004). Unfortunately, the discharge of treated wastewater into natural systems, e.g., rivers and lakes, has also inadvertently resulted in introduction of a complex mixture of organic contaminants to local aquatic ecosystems and, importantly, surface-water derived drinking water supplies. This use of treated wastewater (after mixing with receiving waters) has been referred to as de facto (unplanned) potable reuse by a National Academy of Engineering study (NRC, 2012). It was estimated recently that 50% of 2056 surface water intakes that serve 1210 drinking water treatment plants (DWTPs) across the U.S. are potentially affected by upstream WWTP discharges (Rice and Westerhoff, 2015).
WWTP effluent is one of the main sources of contaminants of emerging concern (CECs) to surface waters (Kolpin et al., 2002). CECs comprise a wide variety of chemicals including pharmaceuticals and personal care products (PPCPs), endocrine disrupting compounds (EDCs), disinfection by-products (DBPs), antibiotics, and pesticides. Many CECs have been reported to cause adverse ecotoxicological effects at environmentally relevant concentrations, raising concerns over their presence in receiving waters (David et al., 2018; Kolpin et al., 2002). Adding to these concerns are the growing number of instances in which these same surface waters are used as drinking water sources (i.e., de facto water reuse). To date, there have been a limited number of studies demonstrating the influence of WWTP effluent on the quality of drinking water sources (Boleda et al., 2009; Guo and Krasner, 2009; Rodayan et al., 2016). For example, under extremely low streamflow conditions, some DWTP intakes could contain almost 100% WWTP discharge, that is, complete de factoreuse (Rice et al., 2013), and the concentrations of some PPCPs at DWTP intakes have been comparable to those from upstream WWTP effluents (Guo and Krasner, 2009). This is potentially concerning given that other studies have shown incomplete (<90%) removal of many CECs during the drinking water treatment process, including benzoylecgonine, 2-ethylidene-1,5-dimethyl-3,3-diphenylpyrrolidine, methadone, cocaine, methylenedioxyamphetamine, ephedrine, and several prescription opioids (Boleda et al., 2009; Rodayan et al., 2016).
The presence of these and many other CECs in streams receiving WWTP effluent, coupled with the potential for their incomplete removal during the drinking water treatment process, poses a signifcant and increasing challenge to chemical risk assessment. Indeed, the vast number of chemicals in wastewater has far exceeded our analytical capability based on individual targeted analysis (NRC, 2007). Moreover, there remains a dearth of information regarding the toxic modes-of-action (MOAs) of most targeted chemicals; this implies an even greater level of uncertainty when unidentified contaminants are considered (Judson et al., 2009). The limited understanding of how individual chemical toxicities are modified when chemicals occur in very complex mixtures at environmental conditions further complicates the risk assessment process (Schwarzenbach et al., 2006). To address these and other limitations of the conventional targeted chemical monitoring approach, a variety of biological effects-based tools have emerged and have increasingly been used in water quality assessment (Escher and Leusch, 2012). These tools include bioassays that target specific biological activities such as estrogenic or androgenic receptor binding (Escher and Leusch, 2012). More recently, “-omics” tools that analyze gene transcripts, proteins, and/or endogenous metabolites have gained popularity for MOA assessments of individual chemicals, as well as complex environmental mixtures (Davis et al., 2017; Skelton et al., 2014; Xia et al., 2017). These effects-based tools are particularly attractive as a complement to conventional chemical monitoring because they are both open-ended (i.e., untargeted), and they provide insights into the biological impacts of complex mixtures even when the individual contaminants cannot be uniquely identified or quantified.
Due to advantages that have been discussed elsewhere (Bundy et al., 2009; Lankadurai et al., 2013), metabolomics has been shown to be a particularly useful “-omics” approach for evaluating comprehensive biological responses to various environmental stressors. For example, this approach has been extensively employed to investigate the MOAs of various xenobiotics, including nanoparticles (Ratnasekhar et al., 2015), PPCPs (Gomez-Canela et al., 2016), and EDCs (Davis et al., 2017), through laboratory-based animal (e.g., rodents, fish, and invertebrates) exposures. In addition, field-based metabolomics assessments using on-site exposure of fish have been used successfully to investigate the biological effect of complex contaminant mixtures discharging from both point (e.g., WWTP effluent) and non-point sources (Davis et al., 2013; Skelton et al., 2014). However, metabolomics studies that employ live animals are labor- and resource-intensive, and typically require lethal sampling to collect the relevant tissues and/or biofluids. In an effort to circumvent these drawbacks, we previously developed a cell culture-based metabolomics method and applied it to investigate the effect(s) of exposure to 17α-ethynylestradiol on zebrafish (Danio rerio) liver (ZFL) cells (Teng et al., 2013). Our findings showed that the cellular responses agreed with those metabolite changes observed in vivo (Teng et al., 2013). This suggested that cell-based metabolomics approaches have the potential of providing valuable ecotoxicological information in a particularly high-throughput and cost-effective manner.
In the current study, the ZFL cell-based metabolomics approach was extended to investigate the biological effect(s) of a WWTP effluent discharged into a stream and further downstream to a DWTP in a de facto potable reuse cycle. Previously, zebrafish has been identified as a model vertebrate for investigating chemical toxicity both in vivo and in vitro and proven highly relevant for both human and ecological health (Dai et al., 2014). The primary goal of this study was to evaluate the effectiveness of cell-based metabolomics for tracking the biological effects of contaminants as they travel from the WWTP to the corresponding downstream DWTP, as well as the final drinking water product. A second goal was to test the applicability and efficacy of cell-based metabolomics for ecological assessments of complex environmental mixtures, and to identify relationships between metabolomics endpoints and conventional contaminant detections.
2. Materials and methods
2.1. Sampling locations
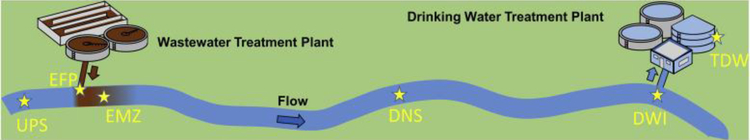
In April 2015, water samples were taken from a stretch of a river in the northeastern USA where a WWTP is located 14.5 km upstream of the intake of a DWTP. The relative locations of all six sampling sites are shown in Fig. 1. Grab samples were collected upstream (UPS) of the WWTP, at the effluent pipe (EFP) of the WWTP, a proximal location downstream of the WWTP outfall (EMZ) where the effluent is well mixed with the river (as demonstrated by conductivity), a distal location downstream of the WWTP outfall (DNS), at the drinking water intake (DWI), and from treated drinking water (TDW) collected from the clear well just prior to discharge to the distribution system. The UPS site was located 1.2 km upstream of EFP, while EMZ, DNS, and DWI were 0.7, 9.1 and 14.5 km, respectively, downstream from EFP. The travel times of water from UPS to: EFP, EMZ, DNS, DWI and TDW (including residence times in the DWTP) were estimated to be 0.8, 1.3, 7.2, 10.9 and 18.9 h, respectively. No precipitation was reported during the period of sampling (Susan Glassmeyer, personal communication, October 24th, 2017). The WWTP effluent flow (1.7 ft3/s) was less than 1% of the flow of river water (1179 ft3/s, average of measurements at three different locations along this segment of river during sample collection), indicating a low magnitude of de factopotable reuse in the study area (Rice and Westerhoff, 2015). All samples were collected in 1-L amber glass bottles, placed immediately on ice, shipped overnight, and stored at −20 °C. Additional descriptions of the WWTP and DWTP are provided in Supporting Information.
Fig. 1.
Schematic diagram (not to scale) showing relative locations of the sampling sites along the river. Site designations are: upstream of effluent (UPS), WWTP effluent discharging point (EFP), a proximal location downstream of the WWTP outfall (EMZ), a distal location downstream of the WWTP outfall (DNS), drinking water intake (DWI), and treated drinking water (TDW).
2.2. Culturing of ZFL cells and exposure experimental design
ZFL cells (CRL-2643, ATCC, Manassas, VA) were cultivated at 28 °C in a humidified atmosphere as monolayer cultures in medium (see the Supporting Information for medium composition) prepared according to ATCC’s instructions (www.atcc.org). To prepare ZFL cells for exposure, four T75 flasks of cells were harvested after they reached 100% confluence. Cells were combined and evenly distributed to 49 culture dishes (there were seven biological replicates for each of six sampling locations and the laboratory control) for precultivation at 28 °C for 48 h. For cell exposure experiments, the cell culture medium was displaced with the prepared mixed exposure medium made from the field-collected grab samples. Specifically, a medium based on the composition as described in the Supporting Information, but at a four-fold higher concentration was prepared. This concentrated medium was mixed in a ratio of 1:3 with a filtered (0.1 μm) water sample from each field site, or with purified water (ultrapure type 1, 18.2 MΩ cm, 0.1-μm filtered, Aqua Solutions, Jasper, GA, USA) that served as the laboratory control (CON). Thus, the final concentrations of environmental samples in the exposure medium were 75% of the concentrations in the initial field-collected water samples. Cells were then incubated at 28 °C for 48 h before quenching. Cytotoxicity of water samples was tested based on a previously published protocol (Feoktistova et al., 2016). No significant (p < 0.05) cytotoxic effects were observed for cells exposed to environmental samples vs. CON (see the Supporting Information for details of the assay).
2.3. Extraction and analysis of intracellular metabolites
A direct quenching method that was developed in our laboratory was applied to quench ZFL cells after the 48-h exposure (Teng et al., 2009). The intracellular metabolites were further extracted using a dual-phase method that was published previously (Teng et al., 2013). A bi-phasic mixture was generated at the end of extraction and further separated into two fractions that contained mainly hydrophilic and lipophilic metabolites, respectively. Both fractions of cell extracts were dried using a vacuum concentrator (Thermo Electron, Waltham, MA, USA). After that, the hydrophilic and lipophilic fractions of cell extracts from individual biological replicates were analyzed with an Inova 600 MHz NMR spectrometer(Agilent, Santa Clara, CA, USA) and a gas chromatograph connected to a single quadrupolemass spectrometer in full scan mode (Thermo Trace 1310 and ISQ, Waltham, MA, USA), respectively. Additional descriptions of metabolite extraction and NMR and GC-MS data acquisitions are provided in the Supporting Information.
2.4. NMR data processing
The NMR spectra were processed with 0.3-Hz apodization. After removing the residual solvent, the spectra were binned at a width of 0.005 ppm within the range of 0.50–10.00 ppm. The binned data were imported into Microsoft® Excel (Microsoft Corporation, Redmond, WA, USA) and normalized to unit total integrated intensity for subsequent analysis.
2.5. GC-MS data processing
The MetAlign software package (Wageningen University, Netherlands) was used to perform baseline correction, background and noise reduction, peak smoothing and selection, and retention time alignment with GC–MS chromatograms (Lommen and Kools, 2012). Peaks with signal-to-noise (S/N) ratios less than three were rejected. Ion groups were assembled into metabolites using the MSClust software package (Wageningen University, Netherlands) and unique ions for each metabolite were used for relative quantification (Tikunov et al., 2012). Artifact peaks arising from the derivatization reagent, column bleed, as well as peaks observed in method blanks were excluded. Only metabolites consistently detected in at least 80% of samples in at least one of the six sites or CON were retained. Metabolites with relative standard deviations (RSD) of greater than 30% in the quality control samples were excluded from the final dataset. Putative annotation of metabolites was conducted by comparing the retention index (Δ of ±10) and EI mass spectrum (similarity ≥ 70%) to those recorded in the NIST05, Wiley 7, and Golm Metabolome Database mass spectral libraries or those obtained from the standards injection. Following these steps, the final dataset contained a total of 107 unique metabolites, 32 of which were annotated (5 at Level I and 27 at Level II as defined by the Metabolomics Standard Initiative (MSI, www.metabolomics-msi.org)). Normalized abundance relative to a constant sum of all detected metabolites was performed for each sample prior to statistical analyses.
2.6. Statistical analysis
Multivariate analyses with NMR and GC-MS data were conducted with SIMCA version 14.1 (Umetrics, Umea, Sweden). The NMR data were mean-centered and pareto-scaled while the GC-MS data were mean-centered and scaled to unit variance. Principal component analysis (PCA) models were built with all data points, and outliers were defined as those points that fell out of the 95% confidence interval of a Hotellings T2 test. ANOVA with Tukey’s honest significant difference (HSD) post-hoc test was performed for multiple-group comparison of PCA scores on the first and second components.
For analysis of the NMR and GC-MS data, a Student’s t-test was performed to compare the normalized abundance of individual metabolites between a given site and the control. Using the results of this analysis, we generated t-test filtered difference spectra by subtracting averaged relative abundances of the control from those of a given site. In addition, we calculated the fold change of lipophilic metabolites that exhibited significant difference between a sampling site and the control. Comparisons were also carried out with a selection of lipophilic metabolites to investigate the abundance change across all sites by applying ANOVA with Tukey’s HSD post-hoc test. Overall, a significance level of 0.05 was set for all statistical tests in this study. More details on generating these difference spectra, including the strategies for controlling false positives with multiple t-tests, and across-site comparisons are provided in Supporting Information. A bioactivity network was built based on significantly changed (p < 0.05) and annotated metabolites using the open-source software Cytoscape (www.cytoscape.org). All data generated or analyzed during this study are included in the main text and Supporting Information.
3. Results and discussion
3.1. Comparison of metabolite profiles of ZFL cells following exposure to water from each site
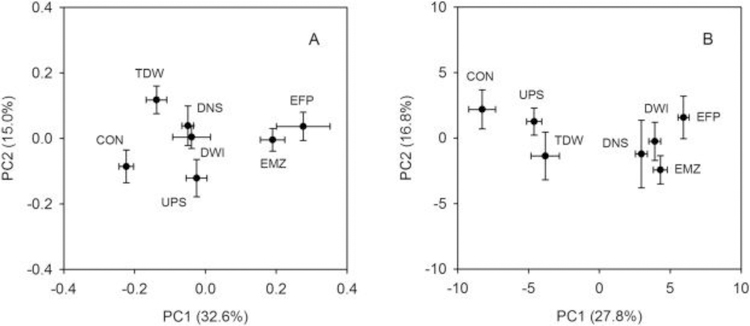
PCA score plots displayed an overall alteration of metabolite profiles of ZFL cells following exposure to field water samples relative to controls (Fig. 2). Distinct separations across several of the sampling sites was achieved in the first component of PCA score plots for both hydrophilic (Fig. 2A) and lipophilic (Fig. 2B) sample extracts. ANOVA with post-hoc (Tukey’s HSD) analysis was applied to evaluate the significance of metabolite profile differences between individual sites using the PCA scores of the first two components (Table S1).
Fig. 2.
Two-component score plots of hydrophilic (A) and lipophilic (B) ZFL cell extracts analyzed with NMR and GC-MS, respectively. Each marker is the mean of the score values for each location with its associated standard error. Site designations are: upstream of effluent (UPS), WWTP effluent discharging point (EFP), a proximal location downstream of the WWTP outfall (EMZ), a distal location downstream of the WWTP outfall (DNS), drinking water intake (DWI), treated drinking water (TDW), and lab control (CON).
Water collected from the UPS site shows discernible differences in metabolome profiles of ZFL cells relative to CON in both score plots (PC1: p = 0.039 in Fig. 2A, p = 0.005 in Fig. 2B). This is perhaps not surprising since there are several known potential sources of contamination (i.e., WWTPs) located upstream of UPS. In addition, as with all field-collected waters, this sample contained natural constituents such as dissolved organic matter (DOM) that were absent in CON. It is reasonable to suspect that these natural constituents were also contributing to the cellular metabolic changes observed at UPS; for example, DOM in natural waters was shown to have a significant effect on the metabolic energy status of Daphnia pulex-pulicaria (Taylor et al., 2016). However, it is notable that in other similar field studies we have seen no separation in PC1 for score plots similarly generated by NMRanalysis of the polar fraction of a relatively pristine field water vs. a similarly prepared lab control (Quincy Teng, personal communication, July 25th, 2018). This suggests that the known potential sources of contamination located upstream from the current study sites are, indeed, contributing to the metabolite marker profiles at UPS.
The water sample collected at EFP reflected the most distinct change in metabolome profile among all sites based on the extent of its separation from CON along PC1 in both hydrophilic and lipophilic score plots. This effect decreased, but not significantly at EMZ, which was proximal below the outfall (PC1: p = 0.77 in Fig. 2A, p = 0.57 in Fig. 2B, EMZ vs.EFP) and where dilution of effluent with river water had taken place. As water flowed further downstream, the ZFL cells exhibited different patterns of change in the hydrophilic metabolome as compared to the lipophilic metabolome. Specifically, the effects of DNS and DWI observed in the hydrophilic extracts significantly decreased relative to the effect of EMZ (Fig. 2A, PC1: p = 0.007 for DNS vs. EMZ and p = 0.007 for DWI vs. EMZ). In fact, the PC1 score values for DNS and DWI were coincident with those of UPS for the score plot generated from the hydrophilic metabolome (Fig. 2A). Conversely, the effects observed in the DNS and DWI lipophilic extracts were not significantly different from that of EMZ (Fig. 2B, PC1: p = 0.799 for DNS vs. EMZ and p = 1 for DWI vs. EMZ) and were significantly greater than that of UPS (PC1: p < 0.001 for DNS vs. UPS and p < 0.001 for DWI vs. UPS). Finally, the effect of TDW on the ZFL metabolome was found to be generally comparable to that of UPS based on PC1 score values in both PCA plots. However, a statistically significant difference was observed when comparing PC1 scores of TDW and CON from the score plot for the lipophilic metabolome (Fig. 2A, p < 0.001), but not in the plot for the hydrophilic metabolome (Fig. 2B, p = 0.78). TDW and CON score values were significantly different along PC2 for the hydrophilic metabolome (p = 0.04), but not for the lipophilic metabolome (p = 0.691). It seems reasonable, although untested, that this difference in metabolite profiles between CON and TDW could be caused by the ubiquitous occurrence of DBPs in treated drinking water (Richardson and Ternes, 2014). For example, similar findings were also reported in an earlier study showing higher baseline toxicity and neurotoxicity in treated drinking water compared with purified water when testing with a variety of bioanalytical tools (Macova et al., 2011).
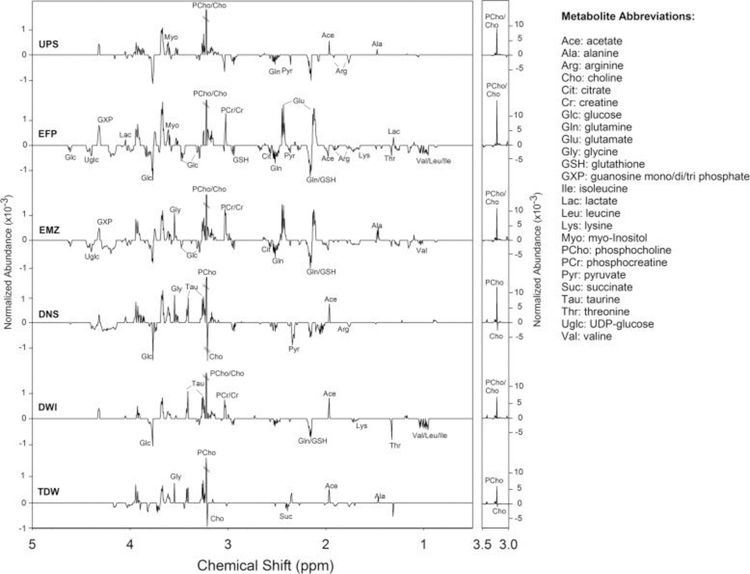
To further investigate the site-specific effects on both the ZFL hydrophilic and lipophilic metabolomes, we focused only on those metabolites that exhibited a statistically significant change in normalized abundance versus that of the controls (p < 0.05). The t-test filtered difference spectra were generated for the hydrophilic extracts (Fig. 3 and S1), as described in the Materials and Methods section. Consistent with the PCA score plot (Fig. 2A), the EFP and EMZ sites (which are directly or indirectly affected by WWTP effluent) exhibited a similar pattern of changes in the difference spectra with respect to the identity of changed hydrophilic metabolites and the magnitude of those changes. The effects at other field sites were less than those observed at EFP and EMZ based on the number and magnitude of altered metabolite peaks.
Fig. 3.
The t-test filtered difference spectra (exposed minus control) generated from binned 1H NMRspectra (0.5–5.0 ppm in chemical shift) of hydrophilic extracts of ZFL cells. The y-axis has been maintained at a constant scale for all spectra to allow for intensity comparisons across sites. For difference spectra showing the full chemical shift range (0.5–10 ppm) see Fig. S1 in Supporting Information.
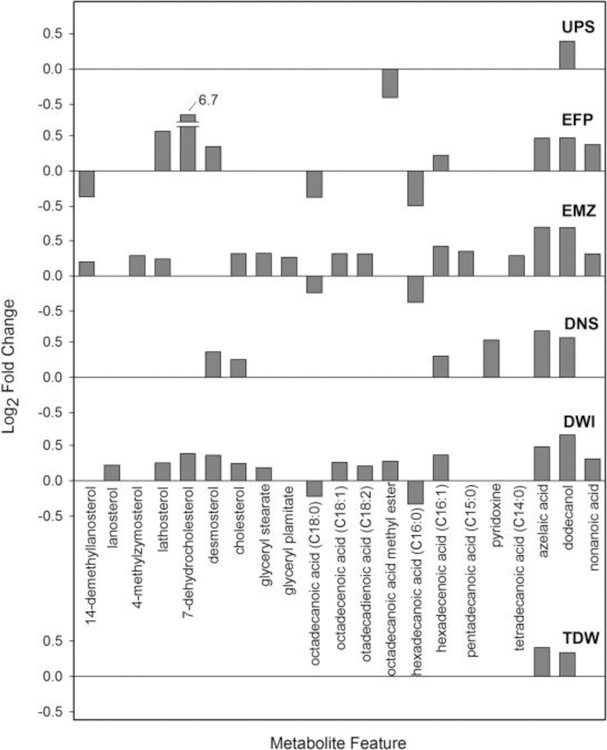
For the lipophilic extracts analyzed with GC-MS, a fold change plot was made for the exposure result for each site versus control (see Fig. 4 for annotated metabolites, Fig. S2 for a version displaying all metabolites). There are 44, 63, 32, and 53 metabolites out of a total of 107 that exhibited significant changes in EFP, EMZ, DNS, and DWI, respectively, versus CON (p < 0.05). In agreement with the PCA score plot, these four sites exhibit similar patterns of metabolite changes (Fig. S2). However, several metabolites exhibited considerably greater increases in normalized abundance for the EFP site than any metabolites at any of the other sites. For the two least impacted sites – UPS and TDW - only 13 and 19 metabolites, respectively, are found to change significantly in normalized abundance versus CON (p < 0.05).
Fig. 4.
Bar chart showing the fold change of 21 annotated metabolites from the lipophilic extracts that exhibited different normalized abundance at one or more of the six sites relative to the control (FDR-corrected p-values<0.05) as measured by GC-MS.
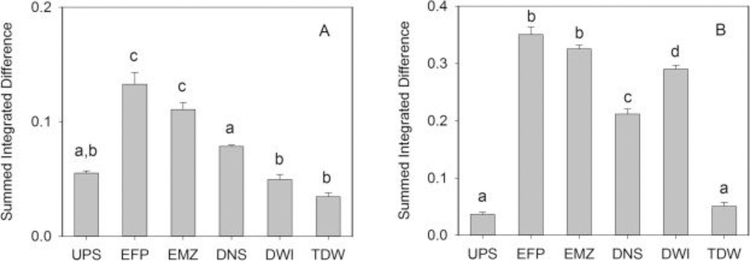
In a further step, we summed the absolute values of intensity differences of all peaks from the t-test filtered difference spectra for ZFL hydrophilic and lipophilic extracts and compared their magnitudes as a proxy for total impact from the exposure for each site (Fig. 5). For both hydrophilic and lipophilic extracts, EFP was found to be the most affected site. In agreement with the PCA score plots (Fig. 2), the level of dilution at the EMZ site did not result in a statistically significant attenuation (p = 0.08 for hydrophilic extracts and p = 0.26 for lipophilic extracts). Still, in general, the effect of effluent from the WWTP gradually decreased along the direction of water flow (particularly in the case of the hydrophilic extract; Fig. 5A). However, we noted a discrepancy between the hydrophilic and lipophilic metabolome at DWI where the integrated impact on hydrophilic metabolites was at the same level as that of UPS (Fig. 5A), while the intensity change of the lipophilic metabolome for DWI was even greater than that observed at DNS (Fig. 5B). (Note that, as discussed above, these trends were also observable in the score plots of Fig. 2.) Inconsistent effects on biological responses when looking at both the hydrophilic and lipophilic metabolites are not uncommon, having also been reported for an in vivo exposure of fathead minnows (Pimephales promelas, FHM) that were caged at various locations near a WWTP (Skelton et al., 2014). However, it serves to reiterate the importance of capturing the greatest extent of the intracellular metabolome as possible, as different classes of metabolites (e.g., lipids, amino acids, carbohydrates) can provide both complementary and disparate types of information.
Fig. 5.
The summed intensity values for significantly changed metabolites from the t-test filtered difference spectra for both hydrophilic (A) and lipophilic (B) extracts across six sampling sites. The intensity value for each site was generated by summing the absolute values of all peaks within each difference spectrum. The error bars represent the standard error of the mean. Bars labeled with different letters indicate significant difference at p < 0.05 based on analysis of variance (ANOVA) with Tukey’s HSD post-hoc test.
An examination of the greater impact on the lipophilic metabolome of the ZFL cells exposed to water from the DWI site suggests an additional source of environmental stressor(s) in the stretch of the river between DNS and DWI that mainly affects lipid metabolism. Regardless of the source of the stressor(s), it appears that they are effectively eliminated during drinking water treatment, based on a comparison of the summed integrated differences of TDW and DWI (Fig. 5B). Indeed, the data in Fig. 5 suggest that, in this system, the response in the ZFL cells from the WWTP effluent is well-controlled downstream through a combination of river dilution, natural attenuation, and existing drinking water treatment technologies.
3.2. Metabolic pathway perturbations observed in the hydrophilic ZFL metabolome
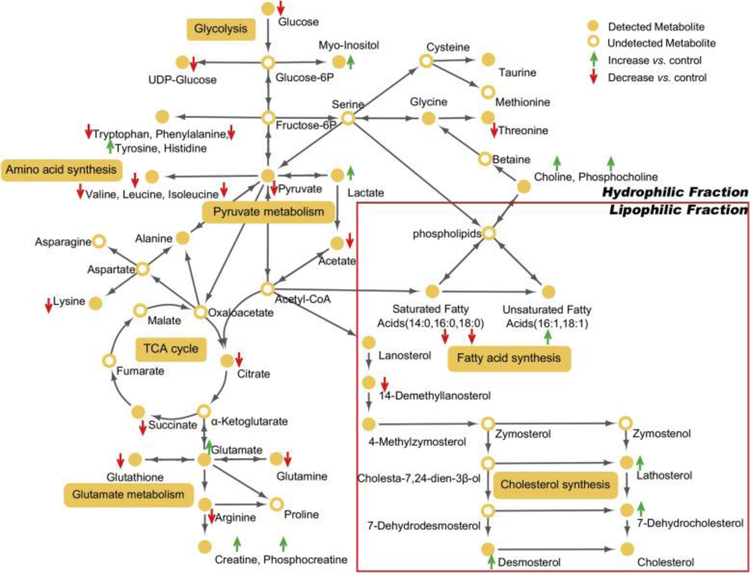
The combined metabolome changes, as depicted in Fig. 2, Fig. 3, Fig. 4, Fig. 5, suggest that WWTP effluent was the primary cause of the responses that we measured in this portion of the river system. Hence, further analyses focused on the perturbed metabolic pathways associated with exposure to the effluent. Using these pathways, a bioactivity network was built using a majority of the annotated metabolites (Fig. 6); the complete list of affected metabolites is presented in Tables S2 and S3.
Fig. 6.
Significantly altered metabolites and proposed perturbed metabolic pathways in ZFL cells after exposure to WWTP effluent (EFP). Green and red arrows indicate significantly increased or decreased metabolites in EFP vs. CON, respectively.
Analysis of the bioactivity network revealed that several metabolites associated with energy metabolism in ZFL were found to change substantially following exposure to the WWTP effluent (EFP). For example, the key entry and exit metabolites for the glycolysis pathway, glucose and pyruvate, respectively, were found to decrease in the ZFL metabolome following exposure. This suggested a greater energetic demand by ZFL cells when exposed to the complex mixtures in the effluent. This hypothesis was further bolstered by findings in the NMR difference spectra of depleted cellular energy molecules AXPs (ATP, ADP, AMP, see Fig. S1 in Supporting Information). In keeping with increased cellular energy consumption, UDP-glucose was also found to decrease following exposure to the EFP site water samples. UDP-glucose is the major form of glucose utilized in the synthesis of polysaccharides (e.g., glycogen) in biological cells. A previous study showed that glucose starvation could result in UDP-glucose deficiency and inactivation of glycogen synthase (Higuita et al., 2004). Thus, it appears that not only the short-term energy balance was affected (e.g., decreased glucose and nucleotides), but potentially so were cellular reserves (e.g., glycogen). Unfortunately, due to the low levels of metabolites characteristic of ZFL cells, we were unable to evaluate effects on glycogen. Regardless, the concomitant decreases in energetic metabolites in the hydrophilic metabolome suggests that glycogen effects may be observed in vivo. Oxidative degradation (i.e., the tricarboxylic acid (TCA) cycle) was also affected by WWTP effluent exposure. Specifically, the abundances of citrateand succinate were found decreased in the EFP-exposed cells compared to CON. Conversely, lactate, the primary by-product of fermentation (i.e., homolactic fermentation), was found to increase in cells that were exposed to water samples collected at the WWTP effluent discharging point.
A number of amino acids in ZFL cells were decreased following wastewater exposure (Fig. 6). Specifically, reductions in valine, leucine, isoleucine, lysine, threonine, glutamine, arginine, tryptophan, and phenylalanine were observed. Given the decreases measured in energetic metabolites (glucose, pyruvate, etc.), the global decrease in amino acids is likely due in part to increased consumption by glycolysis and the TCA cycle (i.e., anaplerosis) to sustain energy production. Exceptions to this apparent global decrease are the increases observed for glutamate and tyrosine. In fact, one of the most prominent metabolite changes following exposure to WWTP effluent is the increase of glutamate (Fig. 3), which appeared to coincide with decreases in glutamine and glutathione. These findings suggest disturbance of the glutamate metabolism pathway, which could have multiple implications for cell viability and function (Newsholme et al., 2003). As the hub of this pathway, glutamate undergoes transformation to numerous other metabolites in order to meet the changing demands of the cell. For example, glutamate can influence the TCA cycle (through deamination to α-ketoglutarate), provide a readily available sink for excess nitrogen (via its conversion to glutamine), and support amino acid homeostasis. Furthermore, as a component of the tripeptide glutathione, glutamate is essential for defense against oxidative stressors (e.g., free radicals, peroxides, and heavy metals) (Newsholme et al., 2003). The reduction in glutathione suggests the presence of components of the WWTP effluent mixture capable of inducing oxidative stress. However, exactly how this decline relates to the observed increase in glutamate is uncertain.
Beyond energetic effects, we also observed a large increase in choline (Cho) and phosphocholine (PCho) in ZFL cells from the EFP site (Fig. 3). Cho and PCho are major components in the biosynthesis of membrane phospholipids that determine the cell membrane fluidity and govern overall cell functions (Li and Vance, 2008). In biological cells, PCho/Cho can either be acquired from the extracellular environment or produced de novovia the action of phospholipase—an enzyme that hydrolyzes phosphatidylcholine (an important component of membrane phospholipids) into fatty acids and other substances (including Cho and PCho) (Li and Vance, 2008). The products from phospholipase action play critical roles in many physiological processes including the generation of signaling lipids as well as regulation of energy metabolism (Li and Vance, 2008). Considering the close relationship between PCho/Cho levels and lipid homeostasis in biological cells, such a large increase of PCho/Cho suggests considerable disturbance of lipid metabolism by the effluent exposure, which we, indeed, observed in the lipophilic ZFL metabolome (Fig. 2, Fig. 4, Fig. 5B).
3.3. Metabolic pathway perturbations observed in the lipophilic ZFL metabolome
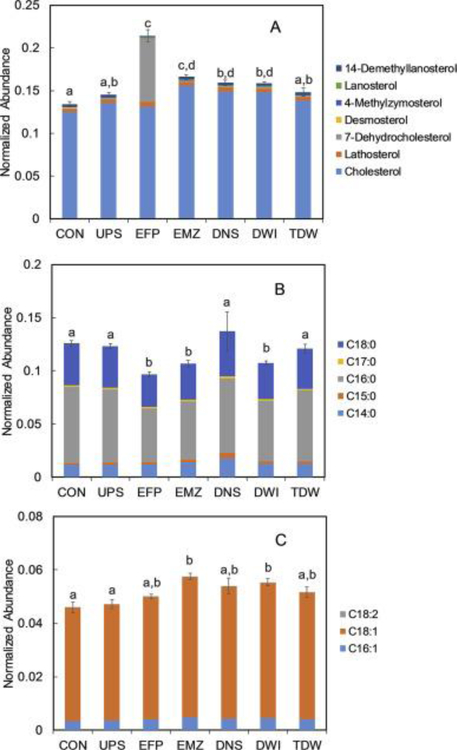
Guided in part by the large change of PCho/Cho levels in the hydrophilic ZFL metabolome (Fig. 3), we further analyzed the GC-MS data to assess which lipid metabolism pathways were affected by exposing ZFL cells to the WWTP effluent. Because 19 of the 21 annotated lipophilic metabolites (Fig. 4) were classified as intermediates or end products in either cholesterol or fatty acid synthesis, we focused our attention on the effects of exposure from the EFP-site water (relative to CON) on these pathways.
Specifically, we first assessed the variation in abundances of cholesterol and its biosynthetic intermediates in the context of a bioactivity network. Cholesterol plays an essential role in vertebrate biochemistry and physiology, maintaining the structural integrity and fluidity of the cell membrane and serving as the precursor for biosynthesis of steroid hormones, vitamin D, and bile acids (van der Wulp et al., 2013). Hence, regulation of cholesterol homeostasis is of paramount importance. In the current study, we found that 14-demethyllanosterol significantly decreased (p = 0.01) while the levels of three other intermediate metabolites, desmosterol, lathosterol, and 7-dehydrocholesterol (7-DHC), were significantly elevated upon exposure to EFP (Fig. 6, p = 0.001 for desmosterol, p = 0.002 for lathosterol, and p < 0.001 for 7-DHC). It should be noted that the normalized abundance of 7-DHC was 102 times that of the CON (see Fig. 4, log2 fold change = 6.7), however, its downstream reaction product, cholesterol, was not significantly changed (p = 0.22). This pattern of metabolite changes has recently been reported with Neuro2a cells that were exposed to over 30 different pharmacologically active compounds that were associated with inhibited conversion of 7-DHC to cholesterol via the enzyme 7-DHC reductase (DHCR7) (Kim et al., 2016). Expression changes (both increases and decreases) in the transcript and/or protein of DHCR7 has frequently been reported in aquatic species exposed to a range of environmental contaminants including Cu (Santos et al., 2010), linuron (Webster et al., 2015), and 17α-ethynylestradiol (Tompsett et al., 2013). Thus, the marked change in 7-DHC observed here is not without some precedent and may represent a candidate marker for exposure to contaminated surface waters.
In regard to fatty acid metabolism, we observed significant decreases in the normalized abundances of two saturated fatty acids (SFA), hexadecanoic acid (C16:0, p < 0.001) and octadecanoic acid (C18:0, p < 0.001), in EFP-exposed ZFL cells relative to control (Fig. 7B). For the monounsaturated fatty acids (MUFA), it was found that hexadecenoic acid (C16:1) abundance increased significantly (p = 0.02) after EFP exposure (Fig. 7C). Another commonly detected MUFA, octadecenoic acid (C18:1) was also found to increase in normalized abundance upon EFP exposure; however, this increase did not meet the threshold for statistical significance (p = 0.12, Fig. 7C). As for the polyunsaturated fatty acids (PUFA), the final dataset for the lipophilic metabolome contained only one metabolite–octadecadienoic acid (C18:2), and there was no significant difference (p = 0.81) in its normalized abundance between EFP and CON. Interestingly, the sum of abundances of all detected fatty acids did not show a significant difference between EFP and CON (p = 0.65). This suggests that the combined pool of fatty acids in the ZFL cells was at a steady state following EFP exposure. However, looking more closely at the selectivity of responses with regard to extent of saturation suggests that exposure to the undiluted wastewater effluent from EFP promoted the biotransformation of SFA to MUFA. MUFA (as opposed to SFA) have been reported to have a protective effect on oxidative stress (Quiles et al., 2006) and cellular inflammation (Liu et al., 2011). Previously, a number of studies have reported elevated MUFA levels across multiple species (fish, rodent, and invertebrate) that were exposed to environmental contaminants including 2,3,7,8-tetrachlorodibenzofuran (Zhang et al., 2015), tributyltin (Janer et al., 2007), and perfluorinated compounds (Wagbo et al., 2012). Considering the fact that WWTP effluent contains a complex mixture of numerous natural and xenobiotic compounds that may exert oxidative stress and/or an inflammatory effect on ZFL cells, it is plausible that the elevated MUFA level (relative to SFA) reflects the physiological status of ZFL cells in combating both types of environmental stress in response to exposure to WWTP effluent.
Fig. 7.
Relative normalized abundances for metabolites identified in the cholesterol and fatty acidsynthesis pathways from lipophilic ZFL extracts. A) Cholesterol synthesis pathway metabolites; B) Saturated fatty acids; and C) Unsaturated fatty acids. The error bars represent the standard error of the mean of summed normalized abundances of all metabolites in each bar. Bars labeled with different letters indicate significant difference at p < 0.05 based on analysis of variance (ANOVA) with Tukey’s HSD post-hoc test.
3.4. Relating metabolite changes with chemicals detected at the WWTP site (EFP)
Data from this study clearly show that the ZFL cell metabolome was impacted to the greatest extent by direct exposure to WWTP effluent (i.e., the EFP site). As discussed above, many of the metabolite changes that were measured provide new or confirmatory knowledge regarding in vitro responses and demonstrate the value of cell-based metabolomics for screening environmental surface waters. Note that water samples collected concurrently at the same study sites were analyzed for over 200 chemical pollutants that cover a wide variety of classes including antibiotics, disinfection byproducts, endocrine disrupting compounds, pharmaceutical compounds and inorganic compounds (results will be published separately, Susan Glassmeyer, personal communication, October 24th, 2017). Using the targeted chemical analysis results, we were able to establish potential links between a selection of chemicals and certain perturbed metabolic pathways that were observed in ZFL cells following exposure to effluent from the EFP site.
As discussed earlier, one of the most distinct metabolite changes in EFP-exposed cells (as opposed to other sites) was the increase in the PCho/Cho level (Fig. 3), which may indicate promoted phospholipase activity. Interestingly, some of the pharmaceuticals detected above their minimum reporting levels only at the EFP site, including acetaminophen and fluoxetine, have been reported to upregulate the activity of phospholipase. For example, acetaminophen was found to increase the expression of mRNA for multiple members of the phospholipase family in mouse liver (Coen et al., 2003); and chronic fluoxetine exposure upregulated activity, protein, and mRNA levels of cytosolic phospholipase A2 in rat frontal cortex (Rao et al., 2006). These studies, along with the occurrence data suggest the potential role of these and related chemicals in the altered levels of PCho/Cho that were observed in the metabolome of ZFL cells exposed to water from EFP.
Data from the present study also show that ZFL cell lipid synthesis pathways were considerably affected by exposure to the WWTP effluent from the EFP site (Fig. 6, Fig. 7). Two pharmaceuticals that were detected above their minimum reporting levels only at the EFP site - bupropion and fluoxetine - have been reported in a recent study to inhibit the activity of DHCR7 (Kim et al., 2016). The detection of these DHCR7 inhibitors in EFP effluent but not at other sites may potentially help explain the considerable accumulation of 7-DHC in cells exposed to the EFP samples (Fig. 6). However, the apparent nonspecific chemical inhibition of DHCR7 does not preclude the role of other compounds in this response, either from additional chemicals that were detected or those that were present but not included in the targeted analytical methods used for compound detection.
A number of other detected chemicals are known to affect certain enzyme activities in fatty acid biosynthetic pathways. For example, the pharmaceutical sitagliptin was reported to inhibit the production of malonyl-CoA and thus the synthesis of SFA (Sujishi et al., 2015). Three detected antidepressant drugs, bupropion, citalopram and fluoxetine, have been shown to induce increased expression of stearoyl-CoA desaturase (SCD), an enzyme that regulates the synthesis of UFA from SFA in human glial cells (Raeder et al., 2006). Conversely, acetaminophen and carbamazepine were found to down-regulate SCD activity in human liver (Elferink et al., 2011) and bronchial epithelial cells (Song et al., 2011), respectively. All of these compounds were detected as part of the complex mixture of chemicals present in the water collected at the EFP site, and none were detected above their minimum reporting levels at any of the other sites. Thus, it seems likely that these and other bioactive chemicals (both detected and undetected) in combination could have resulted in the integrated effects observed for fatty acid metabolism in ZFL cells.
3.5. Trends in metabolite changes and chemical compositions across various sites
In addition to focusing specifically on the EFP site, we also explored the chemical data from the other sites in light of the pattern of ZFL metabolome changes that were observed across these sites. For example, chemical analysis of the EMZ site sample (collected ∼0.7 km downstream of EFP) resulted, as expected, in the detection of substantially fewer and less-concentrated compounds as compared to the EFP site (Susan Glassmeyer, personal communication, October 24th, 2017). Therefore, that we observed no significant difference (p > 0.05) between EFP and EMZ with respect to effect on both the hydrophilic and the lipophilic ZFL metabolome (Fig. 2, Fig. 5) is somewhat surprising. Given the complexity of the chemical mixtures at these sites, there could be multiple factors contributing to this discrepancy. First, an organic farm that cultivates vegetables and fruits was identified in close proximity to EMZ, suggesting the possibility of an additional source of unmeasured bioactive constituents that might have contributed to the overall effect on the ZFL metabolome. Second, given the limited knowledge regarding the transformation products of contaminants in surface waters, it is possible that a variety of unmeasured daughter products were present at EMZ that produced equivalent or greater toxicity to the ZFL cells than the parent compounds. Furthermore, the integrated effects of chemical mixtures on metabolome changes may not produce a linear dose-response relationship, particularly when mixing with river water. Indeed, non-linear dose response curves have been frequently observed in toxicological studies. For example, in numerous studies, the toxicological effect(s) of a given compound appears to follow an inverted U-shaped dose response curve in which increases in dose produce increased effect(s) up to a maximum, followed by a decreasing response despite continuing to increase the dose (Calabrese and Baldwin, 2001). This type of response has been attributed to different types of effects occurring at different dose levels (e.g., stimulatory at low dose and/or inhibitory at high dose) (Calabrese and Baldwin, 2001). Clearly, this relationship between the constituents found in complex environmental mixtures (e.g., various types of contaminants and different dilution factors) and their effects on the ZFL cell metabolome warrant further investigation. Moreover, given the disparity between EFP and EMZ with regard to the number of compounds detected and the intensities of the metabolomic responses observed, the importance of including untargeted biological assessments in conjunction with chemical monitoring efforts should not be underestimated.
Our untargeted metabolomics assessment also suggested that an unidentified source of bioactive compounds (that primarily affect lipid metabolism) may exist in the stretch of river between DNS and DWI. For example, as noted above, the overall effects are greater at DWI than at DNS for the lipophilic, but not for the hydrophilic, metabolome (Fig. 5). Given this unanticipated result, we looked more closely for differences in compound detections at these sites. There were slightly fewer constituents detected at DWI as compared to DNS. But, one steroid hormone, androstenedione, which is often present in human and animal waste, was considerably more abundant at DWI as compared to DNS in spite of the fact that DWI is 5.4 km further downstream from the WWTP. While there is no permitted wastewater discharge between DWI and DNS, a land-use analysis revealed that this segment of river passes through a densely populated area that includes many residential homes, businesses, and a heavily trafficked highway. Thus, some level of additional contamination from these and other sources may be contributing to the lipid response. This may include, for example, stormwater runoff and leaking septic tanks (only 60% of residents in the surrounding area are connected to sewer collection, Susan Glassmeyer, personal communication, October 24th, 2017).
4. Conclusions
The main goal of the current study was to determine the effectiveness of cell-based metabolomics for tracking biological impacts of complex chemical mixtures as they move through aquatic systems from their initial release at a WWTP to a treated water intended for human consumption at a DWTP. The analyses of both the hydrophilic and lipophilic metabolomes revealed a gradient of response intensities, generally diminishing with the distance of the sampling sites downstream from the WWTP and after drinking water treatment. The ability to capture these trends, even when some impacts are relatively small (as in the case of treated drinking water vs. other sites impacted by WWTP effluent), provides a strong impetus to further develop this untargeted in vitro approach for environmental assessments in general, and specifically, for surface-water reuse scenarios. Moreover, these results underscore the importance of including such bioassay methods alongside extensive chemical monitoring for understanding potential sublethal contributions to exposure and risk. The results of this study also emphasize the importance of including lipid analyses alongside more routine metabolomics assessments that are often exclusive of this important class of metabolites. Indeed, as described above, the lipophilic metabolome revealed an unexpectedly large response at the drinking water intake, despite the level of WWTP-contaminant dilution observed at this site. Finally, biochemical pathway analysis of the hydrophilic and lipophilic metabolite changes suggested candidate pathways (energy, amino acids, and lipid metabolism) and potential markers (e.g., 7-DHC, indicative of disrupted cholesterol metabolism) for future investigations of other surface waters.
Highly sensitive, untargeted approaches such as in vitro metabolomics (which can assess effects upon multiple biological pathways) are increasing in importance as the global water crisis places additional focus on the complexity of ecological and human health effects that result from the discharge of natural and xenobiotic compounds into surface waters from WWTPs. However, it is important to note that in vitro experiments, in general, do not fully mimic in vivo exposures. For example, cells used for in vitro exposures typically do not experience the types of intercellular interactions (e.g. exchanges of nutrients/signaling molecules/growth factors) observed in in vivo systems. In addition, xenobiotics from in vitroexposures are introduced directly into the growth medium, while for in vivo experiments the exposure levels of xenobiotics are impacted by a number of factors including substrate uptake and transport. Looking forward, the development and refinement of these tools with regard to linking these cellular level effects to whole organism and population level adverse responses is of considerable importance. Tools such as the adverse outcome pathway framework (Ankley et al., 2010), which has been designed to support such efforts, will be critical for bringing in vitro metabolomics approaches such as the one described here into greater use by the regulatory community.
Supplementary Material
Acknowledgments
H.Z. was supported by the Great Lakes National Program Office and a National Research Council Research Associateship award at the National Exposure Research Laboratory of U.S. Environmental Protection Agency (EPA). The research described in this article has been funded in part by the EPA through Interagency Agreement DW 92401501 to the U.S. Geological Survey (USGS), and through programmatic support of the USGS Toxic Substances Hydrology Program and the EPA’s Office of Research and Development. This document has been reviewed in accordance with EPA and USGS policy and approved for publication. Approval does not signify that the contents reflect the views of the EPA and mention of trade names or commercial products does not constitute endorsement or recommendation for use by EPA. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government. The authors would like to thank the participating DWTPs for their involvement in the project and for their assistance in collecting the samples.
Footnotes
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, Mount DR, Nichols JW, Russom CL, Schmieder PK, Serrrano JA, Tietge JE, Villeneuve DL, 2010. Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem 29 (3), 730e741. [DOI] [PubMed] [Google Scholar]
- Boleda MR, Galceran MT, Ventura F, 2009. Monitoring of opiates, cannabinoids and their metabolites in wastewater, surface water and finished water in Catalonia, Spain. Water Res 43 (4), 1126e1136. [DOI] [PubMed] [Google Scholar]
- Bundy JG, Davey MP, Viant MR, 2009. Environmental metabolomics: a critical review and future perspectives. Metabolomics 5, 3e21. [Google Scholar]
- Calabrese EJ, Baldwin LA, 2001. The frequency of U-Shaped dose responses in the toxicological literature. Toxicol. Sci 62, 330e338. [DOI] [PubMed] [Google Scholar]
- Coen M, Lenz EM, Nicholson JK, Wilson ID, Pognan F, Lindon JC, 2003. An integrated metabonomic investigation of acetaminophen toxicity in the mouse using NMR spectroscopy. Chem. Res. Toxicol 16 (3), 295e303. [DOI] [PubMed] [Google Scholar]
- Dai YJ, Jia YF, Chen N, Bian WP, Li QK, Ma YB, Chen YL, Pei DS, 2014. Zebrafish as a model system to study toxicology. Environ. Toxicol. Chem 33 (1), 11e17. [DOI] [PubMed] [Google Scholar]
- David A, Lange A, Tyler CR, Hill EM, 2018. Concentrating mixtures of neuroactive pharmaceuticals and altered neurotransmitter levels in the brain of fish exposed to a wastewater effluent. Sci. Total Environ 621, 782e790. [DOI] [PubMed] [Google Scholar]
- Davis JM, Collette TW, Villeneuve DL, Cavallin JE, Teng Q, Jensen KM, Kahl MD, Mayasich JM, Ankley GT, Ekman DR, 2013. Field-based approach for assessing the impact of treated pulp and paper mill effluent on endogenous metabolites of fathead minnows (Pimephales promelas). Environ. Sci. Technol 47 (18), 10628e10636. [DOI] [PubMed] [Google Scholar]
- Davis JM, Ekman DR, Skelton DM, LaLone CA, Ankley GT, Cavallin JE, Villeneuve DL, Collette TW, 2017. Metabolomics for informing adverse outcome pathways: androgen receptor activation and the pharmaceutical spironolactone. Aquat. Toxicol 184, 103e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferink MGL, Olinga P, van Leeuwen EM, Bauerschmidt S, Polman J, Schoonen WG, Heisterkamp SH, Groothuis GMM, 2011. Gene expression analysis of precision-cut human liver slices indicates stable expression of ADME-Tox related genes. Toxicol. Appl. Pharmacol 253 (1), 57e69. [DOI] [PubMed] [Google Scholar]
- Escher B, Leusch F, 2012. Bioanalytical Tools in Water Quality Assessment IWA Publishing, London, UK. [Google Scholar]
- Feoktistova M, Geserick P, Leverkus M, 2016. Crystal violet assay for determining viability of cultured cells. Cold Spring Harb. Protoc 4, 343e346. [DOI] [PubMed] [Google Scholar]
- Gomez-Canela C, Miller TH, Bury NR, Tauler R, Barron LP, 2016. Targeted metabolomics of Gammarus pulex following controlled exposures to selected pharmaceuticals in water. Sci. Total Environ 562, 777e788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YC, Krasner SW, 2009. Occurrence of primidone, carbamazepine, caffeine, and precursors for N-Nitrosodimethylamine in drinking water sources impacted by wastewater. J. Am. Water Resour. Assoc 45 (1), 58e67. [Google Scholar]
- Higuita JC, Thelestam M, Katz A, 2004. Glucose starvation results in UDP-glucose deficiency and inactivation of glycogen synthase. Arch. Biochem. Biophys 425 (2), 242e248. [DOI] [PubMed] [Google Scholar]
- Janer G, Navarro JC, Porte C, 2007. Exposure to TBT increases accumulation of lipids and alters fatty acid homeostasis in the ramshorn snail Marisa cornuarietis. Comparative Biochemistry and Physiology C-Toxicology & Pharmacology 146 (3), 368e374. [DOI] [PubMed] [Google Scholar]
- Judson R, Richard A, Dix DJ, Houck K, Martin M, Kavlock R, Dellarco V, Henry T, Holderman T, Sayre P, Tan S, Carpenter T, Smith E, 2009. The toxicity data landscape for environmental chemicals. Environ. Health Perspect 117 (5), 685e695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HYH, Korade Z, Tallman KA, Liu W, Weaver CD, Mirnics K, Porter NA, 2016. Inhibitors of 7-dehydrocholesterol reductase: screening of a collection of pharmacologically active compounds in Neuro2a cells. Chem. Res. Toxicol 29 (5), 892e900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpin DW, Furlong ET, Meyer MT, Thurman EM, Zaugg SD, Barber LB, Buxton HT, 2002. Pharmaceuticals, hormones, and other organic wastewater contaminants in US streams, 1999–2000: a national reconnaissance. Environ. Sci. Technol 36 (6), 1202e1211. [DOI] [PubMed] [Google Scholar]
- Lankadurai BP, Nagato EG, Simpson MJ, 2013. Environmental metabolomics: an emerging approach to study organism responses to environmental stressors. Environ. Rev 21 (3), 180e205. [Google Scholar]
- Li ZY, Vance DE, 2008. Phosphatidylcholine and choline homeostasis. JLR (J. Lipid Res.) 49 (6), 1187e1194. [DOI] [PubMed] [Google Scholar]
- Liu XQ, Strable MS, Ntambi JM, 2011. Stearoyl CoA desaturase 1: role in cellular inflammation and stress. Advances in Nutrition 2 (1), 15e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommen A, Kools HJ, 2012. MetAlign 3.0: performance enhancement by efficient use of advances in computer hardware. Metabolomics 8 (4), 719e726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macova M, Toze S, Hodgers L, Mueller JF, Bartkow M, Escher BI, 2011. Bioanalytical tools for the evaluation of organic micropollutants during sewage treatment, water recycling and drinking water generation. Water Res 45 (14), 4238e4247. [DOI] [PubMed] [Google Scholar]
- Newsholme P, Procopio J, Lima MMR, Pithon-Curi TC, Curi R, 2003. Glutamine and glutamate - their central role in cell metabolism and function. Cell Biochem. Funct 21 (1), 1e9. [DOI] [PubMed] [Google Scholar]
- NRC, 2007. Toxicity Testing in the 21st Century: a Vision and a Strategy The National Academies Press, Washington, D.C. [Google Scholar]
- NRC, 2012. Water Reuse: Expanding the Nation’s Water Supply through Reuse of Municipal Wastewater, Washington, DC. [Google Scholar]
- Quiles JL, Barja G, Battino M, Mataix J, Solfrizzi V, 2006. Role of olive oil and monounsaturated fatty acids in mitochondrial oxidative stress and aging. Nutr.Rev 64 (10), S31eS39. [Google Scholar]
- Raeder MB, Ferno J, Glambek M, Stansberg C, Steen VM, 2006. Antidepressant drugs activate SREBP and up-regulate cholesterol and fatty acid biosynthesis in human glial cells. Neurosci. Lett 395 (3), 185e190. [DOI] [PubMed] [Google Scholar]
- Rao JS, Ertley RN, Lee HJ, Rapoport SI, Bazinet RP, 2006. Chronic fluoxetine upregulates activity, protein and mRNA levels of cytosolic phospholipase A(2) in rat frontal cortex. Pharmacogenomics J 6 (6), 413e420. [DOI] [PubMed] [Google Scholar]
- Ratnasekhar C, Sonane M, Satish A, Mudiam MKR, 2015. Metabolomics reveals the perturbations in the metabolome of Caenorhabditis elegans exposed to titanium dioxide nanoparticles. Nanotoxicology 9 (8), 994e1004. [DOI] [PubMed] [Google Scholar]
- Rice J, Westerhoff P, 2015. Spatial and temporal variation in de facto wastewater reuse in drinking water systems across the USA. Environ. Sci. Technol 49 (2), 982e989. [DOI] [PubMed] [Google Scholar]
- Rice J, Wutich A, Westerhoff P, 2013. Assessment of de facto wastewater reuse across the US: trends between 1980 and 2008. Environ. Sci. Technol 47 (19), 11099e11105. [DOI] [PubMed] [Google Scholar]
- Richardson SD, Ternes TA, 2014. Water analysis: emerging contaminants and current issues. Anal. Chem 86 (6), 2813e2848. [DOI] [PubMed] [Google Scholar]
- Rodayan A, Afana S, Segura PA, Sultana T, Metcalfe CD, Yargeau V, 2016. Linking drugs of abuse in wastewater to contamination of surface and drinking water. Environ. Toxicol. Chem 35 (4), 843e849. [DOI] [PubMed] [Google Scholar]
- Santos EM, Ball JS, Williams TD, Wu HF, Ortega F, Van Aerle R, Katsiadaki I, Falciani F, Viant MR, Chipman JK, Tyler CR, 2010. Identifying health impacts of exposure to copper using transcriptomics and metabolomics in a fish model. Environ. Sci. Technol 44 (2), 820e826. [DOI] [PubMed] [Google Scholar]
- Schwarzenbach RP, Escher BI, Fenner K, Hofstetter TB, Johnson CA, von Gunten U, Wehrli B, 2006. The challenge of micropollutants in aquatic systems. Science 313 (5790), 1072e1077. [DOI] [PubMed] [Google Scholar]
- Skelton DM, Ekman DR, Martinovic-Weigelt D, Ankley GT, Villeneuve DL, Teng Q, Collette TW, 2014. Metabolomics for in situ environmental monitoring of surface waters impacted by contaminants from both point and nonpoint sources. Environ. Sci. Technol 48 (4), 2395e2403. [DOI] [PubMed] [Google Scholar]
- Song M, Kim YJ, Ryu JC, 2011. Phospholipidosis induced by PPAR gamma signaling in human bronchial epithelial (BEAS-2B) cells exposed to amiodarone. Toxicol. Sci 120 (1), 98e108. [DOI] [PubMed] [Google Scholar]
- Sujishi T, Fukunishi S, Ii M, Nakamura K, Yokohama K, Ohama H, Tsuchimoto Y, Asai A, Tsuda Y, Higuchi K, 2015. Sitagliptin can inhibit the development of hepatic steatosis in high-fructose diet-fed ob/ob mice. J. Clin. Biochem. Nutr 57 (3), 244e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NS, Kirwan JA, Yan ND, Viant MR, Gunn JM, McGeer JC, 2016. Metabolomics confirms that dissolved organic carbon mitigates copper toxicity. Environ. Toxicol. Chem 35 (3), 635e644. [DOI] [PubMed] [Google Scholar]
- Teng Q, Ekman DR, Huang WL, Collette TW, 2013. Impacts of 17 alphaethynylestradiol exposure on metabolite profiles of zebrafish (Danio rerio) liver cells. Aquat. Toxicol 130, 184e191. [DOI] [PubMed] [Google Scholar]
- Teng Q, Huang WL, Collette TW, Ekman DR, Tan C, 2009. A direct cell quenching method for cell-culture based metabolomics. Metabolomics 5 (2), 199e208. [Google Scholar]
- Tikunov YM, Laptenok S, Hall RD, Bovy A, de Vos RCH, 2012. MSClust: a tool for unsupervised mass spectra extraction of chromatography-mass spectrometry ion-wise aligned data. Metabolomics 8 (4), 714e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompsett AR, Wiseman S, Higley E, Giesy JP, Hecker M, 2013. Effects of exposure to 17 alpha-ethynylestradiol during larval development on growth, sexual differentiation, and abundances of transcripts in the liver of the wood frog (Litho bates sylvaticus). Aquat. Toxicol 126, 42e51. [DOI] [PubMed] [Google Scholar]
- USDA, 2013. Irrigation & Water Use https://www.ers.usda.gov/topics/farmpractices-management/irrigation-water-use.aspx. accessed Oct 1st, 2017.
- USEPA, 2004. Manual: Guidelines for Water Reuse EPA/625/R-04/108.
- van der Wulp MYM, Verkade HJ, Groen AK, 2013. Regulation of cholesterol homeostasis. Mol. Cell. Endocrinol 368 (1e2), 1e16. [DOI] [PubMed] [Google Scholar]
- Wagbo AM, Cangialosi MV, Cicero N, Letcher RJ, Arukwe A, 2012. Perfluorooctane sulfonamide-mediated modulation of hepatocellular lipid homeostasis and oxidative stress responses in atlantic salmon hepatocytes. Chem. Res. Toxicol 25 (6), 1253e1264. [DOI] [PubMed] [Google Scholar]
- Webster TMU, Perry MH, Santos EM, 2015. The herbicide linuron inhibits cholesterol biosynthesis and induces cellular stress responses in Brown trout. Environ. Sci. Technol 49 (5), 3110e3118. [DOI] [PubMed] [Google Scholar]
- Xia P, Zhang XW, Zhang HX, Wang PP, Tian MM, Yu HX, 2017. Benchmarking water quality from wastewater to drinking waters using reduced transcriptome of human cells. Environ. Sci. Technol 51 (16), 9318e9326. [DOI] [PubMed] [Google Scholar]
- Zhang LM, Hatzakis E, Nichols RG, Hao RX, Correll J, Smith PB, Chiaro CR, Perdew GH, Patterson AD, 2015. Metabolomics reveals that aryl hydrocarbon receptor activation by environmental chemicals induces systemic metabolic dysfunction in mice. Environ. Sci. Technol 49 (13), 8067e8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.