Abstract
A new platform for N-glycoprotein analysis from serum that combines matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI MSI) workflows with antibody slide arrays is described. Antibody Panel Based (APB) N-glycan imaging allows for the specific capture of N-glycoproteins by antibodies on glass slides and N-glycan analysis in a protein-specific and multiplexed manner. Development of this technique has focused on characterizing two abundant and well-studied human serum glycoproteins, alpha-1-antitrypsin and immunoglobulin G. Using purified standard solutions and one microliter samples of human serum, both glycoproteins can be immunocaptured and followed by enzymatic release of N-glycans. N-glycans are detected with a MALDI FT-ICR mass spectrometer in a concentration-dependent manner while maintaining specificity of capture. Importantly, the N-glycans detected via slide-based antibody capture were identical to that of direct analysis of the spotted standards. As a proof of concept, this workflow was applied to patient serum samples from individuals with liver cirrhosis to accurately detect a characteristic increase in an IgG N-glycan. This novel approach to protein-specific N-glycan analysis from an antibody panel can be further expanded to include any glycoprotein for which a validated antibody exists. Additionally, this platform can be adapted for analysis of any biofluid or biological sample that can be analyzed by antibody arrays.
Graphical Abstract
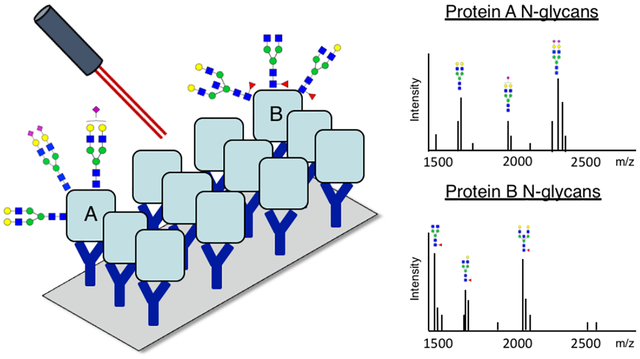
Glycosylation is one of the most common post-translational modifications and often consists of the covalent addition of an oligosaccharide (glycan) to either an asparagine (N-linked) or serine/threonine (O-linked) residue. N-linked glycans have been well-established to change with the progression of cancer and other diseases,1–4 and studies indicate that the N-glycan component of a glycoprotein may act as a specific disease biomarker more than the protein alone.2,5 This has been shown in the success of fucosylated alpha-fetoprotein (AFP) as a biomarker for liver cancer,6,7 yet most N-glycan profiles present on protein biomarkers remain unexplored. Current techniques for analysis of N-glycan profiles and their carrier proteins are often time-consuming or require large amounts of sample,1,3,8,9 which limits the ability to analyze significant numbers of patient samples for the discovery of novel disease biomarkers. Some high throughput methods have utilized differential lectin binding to identify carbohydrate structural motifs,10–12 yet these are limited to the variable and low binding affinities of most lectins, and they cannot be used to report true structural composition or glycan carrier (i.e. N-glycan, O-glycan, or glycosphingolipid) information.10–12
The technology of matrix-assisted laser desorption/ionization (MALDI) mass spectrometry imaging (MSI) has emerged in recent decades to become a powerful technique for analyte detection and localization across tissue sections with high mass accuracy.13–17 This technique creates two-dimensional heat maps of an analyte’s intensity across a tissue sample on a slide. Our lab has previously developed a method for the spatial analysis of released N-glycans across tissue sections18–23 and related workflows have been implemented and adapted by multiple labs.24–27 However, as is common to any method relying on enzymatic release of N-glycans, linking N-glycan signatures to their carrier proteins remains laborious and requires extensive additional analysis.24,28 Leveraging that MALDI MSI can be used to detect N-glycans from the solid surface of a tissue on a slide, we hypothesized that we could extend beyond traditional imaging techniques to detect N-glycan profiles from target glycoproteins captured on a slide-based antibody microarray panel. This would bridge the gap in linking N-glycan signatures to their proteins, as the location of the detected N-glycans along the slide array would be linked with each immunocaptured glycoprotein they were released from. This method would obtain N-glycan profiles for each glycoprotein, rather than other targeted approaches analyzing only certain N-glycan motifs or one protein at a time. Here we report a novel biomarker discovery platform by Antibody Panel Based (APB) N-glycan imaging, which couples the analyte localization of traditional MALDI MSI with the protein capture specificity of an antibody array for use with patient biofluid samples.
Experimental Section
Materials
Nitrocellulose-coated glass microscope slides (PATH microarray slides) and well slide modules (ProPlate Multi-Array Slide System, 24-well) were obtained from Grace Bio-Labs (Bend, OR). Trifluoroacetic acid, α-cyano-4-hydroxycinnamic acid, octyl-β-D-glucopyranoside, human alpha-1-antitrypsin, and stock human serum were obtained from Sigma Aldrich (St. Louis, MO). HPLC grade water, HPLC grade acetonitrile, bovine serum albumin, and phosphate buffered saline were obtained from Fisher Scientific (Hampton, NH). Peptide-N-glycosidase F (PNGase F) Prime ™ was cloned, expressed, and purified in-house as previously described.20 Anti-human A1AT was obtained from Genway Biotech (San Diego, CA). Human immunoglobulin G was obtained from Jackson ImmunoResearch (West Grove, PA) and anti-human IgG from Bethyl Laboratories (Montgomery, TX). Cirrhotic patient serum was obtained from Dr. Amit Singal (University of Texas Southwestern Medical Center, Dallas, TX). Serum samples were obtained via a study protocol approved by the UTSW Institutional Review Board, with written informed consent obtained from each subject. Diagnosis of liver cirrhosis was based on liver histology or clinical, laboratory, and imaging evidence of hepatic decompensation or portal hypertension. Each patient had a normal ultrasound; if serum AFP was elevated, a CT or MRI showed no liver mass. Further patient details regarding these samples are found in our previous publication.29
Antibody array preparation
Nitrocellulose-coated microscope slides were used and wells created by attaching a 24-well slide module. Antibodies were diluted in PBS and manually spotted in wells at 200ng per 1.5μL spot. Spots were then left to adhere overnight at 4°C in a humidity chamber made from a 12×9×3.5cm western blot incubation box lined with a Wypall X 60 paper towel and 2 rolled KimWipes saturated with distilled water. Slides were then air dried at room temperature and rinsed with 0.1% octyl-β-D-glucopyranoside in 1X PBS (referred to as PBS-OGS) to remove any unbound protein from slide. Rinse consisted of 200μL to each well for 1 minute.
Sample capture and N-glycan release
Wells were blocked with 200μL of 1% BSA (prepared in PBS-OGS) for 1 hr with gentle shaking. Wells were then washed with 200μL PBS (3 mins × 2) and 200μL double distilled water (1 min × 1) and let air dry. Samples were added to wells and incubated at room temperature for 2 hours in humidity chamber with gentle shaking. All samples were diluted in PBS, with sample volumes of 100μL added to wells. Slides were then washed with 200μL of PBS-OGS (1 min × 1), 200μL PBS (3 mins × 2), and 200μL double distilled water (1 min × 1) and let air dry. After removing the well module, an additional wash was performed by submerging the slide into 50mL of water for 30 seconds to remove any residual salt. In order to cleave N-glycans from captured proteins, PNGase F Prime™ (0.1μg/μL in HPLC grade water) was applied to the slides using an automated sprayer (M3 TM-Sprayer, HTX Technologies, Chapel Hill, NC) to retain localization with spraying parameters 15 passes at 45°C, 10 psi, flow rate 25μL/min, and 1200mm/min velocity. Slides were incubated overnight at 37°C in humidity chambers made in cell culture dishes with Wypall X 60 paper towels and 2 rolled KimWipes saturated with distilled water.
MALDI MS preparation and imaging
MALDI matrix α-cyano-4-hydroxycinnamic acid (CHCA, 7mg/mL in 50% acetonitrile/0.1% trifluoroacetic acid) was applied to slides using the same automated sprayer. Application parameters were 2 passes at 77°C, 10 psi, 1300 mm/min velocity and flow rate 100 μL/min. Slides were imaged on a solariX Legacy 7T FT-ICR mass spectrometer (Bruker) equipped with a matrix-assisted laser desorption/ionization (MALDI) source. Sampling was done using a SmartBeam II laser operating at 2000 Hz with laser spot size of 25μm. Images were collected using a smartwalk pattern at a 250μM raster with 200 laser shots per pixel. Samples were analyzed in positive ion, broadband mode using a 512k word time domain spanning m/z range 500–5000. An on-slide resolving power of 58,000 at m/z 1501 was calculated.
Data analysis
N-glycan localization and intensity were visualized using FlexImaging v4.1 (Bruker), with data imported at a 0.98 ICR Reduction Noise Threshold. Images were normalized to total ion current and N-glycan peaks were selected manually based on their theoretical mass values. Spectra were recalibrated using Data Analysis 5.0 (Bruker) with an 8-point linear function based on N-glycan theoretical masses. For quantification of peaks at individual spots, spectra were imported into SCiLS Lab software 2017a (Bruker). Each spot was designated a unique region and area under monoisotopic peak values were exported from each region.
HPLC and lectin orthogonal confirmation
HPLC analyses on released labelled N-glycans were performed using a Waters Alliance HPLC system as previously described.30 Lectin analyses on spotted glycoproteins were performed as previously described.31
Results and Discussion
The novel workflow for specific glycoprotein capture and APB N-glycan imaging is illustrated in Figure 1. The workflow is founded upon a similar MALDI mass spectrometry workflow for N-glycan imaging on tissue,20,23 and consists of three major steps. The first (Figure 1A) involves antibody spotting and glycoprotein capture localized to their antibody spots. The second (Figure 1B) consists of PNGase F enzymatic release of N-glycans in a localized manner and matrix coating of the slide, trapping released glycans in area of their release. Figure 1C shows the third step of MALDI MS analysis of the slide, where an overall spectrum is obtained with images correlating to each m/z peak. Images obtained from MS analysis depict the abundance of an N-glycan across a slide with color intensity, creating a heat map for each N-glycan detected. This allows for the visualization of N-glycans released from immunocaptured glycoproteins in an array type format, where N-glycans of interest can be linked back to their protein carriers.
Figure 1.
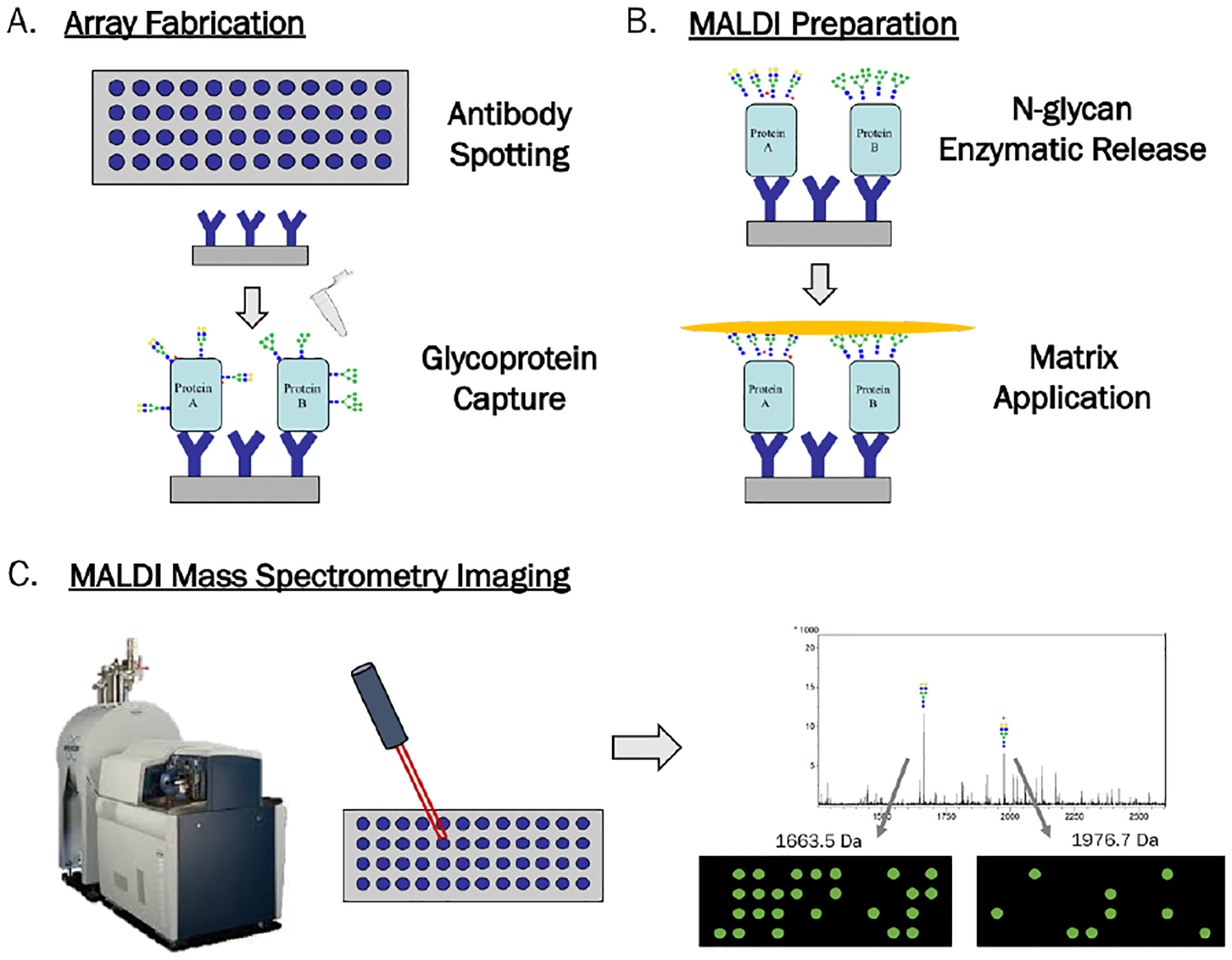
Workflow for Antibody Panel Based N-glycan imaging by MALDI MS. A) Antibodies are spotted and slide blocked with BSA. Sample is added for capture of glycoproteins by their antibodies. B) N-glycans are enzymatically released in a localized manner followed by matrix application. C) Slides are imaged by a MALDI FT-ICR MS to obtain overall spectrum and individual images for each m/z peak, which show the abundance of each N-glycan in two-dimensions across the array.
Initial experiments were done using human alpha-1-antitrypsin (A1AT) and immunoglobulin G (IgG) as they are abundant glycoproteins in human serum with well-characterized N-glycosylation sites.30,32–35 These glycoproteins have distinct N-glycan profiles from each other, as shown in Figure 2 and Figure S1 with orthogonal HPLC and lectin confirmation of unique N-glycan profiles. The N-glycan profiles in Figure 2 were obtained from MALDI MS of A1AT and IgG spotted directly to the slide, and show all detected N-glycans comprising more than 1% of overall glycan signal. Proposed structures for these N-glycans are shown, with the core fucose linkage on IgG N-glycans assigned based on orthogonal HPLC and lectin characterizations of this glycoprotein (Figure S1) as well as other literature sources.34,35 Mass values of all detected N-glycans on A1AT and IgG are listed in Table S1.
Figure 2.
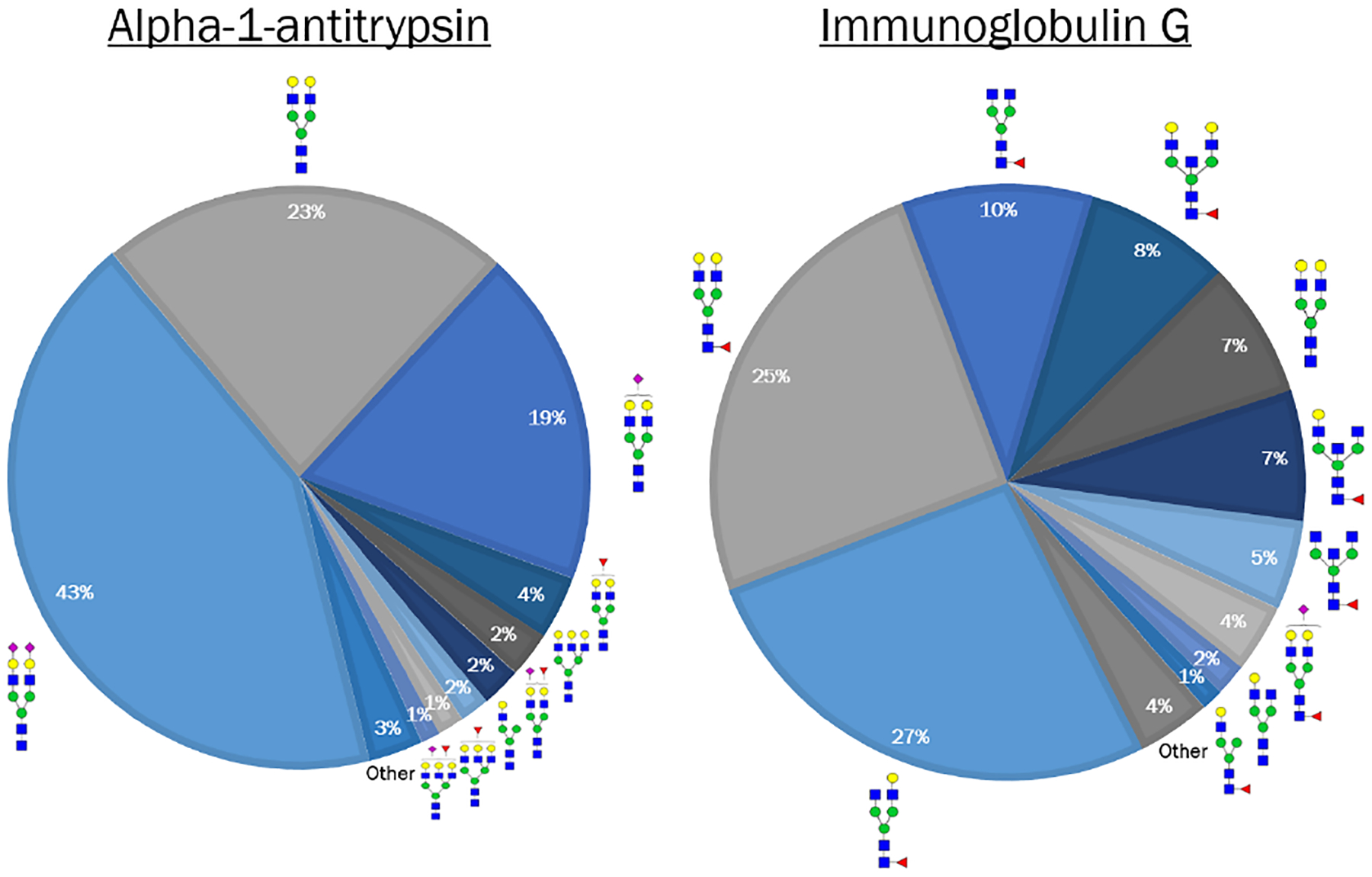
N-glycan profiles observed on human A1AT and IgG by MALDI of spotted proteins. Glycoproteins were spotted (500ng) to a slide and imaged by MALDI FT-ICR MS for detection of N-glycans on each protein. Percentage of each N-glycan specie was calculated by area under the peak divided by the total of all N-glycan peak areas. The proposed structures for all species comprising >1% of each protein’s N-glycome are shown above, and there are clearly distinct N-glycan structures on the two proteins. N-glycan compositions are represented by blue square for N-acetylglucosamine, green circle for mannose, red triangle for fucose, and yellow circle for galactose.
Antibody capture of glycoproteins was performed similar to immunoassay procedures used in other array formats.11,36 Antibodies were manually pipetted at 200ng per spot, each spot a 1.5μL volume. Wells were created with clip-on well modules and antibodies spotted within these wells. Slides were blocked with 1% BSA to prevent nonspecific binding to the slide or other antibodies. Figure 3A illustrates that the slide is sufficiently blocked to prevent A1AT binding when the protein was spotted directly to the slide followed by a wash to remove unbound protein. When A1AT was added to a well as a 100μL sample, capture specificity to its antibody was seen with A1AT N-glycans localized to anti-A1AT but not an adjacent anti-IgG spot, shown in Figure 3B. Circles were added to indicate the position of spotted anti-A1AT (red) and anti-IgG (blue) within the well. Figure 3C contains a dilution series of A1AT added to its antibody, illustrating the successful capture of a glycoprotein and N-glycan detection localized to capture spots. A major N-glycan signature was from m/z 2289.7619 (Hex5HexNAc4NeuAc2+3Na), which is depicted in 3C. This glycan represents approximately 47% of the total glycan pool on A1AT and this peak can be observed at 10 ng of captured protein. This correlates to approximately 3 femtomoles of that glycan, which highlights the sensitivity of this platform. N-glycan signal intensities within each spot were quantified using the area under the peak. As shown in Figure 3D, N-glycan signal from immunocaptured A1AT was detected in a concentration-dependent manner, with a signal plateau observed as the antibodies neared saturation. The profile of the most abundant N-glycans detected on this captured glycoprotein showed extremely strong agreement with that of glycoprotein spotted directly to the slide rather than antibody-captured, Figure 3E. This was done to confirm that the glycoforms of A1AT present in solution are all captured by its antibody. N-glycan of m/z 1809.6923 (Hex5dHex1HexNAc4+Na) was excluded from this analysis as it is highly abundant on the capture antibody and thus would confound the comparison of spotted to captured profiles. An orthogonal analysis of the N-glycan profile of A1AT was performed with HPLC and lectin profiling, Figure S1.
Figure 3.
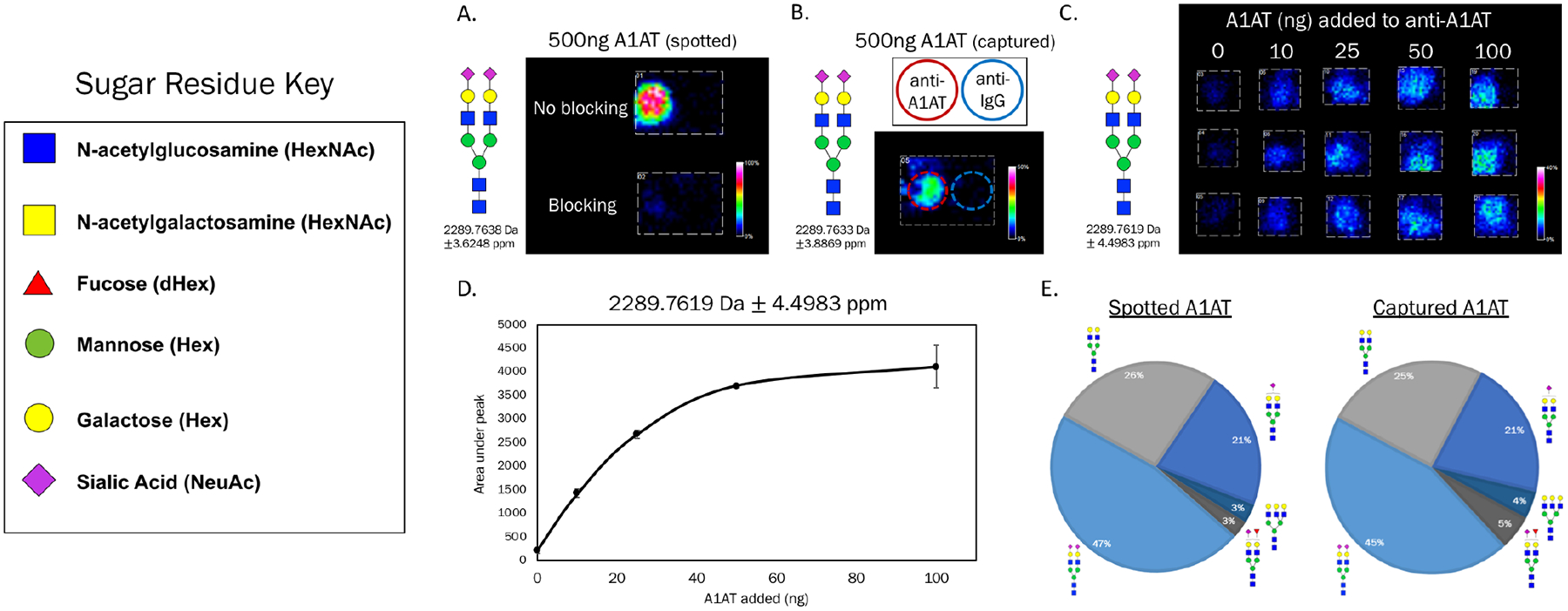
N-glycan detection by MALDI MSI of immunocaptured A1AT. A) A1AT was spotted (1μL) to slide with and without blocking (1% BSA). Imaging data is from the most abundant N-glycan observed on A1AT, which is not seen on the blocked slide. B) A1AT was added in a 100μL volume to a well containing both anti-A1AT and anti-IgG as adjacent spots, red and blue circles added for emphasis of antibody location. C) A dilution series of A1AT standard solutions (100μL) were added in triplicate to anti-A1AT. N-glycan signal is seen at antibody spots with an increase in color intensity observed as more glycoprotein was present. D) Quantifications of imaging data in C were performed by calculating the area under the peak for each sample. Each data point represents the average +/− standard deviation of three samples. E) Comparison of A1AT spotted directly to the slide versus antibody-captured. N-glycan profiles showed strong agreement. Percentages of each N-glycan specie were calculated by area under the peak divided by the total of all N-glycan peak areas. The proposed structures for the most abundant N-glycans on A1AT are shown.
To illustrate the potential of this platform to become a multiplex array for the simultaneous analysis of multiple glycoprotein targets, side-by-side capture of two glycoproteins was tested. Human A1AT and IgG were used and antibodies to the two proteins were spotted side-by-side within each well as depicted in Figure 4A. Mixtures containing both A1AT and IgG standards were added to wells in triplicate 100 μL volumes at the concentrations shown in Figures 4B and 4C, with red and blue circles added to indicate antibody positions. N-glycans were detected from both glycoproteins localized to their individual capture spots, with an N-glycan signature unique to A1AT shown in Figure 4B (m/z 2289.7419, Hex5HexNAc4NeuAc2 +3Na) and unique to IgG in 4C (m/z 1485.5335, Hex3dHex1HexNAc4 +Na). Quantifications to compare the protein signal to antibody background signal for these images are shown in Figure S2. Specificity of capture was observed by the lack of protein-specific N-glycan signals on the opposite antibodies as well as the surrounding slide itself. As the goal of this platform is application to biological samples for biomarker discovery, human serum was also used for side-by-side capture of glycoproteins A1AT and IgG from a more complex mixture. Commercially-obtained human serum was diluted in PBS (1:100) and added to the wells containing both A1AT and IgG antibodies as previously shown. As both glycoproteins are present in serum at concentrations greater than 1mg/mL,37 addition of 1μL of serum per well was more than sufficient to saturate the 200ng of capture antibodies spotted in each well. Figures 4D and 4E depict N-glycan signatures associated with both glycoproteins captured from just 1μL of serum, again showing great specificity of capture.
Figure 4.
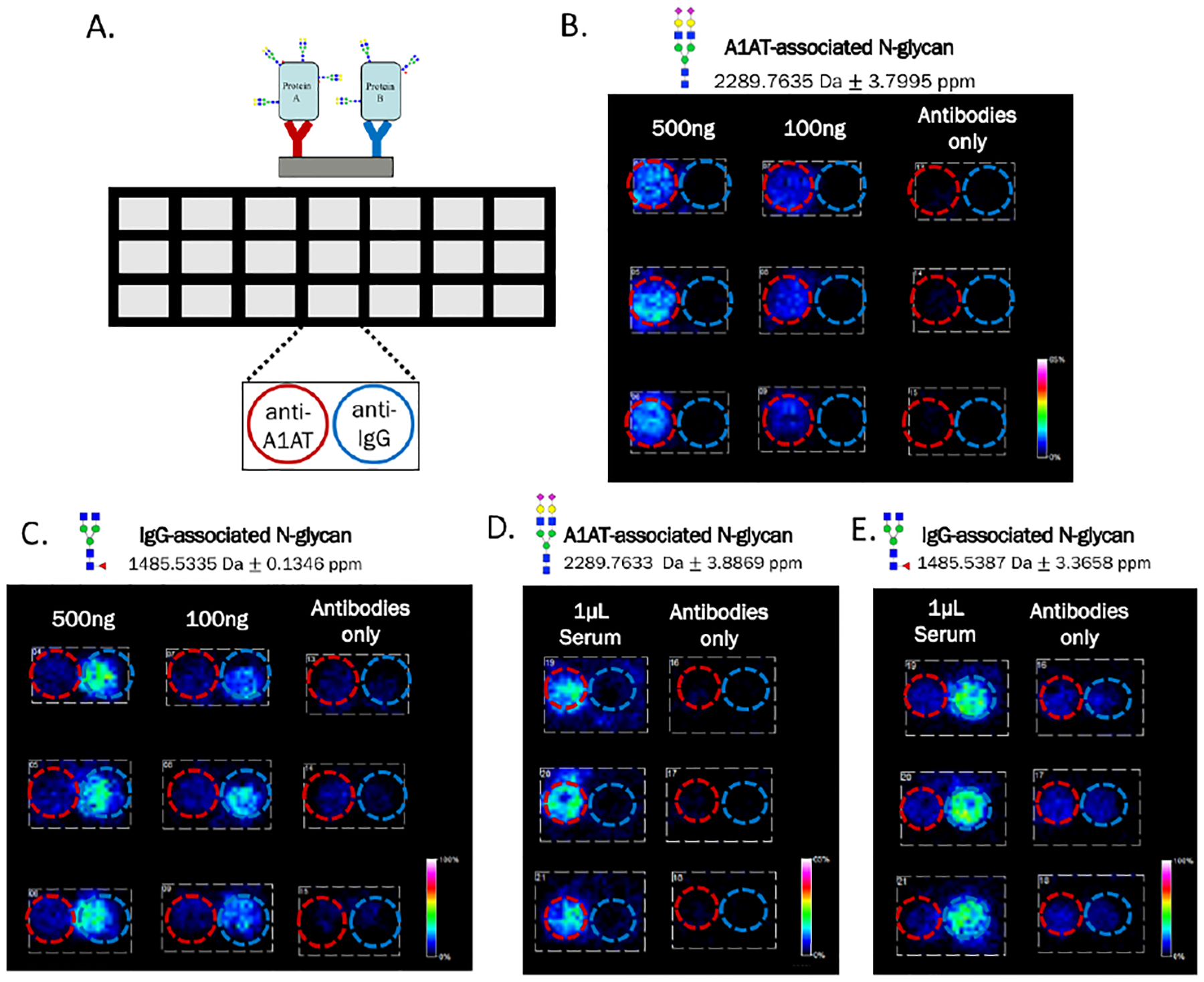
Side-by-side capture of A1AT and IgG from standard solutions and stock human serum. A) Template of the well module used on nitrocellulose-coated microscope slides. Within each well both anti-A1AT and anti-IgG were spotted adjacently at 200ng per 1.5μL spot. B-C) Solutions containing a mixture of both A1AT and IgG standards were added in triplicate to the two antibodies, red and blue circles added to indicate positions of anti-A1AT and anti-IgG, respectively. An A1AT-associated N-glycan was observed localized to the leftof each well, illustrating specific capture of this glycoprotein by anti-A1AT. An IgG-associated N-glycan was observed localized to the right of each well, illustrating specific capture of this glycoprotein by anti-IgG. D-E) Stock human serum was diluted in PBS and added in triplicate to each well at only 1μL of serum per 100μL well. N-glycan signals from both A1AT and IgG were again detected localized to their respective antibodies. The N-glycan compositions are represented by blue square for N-acetylglucosamine, green circle for mannose, red triangle for fucose, and yellow circle for galactose.
An anticipated application for this new platform is the discovery of disease-specific changes in N-glycans on proteins captured from patient biofluid samples. As a proof of concept, serum was pooled from 5 patients with liver cirrhosis, and 1μL of the pooled sample, diluted 1:100 in PBS, was added to the array in the triplicate. Commercially-obtained serum was added in the same manner for comparison. This concentration of serum again allowed for saturating amounts of these very-abundant proteins to be added to the antibodies, which is important to ensure detected glycan changes are not due to variations in protein levels. Glycoproteins were again specifically captured by their antibodies with detectable levels of distinct N-glycans, imaging data and overall N-glycan profiles of IgG shown in Figure 5. Notably, an increase in an IgG nongalactosylated N-glycan m/z 1485.5328 (Hex3dHex1HexNAc4+Na) was observed in the cirrhotic serum compared to stock human serum, Figures 5A–C. This particular N-glycan has been previously described to increase in cirrhotic serum38,39 and this new platform showed agreement with those findings. A subsequent decrease in galactosylated biantennary N-glycans (m/z 1809.6293 Hex5dHex1HexNAc4+Na and 1647.5545 Hex4dHex1HexNAc4+Na) was also observed, illustrating the potential for comparison of glycan ratios that may change in the presence of disease (Figures 5B and 5C). For example, the ratio of glycans m/z 1485.5328 to 1809.6293 increased from 0.35 to 0.68 in cirrhotic patient serum. Intensity value comparisons of these N-glycans are shown in Figure S3. Additional N-glycans were observed on IgG only from cirrhotic samples, which illustrates the potential for greater variance in glycan profiles as clinical samples are used. A list of all N-glycans observed in these samples is found in Table S2. A1AT was also successfully captured from these cirrhotic samples without a loss in binding specificity, as shown in Figure 5D. This experiment illustrates the future potential for this platform to be applied to patient samples for detection of disease-related N-glycan changes for biomarker purposes. Together, these results describe the development of a new APB mass spectrometry platform for protein-specific N-glycan analysis from biofluid samples in a clinically-relevant manner requiring minimal sample consumption.
Figure 5.
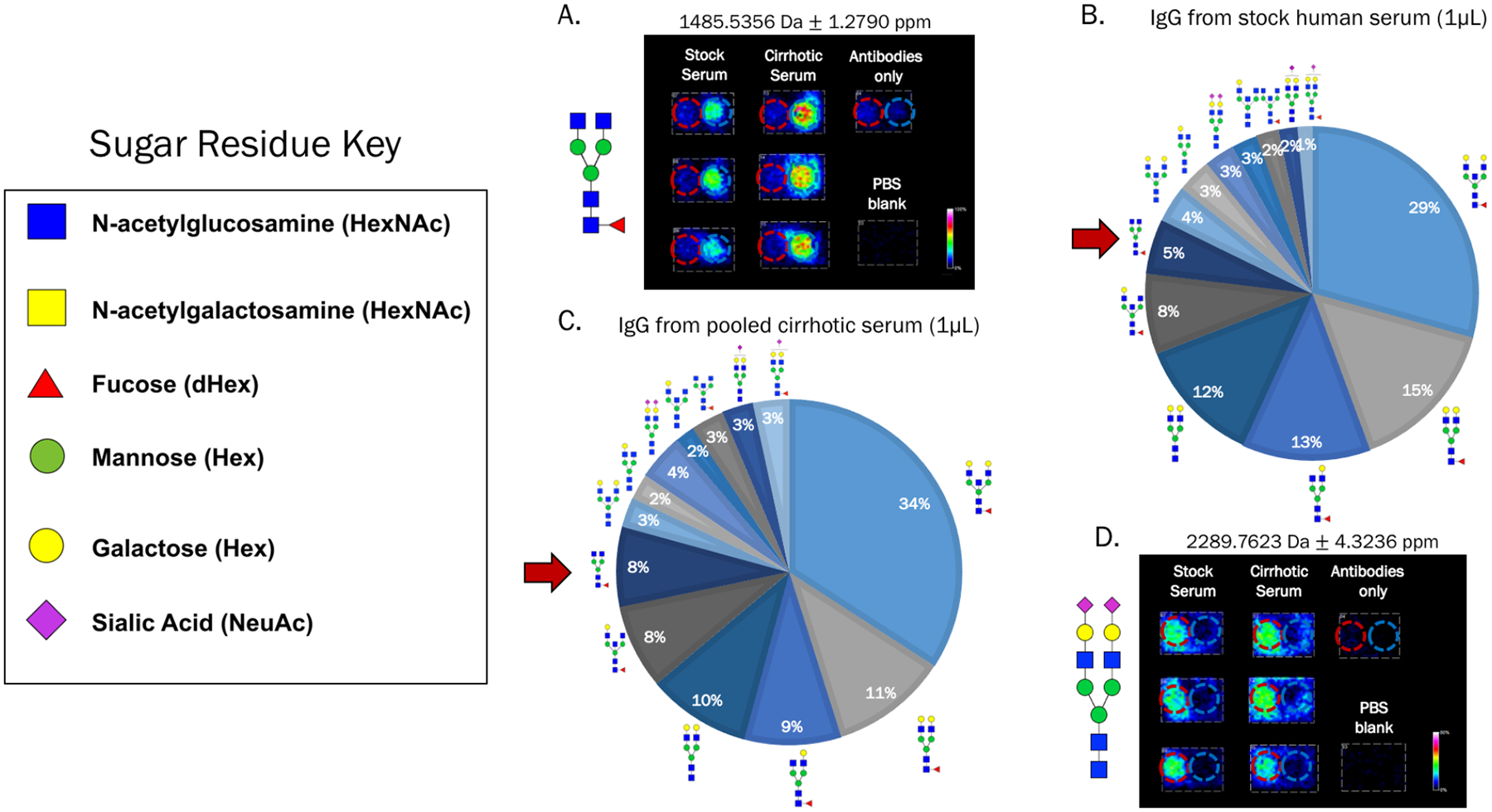
Potential for detection of altered N-glycosylation in patient serum samples. A) Stock human serum and serum pooled from 5 patients with cirrhosis were added in triplicate to wells containing both anti-A1AT and anti-IgG as shown previously. 1μL of serum was diluted in 100μL PBS for the addition to each well. An IgG-associated N-glycan was observed to increase in the cirrhotic samples compared to the stock serum, which has been previously reported. B-C) IgG N-glycan profiles in stock human serum and cirrhotic patient serum illustrating an increase in the nongalactosylated fucosylated biantennary N-glycan from A (red arrow) and a subsequent decrease in the galactosylated fucosylated biantennary N-glycans. Percentages of each N-glycan specie were calculated by area under the peak divided by the total of all N-glycan peak areas. D) An A1AT-associated N-glycan is shown to illustrate that this glycoprotein was also specifically captured out of both stock and cirrhotic patient serum.
Conclusion
Described here is a new antibody panel based mass spectrometry platform for the multiplexed detection of N-glycans in a protein-specific manner from biological samples. The development of this technique was based on a well-established protocol for enzymatic release of N-glycans from tissue sections for MALDI MSI.18,20 The two-dimensional analysis with detection by MSI allows for the mapping of N-glycan signals to their carrier proteins along a slide-based antibody array. In this platform, antibodies are essential for the specific capture of glycoprotein targets from a complex biological mixture, similar to an ELISA. Yet unlike an ELISA, no secondary antibody or lectin is needed for this methodology as mass spectrometry provides sensitive and specific detection of distinct N-glycans. Antibody capture also negates the need for sample clean-up prior to MS analysis, which can be extensive.1,3,40–42 However, as is typical for serum-based assays, evidence for the presence of heterophilic antibodies in cirrhotic serum has been observed with nonspecific binding to some antibodies assayed, which can lead to false positives in clinical assays.29,43 Antibody capture has been previously used to capture a single target protein for MALDI MS analysis,44,45 yet our novel multiplexed technique can be expanded for the analysis of potentially 100s or 1000s of different N-glycoproteins in one imaging run. Each run generates an immense amount of data, as spectra showing potentially hundreds of N-glycan species are gathered localized to each glycoprotein on the array. Therefore, this method has powerful capabilities for the characterization of N-glycosylation across many target proteins simultaneously.
This new method extends the capabilities of existing N-glycan biomarker detection technologies. Lectin microarrays have been used for detection of changes in N-glycans in a biomarker setting,11,46–48 however our new APB N-glycan imaging method significantly increases the amount of information that can be obtained from such analyses. While lectins bind to N-glycan structural motifs, MALDI MS detects the compositions of N-glycans present. The method can be easily adapted to the use of other instrumentation, e.g., ion mobility, which will allow reporting on configuration of N-glycoforms. Additionally, APB glycan imaging obtains a complete mass spectrum for each glycoprotein capture spot, allowing hundreds of potential N-glycan masses to be probed per glycoprotein target as opposed to a select few probed with targeted lectin analysis. Detection of the glycan heterogeneity present on each protein can be used for calculation of glycan ratios, which may represent important alterations in the overall glycosylation of a protein that can be clinically utilized.49,50 Though different glycans will vary in ionization efficiencies, these inherent properties are assumed to remain constant across different samples, thus having a negligible effect on ratio comparison. As previously mentioned, MSI analyses on tissues have been used for elucidating N-glycan changes in the presence of disease.21,22,51,52 While tissue-based analysis is often used for prognosis and pathological examination, it is not as an accessible material for early detection of disease, as is serum or other biofluids. We hypothesize that our new biomarker discovery and validation platform may be used with readily available patient biofluids such as serum or urine.
Further work remains to increase the clinical power of this platform. More antibodies can be added so that more glycoproteins can be probed per analysis. This improvement will be limited by the quality of these antibodies – both in binding affinity and specificity – as is widely recognized in the field of antibody research. The addition of patient samples shown in Figure 5 highlights the importance of antibody quality in specifically capturing target glycoproteins from complex samples that have potential for great heterogeneity. We acknowledge to the reviewer that we understand concerns of capture specificity in clinical samples that will require validation of each antibody used. Observed variance in clinical samples could occur due to poor antibodies, or it possible the sensitivity of our new platform could unveil novel disease-related anomalies not previously recognized by current N-glycan analysis methods. Future work will be performed to remove the N-glycans present on antibodies to limit background signal. Clinical relevance can also be improved by streamlining the workflow, as the current method requires 1.5 days. Additionally, translation of this technique to other mass spectrometry platforms will be important both for achieving complete structural information of the detected N-glycans as well as moving to more clinically-accessible MALDI MS instruments. The MALDI MS instrumentation used in this study does not uncover the full structural configuration of an N-glycan, rather its composition. While the N-glycan profiles of the two glycoproteins used here have been well-characterized by orthogonal methods, future work with less-studied glycoproteins will require additional N-glycan structural characterization or orthogonal confirmation. Mass spectrometry analysis following trypsin digest to observe peptides rather than N-glycans will also be used to confirm glycoprotein binding specificity at each antibody.
The development of this novel platform suggests exciting potential for application in the clinic as both a biomarker discovery tool as well as a new screening platform for a number of diseases in readily available clinical biofluid samples. This platform was able to detect N-glycans on glycoproteins captured from only 1μL of human serum, illustrating its potential for use with very minimal patient sample consumption. N-glycans and their role in disease progression are quickly becoming recognized as an important new frontier for biomedical research. However, the applications of this new technique extend beyond just N-glycans and biofluid samples: this platform could be used with liquids such as cell supernatants or probe other classes of glycans or post-translational modifications.
Supplementary Material
Table S1: All observed N-glycans on A1AT and IgG (PDF)
Table S2: All observed N-glycans in capture from human serum (PDF)
Figure S1: HPLC and lectin N-glycan profiles of A1AT and IgG (PDF)
Figure S2: Quantification of N-glycan peaks in Figure 4 (PDF)
Figure S3: Quantifications of N-glycan peaks observed in normal and cirrhotic serum (PDF)
ACKNOWLEDGMENTS
The authors would like to thank Dr. Amit Singal (UTSW) for providing patient serum samples. This work was supported by funding from South Carolina Clinical & Translational Research Institute, NIH/NCATS grant numbers TL1 TR001451 and UL1 TR001450 (APB), R21 CA225474-01 (ASM and BBH), and the South Carolina Centers of Economic Excellence (ASM and RRD).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
The authors declare no competing financial interest.
REFERENCES
- (1).Kailemia MJ; Park D; Lebrilla CB Glycans and Glycoproteins as Specific Biomarkers for Cancer. Anal. Bioanal. Chem 2017, 409 (2), 395–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Adamczyk B; Tharmalingam T; Rudd PM Glycans as Cancer Biomarkers. Biochim. Biophys. Acta 2012, 1820 (9), 1347–1353. [DOI] [PubMed] [Google Scholar]
- (3).Kuzmanov U; Kosanam H; Diamandis EP The Sweet and Sour of Serological Glycoprotein Tumor Biomarker Quantification. BMC Med. 2013, 11 (31). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ohtsubo K; Marth JD Glycosylation in Cellular Mechanisms of Health and Disease. Cell 2006, 126 (5), 855–867. [DOI] [PubMed] [Google Scholar]
- (5).Meany DL; Chan DW Aberrant Glycosylation Associated with Enzymes as Cancer Biomarkers. Clin. Proteomics 2011, 8 (1), 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Taniguchi N Toward Cancer Biomarker Discovery Using the Glycomics Approach. Proteomics 2008, 8, 3205–3208. [DOI] [PubMed] [Google Scholar]
- (7).Aoyagi Y; Isokawa O; Suda T; Watanabe M; Suzuki Y; Asakura H The Fucosylation Index of Alpha-Fetoprotein as a Possible Prognostic Indicator for Patients with Hepatocellular Carcinoma. Cancer 1998, 83 (10), 2076–2082. [PubMed] [Google Scholar]
- (8).Mariño K; Bones J; Kattla JJ; Rudd PM A Systematic Approach to Protein Glycosylation Analysis: A Path through the Maze. Nature Chemical Biology. 2010. [DOI] [PubMed] [Google Scholar]
- (9).Geyer H; Geyer R Strategies for Analysis of Glycoprotein Glycosylation. Biochim. Biophys. Acta - Proteins Proteomics 2006, 1764 (12), 1853–1869. [DOI] [PubMed] [Google Scholar]
- (10).Reatini BS; Ensink E; Liau B; Sinha JY; Powers TW; Partyka K; Bern M; Brand RE; Rudd PM; Kletter D; et al. Characterizing Protein Glycosylation through On-Chip Glycan Modification and Probing. Anal. Chem 2016, 88, 11584–11592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Chen S; LaRoche T; Hamelinck D; Bergsma D; Brenner D; Simeone D; Brand RE; Haab BB Multiplexed Analysis of Glycan Variation on Native Proteins Captured by Antibody Microarrays. Nat. Methods 2007, 4 (5), 437–444. [DOI] [PubMed] [Google Scholar]
- (12).Hirabayashi J Concept, Strategy and Realization of Lectin-Based Glycan Profiling. J. Biochem 2008, 144 (2), 139–147. [DOI] [PubMed] [Google Scholar]
- (13).Angel PM; Caprioli RM Matrix-Assisted Laser Desorption Ionization Imaging Mass Spectrometry: In Situ Molecular Mapping. Biochemistry 2013, 52 (22), 3818–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Seeley EH; Schwamborn K; Caprioli RM Imaging of Intact Tissue Sections: Moving beyond the Microscope. J. Biol. Chem 2011, 286 (29), 25459–25466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Buchberger AR; DeLaney K; Johnson J; Li L Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights. Anal. Chem 2018, 90 (1), 240–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Balluff B; Schöne C; Höfler H; Walch A MALDI Imaging Mass Spectrometry for Direct Tissue Analysis: Technological Advancements and Recent Applications. Histochem. Cell Biol. 2011, 136 (3), 227–244. [DOI] [PubMed] [Google Scholar]
- (17).Walch A; Rauser S; Deininger S-O; Höfler H MALDI Imaging Mass Spectrometry for Direct Tissue Analysis: A New Frontier for Molecular Histology. Histochem. Cell Biol. 2008, 130 (3), 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Powers TW; Neely BA; Shao Y; Tang H; Troyer DA; Mehta AS; Haab BB; Drake RR; Batra SK MALDI Imaging Mass Spectrometry Profiling of N-Glycans in Formalin-Fixed Paraffin Embedded Clinical Tissue Blocks and Tissue Microarrays. PLoS One 2014, 9 (9), e106255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Drake RR; Powers TW; Jones EE; Bruner E; Mehta AS; Angel PM MALDI Mass Spectrometry Imaging of N-Linked Glycans in Cancer Tissues. Adv. Cancer Res. 2017, 134, 85–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Powers TW; Jones EE; Betesh LR; Romano PR; Gao P; Copland JA; Mehta AS; Drake RR Matrix Assisted Laser Desorption Ionization Imaging Mass Spectrometry Workflow for Spatial Profiling Analysis of N-Linked Glycan Expression in Tissues. Anal. Chem 2013, 85, 9799–9806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).West CA; Wang M; Herrera H; Liang H; Black A; Angel PM; Drake RR; Mehta AS N-Linked Glycan Branching and Fucosylation Are Increased Directly in Hcc Tissue As Determined through in Situ Glycan Imaging. J. Proteome Res. 2018, 17 (10), 3454–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Scott DA; Casadonte R; Cardinali B; Spruill L; Mehta AS; Carli F; Simone N; Kriegsmann M; Del Mastro L; Kriegsmann J; et al. Increases in Tumor N-Glycan Polylactosamines Associated with Advanced HER2 Positive and Triple Negative Breast Cancer Tissues. Proteomics Clin Appl 2018, 13 (1), 1800014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Drake RR; Powers TW; Norris-Caneda K; Mehta AS; Angel PM In Situ Imaging of N-Glycans by MALDI Imaging Mass Spectrometry of Fresh or Formalin-Fixed Paraffin-Embedded Tissue. Curr. Protoc. Protein Sci. 2018, 94 (1), e68. [DOI] [PubMed] [Google Scholar]
- (24).Heijs B; Holst S; Briaire-De Bruijn IH; Van Pelt GW; De Ru AH; Van Veelen PA; Drake RR; Mehta AS; Mesker WE; Tollenaar RA; et al. Multimodal Mass Spectrometry Imaging of N-Glycans and Proteins from the Same Tissue Section. Anal. Chem 2016, 88, 7745–7753. [DOI] [PubMed] [Google Scholar]
- (25).Gustafsson OJR; Briggs MT; Condina MR; Winderbaum LJ; Pelzing M; McColl SR; Everest-Dass AV; Packer NH; Hoffmann P MALDI Imaging Mass Spectrometry of N-Linked Glycans on Formalin-Fixed Paraffin-Embedded Murine Kidney. Anal. Bioanal. Chem 2015, 407 (8), 2127–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Holst S; Heijs B; De Haan Noortje; Van Zeijl RM; Briaire-De Bruijn IH; Van Pelt GW; Mehta AS; Angel PM; Mesker WE; Tollenaar RA; et al. Linkage-Specific in Situ Sialic Acid Derivatization for N-Glycan Mass Spectrometry Imaging of Formalin-Fixed Paraffin-Embedded Tissues. Anal. Chem 2016, 88 (11), 5904–5911. [DOI] [PubMed] [Google Scholar]
- (27).Everest-Dass AV; Briggs MT; Kaur G; Oehler MK; Hoffmann P; Packer NH N-Glycan MALDI Imaging Mass Spectrometry on Formalin-Fixed Paraffin-Embedded Tissue Enables the Delineation of Ovarian Cancer Tissues. Mol. Cell. Proteomics 2016, 15 (9), 3003–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Angel PM; Mehta A; Norris-Caneda K; Drake RR MALDI Imaging Mass Spectrometry of N-Glycans and Tryptic Peptides from the Same Formalin-Fixed, Paraffin-Embedded Tissue Section. In Methods Mol Biol; 2017; pp 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Wang M; Shen J; Herrera H; Singal A; Swindell C; Renquan L; Mehta A Biomarker Analysis of Fucosylated Kininogen through Depletion of Lectin Reactive Heterophilic Antibodies in Hepatocellular Carcinoma. J. Immunol. Methods 2018, 462, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Comunale MA; Rodemich-Betesh L; Hafner J; Wang M; Norton P; di Bisceglie AM; Block T; Mehta A Linkage Specific Fucosylation of Alpha-1-Antitrypsin in Liver Cirrhosis and Cancer Patients: Implications for a Biomarker of Hepatocellular Carcinoma. PLoS One 2010, 5 (8), e12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Klamer Z; Hsueh P; Ayala-Talavera D; Haab B Deciphering Protein Glycosylation by Computational Integration of On-Chip Profiling, Glycan-Array Data, and Mass Spectrometry. Mol. Cell. Proteomics 2019, 18 (1), 28–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Clerc F; Reiding KR; Jansen BC; Kammeijer GSM; Bondt A; Wuhrer M Human Plasma Protein N-Glycosylation. Glycoconj. J 2016, 33 (3), 309–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).McCarthy C; Saldova R; Wormald MR; Rudd PM; McElvaney NG; Reeves EP The Role and Importance of Glycosylation of Acute Phase Proteins with Focus on Alpha-1 Antitrypsin in Acute and Chronic Inflammatory Conditions. J Proteome Res 2014, 13 (7), 3131–3143. [DOI] [PubMed] [Google Scholar]
- (34).Mittermayr S; Bones J; Doherty M; Guttman A; Rudd PM Multiplexed Analytical Glycomics: Rapid and Confident IgG N-Glycan Structural Elucidation. J. Proteome Res 2011, 10, 3820–3829. [DOI] [PubMed] [Google Scholar]
- (35).Saldova R; Stöckmann H; O’Flaherty R; Lefeber DJ; Jaeken J; Rudd PM N-Glycosylation of Serum IgG and Total Glycoproteins in MAN1B1 Deficiency. J. Proteome Res. 2015, 14 (10), 4402–4412. [DOI] [PubMed] [Google Scholar]
- (36).Wang J; Barker K; Steel J; Park J; Saul J; Fest F; Wallstrom G; Yu X; Bian X; Anderson KS; et al. A Versatile Protein Microarray Platform Enabling Antibody Profiling against Denatured Proteins. Proteomics Clin Appl 2014, 7 (0), 378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Anderson NL; Anderson NG The Human Plasma Proteome: History, Character, and Diagnostic Prospects. Mol. Cell. Proteomics 2002, 1 (11), 845–867. [DOI] [PubMed] [Google Scholar]
- (38).Mehta AS; Long RE; Comunale MA; Wang M; Rodemich L; Krakover J; Philip R; Marrero JA; Dwek RA; Block TM Increased Levels of Galactose-Deficient Anti-Gal Immunoglobulin G in the Sera of Hepatitis C Virus-Infected Individuals with Fibrosis and Cirrhosis. J. Virol 2008, 82 (3), 1259–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Lamontagne A; Long RE; Comunale MA; Hafner J; Rodemich-Betesh L; Wang M; Marrero J; Di Bisceglie AM; Block T; Mehta A Altered Functionality of Anti-Bacterial Antibodies in Patients with Chronic Hepatitis C Virus Infection. PLoS One 2013, 8 (6), e64992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Song T; Aldredge D; Lebrilla CB A Method for In-Depth Structural Annotation of Human Serum Glycans That Yields Biological Variations. Anal. Chem 2015, 87, 7754–7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Ruhaak LR; Huhn C; Waterreus W; De Boer AR; Neusu C; Hokke CH; Deelder AM; Wuhrer M; Deelder M Hydrophilic Interaction Chromatography-Based High-Throughput Sample Preparation Method for N-Glycan Analysis from Total Human Plasma Glycoproteins Hydrophilic Interaction Chromatography-Based High-Throughput Sample Preparation Method for N-Glycan Analysis. Anal. Chem 2008, 80 (15), 6119–6126. [DOI] [PubMed] [Google Scholar]
- (42).Reiding KR; Blank D; Kuijper DM; Deelder AM; Wuhrer M High-Throughput Profiling of Protein N-Glycosylation by MALDI-TOF-MS Employing Linkage-Specific Sialic Acid Esterification. Anal. Chem 2014, 86 (12), 5784–5793. [DOI] [PubMed] [Google Scholar]
- (43).Bolstad N; Warren DJ; Nustad K Heterophilic Antibody Interference in Immunometric Assays. Best Pract. Res. Clin. Endocrinol. Metab 2013, 27 (5), 647–661. [DOI] [PubMed] [Google Scholar]
- (44).Darebna P; Spicka J; Kucera R; Topolcan O; Navratilova E; Ruzicka V; Volny M; Novak P; Pompach P Detection and Quantification of Carbohydrate-Deficient Transferrin by MALDI-Compatible Protein Chips Prepared by Ambient Ion Soft Landing. Clin. Chem 2018, 64 (9). [DOI] [PubMed] [Google Scholar]
- (45).Pompach P; Nováková J; Kavan D; Benada O; Růžička V; Volný M; Novák P Planar Functionalized Surfaces for Direct Immunoaffinity Desorption/Ionization Mass Spectrometry. Clin. Chem 2016, 62 (1), 270–278. [DOI] [PubMed] [Google Scholar]
- (46).Yue T; Goldstein IJ; Hollingsworth MA; Kaul K; Brand RE; Haab BB The Prevalence and Nature of Glycan Alterations on Specific Proteins in Pancreatic Cancer Patients Revealed Using Antibody-Lectin Sandwich Arrays. Mol. Cell. Proteomics 2009, 8 (7), 1697–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Nagaraj VJ; Eaton S; Thirstrup D; Wiktor P Piezoelectric Printing and Probing of Lectin NanoProbeArrays for Glycosylation Analysis. Biochem. Biophys. Res. Commun 2008, 375 (4), 526–530. [DOI] [PubMed] [Google Scholar]
- (48).Patwa TH; Zhao J; Anderson MA; Simeone DM; Lubman DM Screening of Glycosylation Patterns in Serum Using Natural Glycoprotein Microarrays and Multi-Lectin Fluorescence Detection. Anal. Chem 2006, 78 (18), 6411–6421. [DOI] [PubMed] [Google Scholar]
- (49).Callewaert N; Van Vlierberghe H; Van Hecke A; Laroy W; Delanghe J; Contreras R Noninvasive Diagnosis of Liver Cirrhosis Using DNA Sequencer-Based Total Serum Protein Glycomics. Nat. Med 2004, 10 (4), 429–434. [DOI] [PubMed] [Google Scholar]
- (50).Verhelst X; Vanderschaeghe D; Castéra L; Raes T; Geerts A; Francoz C; Colman R; Durand F; Callewaert N; Van Vlierberghe H A Glycomics-Based Test Predicts the Development of Hepatocellular Carcinoma in Cirrhosis. Clin. Cancer Res. 2017, 23 (11), 2750–2758. [DOI] [PubMed] [Google Scholar]
- (51).Kunzke T; Balluff B; Feuchtinger A; Buck A; Langer R; Luber B; Lordick F; Zitzelsberger H; Aichler M; Walch A Native Glycan Fragments Detected by MALDI-FT-ICR Mass Spectrometry Imaging Impact Gastric Cancer Biology and Patient Outcome. Oncotarget 2017, 8 (40), 68012–68025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Powers T; Holst S; Wuhrer M; Mehta A; Drake R; Powers TW; Holst S; Wuhrer M; Mehta AS; Drake RR Two-Dimensional N-Glycan Distribution Mapping of Hepatocellular Carcinoma Tissues by MALDI-Imaging Mass Spectrometry. Biomolecules 2015, 5 (4), 2554–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: All observed N-glycans on A1AT and IgG (PDF)
Table S2: All observed N-glycans in capture from human serum (PDF)
Figure S1: HPLC and lectin N-glycan profiles of A1AT and IgG (PDF)
Figure S2: Quantification of N-glycan peaks in Figure 4 (PDF)
Figure S3: Quantifications of N-glycan peaks observed in normal and cirrhotic serum (PDF)


