Abstract
Chaperone-mediated autophagy (CMA) is a selective type of autophagy whereby a specific subset of intracellular proteins is targeted to the lysosome for degradation. These proteins are identified by a chaperone that targets them to lysosomes. There, they are translocated into the organelle lumen through a lysosomal membrane receptor/translocation complex. CMA plays an important role in maintaining cellular proteostasis by eliminating damaged and altered proteins. CMA also participates in the control of the cellular energetic balance through recycling of amino acids resulting from lysosomal proteolysis of the substrate proteins. Lastly, due to the intrinsic protein selectivity of CMA, this type of autophagy exerts regulatory functions by mediating timely degradation of key cellular proteins that participate in processes such as lipid and glucose metabolism, cell cycle, DNA repair, and cellular reprogramming, among others. Dysfunctional CMA occurs with age and has now been described in a growing list of human pathologies such as metabolic disorders, neurodegeneration, cancer, immunodeficiency, and diabetes. In this chapter, we describe current methodologies to quantitatively analyze CMA activity in different experimental models.
Keywords: Chaperones, Lysosomes, Proteolysis, Subcellular fractionation
1. Introduction
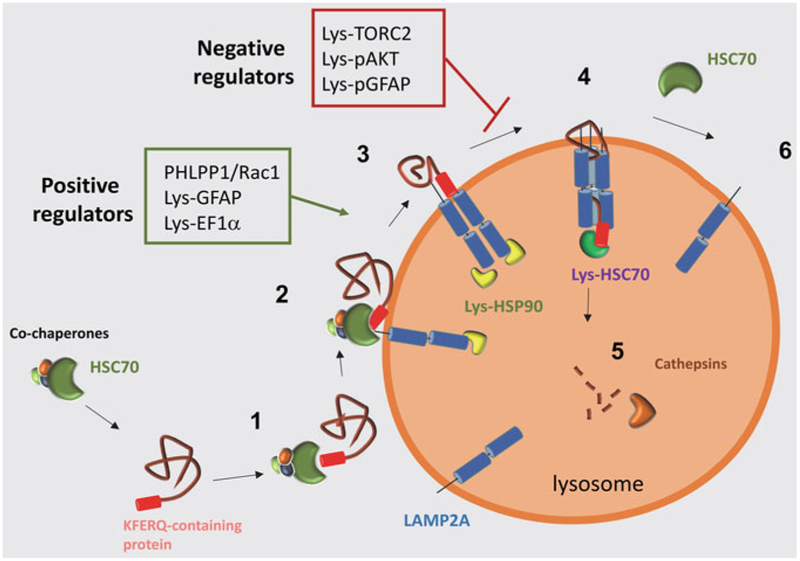
Chaperone-mediated autophagy (CMA) is a multistep process that results in selective degradation of intracellular soluble proteins [1]. Selectivity is driven by the presence of a CMA-targeting motif—a pentapeptide sequence sharing biochemical similarity to KFERQ—in the substrate proteins [2]. This motif is recognized by a constitutively expressed intracellular chaperone, the heat shock-cognate chaperone of 70 kDa (hsc70) (Fig. 1) [3]. Once hsc70 binds the substrate proteins, they are targeted to the surface of a subset of lysosomes active for this autophagic pathway. At the lysosomal membrane, the substrate/chaperone complex docks at the cytosolic tail of a monomeric single-span protein termed lysosome-associated membrane protein type-2A (LAMP2A) [4]. Binding of substrates to LAMP2A initiates its multimerization at the lysosomal membrane, which comprises the basis of the CMA translocation complex [5]. The unfolding of the substrate protein is not required for its binding to the chaperone or to the lysosomal surface, but it is a prerequisite for substrate translocation across the lysosomal membrane. Substrate internalization is mediated by a lysosome-resident hsc70 (lys-hsc70) [6], and it is rapidly followed by complete degradation of the substrate protein into its constitutive amino acids by lysosomal luminal proteases, also known as cathepsins.
Fig. 1.
Chaperone-mediated autophagy: schematic model of the steps in chaperone-mediated autophagy.1. Substrate binding by HSC70 and cochaperones and targeting to lysosomes. 2. Binding of the substrate to LAMP2A at the lysosomal membrane. 3. HSP90 binds to LAMP2A to stabilize it while it organizes into higher molecular weight complexes. 4. Substrate crosses the lysosomal membrane through a LAMP2A-enriched translocation complex, and translocation is complete by the action of luminal HSC70. 5. The substrate is rapidly degraded by luminal proteases (cathepsins). 6. Once substrate translocation is complete, LAMP2A dissociates into monomers in a process dependent on cytosolic HSC70. Red box: negative regulators of CMA at the lysosomal membrane. Green box: positive regulators of CMA at the lysosomal membrane
In addition to these proteins that interact directly with the substrate protein—hsc70 on both side of the lysosomal membrane and LAMP2A—the lysosomal membrane also hosts regulators that modulate CMA activity directly at this compartment. A lysosomal resident form of hsp90 is involved in maintaining the stability of LAMP2A during its multimerization (Fig. 1) [5]. A pair of regulators GFAP/EF1α controls the stability of the LAMP2A translocation complex in a GTP-dependent manner [7]. GFAP exists in the membrane in two forms: unmodified GFAP that associates with the LAMP2A multimer to stabilize the translocation complex and phosphorylated GFAP (pGFAP) that is masked by EF1α [7]. In the presence of GTP, EF1α is released from the membrane allowing pGFAP to become accessible to unmodified GFAP [7]. Affinity of GFAP to form dimers with pGFAP is higher than its binding to LAMP2A; therefore, it dissociates from the translocation complex and binds to pGFAP, resulting in the disassembly of the translocation complex. A second mechanism that controls the dynamics of LAMP2A assembly into the translocation complex involves mTOR complex 2 or TORC2, the kinase AKT, and the phosphatase PHLPP1 [8]. TORC2 provides a continuous inhibitory regulation on CMA through phosphorylation of AKT, that is, the kinase of GFAP at the lysosomal membrane [8]. Continuous phosphorylation of GFAP makes the speed of assembly/disassembly of LAMP2A from the translocation complex constitutively slow (Fig. 1). Whenever maximal activation of CMA is required, this inhibitory effect of TORC2 needs to be released and that is attained through the recruitment of PHLPP1, in a Rac-1-dependent manner, to the lysosomal membrane [8]. PHLPP1 dephosphorylates AKT and that accelerates LAMP2A assembly/disassembly and consequently the rate(s) of substrate(s) uptake (Fig. 1) [8].
In contrast with other types of autophagy, such as macroautophagy or microautophagy that requires formation of vesicles for the uptake of the substrates to be degraded, in CMA, substrate proteins are recognized individually, and they are translocated into the lysosomal lumen independent of vesicular trafficking [1]. Selective targeting of individual proteins for degradation has been reported in a type of microautophagy, known as endosomal microautophagy (e-MI) [9]. In this process, the chaperone hsc70 also recognizes the same KFERQ-like motif in the substrate proteins but delivers them to the surface of late endosomes. There, cargo is internalized in multivesicular bodies (MVB) that form on the surface of this organelle using components of the ESCRT complex [9]. An important difference with CMA is that, despite the abundance of LAMP2A in the late endosomal membrane, e-MI does not use this membrane protein for substrate internalization. Additionally, e-MI substrates do not need to undergo unfolding prior to internalization into MVB luminal vesicles [9], while CMA substrates must be unfolded for internalization into the lysosomal lumen [10].
The selectivity of CMA seems beneficial under conditions in which discrimination between different types of proteins for degradation is required. For example, an increase in CMA activity is observed during prolonged starvation [11]. Degradation of proteins through CMA will provide cells with free amino acids required to sustain protein synthesis under these conditions [11, 12]. Like-wise, activation of CMA during mild oxidative stress or after exposure to compounds that decrease proteostasis allows the selective removal of the proteins damaged or altered under these conditions [13]. In addition, selective removal of proteins through CMA has been shown to exert important regulatory functions in metabolic pathways, DNA repair pathways, and cell cycle, among others [14–16].
Malfunctioning of CMA plays a key role in an increasing amount of severe human disorders [17–20]. In many instances, the mechanisms underlying CMA failure in these pathologies involve perturbations in the functioning of the CMA translocation complex in lysosomes. The interplay of CMA with several neurodegenerative diseases is bidirectional, whereby CMA contributes to the elimination of pathogenic proteins, but it eventually becomes a casualty of the toxic effect of these aberrant proteins [18, 20]. Given this dual role of CMA in neurodegeneration, and the growing number of diseases associated to CMA failure, it has become important to analyze its status in disease conditions to understand if enhancing CMA activity could be a worthwhile endeavor in treating these diseases.
Analysis of the levels of CMA effectors and modulators in lysosomes can yield useful insights into CMA activity in tissues and cells in culture. However, to get an accurate picture of CMA activity, it is necessary to track the targeting and translocation of CMA substrates into lysosomes. To that effect, both in vitro systems with isolated lysosomes and a photoswitchable CMA reporter that works in intact cells are the gold standard methods in the field. In this chapter, we describe the battery of markers that can be used to obtain information on the steady-state status of CMA as well as CMA functional assays and their applicability to different experimental models.
2. Materials
2.1. Isolation of Rat Liver Lysosomes
Wistar rats (200–250 g): Male or female can be selected depending on the purpose of the experiment. To enrich in CMA-active lysosomes and reduce liver glycogen content (as it can interfere with lysosomal isolation), rats can be starved for 24–48 h before liver dissection but should be maintained with water ad libitum.
Tools: Dissection instruments (forceps, scissors, clamps), double cloth gauze, funnel, Teflon/glass homogenizer (for motorized homogenizer).
Centrifugation supplies: Polycarbonate tubes (30 mL), ultraclear tubes for SW41 rotor (Beckman, Fullerton, CA), and SW41 rotor (Beckman).
Homogenization solution: 0.25 M sucrose (American Bioanalytical, Natick, MA) in double-distilled water (ddH2O). Prepare fresh or the day before, and store at 4 °C.
Centrifugation media: Metrizamide (Fitzgerald Industries International, Acton, MA). Prepare as 85.6% (w/v) stock in ddH2O, and adjust to pH 7.2 with NaOH. Store in aliquots at −20 °C protected from light (see Note 1).
Lysosome resuspension buffer: 10 mM 3-(N-Morpholino) propanesulfonic acid (MOPS), 0.25 M sucrose, pH 7.2 (adjusted with NaOH). Store at 4 °C for 1 week maximum.
2.2. Lysosome Purity and Integrity
Millipore multiscreen assay system (Millipore, Bedford, MA), 0.22 μm Durapore filter 96-well plates, vacuum manifold and polystyrene flat-bottom 96-well plates.
Acetate buffer: 0.4 M sodium acetate, pH 4.4.
β-hexosaminidase substrate solution: 4 mM 4-methylumbelli-feryl-N-acetyl-B-D-glucopyranoside in ddH2O. Sonicate to dissolve and store a −20 °C protected from light. Before use, the solution can be sonicated again and kept at 37 °C until use.
10% Triton X-100 (Bio-Rad, Hercules, CA) in ddH2O and store at room temperature.
Reaction mixture: 10 mL acetate buffer, 10 mL β-hexosaminidase substrate solution, 0.5 mL 10% Triton X-100, 19.5 mL ddH2O. Store at −20 °C and thaw by placing in a 37 °C before use.
Stop solution: 0.5 M glycine, 0.5 M Na2CO3, in ddH2O.
Blocking solution: 20 mg/mL bovine serum albumin (BSA) in ddH2O.
0.25 M sucrose (American Bioanalytical, Natick, MA): Prepare fresh in ddH2O.
2.3. Lysosomal Binding/Uptake Assay
Incubation buffer: 10 mM MOPS, 0.25 M sucrose in ddH2O adjusted to pH 7.3. Prepare fresh and store at 4 °C for 1 week maximum.
Chymostatin (Sigma): prepare as 10 mM stock, store at −20 °C, and dissolve in incubation buffer before use.
Proteinase K (Sigma): prepare as 5 mg/mL stock in 10 mM Tris–HCl pH 7.5, 1 mM CaCl2. Store at −20 °C, and dissolve in incubation buffer before use.
4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF, American Analytical): dissolve in ddH2O as 1 mM stock and store at −20 °C.
Reagents for standard SDS-PAGE and immunoblot.
2.4. Protein Degradation Assay
Millipore multiscreen assay system: 0.22 μm Durapore filter 96-well plates, vacuum manifold, and polystyrene flat-bottom 96-well plates.
Proteolysis buffer: 10 mM MOPS pH 7.3, 1 mM DTT, 5.4 μm cysteine, 0.25 M sucrose in ddH2O adjusted to pH 7.3. Prepare fresh and store at 4 °C for 1 week maximum. Cysteine and DTT should be added right before use.
Trichloroacetic acid (America Bioanalytical): 20% in ddH2O and store at room temperature.
BSA dissolved at 20 mg/mL in in ddH2O and stored at 4 °C.
2.5. Dynamics of CMA Translocation Complex
Electrophoresis apparatus: Invitrogen XCell Sure Lock Running Apparatus.
NativePAGE 3–12% Bis-Tris gel (Invitrogen).
Anode buffer: prepare 1 L by adding 50 mL of 20× NativePAGE running buffer (Invitrogen) to 950 mL of ddH2O.
Cathode buffer: prepare 200 mL by adding 10 mL of running buffer, 10 mL of NativePAGE cathode additive, and 180 mL of ddH2O. For a lighter shade of blue, 1 mL of additive can be used instead.
NativePAGE Sample Prep Kit (Invitrogen).
High molecular weight native marker, Amersham (GE Healthcare Life Science).
Solubilizing solution: 20 mM MOPS, 150 mM NaCl, and 1% octylglucoside powder, pH 7.4 in ddH2O. Store at 4 °C.
Coomassie fixing solution: 40% (v/v) methanol, 10% (v/v) acetic acid; fill to 100 mL with ddH2O. Store at room temperature.
Coomassie and BLOT destaining solution: 8% acetic acid in ddH2O. Store at room temperature.
2.6. Cell Immunofluorescence
Cell culture medium: Dulbecco’s modified Eagle’s medium (DMEM) (Sigma) supplemented with 10% newborn calf serum (NCS). The medium should be adapted depending on the cell type requirements.
Microscope cover slips (22 × 22 mm).
Phosphate-buffered saline (PBS): 1.37 M NaCl, 0.03 M KCl, 0.07 M Na2HPO4, 0.11 M K2HPO4 pH 7.4. Store at room temperature.
Methanol fixing solution: 100% methanol placed at −20 °C at least 4 hours prior to fixing cells.
Blocking solution: 0.2 (w/v) powdered nonfat milk, 2% NCS, 0.1 M glycine, 1% BSA, and 0.01% Triton X-100 in PBS. Prepare fresh and maintain at 4 °C until use.
Primary antibodies: See Table 1 for source and dilutions. Please note that IgG rabbit anti-LAMP2A was originally developed in our laboratory [4] and is now available through Invitrogen (cat# 51–2200) (note that most commercial antibodies are developed against the luminal part of LAMP2 and recognize the three isoforms (LAMP2A, B, and C)). For hsc70, we recommend using IgM mouse monoclonal anti-hsc70 antibody clone 13D3 (available through several vendors) because most commercial antibodies recognize both hsp70 and hsc70, but clone 13D3 has been well characterized as specific for hsc70.
Secondary antibodies: Fluorophores are selected depending on the combination of primary antibodies used, but common ones used in these procedures are Alexa Fluor488 goat-conjugated anti-mouse IgM antibody (ThermoFisher Scientific) (for antihsc70) and Alexa Fluro555 goat anti-rabbit IgG (ThermoFisher Scientific) (for anti-LAMP2A).
Mounting media: SlowFade Diamond Antifade Kit with DAPI (ThermoFisher scientific).
Table 1.
Antibodies or CMA-related proteins and recommended dilutions
| Antigen | Type | Source/cat# | Dilution (IB) | Dilution (IF) |
|---|---|---|---|---|
| AKT | Rabbit pAb IgG | Cell signaling [9272] | 1:1000 | 1: 200 |
| pAKT (ser473) | Rabbit pAb IgG | Cell signaling [9271] | 1:1000 | 1: 25 |
| Cath A (A-19) | Goat pAb IgG | Santa Cruz [sc-26049] | 1:500 | N/A |
| Cath D | Goat pAb IgG | Santa Cruz [sc-6486] | 1:500 | N/A |
| EF1α | Mouse mAb IgG | Millipore [05–235] | 1:1000 | N/A |
| GAPDH | Rabbit mAb IgG | Cell signaling [2118] | 1:1000 | 1:100 |
| GFAP | Mouse mAb IgG | Millipore [MAB360] | 1:1000 | 1:200 |
| pGFAP | Rabbit pAb IgG | ABGENT [AP3562a] | 1:1000 | N/A |
| HSC70 (13D3) | Mouse mAb IgM | Novus biological [NB120–2788] | 1:5000 | 1:500 |
| HSP90 | Rat mAb IgG | ENZO | 1:10,000 | N/A |
| LAMP-1 [H4A3] | Mouse mAb IgG | Abcam [ab25630] | 1:3000 | 1:100 |
| LAMP2A | Rabbit pAb IgG | ThermoFisher [51–2200] | 1:1000 | 1:200 |
| mTOR | Rabbit pAb IgG | Cell signaling [2972] | 1:1000 | N/A |
| Rictor | Rabbit mAb IgG | Cell signaling [2114] | 1:1000 | N/A |
| Rac1 | Mouse mAb IgG | Millipore | 1:1000 | N/A |
| Ribonuclease A | Rabbit pAb IgG | Rockland immunochemicals | 1:10,000 | N/A |
pAb polyclonal antibody, mAb monoclonal antibody
2.7. Photoconvertible CMA Reporter
Transfection/transduction reagents: For transient transfection with the plasmid containing the KFERQ-reporter use Lipofec-tamine 2000 (ThermoFisher scientific) and follow manufacturer’s instructions. For lentiviral-mediated stable expression, transduce cells using polybrene/transfection reagent (Sigma) (10 mg/mL stock solution), store at −20 °C, and dilute 1:1000 in culture media before use.
DMEM supplemented with 10% NCS.
Light-emitting diode (LED) at 405 nm wavelength.
Microscope cover slips (22 × 22 mm).
Phosphate-buffered saline (PBS) 1.37 M NaCl, 0.03 M KCl, 0.07 M Na2HPO4, 0.11 M K2HPO4 pH 7.4. Store at room temperature.
Paraformaldehyde fixing solution (PFA): Prepare as 4% PFA in PBS.
Mounting media: SlowFade Diamond Antifade Kit with DAPI (ThermoFisher Scientific).
2.8. Modulation of CMA in Cultured Cells
Serum deprivation: Dulbecco’s modified Eagle’s medium (DMEM) (Sigma) without additions. Washing solution (PBS).
-
Oxidative stress: H2O2 prepared fresh to a final concentration of 100 μM (dilute with culture media); paraquat prepared fresh to a final concentration of 40 μM (dilute with culture media).
Note: final concentration varies depending on the cell type. Concentrations indicated here effectively induce CMA in mouse fibroblasts.
CMA chemical activator: Atypical retinoid 7 (AR7) (originally developed by our laboratory [21] and now commercially available (Sigma). Prepare as 10 mM stock in DMSO, and store at −20 °C until use. Dilute in serum-free DMEM to working solution for a final concentration of 5–20 μM (depending on the cell type).
Inhibitors of lysosomal proteolysis: 2 M NH4Cl prepared fresh in ddH2O for a final concentration of 10–20 mM; 10 mM leupeptin stock solution in ddH2O, store at −20 °C until use, and dilute in culture media to a final concentration of 100–200 μM.
2.9. Measurement of CMA In Vivo
Rats (Wistar or any other strain of interest) or mice (C57BL/6 mice or any other strain of interest).
1 mL TB syringe, slip tip with BD PrecisionGlide Needle (Sigma).
25–30 gauge needles.
Leupeptin prepared in sterile saline (9 g/L NaCl) for a final concentration of 2 mg per 100 g body weight. To avoid injecting large volumes, prepare at a concentration that requires injection of 200–300 μL of solution. Prepare fresh.
3. Methods
The two most common reasons that motivate the study of CMA are (1) the analysis of changes in the activity of this autophagic pathway in different conditions or in response to different interventions and (2) the interest in determining if a specific protein undergoes degradation through this autophagic pathway. In this chapter, we first detail methods to directly assess CMA activity (independently of the substrate degraded), and in the last section, we briefly summarize the array of procedures to test if a protein is a CMA substrate.
3.1. Measuring CMA Activity In Vitro
3.1.1. Isolation of Rat Liver Lysosomes
Rinse the liver from a 24-h-starved rat extensively with 4 °C cold 0.25 M sucrose to remove any residual blood (see Note 2). Weigh the liver, and mince in a 50 mL plastic conical tube, making sure you act quickly and keep the tube on ice to prevent unwanted proteolysis. Add 3 volumes of cold 0.25 M sucrose/g of liver.
Transfer the minced liver into the glass homogenizer, and homogenize using a Teflon pestle and a motorized homogenizer with 8–10 strokes at maximum speed (this is done, if possible, in a cold room or alternatively keep the glass homogenizer inside an ice bucket).
Filter the homogenate through a double cotton gauze, and add 4 volumes/g liver of cold 0.25 M sucrose. Save a small aliquot (100–200 μL) to use as reference of total liver homogenate.
Centrifuge the homogenate at 6800 × g for 5 min at 4 °C, and collect the resulting supernatant into a clean tube (be careful to not collect the white layer above the pellet, as these are mainly heavy mitochondria). Discard the post nuclear pellet that contains unbroken cells, plasma membrane, nuclei, and heavy mitochondria. The best way to collect the supernatant is to directly decant it to the new tube.
Centrifuge the supernatant at 17,000 × g for 10 min at 4 °C, and resuspend the pellet with a “cold finger” (a glass tube with ice inside and dry outside to avoid including water in the sample). Add 3.5 volume/g starting liver of 0.25 M sucrose solution, and centrifuge again at 17,000 × g for 10 min at 4 °C to wash the resuspended pellet (mitochondria/lysosomal fraction), and ensure that any additional cytosolic components incorporated in the initial pellet are released into the supernatant.
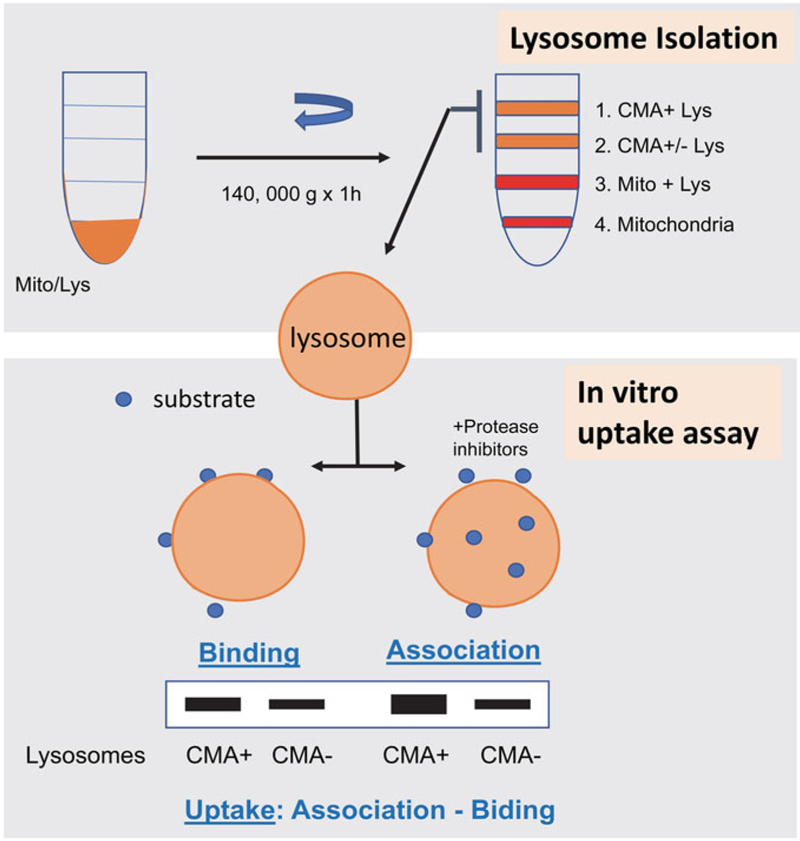
Discard the supernatant from the previous centrifugation. Resuspend the pellet using the “cold finger” and 0.25 M sucrose until the final volume (including the volume of the resuspended pellet) is 1.1 mL. Add 2 volumes of 85.6% (w/v) metrizamide (2.2 mL) to reach a final concentration of 57% metrizamide, and mix everything carefully with a 5 mL plastic pipette. Load the sample at the bottom of a 14 × 95 mm, thin-wall, ultraclear ultracentrifugation tube (make sure to not touch the walls of the tube) (Fig. 2).
Generate a discontinuous metrizamide gradient on top of the 57% metrizamide containing the light mitochondrial/lysosome fraction by overlaying: 2 mL of 32.8% metrizamide, 3.3 mL of 26.3% of metrizamide, and 3.5 mL of 19.8% metrizamide (all diluted in ddH2O). Fill the tube with 0.25 M sucrose up to 2 mm of the top edge of the tube. Generate the gradient while the bottom part of the tube (the one containing the lysosomes) is inside an ice bucket.
Centrifuge in SW 41 swinging bucket rotor at 141,000 × g at 4 °C for 1:09 h (setting acceleration to 4 and deceleration to 9). The time of centrifugation at maximal speed is 1 h, and the 9 min are required for acceleration and deceleration.
After centrifugation, white to light brown material is clearly visible in each of the interphases. Mitochondria are mostly retained at the lowest interphase of the gradient; the following interphase contains a mixture of mitochondria and lysosomes. A mix population of lysosomes is present in the third interphase from the bottom, and lysosomes with high CMA activity migrate to the top interphase. Collect each of the interphases separately (approx. 3–4 mL) with a Pasteur pipet in 30 mL polycarbonate tubes.
Add 5–10 volumes of 0.25 M sucrose solution, and wash by centrifugation at 37,000 × g for 15 min at 4 °C.
Resuspend the pellet coming from the third interphase using a glass rod to avoid lysosomal damage (a Pasteur pipet with the tip blunted with a flame and left to cool down can be used). Once the pellet is disaggregated, add 300 μL of 0.25 M sucrose, pipette up and down with a plastic tip with the end cut to avoid friction during pipetting, and place it into a microfuge tube to centrifuge at 10,000 × g for 5 min at 4 °C.
Use the supernatant obtained from the centrifugation of the third interphase (containing CMA+ lysosomes) to resuspend the pellet coming from the first interphase applying the same procedure (disaggregate the pellet quickly with a glass rod, and then add the supernatant from the third interphase centrifugation and pipette up and down with a cut plastic tip). This will be the final sample enriched in CMA-active lysosomes (CMA+).
Resuspend the pellet from the centrifugation of the third interphase (with a glass rod), and then add 300 μL of 0.25 M sucrose, and pipette up and down with a cut plastic tip. This is the final sample enriched in lysosomes with low CMA activity (CMA-) (Fig. 2).
Fig. 2.
Measurement of CMA in vitro. Top: lysosomes active for CMA can be isolated by flotation in a discontinuous gradient of metrizamide. Bottom: incubation of CMA substrates with intact lysosomes pre-treated or not with inhibitors of lysosomal proteases allows to quantify substrate binding and translocation (uptake) inside the lysosomal lumen
3.1.2. Lysosomal Purity
Prepare a 96-well plastic flat-bottom plate with the homogenate (2 μL) as reference and different volumes of the collected fractions diluted 1:10 in ddH2O (2, 5 and 7 μL). Add ddH2O to a final volume of 25 μL H2O (remember to use a well with only ddH2O as a blank).
Add 100 μL of the pre-warm (37 °C) reaction mixture, and vortex gently. Cover the plate with a plastic lid or Parafilm to avoid evaporation.
Incubate at 37 °C for 30 min.
Stop the assay by adding 75 μL of glycine/carbonate stop solution.
Read in the plate reader at EM: 450 nm/Ex: 370 nm.
Calculate recovery as the percentage of β-hexosaminidase activity of the homogenate recovered in each fraction [(activity fraction × total volume fraction)/(activity homogenate total volume homogenate)] × 100 in the fraction.
Calculate enrichment as the specific β-hexosaminidase activity in the fractions relative to the one in homogenate [(activity fraction/μg of protein in the fraction)/(activity homogenate/μg of protein in homogenate)].
3.1.3. Lysosomal Integrity
Pre-wet the desired wells of the 96-well MultiScreen-Mesh filter plate with a Durapore (PVDF) filter (0.22 μm) by filling with ddH2O (5 min, RT).
Shake the water out, and block with 20 mg/mL BSA in ddH2O (approx. 100 μL) for 30 min at RT.
Filter the blocking solution with the vacuum manifold.
Add 30 μL of 0.25 M sucrose per well.
Add 15 μL of lysosomes (directly from the gradient), and include a blank with only ddH2O.
Apply vacuum for 30 s to 1 min until all the solution is filtered and collected in a 96-well plastic plate.
Take the flow through, and assay for β-hexosaminidase as in the previous section (samples can be stored at −20 °C and assayed for activity later).
3.2. In Vitro Reconstitution of CMA
3.2.1. Lysosomal Binding/Uptake
Incubate freshly isolated rat liver lysosomes with 100 μM chymostatin for 10 min on ice. Depending on the substrate, a protease inhibitor cocktail can be used instead (recommended combination: 100 μM leupeptin, 100 μM AEBSF, 10 μM pepstatin and 1 mM EDTA).
Carry out transport assay in 500 μL microfuge tubes by adding freshly isolated rat liver lysosomes treated or not with protease inhibitor (50–100 μg protein in a volume of 10 μL) and GAPDH or another protein of interest (50 μg) in a final volume of 30 μL of MOPS buffer (see Note 3). Incubate samples for 20 min at 37 °C (Fig. 2).
At the end of the incubation (see Note 4), centrifuge samples at 25,000 × g for 5 min at 4 °C.
Aspirate the supernatants, and wash the pellets with 100 μL of incubation buffer carefully to not disturb the pellets.
Resuspend the pellet in 20 μL of Laemmli sample buffer with protease inhibitors, boil for 5 min at 95 °C, and analyze by SDS-PAGE and immunoblot for GAPDH. A lane with 1/10 of the amount of GAPDH should be included in the immunoblot for quantification purposes.
Perform densitometric analysis on the immunoblots, and use a generated standard curve for each antibody using increasing concentrations of antigen to determine the linear range. These values allow the calculation of (1) binding as the percentage of total added GAPDH associated to lysosomes untreated with protease inhibitors, (2) association as the percentage of GAPDH recovered in lysosomes treated with protease inhibitors, and (3) uptake, calculated as the difference between association and binding.
3.2.2. Intact Lysosomes Protein Degradation Assay
This assay is carried out in 96-well MultiScreen-Mesh filter plate with a Durapore (PVDF) filter (0.22 μm) pre-wet with ddH2O for 10 min at room temperature. After aspirating the water, add to the wells the 20 μL MOPS/DTT proteolysis buffer (see Notes 3 and 5).
Add freshly isolated rat liver lysosomes (25 μg protein) (10 μL of a 1:4 dilution in proteolysis buffer) per well and 10 μL of the radiolabeled protein cocktail (2000 dpm/μL), and adjust the final volume to 60 μL with proteolysis buffer. Incubate for 30 min at 37 °C. One well should contain the same reagents, except for the lysosomes, to determine the amount of spontaneous proteolysis (autolysis) that will be used as a blank.
At the end of the incubation, add 90 μL of 20% TCA and 30 μL of 20 mg/mL BSA to each well to stop the reaction. Incubate at 4 °C for at least 30 min to allow for protein precipitation.
Collect the acid-soluble radioactivity in a polystyrene 96-well plate using the multiscreen vacuum system. Collect the flow through (acid-soluble) from each sample and the filter retained material (acid-precipitable) in separate scintillation vials for dpm counting.
Proteolysis is calculated as the percentage of total protein (radioactivity precipitated by acid at time 0) transformed into amino acids and small dipeptides (radioactivity that remains soluble after adding the acid stop solution) at the end of the incubation. The amount of spontaneous hydrolysis (calculated from the blank well) needs to be discounted following the formula: [((dpm flow through sample – dpm flow through blank)/dpm pellet time 0) × 100] (see Note 6).
3.2.3. Dynamics of the CMA Translocation Complex
Resuspend freshly isolated lysosomes (pellet from the centrifugation at 37,000 × g for 15 min) in 1% octyl glycoside in MOPS pH 7.3 supplemented with protease inhibitors, and incubate on ice for 15 min (see Note 7).
Spin at 16,000 × g for 15 min at 4 °C to collect the solubilized lysosomal membrane complexes in the supernatant.
Discard pellet and add to the supernatant the following components from the NativePAGE Sample Prep Kit: 2.5 μL of NativePAGE 4 × sample buffer, 1 μL of G-250 sample additive, and 10 μL of ddH2O per sample.
Load gel in a cold room (4 °C) and after adding the anode buffer (clear) in the outer chamber and the cathode buffer (blue) in the inner chamber, run the gel at 150 Volts (constant) for ~2.5–3.5 h or until dye collects at the bottom.
Cut out the molecular weight marker lane, and stain with Coomassie Blue (see Note 8).
Remove gel, put it into transfer buffer for at least 2 min, and proceed with the wet transfer into a methanol pre-wet PVDF membrane. Transfer overnight at a constant current of 0.25 Amps.
After removing from the transfer, shake the membrane in 8% acetic acid for 15 min, and wash 3 times for 5 min each time with TBST.
Block with 5% milk in TBST for 1 h. Incubate overnight with the primary antibody against LAMP2A (see Table 1) diluted in 3% BSA in TBST. Incubate with secondary antibody (1:10,000) in 5% milk in TBST for 1 h, wash 3 times (15 min each time) with TBST, and develop using standard chemilumi-nescence detection (see Note 9).
Monomeric LAMP2A is detected as a wide band in the 90–110 KD range (depending on the tissue), and the multimeric complex of LAMP2A shown to participate in substrate uptake into lysosomes via CMA is observed as a well-resolved band of 700 KD. Changes in the percentage of total lysosomal LAMP2A detected in this multimeric complex provide information on CMA activity in lysosomes.
3.3. Measurement of CMA Activity in Intact Cells
3.3.1. Modulation of CMA in Cultured Cells
- CMA activation: CMA can be upregulated in cells in culture by two methods:
-
Inducing stresses known to activate CMA [1]: the stressors used are not selective for CMA and activate other arms of the cellular response to stress but are useful when quantifying the ability of cells to activate CMA. The best characterized are:Serum removal: Starvation is a well-established inducer of CMA [11] and can be mimicked in cells by removing the serum from the culture media [22] as follows:
- Plate cells to 80% confluency in media containing 10% newborn calf serum.
- After 12 h wash the cells extensively with PBS or Hanks solution (×3 washes with half of the volume of the plate or well).
- Add complete medium but without addition of serum for 16–24 h. CMA activation reaches exponential kinetics about 10–16 h (depending on the cell type) and plateaus by 24 h after serum removal.
Oxidative stress: Conditions that induce mild oxidative stress can be used to upregulate CMA [13].- Plate cells to 80% confluency in media containing 10% newborn calf serum.
- After 12 h change media to fresh media containing 10% newborn calf serum and 100 μM of H2O2 or 40 μM of paraquat (see Note 10).
- After 4 h with the oxidizing agent, aspirate the culture media, and replace by fresh media containing 10% newborn calf serum.
- Monitor CMA activity at 12–24 h after the oxidative stress.
Genotoxic stress: CMA is activated during the recovery from stressors inducing double-strand DNA breaks [15].- Plate cells to 80% confluency in media containing 10% newborn calf serum.
- After 12 h change media to fresh media containing 10% newborn calf serum and any of the following agents: 10 μM etoposide, 225 μM MMS, or 20 μM cisplatin.
- After 12 h aspirate the culture media, and replace by fresh media containing 10% newborn calf serum.
- Monitor CMA activity at 12 h after removing the DNA damaging agent.
- Chemical activation of CMA: To date, there is only one type of compound tested and validated to selectively activate CMA [21] (see Note 11).
- Plate cells to 80% confluency in media containing 10% newborn calf serum.
- After 12 h change media to fresh media containing 10% newborn calf serum and 5–10 μM AR7 (CAS number 80306–38–3; #SML0921, Sigma) (see Note 11).
- Monitor CMA activity at 8–16 h after adding AR7.
-
- CMA inhibition:
- Genetic downregulation of CMA: The most selective way to block CMA is through genetic knockdown of LAMP2A, [23, 24] since other components involved in CMA are also shared by other cellular processes. To knock down LAMP2A using lentivirus-mediated shRNA in cultured cells:
- Plate approximately 500,000 cells/well in a 6-well plate a day before transduction.
- Replace the media with 0.5 mL of culture media containing 200 μg/mL of polybrene, and add 0.5 mL of lentiviral particles containing shRNA against LAMP2A (the original construct is in a backbone vector that also expresses GFP) [24].
- After 24 h add an additional 1 mL of serum-supplemented media.
- After 48–72 h, check for GFP expression with a fluorescence microscope to determine transduction efficiency.
- Check for LAMP2A knockdown efficiency by immunoblotting after 7–10 days (half-life of LAMP2A protein in most cell types under basal conditions is about3.5 days).
- Chemical downregulation: To date, there is no chemical compound capable to selectively inhibit CMA without affecting other lysosomal degradation pathways. Compounds that block degradation in the lysosomal lumen (either by increasing the lysosomal pH or by inhibiting the catalytic activity of lysosomal proteases) will also block protein degradation dependent on CMA.
3.3.2. Measuring Protein Degradation by CMA
Degradation of proteins in lysosomes via CMA in cultured cells can be determined using metabolic labeling and tracking degradation of intracellular proteins as a pool as follows (see Note 12):
Plate cells in 12-well plate to 60% of confluency, and supplement the media with 10% serum and 3H-leucine to 2 μCi/ml.
After 48 h wash cells extensively, and add 300 μL of fresh media supplemented with an unlabeled excess of leucine (for cells growing in standard DMEM, add 2.8 mM leucine).
After 1 h aspirate the media, and replace with fresh media with an excess of unlabeled leucine as above (this allows eliminating short half-life proteins usually degraded by the proteasome). Leave triplicate wells without additions, and supplement 3 wells with 20 mM NH4Cl and 100 μM leupeptin (to block all lysosomal degradation) and 3 wells with 10 mM 3-methyladenine (to block macroautophagy).
Take aliquots of 35 μL of the media every 6 h for 24 h, and precipitate them with TCA and BSA as described in the previous sections.
At the last time point, aspirate the media and add 500 μL of solubilizing buffer. After 24 h count the radioactivity in 20 μL of the solubilized wells.
Proteolysis is calculated as the percentage of total protein labeled at time 0 converted into free radiolabeled amino acids at each time point. Lysosomal proteolysis is calculated by discounting the proteolysis remaining in the wells treated with lysosomal proteolysis inhibitors to the total proteolysis. CMA-dependent degradation is calculated as the percentage of lysosomal proteolysis insensitive to 3-methyladenine (see Note 13).
3.3.3. Cell Immunofluorescence
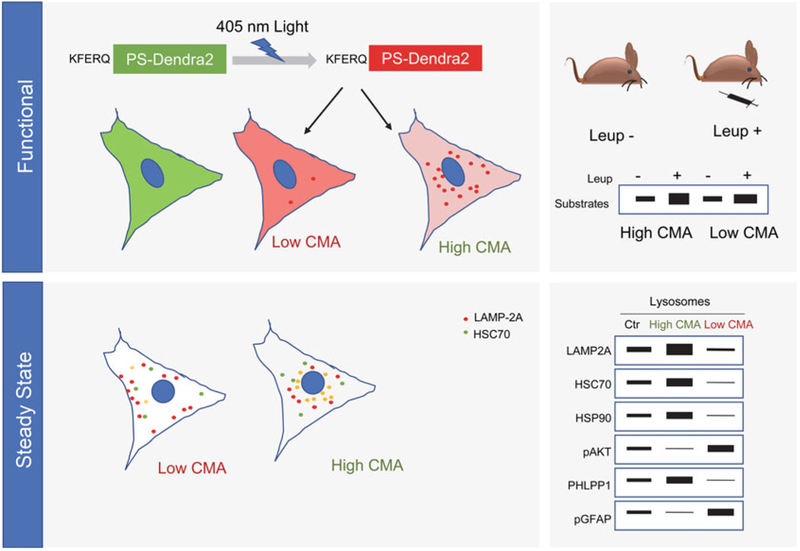
Quantification of changes in the number of lysosomes active for CMA can be used as a complementary measurement of CMA (Fig. 3). Lysosomes with the capability to perform CMA contain HSC70 in their lumen and can be identified as HSC70- and LAMP2A-positive vesicles as follows:
Fig. 3.
Measurement of CMA in cultured cells and in vivo. Top: functional assays, expression of photoswitchable reporters (left) allows quantification of CMA in cultured cells as the number of fluorescent puncta (lysosomes) per cell. Injection of leupeptin in vivo (to block lysosomal proteolysis) and immunoblot for CMA substrate of lysosomes from injected and not injected mice allows quantification of substrate flux by CMA (right). Bottom: steady-state assays, quantification of number of lysosomes positive for LAMP2A and HSC70 allows for detecting changes in lysosomes capable of performing CMA (left). Immunoblot of isolated lysosomes for well-characterized CMA activator and inhibitor proteins. Profile of differences in specific protein levels in lysosomes with upregulated and downregulated CMA activity are shown
Plate cells on sterile cover slips at the bottom of a 24-well plate until they reach semi-confluence in media with 10% NCS.
Warm serum-free media and PBS at 37 °C.
Remove culture medium from cells, and wash once with the media and a second time with PBS.
Remove PBS and add −20 °C chilled 100% methanol for 4 min to extract the fraction of soluble HSC70 and preserve the one associated to membranes.
Wash 3 times with PBS to remove methanol.
Block/permeabilize for 30 min at room temperature.
Wash 3 times with PBS, and add the two primary antibodies for 1 h.
Wash 3 times with PBS, and add both secondary antibodies.
Wash off secondary antibodies with PBS and mount for imaging as usual to visualize in a fluorescent microscope and determine the number of lysosomes capable to perform CMA as the percentage of LAMP2A-positive vesicles that also label for HSC70.
3.3.4. Photoconvertible CMA Reporter
CMA activity can be measured in cells in culture using photoswitchable (PS) artificial substrates. Three different versions have been generated by our group and tested and validated by other groups: KFERQ-PS-CFP [25], KFERQ-photoactivable (PA)-mCherry [25], and KFERQ-PS-Dendra (unpublished). These differ in the color they fluoresce but follow the same principle and can be used indistinguishably. When exposed to a 405 nm LED light, KFERQ-PS-CFP protein changes from blue to green fluorescence, KFERQ-PA-mCherry that normally does not fluoresce changes to red fluorescence, and KFERQ-PS-Dendra changes from green to red fluorescence. Change in color allows performing pulse/chase type experiments where the switched fluorescence can be followed as it is delivered to lysosomes (Fig. 3) [25].
Transduce cells with lentivirus carrying the reporter following the same steps as for shRNA delivery (see previous section).
Twenty-four hour after transduction, the cells can be photo-activated by exposure to a 3.5 mA (current constant) and 90 V light-emitting diode (LED: Norlux, 405 nm) for 4 min.
Split photoswitched cells, and plate on cover slip at the bottom of 24-well plates.
Once attached (2–3 h) add the desired treatments (physiological stimuli, drugs, etc.).
Fix cells at different times or at a single end time point (16 h after treatment) with 4% paraformaldehyde in PBS for 15 min.
Capture images using a fluorescent microscope. CMA can be quantified as the number of fluorescent puncta per cell (see Note 14).
3.4. Measurement of CMA Activity In Vivo
3.4.1. Changes in CMA Components
Using CMA-active lysosomes isolated from mice, changes in CMA mediators and modulators can be measured via immunoblotting (Fig. 3). When possible, two to three mice livers can be pooled to get enough material for multiple immunoblots. Increase in LAMP2A, HSC70, PHLPP1, and GFAP in lysosomes is supportive of CMA activation; increase in phospho-AKT and phospho-GFAP is supportive of CMA inactivation. Increase in levels of known CMA substrates such as GAPDH, ribonuclease A, alpha-synuclein in the isolated lysosomes is also a good indication of CMA activation.
3.4.2. Substrate Degradation
Degradation of substrate proteins in lysosomes (flux) can be measured upon blocking their proteolysis inside the lysosomal lumen as follows (Fig. 3):
Inject leupeptin prepared in saline intraperitoneally to mice or rats (2 mg/100 g of body weight) or only saline to the control animals.
After 3 h isolate lysosomes active for CMA following the procedures described in the previous section.
Immunoblot lysosomes for well-known CMA substrates such as GAPDH, ribonuclease A, alpha-synuclein (for a more complete list see [1]). Calculate CMA flux as the increase in levels of the substrates in lysosomes isolated from leupeptin-treated animals relative to levels in those from untreated animals.
4. Determining if a Protein Is a CMA Substrate
Investigators interested in testing whether a protein is a bona fide CMA substrate should perform as many of the following tests as possible in their system of interest:
Sequence analysis: every CMA substrate protein contains in its amino acid sequence a KFERQ-like pentapeptide motif that is both necessary and sufficient for its targeting to the lysosome [2]. The initial step in determining if a protein is a CMA substrate is to identify the presence of this motif. This consensus motif has a very specific amino acid composition, and any variation from the original set of criteria needs to be experimentally confirmed as a bona fide targeting motif. Briefly, the motif is based on the physical properties of the amino acid residues which have been identified as a combination of one or two of the positively charged residues K, R; one of the negatively charged residues D, E; and one or two of the hydrophobic residues F, L, V, I and a glutamine (Q) that either initiates or terminates the sequence [2] (see Note 15).
Association with lysosomes, preferentially with those that have higher CMA activity (positive for lys-HSC70) [26]: this can be determined using co-localization by immunofluorescence (see Subheading 3.3) or by immunoblot in isolated lysosomes (see Subheading 3.1).
Degradation in lysosomes: detected as an increase in the cellular levels of the protein of interest upon treatment of cells with inhibitors of lysosomal proteolysis (20 mM NH4Cl/100 μM leupeptin) for 12 h. Note that this property is also shared with proteins degraded by other autophagic pathways (macro- and microautophagy) and by endocytosis.
Interaction with CMA components: detectable by co-immunoprecipitation of the protein of interest with HSC70 in cytosolic fractions [3] or with the cytosolic tail of LAMP2A in isolated lysosomes [4]. Note that because protein interaction with HSC70 also occurs in many other cellular pathways, confidence of a relation with CMA degradation will be higher if the HSC70/protein interaction can be completed by addition of KFERQ-containing proteins (i.e., ribonuclease A (RNase A)). Because KFERQ-dependent binding to HSC70 also mediates degradation of proteins by e-MI, additional criteria need to be used to differentiate between both autophagic pathways.
Changes in intracellular levels upon CMA modulation: increase in the intracellular levels of the candidate protein in cells that lack LAMP2A (although activation of other autophagy pathways to compensate for reduced CMA prevents substrate accumulation in many instances).
Capability to directly translocate into isolated lysosomes: this is the most definitive evidence of a protein being a CMA substrate; as in this type of in vitro assay, there is no contribution of any other proteolytic system to the observed lysosomal translocation and degradation (see Subheading 3.2, for details).
Mutagenesis of the targeting motif: given that the KFERQ-like targeting motif is both a necessary and sufficient sequence that is required for HSC70 to target substrate proteins to the lysosomal membrane, alterations in this motif can be utilized to support the involvement of this chaperone in their degradation. Mutation of the Q residue in the motif to alanine (A) disrupts the interaction of the protein with HSC70 and its subsequent targeting to lysosomes. Elimination or alteration of the targeting motif will result in a decreased association of the substrate with lysosomes in the case of CMA or with late endosomes/MVB in the case of e-MI. Furthermore, in some instances, interaction with HSC70 persists even in the absence of the CMA-targeting motif. HSC70 binding in these cases often has switched to another region in the protein, due to the ability of HSC70 to bind hydrophobic protein patches. However, that type of interaction does not target the protein for CMA degradation and can be easily identified by demonstrating lack of competition for HSC70 binding with other proteins bearing the CMA-targeting motif such as RNase A.
5. Notes
Metrizamide is light sensitive and needs to be dissolved in the dark (beaker wrapped with aluminum foil) and slowly to avoid solid clump formation. Best to start with half of the final volume of water and slowly add metrizamide powder while stirring. To adjust the pH of the solution use 0.01 M NaOH once the metrizamide is dissolved.
Liver perfusion is not strictly necessary. Extensive washing of the livers with sucrose removes most of the blood remaining in the tissue.
In the transport and degradation assays with intact lysosomes, supplementation with HSC70 and ATP may be necessary or not depending on the origin of the lysosomes. In our experience, lysosomes from rat liver, when prepared following this protocol, contain enough HSC70 associated to the cytosolic side of their membrane, and supplementation is not required. However, lysosomes from cultured cells or from some tissues need to be supplemented with additional recombinant HSC70 (10 μg/mL final concentration) and ATP (5 mM) to maximize uptake. Although we recommend an incubation of 30 min at 37 °C to measure proteolysis in intact lysosomes, in those instances in which lysosomal fragility is suspected, the incubation can be performed instead for 45–60 min at room temperature (25 °C) to reduce lysosomal breakage. In this case, it is advisable to use an ATP regenerating system in the incubation media [ATP (10 mM), MgCl2 (10 mM), phosphocreatine (2 mM) and creatine phosphokinase (50 pg/mL)].
In some instances, uptake of substrates by lysosomes can also be determined using proteinase K treatment to remove the protein associated with the cytosolic side of the lysosomal membrane. In that case, after incubation with the substrate, all samples are cooled down on ice (1 min), and the samples treated with chymostatin are exposed to proteinase K (5 μL of 1 mg/mL solution). Samples are incubated on ice for 10 min, and the reaction stopped by adding 5 μL AEBSF (100 μM). All samples are collected by centrifugation at 25,000 × g for 5 min at 4 °C. Uptake is determined as the percentage of GAPDH added that is associated with the proteinase K-treated samples.
Presence of DTT and cysteine in the proteolysis buffer is absolutely necessary to obtain maximal degradation. DTT needs to be added fresh to the solution.
Although the use of multiple technical replicates is always encouraged, it is especially important in this assay since due to the small volume of lysosomes added to the reaction and the fact that lysosomes are in suspension, there are higher chances of variability in the total amount of lysosomes added per well. The short half-life of lysosomes once purified (membrane breakage and loss of luminal pH start to increase exponentially 1 h after isolation) makes it not possible to determine the concentration of lysosomal protein added per well. When working with lysosomes of different origins, it is necessary to save a small amount frozen to determine both protein concentration and β-hexosaminidase activity later and correct proteolysis rates.
If the sample subjected to solubilization is in solution (instead of as a pellet), adjust the final concentration of detergent in the solution using a concentrated stock.
For fast Coomassie staining of the molecular weight marker, place the strip of gel in Coomassie fixing solution, and micro-wave for 45 s. Shake 15 min in this same solution. Remove solution, and place gel in destaining solution. Microwave for 45 s, and shake in destaining overnight at room temperature. The next day, remove destaining, and store in water.
We have found that using concentrations of LAMP2A antibody higher than recommended in Table 1 has a negative impact on the detection of the 700 KD multimeric complex of LAMP2A and therefore strongly recommend using a 1:5000 dilution (instead of the standard 1:1000 used for regular immunoblot as indicated in Table 1). In some instances, in which only a small fraction of LAMP2A is organized into the multimeric complex, the high abundance of monomers in lysosomes could make detection of the multimers challenging when developing the membrane. This problem can be resolved by doing a short exposure of the membrane (to identify the LAMP2A monomer) and then proceed to a long exposure covering the bottom of the membrane containing the LAMP2A monomer with foil paper to eliminate this signal that may otherwise cover the considerably weaker signal of the multimer.
Concentrations of H2O2 and paraquat need to be optimized for each cell type. These concentrations work well in mouse and human fibroblasts in culture and were determined after doing viability assays. In these cells, we have found that concentrations of paraquat up to 100 μM result in less than 10% cell death. Concentrations inducing more than 15% cell death in the cell type of interest should be avoided. Paraquat should be handled carefully with appropriate safety personal protective equipment (mask, gloves) as it is highly toxic when in powder form.
There has been some level of confusion regarding methods to chemically activate CMA due to the misinterpretation of a study from the late James Dice group [27]. In this study, it was described how compounds that modify other biological processes such as anisomycin or cycloheximide (that inhibit protein synthesis) or 6-aminonicotinamide (that inhibits pentose phosphate metabolism) lead to activation of CMA (among many other pathways, including macroautophagy). Consequently, the use of these drugs for selective activation of CMA is highly discouraged, and they should be used more under the category of cellular stressors that elicit a CMA response. Activation of other cellular stress pathways, including other proteolytic systems, by these drugs, makes interpretation of findings complex. Use of AR7 or related derivatives [21] is recommended as for these compounds; lack of effect on proteasomal or lysosomal degradation pathways (other than CMA) has been confirmed. Concentrations of AR7 resulting in maximal activation of CMA should be determined experimentally for each cell type. The range indicated here has been proven effective in mouse fibroblasts, astrocytes, macrophages, T cells, and kidney epithelial cells.
It is also possible to determine CMA activity in cells in culture by tracking the degradation of previously well-characterized substrates by immunoblot after addition of the inhibitors in the same conditions. However, because degradation of specific proteins by CMA may depend on the cellular conditions, following a pool of proteins provides a better representation of CMA activity than a single protein.
Other inhibitors of macroautophagy can be used, but times and conditions in this protocol have been optimized for 3-methyladenine. Acute chemical blockage of macroautophagy is preferred to genetic blockage for these experiments as that will prevent activation of compensatory mechanisms.
Using PS reporters allows measuring degradation of the switched proteins without having to inhibit protein synthesis. When using the fluorescent reporters, it is important to use all the controls as described in [25] to confirm that the fluorescent puncta are indeed lysosomes and not protein aggregates.
Note that the KFERQ-like motif is used for interaction with HSC70, and consequently the presence of the motif is not sufficient to differentiate CMA substrates from e-MI substrates [9].
References
- 1.Kaushik S, Cuervo AM (2012) Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol 22:407–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dice JF (1990) Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci 15:305–309 [DOI] [PubMed] [Google Scholar]
- 3.Chiang H et al. (1989) A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 246:382–385 [DOI] [PubMed] [Google Scholar]
- 4.Cuervo AM, Dice JF (1996) A receptor for the selective uptake and degradation of proteins by lysosomes. Science 273:501–503 [DOI] [PubMed] [Google Scholar]
- 5.Bandyopadhyay U et al. (2008) The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol 28:5747–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agarraberes F, Terlecky S, Dice J (1997) An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol 137:825–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bandyopadhyay U et al. (2010) Identification of regulators of chaperone-mediated autophagy. Mol Cell 39:535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arias E et al. (2015) Lysosomal mTORC2/PHLPP1/Akt Regulate Chaperone-Mediated Autophagy. Mol Cell 59:270–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahu R et al. (2011) Microautophagy of cytosolic proteins by late endosomes. Develop Cell 20:131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salvador N et al. (2000) Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. J Biol Chem 275:27447–27456 [DOI] [PubMed] [Google Scholar]
- 11.Cuervo AM et al. (1995) Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Phys 269: C1200–C1208 [DOI] [PubMed] [Google Scholar]
- 12.Wing SS et al. (1991) Proteins containing peptide sequences related to KFERQ are selectively depleted in liver and heart, but not skeletal muscle, of fasted rats. Biochem J 275:165–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiffin R et al. (2004) Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell 15:4829–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider JL, Suh Y, Cuervo AM (2014) Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab 20:417–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park C, Suh Y, Cuervo AM (2015) Regulated degradation of Chk1 by chaperone-mediated autophagy in response to DNA damage. Nat Commun 6:6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valdor R et al. (2014) Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat Immunol 15:1046–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuervo AM, Dice JF (2000) Age-related decline in chaperone-mediated autophagy. J Biol Chem 275:31505–31513 [DOI] [PubMed] [Google Scholar]
- 18.Cuervo AM et al. (2004) Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 305:1292–1295 [DOI] [PubMed] [Google Scholar]
- 19.Kiffin R et al. (2007) Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci 120:782–791 [DOI] [PubMed] [Google Scholar]
- 20.Orenstein SJ et al. (2013) Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci 16:394–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anguiano J et al. (2013) Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat Chem Biol 9:374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger JJ, Dice JF (1985) Effect of serum deprivation and replacement on proteolysis in cultured human fibroblasts. Prog Clin Biol Res 180:479–481 [PubMed] [Google Scholar]
- 23.Massey AC et al. (2006) Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A 103:5905–5910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massey AC et al. (2008) Early cellular changes after blockage of chaperone-mediated autophagy. Autophagy 4:442–456 [DOI] [PubMed] [Google Scholar]
- 25.Koga H et al. (2011) A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nat Commun 2:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuervo AM, Dice JF, Knecht E (1997) A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem 272:5606–5615 [DOI] [PubMed] [Google Scholar]
- 27.Finn P et al. (2005) Effects of small molecules on chaperone-mediated autophagy. Autophagy 1:141–145 [DOI] [PubMed] [Google Scholar]