Abstract
Sine oculis homeobox homolog 4 (SIX4), a member of the SIX family, play important role in the development and construction of vertebrate tissues and organs. There is very little known about the function of SIX4 in cancer cells. Herein, we investigated whether SIX4 promote cancer metastasis in addition to its direct role in breast cancer cells. Our study showed that the expression of SIX4 was profoundly increased in breast cancer tissues, and the high expression of SIX4 correlated strongly with distant metastasis and poor prognosis. Functional experiments demonstrated that SIX4 obviously promoted the cell migration and invasion of breast cancer cells, and up-regulated the expression of EMT mesenchymal marker, down-regulated the epithelial molecules by Snai1 induction in vitro. Moreover, SIX4 knockdown significant suppressed breast cancer lung metastasis in vivo. Mechanistically, SIX4 directly interacted with STAT3 and promoted the phosphorylated STAT3 nuclear translocation, thereby inducing EMT program activation. Collectively, our findings demonstrated that SIX4 may be a promising target for intervention to prevent cancer metastasis.
Keywords: SIX4, metastasis, STAT3, breast cancer
Introduction
The sine oculis homeobox (SIX) protein family was initially identified as the key determinants of Drosophila retinal development, play important role in the development and construction of tissues and organs in vertebrates. A lot of evidence indicate that SIX have disorder of transcription or post-translational modifications in a variety of primary tumor and metastatic lesions [1]. The SIX family is comprised of six members (SIX1-SIX6), of which SIX1 is most studied for its roles in cancer initiation and progression [2]. In contrast, little is known about the other SIX family members in tumorigenesis and progression. Few studies have implicated SIX4 in cancer. For example, SIX4 promotes metastasis in colorectal cancer [3] and correlates with higher TNM stage in non-small cell lung cancer [4]. SIX4 was identified herein as both an oncogene and miR-203a target [5]. These data suggest that SIX4 can promote cancer progression. However, the signalling mechanisms that induce and promote tumorigenesis and metastasis by SIX4 have not yet been elucidated.
The epithelial-mesenchymal transition (EMT) process has been an important explanation for the distant metastases of epithelial malignancies, including breast cancer [6]. EMT is driven by pleiotropic signalling factors such as EMT-inducing transcription factors (ZEB1, ZEB2, Snail, Slug, Twist etc.), epigenetic and post-translational regulators [7]. EMT is usually characterized by the down-regulation of epithelial molecules, including E-cadherin, zona occludens-1, cytokeratin, claudins, and up-regulation in mesenchymal markers such as N-cadherin, vimentin, fibroblast specific protein-1, α-smooth muscle actin, fibronectin and so on [8]. These cells acquire the ability to reconstitute metastatic colonies in the distant sites.
Recently, emerging evidence suggests that STAT3 signalling pathway plays a significant role in breast cancer angiogenesis, proliferation, invasion, metastasis, and also leads to cancer stem cell self-renewal traits, induction of EMT [9]. In a previous study, it was reported that STAT3 induces EMT in cancer cells through JAK/STAT3/Twist-1 signal pathway in breast cancer [10]. Silencing of the STAT3 signalling pathway also suppresses EMT progression via the down-regulation of Snail expression in breast cancer [11]. In addition, IL-6/STAT3 mediated repression of MIR34A promotes EMT, invasiveness, and metastasis in colorectal cancer [12]. In this study, we confirmed that the expression of SIX4 was significantly increased in breast cancer tissues, and the high expression of SIX4 correlated strongly with distant metastasis and poor prognosis. In vitro, we found that SIX4 obviously promoted the cell migration and invasion of breast cancer cells, and up-regulated the expression of EMT mesenchymal marker, down-regulated the epithelial molecules. In vivo, we also found that SIX4 knockdown obviously suppressed breast cancer lung metastasis. Mechanistically, SIX4 directly interacted with STAT3 and promoted the phosphorylated STAT3 nuclear translocation, thereby inducing EMT program activation. Collectively, our findings demonstrated that SIX4 may be a promising target for intervention to prevent cancer metastasis.
Materials and methods
Cell culture and reagents
MDA-MB-231, MDA-MB-468, ZR-75-1, BT-549, MCF-7, MCF-10A and HEK293T cells were obtained from Shanghai GENECHEM, Co., Ltd. The MDA-MB-231, MDA-MB-468 cells were cultured in L-15 medium (HyClone, GE Healthcare). ZR-75-1, BT-549 were cultured in RPMI-1640 (HyClone, GE Healthcare). The MCF-7 and HEK293T cells were cultured in DMEM (HyClone, GE Healthcare). MCF-10A cells were cultured in MEBM basal medium (Lonza, MD, USA) supplemented with the MEGM SingleQuots Kit (#CC-4136, Lonza). Add 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc) and 1% penicillin/streptomycin (KeyGEN; KeyGEN Bio TECH Corp., Ltd) to L15, RPMI-1640 and DMEM culture medium. According to the ATCC instructions, MDA-MB-23 and MDA-MB-468 cells were cultured at 37°C in 100% air incubator, while other cells were cultured at 37°C in 5% CO2 incubator. STAT3 inhibitor AG-490 and AKT inhibitor AZD5363 were obtained from MedChemExpress (MCE, NJ, USA). Mouse anti-SIX4 (sc-81984), p-JAK2 (sc-21870), HA (sc-7392), Flag (sc-166355), GAPDH (sc-137179) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibodies for JAK2 (#3230), Histone 3 (#4499), HA-Tag (#3724), STAT3 (#12640), phosho-STAT3 (Tyr705) (#9145), E-cadherin (#14472), N-cadherin (#13116), Vimentin (#5741), Snai1 (#3879), Slug (#9585), Zeb1 (#3396), Twist (#46702) were obtained from Cell Signaling Technology (Danvers, MA, USA).
Construction of lentiviral-infected cell lines
The three SIX4-shRNAs and the control RNA packed lentivirus were purchased from GENECHEM (Shanghai, China). The lentivirus was used to infect MDA-MB-231 and MDA-MB-468. Briefly, approximately 5 × 104 cells were seeded on six-well plates in L15 medium and incubated 24 h. Lentivirus (volume = multiplicity of infection × cells number/lentivirus titer) mixed with 5 μg/ml polybrene was added to cells. At 12 h following infection, the culture medium was replaced with fresh L15 medium. The medium was replaced with fresh medium with 2 μg/ml puromycin (Sangon Biotech Co., Ltd., Shanghai, China) after 3 days of infection. Next this medium containing puromycin was replaced every two days until the control cells were completely dead. We used MD2-G, PAX, pLKO-SIX4 three-pack system for high-expression virus packaging, the packaged lentivirus immediately infected MCF-7 and MDA-MB-231 cells. The effect of knockdown and overexpression of SIX4 were confirmed by Western blot.
Wound healing and transwell assays
Briefly, for wound healing assay, approximately 1 × 106 MDA-MB-231, MCF-7 cells were uniformly seeded on six-well plates. When the cell density reached approximately 100%, scratch-wound assay was carried out using a 10-μl pipette tip. The medium was replaced by L15 or DMEM without serum, including 1 μM 5-fluouracil (Sigma) to block cell proliferation after scratch wounding. Record the scratches after 0 and 24 h with a microscope.
For transwell migration assays, approximately 104 cells were seeded on the upper chamber membranes of 24-well plates. Then, 650 μl of L15 or DMEM culture medium supplemented with 10% FBS was added to the lower chamber. 24 h later, the membrane was fixed with 4% paraformaldehyde, sequentially stained with 0.1% crystal violet. The inside of the membrane was gently wiped with cotton swab, At last, quantify the number of cells with a microscope.
Quantitative real-time PCR (qRT-PCR)
Total RNAs were prepared using TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. Reverse transcription was performed using an RT-PCR kit (TaKaRa). PCR and data collection were performed using the ABI 7300 Fast real-time PCR system (Applied Biosystems). The gene expression fold-change was calculated using the 2-ΔΔCT method compared with GAPDH. The qPCR primer sequences of SIX4, Snai1, Slug, Twist, Zeb1, E-cadherin, N-cadherin, Vimentin and GAPDH were as follows: SIX4 (forward primer: 5’-AGCAGCTCTGGTACAAGGC-3’, reverse primer: 5’-CTTGAAACAATACACCGTCTCCT-3’); Snai1 (forward primer: 5’-ACTGCAACAAGGAATACCTCAG-3’, reverse primer: 5’-GCACTGGTACTTCTTGACATCTG-3’); Slug (forward primer: 5’-CGAACTGGACACACATACAGTG-3’, reverse primer: 5’-CTGAGGATCTCTGGTTGTGGT-3’); Twist (forward primer: 5’-GTCCGCAGTCTTACGAGGAG-3’, reverse primer: 5’-GCTTGAGGGTCTGAATCTTGCT-3’); Zeb1 (forward primer: 5’-GATGATGAATGCGAGTCAGATGC-3’, reverse primer: 5’-ACAGCAGTGTCTTGTTGTTGT-3’); E-cadherin (forward primer: 5’-CGAGAGCTACACGTTCACGG-3’, reverse primer: 5’-GGGTGTCGAGGGAAAAATAGG-3’); N-cadherin (forward primer: 5’-TCAGGCGTCTGTAGAGGCTT-3’, reverse primer: 5’-ATGCACATCCTTCGATAAGACTG-3’); Vimentin (forward primer: 5’-TGCCGTTGAAGCTGCTAACTA-3’, reverse primer: 5’-CCAGAGGGAGTGAATCCAGATTA-3’); GAPDH (forward primer: 5’-GGAGCGAGATCCCTCCAAAAT-3’, reverse primer: 5’-GGCTGTTGTCATACTTCTCATGG-3’).
Immunofluorescence (IF)
The right number of cells was seeded on 12 mm coverslips cultured in 24-well plates. 24 h later, the coverslips were fixed with 4% paraformaldehyde and washed with PBS three times. Next, the cells permeabilized with 0.1% Triton X-100/PBS. Blocked with 1% bovine serum albumin for 2 h. Subsequently, the cells were incubated with the primary antibodies overnight at 4°C. Next day, the cells were washed with PBS and incubated with the fluorescently labelled secondary antibody for 2 h at room temperature in the dark. Nuclei was strained by DAPI for 10 min, Samples were imaged using confocal microscopy.
Western blot
Total protein was extracted with NP40 lysates with the addition pf protein phosphatase inhibitors. Extraction of cytoplasmic proteins and nuclear proteins using the cytosolic nucleus isolation kit (BioVision). Protein samples were separated on 10% SDS-PAGE and then transferred to 0.45 um PVDF membranes (Bio-Rad). After blocking in 5% non-fat milk for 1 h at room temperature, the membranes were incubated with primary antibodies overnight at 4°C. After washing three times, the membranes were incubated with secondary antibodies (Thermo Fisher Scientific) for 2 h at room temperature. Finally, the membranes were washed three times and were detected with ECL regents (Thermo Fisher Scientific).
Immunoprecipitation assay
For immunoprecipitation assays, the cells were harvested and lysed in NP40 for 30 min at 4°C. After centrifugal, the supernatants were precleared with 10 μl of protein A/G agarose beads for 2 h. The primary antibody was separately added to the supernatants, and EP tube was placed on the suspension and shaken overnight at 4°C. Next day, the supernatants mixed with 60 μl of protein A/G agarose beads for 4 h at 4°C. After the agarose beads were boiled with 1 × loading buffer, the protein samples were prepared for the western blot.
Tissue samples and ethical approval
Tissues samples used here were obtained from First Affiliated Hospital, School of Medicine, Shihezi University. All patients have been clinically and histopathologically diagnosed at First Affiliated Hospital. Written informed consent was obtained from individual prior to this experiment and approved by the research medical ethics committee of First Affiliated Hospital, School of Medicine, Shihezi University (approval number: 2017-099-01).
Immunohistochemistry (IHC)
For IHC, formalin-fixed paraffin-embedded tissues were cut into 4-mm sections. The sides were stained with antibody against SIX4, p-STAT3. Protein SIX4 staining was mainly stained in the cytoplasm and partially stained in the nucleus. Protein p-STAT3 staining was stained in the nucleus. Two experienced pathologists examined immunostaining results. The expression level was calculated by multiplying an intensity score and a proportion score, and was categorized as low expression (score 0-3), high expression (score more than 4). The proportion score revealed the fraction of positive-stained cells (0: <10%; 1: 10% to ≤30%; 2: 31% to ≤70%; 4: >70%), the staining intensity score (0: no staining, 1: weak, 2: intermediate, 3: strong).
Animal experiment
We examined the ability of SIX4 on metastasis by using lung metastasis model. This experiment was maintained according to guidelines approved by Institutional Animal Care and Use Committee, Huazhong University of Science and Technology (Hubei China; IACUC ID: 2018S2183). We used 4 weeks old female BABL/C NOD mice for follow-up experiment. MDA-MB-231 Sh-NC, Sh-SIX4 cells were harvested and suspended in serum-free medium, injected tail vein 105 of cancer cells for experimental lung metastases. After eight weeks, mice were sacrificed and wet lungs were dissociated to weigh and analysis. Then fixed with formaldehyde and sent to HE and IHC staining.
Statistical analysis
All data were analyzed using SPSS20.0 software. The expression of SIX4 and clinicopathological features was compared using the Chi-square test. A Kaplan-Meier analysis was used to obtain the survival curves, and differences were assessed with the Cox regression analysis. The differences between groups in cell experiments were tested by Student’s t-test. The differences were considered statistically significant at P<0.05.
Results
SIX4 is overexpressed in breast cancer and associated with metastasis and poor prognosis
To detect the potential role of SIX4 in breast cancer development, we examined the expression of SIX4 in normal tissues and breast cancer samples with the Oncomine database. As shown in Figure 1A, the expression of SIX4 was significant overexpressed in breast cancer samples compared with normal tissues. To confirm this finding, we further analysed the expression of SIX4 in our clinical breast cancer samples by IHC staining, we found that the expression of SIX4 was higher in the lung metastases than in the primary tumor, and the primary tumor was significantly higher than normal tissue (Figure 1B, 1C). Further chi-square test showed that the expression of SIX4 was positive correlated with lymph node metastasis, TNM stage, histological differentiation, HER-2-/ER-/PR- in patients with breast cancer (Table 1). The data in Next, the multivariate analysis by Cox regression showed that SIX4 expression, N stage, HER-2-/ER-/PR-(TNBC) were the independent prognostic factor (Figure 1D). In addition, Kaplan-Meier analysis presented that in tumor tissues, the high expression of SIX4 was associated with a lower overall survival rate and disease free survival rate (Figure 1E, 1F). In collection, all these finding suggest that SIX4 may play an important role in the progression and metastasis of breast cancer.
Figure 1.
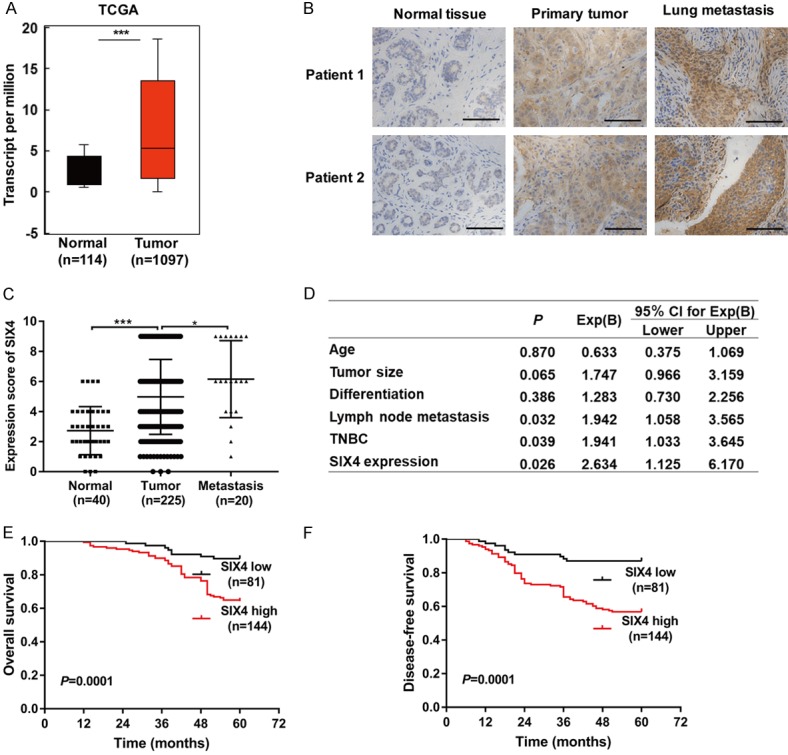
SIX4 is overexpressed in breast cancer and associated with metastasis and poor prognosis. A. Analysis of SIX4 levels in breast cancer tissues compared to those in normal samples using Oncomine databases. B. Analysis of SIX4 protein levels in the normal tissue, primary tumor, and lung metastases tissues. Scale bar, 100 μm. C. Quantification of the protein levels of SIX4 in normal tissue, primary tumor, and lung metastases tissue. D. Cox regression analysis to evaluate the significance of the association between SIX4 expression and breast cancer patients prognosis in the presence of clinical variables. E, F. Kaplan-Meier analysis of OS (overall survival) rate and DFS (disease free survival) rate related to the expression of SIX4 in 225 breast cancer patients. ***P<0.001. *P<0.05.
Table 1.
The correlations between the SIX4 expression levels and the clinicopathological features of patients with breast cancer
| Clinicopathological features | Cases | SIX4 | P-value | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Age (years) | ||||
| ≤45 | 96 | 34 | 62 | 0.745 |
| >45 | 129 | 43 | 86 | |
| Tumor size (cm) | ||||
| ≤2 | 94 | 38 | 56 | 0.097 |
| >2 | 131 | 39 | 92 | |
| Differentiation | ||||
| Well | 18 | 11 | 7 | 0.001 |
| Moderate | 160 | 61 | 99 | |
| Poor | 47 | 5 | 42 | |
| Lymph node metastasis | ||||
| No | 120 | 63 | 57 | 0.001 |
| Yes | 105 | 14 | 91 | |
| TNM stage | ||||
| I | 64 | 35 | 29 | 0.001 |
| II | 105 | 31 | 74 | |
| III | 56 | 11 | 45 | |
| TNBC | ||||
| No | 180 | 71 | 109 | 0.031 |
| Yes | 45 | 10 | 35 | |
SIX4 expression significantly impacts breast cancer cells migration and invasion in vitro
Previous results demonstrate that SIX4 was involved in the progression and metastasis of breast cancer. We designed in vitro experiment to validate this result.
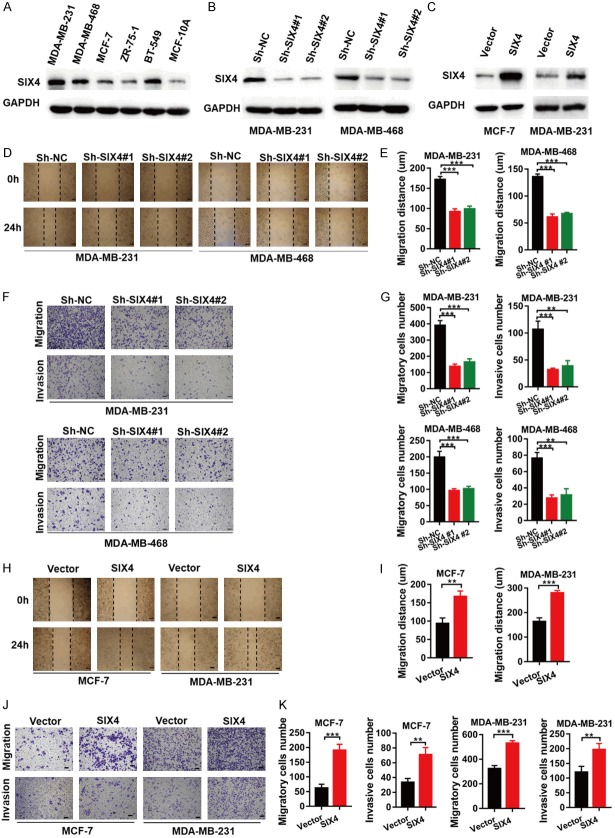
We first detected the expression of SIX4 in normal breast cell lines and breast cancer cell lines. SIX4 showed high expression in MDA-MB-231, MDA-MB-468 and BT549 cells and low expression in the normal cell line MCF-10A and other breast cancer cell lines (Figure 2A). We used lentivirus technology to silence SIX4 expression in MDA-MB-231, MDA-MB-468 and improve its expression in MCF-7, MDA-MB-231 cells. As shown in Figure 2B and 2C, the expression of SIX4 were analysed by western blot analysis. To test the effect of SIX4 on wound healing, migration, and invasion of breast cancer cells, we used MDA-MB-231, MAD-MB-468 cells silenced with Sh-NC or SIX4 Sh-RNA and MCF-7, MDA-MB-231 cells overexpressed with Vector or SIX4 in subsequent experiments. Wound healing assays showed that the healing capability of MDA-MB-231 and MDA-MB-468 Sh-SIX4 cells were significantly weakened compared with Sh-NC cells (Figure 2D, 2E). Transwell migration and Matrigel invasion assays demonstrated that SIX4 knockdown obviously reduced the migratory ability of MDA-MB-231 and MDA-MB-468 cells compared with negative control cells (Figure 2F, 2G). On the contrary, we found that overexpression of SIX4 in MCF-7 and MDA-MB-231 cells enhanced the migration and invasion ability (Figure 2H-K). These results suggest that SIX4 expression significantly impacts cell migration and invasion in vitro.
Figure 2.
SIX4 expression significantly impacts breast cancer cells migration and invasion in vitro. A. The whole-cell extracts from five breast cancer cell lines and a normal breast cell line, they well then collected and subjected to IB analysis. B. MDA-MB-231 and MDA-MB-468 cells were silenced with control (Sh-NC) or SIX4 Sh-RNA (no. 1 and 2), and whole-cell extracts were collected for IB analysis. C. MCF-7 and MDA-MB-231 cells were overexpressed with Vector or SIX4, and whole-cell extracts were collected for IB analysis. D, E. Wound healing assays in MDA-MB-231 cells and MDA-MB-468 cells silenced with control (Sh-NC) or SIX4 Sh-RNA (no. 1 and 2). F, G. Migration and invasion assays in MDA-MB-231 cells and MDA-MB-468 cells silenced with control (Sh-NC) or SIX4 Sh-RNA (no. 1 and 2). H, I. Wound healing assays in MCF-7 cells and MDA-MB-231 cells overexpressed with Vector or SIX4. J, K. Migration and invasion assays in MCF-7 cells and MDA-MB-231 cells overexpressed with Vector or SIX4. Each bar represents the mean ± SD of three independent experiments. Scale bar, 100 μm. ***P<0.001, **P<0.01.
SIX4 regulates invasion-metastasis cascade through EMT program
It has been generally believed that EMT program may promote breast cancer metastasis. We speculated that the acquisition of metastatic potential by SIX4 might depend on induction of EMT process. To test this hypothesis, we first detected whether SIX4 affect EMT-related transcription (EMT-TFs) and markers in breast cancer. The results showed that only Snai1 was significantly decreased in MDA-MB-231 ShSIX4 cells compared to Sh-NC cells, while other tested TFs was not changed.
Correspondingly, E-cadherin increased, while N-cadherin and Vimentin decreased both in mRNA and protein level in MDA-MB-231 Sh-SIX4 cells (Figure 3A, 3B). In contrast, overexpression of SIX4 increased the expression of transcription factor Snai1, which in turn decreased E-cadherin, while elevated N-cadherin and Vimentin in MCF-7 cells (Figure 3C, 3D). In addition, similar results were obtained by immunofluorescence (IF) assay (Figure 3E, 3F). Taken together, those data indicate that overexpression of SIX4 gain the mesenchymal characteristics and lose epithelial properties.
Figure 3.
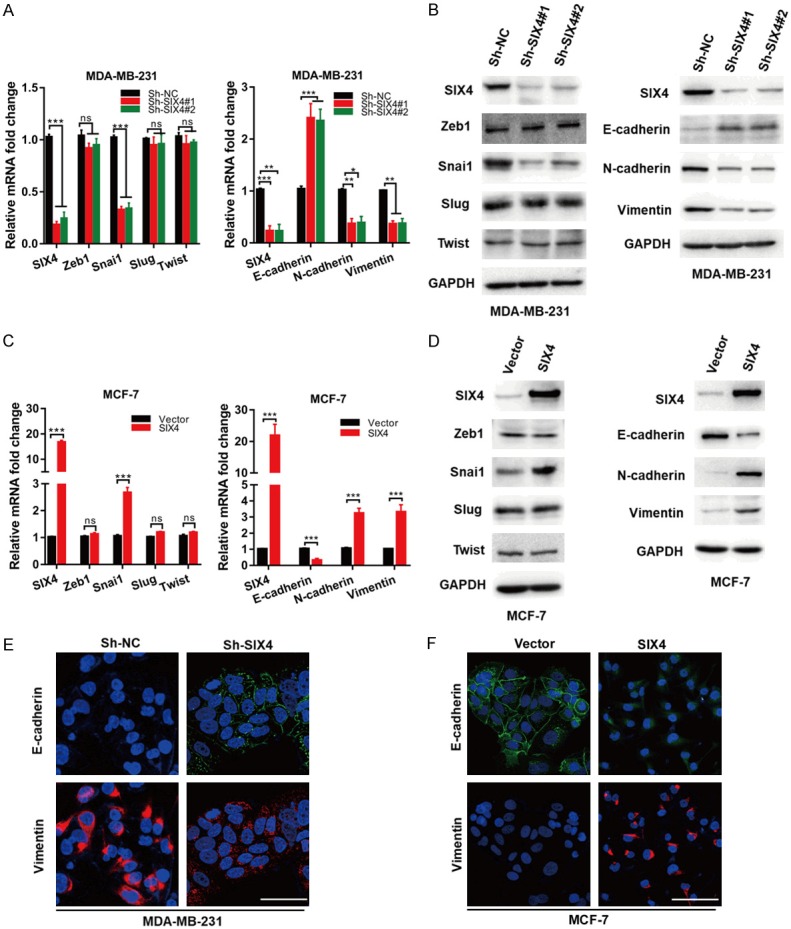
SIX4 regulates invasion-metastasis cascade through EMT program. A. The mRNA expression of EMT-TFs and EMT markers were determined in MDA-MB-231 cells silenced with control (Sh-NC) or SIX4 Sh-RNA (no. 1 and 2). B. MDA-MB-231 cells were silenced with control (Sh-NC) or SIX4 Sh-RNA (no. 1 and 2), and whole-cell extracts were collected for IB analysis. C. The mRNA expression of EMT-TFs and EMT markers were determined in MCF-7 cells overexpressed with Vector or SIX4. D. MCF-7 cells were overexpressed with Vector or SIX4, and whole-cell extracts were collected for IB analysis. E. Immunofluorescence staining of E-cadherin and Vimentin in paired MDA-MB-231/Sh-NC, MDA-MB-231/Sh-SIX4 cells. F. Immunofluorescence staining of E-cadherin and Vimentin in paired MCF-7/Vector and MCF-7/SIX4 cells. Nuclei were visualized with DAPI staining (blue). Each bar represents the mean ± SD of three independent experiments. Scale bar, 50 μm. ***P<0.001, **P<0.01, *P<0.05.
SIX4 promotes the activation of the STAT3 signalling pathway in breast cancer
Previous reports have shown the important role of the STAT3 signalling pathway in cancer EMT. For example, STAT3 is involved in mediating the EMT program and is also inextricably linked to EMT transcription factors. Moreover, STAT3 crosstalk with other signalling pathways leading to EMT [13]. We hypothesized that SIX4 regulates EMT by the STAT3 signalling pathway, to confirm this opinion, we detected the total and phosphorylation protein levels of JAK2, STAT3 in breast cancer cell lines. The western blot results showed that SIX4 could notably increase the phosphorylation of STAT3, but the expression of total JAK2, STAT3 and phosphorylation JAK2 were not changed in breast cancer cell lines (Figure 4A, 4B). It well known that phosphorylated STAT3 acts as a transcription factor by translocating to the nucleus to activate downstream target genes, including EMT related genes. We examined whether SIX4 affect the STAT3 nuclear translocation. As shown in Figure 4C and 4D, knockdown of SIX4 strongly decreased the level of STAT3 in the nucleus, while overexpression of SIX4 enhanced the level of STAT3 in the nucleus. To elucidate the potential role of SIX4 in STAT3 activation, we examined whether SIX4 induces STAT3 through SIX4-STAT3 complex formation. Flag-SIX4 and HA-STAT3 plasmids were co-transfected into HEK293T cells and detected by exogenous co-immunoprecipitation test, the western blot assay showed that Flag-SIX4 and HA-STAT3 bind to each other (Figures 4E, S1A). In agreement with this finding, immunoprecipitation assay (Figure 4F) and immunofluorescence assay (Figure 4G) showed that the endogenous SIX4 could bind to endogenous STAT3 in MDA-MB-231 cells. Furthermore, we demonstrated that knockdown of SIX4 weaken the combination of JAK2 and STAT3 by co-immunoprecipitation assay, whereas, the total level of JAK2 protein was not affected in MDA-MB-231 Sh-SIX4 cells compared with Sh-NC cells (Figure 4H). To further validate this, we found that knockdown of SIX4 didn’t affect the combination of STAT3 and EGFR or SRC by co-immunoprecipitation assay (Figure S1B). These data indicate that SIX4 may enhance phosphorylated STAT3 via interfering with the interaction between JAK2 and STAT3 thereby inhibit the EMT program of breast cancer.
Figure 4.
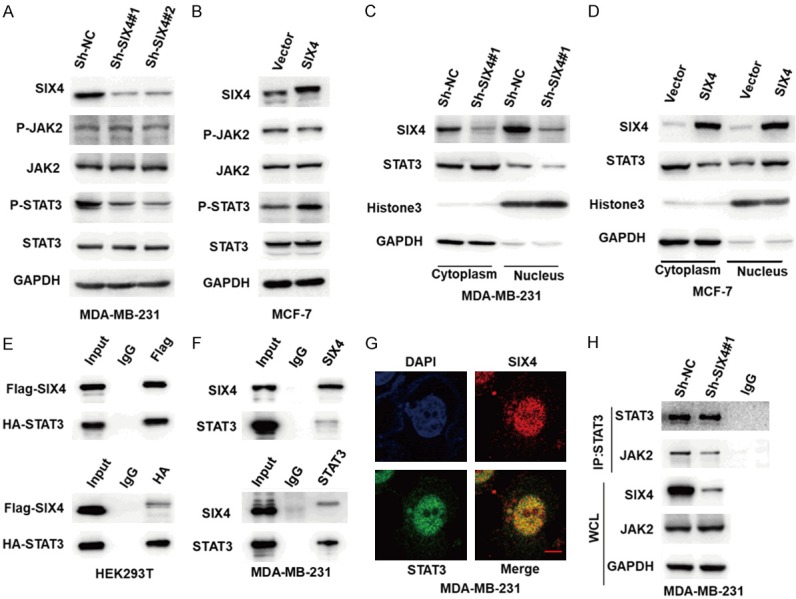
SIX4 promotes the activation of the STAT3 signalling pathway in breast cancer. A. MDA-MB-231 cells were silenced with control (Sh-NC) or SIX4 Sh-RNA (no. 1 and 2), and whole-cell extracts were collected for IB analysis of the indicated proteins. B. MCF-7 cells were overexpressed with Vector or SIX4, and whole-cell extracts were collected for IB analysis of the indicated proteins. C, D. IB analysis showed nuclear localization of total STAT3 in those indicated cells. E. HEK293T cells were co-transfected with Flag-SIX4 and HA-STAT3 plasmids, and whole-cell extracts were collected for IP with Flag or HA antibody, followed by IB analysis. F. MDA-MB-231 cells were lysed, then whole-cell extracts were collected for IP with SIX4 or STAT3 antibody, followed by IB analysis. G. Immunofluorescence staining showing co-localization of SIX4 (red) and STAT3 (green) in MDA-MB-231 cells. Scale bar: 10 μm. H. MDA-MB-231 cells were silenced with control (Sh-NC) or SIX4 Sh-RNA#1, whole-cell extracts were collected for IP with STAT3 or lgG antibody, followed by IB analysis of the indicated proteins.
The effect of SIX4 on EMT depends on the STAT3 signalling pathway in breast cancer
To further investigate the role of STAT3 in SIX4-induced EMT process, we used STAT3 siRNA and inhibitor to perform the reverse experiment. Interestingly, we found that siSTAT3 almost completely reversed the increase in Snai1 mRNA and protein levels mediated by SIX4 overexpression (Figure 5A, 5B). Furthermore, we obtained the same result in mRNA and protein levels of EMT marker (Figure 5C, 5D). To further confirm these results, we used siSTAT3 to perform migration and invasion reversal experiments. We found that siSTAT3 could suppress the regulatory effects of SIX4 both in migration and invasion (Figure 5E, 5F). Correspondingly, when STAT3 pathway was blocked by inhibitor AG-490, the SIX4 overexpressing failed to facilitate MCF-7 cells migration and invasion (Figure 5G). However, it had been reported that PI3K/Akt was regulated by SIX4 in colorectal cancer [3]. In this study, we demonstrated that when AKT pathway was blocked by inhibitor AZD5363, the SIX4 overexpressing could still facilitate migration and invasion in MCF-7 cells (Figure S1C, S1D). Collectively, these data suggest that SIX4 promotes EMT depends on the STAT3 signalling pathway in breast cancer.
Figure 5.
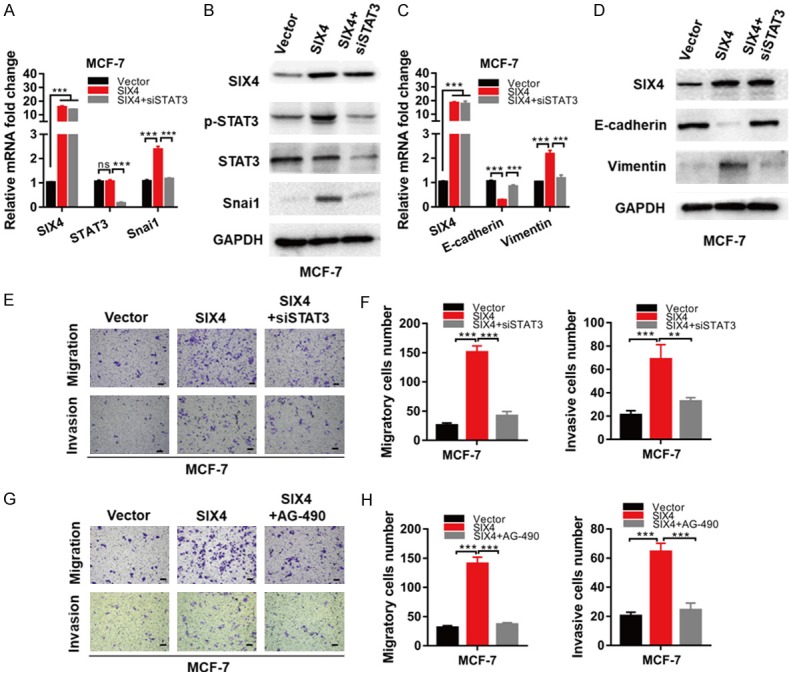
The effect of SIX4 on EMT depends on the STAT3 signalling pathway in breast cancer. A. MCF-7 cells were overexpressed with Vector, SIX4 or SIX4 plus siSTAT3, mRNA extracts were collected for qRT-PCR analysis of indicated markers. B. MCF-7 cells were overexpressed with Vector, SIX4 or SIX4 plus siSTAT3, whole-cell extracts were collected for IB analysis of the indicated proteins. C. MCF-7 cells were overexpressed with Vector, SIX4 or SIX4 plus siSTAT3, mRNA extracts were collected for qRT-PCR analysis of indicated markers. D. MCF-7 cells were overexpressed with Vector, SIX4 or SIX4 plus siSTAT3, whole-cell extracts were collected for IB analysis of the indicated proteins. E, F. Migration and invasion assays in MCF-7 cells overexpressed with Vector, SIX4 or SIX4 plus siSTAT3. G, H. Migration and invasion assays in MCF-7 cells overexpressed with Vector, SIX4 or SIX4 plus STAT3 inhibitor. Each bar represents the mean ± SD of three independent experiments. Scale bar, 100 μm. ***P<0.001, **P<0.01.
Knockdown of SIX4 suppresses breast cancer metastasis in vivo
In an effort to investigate the effect of SIX4 on metastasis in vivo, MDA-MB-231 Sh-NC, Sh-SIX4 cells were respectively injected into caudal veins of female nude mice. At eight weeks after injection, all mice were sacrificed and the lungs were harvested. We found that the lungs of the Sh-SIX4 mice displayed less macroscopically visible metastases than Sh-NC mice (Figure 6A, 6B). We also checked the wet lung weight between two groups, data indicated that the wet lung weight of Sh-SIX4 groups was lighter than that of Sh-NC groups (Figure 6C). Moreover, mice lungs embedded in and sectioned for H&E staining, the result showed that Sh-SIX4 significantly suppressed the migration nodules (Figure 6D). We also tested the association between SIX4 and p-STAT3 protein expression in lung metastatic nodules by IHC staining, the data showed that expression of p-STAT3 was weakened with knockdown of SIX4 in vivo (Figure 6E). Taken together, our study reveals that SIX4 plays a potential role in breast cancer metastasis.
Figure 6.
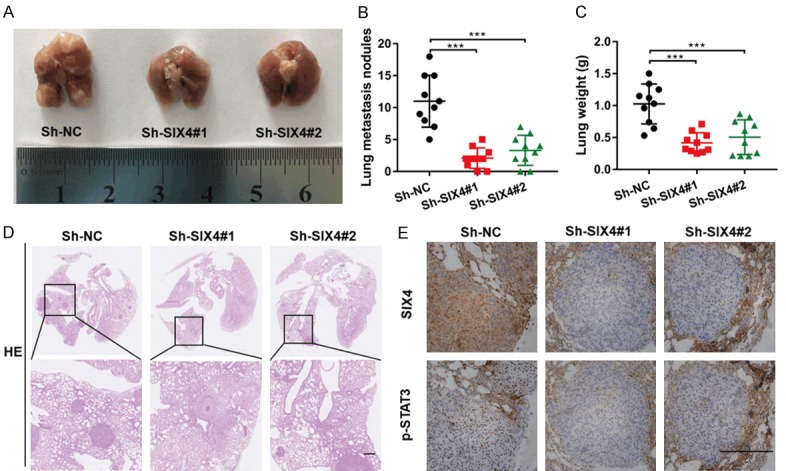
Knockdown of SIX4 suppresses breast cancer metastasis in vivo. (A) Nude mice were randomly separated three groups intravenously tail-vein injected with MDA-MB-231 cells expressing Sh-NC, Sh-SIX4#1 and Sh-SIX4#2. Representative images of lung showing metastatic foci generated from MDA-MB-231 cells and (B) quantitative analysis of the average number of metastatic foci and (C) the wet weight of lung in three groups. ***P<0.001. (D) Representative HE and (E) Representative IHC pictures of SIX4 and p-STAT3 expression in MDA-MB-231 cells lung metastatic foci. Scale bar: 100 μm.
Discussion
There is increasing evidence supporting an important role of SIX family in male gonadal differentiation, myogenesis, neurogenesis, development of organs and tumor [14-17]. As a member of the SIX family, SIX4 is rarely seen in research reports on tumors. Wei Q et al. [18] demonstrated SIX1 and SIX4 are highly expressed in esophageal squamous cell carcinoma. LI G et al. [3] found that SIX4 was up-regulated in GEO cohorts and Cancer Genome Atlas CRC cohort. X.Y. Na et al. [5] reported that MiR-203a plays a role in tumor suppression by targeting down-regulation of SIX4 in bladder cancer. However, the mechanism by which SIX4 contributes to above carcinoma is unclear. In addition, a role for SIX4 in other cancers has not been examined. In this study, we demonstrated that SIX4 was higher in the lung metastases than the primary tumor. SIX4 expression was positive correlated with lymph node metastasis, TNM stage, histological differentiation, HER-2-/ER-/PR- in patients with breast cancer patients. Breast cancer is the most common cancer in women and ranks second among female cancer deaths. The occurrence of distant metastasis is the leading cause of death in breast cancer patients. Triple-negative breast cancer (TNBC) accounts for about 10-20% of all breast cancers. It is characterized by highly invasive, early recurrence and distant metastasis, and poor overall survival [19]. In present study, we demonstrate that the expression of SIX4 in TNBC was higher, and it was the independent prognostic factor and associated with poor survival. The above data suggested that SIX4 may play an important role in breast cancer metastasis. Subsequently, we designed some experiments in vitro and in vivo to explore.
To date, the EMT process has been an important cause of distant metastases for epithelial cancers, including breast cancer [20]. The EMT is triggered by so-called EMT-TFs such as Zeb1, Twist, Snai1, Slug and so on. In our study, we found that SIX4 impacted snai1 mRNA and protein expression, while that of the other tested EMT-TFs was not changed. SIX4 further affected the EMT markers by affecting transcription factors. Therefore, our observations suggest that SIX4 impacted breast cancer cells migration and invasion, and plays an important role in the promotion of EMT.
EMT is closely related to STAT3 signalling pathways [21]. In the current study, we demonstrated knockdown of SIX4 notably decreased the phosphorylation of STAT3 translocated to nuclear, which in turn suppressed the EMT process. In contrast, the overexpressing of SIX4 significantly promoted the EMT process. For further study, we explored that SIX4 impacted the combination of JAK2 and STAT3, which in turn affected STAT3 signalling pathway. Moreover, EMT up-regulation by SIX4 overexpression can also be reversed by siSTAT3 or STAT3 inhibitor. These results explained the possible mechanism by which SIX4 affects STAT3 and thus induced EMT, leading to distant metastasis. In addition, in vivo experiment, our result indicated a marked reduction in metastases in MDA-MB-231 Sh-SIX4 compared with Sh-NC mice, and that STAT3 phosphorylation was positively associated with the expression of SIX4.
In summary, we identified a new molecular and functional network present in breast cancer metastasis that regulates and coordinates SIX4. The result suggested that SIX4 seems to represent a novel potential target for breast cancer therapeutics.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81773113, No. 81572725, No. 81874186, No. 31670891 and No. 81874148).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Adv Cancer Res. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- 2.Wu W, Ren Z, Li P, Yu D, Chen J, Huang R, Liu H. Six1: a critical transcription factor in tumorigenesis. Int J Cancer. 2015;136:1245–1253. doi: 10.1002/ijc.28755. [DOI] [PubMed] [Google Scholar]
- 3.Li G, Hu F, Luo X, Hu J, Feng Y. SIX4 promotes metastasis via activation of the PI3K-AKT pathway in colorectal cancer. PeerJ. 2017;5:e3394. doi: 10.7717/peerj.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Q, Li A, Tian Y, Liu Y, Li T, Zhang C, Wu JD, Han X, Wu K. The expression profile and clinic significance of the SIX family in non-small cell lung cancer. J Hematol Oncol. 2016;9:119. doi: 10.1186/s13045-016-0339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Na XY, Shang XS, Zhao Y, Ren PP, Hu XQ. MiR-203a functions as a tumor suppressor in bladder cancer by targeting SIX4. Neoplasma. 2019;66:211–221. doi: 10.4149/neo_2018_180512N312. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 8.Wu Y, Sarkissyan M, Vadgama JV. Epithelial-mesenchymal transition and breast cancer. J Clin Med. 2016;5 doi: 10.3390/jcm5020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 10.Kim MS, Lee HS, Kim YJ, Lee DY, Kang SG, Jin W. MEST induces Twist-1-mediated EMT through STAT3 activation in breast cancers. Cell Death Differ. 2019;26:2594–2606. doi: 10.1038/s41418-019-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saitoh M, Endo K, Furuya S, Minami M, Fukasawa A, Imamura T, Miyazawa K. STAT3 integrates cooperative Ras and TGF-beta signals that induce Snail expression. Oncogene. 2016;35:1049–1057. doi: 10.1038/onc.2015.161. [DOI] [PubMed] [Google Scholar]
- 12.Rokavec M, Öner MG, Li H, Jackstadt R, Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S, Slotta-Huspenina J, Bader FG, Greten FR, Hermeking H. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124:1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takata N, Abbey D, Fiore L, Acosta S, Feng R, Gil HJ, Lavado A, Geng X, Interiano A, Neale G, Eiraku M, Sasai Y, Oliver G. An eye organoid approach identifies Six3 suppression of R-spondin 2 as a critical step in mouse neuroretina differentiation. Cell Rep. 2017;21:1534–1549. doi: 10.1016/j.celrep.2017.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimoto Y, Tanaka SS, Yamaguchi YL, Kobayashi H, Kuroki S, Tachibana M, Shinomura M, Kanai Y, Morohashi K, Kawakami K, Nishinakamura R. Homeoproteins Six1 and Six4 regulate male sex determination and mouse gonadal development. Dev Cell. 2013;26:416–430. doi: 10.1016/j.devcel.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 15.Santolini M, Sakakibara I, Gauthier M, Ribas-Aulinas F, Takahashi H, Sawasaki T, Mouly V, Concordet JP, Defossez PA, Hakim V, Maire P. MyoD reprogramming requires Six1 and Six4 homeoproteins: genome-wide cis-regulatory module analysis. Nucleic Acids Res. 2016;44:8621–8640. doi: 10.1093/nar/gkw512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Liu M, Zhao L, Li Y, Zhang M, Jin Y, Xiong Q, Liu X, Zhang L, Jiang H, Chen Q, Wang C, You Z, Yang H, Cao C, Dai Y, Li R. Disabling of nephrogenesis in porcine embryos via CRISPR/Cas9-mediated SIX1 and SIX4 gene targeting. Xenotransplantation. 2019;26:e12484. doi: 10.1111/xen.12484. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi M, Tamura M, Sato S, Kawakami K. Mice doubly deficient in Six4 and Six5 show ventral body wall defects reproducing human omphalocele. Dis Model Mech. 2018;11 doi: 10.1242/dmm.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei Q, Yu WW, Zhao KL, Fu XL, Zhu ZF, Qin GQ, Chen H, Zhang ZX, Gu YZ, Xiang JQ, Chen HQ, Du X, Sun MH. Expression of Six1 and Six4 in esophageal squamous cell carcinoma and their correlation with clinical prognosis. Zhonghua Bing Li Xue Za Zhi. 2013;42:446–450. doi: 10.3760/cma.j.issn.0529-5807.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khaled N, Bidet Y. New insights into the implication of epigenetic alterations in the EMT of triple negative breast cancer. Cancers (Basel) 2019;11 doi: 10.3390/cancers11040559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balanis N, Wendt MK, Schiemann BJ, Wang Z, Schiemann WP, Carlin CR. Epithelial to mesenchymal transition promotes breast cancer progression via a fibronectin-dependent STAT3 signaling pathway. J Biol Chem. 2013;288:17954–17967. doi: 10.1074/jbc.M113.475277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.