Abstract
Long non-coding RNAs (lncRNAs) have been highlighted to play key roles in the gene regulatory network, and the dysregulation of lncRNAs has also been implicated in various malignancies. However, little is known regarding the expression of lncRNA and their functions in the progression of nephroblastoma. Thus, the present study aimed to explore the potential role of homeobox A11 (HOXA11)-AS in nephroblastoma. Microarray-based analysis was initially applied to screen the differentially expressed lncRNAs, and HOXA11-AS was selected as the candidate. The HFWT cells were performed with gain- and loss-of function test to evaluate the role of HOXA11-AS in cell cycle and apoptosis in nephroblastoma using flow cytometry and Western blots. Moreover, the relationship between HOXA11-AS and forkhead box P2 (FOXP2) was verified by Cross-linking RIP, and the direct interaction between HOXA11-AS and Cyclin D2 (CCND2) was detected using a dual luciferase reporter gene assay. Tumor formation in nude mice was used to investigate the effect of HOXA11-AS in vivo. HOXA11-AS was found to be highly expressed in the nephroblastoma. Furthermore, the silencing of HOXA11-AS promoted apoptosis and cell cycle arrest at the G1/S phase in nephroblastoma through the transcription factor FOXP2 to downregulate the expression of CCND2. Consistently, the tumor formation data in nude mice verified the results in vivo. Taken together, silencing of HOXA11-AS promotes apoptosis and inhibits the cell cycle entry in nephroblastoma by recruiting the transcription factor FOXP2 to downregulate the expression of CCND2, highlighting a promising novel direction for future nephroblastoma treatment.
Keywords: Long non-coding RNA HOXA11-AS, forkhead box P2, cyclin D2, nephroblastoma, cell apoptosis, cell cycle
Introduction
As a common primary renal tumor, nephroblastoma commonly occurs during childhood and is widely considered to be an embryonic neoplasm arising from the metanephric blastema [1]. Nephroblastoma has significant metastatic potential, often spreading to other organs. Surgical resection with adjuvant chemotherapy is a treatment approach for all stages of nephroblastoma arising from a teratoma, with the exception of primary extra-renal nephroblastoma [2]. Classically, the tumor often exhibits a triphasic combination of blastemal, stromal and epithelial components, although the percentage of each component has been shown to vary considerably [3]. The increase in abdominal mass or circumference is the most typical symptoms of nephroblastoma [4]. Currently, long non-coding RNAs (lncRNAs) have been demonstrated to be involved in the development of nephroblastoma [5].
LncRNAs are longer than 200 nt in length, and have been found to participate in various biological processes, including the regulation of transcription, translation, and protein modification [6]. Interestingly, lncRNAs can promote tumor formation, progression, and metastasis in many human malignancies, including kidney cancer [7]. For instance, SOX21-AS1 has been found to modulate cell proliferation in nephroblastoma [8]. Interestingly, a newly identified homeobox (HOX) A11 antisense (HOXA11-AS), 1,628 bp in length, has been highlighted to be a vital initiator and facilitator in proliferation and metastasis of malignant tumors [9]. Moreover, cyclin D (CCND) family members are dysregulated in different types of cancers, highlighting their potential as therapeutic targets for future cancer therapy [10]. Faussillon et al. indicate that CCND2 is upregulated in nephroblastoma [11]. A previous study revealed that lnc00598 could modulate the transcription of CCND2 and regulate the G1 checkpoint [12].
The forkhead box (Fox) genes have been linked with numerous human diseases, including cancer, glaucoma and language disorders, and act as transcription factors with DNA-binding ability [13]. FOXP2 is involved in the process of nucleic acid-binding and protein oligomerization, and also functions in embryonic development of various tissues, such as the brain, lung, intestine and heart tissues [14].
SNHG1 has been demonstrated to promote the malignant biological behavior of glioma cells by participating in the miR-154-5p/miR-376b-3p-FOXP2-KDM5B axis with a positive feedback loop [15]. SOX2OT has been reported to promote the carcinogenesis of nephroblastoma by regulating the expression and function of miR-363/FOXP4 [16]. Based on the aforementioned findings, we hypothesized that lncRNA could regulate the development of nephroblastoma via the CCND2 and FOXP2. Therefore, HOXA11-AS was selected as the study focus of our investigation in order to investigate this hypothesis and potentially elucidate a novel treatment strategy for nephroblastoma.
Materials and methods
Ethics statement
Written consent was obtained from all participating patients prior to the study. The protocols of this study were approved by the Ethic Committee of Guangzhou Women and Children’s Medical Center, Guangzhou Medical University and performed in strict accordance with the ethical principles for medical research involving human subjects of the Declaration of Helsinki. All animal experiments were performed in strict adherence with the Guide for the Management and Use of Laboratory Animals issued by the National Institutes of Health. The protocol of animal experiments was approved by the Institutional Animal Care and Use Committee of Guangzhou Women and Children’s Medical Center, Guangzhou Medical University.
Microarray-based analysis
Nephroblastoma gene expression datasets, including GSE11151, GSE19249 and GSE73209 were downloaded from the Gene Expression Omnibus database. The background normalization and standardized preprocessing of gene expression datasets were performed by the affy package in R, with the differential expression analysis performed using the “limma” package in R. The heatmap was subsequently constructed using the “pheatmap” package.
Sample collection and cell culture
This study included samples from 28 patients diagnosed with nephroblastoma who underwent surgery at the Guangzhou Women and Children’s Medical Center, Guangzhou Medical University between April 2017 and July 2018. All collected samples were classified in accordance with the World Health Organization criteria for nephroblastoma.
The nephroblastoma cell line HFWT was purchased from RIKEN BioResource Research Center (RCB0665, Tsukuba, Japan) and cultured in Ham F12 medium containing 15% fetal bovine serum (FBS), 100 ug/mL streptomycin and 100 U/mL penicillin. Another nephroblastoma cell line SK-NEP-1 was purchased from American Type Culture Collection (HTB-48, Manassas, VA, USA) and cultured in McCoy’s 5a medium modified (30-2007) containing 15% FBS. Normal human embryonic kidney cells HEK-293A were purchased from the European Collection of Animal Cell Cultures and cultured in Multi Experiment Matrix (Gibco, Carlsbad, CA, USA) medium containing 15% FBS, 100 ug/mL streptomycin and 100 U/mL penicillin. All cells were grown at 37°C in a humidified 5% CO2 atmosphere.
Transfection
Nephroblastoma HFWT cells were transfected with overexpressed (oe)-negative control (NC) + small hairpin (sh)-NC, oe-HOXA11-AS + sh-NC, oe-NC + sh-HOXA11-AS, oe-NC, oe-HOXA11-AS, sh-NC, sh-HOXA11-AS, oe-HOXA11-AS + sh-FOXP2, oe-HOXA11-AS + oe-FOXP2 + sh-NC, and oe-HOXA11-AS + oe-FOXP2 + sh-CCND2 plasmids. The overexpressed vector was pcDNA3.1, and the RNAi vector was pRNAT-U6.1/neo, all of which were designed and constructed by GenePharma (Shanghai, China). The transfection was performed using the Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted by Trizol (15596026, Invitrogen, Carlsbad, CA, USA), and subsequently reversely transcribed into cDNA using a reverse transcription kit (RR047A, Takara, Japan). The reaction was conducted using a SYBR Premix EX Taq kit (RR420A, Takara Bio Inc., Otsu, Shiga, Japan) in a real-time PCR instrument (ABI7500, ABI, Foster City, CA, USA). Three replicates were considered to be an experiment set. The primers (Table 1) were synthesized by Shanghai Sangon Biotechnology Co., Ltd. (Shanghai, China). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was regarded as an internal control. The relative transcript level of the product was calculated by the 2-ΔΔCt method [17].
Table 1.
RT-PCR primer sequences
| Primer sequences | |
|---|---|
| HOXA11-AS | F: 5’-TGCCAAGTTGTACTTACTACGTC-3’ |
| R: 5’-GTTGGAGGAGTAGGAGTATGTA-3’ | |
| CCND2 | F: 5’-TGGAGCTGCTGTGCCACG-3’ |
| R: 5’-GTGGCCACCATTCTGCGC-3’ | |
| Ki67 | F: 5’-ACGCCTGGTTACTATCAAAAGG-3’ |
| R: 5’-CAGACCCATTTACTTGTGTTGGA-3’ | |
| caspase-3 | F: 5’-TGTGGCATTGAGACAGAC-3’ |
| R: 5’-CACTTGCCATACAAACTA-3’ | |
| GAPDH | F: 5’-GGGCTGCTTTTAACTCTGGT-3’ |
| R: 5’-TGGCAGGTTTTTCTAGACGG-3’ |
Note: HOXA11-AS, homeobox A11 antisense RNA; CCND2, cyclin D2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; F, forward; R, reverse.
Fluorescence in situ hybridization (FISH)
The subcellular localization of HOXA11-AS was identified by FISH. According to the instructions of RiboTM lncRNA FISH Probe Mix (Red) (C10920, RiboBio company, Guangzhou, Guangdong, China), FISH was performed as follows: slides were placed in a 24-well culture plate, and HFWT cells were seeded at 6 × 104 cells/well and fixed with 1 mL 4% paraformaldehyde at room temperature for 10 min. Pre-hybrid solution (200 uL) was added to each well for 30 min at 37°C. Next, anti-HOXA11-AS nucleotide probe (Wuhan GeneCreate Biological Engineering Co., Ltd., Wuhan, Hubei, China) were added and incubated overnight at 37°C under conditions void of light. The following day, the slices were washed three times (5 min each) at 42°C with 0.1% Tween-20 in 4 × sodium citrate buffer (SSC), 2 × SSC and 1 × SSC under dark conditions. After rinsing with 1 × PBS for three times (5 min each) at room temperature, the slices were stained with 4’6-diamidino-2-phenylindole (1:800) for 10 min. A fluorescent microscope (Olympus, Tokyo, Japan) was employed to photograph and analyze the slices under five different fields of vision.
Native RNA immunoprecipitation (RIP)
The RIP kit (Milipore Corporation, Bedford, MA, USA) was employed to detect the binding of HOXA11-AS to the transcription factor FOXP2 protein. The cell lysate was co-precipitated with the magnetic bead-antibody complex. The antibody used for RIP was FOXP2 (#5337, 1:200, Cell Signaling Technology, Beverly, MA, USA). The antibody was mixed at room temperature for 30 min, and immunoglobulin G (IgG; ab172730, 1:100, Abcam Inc., Cambridge, UK) was regarded as the NC. Finally, RNA extraction was performed for qPCR analysis.
Cross-linking RIP
The HFWT cells transfected with different plasmids for 48 h were cross-linked using 0.75% formaldehyde. Chromatin was then sheared by sonication. The FOXP2 antibody (#5337, 1:200, Cell Signaling Technology, Beverly, MA, USA) was subsequently added to the nucleic acid extract and incubated overnight at 4°C. Protein A/G Magnetic Beads (88803, Thermo Fisher Scientific Inc., Waltham, MA, USA) were then added and incubated for 4 h at 4°C. Following proteinase K treatment, the total RNA was isolated from lysates for reverse transcription into cDNA and RT-qPCR analysis.
RNA pull-down
Biotinylated HOXA11-AS and U6 RNA were mixed with proteins from nuclear extracts of HFWT cells. A complex of biotinylated HOXA11-AS and protein was isolated using streptavidin agarose beads (Thermo Fisher Scientific, Waltham, MA, USA). The protein was then eluted from the RNA-protein complex. Finally, immunoblotting with FOXP2 antibody was applied in order to detect and evaluate the interaction.
Western blot analysis
Total protein in the tissues or cells was isolated using RIP assay lysis buffer containing phenylmethylsulfonyl fluoride. The protein (50 μg) was subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis gel electrophoresis, and transferred onto a polyvinylidene fluoride membrane by a wet transfer method. The membrane was blocked with 5% skim milk at room temperature for 1 h, and then incubated with diluted rabbit antibodies to CCND2 (ab230883, 1:1000), Ki67 (ab16667, 1:1000), or Cleaved-caspase-3 (ab2302, 1:1000) overnight at 4°C. GAPDH (ab9485, 1:2500) was employed as the internal reference. Next, the membrane was incubated with horseradish peroxidase-labeled secondary goat anti-rabbit antibody to IgG H&L (ab97051, 1:2000) for 1 h. All the aforementioned antibodies were purchased from Abcam (Cambridge, UK). The signal was developed by enhanced chemiluminescence fluorescence detection Kit (BB-3501, Amersham, Little Chalfont, UK) and exposed in the Bio-Rad image analysis system (Bio-Rad, Richmond, Cal., USA). Quantity One v4.6.2 software was used for densitometry analysis. The relative protein content was determined by the ratio between the target protein and GAPDH.
Dual luciferase reporter gene assay
Plasmids oe-NC, oe-HOXA11-AS, sh-NC, or sh-HOXA11-AS were co-transfected into HFWT cells with CCND2-2Kb luciferase reporter plasmid respectively to evaluate the effects associated with HOXA11-AS on the CCND2 promoter activity. Renilla luciferase was considered as an internal reference. After a 48-h transfection, the cells were harvested and lysed using a luciferase assay kit (K801-200, BioVision, Milpitas, CA, USA). The luciferase reporter assay was performed using a dual luciferase reporter analysis system (Promega, Madison, WI, USA). The ratio of firefly luciferase activity to Renilla luciferase activity was used as the relative luciferase activity.
Based on data obtained from the University of California Santa Cruz (UCSC; http://genome.ucsc.edu/) and JASPAR (http://jaspar.genereg.net/), we were able to confirm that the FOXP2 protein was most likely to bind to two sites of CCND2 DNA. The recombinant luciferase reporter vector was co-transfected into HFWT cells with FOXP2 expression vector for dual luciferase reporter assay to verify the specific sites of FOXP2 protein binding to CCND2 DNA.
Chromatin immunoprecipitation (ChIP)
The proteins were cross-linked to the DNA by adding formaldehyde for 10 min, after which the DNA was sheared to an average fragment size of 300-500 bp by sonication. After centrifugation at 12000 g for 10 min at 4°C, the supernatant was collected into three separate tubes, one of which was used as the Input. The remaining two tubes were incubated with NC rabbit antibody to IgG (ab109489, 1:300, Abcam Inc., Cambridge, UK) or antibody to FOXP2 (Upstate Biotechnology, Lake Placid, NY, USA) at 4°C overnight. The protein and DNA complex was precipitated with protein A/G Magnetic Beads (88803, Thermo, Waltham, MA, USA) at 65°C overnight, with the DNA fragments precipitated and purified via the phenol/chloroform method. The primer was designed to amplify a binding site between FOXP2 and the promoter region of CCND2 (Forward: 5’-GGGTGACGGGAGGAAGGAGGTGA-3’, Reverse: 5’-GGGAGGGGGCGAGTGAGGGATTA-3’). NC primer set was designed to amplify the sequence away from the CCND2 DNA promoter region (Forward: 5’-CCAGCAATCATAAAACGAAA-3’, Reverse: 5’-GTAGGAATCAGATGCAGAAA-3’). The purified DNA fragment was employed as a template for RT-qPCR using a site1 primer and Distal primer (control) to verify whether FOXP2 could bind to the site 1 of CCND2 DNA.
Similarly, the same procedure was conducted in order to identify the point at which HOXA11-AS was knocked down.
Cell counting Kit-8 (CCK-8)
Cell proliferation was measured using a CCK-8 kit (Dojindo Laboratories, Kumamoto, Japan). Briefly, HFWT cells were seeded into 96-well plates. After 48-h transfection with indicated siRNA or plasmids, the CCK-8 reagent was added to each well and further incubated for 1 h. The optical density (OD) value at a wavelength of 450 nm was detected using a microplate reader.
Flow cytometry
After 48-h transfection of indicated siRNA or plasmids, cells were harvested for cell cycle analysis. The cells were fixed with 70% cold absolute ethanol at 4°C for 2 h and subsequently stored at -20°C. The fixed cells were washed and then incubated with propidium iodide (PI, Sigma-Aldrich, St Louis, MO, USA) in the presence of RNase A for 30 min at room temperature. The cell cycle distribution was immediately analyzed by flow cytometry using Cell Quest software (FACSCalibur, BD Biosciences, Franklin Lakes, NJ, USA). Apoptosis was measured using the FITC-Annexin V apoptosis detection kit (BD Biosciences, Franklin Lakes, NJ, USA).
Tumor formation in nude mice
Twenty BALB/C nude mice, aged 4-6 weeks, weighing 19-22 g, were randomly divided into 2 groups, with 10 mice placed in each group. HFWT cells transfected with sh-HOXA11-AS or sh-NC were injected subcutaneously into the thigh of each nude mouse. The tumor volume was examined weekly and calculated in accordance with the following formula: length × width2/2. The growth curve was recorded based on the volume of the transplanted tumor. After six-week injections, the nude mice were euthanized, after which their respective tumors were collected and weighed.
Statistical analysis
All data analysis was performed using SPSS 22.0 (IBM Corp. Armonk, NY, USA) statistical software. Quantitative data was expressed as mean ± standard deviation. Paired t test was applied to compare the paired data of the paired experiments with normal distributions, while an unpaired t test was employed to compare the two sets of data in a non-paired design following normal distributions. Comparisons among multiple groups were analyzed using one-way analysis of variance (ANOVA) with Tukey’s post hoc tests. Comparisons among groups at different time points were measured using repeated measures ANOVA with Bonferroni post hoc tests. Pearson correlation analysis was performed to evaluate the relationship between HOXA11-AS and CCND2 expression in nephroblastoma tissues. Kaplan-Meier estimator was applied to calculate the overall survival curve of patients with nephroblastoma, while a log-rank was used to analyze the survival difference among patients. P < 0.05 was considered to be indicative of statistical significance.
Results
Highly expressed HOXA11-AS is associated with malignancy of nephroblastoma
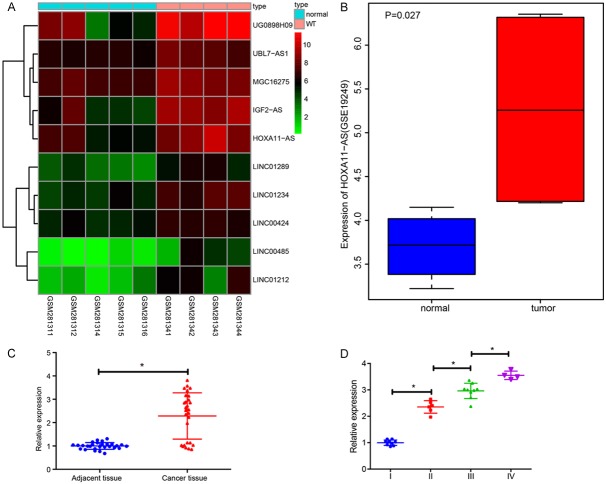
In order to elucidate the correlation between HOXA11-AS expression and nephroblastoma, differentially expressed lncRNAs were analyzed based on the datasets GSE11151 and GSE19249. A heatmap of the top 10 upregulated lncRNAs in dataset GSE11151 is depicted in Figure 1A. Moreover, HOXA11-AS was upregulated in nephroblastoma tumor tissues compared with normal tissues in the dataset GSE19249 (Figure 1B), suggesting that HOXA11-AS was more likely to be associated with nephroblastoma.
Figure 1.
HOXA11-AS is highly expressed in nephroblastoma and may be involved in the pathogenesis of nephroblastoma. A. The heatmap of top 10 up-regulated differentially expressed lncRNAs in dataset GSE11151; the X axis indicating the sample number and the Y axis indicating the differentially expressed lncRNA; the upper right histogram represents the color scale, and each rectangle in the figure corresponds to the sample expression value; B. The expression of HOXA11-AS in gene expression dataset GSE19249; C. RT-qPCR was used to detect HOXA11-AS expression in nephroblastoma tissues and corresponding adjacent normal tissues obtained from patients, n = 28, *P < 0.05 vs. the adjacent normal tissues. D. RT-qPCR was used to detect the expression of HOXA11-AS in different grades of nephroblastoma tumor tissues (9 for grade I, 6 for grade II, 9 for grade III, and 4 for grade IV), *P < 0.05 vs. grade I tissues. The values in the figures were quantitative data, and expressed as mean ± standard deviation. Paired t test was used to compare the paired data of the paired design with normal distribution and variance, and unpaired t test was used to compare the two sets of data in a non-paired design that obeyed the normal distribution and the variance. One-way ANOVA was used for the data comparison between multiple groups with Tukey’s post hoc test.
Next, to further validate the bioinformatics results, we examined the expression of HOXA11-AS in 28 patients with nephroblastoma. HOXA11-AS expression was detected in the nephroblastoma tissues obtained from the patient and the adjacent normal tissues by RT-qPCR (Figure 1C). HOXA11-AS expression was upregulated in nephroblastoma tumor tissues compared with the adjacent normal tissues (P < 0.05). The expression of HOXA11-AS was higher in the advanced stage of nephroblastoma tumor tissues compared with early stage of tumor tissues (P < 0.05; Figure 1D). Taken together, highly expressed HOXA11-AS was associated with high-grade and unfavorable histology in nephroblastoma.
HOXA11-AS inhibits apoptosis and arrests cell cycle progression of nephroblastoma in vitro
In order to further analyze the effect of HOXA11-AS on nephroblastoma tumor cells, we initially detected the expression of HOXA11-AS in the normal embryonic renal cells (HEK-293A) and nephroblastoma cell lines HFWT and SK-NEP-1. The results obtained revealed that HOXA11-AS was significantly higher in nephroblastoma cells than that of HEK-293A cells (Figure 2A). Hence, we selected HFWT cells with higher HOXA11-AS expression and SK-NEP-1 cells with lower HOXA11-AS expression for subsequent experiments. Next, we designed three sh-HOXA11-AS plasmids, which were respectively transfected into the HFWT cells for silencing HOXA11-AS. The results obtained indicated that sh-HOXA11-AS-3 was the most efficient plasmid for inhibiting the expression of HOXA11-AS (Figure 2B). Hence, sh-HOXA11-AS-3 was employed for subsequent experiments. Next, the HFWT cells were transfected with oe-NC + sh-NC, oe-HOXA11-AS + sh-NC, and oe-NC + sh-HOXA11-AS plasmids. The cell proliferation of HFWT in each transfected group was detected by CCK-8 (Figure 2C), which revealed that the overexpression of HOXA11-AS enhanced cell viability, while the silencing of HOXA11-AS was found to attenuate cell viability (P < 0.05). The apoptosis and cell cycle distribution of the HFWT cells in each group were detected by flow cytometry (Figure 2D, 2E), the results of which indicated that the overexpression of HOXA11-AS inhibited apoptosis and G1/S cell cycle arrest, while silencing of HOXA11-AS significantly promoted apoptosis and G1/S cell cycle arrest (P < 0.05). The expression of Ki67 and cleaved-caspase-3 in HFWT cells was detected by RT-qPCR and Western blot analysis (Figure 2F). Overexpressed HOXA11-AS was found to promote the expression of Ki67 and inhibit the expression of cleaved-caspase-3. Conversely, HOXA11-AS silencing was found to repress the expression of Ki67 while inducing the expression of cleaved-caspase-3 (P < 0.05). In conclusion, HOXA11-AS can inhibit apoptosis and cycle arrest of nephroblastoma cells in vitro.
Figure 2.
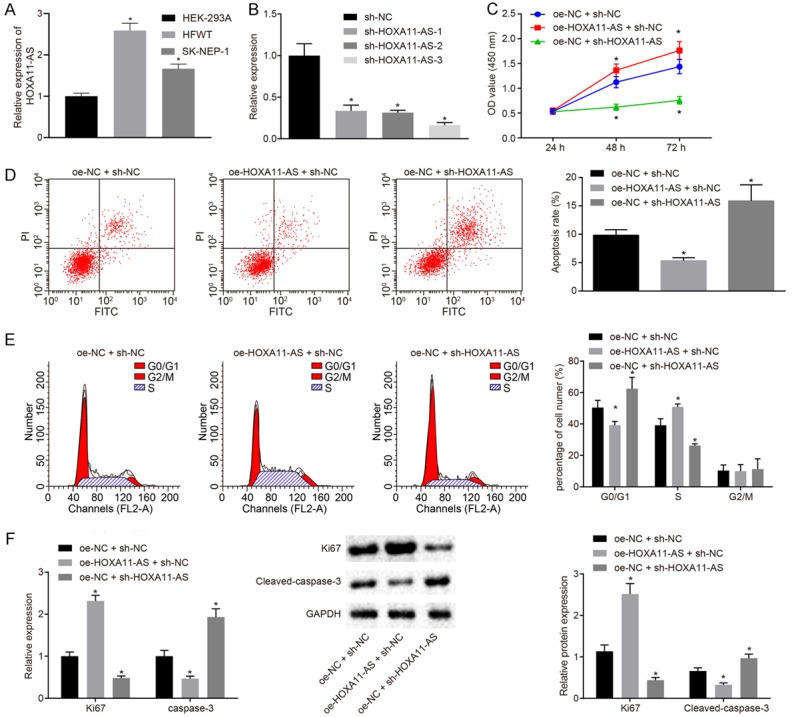
HOXA11-AS inhibits apoptosis and promotes cell cycle progression of nephroblastoma in vitro. A. RT-qPCR was used to detect the HOXA11-AS expression in HEK-293A, HFWT and SK-NEP-1 cells; B. RT-qPCR was used to detect the interference efficiency of three sh-HOXA11-AS plasmids; C. CCK-8 was used to detect the viability of HFWT cells; D. Flow cytometry was used to detect the apoptosis of HFWT cells in each group; E. Flow cytometry was used to detect the cell cycle distribution of HFWT cells in each group; F. RT-qPCR and Western blot analysis were used to detect the expression of Ki67 and cleaved-caspase-3 in HFWT cells. The values in the figures were quantitative data, and expressed as mean ± standard deviation. One-way ANOVA was used for the data comparison among multiple groups, with Tukey’s post hoc test. Repeated measures ANOVA was used to the data comparison between groups at different time points and Bonferroni was used for post hoc test. *P < 0.05 vs. the HFWT treated with oe-NC + sh-NC. The experiment was repeated for three times.
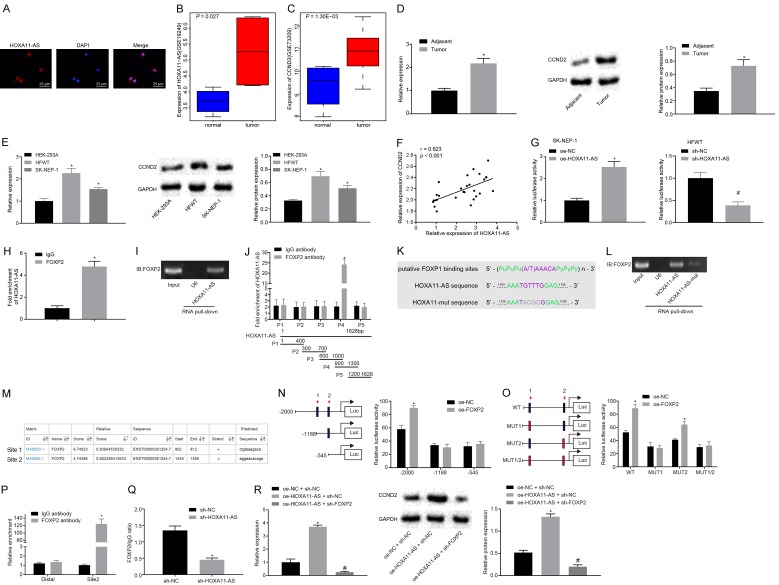
HOXA11-AS upregulates the expression of CCND2 by recruiting the transcription factor FOXP2
The micro-location of HOXA11-AS in HFWT cells was detected by FISH (Figure 3A). The results showed of which that revealed that HOXA11-AS was mainly predominately distributed in the nucleus of HFWT cells, suggesting that HOXA11-AS could potentially play a role in the regulation of gene expression in the nucleus. It was predicted in the LncMAP database (http://bio-bigdata.hrbmu.edu.cn/LncMAP/) that HOXA11-AS may regulate CCND2 by the transcription factor FOXP2. The expression of CCND2 in tumor tissues was drastically higher compared with normal tissues in nephroblastoma gene expression dataset GSE11151 (Figure 3B) and GSE73209 (Figure 3C). RT-qPCR and Western blot analysis were performed in order to detect the expression of CCND2 in nephroblastoma tumor tissues as well as the HEK-293A, HFWT and SK-NEP-1 cells (Figure 3D, 3E). The results showed that CCND2 was highly expressed in the nephroblastoma tumor tissues and HFWT and SK-NEP-1 cells compared with their control counterparts respectively (P < 0.05). In addition to the in vitro study, the potential relationship between the data and our clinical samples was subsequently investigated. The expression of HOXA11-AS and CCND2 among the 28 patients with nephroblastoma was analyzed (Figure 3F). The results revealed that the expression of HOXA11-AS was positively correlated with CCND2 among the clinical nephroblastoma cases (P < 0.05).
Figure 3.
HOXA11-AS up-regulates the expression of CCND2 by recruiting the transcription factor FOXP2 in nephroblastoma cells. A. The micro-location of HOXA11-AS in HFWT cells detected by RNA-FISH (× 400); B and C. The expression of CCND2 in gene expression datasets GSE11151 and GSE73209; D. The expression of CCND2 in nephroblastoma tissues detected by RT-qPCR and Western blot analysis, *P < 0.05 vs. adjacent normal tissues; E. The expression of CCND2 in HFWT cells detected by RT-qPCR and Western blot analysis, *P < 0.05 vs. HEK-293A cells; F. Pearson correlation analysis of HOXA11-AS and CCND2 expression in 28 nephroblastoma tissues and adjacent tissues; G. The effect of HOXA11-AS on CCND2 promoter activity examined by a dual luciferase reporter gene assay, *P < 0.05 vs. cells treated with oe-NC, #P < 0.05 vs. cells treated with sh-NC; H. HOXA11-AS binding to transcription factor FOXP2 confirmed by RIP, *P < 0.05 vs. IgG control; I. Transcription factor FOXP2 combining with HOXA11-AS verified by RNA pull-down; J. The specific binding of FOXP2 protein to HOXA11-AS detected by formaldehyde cross-linking RIP; K. Predicted specific binding sites of FOXP2 and HOXA11-AS; L. The binding of transcription factor FOXP2 to mut-HOXA11-AS in HFWT cells detected by RNA pull-down; M. Website analysis showed that the transcription factor FOXP2 was most likely to bind to two sites of the CCND2 DNA promoter region; N. The truncated CCND2 recombinant luciferase reporter gene vector and FOXP2 expression vector were co-transfected into SK-NEP-1 cells for a dual luciferase reporter gene assay, *P < 0.05 vs. cells treated with oe-NC; O. The constructed CCND2 recombinant luciferase reporter gene vector and FOXP2 expression vector were co-transfected into SK-NEP-1 cells for a dual luciferase reporter gene assay, *P < 0.05 vs. cells treated with oe-NC. P. The binding ability of FOXP2 at binding site 2 of CCND2 promoter region detected by ChIP, *P < 0.05 vs. IgG control; Q. The enrichment of CCND2 by FOXP2 after silencing HOXA11-AS in HFWT cells detected by ChIP, *P < 0.05 vs. cells treated with sh-NC; R. The expression of CCND2 in transfected SK-NEP-1 cells detected by RT-qPCR and Western blot analysis, *P < 0.05 cells treated with oe-NC + sh-NC, #P < 0.05 vs. cells treated with HOXA11-AS + sh-NC. The values in the figures were quantitative data, and expressed as mean ± standard deviation. Unpaired t test was used to compare the two sets of data in a non-paired design that obeyed the normal distribution and the variance. Data comparisons among groups were measured using one-way ANOVA with Tukey’s post hoc test used. Data comparison among groups at different time points was measured by repeated measures ANOVA, and Bonferroni was used for post hoc test. The experiment was repeated for three times.
Dual luciferase reporter gene assay was performed to verify the interaction of HOXA11-AS with CCND2 promoter region (Figure 3G). The results demonstrated the cells treated with oe-HOXA11-AS induced higher activity in CCND2 promoter than the cells treated oe-NC (P < 0.05). The activity of CCND2 promoter in the cells treated with sh-HOXA11-AS was lower than that in the cells treated with sh-NC (P < 0.05), indicating that HOXA11-AS positively regulated the expression of CCND2. In order to further investigate the mechanism by which HOXA11-AS positively regulates CCND2 expression, RIP was performed to understand whether HOXA11-AS bound to the transcription factor FOXP2 (Figure 3H). The results showed that enrichment of HOXA11-AS was increased by FOXP2 compared with the IgG control (P < 0.05), indicating that HOXA11-AS could specifically bind to FOXP2 protein. Meanwhile, RNA pull-down assay elucidated a similar tendency of the binding relation between FOXP2 and HOXA11-AS (Figure 3I).
In order to identify the specific binding sites of FOXP2 with HOXA11-AS, we designed 5 pairs of primers to detect various fragments of HOXA11-AS based on the formaldehyde cross-linking RIP assay. Our data indicated that the specific binding sites of HOXA11-AS interacting with FOXP2 located at 900-1300 nt (Figure 3J). Additionally, a bioinformatics tool namely JASPAR (http://jaspar.genereg.net/) was used to predict the binding site between these two genes and 1127 nt-1132 nt region of HOXA11-AS was predicted to interact with FOXP2 (Figure 3K). We then mutated the site on HOXA11-AS 1127 nt-1132 nt and performed an RNA pull-down assay in the HFWT cells to determine the binding ability of mut-HOXA11-AS to FOXP2 (Figure 3L). The results showed that the transcription factor FOXP2 could not bind to the mutated HOXA11-AS, indicating that the 1127 nt-1132 nt site (UGUUUG) of HOXA11-AS was the specific site for the interaction with FOXP2.
Furthermore, we predicted the two most possible specific binding sites in the promoter region of CCND2 with FOXP2 with two databases UCSC (http://genome.ucsc.edu/) and JASPAR (http://jaspar.genereg.net/; Figure 3M). In order to confirm this observation, a dual luciferase reporter assay was performed (Figure 3N, 3O), which showed that FOXP2 specifically associated with the binding site 1 in CCND2 promoter region. Next, the binding ability of the binding site 2 in CCND2 promoter region with FOXP2 was examined in the HFWT cells using ChIP (Figure 3P). The data illustrated that an elevated amount of amplification products was obtained using primers specific for binding site 2 between cells treated with FOXP2 antibodies relative to the IgG control (P < 0.05), while no significant difference was detected using the Distal primer between these two groups (P > 0.05), indicating that the site 1 of the CCND2 promoter region (CTGTAAAGACA) was indeed the binding site of the transcription factor FOXP2. Next, to verify the critical role of HOXA11-AS in the regulation of transcription factors FOXP2 and CCND2 DNA, we silenced HOXA11-AS in the HFWT cells (Figure 3Q). Following enrichment by applying FOXP2 antibody in the cell lysates, the cells transfected with sh-HOXA11-AS exhibited diminished amplification products when compared with the cells treated with sh-NC based on the data in RT-qPCR using CCND2 site2 primer (P < 0.05).
Next, in order to further understand whether HOXA11-Az regulated the expression of the CCND2 by recruiting the binding transcription factor FOXP2, the cells were treated with oe-NC + sh-NC, oe-HOXA11-AS + sh-NC, and oe-HOXA11-AS + sh-FOXP2. The expression of CCND2 in each group was detected by RT-qPCR and Western blot analysis (Figure 3R). The results showed that the expression of CCND2 was increased in cells treated with oe-HOXA11-AS + sh-NC compared with the cells treated with oe-NC + sh-NC (P < 0.05), while a decreased trend was observed in cells treated with oe-HOXA11-AS + sh-FOXP2 compared with cells treated with oe-HOXA11-AS + sh-NC (P < 0.05). These results indicate that HOXA11-AS recruits the transcription factor FOXP2 to upregulate CCND2 expression.
HOXA11-AS upregulates the expression of CCND2 to inhibit the apoptosis and affect cell cycle distribution in nephroblastoma by recruiting the transcription factor FOXP2
Next, we set out to explore the effects of the HOXA11-AS/FOXP2/CCND2 axis on the biological characteristics of nephroblastoma. The cells were then treated with oe-NC + sh-NC, oe-HOXA11-AS + sh-NC, oe-HOXA11-AS + oe-FOXP2 + sh-NC, and oe-HOXA11-AS + oe-FOXP2 + sh-CCND2 plasmids. The viability of the HFWT cells in each group was detected by CCK-8 (Figure 4A). The results showed that the cell viability was higher in those treated with oe-HOXA11-AS + sh-NC and oe-HOXA11-AS + oe-FOXP2 + sh-NC than that of the cells treated with oe-NC + sh-NC. In contrast, the cell viability of the cells treated with oe-HOXA11-AS + oe-FOXP2 + sh-CCND2 was lower than that of the cells treated with oe-HOXA11-AS + oe-FOXP2 + sh-NC.
Figure 4.
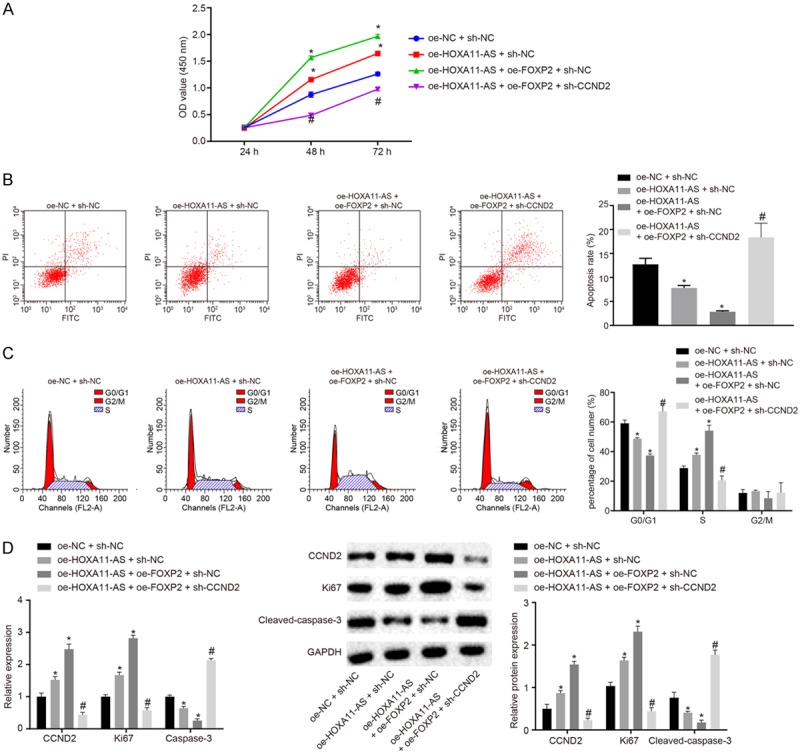
The cancer oncogene effect of CCND2 was mediated by HOXA11-AS in nephroblastoma cells. A. The viability of HFWT cells in each group detected by CCK-8; B. The apoptosis of HFWT cells in each group detected by flow cytometry; C. The cell cycle distribution of HFWT cells in each group detected by flow cytometry; D. The expressions of Ki67, CCND2 and cleaved-caspase-3 in HFWT cells detected by RT-qPCR and Western blot analysis. The values in the figures were quantitative data, and expressed as mean ± standard deviation. Unpaired t test was used for the comparison between groups. *P < 0.05 vs. cells treated with oe-NC + sh-NC, #P < 0.05 vs. cells treated with oe-HOXA11-AS + oe-FOXP2 + sh-NC. One-way ANOVA was used for the data comparison among groups with Tukey’s post hoc test. Repeated measures ANOVA was used to data comparison among groups at different time points, with Bonferroni post hoc test. The experiment was repeated for three times.
The apoptosis and cell cycle distribution of HFWT cells in each group were detected by flow cytometry methods (Figure 4B, 4C). The results showed that the apoptosis rate of cells treated with oe-HOXA11-AS + sh-NC and oe-HOXA11-AS + oe-FOXP2 + sh-NC was lower than cells treated with oe-NC + sh-NC. Likewise, the percentage of cells in G0/G1 phase decreased and the percentage of S phase increased. On the other hand, the apoptosis rate of cells treated with oe-HOXA11-AS + oe-FOXP2 + sh-CCND2 was higher than that of the cells treated with oe-HOXA11-AS + oe-FOXP2 + sh-NC, with an increased percentage of cells at the G0/G1 phase and a diminished percentage of cells at the S phase.
The expression of Ki67 and cleaved-caspase-3 in HFWT cells was detected by RT-qPCR and Western blot analysis (Figure 4D). The results revealed that the expression of Ki67 and CCND2 was increased and the expression of cleaved-caspase-3 was decreased in cells treated with oe-HOXA11-AS + sh-NC and oe-HOXA11-AS + oe-FOXP2 + sh-NC compared with cells treated with oe-NC + sh-NC. Reversely, the expression of Ki67 and CCND2 in cells treated with oe-HOXA11-AS + oe-FOXP2 + sh-CCND2 was lower compared with cells treated with oe-HOXA11-AS + oe-FOXP2 + sh-NC, and the expression of cleaved-caspase-3 was increased. These results support the hypothesis that HOXA11-AS inhibits apoptosis and affects cell cycle distribution via its interaction with CCND2 through FOXP2 in nephroblastoma cells.
Silencing of HOXA11-AS inhibits the development of nephroblastoma in vivo
In order to further investigate the effect of HOXA11-AS on the development of nephroblastoma in vivo, the cells were treated with sh-NC and sh-HOXA11-AS plasmids, and stably transfected cell lines were constructed and injected into nude mice for tumor formation. The visualized images of the obtained transplanted tumors are depicted in Figure 5A. The measurements of the transplanted tumors, such as size and weight, are illustrated in Figure 5B, 5C, respectively. Compared with the mice treated with sh-NC, the mean volume of xenografts and the weight in mice treated with sh-HOXA11-AS were reduced (P < 0.05). TUNEL was applied to detect the apoptosis in each group (Figure 5D). The results revealed that the ratio of apoptosis in mice treated with sh-HOXA11-AS was higher than mice treated with sh-NC. RT-qPCR and Western blot analysis methods were performed to detect the expression of CCND2, Ki67 and cleaved-caspase-3 in tumor tissues (Figure 5E). The result showed that the expression of CCND2 and Ki67 was notably decreased with the expression of cleaved-caspase-3 increased in the cells treated with sh-HOXA11-AS when compared to the cells treated with sh-NC (P < 0.05). Taken together, the above-mentioned key findings revealed that silencing of HOXA11-AS could inhibit the development of nephroblastoma in vivo.
Figure 5.
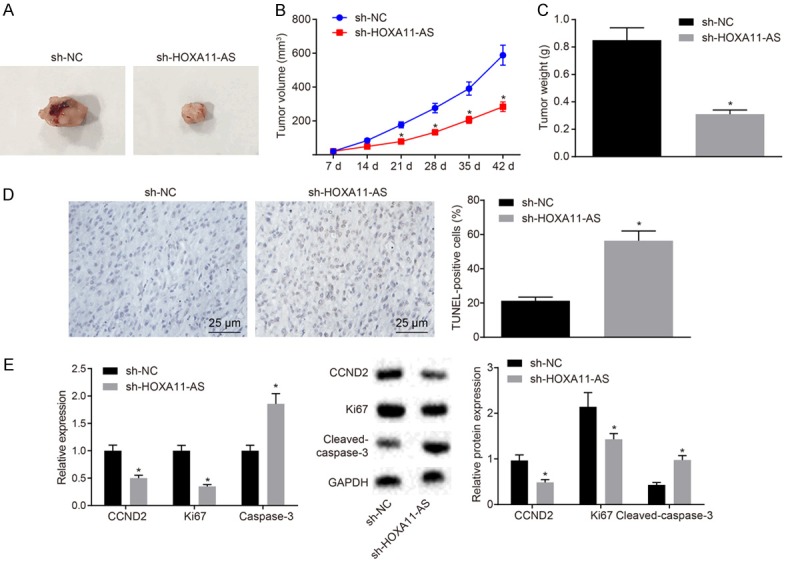
HOXA11-AS silencing suppresses progression of nephroblastoma tumor. A. Representative photographs of tumor images; B. Growth curve of transplanted tumors in each group; C. Tumor weight of transplanted tumors in each group; D. The apoptosis of cells in transplanted tumor sections detected by TUNEL; E. The expression of CCND2, Ki67 and cleaved-caspase-3 in the transplanted tumor tissues detected by RT-qPCR and Western blot analysis. The values in the figures were quantitative data, n = 10. *P < 0.05 vs. oe-NC + sh-NC. Quantitative data were expressed as means ± standard deviation, and verified by unpaired t test. Repeated measures ANOVA was used to data comparison among groups at different time points, and Bonferroni was used for post hoc test.
Discussion
As one of the most commonly diagnosed urinary system malignancy, nephroblastoma is widely considered to occur during childhood [4]. Existing clinical literature has highlighted a high incidence of extra renal nephroblastoma; hence, timely treatment of this disease is of urgent need [2]. Recently, lncRNAs have been identified as participants in the development of nephroblastoma [5]. Hence, during the current study we set out to explore the specific mechanism by which lncRNA HOXA11-AS influences the development of nephroblastoma. Our central findings demonstrated that silencing of HOXA11-AS could promote apoptosis and arrest the cell cycle at the G1/S phase.
Initially, we demonstrated that the expression of HOXA11-AS and CCND2 was upregulated in nephroblastoma. CCND2 was identified as a target gene of HOXA11-AS. The involvement of lncRNAs has been reported in multiple human diseases, particularly tumors [18]. Interestingly, a previous study has proved that CCND2 was highly expressed in nephroblastoma [11]. Another study also further concluded that HOXA11 is overexpressed in nephroblastoma [19]. Buschges et al. demonstrated the ability of CCND2 to promote the development of cell cycle from G1 to S phase [20]. In another case, PDIA3P was reported to bind to miR-185-5p and regulate the development of oral squamous cell carcinoma by targeting CCND2 [21]. LINC00598, a lncRNA that has been implicated in the regulation of numerous cellular functions, capable of modulating CCND2 transcription and the G1 checkpoint [12]. In line with our study, a previous study verifies that nephroblastoma patients with larger tumor size, advanced National Wilms Tumor Study stage or unfavorable histopathological type exhibited higher SOX21-AS1 expression [8]. SOX2OT has been shown to promote the carcinogenesis of nephroblastoma by regulating the axis of miR-363/FOXP4 axis [15]. In conclusion, many lncRNAs have been shown to regulate CCND2 expression and we also found that HOXA11-AS could bind to CCND2.
In addition, we also found that silencing of HOXA11-AS triggered a decline in cell proliferation and elevations in cell apoptosis, as evidenced by downregulated Ki67 and upregulated cleaved-caspase-3 both in vitro and in vivo. Ki-67 is a tumor proliferation marker acting as an essential prognostic factor in multiple cancers, and it has been demonstrated that a higher level of blastemal Ki-67 proliferation index can be found in nephroblastoma with metastases, indicating that Ki-67 can be an unfavorable indicator for tumor treatment [22]. Cleaved caspase-3 is biological marker related to apoptosis, and caspase-3 is cleaved at two sites and divided into three fragments; the prodomain, the large subunit, and the small subunit [23]. Similarly, the downregulation of HOXA11-AS has been reported to promote an increase in cell apoptosis and caspase-3 activities in oral squamous cell carcinoma cells resistant to cisplatin [24]. In colorectal cancer cells, HOXA11-AS overexpression induces propofol-suppressed cell proliferation and inhibits caspase-3 activity as well as cell apoptosis induced by propofol [25].
Depleted HOXA11-AS has been shown to stimulate apoptosis and inhibit cell cycle entry in nephroblastoma by recruiting FOXP2 to downregulate CCND2. FOXP2 is a transcription factor associated with cognitive function in humans, and FOXP2 gene heterozygous mutations induce a monogenic speech and language disorder, which can affect the development of embryonic brain [26]. A previous report proved that SNHG1 promotes glioma cell malignant biological behaviors through the positive feedback loop characterized by the association among miR-154-5p, miR-376b-3p, FOXP2 and KDM5B [15]. Our study indicated that there was a transcriptional regulation relationship between CCND2 and FOXP2. FOXP2 has been demonstrated to regulate the cell growth in osteosarcoma [27]. CCND2, a cell cycle factor, has been proved to promote the cell cycle progression [28]. LOXL1-AS1 has been demonstrated to modulate cell proliferation and cell cycle progression in prostate cancer by targeting miR-541-3p and CCND1 [29]. A previous study concluded that up-regulated FOXP3 in lung adenocarcinoma helps to maintain malignant performance by up-regulating the expression of G1/S transition gene CCND1 [30].
In summary, our findings demonstrated the role of HOXA11-AS in nephroblastoma. More specifically, the depletion of HOXA11-AS can promote apoptosis and inhibit cell cycle distribution in nephroblastoma by recruiting the transcription factor FOXP2 to down-regulate the expression of CCND2 (Figure 6). The aforementioned findings present a novel therapeutic target for the treatment of nephroblastoma. Future investigations are required as we are still at the early stages of evaluating the specific role of HOXA11-AS in nephroblastoma. Further studies are expected to validate its applicable value from a clinical practice perspective.
Figure 6.
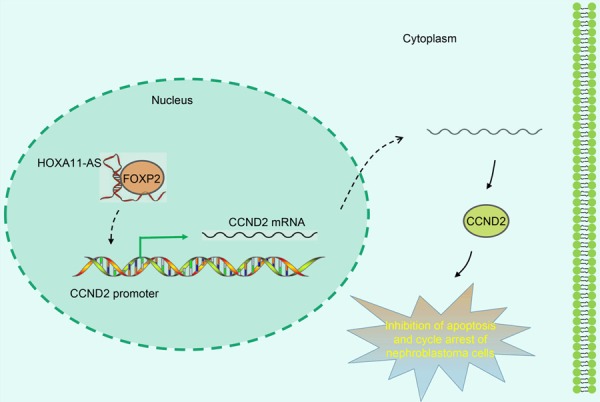
HOXA11-AS positively regulate Cyclin D2 to inhibit apoptosis and promote cell cycle progression in nephroblastoma by recruiting FOXP2.
Acknowledgements
We would like to give our sincere appreciation to the reviewers for their helpful comments on this article. This work was supported by grants from the Science and Technology Planning Project of Guangdong Province (No: 2016A020215009), and Guangzhou Institute of Pediatrics/Guangzhou Women and Children’s Medical Center (No: pre-NSFC-2018-016; YIP-2018-021).
Disclosure of conflict of interest
None.
References
- 1.Li Y, Lei C, Xiang B, Li F, Wang C, Wang Q, Chen S, Ji Y. Extrarenal teratoma with nephroblastoma in the retroperitoneum: case report and literature review. Medicine (Baltimore) 2017;96:e8670. doi: 10.1097/MD.0000000000008670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sindhu II, Saeed H, Wali R, Mehreen A. Primary extra-renal wilms’ tumor in urinary bladder: rare presentation of a common pediatric malignancy. J Coll Physicians Surg Pak. 2019;29:S31–S33. doi: 10.29271/jcpsp.2019.06.S31. [DOI] [PubMed] [Google Scholar]
- 3.Sinha A, Phukan JP, Bandyopadhyay G, Mukherjee S. Teratoid Wilms’ tumor in a child: a report of a rare case. Int J Appl Basic Med Res. 2013;3:72–74. doi: 10.4103/2229-516X.112248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu S, Sun N, Zhang WP, Song HC, Huang CR. Management of Wilms tumor with intravenous thrombus in children: a single center experience. World J Pediatr. 2019;15:476–482. doi: 10.1007/s12519-019-00272-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu S, Fu W, Zhang L, Fu K, Hu J, Jia W, Liu G. LINC00473 antagonizes the tumour suppressor miR-195 to mediate the pathogenesis of Wilms tumour via IKKα. Cell Prolif. 2018;51 doi: 10.1111/cpr.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65:1140–1151. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Hou T, Qi X, Wang J, Sun X. SOX21-AS1 is associated with clinical stage and regulates cell proliferation in nephroblastoma. Biosci Rep. 2019;39 doi: 10.1042/BSR20190602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue JY, Huang C, Wang W, Li HB, Sun M, Xie M. HOXA11-AS: a novel regulator in human cancer proliferation and metastasis. Onco Targets Ther. 2018;11:4387–4393. doi: 10.2147/OTT.S166961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pei Y, Singh RK, Shukla SK, Lang F, Zhang S, Robertson ES. Epstein-barr virus nuclear antigen 3C facilitates cell proliferation by regulating cyclin D2. J Virol. 2018;92 doi: 10.1128/JVI.00663-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faussillon M, Monnier L, Junien C, Jeanpierre C. Frequent overexpression of cyclin D2/cyclin-dependent kinase 4 in Wilms’ tumor. Cancer Lett. 2005;221:67–75. doi: 10.1016/j.canlet.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Jeong OS, Chae YC, Jung H, Park SC, Cho SJ, Kook H, Seo S. Long noncoding RNA linc00598 regulates CCND2 transcription and modulates the G1 checkpoint. Sci Rep. 2016;6:32172. doi: 10.1038/srep32172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 2009;10:233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haussermann K, Young G, Kukura P, Dietz H. Dissecting FOXP2 oligomerization and DNA binding. Angew Chem Int Ed Engl. 2019;58:7662–7667. doi: 10.1002/anie.201901734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Xue Y, Ma J, Shao L, Wang D, Zheng J, Liu X, Yang C, He Q, Ruan X, Li Z, Liu Y. SNHG1 promotes malignant biological behaviors of glioma cells via microRNA-154-5p/miR-376b-3p-FOXP2-KDM5B participating positive feedback loop. J Exp Clin Cancer Res. 2019;38:59. doi: 10.1186/s13046-019-1063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma L, Sun X, Kuai W, Hu J, Yuan Y, Feng W, Lu X. Retracted: long noncoding RNA SOX2OT accelerates the carcinogenesis of wilms’ tumor through ceRNA through miR-363/FOXP4 axis. DNA Cell Biol. 2018 doi: 10.1089/dna.2018.4420. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 17.Ayuk SM, Abrahamse H, Houreld NN. The role of photobiomodulation on gene expression of cell adhesion molecules in diabetic wounded fibroblasts in vitro. J Photochem Photobiol B. 2016;161:368–374. doi: 10.1016/j.jphotobiol.2016.05.027. [DOI] [PubMed] [Google Scholar]
- 18.Sheng SR, Wu JS, Tang YL, Liang XH. Long noncoding RNAs: emerging regulators of tumor angiogenesis. Future Oncol. 2017;13:1551–1562. doi: 10.2217/fon-2017-0149. [DOI] [PubMed] [Google Scholar]
- 19.Li CM, Guo M, Borczuk A, Powell CA, Wei M, Thaker HM, Friedman R, Klein U, Tycko B. Gene expression in Wilms’ tumor mimics the earliest committed stage in the metanephric mesenchymal-epithelial transition. Am J Pathol. 2002;160:2181–2190. doi: 10.1016/S0002-9440(10)61166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buschges R, Weber RG, Actor B, Lichter P, Collins VP, Reifenberger G. Amplification and expression of cyclin D genes (CCND1, CCND2 and CCND3) in human malignant gliomas. Brain Pathol. 1999;9:435–442. doi: 10.1111/j.1750-3639.1999.tb00532.x. discussion 432-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun CC, Zhang L, Li G, Li SJ, Chen ZL, Fu YF, Gong FY, Bai T, Zhang DY, Wu QM, Li DJ. The lncRNA PDIA3P interacts with miR-185-5p to modulate oral squamous cell carcinoma progression by targeting cyclin D2. Mol Ther Nucleic Acids. 2017;9:100–110. doi: 10.1016/j.omtn.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juric I, Pogorelic Z, Kuzmic-Prusac I, Biocic M, Jakovljevic G, Stepan J, Zupancic B, Culic S, Kruslin B. Expression and prognostic value of the Ki-67 in Wilms’ tumor: experience with 48 cases. Pediatr Surg Int. 2010;26:487–493. doi: 10.1007/s00383-010-2588-3. [DOI] [PubMed] [Google Scholar]
- 23.Crowley LC, Waterhouse NJ. Detecting cleaved caspase-3 in apoptotic cells by flow cytometry. Cold Spring Harb Protoc. 2016;2016 doi: 10.1101/pdb.prot087312. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Li H, Shi J. LncRNA HOXA11-AS promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by suppression of miR-214-3p expression. Biomed Res Int. 2019;2019:8645153. doi: 10.1155/2019/8645153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren YL, Zhang W. Propofol promotes apoptosis of colorectal cancer cells via alleviating the suppression of lncRNA HOXA11-AS on miRNA let-7i. Biochem Cell Biol. 2019 doi: 10.1139/bcb-2018-0235. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 26.Vernes SC, Oliver PL, Spiteri E, Lockstone HE, Puliyadi R, Taylor JM, Ho J, Mombereau C, Brewer A, Lowy E, Nicod J, Groszer M, Baban D, Sahgal N, Cazier JB, Ragoussis J, Davies KE, Geschwind DH, Fisher SE. Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genet. 2011;7:e1002145. doi: 10.1371/journal.pgen.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gascoyne DM, Spearman H, Lyne L, Puliyadi R, Perez-Alcantara M, Coulton L, Fisher SE, Croucher PI, Banham AH. The forkhead transcription factor FOXP2 is required for regulation of p21WAF1/CIP1 in 143B osteosarcoma cell growth arrest. PLoS One. 2015;10:e0128513. doi: 10.1371/journal.pone.0128513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li YL, Wang J, Zhang CY, Shen YQ, Wang HM, Ding L, Gu YC, Lou JT, Zhao XT, Ma ZL, Jin YX. MiR-146a-5p inhibits cell proliferation and cell cycle progression in NSCLC cell lines by targeting CCND1 and CCND2. Oncotarget. 2016;7:59287–59298. doi: 10.18632/oncotarget.11040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long B, Li N, Xu XX, Li XX, Xu XJ, Liu JY, Wu ZH. Long noncoding RNA LOXL1-AS1 regulates prostate cancer cell proliferation and cell cycle progression through miR-541-3p and CCND1. Biochem Biophys Res Commun. 2018;505:561–568. doi: 10.1016/j.bbrc.2018.09.160. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Li D, Yang W, Fu H, Liu Y, Li Y. Overexpression of the transcription factor FOXP3 in lung adenocarcinoma sustains malignant character by promoting G1/S transition gene CCND1. Tumour Biol. 2016;37:7395–7404. doi: 10.1007/s13277-015-4616-3. [DOI] [PubMed] [Google Scholar]