Abstract
PD-1/PD-L1 immune checkpoint blockade therapy has become an effective method for the treatment of cancers in the clinic. It has great clinical advantages and therapeutic effects in the treatment of various cancers. However, a considerable number of cancer patients currently have relatively low response rates and drug resistance to PD-1/PD-L1 immunotherapy. Therefore, an in-depth understanding of the regulatory mechanism of PD-L1 expression in tumor cells will provide new insights into PD-1/PD-L1 immunotherapy. This review will systematically review the regulatory mechanisms of PD-L1 including genomic amplification, epigenetic regulation, transcriptional regulation, translational regulation and posttranslational modification. We will also discuss PD-L1 expression regulation in clinical applications. Finally, we hope to provide new routes for PD-1/PD-L1 immunotherapy in the clinic.
Keywords: Immune checkpoint blockade therapy, PD-1/PD-L1, gene expression, regulatory mechanism
Introduction
In recent years, immunotherapy has become a new method of cancer treatment. Currently, immune checkpoint blockade therapy is one of the most widely used methods of tumor immunotherapy. The pathway involving programmed death protein 1 (PD-1) and its ligand (PD-L1) is a well-characterized immune checkpoint and has been applied in the clinical treatment of various cancers. Antibodies targeting the PD-1/PD-L1 pathway have been approved for various cancers, including melanoma, non-small cell lung cancer (NSCLC), Hodgkin’s lymphoma, bladder cancer, renal cell carcinoma (RCC), head and neck squamous cell carcinoma (HNSCC), breast cancer, Merkel cell carcinoma, hepatocellular carcinoma (HCC) and gastric cancer (GC) [3]. However, these antibodies are only efficacious in a small portion of patients with certain cancers.
At present, the understanding of the resistance mechanism of immune checkpoint blockade therapy and the regulation of PD-L1 expression is quite limited. To develop a more effective and lasting immune checkpoint blocking therapy strategy, it is necessary to gain insights into the multiple roles and complex regulatory mechanisms of PD-L1 in cancers. In this review, we will discuss the molecular mechanisms of PD-L1 expression in cancer cells at the levels of genomic amplification, epigenetic regulation, transcriptional regulation, posttranscriptional regulation, translational regulation, and posttranslational modification. These findings may provide new insights into targeting tumor immune escape after immunotherapy in the clinic.
Classification of PD-L1 expression in tumor cells
The expression of PD-L1 can be divided into constitutive expression and inducible expression depending on the extrinsic or intrinsic stimuli (Figure 1). Constitutive expression of PD-L1 in tumor cells is induced by dysregulation of oncogenic or tumor suppressor gene signaling pathways, by activation of abnormal transcription factors, or by genomic aberrations or gene amplifications. Many oncogenic transcription factors have been found to directly regulate PD-L1 expression.
Figure 1.
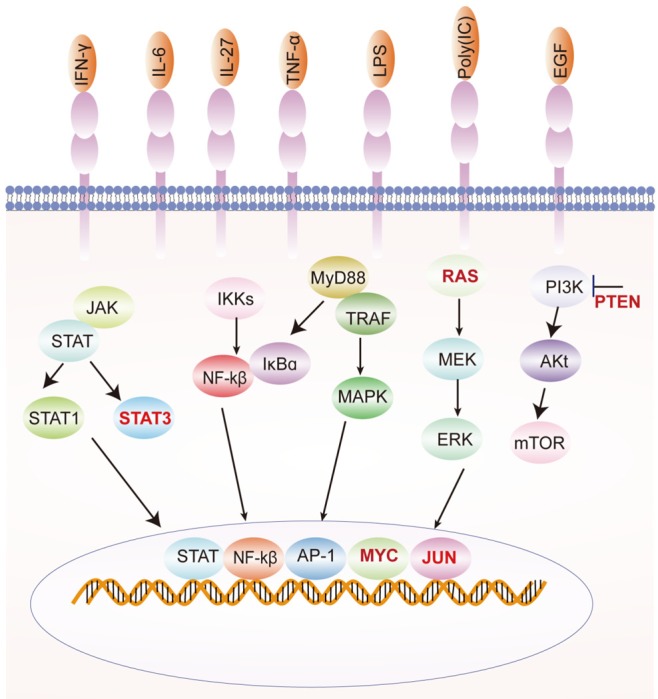
Classification of PD-L1 expression. PD-L1 expression can be divided into constitutive expression and inducible expression. Constitutive expression is induced by dysregulation of signal transduction components in tumor cells. Inducible expression is induced by a number of inflammatory cytokines.
The oncogenic transcription factor MYC is abnormally expressed in many cancer patients [1,2]. Inhibition of MYC gene expression in mouse or human tumor cells can reduce the expression of PD-L1 at both the gene and protein levels [3-6]. Further studies showed that MYC could bind to the promoter region of PD-L1 and regulate the expression of PD-L1 [3]. Approximately 41% of NSCLC patients show overexpression of MYC [7]. Immunostaining of NSCLC tissues revealed that MYC expression significantly correlated with PD-L1 expression in non-small cell lung cancer [8]. PD-L1 expression was up-regulated by a KRAS mutation and through p-ERK signaling in lung adenocarcinoma [9]. Other studies have shown that oncogenic RAS signaling can drive PD-L1 expression through the RAS-MEK signaling pathway [10]. STAT3 has also been found to act on the PD-L1 promoter to regulate PD-L1 expression [4,11] (Figure 1).
Inducible expression refers to the expression of PD-L1-controlled inflammatory signals from tumor cells or other immune cells, such as APCs and T cells, in the tumor microenvironment. A number of inflammatory cytokines have been found to induce the expression of PD-L1. These inflammatory factors include IFN-γ, TNF-α, IL-17, IL-27, IL-10, IL-4, IL-2 and IL-10 [12,13] (Table 1).
Table 1.
Classification of PD-L1 expression
| Type | Inducer | Type of cancers | Ref |
|---|---|---|---|
| Constitutive expression | MYC | NSCLC, lymphoma, HCC, melanoma | [3-5,8] |
| KRAS | NSCLC, lung cancer | [9,10,35,71] | |
| STAT3 | HNSC, lymphoma, melanoma | [4,11,72,73] | |
| JUN | Lymphoma, melanoma, medulloblastoma | [53,72,74] | |
| PTEN | Glioma, colorectal cancer, melanoma, breast cancer | [72,75-78] | |
| EGFR | Head and neck cancer, breast cancer, NSCLC | [10,61,79] | |
| MEK-ERK | Melanoma, lymphoma, multiple myeloma | [67,80,81] | |
| Inducible expression | IFN-γ | Pancreatic cancer, colon cancer, HCC, melanoma, lung cancer, gastric cancers | [82-86] |
| IL-6 | HCC, lung cancer, prostate cancer | [87-89] | |
| IL-27 | Lung cancer, epithelial ovarian cancer | [88,90] | |
| TNF-α | Breast cancer, HCC, prostate and colon cancer cells | [52,83,91] | |
| LPS | Gastric cancers | [92] | |
| EGF | NSCLC, breast cancer | [10,61,71,93] | |
| IL-8 | Gastric cancers, NSCLC, melanoma | [94,95] |
Regulation of PD-L1 expression by genomic amplification
PD-L1 and PD-L1 are located on chromosome 9p24.1. The amplification of the 9p24.1 region is closely related to an increase in PD-L1 levels in a wide range of cancers [14].
It has been found that copy number alterations (CNAs) of PD-L1 occur in various types of tumors, which lead directly to up-regulation of PD-L1 expression [15].
The highest frequency of CNAs of PD-L1 has been found in primary mediastinal B-cell lymphoma (PMBCL), classical Hodgkin lymphoma (cHL), and triple-negative breast cancer (TNBC), at 63% [16], 40% [17] and 29% [18], respectively. However, in GC, small cell lung cancers, NSCLCs and diffuse large B-cell lymphoma (DLBCL), the CNAs were much lower, with frequencies of 15% [19], 1.9% [20], 5.3% [21] and 3% [22], respectively. In general, the increase in CNAs is positively correlated with PD-L1 protein levels [23] (Figure 2).
Figure 2.
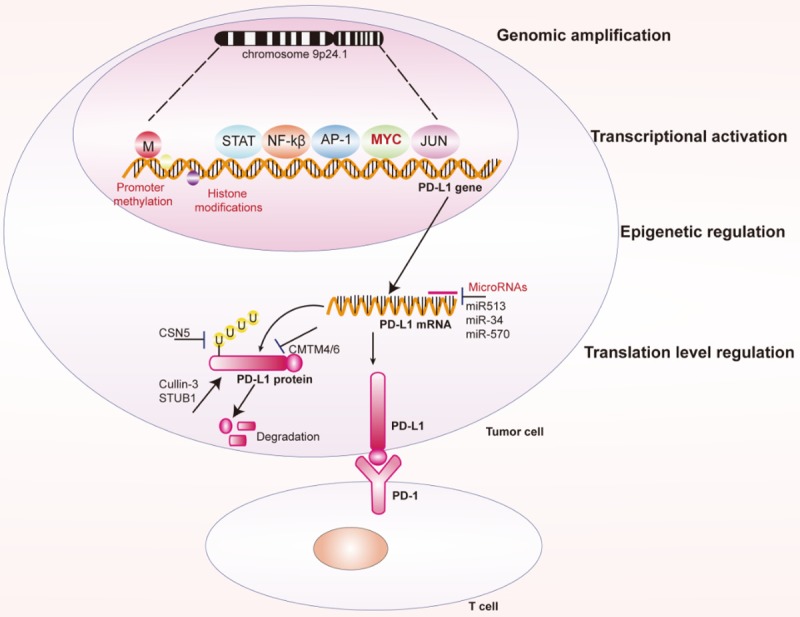
Regulation of PD-L1 expression in cancer cells at different levels. PD-L1 expression can be regulated by genomic amplification, transcriptional regulation, epigenetic regulation and transcriptional regulation.
Epigenetic regulation of PD-L1 expression
Epigenetic modifications, such as microRNAs (miRNAs), promoter DNA methylation and histone modifications, can regulate the recognition and binding of transcription factors to DNA elements without affecting DNA sequences, thereby altering chromatin structure and regulating PD-L1 expression [24] (Figure 2).
miRNAs are a class of non-coding single-stranded RNAs that contain 22-24 nucleotides. miRNAs inhibit translation or degradation of target mRNA by binding to the 3’untranslated region (3’UTR) of the target mRNA. A number of miRNAs have been found to regulate PD-L1 expression in different types of cancers [24]. They can regulate PD-L1 expression directly or indirectly.
Direct effectors regulate PD-L1 expression primarily by binding to PD-L1 mRNA. miRNAs that directly regulate PD-L1 expression include miR513 [25], miR-34 [26], miR-570 [27,28], miR-152 [29], miR-200 [30], miR-138 [31], miR-142-5p [32], miR-424 [33], miR-193a [34] and miR-140/142/340/383 [35]. Indirect effects mainly occur through affecting the expression of other PD-L1 regulators. miRNAs that indirectly regulate PD-L1 expression include miR-20b, miR-21, miR-130b [36], and miR-197 [37].
Recently, it was found that the promoter methylation of PD-L1 was negatively correlated with PD-L1 expression in a number of cancers [38-42]. PD-L1 promoter methylation has been found in many cancers, including acute myeloid leukemia [38], HNSCC [43-45], glioblastoma [41], glioma [42,43], colorectal cancer [40], and prostate cancer [46]. Analysis of PD-L1 promoter methylation has clinical significance for predicting the outcome of PD-1/PD-L1 immune checkpoint blockade therapies. In PD-1/PD-L1 targeted drug-treated patients, increased PD-L1 promoter methylation is associated with overall patient survival and recurrence-free survival [40].
In addition, histone modifications, including methylation, acetylation, phosphorylation, adenylation, ubiquitination, and ADP ribosylation, can also regulate PD-L1 gene expression [24]. The histone acetylation of the promoter region of the PD-L1 gene is essential for the expression of PD-L1 [24].
Transcriptional activation of PD-L1 expression
A number of transcription factors have been found to regulate PD-L1 transcriptional activation. These transcription factors include MYC, STAT3, NF-kβ, AP1, and HIF-1 (Figure 2).
The oncogene MYC is a transcription factor that is overexpressed and activated in a variety of tumors and involved in tumorigenesis [47]. However, there is controversy about the regulation of PD-L1 expression by MYC. Casey et al. found that inhibition of MYC in tumor cells resulted in a decrease in PD-L1 mRNA and protein expression. MYC can bind directly to the promoters of PD-L1 and enhance the anti-tumor immune response [3]. In contrast, Hogg and Durand-Panteix et al. reported that MYC transcriptional levels inhibited PD-L1 mRNA expression [48,49]. Future research is also needed to clarify these discrepancies.
STAT3 is another reported transcription factor that is involved in the regulation of PD-L1 expression. In chimeric nucleophosmin (NPM)/ALK-carrying T cell lymphoma, STAT3 upregulates PD-L1 expression by binding to the PD-L1 promoter. This effect can be suppressed by silencing STAT3 with siRNA [49]. It was also reported that latent membrane protein-1 (LMP1) of the Epstein-Barr virus can induce PD-L1 expression through inducing the phosphorylation of STAT3 [50].
NF-kβ is a nuclear transcription factor that also regulates PD-L1 expression. However, the mechanism of regulation is still unclear. In natural killer/T-cell lymphoma (NKTCL), inhibition of the NF-kβ signaling pathway reduces PD-L1 expression [51]. Recently, Lim et al. found that the inflammatory factor TNF-α activates the NF-kβ signaling pathway and activates COP9 signalosome 5 (CSN5) to inhibit ubiquitination and degradation of PD-L1 protein [52].
The transcription factor AP-1 is a dimeric complex composed of c-Jun, FOS, MAF, or ATF. Expression of PD-L1 in Hodgkin’s lymphoma is induced by AP1 via binding to the enhancer region of the first intron of the PD-L1 gene [53].
Hypoxia-inducible factor 1α (HIF-1α) is another important carcinogenic factor and has clinical significance in regulating the expression of PD-L1 in tumor cells [54]. Binding of HIF-1α to the PD-L1 proximal promoter stimulates transcription of PD-L1. Overexpression of HIF-1α induces an increase in PD-L1 levels [54,55].
Translation-level regulation of PD-L1
It has been found that ubiquitination, deubiquitination, glycosylation and phosphorylation can affect the stability of PD-L1 protein in cancer cells, thereby regulating the expression of PD-L1 protein (Figure 2).
Several proteins were reported to regulate the stability of the PD-L1 protein through ubiquitination. CSN5 is the fifth component of the CSN complex, which contains a conserved JAMM motif. CSN5 has deubiquitination activity through the JAMM motif and plays an important role during tumorigenesis. Lim et al. found that macrophages secrete TNF-α to activate NF-kβ and then induce transactivation of CSN5. Activation of CSN5 results in deubiquitination of PD-L1 in breast cancer cells and enhances the stability of PD-L1 [52]. Cyclin-dependent kinase 4/6 is a key regulator of the cell cycle. Cyclin D-CDK4 induces ubiquitination degradation of PD-L1 via cullin 3-SPOP to control therapeutic efficacy in human cancers [56]. CMTM6 was a recently identified type 3 transmembrane protein involved in regulating PD-L1 expression [57,58]. A genome-wide CRISPR-Cas9 screening technology revealed that CMTM6 inhibits ubiquitination and inhibits lysosomal-mediated degradation of PD-L1 by interacting with PD-L1 on the surface of tumor cells [57]. In addition to CMTM6, its closest family member, CMTM4, has similar functions [58]. Epidermal growth factor (EGF) treatment also induces ubiquitination of PD-L1 and regulates PD-L1 protein expression [59].
Glycosylation is an important posttranslational modification of proteins. N-linked glycosylation is a key protein modification that determines the structure and function of proteins and plays an important role in regulating membrane proteins. N-linked glycosylation of PD-L1 was shown to stabilize the PD-L1 protein and prevent degradation by the 26S proteasome [60,61]. In triple-negative breast cancer, β-1,3-N-acetylglucosaminyl transferase (B3GNT3) was required for the interaction between PD-L1 and PD-1 [60].
Clinical application of PD-L1 expression regulation
Due to tumor heterogeneity and genetic differences between individuals, there are significant defects in the therapeutic effects of targeting the PD-1/PD-L1 pathway alone. Recent studies have found that combining PD-L1/PD1 immunotherapy with targeted therapy significantly improves therapeutic effects by regulating PD-L1 at a very low level [62]. This strategy inhibits PD-L1 expression by regulating key proteins in the signaling pathway, and it combines with the immunotherapy of PD-L1 or PD-1 antibody to achieve a greater therapeutic effect.
In NSCLC, EGFR mutations can induce PD-L1 expression. The combination of osimertinib and durvalumab in the treatment of NSCLC patients with EGFR mutations showed significant efficacy and an overall response rate (ORR) of up to 70% [63-65]. Patients with advanced NSCLC treated with nivolumab in combination with erlotinib for EGFR mutations showed a durable clinical benefit [66]. The use of the KRAS/MEK inhibitor trametinib in combination with anti-PD-1 antibodies also significantly reduced PD-L1 expression and showed better therapeutic effects than individual treatments in NSCLC [67,68].
On the other hand, the expression of PD-L1 is also regulated by MAPK and PI3K/Akt signaling pathways, and inhibition of these pathways also reduces PD-L1 expression [69]. Inhibition of these signaling pathways can inhibit cell proliferation and regulate PD-L1 expression. Clinical studies have found that receptor tyrosine kinase inhibitors have a better therapeutic effect in lung cancers with high PD-L1 expression [70].
Conclusions and future challenges
Immunotherapies are a new direction in cancer therapy and have many advantages over traditional treatments. Currently, immunotherapy that targets the PD-1/PD-L1 axis has been clinically approved in many countries for the treatment of various human cancers. It has shown unprecedented efficacy in the treatment of a wide range of human cancers. However, only a small proportion of patients show an effect with PD-1/PD-L1 immune checkpoint blockade therapy. The expression of PD-L1 varies greatly in tumor tissues. At present, methods to detect PD-L1 expression in tumor tissues include immunostaining, Western blotting, qPCR and microarray. However, these methods for detecting the expression of PD-L1 vary greatly. An in-depth understanding of the regulatory mechanism of PD-L1 expression has been very helpful for PD-1/PD-L1 immunotherapy in the clinic. Although the regulatory mechanism of PD-L1 expression has been investigated to some extent, there are still many questions that need to be solved. For example, new mechanisms that regulate PD-L1 expression need to be investigated in future studies.
The expression of PD-L1 can be regulated at different levels; however, it is necessary to study which regulatory mechanism plays a critical role in certain types of cancer. A number of transcription factors that regulate the expression of PD-L1 regulate it by binding to the PD-L1 promoter, but the transcription factors that play key roles in certain types of cancer also need to be identified. In addition, the expression of PD-L1 varies greatly in different stages of tumor development, such as in primary cancer and metastatic cancer. In addition to antibody drugs, it is also necessary to develop a small molecule inhibitor of PD-L1 for treatment of cancer patients. Finally, these studies will provide new ideas for immunological checkpoint blocking therapy.
The understanding of the regulatory mechanism of PD-L1 expression will continue to deepen and will finally provide more choices and more effective methods for tumor immunotherapy of the PD-1/PD-L1 pathway.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81502621 and 81502088), the China Postdoctoral Science Special Foundation (2017M5654), medical clinical science and technology development fund of Jiangsu University (JLY20180033).
Disclosure of conflict of interest
None.
References
- 1.Hsieh AL, Walton ZE, Altman BJ, Stine ZE, Dang CV. MYC and metabolism on the path to cancer. Semin Cell Dev Biol. 2015;43:11–21. doi: 10.1016/j.semcdb.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, Gouw AM, Baylot V, Gütgemann I, Eilers M, Felsher DW. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atsaves V, Tsesmetzis N, Chioureas D, Kis L, Leventaki V, Drakos E, Panaretakis T, Grander D, Medeiros LJ, Young KH, Rassidakis GZ. PD-L1 is commonly expressed and transcriptionally regulated by STAT3 and MYC in ALK-negative anaplastic large-cell lymphoma. Leukemia. 2017;31:1633–1637. doi: 10.1038/leu.2017.103. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Jia Y, Zhao S, Zhang X, Wang X, Han X, Wang Y, Ma M, Shi J, Liu L. BIN1 reverses PD-L1-mediated immune escape by inactivating the c-MYC and EGFR/MAPK signaling pathways in non-small cell lung cancer. Oncogene. 2017;36:6235–6243. doi: 10.1038/onc.2017.217. [DOI] [PubMed] [Google Scholar]
- 6.Casey SC, Baylot V, Felsher DW. MYC: master regulator of immune privilege. Trends Immunol. 2017;38:298–305. doi: 10.1016/j.it.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lorenz J, Friedberg T, Paulus R, Oesch F, Ferlinz R. Oncogene overexpression in non-small-cell lung cancer tissue: prevalence and clinicopathological signifi cance. Clin Investig. 1994;72:156–163. doi: 10.1007/BF00184595. [DOI] [PubMed] [Google Scholar]
- 8.Kim EY, Kim A, Kim SK, Chang YS. MYC expression correlates with PD-L1 expression in non-small cell lung cancer. Lung Cancer. 2017;110:63–67. doi: 10.1016/j.lungcan.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Chen N, Fang W, Lin Z, Peng P, Wang J, Zhan J, Hong S, Huang J, Liu L, Sheng J, Zhou T, Chen Y, Zhang H, Zhang L. KRAS mutation-induced upregulation of PD-L1 mediates immune escape in human lung adenocarcinoma. Cancer Immunol Immunother. 2017;66:1175–1187. doi: 10.1007/s00262-017-2005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coelho MA, de Carné Trécesson S, Rana S, Zecchin D, Moore C, Molina-Arcas M, East P, Spencer-Dene B, Nye E, Barnouin K, Snijders AP, Lai WS, Blackshear PJ, Downward J. Oncogenic RAS signaling promotes tumor immunoresistance by stabilizing PD-L1 mRNA. Immunity. 2017;47:1083–1099. e6. doi: 10.1016/j.immuni.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marzec M, Zhang Q, Goradia A, Raghunath PN, Liu X, Paessler M, Wang HY, Wysocka M, Cheng M, Ruggeri BA, Wasik MA. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1) Proc Natl Acad Sci U S A. 2008;105:20852–20857. doi: 10.1073/pnas.0810958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48:434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Y. Regulatory mechanisms of PD-L1 expression in cancer cells. Cancer Immunol Immunother. 2018;67:1481–1489. doi: 10.1007/s00262-018-2226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Teng F, Kong L, Yu J. PD-L1 expression in human cancers and its association with clinical outcomes. Onco Targets Ther. 2016;9:5023–5039. doi: 10.2147/OTT.S105862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Budczies J, Bockmayr M, Denkert C, Klauschen F, Gröschel S, Darb-Esfahani S, Pfarr N, Leichsenring J, Onozato ML, Lennerz JK, Dietel M, Fröhling S, Schirmacher P, Iafrate AJ, Weichert W, Stenzinger A. Pan-cancer analysis of copy number changes in programmed death-ligand 1 (PD-L1, CD274)-associations with gene expression, mutational load, and survival. Genes Chromosomes Cancer. 2016;55:626–639. doi: 10.1002/gcc.22365. [DOI] [PubMed] [Google Scholar]
- 16.Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O’Donnell E, Chapuy B, Takeyama K, Neuberg D, Golub TR, Kutok JL, Shipp MA. Integrative analysis reveals selective 9p24.1 amplifi cation, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood. 2010;116:3268–3277. doi: 10.1182/blood-2010-05-282780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, Schuster SJ, Millenson MM, Cattry D, Freeman GJ, Rodig SJ, Chapuy B, Ligon AH, Zhu L, Grosso JF, Kim SY, Timmerman JM, Shipp MA, Armand P. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372:311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett MT, Anderson KS, Lenkiewicz E, Andreozzi M, Cunliffe HE, Klassen CL, Dueck AC, McCullough AE, Reddy SK, Ramanathan RK, Northfelt DW, Pockaj BA. Genomic amplifi cation of 9p24.1 targeting JAK2, PD-L1, and PD-L2 is enriched in high-risk triple negative breast cancer. Oncotarget. 2015;6:26483–26493. doi: 10.18632/oncotarget.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.George J, Saito M, Tsuta K, Iwakawa R, Shiraishi K, Scheel AH, Uchida S, Watanabe SI, Nishikawa R, Noguchi M, Peifer M, Jang SJ, Petersen I, Büttner R, Harris CC, Yokota J, Thomas RK, Kohno T. Genomic amplifi cation of CD274 (PD-L1) in small-cell lung cancer. Clin Cancer Res. 2017;23:1220–1226. doi: 10.1158/1078-0432.CCR-16-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda S, Okamoto T, Okano S, Umemoto Y, Tagawa T, Morodomi Y, Kohno M, Shimamatsu S, Kitahara H, Suzuki Y, Fujishita T, Maehara Y. PD-L1 is upregulated by simultaneous amplifi cation of the PD-L1 and JAK2 genes in non-small cell lung cancer. J Thorac Oncol. 2016;11:62–71. doi: 10.1016/j.jtho.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Georgiou K, Chen L, Berglund M, Ren W, de Miranda NF, Lisboa S, Fangazio M, Zhu S, Hou Y, Wu K, Fang W, Wang X, Meng B, Zhang L, Zeng Y, Bhagat G, Nordenskjöld M, Sundström C, Enblad G, Dalla-Favera R, Zhang H, Teixeira MR, Pasqualucci L, Peng R, Pan-Hammarström Q. Genetic basis of PD-L1 overexpression in diffuse large B-cell lymphomas. Blood. 2016;127:3026–3034. doi: 10.1182/blood-2015-12-686550. [DOI] [PubMed] [Google Scholar]
- 23.Zerdes I, Matikas A, Bergh J, Rassidakis GZ, Foukakis T. Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: biology and clinical correlations. Oncogene. 2018;37:4639–4661. doi: 10.1038/s41388-018-0303-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S, Sharawat SK. Epigenetic regulators of programmed death-ligand 1 expression in human cancers. Transl Res. 2018;202:129–145. doi: 10.1016/j.trsl.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 25.Gong AY, Zhou R, Hu G, Li X, Splinter PL, O’Hara SP, LaRusso NF, Soukup GA, Dong H, Chen XM. MicroRNA-513 regulates B7-H1 translation and is involved in IFN-gamma-induced B7-H1 expression in cholangiocytes. J Immunol. 2009;182:1325–1333. doi: 10.4049/jimmunol.182.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Li J, Dong K, Lin F, Long M, Ouyang Y, Wei J, Chen X, Weng Y, He T, Zhang H. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal. 2015;27:443–452. doi: 10.1016/j.cellsig.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Wang W, Li F, Mao Y, Zhou H, Sun J, Li R, Liu C, Chen W, Hua D, Zhang X. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum Genet. 2013;132:641–648. doi: 10.1007/s00439-013-1275-6. [DOI] [PubMed] [Google Scholar]
- 28.Wu L, Chen Z, Zhang J, Xing Y. Effect of miR-513a-5p on etoposide-stimulating B7-H1 expression in retinoblastoma cells. J Huazhong Univ Sci Technolog Med Sci. 2012;32:601–606. doi: 10.1007/s11596-012-1004-8. [DOI] [PubMed] [Google Scholar]
- 29.Xie G, Li W, Li R, Wu K, Zhao E, Zhang Y, Zhang P, Shi L, Wang D, Yin Y, Deng R, Tao K. Helicobacter pylori promote B7-H1 expression by suppressing miR-152 and miR-200b in gastric cancer cells. PLoS One. 2017;12:e0168822. doi: 10.1371/journal.pone.0168822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, Zhang X, Yi X, Dwyer D, Lin W, Diao L, Wang J, Roybal J, Patel M, Ungewiss C, Peng D, Antonia S, Mediavilla-Varela M, Robertson G, Suraokar M, Welsh JW, Erez B, Wistuba II, Chen L, Peng D, Wang S, Ullrich SE, Heymach JV, Kurie JM, Qin FX. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int. 2017;41:1056–1064. doi: 10.1002/cbin.10826. [DOI] [PubMed] [Google Scholar]
- 32.Jia L, Xi Q, Wang H, Zhang Z, Liu H, Cheng Y, Guo X, Zhang J, Zhang Q, Zhang L, Xue Z, Li Y, Da Y, Zhao P, Zhang R. miR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem Biophys Res Commun. 2017;488:425–431. doi: 10.1016/j.bbrc.2017.05.074. [DOI] [PubMed] [Google Scholar]
- 33.Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang T, Song W, Chen Y, OuYang J, Chen J, Kong F, Dong Y, Jiang SW, Li W, Wang P, Yuan Z, Wan X, Wang C, Li W, Zhang X, Chen K. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat Commun. 2016;7:11406. doi: 10.1038/ncomms11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kao SC, Cheng YY, Williams M, Kirschner MB, Madore J, Lum T, Sarun KH, Linton A, McCaughan B, Klebe S, van Zandwijk N, Scolyer RA, Boyer MJ, Cooper WA, Reid G. Tumor suppressor microRNAs contribute to the regulation of PD-L1 expression in malignant pleural mesothelioma. J Thorac Oncol. 2017;12:1421–1433. doi: 10.1016/j.jtho.2017.05.024. [DOI] [PubMed] [Google Scholar]
- 35.Dong P, Xiong Y, Yu J, Chen L, Tao T, Yi S, Hanley SJB, Yue J, Watari H, Sakuragi N. Control of PD-L1 expression by miR-140/142/340/383 and oncogenic activation of the OCT4-miR-18a pathway in cervical cancer. Oncogene. 2018;37:5257–5268. doi: 10.1038/s41388-018-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu J, Chen L, Zou L, Yang P, Wu R, Mao Y, Zhou H, Li R, Wang K, Wang W, Hua D, Zhang X. MiR-20b, -21, and -130b inhibit PTEN expression resulting in B7-H1 over-expression in advanced colorectal cancer. Hum Immunol. 2014;75:348–353. doi: 10.1016/j.humimm.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Fujita Y, Yagishita S, Hagiwara K, Yoshioka Y, Kosaka N, Takeshita F, Fujiwara T, Tsuta K, Nokihara H, Tamura T, Asamura H, Kawaishi M, Kuwano K, Ochiya T. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol Ther. 2015;23:717–727. doi: 10.1038/mt.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goltz D, Gevensleben H, Grunen S, Dietrich J, Kristiansen G, Landsberg J, Dietrich D. PD-L1 (CD274) promoter methylation predicts survival in patients with acute myeloid leukemia. Leukemia. 2017;31:738–743. doi: 10.1038/leu.2016.328. [DOI] [PubMed] [Google Scholar]
- 39.Franzen A, Vogt TJ, Muller T, Dietrich J, Schröck A, Golletz C, Brossart P, Bootz F, Landsberg J, Kristiansen G, Dietrich D. PD-L1 (CD274) and PD-L2 (PDCD1LG2) promoter methylation is associated with HPV infection and transcriptional repression in head and neck squamous cell carcinomas. Oncotarget. 2018;9:641–650. doi: 10.18632/oncotarget.23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goltz D, Gevensleben H, Dietrich J, Dietrich D. PD-L1 (CD274) promoter methylation predicts survival in colorectal cancer patients. Oncoimmunology. 2017;6:e1257454. doi: 10.1080/2162402X.2016.1257454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heiland DH, Haaker G, Delev D, Mercas B, Masalha W, Heynckes S, Gäbelein A, Pfeifer D, Carro MS, Weyerbrock A, Prinz M, Schnell O. Comprehensive analysis of PD-L1 expression in glioblastoma multiforme. Oncotarget. 2017;8:42214–42225. doi: 10.18632/oncotarget.15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rover LK, Gevensleben H, Dietrich J, Bootz F, Landsberg J, Goltz D, Dietrich D. PD-1 (PDCD1) promoter methylation is a prognostic factor in patients with diffuse lower-grade gliomas harboring isocitrate dehydrogenase (IDH) mutations. EBioMedicine. 2018;28:97–104. doi: 10.1016/j.ebiom.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marwitz S, Scheufele S, Perner S, Reck M, Ammerpohl O, Goldmann T. Epigenetic modifi cations of the immune-checkpoint genes CTLA4 and PDCD1 in non-small cell lung cancer results in increased expression. Clin Epigenetics. 2017;9:51. doi: 10.1186/s13148-017-0354-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Xiang C, Wang Y, Duan Y, Liu C, Zhang Y. PD-L1 promoter methylation mediates the resistance response to anti-PD-1 therapy in NSCLC patients with EGFR-TKI resistance. Oncotarget. 2017;8:101535–101544. doi: 10.18632/oncotarget.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goltz D, Gevensleben H, Dietrich J, Schroeck F, de Vos L, Droege F, Kristiansen G, Schroeck A, Landsberg J, Bootz F, Dietrich D. PDCD1 (PD-1) promoter methylation predicts outcome in head and neck squamous cell carcinoma patients. Oncotarget. 2017;8:41011–41020. doi: 10.18632/oncotarget.17354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gevensleben H, Holmes EE, Goltz D, Dietrich J, Sailer V, Ellinger J, Dietrich D, Kristiansen G. PD-L1 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncotarget. 2016;7:79943–79955. doi: 10.18632/oncotarget.13161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lancho O, Herranz D. The MYC enhancer-ome: long-range transcriptional regulation of MYC in cancer. Trends Cancer. 2018;4:810–822. doi: 10.1016/j.trecan.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hogg SJ, Vervoort SJ, Deswal S, Ott CJ, Li J, Cluse LA, Beavis PA, Darcy PK, Martin BP, Spencer A, Traunbauer AK, Sadovnik I, Bauer K, Valent P, Bradner JE, Zuber J, Shortt J, Johnstone RW. BET-bromodomain inhibitors engage the host immune system and regulate expression of the immune checkpoint ligand PD-L1. Cell Rep. 2017;18:2162–2174. doi: 10.1016/j.celrep.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Durand-Panteix S, Farhat M, Youlyouz-Marfak I, Rouaud P, Ouk-Martin C, David A, Faumont N, Feuillard J, Jayat-Vignoles C. B7-H1, which represses EBV-immortalized B cell killing by autologous T and NK cells, is oppositely regulated by c-Myc and EBV latency III program at both mRNA and secretory lysosome levels. J Immunol. 2012;189:181–190. doi: 10.4049/jimmunol.1102277. [DOI] [PubMed] [Google Scholar]
- 50.Fang W, Zhang J, Hong S, Zhan J, Chen N, Qin T, Tang Y, Zhang Y, Kang S, Zhou T, Wu X, Liang W, Hu Z, Ma Y, Zhao Y, Tian Y, Yang Y, Xue C, Yan Y, Hou X, Huang P, Huang Y, Zhao H, Zhang L. EBV-driven LMP1 and IFN-gamma up-regulate PD-L1 in nasopharyngeal carcinoma: implications for oncotargeted therapy. Oncotarget. 2014;5:12189–12202. doi: 10.18632/oncotarget.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bi XW, Wang H, Zhang WW, Wang JH, Liu WJ, Xia ZJ, Huang HQ, Jiang WQ, Zhang YJ, Wang L. PD-L1 is upregulated by EBV-driven LMP1 through NF-kappaB pathway and correlates with poor prognosis in natural killer/T-cell lymphoma. J Hematol Oncol. 2016;9:109. doi: 10.1186/s13045-016-0341-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu Y, Chang SS, Lin WC, Hsu JM, Hsu YH, Kim T, Chang WC, Hsu JL, Yamaguchi H, Ding Q, Wang Y, Yang Y, Chen CH, Sahin AA, Yu D, Hortobagyi GN, Hung MC. Deubiquitination and stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30:925–939. doi: 10.1016/j.ccell.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green MR, Rodig S, Juszczynski P, Ouyang J, Sinha P, O’Donnell E, Neuberg D, Shipp MA. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clin Cancer Res. 2012;18:1611–1618. doi: 10.1158/1078-0432.CCR-11-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noman MZ, Desantis G, Janji B, Hasmim M, Karray S, Dessen P, Bronte V, Chouaib S. PD-L1 is a novel direct target of HIF-1alpha, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J Exp Med. 2014;211:781–790. doi: 10.1084/jem.20131916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shehade H, Oldenhove G, Moser M. Hypoxia in the intestine or solid tumors: a benefi cial or deleterious alarm signal? Eur J Immunol. 2014;44:2550–2557. doi: 10.1002/eji.201444719. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, Tan Y, Ci Y, Wu F, Dai X, Guo J, Huang YH, Fan C, Ren S, Sun Y, Freeman GJ, Sicinski P, Wei W. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553:91–95. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, Lam EYN, Henderson MA, Bell CC, Stolzenburg S, Gilan O, Bloor S, Noori T, Morgens DW, Bassik MC, Neeson PJ, Behren A, Darcy PK, Dawson SJ, Voskoboinik I, Trapani JA, Cebon J, Lehner PJ, Dawson MA. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–105. doi: 10.1038/nature23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, Logtenberg MEW, Slagter M, Rozeman EA, Hofland I, Broeks A, Horlings HM, Wessels LFA, Blank CU, Xiao Y, Heck AJR, Borst J, Brummelkamp TR, Schumacher TNM. Identifi cation of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549:106–110. doi: 10.1038/nature23669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horita H, Law A, Hong S, Middleton K. Identifying regulatory posttranslational modifi cations of PD-L1: a focus on monoubiquitinaton. Neoplasia. 2017;19:346–353. doi: 10.1016/j.neo.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li CW, Lim SO, Chung EM, Kim YS, Park AH, Yao J, Cha JH, Xia W, Chan LC, Kim T, Chang SS, Lee HH, Chou CK, Liu YL, Yeh HC, Perillo EP, Dunn AK, Kuo CW, Khoo KH, Hsu JL, Wu Y, Hsu JM, Yamaguchi H, Huang TH, Sahin AA, Hortobagyi GN, Yoo SS, Hung MC. Eradication of triple-negative breast cancer cells by targeting glycosylated PD-L1. Cancer Cell. 2018;33:187–201. e10. doi: 10.1016/j.ccell.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, Khoo KH, Chang SS, Cha JH, Kim T, Hsu JL, Wu Y, Hsu JM, Yamaguchi H, Ding Q, Wang Y, Yao J, Lee CC, Wu HJ, Sahin AA, Allison JP, Yu D, Hortobagyi GN, Hung MC. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016;7:12632. doi: 10.1038/ncomms12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moya-Horno I, Viteri S, Karachaliou N, Rosell R. Combination of immunotherapy with targeted therapies in advanced non-small cell lung cancer (NSCLC) Ther Adv Med Oncol. 2018;10:1758834017745012. doi: 10.1177/1758834017745012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li X, Lian Z, Wang S, Xing L, Yu J. Interactions between EGFR and PD-1/PD-L1 pathway: implications for treatment of NSCLC. Cancer Lett. 2018;418:1–9. doi: 10.1016/j.canlet.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 64.Chih-Hsin Yang J, Shepherd FA, Kim DW, Lee GW, Lee JS, Chang GC, Lee SS, Wei YF, Lee YG, Laus G, Collins B, Pisetzky F, Horn L. Osimertinib plus durvalumab versus osimertinib monotherapy in EGFR T790M-positive NSCLC following previous EGFR TKI therapy: CAURAL brief report. J Thorac Oncol. 2019;14:933–939. doi: 10.1016/j.jtho.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 65.Ahn MJ. Combination of osimertinib with durvalumab in epidermal growth factor receptor-mutant non-small cell lung cancer: is there room for reinvestigation? J Thorac Oncol. 2019;14:766–767. doi: 10.1016/j.jtho.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 66.Neoadjuvant PD-1 Blockade in Resectable Lung Cancer; Nivolumab and Ipilimumab in Advanced Melanoma; Overall Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma; Prolonged Survival in Stage III Melanoma with Ipilimumab Adjuvant Therapy; Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma; Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma; Nivolumab and Ipilimumab versus Ipilimumab in Untreated Melanoma; Rapid Eradication of a Bulky Melanoma Mass with One Dose of Immunotherapy; Genetic Basis for Clinical Response to CTLA-4 Blockade; Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma; Nivolumab plus Ipilimumab in Advanced Melanoma; Safety and Tumor Responses with Lambrolizumab (Anti-PD-1) in Melanoma; Hepatotoxicity with Combination of Vemurafenib and Ipilimumab. N Engl J Med. 2018;379:2185. doi: 10.1056/NEJMx180040. [DOI] [PubMed] [Google Scholar]
- 67.Liu L, Mayes PA, Eastman S, Shi H, Yadavilli S, Zhang T, Yang J, Seestaller-Wehr L, Zhang SY, Hopson C, Tsvetkov L, Jing J, Zhang S, Smothers J, Hoos A. The BRAF and MEK inhibitors dabrafenib and trametinib: effects on immune function and in combination with immunomodulatory antibodies targeting PD-1, PD-L1, and CTLA-4. Clin Cancer Res. 2015;21:1639–1651. doi: 10.1158/1078-0432.CCR-14-2339. [DOI] [PubMed] [Google Scholar]
- 68.Gettinger S, Hellmann MD, Chow LQM, Borghaei H, Antonia S, Brahmer JR, Goldman JW, Gerber DE, Juergens RA, Shepherd FA, Laurie SA, Young TC, Li X, Geese WJ, Rizvi N. Nivolumab plus erlotinib in patients with EGFR-mutant advanced NSCLC. J Thorac Oncol. 2018;13:1363–1372. doi: 10.1016/j.jtho.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 69.Lastwika KJ, Wilson W 3rd, Li QK, Norris J, Xu H, Ghazarian SR, Kitagawa H, Kawabata S, Taube JM, Yao S, Liu LN, Gills JJ, Dennis PA. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res. 2016;76:227–238. doi: 10.1158/0008-5472.CAN-14-3362. [DOI] [PubMed] [Google Scholar]
- 70.Lin C, Chen X, Li M, Liu J, Qi X, Yang W, Zhang H, Cai Z, Dai Y, Ouyang X. Programmed death-ligand 1 expression predicts tyrosine kinase inhibitor response and better prognosis in a cohort of patients with epidermal growth factor receptor mutation-positive lung adenocarcinoma. Clin Lung Cancer. 2015;16:e25–35. doi: 10.1016/j.cllc.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 71.Lastwika KJ, Wilson W 3rd, Li QK, Norris J, Xu H, Ghazarian SR, Kitagawa H, Kawabata S, Taube JM, Yao S, Liu LN, Gills JJ, Dennis PA. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res. 2016;76:227–238. doi: 10.1158/0008-5472.CAN-14-3362. [DOI] [PubMed] [Google Scholar]
- 72.Jiang X, Zhou J, Giobbie-Hurder A, Wargo J, Hodi FS. The activation of MAPK in melanoma cells resistant to BRAF inhibition promotes PD-L1 expression that is reversible by MEK and PI3K inhibition. Clin Cancer Res. 2013;19:598–609. doi: 10.1158/1078-0432.CCR-12-2731. [DOI] [PubMed] [Google Scholar]
- 73.Bu LL, Yu GT, Wu L, Mao L, Deng WW, Liu JF, Kulkarni AB, Zhang WF, Zhang L, Sun ZJ. STAT3 induces immunosuppression by upregulating PD-1/PD-L1 in HNSCC. J Dent Res. 2017;96:1027–1034. doi: 10.1177/0022034517712435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dorand RD, Nthale J, Myers JT, Barkauskas DS, Avril S, Chirieleison SM, Pareek TK, Abbott DW, Stearns DS, Letterio JJ, Huang AY, Petrosiute A. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science. 2016;353:399–403. doi: 10.1126/science.aae0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parsa AT, Waldron JS, Panner A, Crane CA, Parney IF, Barry JJ, Cachola KE, Murray JC, Tihan T, Jensen MC, Mischel PS, Stokoe D, Pieper RO. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med. 2007;13:84–88. doi: 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- 76.Song M, Chen D, Lu B, Wang C, Zhang J, Huang L, Wang X, Timmons CL, Hu J, Liu B, Wu X, Wang L, Wang J, Liu H. PTEN loss increases PD-L1 protein expression and affects the correlation between PD-L1 expression and clinical parameters in colorectal cancer. PLoS One. 2013;8:e65821. doi: 10.1371/journal.pone.0065821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Atefi M, Avramis E, Lassen A, Wong DJ, Robert L, Foulad D, Cerniglia M, Titz B, Chodon T, Graeber TG, Comin-Anduix B, Ribas A. Effects of MAPK and PI3K pathways on PD-L1 expression in melanoma. Clin Cancer Res. 2014;20:3446–3457. doi: 10.1158/1078-0432.CCR-13-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mittendorf EA, Philips AV, Meric-Bernstam F, Qiao N, Wu Y, Harrington S, Su X, Wang Y, Gonzalez-Angulo AM, Akcakanat A, Chawla A, Curran M, Hwu P, Sharma P, Litton JK, Molldrem JJ, Alatrash G. PD-L1 expression in triple-negative breast cancer. Cancer Immunol Res. 2014;2:361–370. doi: 10.1158/2326-6066.CIR-13-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Concha-Benavente F, Srivastava RM, Trivedi S, Lei Y, Chandran U, Seethala RR, Freeman GJ, Ferris RL. Identifi cation of the cell-intrinsic and -extrinsic pathways downstream of EGFR and IFNgamma that induce PD-L1 expression in head and neck cancer. Cancer Res. 2016;76:1031–1043. doi: 10.1158/0008-5472.CAN-15-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu J, Hamrouni A, Wolowiec D, Coiteux V, Kuliczkowski K, Hetuin D, Saudemont A, Quesnel B. Plasma cells from multiple myeloma patients express B7-H1 (PD-L1) and increase expression after stimulation with IFN-γ and TLR ligands via a MyD88-, TRAF6-, and MEK-dependent pathway. Blood. 2007;110:296–304. doi: 10.1182/blood-2006-10-051482. [DOI] [PubMed] [Google Scholar]
- 81.Yamamoto R, Nishikori M, Tashima M, Sakai T, Ichinohe T, Takaori-Kondo A, Ohmori K, Uchiyama T. B7-H1 expression is regulated by MEK/ERK signaling pathway in anaplastic large cell lymphoma and Hodgkin lymphoma. Cancer Sci. 2009;100:2093–2100. doi: 10.1111/j.1349-7006.2009.01302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Imai D, Yoshizumi T, Okano S, Itoh S, Ikegami T, Harada N, Aishima S, Oda Y, Maehara Y. IFN-γ promotes epithelial-mesenchymal transition and the expression of PD-L1 in pancreatic cancer. J Surg Res. 2019;240:115–123. doi: 10.1016/j.jss.2019.02.038. [DOI] [PubMed] [Google Scholar]
- 83.Li N, Wang J, Zhang N, Zhuang M, Zong Z, Zou J, Li G, Wang X, Zhou H, Zhang L, Shi Y. Cross-talk between TNF-alpha and IFN-gamma signaling in induction of B7-H1 expression in hepatocellular carcinoma cells. Cancer Immunol Immunother. 2018;67:271–283. doi: 10.1007/s00262-017-2086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X, Parisi G, Saus CP, Torrejon DY, Graeber TG, Comin-Anduix B, Hu-Lieskovan S, Damoiseaux R, Lo RS, Ribas A. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017;19:1189–1201. doi: 10.1016/j.celrep.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang X, Zeng Y, Qu Q, Zhu J, Liu Z, Ning W, Zeng H, Zhang N, Du W, Chen C, Huang JA. PD-L1 induced by IFN-gamma from tumor-associated macrophages via the JAK/STAT3 and PI3K/AKT signaling pathways promoted progression of lung cancer. Int J Clin Oncol. 2017;22:1026–1033. doi: 10.1007/s10147-017-1161-7. [DOI] [PubMed] [Google Scholar]
- 86.Moon JW, Kong SK, Kim BS, Kim HJ, Lim H, Noh K, Kim Y, Choi JW, Lee JH, Kim YS. IFNgamma induces PD-L1 overexpression by JAK2/STAT1/IRF-1 signaling in EBV-positive gastric carcinoma. Sci Rep. 2017;7:17810. doi: 10.1038/s41598-017-18132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chan LC, Li CW, Xia W, Hsu JM, Lee HH, Cha JH, Wang HL, Yang WH, Yen EY, Chang WC, Zha Z, Lim SO, Lai YJ, Liu C, Liu J, Dong Q, Yang Y, Sun L, Wei Y, Nie L, Hsu JL, Li H, Ye Q, Hassan MM, Amin HM, Kaseb AO, Lin X, Wang SC, Hung MC. IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J Clin Invest. 2019;129:3324–3338. doi: 10.1172/JCI126022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carbotti G, Nikpoor AR, Vacca P, Gangemi R, Giordano C, Campelli F, Ferrini S, Fabbi M. IL-27 mediates HLA class I up-regulation, which can be inhibited by the IL-6 pathway, in HLA-defi cient Small Cell Lung Cancer cells. J Exp Clin Cancer Res. 2017;36:140. doi: 10.1186/s13046-017-0608-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu L, Chen X, Shen M, Yang DR, Fang L, Weng G, Tsai Y, Keng PC, Chen Y, Lee SO. Inhibition of IL-6-JAK/Stat3 signaling in castration-resistant prostate cancer cells enhances the NK cell-mediated cytotoxicity via alteration of PD-L1/NKG2D ligand levels. Mol Oncol. 2018;12:269–286. doi: 10.1002/1878-0261.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carbotti G, Barisione G, Airoldi I, Mezzanzanica D, Bagnoli M, Ferrero S, Petretto A, Fabbi M, Ferrini S. IL-27 induces the expression of IDO and PD-L1 in human cancer cells. Oncotarget. 2015;6:43267–43280. doi: 10.18632/oncotarget.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang X, Yang L, Huang F, Zhang Q, Liu S, Ma L, You Z. Inflammatory cytokines IL-17 and TNF-alpha up-regulate PD-L1 expression in human prostate and colon cancer cells. Immunol Lett. 2017;184:7–14. doi: 10.1016/j.imlet.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li H, Xia JQ, Zhu FS, Xi ZH, Pan CY, Gu LM, Tian YZ. LPS promotes the expression of PD-L1 in gastric cancer cells through NF-kappaB activation. J Cell Biochem. 2018;119:9997–10004. doi: 10.1002/jcb.27329. [DOI] [PubMed] [Google Scholar]
- 93.Chen N, Fang W, Zhan J, Hong S, Tang Y, Kang S, Zhang Y, He X, Zhou T, Qin T, Huang Y, Yi X, Zhang L. Upregulation of PD-L1 by EGFR activation mediates the immune escape in EGFR-driven NSCLC: implication for optional immune targeted therapy for NSCLC patients with EGFR mutation. J Thorac Oncol. 2015;10:910–923. doi: 10.1097/JTO.0000000000000500. [DOI] [PubMed] [Google Scholar]
- 94.Sun L, Wang Q, Chen B, Zhao Y, Shen B, Wang H, Xu J, Zhu M, Zhao X, Xu C, Chen Z, Wang M, Xu W, Zhu W. Gastric cancer mesenchymal stem cells derived IL-8 induces PD-L1 expression in gastric cancer cells via STAT3/mTOR-c-Myc signal axis. Cell Death Dis. 2018;9:928. doi: 10.1038/s41419-018-0988-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sanmamed MF, Perez-Gracia JL, Schalper KA, Fusco JP, Gonzalez A, Rodriguez-Ruiz ME, Oñate C, Perez G, Alfaro C, Martín-Algarra S, Andueza MP, Gurpide A, Morgado M, Wang J, Bacchiocchi A, Halaban R, Kluger H, Chen L, Sznol M, Melero I. Changes in serum interleukin-8 (IL-8) levels reflect and predict response to anti-PD-1 treatment in melanoma and non-small-cell lung cancer patients. Ann Oncol. 2017;28:1988–1995. doi: 10.1093/annonc/mdx190. [DOI] [PMC free article] [PubMed] [Google Scholar]


