Abstract
Small extracellular vesicles (sEVs) mediate the interaction between tumor and tumor-associated macrophages (TAMs). This study aims to demonstrate that the pancreatic ductal adenocarcinoma (PDAC)-derived sEV Ezrin (sEV-EZR) could modulate macrophage polarization and promote PDAC metastasis. We isolated PDAC-derived sEVs and plasma sEVs from PDAC patients. Human blood mononuclear cell (PBMC)-derived macrophages were treated with PDAC-derived sEVs or the counterpart depleted Ezrin (EZR) with shRNA-mediated knockdown. We used enzyme-linked immunosorbent assays and flow cytometry to monitor macrophages polarization. NOD/SCID/IL2Rγnull mice were treated with sEVs to study PDAC liver metastasis. The plasma sEV-EZR levels of 165 PDAC patients and 151 high-risk controls were analyzed. The EZR levels are higher in sEVs derived from PDAC cells and PDAC-patient plasma than that of the normal controls. PDAC-derived sEVs modulate the polarization of macrophages to M2 phenotype, while PDAC-shEZR-derived sEVs polarize macrophages into M1 phenotype. We found an increase in M1 TAMs and a decrease in M2 TAMs in orthotropic tumors treated with PDAC-shEZR-derived sEVs. The amount of liver metastasis in PDAC-shEZR-derived sEVs-treated mice was observed to be smaller than that of controls. The mean plasma sEV-EZR levels from PDAC patients were significantly higher than those from the controls (32.43±20.78 vs. 21.88±11.43 pg/ml; P<0.0001). The overall survival in the high-plasma sEV-EZR patients was significantly shorter than that in the low-EZR group (6.94±15.25 vs. 9.63±15.11 months; P=0.0418). sEV-EZR could modulate macrophage polarization and promote metastasis in PDAC. Targeting sEV-EZR can be considered a promising therapeutic strategy to inhibit PDAC metastasis.
Keywords: Pancreatic cancer, extracellular vesicles, macrophage polarization, Ezrin, metastasis
Introduction
The tumor microenvironment (TME) of pancreatic ductal adenocarcinoma (PDAC), which comprises extracellular matrix, fibroblasts, endothelial cells and immune cells together with a minority of malignant cells, characterizes prominent desmoplastic change and plays a vital role in cancer development and chemoresistance [1]. Therapeutic failures of chemotherapy, targeted therapy, and immunotherapy have all been attributed to the PDAC microenvironment [2,3]. Macrophages are one of the major components of the TME involving tumor progression [4,5]. Tumor-associated macrophages (TAMs) are observed to secrete cytokines and inflammatory mediators that provide a favorable milieu for cancer cell [6,7]. According to their polarization states, macrophages are categorized into two types: classically activated type 1 (M1 macrophages), and alternatively activated type 2 (M2 macrophages) [8]. M1 macrophages, characterized by the expression of the inducible-type nitric oxide synthase (iNOS), are pro-inflammatory and develop in response to lipopolysaccharides (LPS) or interferon-γ (IFN-γ) [9]. M2 macrophages, or anti-inflammatory macrophages, develop in response to interleukin (IL)-4, IL-13 or glucocorticoids, and are characterized by the secretion of anti-inflammatory mediators, including transforming growth factor-β1 (TGF-β1) and IL-10 to promote extracellular matrix remodeling and angiogenesis [10,11]. M2 TAMs are related to pro-tumor features, whereas M1 macrophages exert anti-tumor functions [12]. Abundant M2-related markers (e.g., CD163 and CD206) in tumor tissues correlate negatively with the survival of cancer patients, including those with PDAC [5,13].
Extracellular vesicles (EVs) constitute a heterogeneous family of cell-released fluid-filled sacs bounded by a phospholipid bilayer without functional nucleus [14]. Small EVs (sEVs; less than 200 nm) or exosomes are the principal families of EVs with multiple biological functions participating in physiological and pathological processes from aging to cancer, inflammation, immune signaling, infectious disease and obesity [15,16]. sEVs originating from tumor cells contain substantial proteomic and genetic information for disease diagnostics and for monitoring cancer progression, metastasis and drug efficacy [17]. In addition, sEVs are present in diverse biofluids such as plasma [18,19], breast milk [19], urine [20] and ascites [21]. Also, sEVs have been shown to regulate interaction between tumor and immune cells, including the regulation of TAMs, contributing to the pro- or anti-tumor responses [22].
EZR, a member of the Ezrin-radixin-moesin (ERM) family, regulates cell proliferation, morphogenesis, migration and adhesion, and modulates plasma membrane signaling transduction [23]. EZR is preferentially produced in epithelial cells, to whose apical surface it localizes [24]. EZR expression was up-regulated in PDAC and was associated with tumor progression [25,26]. Little is known about the role of small extracellular vesical Ezrin (sEV-EZR) involved in TAM regulation in PDAC. This study aims to demonstrate that the PDAC-derived sEV-EZR could regulate the macrophage polarization and promote PDAC metastasis, and that sEV-EZR is significantly associated with PDAC patient survival.
Materials and methods
Culture of cell lines
Human pancreatic duct epithelial cell (HPDE) was cultured in keratinocyte serum-free (KSF) medium supplemented by epidermal growth factor and bovine pituitary extract (Life Technologies, Inc., Grand Island, NY). PANC-1 cells were cultured in DMEM and BXPC-3, PDAC patient-derived xenograft PC080, and PC084 cells were cultured in RPMI1640 (Gibco, Grand Island, NY, USA) supplemented with 10% exosome-depleted fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA), 1 mM sodium pyurvate and 1% non-essential amino acids (Gibco, Grand Island, NY, USA). THP-1 and U937 cells, a pro-monocytic cell line, were cultured in RPMI 1640 supplemented with 10% fetal bovine serum. All cells were cultured at 37°C in a 5% CO2 atmosphere and maintained within 3 months of resuscitation from the frozen aliquots, with less than 20 passages for each experiment.
Patient samples and tissue collection
Between January 2005 and December 2017, peripheral blood and surgical tissues were collected at the National Taiwan University Hospital from 165 patients with cytologically and/or pathologically confirmed PDAC after their written informed consents were obtained. All the patients’ demographic data, including age, sex, serological study results, image study results, survival data, and clinical manifestations were collected. Peripheral blood was collected from 151 controls at high risk for PDAC (high-risk controls; HRCs) who had a family history of PDAC and participated in the pancreatic cancer screening program at the National Taiwan University Hospital between January 2005 and December 2015 [27]. All control subjects underwent a detailed history assessment and physical examination, family history collection, personal and family health history collection, magnetic resonance imaging/magnetic resonance cholangiopancreatography (MRI/MRCP) examination and blood testing. All controls were followed up for more than 2 years and were free of pancreatic malignancy. The study was reviewed and approved by the Institutional Review Board of the National Taiwan University Hospital.
Separation and characterization of sEVs
Separation of sEVs from cells
For separation of sEVs from HPDE, BXPC-3, PANC-1, PC080 and PC084 cell-conditioned medium, 160 ml of medium after incubation with each cell line was harvested, and the cells were removed from the conditioned media by centrifugation at 500 × g for 10 min. The supernatants were successively centrifuged at 2000 × g for 20 min, filtered through a 0.22-um filter, and ultra-centrifuged at 100,000 × g for 1.5 hours (Beckman 70Ti rotor). Pellets were resuspended in 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) and loaded on top of sucrose gradients with the indicated compositions (from bottom to top, 1.2 ml of 2 M, 2 ml of 1.3 M, 2 ml of 1.16 M, 1.8 ml of 0.8 M, 1.8 ml of 0.5 M, and 1.2 ml of 0.25 M with 20 mM Tris, pH 7.4). Ultra-centrifugation was performed at 100,000 × g for 16 hours (Beckman SW41 rotor), and 0.5 ml from each fraction was collected. The isolated EVs were washed 3 times with PBS through Amicon-0.5 ml centrifugal filters with a cut-off of 100 kD (Millipore, Billerica, MA, USA) for further analysis.
Separation of sEVs from human plasma samples
Ten ml of peripheral venous blood was collected into a plasma separator tube (Plastic K2EDTA tube 10 ml with Lavender Hemogard Closure, BD Vacutainer Systems, Franklin Lakes, NJ, USA) from each patient and control under fasting conditions, and then the plasma was immediately separated by centrifugation at 3000 × g for 10 min in a refrigerated centrifuge. The plasma was stored at -80°C for further analysis. Size-exclusion chromatography (SEC) was performed to isolate sEVs from plasma. In brief, 20 ml of Sepharose CL-4B (Sigma Aldrich, St. Louis, MO, USA) was stacked in a 27 ml syringe (BD PlasticpakcTM, San Jose, CA), washed and equilibrated with PBS. Then, 1.2 ml of plasma was diluted 1:1 with PBS, filtered through a 0.22-µm filter and centrifuged at 13,000 × g for 15 min. Finally, 2 ml of supernatant was loaded onto the column, followed immediately by fraction collection (0.5 ml per fraction and a total of 20 fractions were collected) using PBS as the elution buffer and EV concentration (30-kDa MWCO Amicon Ultra 0.5 ml, Millipore, Billicera, MA, USA).
sEV size analysis
Size distribution and concentration of isolated vesicles were measured using the NanoSight NS300 instrument (Malvern Instruments Ltd, Malvern, UK) equipped with a 488 nm laser and a CCD camera (model NS-300). Data were analyzed using the Nanoparticle Tracking Analysis (NTA) software (versions 3.1 build 3.1.46). Samples processed by SEC were diluted 90-100 times using sterile and filtered phosphate buffered saline (PBS) to reduce the number of particles in the field of view below 100/frame. Readings were taken in a single capture during 60 s at 30 frames per second (fps), with the camera level set to 15 and manual monitoring of temperature.
Immunoblot analysis of sEV proteins
Cells or sEVs were lysed in RIPA lysis buffer (containing 50 mM Tris-HCl, 1% NP-40, 150 mM NaCl, 0.1% SDS, 1 mM PMSF, 1 mM Na3VO4) with complete protease and protease inhibitor cocktail (1:1000) (Sigma-Aldrich, Inc.). Protein concentrations were then determined using the BCA assay kit (Thermo, USA) in accordance with the manufacturer’s instructions. Then, 5 μg of protein lysates from cells and 0.5 μg of protein lysates from sEVs were loaded onto 10% SDS polyacrylamide gels and subjected to electrophoresis, followed by transferring the proteins to poly-vinylidene fluoride membranes (Pall Life Sciences, Glen Cove, NY). The membranes were then probed with antibodies specific for sEV-enriched markers, Alix (ab186429), Flotillin-1 (ab133497), TSG101 (ab125011), and CD9 (ab92726) (Abcam, Cambridge, UK); CD81 (se-166029) (Santa Cruz, Dallas, TX); antibody against Calnexin (#22595) (Cell Signaling, Danvers, Massachusetts, USA); antibodies against Annexin A11 (ab137424); antibodies against Ezrin (ab41672) (Abcam, Cambridge, UK); antibodies against Moesin (GTX101708) and antibodies against GPRC5A (GTX108135) (GeneTex, Irvine, CA, USA). Signals from HRP-coupled secondary antibodies were visualized using the enhanced chemiluminescence (ECL) detection system (PerkinElmer, Waltham, MA). The membrane was exposed using enhanced chemiluminescence reagents and analyzed using the BioSpectrum 810 Imaging Systems (UVP, Upland, CA).
Transmission electron microscope
Formvar/carbon-coated copper grids (Ted Pella, Inc., Redding, CA, USA) were glow-discharged before the purified sEV samples (0.1 μg) were loaded. The grids and samples were incubated for 1 min and were contrasted in 1% uranyl acetate. The preparations were examined using a transmission electron microscope (TEM) (Tecnai G2 F20 X-TWIN, FEI, USA).
shRNA-mediated downregulation of sEV-EZR
For gene knockdown experiments, shRNA clones for EZR and their control pLKO.1-scramble (ASN0000000004) were obtained from the National RNAi Core Facility (Academia Sinica, Taiwan). The pcDNA3.1-EZR plasmid was generated by inserting full-length cDNA (Ezrin: NM_003379) into the pcDNA3.1+ vector. For transfection of the other plasmids, cells were transiently transfected with 5 μg of plasmids using Lipofectamine 3000 from Invitrogen (CA, USA) according to the manufacturer’s protocol.
Flow cytometry: macrophage differentiation and polarization
Mononuclear cells were isolated from human whole blood by standard density gradient centrifugation with lymphoprep (StemCell Technologies Inc., Vancouver, Canada). CD14+ cells were subsequently purified from peripheral mononuclear cells by high-gradient magnetic sorting using anti-CD14 microbeads (EasySep™ Human CD14 Positive Selection Kit II, StemCell Technologies Inc., Vancouver, Canada). To prepare human blood mononuclear cell (PBMC)-derived macrophages, CD14+ cells were cultured in RPMI media containing 10% FBS and 50 ng/ml GM-CSF or M-CSF (R&D, Minneapolis, MN, USA) for 6 days. Then, M0 cells were polarized into M1 using 20 ng/ml IFN-γ (R&D, Minneapolis, MN, USA) and 100 ng/ml LPS (Sigma-Aldrich, USA) or polarized into M2 using IL-4 and IL13 (50 ng/ml, R&D, Minneapolis, MN, USA), both co-incubated with 5 μg/ml cell-derived sEVs for 72 h. The cells were harvested, washed twice with PBS, fixed in 1% paraformaldehyde (PFA) overnight at 4°C, washed again, and resuspended in flow cytometry buffer (1 × PBS buffer containing 1% FSA). To determine M1 or M2 phenotypes, these cells were labeled with the surface fluorchrome-conjugated antibodies: CD80 and CD86 for M1, and CD163 and CD206 for M2 (BD Biosciences, Franklin Lakes, NJ) for 60 min on ice in the dark. The stained cells were analyzed in a BD FACSCantor™ system (BD Biosciences, Franklin Lakes, NJ) using the CELLQuest Analysis software (BD Biosciences, Franklin Lakes, NJ).
Enzyme-linked immunosorbent assay (ELISA)
Prior to ELISA, 2X RIPA lysis buffer was added to sEVs, followed by 10-min sonication to release proteins and 10-min centrifugation at 13000 × g to remove debris. Plasma sEV-EZR was assayed via a quantitative sandwich ELISA according to the manufacturer’s (elabscience, Selangor, Malaysia) protocol. Following treatment of PBMC CD14+ macrophages with sEVs only, with sEVs and LPS+INF-γ, or with sEVs and IL-4+IL-13 (R&D Systems, Inc., Minneapolis, MN, USA), cell supernatants were collected and processed using R&D Duoset ELISA kits (R&D Systems, Inc, Minneapolis, MN, USA) according to the manufacturer’s instructions in detection of cell secretion of IL-10 (cat# DY217B, CCL-18 (cat# DY394), TNF-α (cat# DY210), and IL-1β (cat# DY201).
Animal studies
All animals were monitored for abnormal tissue growth or ill effects according to AAALAS guidelines and euthanized if excessive deterioration of animal health was observed. No statistical method was employed to pre-determine sample size. No method of randomization was adopted to allocate animals into experimental groups. The investigators were not blinded to allocation during experiments and outcome assessment. Mice that died before the predetermined end of the experiment were excluded from the analysis. To study liver metastasis and tumor size, 6-week NOD/SCID/IL2Rγnull (NSG) male mice (kindly provided by the Dr. Michael Hsiao, Genomic Research Center, Academia Sinica, Taipei) were retro-orbitally injected with sEVs (corresponding to a total protein content of 5 µg) for 1 week. Then, 1 × 106 luciferase-positive PC080 cancer cells, which were resuspended in 25 µl matrigel (Corning), were injected orthotopically into pancreas. After cancer cell injection, the mice were injected with sEVs (corresponding to a total protein content of 5 µg) every other day for 1 week. After 21 days, tumors at pancreas of each mouse were analyzed by weight and size, and metastases of liver were measured using bioluminescence IVIS imaging (PerkinElmer, Waltham, MA, USA).
Immunohistochemical (IHC) staining
Formalin-fixed paraffin-embedded primary tumor tissue sections were used for IHC. For heat-induced antigen retrieval, slides were soaked in citric acid buffer (pH 6.0) and heated till 121°C for 30 min. After quenching endogenous peroxidase activity with 3% H2O2, sections were blocked. Mouse tissues were then incubated with anti-CD206 antibody (Ab64693, 1:100) and anti-iNOS antibody (Ab15323, 1:50) (Abcam, Cambridge, MA), while human tissues were incubated with anti-CD68 antibody (M0814, 1:3000) (DakoCytomation, CA, USA) and anti-CD206 antibody (#91992, 1:200) (Cell Signaling, Danvers, Massachusetts, USA) at 4°C overnight. Specific signals were then developed with LSAB+ kit (DakoCytomation, CA, USA) using diaminobenzidine (Biocare) as chromogen. Sections were then counterstained with hematoxylin and observed under light microscope. Slides were scanned using Leica Aperio AT2 (Aperio, Leica Microsystems, Vista, CA) with 40 × magnification. Immunohistochemical expression was quantified in immunohistochemistry digital slides with Leica Aperio positive pixel count algorithm using whole slide analysis (Aperio ImageScope v12.3).
Statistical analysis
The plasma sEV-EZR level was summarized as the mean and standard deviations, and was then compared across patient subgroups on the basis of tumor stages using the Mann-Whitney-Wilcoxon test. Survival curves were plotted using the Kaplan-Meier method and then compared with the log-rank test. Receiver operating characteristic (ROC) curves and areas under the ROC curves (AUCs) were established for discriminating PDAC patients from controls. The cutoff values for plasma sEV-EZR levels were determined from the ROC curves by calculating the Youden index. The sensitivity (SN) and specificity (SP) were calculated for significant variables using the optimal cutoff as determined by the ROC analysis. The data of macrophage polarization experiment were expressed as the mean ± standard deviation (SD) from at least three independent experiments. Differences of various treatment groups were assessed using the Student’s t test or ANOVA when appropriate. Differences were considered significant at P<0.05. Data analyses were performed using GraphPad Prism Ver. 5.02 (San Diego, CA) and MedCalc v.5 software (Mariakerke, Belgium).
Results
PDAC-derived sEVs regulate the polarization of PBMC and THP-1/U937-derived macrophages to M2 phenotype
We have applied the ultracentrifugation (UC) followed with sucrose density gradient (SDG) to purify sEVs from the culture medium. Purified sEV (exosomes) have been characterized by Western Blot (exosome-enriched markers including Alix, TSG 101, Flotillin-1, and CD81) (Figure 1A); NTA (particle-size confirmation) (Figure 1C); TEM (morphological detection) (Figure 1D).
Figure 1.
Separation and Characterization of sEVs from conditioned medium via ultracentrifugation (UC) and a sucrose density gradient (SDG). A. Western blots of sEV-enriched markers. Abundant Flotillin-1, Alix, TSG101, and CD81 are detected in sEVs isolated from conditioned medium in fraction 7-10 by UC-SDG. B. RNA, protein (up panel), and EVs concentrations (down panel) in fraction 7-10. C. Nanoparticle tracking analysis by NasoSight 300 of purified EVs in fraction 7-9. D. PDAC cell-derived sEVs were observed by TEM. Scale bars are 50 nm.
The following cytokines were measured by ELISA to assess macrophage phenotypes: TNF-α/IL-1β for M1 and IL-10/CCL18 for M2. The surface markers were detected by flow cytometry to characterize macrophage phenotypes: CD80/CD86 for M1 and CD163/CD206 for M2. Freshly isolated CD14+ PBMCs-derived macrophages were incubated with sEVs for 72 hr. The amount of M1 cytokines IL-1β and TNF-α treated with PDAC cell (PC080 and PC084)-derived sEVs is found to be less than HPDE cell-derived sEVs (Figure 2A). A significant increase was found in M2 cytokines CCL18 and IL-10 treated with PDAC cell-derived sEVs, compared with HPDE cell-derived sEVs (Figure 2A). Flow cytometry showed the increased macrophages with M2 markers (CD163 and CD206) and reduced macrophages with M1 markers (CD80 and CD86) in CD14+ PBMCs-derived macrophages treated with PDAC cell-derived sEVs, compared with the macrophages treated with HPDE cell-derived sEVs (Figure 2B). Also, the role of PDAC sEVs in macrophage polarization was further explored.
Figure 2.
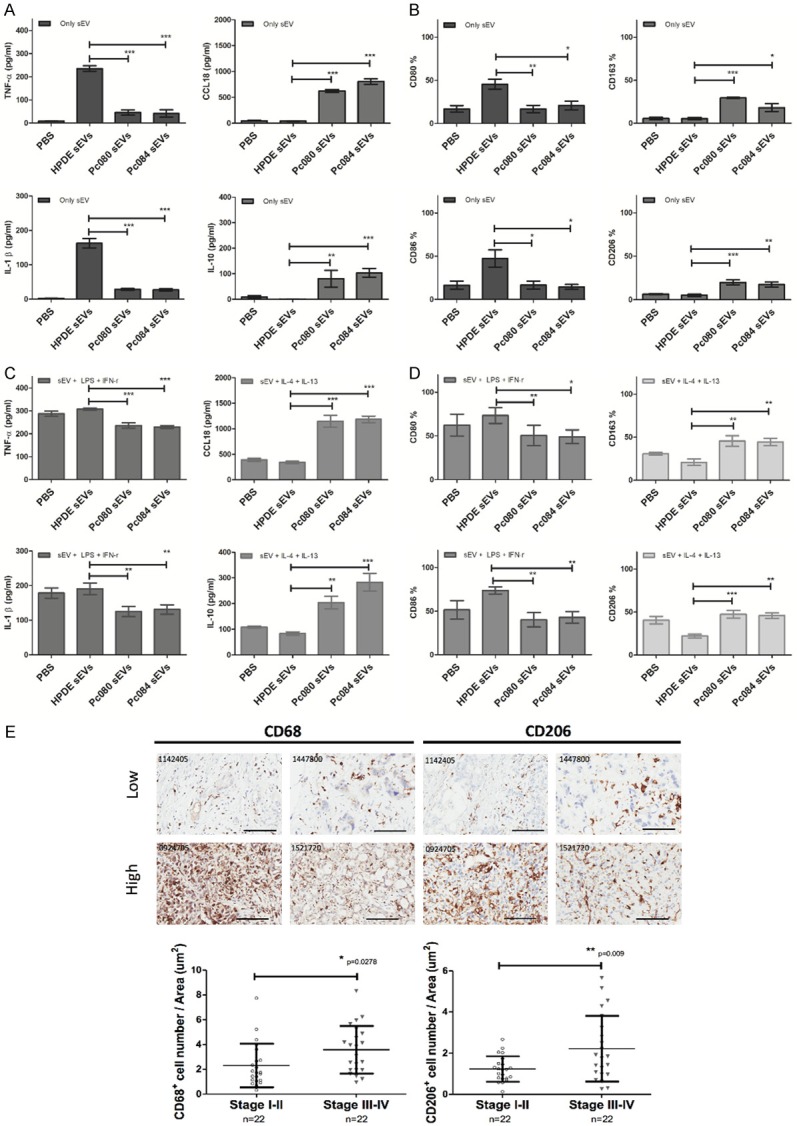
PDAC cell-derived sEVs promote M2 polarization in CD14+ PBMC-derived macrophages. A. ELISA analysis of M1 cytokines (TNF-α and IL-1-β, left panel) and M2 cytokines (CCL18 and IL10, right panel) in CD14+ PBMCs-derived macrophages incubated with PDAC cells (PC080 and PC084)-derived sEVs or HPDE cell-derived sEVs for 72 hrs. B. Flow cytometry analysis of M1 cell surface markers (CD80 and CD86 left panel) and M2 (CD163 and CD206, right panel) cell surface markers on CD14+ PBMCs-derived macrophages incubated with PDAC cells (PC080 and PC084)-derived sEVs or HPDE cell-derived sEVs for 72 hrs. C. ELISA analysis of M1 and M2 cytokines in CD14+ PBMCs-derived macrophages treated with LPS+INF-γ (M1 induction cytokine, left panel) or IL-4+IL-13 (M2 induction cytokine, right panel) in combination with sEVs (derived from either PDAC cell or HPDE cells) for 72 hrs. D. Flow cytometry analysis of M1 and M2 cytokines in CD14+ PBMCs-derived macrophages treated with LPS+INF-γ (M1 induction cytokine, left panel) or IL-4+IL-13 (M2 induction cytokine, right panel) in combination with sEVs (derived from either PDAC cell or HPDE cells) for 72 hrs. Above data represented means ± SD from 4 independent experiments. E. Representative images of CD68 and CD206 (M2 macrophage marker) staining by immunohistochemistry (IHC) in human PDAC tissues (upper panel). The bar chart showed the percentages of CD68+ macrophage and CD206+ macrophage of total cells in IHC images. Each dot represented the datum from one patient. Scale bar, 100 μm. 40 × magnification. Level of significance was determined using Student’s t-test. *P<.05, **P<.005, ***P<0.001.
In earlier research, lipopolysaccharides (LPS) and interferon-γ (IFN-γ) can polarize macrophages into M1 macrophages [9], while interleukin (IL)-4 and IL-13 can polarize macrophages into M2 macrophages [11,28,29]. The amount of M1 cytokines IL-1β and TNF-α in CD14+ PBMCs-derived macrophages treated with LPS, IFN-γ, and PDAC cell-derived sEVs is found to be smaller than that in CD14+ PBMCs-derived macrophages treated with LPS, IFN-γ, and HPDE cell-derived sEVs (Figure 2C). A significant increase of M2 cytokines CCL18 and IL-10 in CD14+ PBMCs-derived macrophage treated with IL-4, IL-13, and PDAC cell-derived sEVs is observed in contrast to CD14+ PBMCs-derived macrophage treated with IL-4 and IL-13, and HPDE cell-derived sEVs (Figure 2C). Similarly, flow cytometry showed the increased macrophages with M2 markers (CD163 and CD206) in CD14+ PBMCs-derived macrophages treated with IL-4, IL-13, and PDAC cell-derived sEVs, compared with the macrophages treated with IL-4, IL-13 and HPDE cell-derived sEVs. Macrophages with M1 markers (CD80 and CD86) were reduced in CD14+ PBMCs-derived macrophages treated with LPS, IFN-γ, and PDAC cell-derived sEVs, compared with the macrophages treated with LPS, IFN-γ, and HPDE cell-derived sEVs (Figure 2D).
Similar patterns in terms of sEV-induced macrophage polarization can be found in human THP-1 and U937 cell lines (Figure 3). M2 TAMs have been reported to be associated with rapid progression of PDAC [5,13,30]. We also observed the increased staining of M2 marker CD206 as well as CD68 by immunohistochemistry in PDAC tissues, and that the increase in both markers was correlated with the cancer stages (Figure 2E).
Figure 3.
PDAC cell-derived sEVs promotes M2 polarization in THP-1/U937 derived macrophages. A. ELISA analysis of M1 cytokines (TNF-α and IL-1-β, left-panel) and M2 cytokines (CCL18 and IL10, right panel) in THP-1/U937 derived macrophages incubated with PDAC cells (PC080 and PC084)-derived sEVs or HPDE cell-derived sEVs for 72 hrs. B. Flow cytometry analysis of M1 cell surface markers (CD80 and CD86, left-panel) and M2 (CD163 and CD206, right panel) cell surface markers on THP-1/U937 derived macrophages incubated with PDAC cells (PC080 and PC084)-derived sEVs or HPDE cell-derived sEVs for 72 hrs. C. ELISA analysis of M1 and M2 cytokines in THP-1/U937 derived macrophages treated with LPS+INF-γ (M1 induction cytokine, left panel) or IL-4+IL-13 (M2 induction cytokine, right panel), co-incubated with sEVs (sEVs derived from either PDAC cell or HPDE cells) for 72 hrs. D. Flow cytometry analysis of M1 and M2 cytokines in THP-1/U937 derived macrophages treated with LPS+INF-γ (M1 induction cytokine, left panel) or IL-4+IL-13 (M2 induction cytokine, right panel), co-incubated with sEVs (sEVs derived from either PDAC cell or HPDE cells) for 72 hrs. Data represented means ± SD from 3 independent experiments. All results are averaged from 3 independent experiments. Data represents means ± S.D. *P<.05, **P<.005, ***P<0.001 by Student’s t-test.
EZR level higher in sEVs derived from PDAC cells and PDAC-patient plasma
To investigate the content of sEVs for this biological function, we purified sEVs from human PDAC cell lines (PANC-1, BXPC-3, and PC080) and HPDE cells (control cell line) analyzed by a liquid chromatography (LC)-tandem mass spectrometry (MS/MS) analysis. Four polypeptides, Annexin A11 (ANXA11), Ezrin (EZR), Moesin (MSN), and Retinoic acid-induced protein (GPRC5A), were identified with 1.5 fold high level in sEVs derived from BXPC-3, PANC-1, and pc080 cell sEVs, compared with those from HPDE (Figure 4A). Western blotting showed that the EZR level appears to be consistently higher in sEVs derived from PDAC cell lines than in those from HPDE cells (Figure 4B). We used size-exclusion chromatography (SEC) to purify sEVs from 1 ml aliquots of plasma from patients with PDAC and controls (Figure 5A). Purified sEVs from plasma have also been characterized by NTA (particle-size confirmation) (Figure 5B); NanoDrop spectrophotometer (protein and RNA concentrations within fractions of 6 to 8) (Figure 5C); Western Blotting (exosome-enriched markers including Alix, Flotillin-1, CD9, CD63, and CD81) (Figure 5D); TEM (morphological detection) (Figure 5E). Western blotting also showed that the EZR level appears to be consistently higher in sEVs derived from PDAC-patient plasma than in those from non-cancer controls (Figure 4C).
Figure 4.
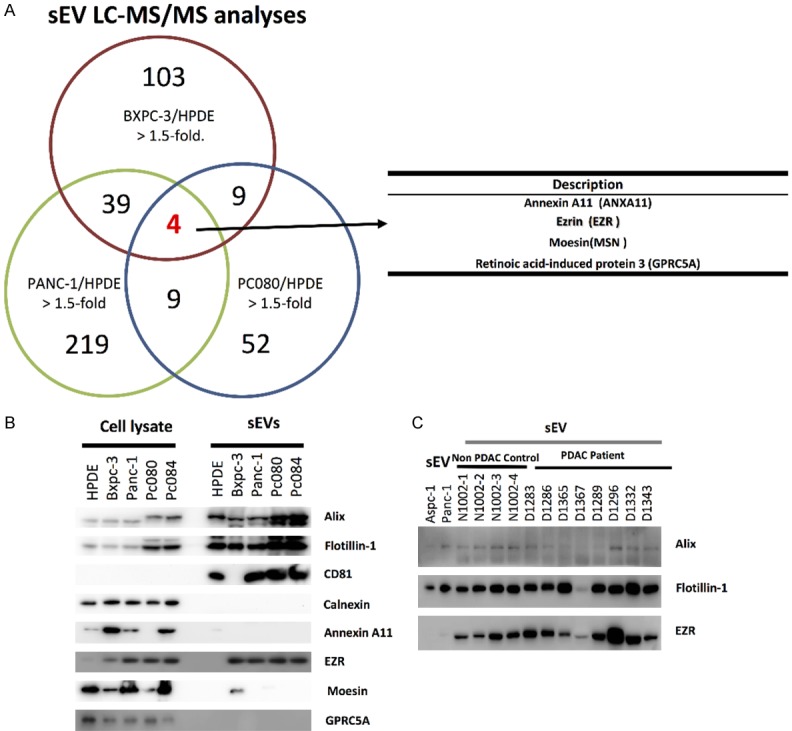
Ezrin found in PDAC cell-derived sEVs and PDAC patient plasma-derived sEVs. A. Identification of sEV peptides from LC-MS/MS analysis. Venn diagram indicates the intersection of sEV peptides across three different PDAC cell lines (BXPC-3, PANC-1 and PC080). B. Ezrin (EZR) is overexpression in PDAC cell-derived sEVs. C. The EZR in sEVs from plasmas of PDAC patients.
Figure 5.
Separation and Characterization of sEVs derived from PDAC-patient plasma. A. Schema for representing the experiment procedures of size-exclusion chromatography (SEC)-based purification of sEVs from human plasma. B. Nanoparticle tracking analysis by NasoSight 300 of purified EVs in fraction 6-8. C. RNA, protein (up panel), and EVs concentrations (down panel) in fraction 5-9. D. Western blots of sEV-enriched markers. Abundant Flotillin-1, Alix, CD63, CD9, and CD81 are detected in sEVs isolated from plasma of human in fraction 6-8 by SEC. E. Human plasma sEVs observed by TEM. Scale bars are 100 nm.
High plasma sEV-EZR level correlates with poor prognosis of PDAC patients
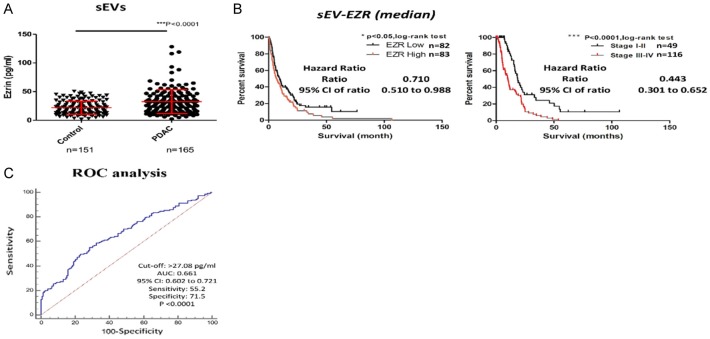
To explore the potential clinic significance of sEV-EZR, we analyzed sEVs from plasma of 165 PDAC patients and 151 controls. The demographic data of study subjects in this study was shown in Table 1. The mean EZR levels were significantly higher in plasma sEVs from PDAC patients than those from the controls (32.43±20.78 vs. 21.88±11.43 pg/ml; P<0.0001; Figure 6A). However, the EZR level in plasma sEVs did not correlate with the tumor stage (P=0.1914; Figure 7A). We observed no differences in EZR levels between PDAC plasma and control plasma (Figure 7B). In this study, PDAC patients were stratified into low-EZR (<28.89 pg/ml) and high-EZR (≥28.89 pg/ml) groups on the basis of the plasma sEV-EZR level (Table 2). The OS in the high-EZR group was statistically significant shorter than that in the low-EZR group (6.94±15.25 vs. 9.63±15.11 months; P=0.0418) (Figure 6B). Multivariate analysis, including age, sex, tumor stage, and plasma sEV-ZER level, showed that plasma sEV-ZER level is an independent prognostic marker in PDAC patients (Table 3).
Table 1.
Baseline characteristics of the study subjects
| PDAC patients (n=165) | Controls (n=151) | |
|---|---|---|
| Sex (M/F) (%) | 70 (42.4%)/95 (57.6%) | 85 (56.3%)/66 (43.7%) |
| Age, mean (SD), years* | 63.62 (12.49) | 47.74 (14.74) |
| CA19-9, mean (SD), U/ml* | 4330.5 (8980.90) | 15.82 (21.38) |
| sEV-EZR, mean (SD), pg/ml* | 32.43 (20.78) | 21.88 (11.43) |
| sEV count, mean (SD), Cal. Conc. (particles/ml plasma) | 2.34E+12 (2.40E+12) | 2.20E+12 (2.34E+12) |
| sEV particle size | ||
| Mean size, mean (SD), nm* | 90.92 (18.43) | 80.057 (8.25) |
| Mode size, mean (SD), nm* | 70.38 (10.96) | 63.63 (6.98) |
| TNM stage (%) | ||
| I-II | 49 (29.7%) | |
| III-IV | 116 (70.3%) | |
| Overall survival, median (SD), months | 7.96 (15.20) | |
| TNM stages I-II | 15.75 (20.47) | |
| TNM stages III-IV** | 5.28 (10.54) |
P<.0001 between PDAC patients and high-risk controls.
P<.0001 between stages I-II and stages III-IV PDAC patients.
PDAC, pancreatic ductal adenocarcinoma; M, male; F, female.
Figure 6.
A. Plasma sEV-EZR levels in PDAC patients and controls. B. Kaplan-Meier analysis of overall survival in PDAC patients with a low or high plasma sEV-EZR (left panel, P=0.0418, log-rank test). Kaplan-Meier analysis of overall survival in patients with early-stage and advanced PDAC (right panel, P<.0001, log-rank test). C. Receiver operating characteristic (ROC) curves of sEV-EZR concentrations in plasma from patients with all stages of PDAC versus concentrations in controls patients (PDAC, n=165; controls, n=151).
Figure 7.

EZR level in plasma sEVs did not correlate with the tumor stage. A. sEV-EZR level in plasma from different PDAC stages (one-way ANOVA, P<0.0001). B. Plasma EZR levels in patients with PDAC (n=64) and controls (n=15).
Table 2.
Comparisons between low and high plasma sEV-EZR groups of PDAC patients
| Low-EZR n=82 | High-EZR n=83 | |
|---|---|---|
| Age, mean (SD), years | 62.32 (12.15) | 64.90 (12.87) |
| Sex (M/F) | 37/45 | 33/50 |
| sEV count, mean (SD), Cal. Conc. (particles/ml plasma) | 2.28E+12 (2.51E+12) | 2.36E+12 (2.32E+12) |
| sEV particle size | ||
| Mean size, mean (SD), nm | 87.46 (17.13) | 94.33 (18.92) |
| Mode size, mean (SD), nm | 68.94 (10.48) | 71.80 (11.14) |
| Stage | ||
| I-II | 21 | 28 |
| III-IV | 61 | 55 |
| CA19-9, mean (SD), U/ml | 4367.96 (9499.22) | 4293.55 (8641.50) |
| sEV-EZR levels, mean (SD), pg/ml** | 17.62 ( 6.45) | 47.06 (19.82) |
| OS, median (SD), months* | 9.63 (15.11) | 6.94 (15.25) |
P<.05 between low- and high-sEV-EZR PDAC patients;
P<.0001 between low- and high-sEV-EZR PDAC patients.
Table 3.
Multivariate analysis of overall survival in PDAC patients
| HR | 95% CI | Multivariate P-value | |
|---|---|---|---|
| Age | 0.640 | 0.453-0.905 | 0.011* |
| Sex | 0.759 | 0.539-1.070 | 0.115 |
| PDAC stage (I-II vs. III-IV) | 0.402 | 0.271-0.597 | 0.000* |
| sEV-EZR, pg/ml (cutoff ≥28.89) | 0.670 | 0.480-0.936 | 0.019* |
| CA19-9, U/ml (cutoff ≥37.0) | 0.549 | 0.347-0.868 | 0.010* |
P<.05.
sEV-EZR, small extracellular vesicles-Ezrin; PDAC, pancreatic ductal adenocarcinoma; HR, hazard ratio; CI, confidence interval; CA19-9, carbohydrate antigen 19-9.
The ROC curve analysis was performed to figure out the SN and the SP of plasma sEV-EZR as a diagnostic biomarker for PDAC. Based on the Youden index calculation, the optimal cut-off value was ascertained for the ability of plasma sEV-EZR to distinguish PDAC patients from controls. At the cut-off value of 27.08 pg/ml, plasma sEV-EZR yielded an SN of 55.2%, an SP of 71.5%, and an AUC of 0.661 [95% confidence interval: 0.602-0.721] for distinguishing PDAC patients from controls (Figure 6C).
Knockdown of PDAC-derived sEV-EZR polarizes PBMC and THP-1/U937-derived macrophages toward M1 phenotype
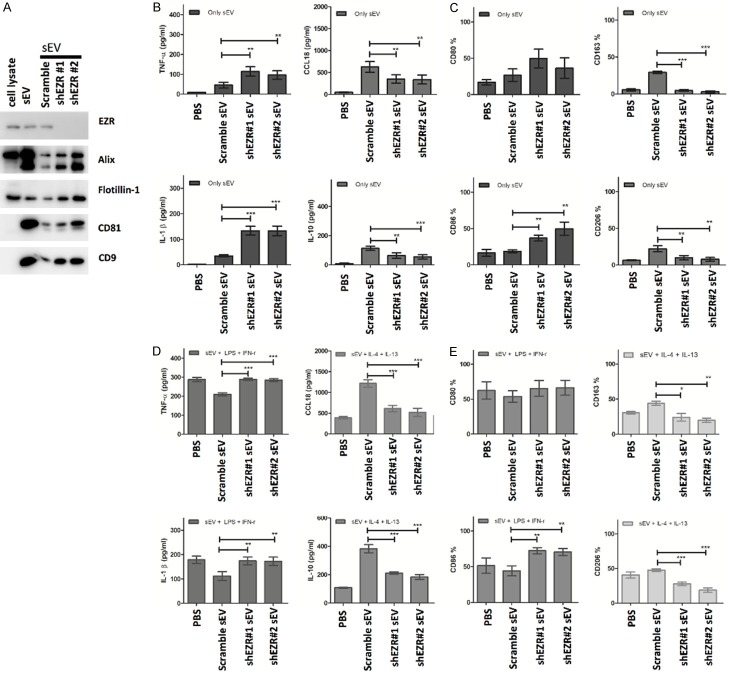
We next determined the role of PDAC-derived sEV-EZR in the polarization of M1/M2 phenotype macrophages. EZR expression in PC084 cells was knocked down with shRNA (PC084-shEZR). Afterward, the EZR levels in sEVs derived from PC084 cells transfected with EZR shRNA were verified by Western blotting (Figure 8A).
Figure 8.
sEV-EZR regulates macrophage polarization. A. Western blot analysis of expression EZR in PC084-derived sEVs with knockdown EZR by shRNA (#1 and #2). B. ELISA analysis of M1 (TNF-α and IL-1-β, left-panel) and M2 cytokines (CCL18 and IL10, right panel) in CD14+ PBMCs-derived macrophages incubated with PC084-shEZR-derived sEVs or PC084-scramble controls-derived sEVs for 72 hrs. Data represented means ± SD from 4 independent experiments (each experiment contains 2 technical replicates). C. Flow cytometry analysis of M1 (CD80 and CD86, left-panel) and M2 (CD163 and CD206, right panel) cell surface markers in CD14+ PBMCs-derived macrophages incubated with PC084-shEZR-derived sEVs or PC084-scramble controls-derived sEVs for 72 hrs. Data represented means ± SD from 4 independent experiments. D. ELISA analysis of M1 or M2 cytokines in CD14+ PBMCs-derived macrophages treated with LPS+INF-γ (M1 induction cytokine, left panel) or IL-4+IL-13 (M2 induction cytokine, right panel) co-incubated with sEVs for 72 hrs. Data represented means ± SD from 4 independent experiments. E. Flow cytometry analysis of M1 and M2 surface markers on CD14+ PBMCs-derived macrophages treated with LPS+INF-γ (M1 induction cytokine, left panel) or IL-4+IL-13 (M2 induction cytokine, right panel) co-incubated with sEVs for 72 hrs. Data represented means ± SD from 4 independent experiments. Level of significance was determined using Student’s t-test. *P<.05, **P<.005, ***P<0.001.
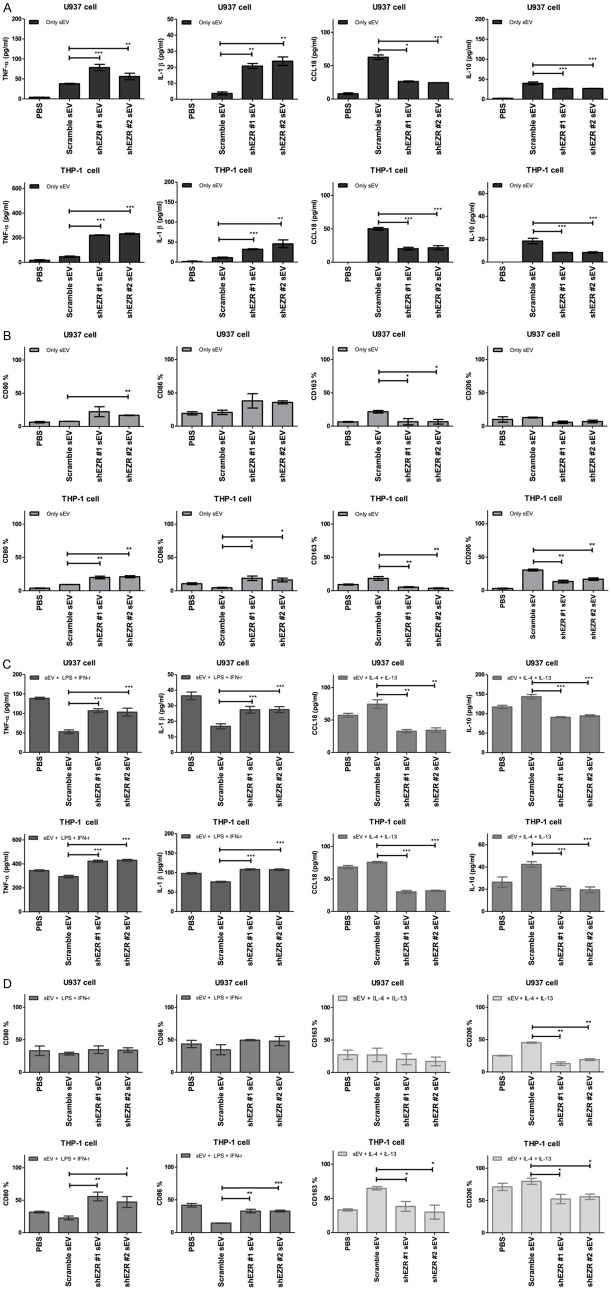
Freshly isolated CD14+ PBMCs-derived macrophages were incubated with the PC084-shEZR-derived sEVs for 72 hr. The amount of M1 cytokines IL-1β and TNF-α treated with PC084-shEZR-derived sEVs is found to be higher than that of PC084-scramble controls (Figure 8B). A significant decrease was found in M2 cytokines CCL18 and IL-10 treated with PC084-shEZR-derived sEVs, compared with PC084-scramble controls (Figure 8B). Flow cytometry showed the decreased macrophages with M2 markers (CD163 and CD206) and increased macrophages with M1 markers (CD80 and CD86) in CD14+ PBMCs-derived macrophages treated with PC084-shEZR-derived sEVs, compared with the macrophages treated with PC084-scramble controls (Figure 8C). The amount of M1 cytokines IL-1β and TNF-α in CD14+ PBMCs-derived macrophages treated with LPS, IFN-γ, and PC084-shEZR-derived sEVs is found to be greater than that in CD14+ PBMCs-derived macrophages treated with LPS, IFN-γ, and PC084-scramble controls (Figure 8D). A significant decrease of M2 cytokines CCL18 and IL-10 in CD14+ PBMCs-derived macrophage treated with IL-4, IL-13, and PC084-shEZR-derived sEVs is observed in contrast to CD14+ PBMCs-derived macrophage treated with IL-4 and IL-13, and PC084-scramble controls (Figure 8E). Similarly, flow cytometry showed the decreased macrophages with M2 markers (CD163 and CD206) in CD14+ PBMCs-derived macrophages treated with IL-4, IL-13, and PC084-shEZR-derived sEVs, compared with the macrophages treated with IL-4, IL-13 and PC084-scramble controls. Macrophages with M1 markers (CD80 and CD86) were increased in CD14+ PBMCs-derived macrophages treated with LPS, IFN-γ, and PC084-shEZR-derived sEVs, compared with the macrophages treated with LPS, IFN-γ, and PC084-scramble controls (Figure 8E). Similar patterns in terms of sEV-induced macrophage polarization can be found in human THP-1 and U937 cell lines (Figure 9).
Figure 9.
sEV-EZR suppresses M1 polarization and induce M2 polarization in THP-1/U937 derived macrophages. A. ELISA analysis of M1 (TNF-α and IL-1-β, left panel) and M2 cytokines (CCL18 and IL10, right panel) in THP-1/U937 derived macrophages which incubated with PC084-shEZR-derived sEVs or PC084-scramble-derived sEVs for 72 hrs. Data represented means ± SD from 3 independent experiments (each experiment contains 2 technical replicates). B. Flow cytometry analysis of M1 (CD80 and CD86, left panel) and M2 (CD163 and CD206, right panel) cell surface markers in THP-1/U937 derived macrophages which incubated with PC084-shEZR-derived sEVs or PC084-scramble-derived sEVs for 72 hrs. Data represented means ± SD from 3 independent experiments. C. ELISA analysis of M1 or M2 cytokines in THP-1/U937 derived macrophages treated with LPS+INF-γ (M1 induction cytokine, left panel) or IL-4+IL-13 (M2 induction cytokine, right panel), co-incubated with sEVs (sEVs derived from PC084-shEZR or PC084-scramble cells) for 72 hrs. Data represented means ± SD from 3 independent experiments (each experiment contains two technical replicates). D. Flow cytometry analysis of M1 and M2 surface markers on THP-1/U937 derived macrophages which treated with LPS+INF-γ (M1 induction cytokine, left panel) or IL-4+IL-13 (M2 induction cytokine, right panel), co-incubated with sEVs (sEVs derived from PC084-shEZR or PC084-scramble cells) for 72 hrs. Data represented means ± SD from 3 independent experiments. Level of significance was determined using Student’s t-test. *P<.05, **P<.005, ***P<0.001.
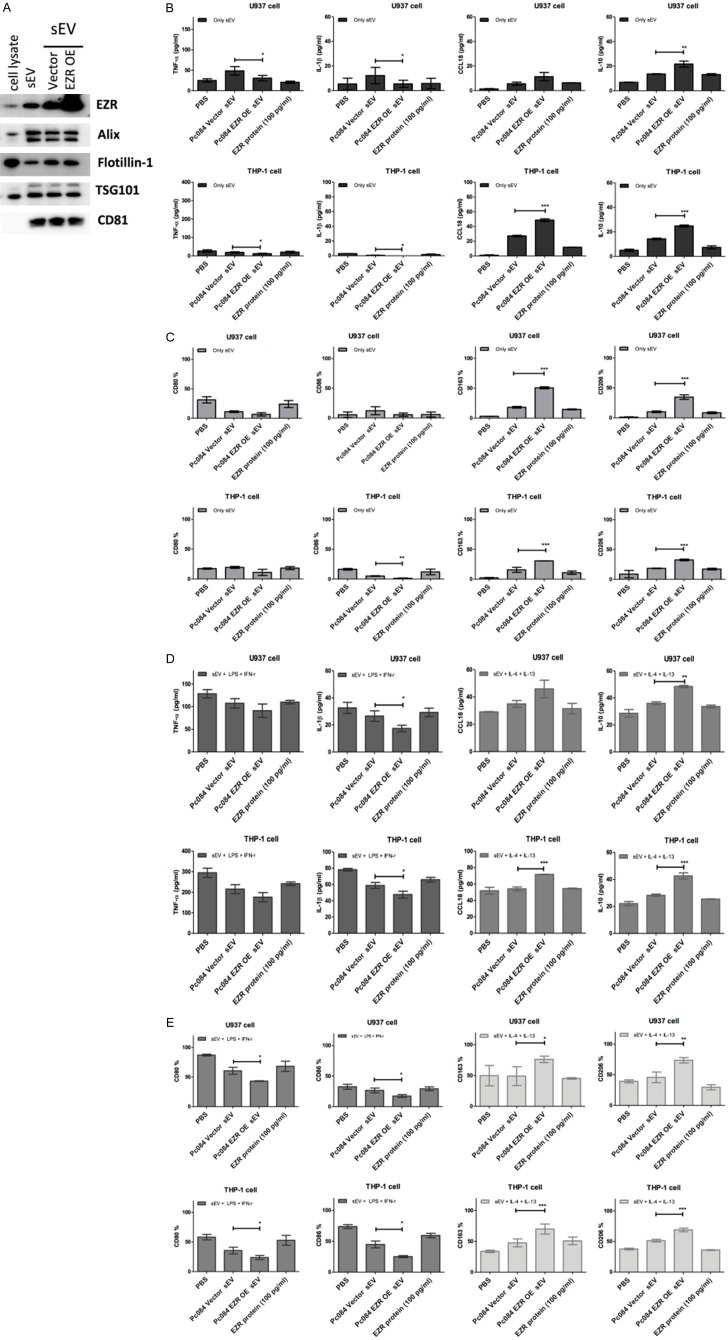
Note that overexpression EZR in PC084-derived sEVs showed the contrary results in terms of the amounts of M1/M2 cytokines and the amounts of macrophages with M1/M2 markers, respectively, when compared with PC084-derived sEVs transfected with vector only (Figure 10).
Figure 10.
PC084 EZR-overexpressed sEVs induce M2 macrophage polarization in THP-1/U937 derived macrophages. A. Western blot analysis of expression of EZR in sEVs derived from PC084-EZR or PC084-Vector. B. ELISA analysis of M1 (TNF-α and IL-1-β, left-panel) and M2 cytokines (CCL18 and IL10, right panel) in THP-1/U937 derived macrophages which incubated with PC084-EZR-derived sEVs, PC084-Vector-derived sEVs, or 100 pg/ml EZR recombinant protein for 72 hrs. Data represented means ± SD from 3 independent experiments (each experiment contains 2 technical replicates). C. Flow cytometry analysis of M1 (CD80 and CD86, left-panel) and M2 (CD163 and CD206, right panel) cell surface markers on THP-1/U937 derived macrophages co-incubated with PC084-EZR-derived sEVs, PC084-Vector-derived sEVs, or 100 pg/ml EZR recombinant protein for 72 hrs. Data represented means ± SD from 3 independent experiments. D. ELISA analysis of M1 or M2 cytokines in THP-1/U937 derived macrophages treated with LPS+INF-γ (M1 induction cytokine, left panel) or IL-4+IL-13 (M2 induction cytokine, right panel) co-incubated with 100 pg/ml EZR recombinant protein, sEVs derived from PC084-EZR or PC084-Vector for 72 hrs. Data represented means ± SD from 3 independent experiments (each experiment contains two technical replicates). E. Flow cytometry analysis of M1 and M2 surface markers on THP-1/U937 derived macrophages which treated with LPS+INF-γ (M1 induction cytokine, left panel) or IL-4+IL-13 (M2 induction cytokine, right panel) co-incubated with 100 pg/ml EZR recombinant protein or sEVs derived from PC084-EZR or PC084-Vector for 72 hrs. Data represented means ± SD from 3 independent experiments. Level of significance was determined using Student’s t-test. *P<.05, **P<.005, ***P<0.001.
PDAC-derived sEV-EZR promotes PDAC metastasis to the liver
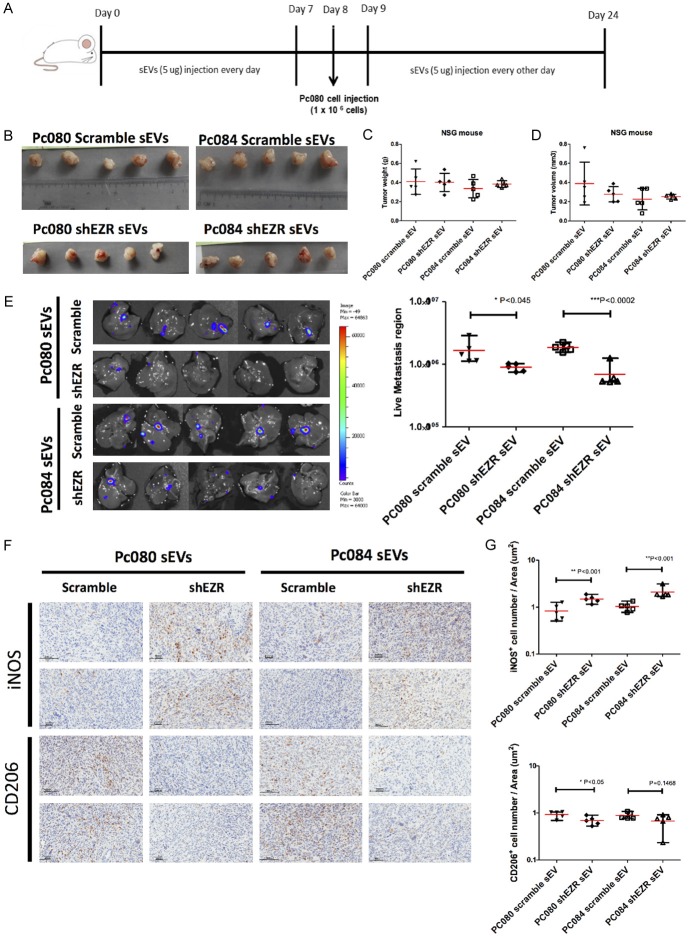
To test the potential roles of sEV-EZR in tumor metastasis, we injected 5 mg of PC084- and PC080-shEZR-derived sEVs into immunocompromised NSG mice once daily for 7 days. On day 8, we implanted 1 × 106 PC080 cells into the pancreas of each NSG mouse, which developed into aggressive orthotropic tumors that metastasized to the liver. On day 9, mice were treated with PC080-shEZR-derived sEVs, PC084-shEZR-derived sEVs, PC080-scramble controls, or PC084-scramble controls. Subsequent injections were administered every other day for a total of 7 injections. The metastasis burden was determined using non-invasive imaging of bioluminescence (IVIS). On day 21, all mice were euthanized, and the xenograft tumors were collected (Figure 11A). Whichever sEVs, PC080-shEZR-derived sEVs, PC084-shEZR-derived sEVs, PC080-scramble controls, or PC084-scramble controls, similar sizes and weights of the orthotropic tumors have been observed (Figure 11B-D). The amount of liver metastasis in PC080-shEZR-derived sEVs and PC084-shEZR-derived sEVs-treated mice was observed to be smaller than that of PC080-scramble controls and PC084-scramble controls (Figure 11E). We found an increase in M1 TAM and a decrease in M2 TAM in orthotropic tumors treated with PC080-shEZR-derived sEVs and PC084-shEZR-derived sEVs, compared with PC080-scramble controls and PC084-scramble controls (Figure 11F, 11G).
Figure 11.
PDAC-derived sEV-EZR promotes PDAC metastasis and increases M2 macrophages in animal models. (A) Schematic illustration of animal study setup and time course. PDAC-derived sEVs (PC080 or PC084 scramble sEVs and shEZR-sEVs) were administered every day for 1 week before PC080 cells injection in NSG mice then were administered every other day until day 24. (B) At day 24, PC080 tumors were harvested and measured by tumor weight (C) and tumor volume (D). Data analyzed by using the Student t-test (E) IVIS images showed liver metastasis at day 24. The bioluminescence photon counts in the region of liver metastasis (n=5) measured by IVIS software. (F) Representative IHC images and (G) the bar chart showed the percentage of iNOS+ (M1 macrophage marker) and CD206+ (M2 macrophage marker) cells in mouse pancreatic cancer tissues. Each dot represents the datum from one mouse. Scale bar, 100 μm. 40 × magnification. Data represented means ± SD. Level of significance was determined using Student’s t-test. *P<.05, **P<.005, ***P<0.001, or using ANOVA test. *P<0.05.
Discussion
sEVs or exosomes are considered as key components in intercellular communication in cancer and play an important role in immune modulation [31,32]. It has been demonstrated that in tumor microenvironment, TAMs which mostly express M2 markers and participate in tumor metastasis, are the most effective cells for tumor progression [33]. In this study, we have shown that PDAC-derived sEV-EZR regulated macrophage polarization into M2 phenotype and promoted PDAC metastasis. In addition, the plasma level of sEV-EZR is a prognostic marker for PDAC patients. We observed that the overall survival is shorter in the high plasma sEV-EZR PDAC patients than that in the low sEV-EZR patients. Our study proposed that PDAC cells secrete EZR-rich sEVs into the tumor microenvironment, which in turn polarizes macrophages that promote tumor metastasis in PDAC (Figure 12). Hence, targeting the interaction between PDAC-derived sEV-EZR and macrophages might be a potential therapeutic strategy for PDAC.
Figure 12.

A proposed model for PDAC cell-derived sEV-EZR in regulating macrophages polarization and promoting liver metastasis. Cancer cells (blue) sEV-EZR (red), macrophages (purple).
Cancer cells could promote tumor progression by evading immune check through intercellular interactions within the tumor microenvironment. Tumor-derived sEVs are being recognized as essential mediators of intercellular communication between cancer and immune cells. They can direct immune cells toward a tumor-promoting phenotype, and significantly contribute to different aspects of tumor progression, including invasion of the surrounding tissues, angiogenesis, formation of pre-metastatic niches, and metastatic dissemination [34-36]. Tumor-derived sEVs elicit immune-evasion effects through several mechanisms. sEV miRNAs including miR-155, miR-125, and miR-21 has been reported on the regulation of macrophage activation [37]. These results suggest the significant role of sEVs in the regulation of TAMs. TAMs are the prominent component of stromal cells in many solid tumors including PDAC [38]. TAMs can pave the way for cancer-cell dissemination from the primary tumor site by producing proteolytic enzymes that digest the extracellular matrix, thereby contributing to metastasis [39]. The presence of TAMs in many solid tumors is associated with poor prognosis [40]. Two major polarization states have been described for macrophages phenotype: M1 and M2. The phenotype of macrophages within tumor microenvironment switches from anti-tumor M1 to pro-tumor M2 types [41]. In the early stage of lung cancer development, TAMs show M1 polarization and exert their antitumor effects, but in the late stage TAMs switch to the M2-like state to participate in tumor progression [33]. Su et al. had reported that PDAC cell line-derived sEVs containing miR-155 had a dose-dependent effect on macrophage polarization [42]. In this study, we have shown that PDAC-derived sEVs can polarize the macrophage into M2 phenotype, and that the M2 macrophages increased in advanced stages of PDAC tissues. These results confirm the role of sEVs on the regulation of macrophage polarization in PDAC, and are consistent with the findings from previous studies. Regarding how PDAC-derived sEVs regulate the macrophage polarization, we found that EZR in PDAC-derived sEVs plays an important role in regulation of the macrophage polarization into pro-tumor M2 phenotype, and that knockdown EZR in PDAC-derived sEVs attenuates the potential of polarizing macrophages into M2 phenotype. Previous studies have showed that STAT3 and STAT6 were involved in the polarization of M1 to M2 macrophage phenotypes [43] and that EZR overexpression could regulate STAT3 activation [44]. The mechanisms of sEV-EZR regulating the macrophage polarization via STAT3 or STAT6 remain to be elucidated.
EZR is a member of the ERM family, which regulates membrane remodeling [38]. ERM proteins have been reported to serve as intracellular scaffolds, which facilitate signal transduction and regulate B- and T-cell activation [45,46]. Previous studies have shown that EZR expression is upregulated in PDAC and lung cancer and is related to colon cancer progression [25,26,47,48]. In this study, we initially found that the EZR level is higher in sEVs derived from PDAC cell lines than in those from HPDE cells by LC-MS/MS analysis. Next, we observed that the EZR level appears to be higher in sEVs derived from PDAC-patient plasma than in those from non-cancer controls. Although the EZR level in plasma sEVs did not correlate with the tumor stage, the OS in the high plasma-sEV-EZR PDAC patients was statistically significant shorter than that in the low plasma-sEV-EZR patients. In addition, we found an increase in M1 TAMs, a decrease in M2 TAMs in orthotropic tumors, and a decrease in the amount of liver metastasis treated with PDAC-derived sEVs whose EZR was knocked down in the PDAC animal model. These results were the first to unveil the roles of sEV-EZR in the regulation of macrophage polarization and in the tumor progression of PDAC.
The discovery of reliable biomarkers for the early detection of PDA is the ‘holy grail’ to improve the dismal outcomes of PDAC. Circulating sEVs in blood containing a reliable source of cancer-associated molecule have clinical utility in cancer diagnosis and management. It is reported that circulating sEV-glypican-1 potentially enables the detection of PDAC with an accuracy of 100% [49]. However, we were not able to confirm did not observe the overexpression of glypican-1 in the PDAC-derived sEVs or in the plasma sEVs from the PDAC patients. Instead, we have analyzed the plasma sEV-EZR as a diagnostic biomarker for PDAC. At the cut-off value of 27.08 pg/ml, plasma sEV-EZR yielded an SN of 55.2%, an SP of 71.5%, and an AUC of 0.661 for distinguishing PDAC patients from controls. As a result, the plasma sEV-EZR is not an ideal diagnostic biomarker for PDAC, but it can be a prognostic marker.
There is no universally accepted approach to isolating and characterizing sEVs, especially from human blood. Various international professional societies have developed guidelines for standardizing protocols for the study of EVs and their biomarker development [50]. Varying methodologies for isolating sEVs include ultracentrifugation, density gradient centrifugation, antibody affinity columns, and precipitation-ultracentrifugation techniques. Of these methods, ultracentrifugation plus density gradient centrifugation appears to be one of the most commonly used technique worldwide but are both time- and labor-intensive. Density gradient centrifugation requires substantial biofluid volume therefore limiting point-of-care clinical translation. We have used simple self-made size-exclusion chromatography (SEC) columns to rapidly isolate sEVs from small input quantities of human plasma with high purity and reproducibility. Further efforts are needed to standardize the isolation and characterization of sEVs from human plasma for clinical uses.
In conclusion, this is the first study to show that sEV-EZR could regulate macrophage polarization and promote metastasis in PDAC. The sEV-EZR polarizes macrophages into a pro-tumor, instead of anti-tumor, phenotype, and hence, has a significant impact on the survival of PDAC patients. Thus, targeting sEV-EZR to inhibit its detrimental effects on macrophages and manipulating sEV content to reprogram TAMs toward M1 phenotype can be considered a promising therapeutic strategy to curb PDAC metastasis.
Disclosure of conflict of interest
None.
References
- 1.Erkan M. Understanding the stroma of pancreatic cancer: co-evolution of the microenvironment with epithelial carcinogenesis. J Pathol. 2013;231:4–7. doi: 10.1002/path.4213. [DOI] [PubMed] [Google Scholar]
- 2.Zhan HX, Zhou B, Cheng YG, Xu JW, Wang L, Zhang GY, Hu SY. Crosstalk between stromal cells and cancer cells in pancreatic cancer: new insights into stromal biology. Cancer Lett. 2017;392:83–93. doi: 10.1016/j.canlet.2017.01.041. [DOI] [PubMed] [Google Scholar]
- 3.Feig C, Gopinathan A, Neesse A, Chan DS, Cook N, Tuveson DA. The pancreas cancer microenvironment. Clin Cancer Res. 2012;18:4266–4276. doi: 10.1158/1078-0432.CCR-11-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams CB, Yeh ES, Soloff AC. Tumor-associated macrophages: unwitting accomplices in breast cancer malignancy. NPJ Breast Cancer. 2016:2. doi: 10.1038/npjbcancer.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye H, Zhou Q, Zheng S, Li G, Lin Q, Wei L, Fu Z, Zhang B, Liu Y, Li Z, Chen R. Tumor-associated macrophages promote progression and the Warburg effect via CCL18/NF-kB/VCAM-1 pathway in pancreatic ductal adenocarcinoma. Cell Death Dis. 2018;9:453. doi: 10.1038/s41419-018-0486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meng F, Li W, Li C, Gao Z, Guo K, Song S. CCL18 promotes epithelial-mesenchymal transition, invasion and migration of pancreatic cancer cells in pancreatic ductal adenocarcinoma. Int J Oncol. 2015;46:1109–1120. doi: 10.3892/ijo.2014.2794. [DOI] [PubMed] [Google Scholar]
- 7.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chávez-Galán L, Olleros ML, Vesin D, Garcia I. Much more than M1 and M2 macrophages, there are also CD169(+) and TCR+ macrophages. Front Immunol. 2015;6:263. doi: 10.3389/fimmu.2015.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarique AA, Logan J, Thomas E, Holt PG, Sly PD, Fantino E. Phenotypic, functional, and plasticity features of classical and alternatively activated human macrophages. Am J Respir Cell Mol Biol. 2015;53:676–688. doi: 10.1165/rcmb.2015-0012OC. [DOI] [PubMed] [Google Scholar]
- 10.Helm O, Held-Feindt J, Schäfer H, Sebens S. M1 and M2: there is no “good” and “bad”-How macrophages promote malignancy-associated features in tumorigenesis. Oncoimmunology. 2014;3:e946818. doi: 10.4161/21624011.2014.946818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoger JL, Gijbels MJ, van der Velden S, Manca M, van der Loos CM, Biessen EA, Daemen MJ, Lutgens E, de Winther MP. Distribution of macrophage polarization markers in human atherosclerosis. Atherosclerosis. 2012;225:461–468. doi: 10.1016/j.atherosclerosis.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 12.Genard G, Lucas S, Michiels C. Reprogramming of tumor-associated macrophages with anticancer therapies: radiotherapy versus chemo- and immunotherapies. Front Immunol. 2017;8:828. doi: 10.3389/fimmu.2017.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu H, Hang JJ, Han T, Zhuo M, Jiao F, Wang LW. The M2 phenotype of tumor-associated macrophages in the stroma confers a poor prognosis in pancreatic cancer. Tumour Biol. 2016;37:8657–8664. doi: 10.1007/s13277-015-4741-z. [DOI] [PubMed] [Google Scholar]
- 14.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Toro J, Herschlik L, Waldner C, Mongini C. Emerging roles of exosomes in normal and pathological conditions: new insights for diagnosis and therapeutic applications. Front Immunol. 2015;6:203. doi: 10.3389/fimmu.2015.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah R, Patel T, Freedman JE. Circulating extracellular vesicles in human disease. N Engl J Med. 2018;379:958–966. doi: 10.1056/NEJMra1704286. [DOI] [PubMed] [Google Scholar]
- 17.Subramanian A, Gupta V, Sarkar S, Maity G, Banerjee S, Ghosh A, Harris L, Christenson LK, Hung W, Bansal A, Banerjee SK. Exosomes in carcinogenesis: molecular palkis carry signals for the regulation of cancer progression and metastasis. J Cell Commun Signal. 2016;10:241–249. doi: 10.1007/s12079-016-0338-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 19.Lässer C, Alikhani VS, Ekström K, Eldh M, Paredes PT, Bossios A, Sjöstrand M, Gabrielsson S, Lötvall J, Valadi H. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011;9:9. doi: 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raj DA, Fiume I, Capasso G, Pocsfalvi G. A multiplex quantitative proteomics strategy for protein biomarker studies in urinary exosomes. Kidney Int. 2012;81:1263–1272. doi: 10.1038/ki.2012.25. [DOI] [PubMed] [Google Scholar]
- 21.Dai S, Wei D, Wu Z, Zhou X, Wei X, Huang H, Li G. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol Ther. 2008;16:782–790. doi: 10.1038/mt.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooks T, Pateras IS, Jenkins LM, Patel KM, Robles AI, Morris J, Forshew T, Appella E, Gorgoulis VG, Harris CC. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun. 2018;9:771. doi: 10.1038/s41467-018-03224-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arpin M, Chirivino D, Naba A, Zwaenepoel I. Emerging role for ERM proteins in cell adhesion and migration. Cell Adh Migr. 2011;5:199–206. doi: 10.4161/cam.5.2.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berryman M, Franck Z, Bretscher A. Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. J Cell Sci. 1993;105:1025–1043. doi: 10.1242/jcs.105.4.1025. [DOI] [PubMed] [Google Scholar]
- 25.Piao J, Liu S, Xu Y, Wang C, Lin Z, Qin Y, Liu S. Ezrin protein overexpression predicts the poor prognosis of pancreatic ductal adenocarcinomas. Exp Mol Pathol. 2015;98:1–6. doi: 10.1016/j.yexmp.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Jin T, Jin J, Li X, Zhang S, Choi YH, Piao Y, Shen X, Lin Z. Prognostic implications of ezrin and phosphorylated ezrin expression in non-small cell lung cancer. BMC Cancer. 2014;14:191. doi: 10.1186/1471-2407-14-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang MC, Wu CH, Yang SH, Liang PC, Chen BB, Jan IS, Chang YT, Jeng YM. Pancreatic cancer screening in different risk individuals with family history of pancreatic cancer-a prospective cohort study in Taiwan. Am J Cancer Res. 2017;7:357–369. [PMC free article] [PubMed] [Google Scholar]
- 28.Kittan NA, Allen RM, Dhaliwal A, Cavassani KA, Schaller M, Gallagher KA, Carson WF 4th, Mukherjee S, Grembecka J, Cierpicki T, Jarai G, Westwick J, Kunkel SL, Hogaboam CM. Cytokine induced phenotypic and epigenetic signatures are key to establishing specific macrophage phenotypes. PLoS One. 2013;8:e78045. doi: 10.1371/journal.pone.0078045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quatromoni JG, Eruslanov E. Tumor-associated macrophages: function, phenotype, and link to prognosis in human lung cancer. Am J Transl Res. 2012;4:376–389. [PMC free article] [PubMed] [Google Scholar]
- 30.Almatroodi SA, McDonald CF, Darby IA, Pouniotis DS. Characterization of M1/M2 tumour-associated macrophages (TAMs) and Th1/Th2 cytokine profiles in patients with NSCLC. Cancer Microenviron. 2016;9:1–11. doi: 10.1007/s12307-015-0174-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McAndrews KM, Kalluri R. Mechanisms associated with biogenesis of exosomes in cancer. Mol Cancer. 2019;18:52. doi: 10.1186/s12943-019-0963-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14:195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaynagetdinov R, Sherrill TP, Polosukhin VV, Han W, Ausborn JA, McLoed AG, McMahon FB, Gleaves LA, Degryse AL, Stathopoulos GT, Yull FE, Blackwell TS. A critical role for macrophages in promotion of urethane-induced lung carcinogenesis. J Immunol. 2011;187:5703–5711. doi: 10.4049/jimmunol.1100558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, Xia H, Man Q, Zhong W, Antelo LF, Wu B, Xiong X, Liu X, Guan L, Li T, Liu S, Yang R, Lu Y, Dong L, McGettigan S, Somasundaram R, Radhakrishnan R, Mills G, Lu Y, Kim J, Chen YH, Dong H, Zhao Y, Karakousis GC, Mitchell TC, Schuchter LM, Herlyn M, Wherry EJ, Xu X, Guo W. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh CH, Tai SK, Yang MH. Snail-overexpressing cancer cells promote M2-like polarization of tumor-associated macrophages by delivering miR-21-abundant exosomes. Neoplasia. 2018;20:775–788. doi: 10.1016/j.neo.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li ZL, Lv LL, Tang TT, Wang B, Feng Y, Zhou LT, Cao JY, Tang RN, Wu M, Liu H, Crowley SD, Liu BC. HIF-1 alpha inducing exosomal microRNA-23a expression mediates the cross-talk between tubular epithelial cells and macrophages in tubulointerstitial inflammation. Kidney Int. 2019;95:388–404. doi: 10.1016/j.kint.2018.09.013. [DOI] [PubMed] [Google Scholar]
- 37.Squadrito ML, Etzrodt M, De Palma M, Pittet MJ. MicroRNA-mediated control of macrophages and its implications for cancer. Trends Immunol. 2013;34:350–359. doi: 10.1016/j.it.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melstrom LG, Salazar MD, Diamond DJ. The pancreatic cancer microenvironment: a true double agent. J Surg Oncol. 2017;116:7–15. doi: 10.1002/jso.24643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14:399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang QW, Liu L, Gong CY, Shi HS, Zeng YH, Wang XZ, Zhao YW, Wei YQ. Prognostic significance of tumor-associated macrophages in solid tumor: a meta-analysis of the literature. PLoS One. 2012;7:e50946. doi: 10.1371/journal.pone.0050946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang H, Declerck YA. Targeting the tumor microenvironment: from understanding pathways to effective clinical trials. Cancer Res. 2013;73:4965–4977. doi: 10.1158/0008-5472.CAN-13-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su MJ, Aldawsari H, Amiji M. Pancreatic cancer cell exosome-mediated macrophage reprogramming and the role of microRNAs 155 and 125b2 transfection using nanoparticle delivery systems. Sci Rep. 2016;6:30110. doi: 10.1038/srep30110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi JW, Kwon MJ, Kim IH, Kim YM, Lee MK, Nam TJ. Pyropia yezoensis glycoprotein promotes the M1 to M2 macrophage phenotypic switch via the STAT3 and STAT6 transcription factors. Int J Mol Med. 2016;38:666–674. doi: 10.3892/ijmm.2016.2656. [DOI] [PubMed] [Google Scholar]
- 44.Ghaffari A, Hoskin V, Szeto A, Hum M, Liaghati N, Nakatsu K, LeBrun D, Madarnas Y, Sengupta S, Elliott BE. A novel role for ezrin in breast cancer angio/lymphangiogenesis. Breast Cancer Res. 2014;16:438. doi: 10.1186/s13058-014-0438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaffer MH, Dupree RS, Zhu P, Saotome I, Schmidt RF, McClatchey AI, Freedman BD, Burkhardt JK. Ezrin and moesin function together to promote T cell activation. J Immunol. 2009;182:1021–1032. doi: 10.4049/jimmunol.182.2.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Treanor B, Depoil D, Bruckbauer A, Batista FD. Dynamic cortical actin remodeling by ERM proteins controls BCR microcluster organization and integrity. J Exp Med. 2011;208:1055–1068. doi: 10.1084/jem.20101125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang HJ, Zhu JS, Zhang Q, Sun Q, Guo H. High level of ezrin expression in colorectal cancer tissues is closely related to tumor malignancy. World J Gastroenterol. 2009;15:2016–2019. doi: 10.3748/wjg.15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan D, Wang S, Ye H, Xu S, Ye G. Ezrin expression in the primary hepatocellular carcinoma patients and associated with clinical, pathological characteristics. J Cancer Res Ther. 2016;12:C291–C294. doi: 10.4103/0973-1482.200761. [DOI] [PubMed] [Google Scholar]
- 49.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borràs FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MÁ, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Górecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzás EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D’Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekström K, El Andaloussi S, Elie-Caille C, Erdbrügger U, Falcón-Pérez JM, Fatima F, Fish JE, Flores-Bellver M, Försönits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gámez-Valero A, Gardiner C, Gärtner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Görgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ 2nd, Kornek M, Kosanović MM, Kovács ÁF, Krämer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lässer C, Laurent LC, Lavieu G, Lázaro-Ibáñez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Linē A, Linnemannstöns K, Llorente A, Lombard CA, Lorenowicz MJ, Lörincz ÁM, Lötvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG Jr, Meehan KL, Mertens I, Minciacchi VR, Möller A, Møller Jørgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-’t Hoen EN, Noren Hooten N, O’Driscoll L, O’Grady T, O’Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Østergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saá P, Sahoo S, Salas-Huenuleo E, Sánchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schøyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder OL 2nd, Soares RP, Sódar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ Jr, Veit TD, Vella LJ, Velot É, Verweij FJ, Vestad B, Viñas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yáñez-Mó M, Yin H, Yuana Y, Zappulli V, Zarubova J, Žėkas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]