Abstract
Introduction
Neurofibromatosis type 1 (NF1) is an autosomal dominantly inherited tumor predisposition syndrome with an incidence of one in 3000–4000 individuals with no currently effective therapies. The NF1 gene on chromosome 17 encodes neurofibromin, which functions as a negative regulator of RAS. NF1 is a chronic multi-system disorder affecting many different tissues. Due to cell-specific complexities of RAS signaling, therapeutic approaches for NF1 will likely have to focus on a particular tissue and manifestation of the disease.
Areas covered
In this review, we discuss the multi-system nature of NF1 and the signaling pathways affected due to deficiency of neurofibromin. We explore the cell/tissue-specific molecular and cellular consequences of aberrant RAS signaling in NF1 and speculate on their potential as therapeutic targets for the disease. We discuss recent genomic, transcriptomic and proteomic studies combined with molecular, cellular and biochemical analyses which have identified several targets for specific NF1 manifestations. We also consider the possibility of patient-specific gene therapy approaches to tackle NF1.
Expert opinion
The emergence of NF1 genotype-phenotype correlations, characterization of cell-specific signaling pathways affected in NF1, identification of novel biomarkers, and the development of sophisticated animal models accurately reflecting human pathology will continue to provide opportunities to develop therapeutic approaches to combat this multi-system disorder.
Keywords: Neurofibromas, neurofibromatosis type 1, peripheral nerve sheath tumors, RAS-MEK-ERK signaling, RASopathies
1. Introduction
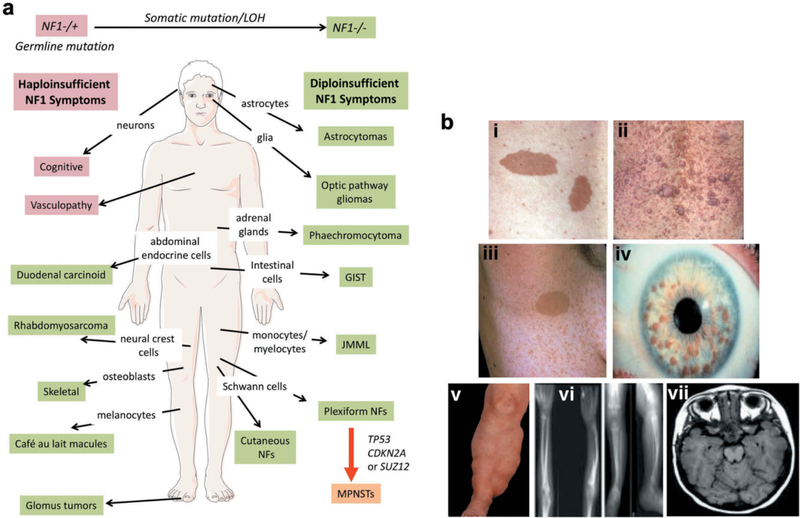
Neurofibromatosis type 1 (NF1: Online Mendelian Inheritance in Man (OMIM), #162200) is an autosomal dominantly inherited tumor predisposition syndrome first described by Friedrich Daniel von Recklinghausen in 1882. Affecting 1/3000–4000 individuals worldwide, it results from constitutional germline mutations of the NF1 gene which is located on the long arm of chromosome 17 [1]. Although the disease is usually fully penetrant by age 5, there is a high degree of variability and unpredictability in disease outcome, even between closely related family members. The major defining features of NF1 include café-au-lait macules (CALMs), cutaneous neurofibromas (CNF), plexiform neurofibromas (PNF), axillary freckling, optic pathway gliomas (OPG), Lisch nodules of iris and skeletal abnormalities [2] (Fig 1A). Patients are predisposed to developing many symptoms (Fig. 1B), including near universal benign, but often disfiguring, peripheral nerve associated tumors known as neurofibromas. About 10% of NF1 patients develop malignant peripheral nerve sheath tumors (MPNSTs) which carry a poor prognosis and are often fatal [3, 4]. NF1 patients are at increased risk to develop a variety of other tumors including optic pathway gliomas (OPG), astrocytic neoplasms, juvenile myelomonocytic leukemias (JMML), gastrointestinal stromal tumors (GIST), breast cancers, phaeochromocytomas, duodenal carcinoids, glomus tumor, juvenile xanthogranuloma and rhabdomyosarcomas [1]. While the tumor suppressor role of NF1 is well known, NF1 is a chronic multi-system disorder whose non-tumor symptoms contribute significantly to its morbidity. These include pigmentary lesions, vascular, skeletal abnormalities, and cognitive deficits [5]. No effective therapy for any NF1 symptom yet exists.
Figure 1. Neurofibromatosis type 1 (NF1) is a multisystem disorder.
A. NF1 patients are predisposed to developing symptoms affecting multiple cells of origin and tissues. Some manifestations associated with NF1, such as cognitive and vascular problems, result from haploinsufficiency of NF1. In contrast, other symptoms are triggered by somatic NF1 mutation/loss of heterozygosity (LOH) resulting in biallelic NF1 inactivation. Further, transformation of plexiform neurofibromas (NFs) into malignant peripheral nerve sheath tumors (MPNSTs) involves additional genetic events. Abbreviations: juvenile myelomonocytic leukemias (JMML) and gastrointestinal stromal tumors (GIST). B. The major defining features of NF1 include: (i) Café-au-lait macules, (ii) cutaneous neurofibromas, (iii) axillary freckling, (iv) Lisch nodules, (v) plexiform neurofibromas, (vi) thinning of long bone cortex and (vii) optic pathway glioma. Adapted from [2].
The NF1 gene encodes neurofibromin, a ubiquitously expressed protein, which functions as a RAS-GTPase Activating Protein (RAS-GAP), a negative regulator of RAS activity (Fig. 2). Consequently, neurofibromin deficiency leads to increased RAS signaling which is assumed to be the root cause of NF1 pathology. NF1 is classified as a RASopathy – a group of clinically related disorders which include Noonan syndrome, Noonan with multiple lentigines, Costello, cardiofacio-cutaneous, capillary malformation-arteriovenous malformation, gingival fibromatosis, and autoimmune lymphoproliferative syndrome that arise due to mutations in multiple genes encoding components of the RAS-MEK–ERK pathway [6].
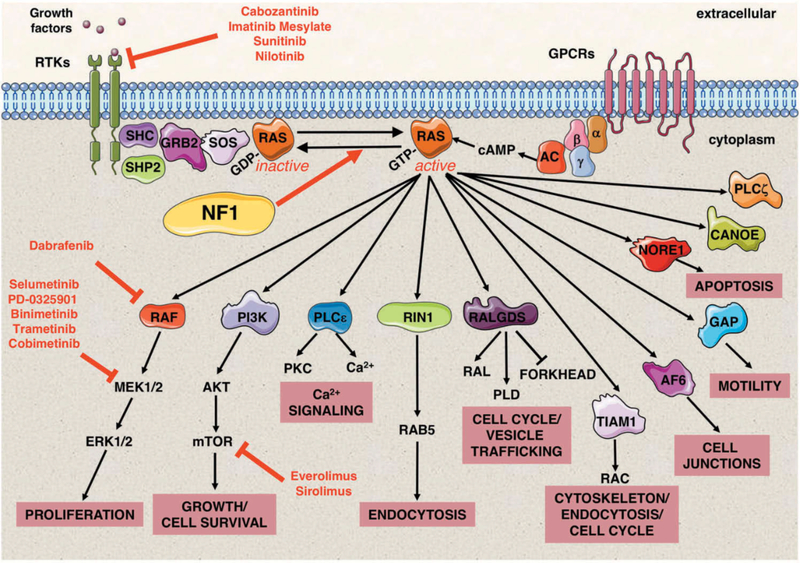
Figure 2. Neurofibromin is a negative regulator of the RAS signaling pathway.
RAS proteins function as fundamental signaling switches controlling a multitude of cellular processes including normal cell growth and differentiation. RAS proteins exist in two cellular states: an inactive GDP-bound form and an active GTP-bound form. Although RAS has a low intrinsic GTPase-activity, RAS-GTPase activating proteins (GAPs) stimulate this activity and consequently help to hydrolyze bound GTP back to GDP. Conversely, guanine nucleotide exchange factors (GEFs), e.g. SOS, convert RAS to its active GTP-bound form. NF1 encodes neurofibromin, a RAS-GAP, which is able to negatively regulate HRAS, KRAS, NRAS, MRAS, RRAS, and RRAS2 (TC21). Neurofibromin deficiency therefore results in elevated RAS signaling. RAS signaling transduces extracellular signals from ligand-activated receptors, both receptor tyrosine kinases (RTKs) and G protein-coupled receptors (GPCRs) at the cell surface through multiple pathways. RAS-GTP proteins activate a multitude of effector proteins, including RAF proteins (ARAF, BRAF, and CRAF) and the MEK–ERK signaling cascade, RalGDS, and the PI3 kinase family. The molecular and cellular events controlled by normal and aberrantly regulated RAS signaling are cell-type specific, depending on the expression of different RAS isoforms and their relative engagement of particular RAS effector proteins. Small molecule inhibitors to upstream regulators of RAS and its downstream effectors in NF1 clinical trials are shown in red. Abbreviations: Growth factor receptor-bound protein 2 (GRB2), src homology 2 domain (SHC), adenyl cyclase (AC), cyclic adenosine monophosphate (cAMP), α, β and γ represent the subunits of a heterotrimeric G protein complex.
In this review we focus on the molecular mechanisms and the signaling pathways that are affected by loss of neurofibromin and that result in NF1 disease progression. We explore potential targets for devising therapeutic strategies that may be useful in combating the variety of symptoms in NF1, including gene therapy approaches.
2. Neurofibromatosis type 1 (NF1) – a multisystem disorder
The occurrence of multiple neurofibromas is generally considered to be the hallmark of NF1. Neurofibromas are benign peripheral nerve sheath tumors and are a major cause of morbidity in NF1 patients. These tumors arise due to bi-allelic inactivation of the NF1 gene in a subpopulation of Schwann cells (SCs) and can be subdivided into five subtypes: cutaneous, subcutaneous, nodular or diffuse plexiform, spinal and atypical neurofibromatous neoplasms of uncertain biologic potential (ANNUBP). CNFs are present in almost all adult patients in the dermis and are associated with single nerve endings. Subcutaneous neurofibromas are located in epidermis and are evident on palpation of skin. Diffuse PNFs are found in 30–50% of NF1 patients, although MRI has revealed internal PNFs in over 50% of NF1 young adults [7]. PNFs are usually congenital and generally much larger than CNFs and are associated with major nerve trunks and nerve plexi and can be associated with major disfigurement. Further complications from large PNFs can arise with compression of vital organs and can be debilitating when their overgrowth involves entire limbs or body segments. PNFs like CNFs, comprise a mixture of SCs, perineurial-like cells, mast cells, macrophages, fibroblasts, blood vessels and extracellular matrix and have a highly infiltrative nature, complicating surgery (Fig. 3). Spinal neurofibromas can occur at single or multiple nerve roots and are associated with both sensory and motor deficits. ANNUBPs (atypical neurofibromas) exhibit hypercellularity and atypical nuclei but few mitoses and no necrosis. They reveal molecular changes that are also found in malignant peripheral nerve sheath tumors (MPNSTs) and hence referred to as pre-malignant [8].
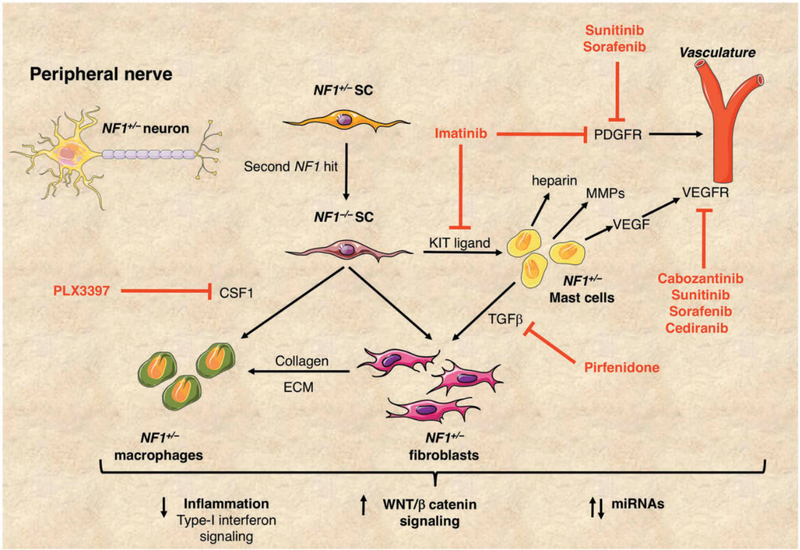
Figure 3. Plexiform neurofibromas in NF1.
Plexiform neurofibromas (PNFs) develop from NF1+/− Schwann cells (SC) with a second hit resulting in NF1 biallelic inactivation (NF1−/−). These tumors comprise a mixture of SCs, mast cells, macrophages and fibroblasts intrinsic to the peripheral nerve. The growth of PNFs depends on the complex interplay between these cell types. KIT ligand is secreted by NF1−/− SCs and acts as a chemo-attractant for NF1+/− mast cells. In turn, NF1+/− mast cells produce TGFβ, stimulating NF1+/− fibroblasts to increase collagen production and other extracellular matrix (ECM) proteins. NF1+/− mast cells also produce heparin, vascular endothelial growth factor (VEGF) and matrix metalloproteinases (MMPs) promoting tumor vascularization and tumor invasiveness. NF1−/− SCs secrete colony-stimulating factor (CSF1) thereby recruiting macrophages, aiding tumor progression. Small molecule inhibitors in NF1 clinical trials are shown in red. Recent reports have highlighted the importance of inflammation, increased signaling through the WNT/β-catenin pathway and misregulated miRNAs in PNFs, all of which may represent potential therapeutic targets.
Critically, 10–15% of benign PNFs develop into aggressive MPNSTs, via atypical or deep-seated neurofibromas [9]. In contrast, CNFs rarely undergo malignant transformation. MPNSTs are the main cause of mortality in NF1 [3] and surgery is the primary means of treating these highly aggressive sarcomas, while adjuvant radiation therapy and/or chemotherapy show no clear survival benefits.
The most common non-neoplastic manifestations in NF1 are pigmentary features, including CALMs which are dense populations of melanocytes observed in 99% of NF1 patients and are used in early childhood diagnosis. Cognitive and behavioural deficits occur in 65–80% of children with NF1. Specific learning disabilities affecting performance in visuospatial tasks, literacy skills, and oral and written expression often lead to poor academic performance [10]. Further, attention-deficit hyperactivity disorder (ADHD) symptoms such as decreased impulse control and planning ability have also been observed [11]. Other non-tumor symptoms in NF1 include skeletal abnormalities, iris hamartomas (Lisch nodules), and reduced overall growth. NF1 patients are also at increased risk of developing neurofibromatous neuropathies [12]. Cardiovascular manifestations of NF1 include vasculopathy, hypertension and congenital heart defects. Patients with constitutional mismatch repair deficiency (CMMRD) and Legius syndrome (caused by germline SPRED1 mutation) may have overlapping clinical symptoms, underlying the need for molecular diagnosis [13].
3. Genetic alterations causing NF1
About one half of NF1 cases are sporadic caused by de novo NF1 mutations. 5% of NF1 patients have segmental NF1 involving one segment of the body, such as a single limb. Such cases are the result of genetic mosaicism with NF1 mutations occurring in the developing embryo post-fertilization [14]. Currently, over 3000 different inherited mutations in NF1 have been reported in the Human Gene Mutation Database (HGMD®) as a cause of NF1 (www.hgmd.cf.ac.uk). Mutational analysis of the NF1 gene can identify 95% causative mutations in patients with classic NF1 [15]. Mutations vary in size from large genomic deletions spanning several megabases, to single base-pair substitutions (missense/nonsense) altering an encoded amino acid or splice site function. The majority (>80%) of germline NF1 mutations are predicted to be inactivating, resulting in almost complete absence of transcript or protein [16].
While some manifestations associated with NF1, such as cognitive problems, result from haploinsufficiency of NF1, others require a somatic mutation resulting in biallelic NF1 inactivation (the “two-hit” hypothesis, first proposed by Knudson) – as seen in the development of CALMs, neurofibromas, GIST, glomus tumors, JMML, bone abnormalities and phaechromocytoma [1] (Fig. 1B). Furthermore, additional genetic alterations such as mutation of TP53, CDKN2A or SUZ12, are required for the progression of plexiform neurofibromas into MPNSTs [9, 17, 18, 19].
The identification of somatic NF1 mutations in many non-NF1-associated sporadic cancers have been reported, including melanoma, glioblastoma, neuroblastoma, breast cancer, ovarian serous carcinoma, paraganglioma and phaeochromocytoma, lung adenocarcinoma, lung squamous cell carcinoma, bladder and colorectal cancer, and leukaemia (reviewed in [20]. Neurofibromin is therefore likely to play both a critical and a general role in cancer, far beyond that evident in NF1. Elucidation of the role of NF1 mutations in the initiation and progression of these tumors, including how they confer resistance or sensitivity to a therapeutic intervention, may provide insights into the mechanisms underlying tumor development in NF1.
The complex natural history of NF1 and the relatively small number of recurrent mutations has hindered correlating NF1 genotype and disease outcome. So far only four definite genotype-phenotype correlations are reported. The clearest genotype-phenotype correlation is that of large constitutional NF1 deletions (1.4 Mb), encompassing the NF1 gene and 14 adjacent genes, which occur in 5 – 10% of NF1 cases. These deletions are associated with a more severe phenotype including learning disabilities and increased susceptibility to MPNSTs [21]. SUZ12, one of the 14 co-deleted genes, encodes a subunit of the chromatin remodelling complex Polycomb repressive complex 2 (PRC2) has been implicated as the responsible cooperating tumor suppressor [18, 19, 22]. Two germline mutations are associated with milder disease outcomes in NF1: individuals with deletion of Met 992 or missense mutation of Arg 1809 lack plexiform or cutaneous neurofibromas [23, 24, 25]. Furthermore, individuals with missense mutations of codons 844–848 have a high prevalence of a severe phenotype, including plexiform and symptomatic spinal neurofibromas, symptomatic optic pathway gliomas, other malignant neoplasms, as well as bone abnormalities [26]. Such genotype-phenotype correlations help to highlight amino acid residues or regions of neurofibromin with important functions that may be restricted to particular tissue/cell types and thereby only result in certain clinical aspects of NF1 when mutated.
The extraordinary variation between disease outcome in individuals with the same germline NF1 mutation points toward the strong effects of modifier genes. Studies of twins with NF1 have revealed that each major symptom associated with NF1 is likely to be affected by distinct genetic modifiers [27, 28, 29]. Knowledge of these modifiers could potentially implicate pathways that impinge significantly on NF1 deficiency; however, the search for definite modifiers continues.
4. Neurofibromin - regulator of RAS signaling
The NF1 gene encodes neurofibromin, a 2818 amino acid protein, ubiquitously expressed but with its highest levels found in cells of the central nervous system (CNS). Neurofibromin functions as 1 of at least 12 human RAS-GTPase activating protein (GAP)-related proteins to negatively regulate RAS [30]. All RAS proteins exist in two cellular states; while the majority of RAS is found in its inactive GDP-bound form, only a very small fraction is present in its active GTP-bound form. Neurofibromin is able to down-regulate activated GTP-bound RAS by stimulating the low intrinsic GTPase-activity of the RAS proteins themselves, thereby promoting their conversion into the inactive RAS-GDP state (Fig. 2). Consequently, mutations in NF1 which compromise neurofibromin function result in sustained intracellular levels of active RAS-GTP, leading to prolonged activation of downstream signaling pathways. RAS signaling transduces extracellular signals from ligand-activated receptors at the cell surface through multiple pathways that ultimately result in post-translational modifications of proteins (e.g. phosphorylation) or transcriptional alterations, mediating signals responsible for proliferation and survival [31].
The predicted outcome of NF1 deficiency in a particular cell type is confounded due to multiple contextual aspects of RAS signaling [31]. Firstly, neurofibromin has been demonstrated to not only negatively regulate HRAS, KRAS, and NRAS, but also MRAS, RRAS, and RRAS2 (TC21) [32]. RAS-GTP proteins are able to activate a multitude of effector proteins, including RAF proteins (ARAF, BRAF, and CRAF), RalGDS, and the PI3 kinase family (Fig. 2). Differences in the expression and engagement of specific RAS effector proteins between cell types and their relative importance on specific signaling pathways in a particular tissue adds further complexity when determining the outcome of neurofibromin deficiency.
Interaction between RAS and RAF results in the re-localisation of RAF to the plasma membrane where it activates the MEK–ERK signaling cascade to control proliferation. Activated RAS-GTP also stimulates PI3K/AKT signaling which protects cells from apoptosis. In the absence of functional neurofibromin, these pathways can become constitutively activated resulting in an increase in cell proliferation and survival. The RAF/MEK/ERK and PI3K/AKT pathways both activate mTOR signaling, a pathway found to be highly active in neurofibromas. Activation of the mTOR pathway has been shown to occur in the absence of growth factors, in both NF1 tumors and neurofibromin-deficient cultured cells [33]. Increased RAS activity in neurons, caused by NF1 haploinsuficiency, has been implicated in NF1-related learning deficiencies, possibly due to increased γ-aminobutyric acid (GABA)-mediated inhibitory neurotransmission [34].
To date the only well-established function of neurofibromin is the downregulation of RAS as a RAS-GAP. This activity is contained within its most highly conserved region - the centrally located GAP-related domain (GRD). However, many pathogenic NF1 missense mutations affect residues located outside of the GRD, suggesting other unknown functions of neurofibromin or important regulatory interactions either within the protein itself, or with other proteins. For example, mutated residues in neurofibromin flanking the GRD have been shown to be important for mediating its interaction with SPRED1 (Sprouty-related EVH1-domain containing protein) [35]. Interaction with SPRED1 is required to translocate neurofibromin to the plasma membrane, although it does not appear to affect GAP activity. Neurofibromin has been shown to associate with a large number of proteins and the functional characterization of these interactions may reveal new avenues for targeting NF1 [2].
5. Possible Therapeutic Approaches for NF1
The majority of drugs currently employed for treatment of NF1 are repurposed agents used against cancer (clinicaltrials.gov, clinicaltrialsregister.eu and Table 1). However, novel therapeutics are also being developed based upon unique aspects of NF1 pathology. High-throughput animal and human-derived genomic, transcriptomic, proteomic and methylome studies combined with molecular, cellular and biochemical analyses of signaling pathways have identified several targets for NF1 manifestations. Below we discuss possible gene therapy strategies to replace or restore the function of defective neurofibromin, small molecule inhibitor approaches to target generic RAS signaling components, as well as more specific pathways involved in NF1 that may represent targets for different manifestations of the disease.
Table 1.
Clinical Trials for NF1.
| Target | Therapeutic | Children/Adults | Clinical Trial Identifier | Sponsor | Phase | Active/Recruiting/Completed | Results Posted/Reference | Study Start Date |
|---|---|---|---|---|---|---|---|---|
| Plexiform neurofibromas | ||||||||
| MEK | Selumetinib | 2Y-18Y | NCI | II | Recruiting | 2011 | ||
| >18Y | NCI | I/II | Recruiting | 2015 | ||||
| >18Y | NCI | II | Recruiting | 2015 | ||||
| MEK | PD-0325901 | >16Y | UAB | II | Active | 2014 | ||
| MEK | Binimetinib | >1Y | UAB | II | Recruiting | 2017 | ||
| MEK/B-RAF | Trametinib + Dabrafenib | 1mth-17Y | Novartis | I | Recruiting | 2015 | ||
| mTOR | Everolimus (RAD001) | 18Y-60Y | Novartis | II | Completed | 2011 | ||
| >6Y | Novartis | II | Terminated | Yes | 2012 | |||
| 2Y-65Y | EUCTR-2016–001563–36 | Novartis | II | Recruiting | 2016 | |||
| mTOR | Sirolimus | 3Y-75Y | UAB | II | Completed | [133] | 2008 | |
| c-MET, VEGFR2 | Cabozantinib | >16Y | UAB | II | Active | 2014 | ||
| KIT | Pexidartinib | 3Y-35Y | NCI | I/II | Suspended | 2015 | ||
| ABL, c-KIT, PDGFR | Imatinib Mesylate | 3Y-65Y | IU | II | Completed | [65] | 2006 | |
| <18Y | EUCTR-2009–016922–15 | HSJD | II | Active | 2009 | |||
| 2Y-65Y | EUCTR-2012–000869–21 | IRCCS | I | Active | 2012 | |||
| 3Y-65Y | IU School of Medicine | I/II | Completed | Yes | 2012 | |||
| 2Y-21Y | St. Justine’s Hospital | II | Recruiting | 2014 | ||||
| RTK: PDGFR, VEGFR, RET, CSF-1R, Flt3 | Sunitinib | 3Y-65Y | IU | II | Suspended | Yes | 2012 | |
| RTK | Nilotinib | >18Y | IU | I | Completed | Yes | 2011 | |
| JAK/Stat | Peg-Interferon alpha-2b | 2Y-30Y | SHH | II | Active | Yes | 2008 | |
| 18 mth-21Y | Univ. of Pittsburgh | II | Completed | 2006 | ||||
| TGF-beta | Pirfenidone | 18Y-70Y | Mayo Clinic | II | Completed | [134] | 2000 | |
| 3Y-21Y | NCI | I | Completed | [135] | 2002 | |||
| RAF, VEGFR, PDGFR | Sorafenib | 3Y-18Y | NCI | I | Completed | [136] | 2008 | |
| VEGFR | Cediranib | >18Y | NCI | II | Terminated | Yes | 2006 | |
| RAS | Tipifarnib | 3Y-25Y | NCI | II | Completed | [137] | 2001 | |
| DHFR inhibitor/mitosis inhibitor | Methotrexate + Vinblastine | -25Y | CHoP | II | Completed | 2001 | ||
| Photosensitizer | Talaporfin sodium (LS11) | 3Y-21Y | CHoP | I | Terminated | 2008 | ||
| 14Y-30Y | MCW | II | Recruiting | 2016 | ||||
| Cutaneous neurofibromas | ||||||||
| MEK | Selumetinib | >18Y | NCI | II | Recruiting | 2017 | ||
| VEGF-A | Ranibizumab | >18Y | MGH | I | Completed | 2008 | ||
| mTOR | Everolimus | >18Y | UTHealth | II | Completed | Yes | 2015 | |
| mTOR | Sirolimus (rapamycin) | >13Y | UTHealth | I | Completed | 2009 | ||
| Photosensitizer | Levulan (5-aminolevulinic acid) | 18Y-90Y | MCW | I | Active | 2011 | ||
| Immune response modifier | Imiquimod | >18Y | MGH | I | Completed | 2009 | ||
| MPNSTs | ||||||||
| EGFR | Erlotinib | >18Y | NCI | II | Completed | [138] | 2003 | |
| ABL, c-KIT, PDGFR | Imatinib Mesylate | >10Y | NCI | II | Completed | [139] | 2002 | |
| RAF/VEGFR | Sorafenib | >18Y | NCI | II | Completed | [140] | 2005 | |
| RTK | Dasatinib | >13Y | SARC | II | Active | [141] | 2007 | |
| AURKA | Alisertib | >18Y | NCI | II | Completed | [142] | 2012 | |
| Granulocyte - colony stimulating factor (G-CSF) | Filgrastim | All ages | NCI | II | Completed | 2005 | ||
| BET | CPI-0610 | >18Y | UTSW | II | Recruiting | 2017 | ||
| RTK, mRTOR | PLX3397 + Sirolimus | >18Y | Columbia Univ. | I/II | Recruiting | 2015 | ||
| mTOR/VEGF-A | Everolimus + Bevacizumab | >18Y | SARC | II | Active | [143] | 2012 | |
| mTOR/HSP90 | Sirolimus + Ganetespib | >16Y | SARC | I | Active | 2013 | ||
| Anti-PD-1 immunotherapy | Pembrolizumab | >18Y | EUCTR-2015–004747–39/ | Oslo Univ. Hospital | II | Recruiting | 2015/2016 | |
| Gastrointestinal Stromal Tumors (GIST) | ||||||||
| MEK | Selumetinib | >18Y | NCI | II | Recruiting | 2017 | ||
| All tumors | ||||||||
| MEK | Binimetinib | >18Y | Array BioPharma | II | Completed | 2013 | ||
| MEK | Cobimetinib | 6 mth-30Y | Hoffmann-La Roche | I/II | Recruiting | 2016 | ||
| Neuroendocrine tumors | ||||||||
| RTK PDGFR, VEGFR, RET, CSF-1R, Flt3/mTOR | Sunitinib + Everolimus | >18Y | NCI | II | Recruiting | 2015 | ||
| Optic Pathway Glioma | ||||||||
| Anti-angiogenic | Lenalidomide | <21Y | NCI | II | Active | 2012 | ||
| Anti-angiogenic and anti-myeloma cell growth | Pomalidomide | 3Y-20Y | NCI | I | Active | 2015 | ||
| Low-Grade Gliomas | ||||||||
| RAF, VEGFR, PDGFR | Sorafenib | >2Y | NYUSM | II | Terminated | [144] | 2011 | |
| MEK | Selumetinib | 3Y-21Y | NCI | I/II | Recruiting | 2010 | ||
| MEK | Binimetinib | 1Y-18Y | CHLA | I/II | Recruiting | 2016 | ||
| EGFR/mTOR | Erlotinib + Sirolimus | <21Y | CRI | I | Completed | 2007 | ||
| mTOR | Everolimus | 1Y-21Y | UAB | II | Active | 2011 | ||
| Cytotoxic | Carboplatin + Vincristine | <9Y | NCI | III | Completed | [145] | 1997 | |
| Cytotoxic | Carboplatin + Vinblastine | <21Y | COG | I | Completed | 2006 | ||
| Mitosis inhibitor/RTK | Vinblastine + Nilotnib | <65Y | EUCTR-2012–003005–10 | Gustave Roussy | I/II | Active | 2012 | |
| Juvenile Myelomonocytic Leukemia | ||||||||
| TNF-alpha | Etanercept | 6mth-18Y | M.D. Anderson | II | Terminated | Yes | 2004 | |
| Cognition | ||||||||
| Dopamine–norepinephrine reuptake inhibitor | Methylphenidate | 7Y-12Y | Hospices Civils de Lyon | IV | Completed | [146] | 2004 | |
| HMG-CoA reductase inhibitor | Lovastatin | 8Y-15Y | UAB | II | Active | [120] | 2009 | |
| HMG-CoA reductase inhibitor | Simvastatin | <18Y | EUCTR-2009–010965–22 | Erasmus MC | III | Completed | [147, 148] | 2009 |
| Na-channel blocker | Lamotrigine | <18Y | EUCTR-2013–003405–26 | Erasmus MC | II | Active | 2013 | |
| 12Y-18Y | Erasmus MC | II | Recruiting | 2014 | ||||
| Autism | ||||||||
| HMG-CoA reductase inhibitor | Simvastatin | <18Y | EUCTR-2012–005742–38 | CMFT | II | Completed | [149] | 2012 |
| Bone deficits | ||||||||
| Vitamin D supplement | Cholecalciferol | 25Y-40Y | University of Utah | II | Recruiting | 2017 | ||
| Recombinant human bone morphogenetic protein-2 | rhBMP-2 | 2Y-18Y | UAB | II | Recruiting | 2016 | ||
| <18Y | EUCTR-2007–003835–22 | HCL | III | Active | 2008 |
Drug therapies in clinical trials are listed for specific NF1 complications including their molecular targets. Children/Adults refers to age of participants eligible for trial. The clinical trial identifier refers to either that of either NIH ClinicalTrials.gov or the European Clinical Trials Database (EudraCT). Sponsor abbreviations: University of Alabama at Birmingham (UAB), Massachusetts General Hospital (MGH), Children’s Hospital of Philadelphia (CHoP), Medical College of Wisconsin (MCW), Indiana University (IU), Spectrum Health Hospitals (SHH), The University of Texas Health Science Center at Houston (UTHealth), University of Texas Southwestern Medical Center (UTSW), Sarcoma Alliance for Research through Collaboration (SARC), NYU School of Medicine (NYUSM), The Foundation of the Carlo Besta Neurological Institute, IRCCS, Italy (IRCCS), Hospital Sant Joan de Deu (HSJD), Erasmus Medical Center (Erasmus MC), Children’s Hospital Los Angeles (CHLA), Children’s Research Institute (CRI), Novartis Pharmaceuticals (Novartis), Children’s Oncology Group (COG), MD Anderson Cancer Center (MD Anderson), Central Manchester University Hospitals NHS Foundation Trust (CMFT), Hospices Civils de Lyon (HCL).
5.1. Restoring neurofibromin function by targeting the NF1 gene
While a strategy to restore neurofibromin levels within a cell is not trivial, it is possible that even a modest decrease in RAS signaling could be beneficial for NF1 patients. As noted above, while some symptoms, such as cognitive deficits, result from NF1 haploinsufficiency, others, including tumor formation, result from NF1 biallelic inactivation. Due to the diverse spectrum of constitutional NF1 mutations and unpredictable nature of somatic mutations, a patient-specific approach to gene therapy may have to be employed, depending on the exact nature of the NF1 germline lesion. Approaches range from restoring neurofibromin function by overcoming the effect of NF1 nonsense or splicing mutations to gene replacement requiring efficient in vivo transduction to deliver functional NF1 to affected tissues.
5.1.1. Nonsense suppression
At least 20% of NF1 cases are caused by nonsense germline mutations leading to a premature termination codon (PTC) resulting in truncated neurofibromin protein [16]. Nonsense suppression therapy would aim to suppress translation termination at in-frame PTCs to restore deficient neurofibromin function. Strategies include drugs promoting readthrough, suppressor tRNAs, PTC pseudouridylation, and inhibition of nonsense-mediated mRNA decay [36]. Aminoglycoside antibiotics have been shown to cause a read-through of nonsense mutations and restore functional protein in short-term studies using mouse models of various neurological diseases, whereas pilot clinical trials have produced variable results [36, 37].
5.1.2. Exon skipping
Splicing mutations account for 27% of NF1 germline mutations [16]. While many of these mutations result in exon skipping, a small subset (2%) are deep intronic single nucleotide changes creating either a novel 5′ or 3′ site and thereby increase pseudoexon inclusion into mature mRNA. Splice-blocking antisense oligonucleotide (ASO) have been shown to be effective in skipping mutant exons in cultured cells harboring NF1 deep intronic mutations and restoring expression of neurofibromin [38].
5.1.3. Gene therapy
Precise targeting and efficient delivery of therapeutic payloads for gene therapy treatments is still in its infancy. Gene therapy for NF1 without genome editing could involve introduction of the full length normal NF1 gene using a recombinant adeno-associated virus (rAAV) containing an expression cassette, to replace mutant alleles and thereby restore neurofibromin levels and function. However, due to the large size of NF1 cDNA (8.5 kb), full gene replacement will require increasing the cargo capacities of available vectors or developing novel delivery systems. Alternatively, a truncated version of the NF1 gene retaining an appropriate level of function could be employed. For symptoms caused by NF1 haploinsufficiency, advances in targeting gene activation, including the engineering of transcription factors based on zinc finger proteins and transcription activator-like effectors (TALE) proteins could potentially be used to increase transcription from the remaining NF1 wild-type allele [39]. Another therapeutic strategy could make use of modified ASOs designed to bind upstream open reading frames (uORFs) in the 5ʹ-UTR region of mRNAs that have been shown to increase translation from the downstream primary ORF (pORF) [40]. The 5ʹ-UTR of human NF1 mRNA contains several such uORFs, although it is unknown whether these normally affect translation.
Genome editing technologies, such as CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats CRISPR- associated 9), have the potential to revolutionize the treatment of genetic disorders [41]. Permanent correction of NF1 missense and nonsense mutations could produce long-term and even lifelong restoration of neurofibromin deficiency. However, due to the thousands of unique NF1 gene mutations, personalized therapeutic approaches would be required. This approach is further complicated by the fact that, as mentioned before, about half of NF1 cases reflect de novo mutations. Routine use of CRISPR/Cas9 gene editing in the clinic will require further developments in efficient delivery, precision of editing and resolving safety issues.
5.2. Biomarkers for early detection of MPNSTs
The identification and development of assays for biomarkers of MPNSTs will enable the early detection of plexiform/atypical neurofibromas transition into these highly aggressive tumors [42]. Although surgery is currently the only effective treatment option available for MPNSTs, biomarkers could potentially be exploited together with gene delivery technologies to control and potentially kill malignant tumors. Synthetic lethal screens (e.g. using siRNA and/or CRISPR libraries) may be useful to identify genetic vulnerabilities of MPNSTs that could be exploited to devise therapies to specifically kill NF1-deficient tumors.
Telomere length may play a role in driving genomic instability and clonal progression in NF1-associated MPNSTs [43]. Furthermore, an increase in telomerase activity in high grade MPNSTs, but not detectable in diffuse PNFs or CNFs, is consistent with a role for telomere dysfunction during the progression of malignancy [44].
5.3. Inhibiting RAS signaling in NF1-deficient cells
As previously mentioned, the only established function of neurofibromin is that of a RAS-GAP (Figure 2). If the only consequence of neurofibromin loss is elevated RAS-GTP, correcting NF1 symptoms should be easier than dealing with the constitutively active RAS alleles found in ~30% of malignant tumors. However, the development of RAS inhibitors has been challenging due to the lack of well-defined druggable pockets and cavities on the surface of RAS [31]. Approaches have focused on the post-translational modification of RAS isoforms by prenylation (addition of farnesyl or geranyl–geranyl moieties) required for their correct localization to the plasma membrane. The farnesyltransferase inhibitors (FTIs) to farnesyl- transferase (FTase), the enzyme catalyzing the prenylation of HRAS have been developed to combat the transforming activities of oncogenic RAS [45]. While showing high preclinical anti-cancer activity in tumors driven by H-RAS, FTIs are not effective against N-RAS and K-RAS. This reflects at least in part that in the absence of active FTase, RAS can also undergo alternative prenylation by geranylgeranyltransferase. Moreover, RAS proteins are not the only substrates for FTase [46]. Tipifarnib, an FTI, has been tested in NF1 patients with PNFs and although sufficiently tolerated, failed to be effective [47]. Progress in developing small molecule inhibitors that bind directly to RAS and prevent guanine nucleotide exchange factor (GEF)-mediated GDP/GTP exchange [48] could be used to target wild-type RAS in NF1-deficient cells. Despite these advances, chronically blocking RAS may not be an appropriate strategy for treating the many non-life-threatening NF1 symptoms due to the dose-limiting toxic effects of FTIs and pan- RAS inhibitors.
5.4. Targeting RAS effectors
5.4.1. The RAS-MEK-ERK Pathway
An alternative approach to deal with increased levels of activated RAS in NF1-deficient cells, is the ongoing development of drugs targeting RAS effectors. Small molecules blocking RAS-binding domains (RBDs), the protein–protein interfaces of RAS-effector complexes, have been developed.
Although the first RAF kinase inhibitor to enter clinical trials, sorafenib, showed no benefit in KRAS-driven cancers, it is currently in trials for both PNFs and low-grade gliomas (Table 1). However, although RAF inhibitors are employed for tumors expressing mutant BRAF, their usefulness is limited by toxicity caused by their paradoxical enhancement of ERK signaling in BRAF wild-type cells (reviewed in [49]). Another potential avenue is provided by rigosertib, a first-in-class small molecule RAS-mimetic, currently in clinical trials for the treatment of chronic myelomonocytic leukemia. Rigosertib interacts with the RBDs of not only RAF kinases, but also of Ral-GDS and PI3Ks, results in the inability of these proteins to bind RAS, leading to the disruption of RAF activation, and inhibition of the RAS-RAF-MEK pathway [50].
Once activated, RAF phosphorylates MEK, which in turn phosphorylates and activates ERK, resulting in its translocation to the nucleus where it mediates transcription factor activation. MEK inhibitors target their conserved allosteric binding pocket, blocking phosphorylation, and subsequent activation. The potential of MEK inhibitors for NF1 treatment is demonstrated by selumetinib, which appears to benefit children with inoperable PNFs with tolerable side effects [51]. Another MEK inhibitor, PD0325901 appears to be promising in preclinical assessments of NF1 murine models of both PNFs [52,53] and JMML [54] and this drug is being evaluated in ongoing clinical trials for PNFs. Table 1 lists the usage of other MEK inhibitors for the NF1-OPG and other low-grade gliomas.
However, targeting MEK in NF1 has potential serious drawbacks. The toxicity of MEK inhibitors may not be suitable for long-term NF1 administration. The MEK/ERK signaling pathway is characterized by the presence of multiple nodes with feedback loops that regulate their activation [55]. Patients who respond well to MEK inhibitors often become resistant after several months due to re-activation of signal flux through the ERK pathway or acquiring MEK mutations. In addition, the PI3K and MEK-ERK pathways interact in multiple ways, by co-regulating their functions. Disruption of these regulatory loops can cause upregulation of pathway components and/or activation of parallel circuits.
The problems of toxicity and acquired resistance of tumors to BRAF and MEK inhibitors in cancer treatment have motivated the development of novel, selective ERK inhibitors, such as SCH772984. In addition to its direct effect on ERK activity, SCH772984 also inhibits MEK-mediated phosphorylation of ERK and could be considered for treatment of NF1 [56].
5.4.2. The PI3K/AKT/mTOR signaling pathway
Activation of the PI3K/AKT/mTOR pathway downstream of RAS has been shown to be important in the development of tumors in NF1 [33]. Both sirolimus (rapamycin) and everolimus (RAD001) bind FKBP12, specifically inhibiting mTORC1 and have shown varied efficacy in preclinical studies in genetically engineered mouse models (GEMM) of tumors and human NF1-associated tumor explants [33,57,58]. Several inhibitors of mTORC1 are currently in clinical trials for NF1-associated tumors (Table 1).
Since the effects of rapamycin and rapalogs are cytostatic and not cytotoxic, co-targeting mTOR and MEK-ERK pathways is likely to be more effective in combating MPNSTs than using a single agent. Everolimus and PD0325901 have been shown to synergistically inhibit proliferation and induce apoptosis in multiple MPNST cell lines, as well as reduce tumor burden in a GEMM, although long-term treatment results in the development of drug resistance, with reactivation of the target pathways [59].
A study identifying p110α as the specific isoform of PI3K catalytic subunit responsible for pro-proliferative PI3K signaling in MPNSTs [60], suggests potentially less toxic, specific p110α inhibitors could be useful against NF1 tumors [61]. This study also demonstrated that mTORC1 is the key PI3K effector in NF1 tumors, while mTORC2 and AKT are dispensable in NF1-deficient tumors [60].
5.5. Receptor tyrosine kinases (RTK) as therapeutic targets for NF1 tumors
While tumors expressing oncogenic RAS are largely resistant to therapies targeting RTKs, since NF1-deficient cells contain wild-type RAS proteins, upstream regulators of the RAS pathway represent potential therapeutic targets for NF1 (Figure 2). The transition from benign neurofibromas to MPNSTs is characterized by gene amplification of several RTKs (i.e. EGFR, PDGFR, MET) [62]. Several multi-kinase inhibitors and general RTK inhibitors approved for the treatment of various malignant cancers have been commandeered for NF1 clinical trials. However, inhibitors with relaxed specificity can cause harsh side effects and are unlikely to be suitable for long-term use [47]. Moreover, development of drug resistance is a major issue when using agents against RTK due in part to compensatory signaling pathways activated in response to RTK inhibition. Despite this, several RTK inhibitors including those against c-KIT, PDGFR, EGFR and c-MET, either alone or in combination, are currently used in clinical trials for neurofibromas and MPNSTs (Table 1).
Imatinib mesylate, a specific inhibitor of tyrosine kinase domains in ABL, c-KIT, and PDGF receptors was originally pinpointed for treating NF1 based on its ability to inhibit conditioned media from NF1−/− SCs to hyper-stimulate proliferation of cultured heterozygous (NF1+/−) mast cells in vitro, as discussed further in section 5.9 [63]. Although attenuation of c-KIT signaling was assumed to be the target of imatinib, an alternative mechanism of inhibiting the PDGF signaling pathway required for neovascularization of tumors has also been suggested [64]. Imatinib was shown to be effective in reducing the volume of PNF in a small-scale phase II trial [65] and additional clinical studies are ongoing (Table 1).
Neurofibromas appear to require an ‘angiogenic switch’ for their malignant transformation to microvascular blood vessel containing MPNSTs [66]. Several multi-target RTK inhibitors with activity against vascular endothelial growth factor (VEGF) RTKs are included in ongoing clinical trials for NF1- associated plexiform, cutaneous, and paraspinal neurofibromas and low-grade astrocytomas (Table 1). The signal transducer and activator of transcription 3 (STAT3) has been suggested to act as a signaling nexus required for the angiogenic response by regulating hypoxiainducible factor (HIFα) and VEGF-A expression in MPNSTs [67]. Since STAT3 is a common point of convergence of several RTKs, it may represent a better therapeutic targeting option for anti-angiogenic therapy than RTK inhibition with its potential for acquired drug resistance.
5.6. WNT/β-catenin signaling in NF1-associated tumors
Several studies have shown the importance of WNT/β-catenin pathway activation in NF1 tumorigenesis with expression of WNT genes being significantly deregulated in both PNFs and MPNSTs [68]. In some MPNSTs, the chemokine receptor CXCR4 and its ligand, CXCL12, has been shown to stabilize β-catenin through activation of PI3K-AKT-glycogen synthase kinase 3β (GSK3β), with β-catenin promoting tumor growth by stimulating cyclin D1 expression and cell cycle progression [69]. An insertional mutagenesis screen to identify genes and signaling pathways that initiate neurofibroma formation provided an alternative mechanism in which P-STAT3 activation represses GSK3β as well as the SWI/SNF gene Arid1b to increase β-catenin [70]. This report showed that P-STAT3 represses Arid1b through histone modification in a BRG1-dependent manner, suggesting epigenetic modification plays a role in early tumorigenesis. Together these studies support WNT/β-catenin and JAK/ STAT signaling as possible therapeutic targets in NF1-driven tumors.
5.7. Novel therapeutic targets in MPNSTs
While MEK-ERK and AKT/mTORC1 signaling are known to be critical for both neurofibroma and MPNST growth, investigations of the molecular events occurring during the malignant transformation of benign PNFs have identified promising new therapeutic targets for MPNSTs [4,71,72].
Combination therapy is a cornerstone of cancer therapy and recent efforts have focused on strategies to co-target multiple pathways in cell and animal MPNST models. For example, increasing proteotoxic stress using the HSP90 inhibitor IPI-504, in combination with inhibiting the mTOR pathway has been shown to cause MPNST regression in mice [73]. More recently, mTOR and HDAC inhibitors have been used together to promote catastrophic oxidative stress and regression of MPNSTs, with both inhibitors causing upregulation of thioredoxin-interacting protein (TXNIP) to trigger cell death by inhibiting thioredoxin and activating apoptosis signal-regulating kinase 1 (ASK1) [74].
As mentioned previously, the polycomb group gene SUZ12 has been identified as a tumor suppressor in NF1-associated MPNSTs and exacerbates the effects of NF1 loss by amplifying RAS-driven transcription through its effects on chromatin [18,19,22]. Inactivation of SUZ12 results in an epigenetic switch with an upregulation of BRD4 [75]. BRD4 is a BET (bromodomain and extra-terminal) family member that regulates expression of mitotic genes required for cell cycle progression. The subsequent finding that MPNSTs are sensitized to BET bromodomain inhibitors, which induce the proapoptotic BIM, offers a new approach for future therapies [18,75]. Combinatorial therapy involving BRD4 and MEK1 or mTOR inhibitors has proved to be more effective [18,76].
Transcriptome analysis has revealed that Aurora kinase A (AURKA) is dramatically overexpressed and genomically amplified in MPNSTs [77], suggesting that AURKA could be a potential MPNST therapeutic target. An AURKA selective inhibitor (MLN8237) stabilized tumor volume and significantly increased survival of mice with MPNST xenografts [77]. Recently, the same group used a lentiviral short hairpin RNA screen to reveal the homeobox protein MEIS1 as a MPNST driver gene [78]. MEIS1 promotes cell growth/survival, via the transcription factor ID1, by downregulating expression of a pro-cell death protein p27Kip, suggesting cell cycle inhibitors may have therapeutic value for MPNSTs.
Group I p21-activated kinases (Group I PAKs, PAK1/2/3) are important effectors of Rho family small GTPases RAC1 and CDC42 and are able to modulate signaling through MEK-ERK, Akt/mTORC1, and WNT/β-catenin signaling pathways [79]. Inhibition of PAK1/2/3 (Frax1036) and MEK (PD0325901) showed synergistic restriction of MPNST cell growth in vitro and dramatically decreased local and metastatic MPNST growth in mouse models [80].
Retinoids regulate specific target genes mediated by retinoid acid receptors (RARs) and are known to play a role in tumor growth, angiogenesis, and metastasis. Recent studies have shown that cellular retinoic acid-binding protein 2 (CRABP2), a transcriptional co-activator of retinoic acid signaling, is upregulated in MPNSTs and promotes their survival [81]. The viability and proliferation of MPNST cell lines is reduced by either knockdown of CRABP2 or by combined application of all-trans retinoic acid (ATRA) and MEK inhibitors, suggesting targeting CRABP2 overexpression could represent a novel approach for MPNST treatment [82].
Integrated proteomics has identified a novel neurofibromin-associated protein, translationally controlled tumor protein (TCTP) as being up-regulated in NF1-associated tumors via MEK-ERK and mTOR signaling [83]. Artesunate, a semisynthetic derivative of the antimalarial drug artemisinin, which binds to and degrades TCTP, significantly suppressed the viability of MPNST cells but not normal SCs, while combinational use of artesunate and rapamycin enhanced the cytotoxic effect on MPNST cells [83].
Activation of the HIPPO signaling pathway has been implicated in the transformation and progression of MPNSTs. The kinases LATS1 and LATS2 negatively regulate the HIPPO path way by phosphorylating TAZ/YAP, thereby preventing their translocation to the nucleus where they interact with transcriptional partners to regulate cell growth [84]. Genomic profiling of patients with peripheral nerve sheath tumors has revealed mutations in LATS1, loss of a copy of either LATS1 or LATS2, or hypermethylation of their promoters [85,86]. A recent study has shown changes in gene expression suggestive of HIPPO-TAZ/YAP hyperactivity in human MPNSTs [87]. Ablation of LATS1/2 in mouse SCs results in hyperactivation of TAZ/YAP and induces high-grade nerve-associated tumors. Elevated TAZ/YAP activity in SCs was also shown to activate oncogenic programs, including PDGF signaling. Pharmacological co-targeting of TAZ/YAP (verteporfin) and PDGFR (sorafenib) inhibits tumor growth, suggesting a novel therapeutic strategy for MPNSTs [87].
As can be seen, the implication of novel pathways involved in MPNST transformation and viability, as well as uncovering their vulnerabilities to existing compounds has led to the identification of numerous therapeutic targets (Figure 4). A contrasting approach, of screening a library of 200,000 small molecules on mouse Nf1 mutant MPNST cells has identified a novel inducer of apoptosis, which may be effective against MPNSTs [88]. Compound 21 (Cpd21) inhibits growth and leads to apoptosis of all available in vitro models of MPNSTs and human MPNST cell lines, while remaining nontoxic to normally dividing SCs. Another study involving a screen of plant-derived compounds revealed silvestrol, an eIF4A inhibitor, as being able to induce G2/M arrest in MPNST cell lines, decreasing the levels of multiple cyclins, Aurora A, AKT, and ERK and dramatic suppression of tumor growth inNF1 MPNST mouse models [89].
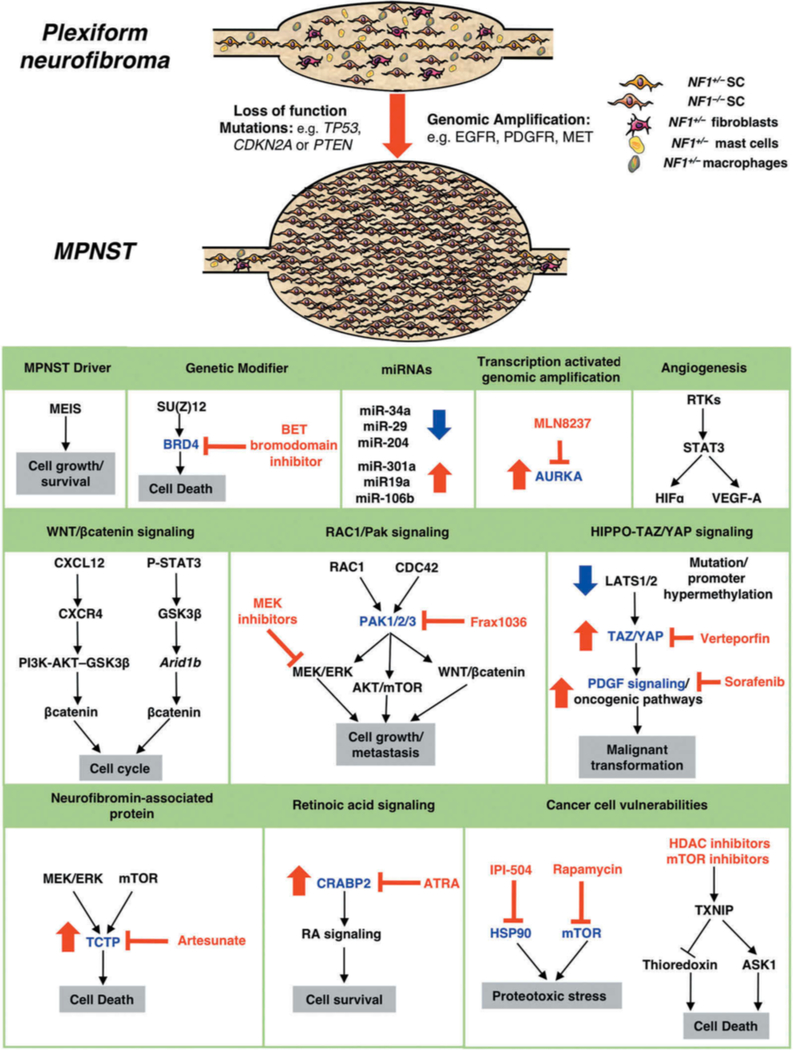
Figure 4. Progression of plexiform neurofibromas to malignant peripheral nerve sheath tumors.
MPNSTs are highly invasive soft tissue sarcomas that frequently metastasize and arise from benign PNFs harboring biallelic NF1 inactivation. Additional genetic changes leading to the transition from PNFs to MPNSTs include loss-of-function mutations in tumor suppressors such as TP53, CDKN2A and PTEN, or genomic amplification of RTKs or signaling factors. NF1−/− SCs consequently have dramatically increased proliferation rates compared to those in benign PNFs. Potential therapeutic targets for MPNSTs are highlighted. Target proteins implicated in the malignant progression of MPNSTs are shown in blue (p21-activated kinases (PAK1/2/3), bromodomain-containing protein 4 (BRD4), Cellular Retinoic Acid Binding Protein 2 (CRABP2), heat shock protein 90 (HSP90), mTOR, Translationally controlled tumor protein (TCTP) and Aurora kinase A (AURKA)) and small molecule inhibitors (BET bromodomain inhibitors, FRAX1036, IPI-504, rapamycin, HDAC inhibitors, artesunate, MLN8237, all-trans retinoic acid (ATRA)) in red. Increase in level of protein/miRNA in MPNSTs is denoted by red arrow, blue arrow denotes decrease.
5.8. miRNAs as targets for MPNSTs
Numerous studies have implicated miRNAs (miRs) as playing a role in malignant transformation of neurofibromas, which could potentially represent therapeutic targets for MPNSTs [90]. Comparison of miR expression profiles between PNFs and MPNSTs have identified both down- and upregulation of various miRs. Downregulated miRs in MPNSTs include miR-34a, miR-29c and miR-204 [91–93]. Restoring expression of these mIRs in MPNST cell lines and/or mouse models result in apoptosis [91], reduced cell invasion [93] or decreased tumor growth/malignant progression in vivo [92] respectively. In contrast, the upregulation of a group of miRs (miR-301a, miR19a and miR-106b) involved in PTEN repression has also been implicated in MPNST development [94].
5.9. Importance of the microenvironment and role of immune system in NF1 tumors
Studies with conditional knockout mice and genetic analysis of cells cultured from human neurofibromas have collectively shown that while bi-allelic inactivation of the NF1 gene is a prerequisite for neurofibroma formation, tumorigenesis can occur only in a NF1+/− background [95]. Thus, non-neoplastic cells in the tumor microenvironment play an essential role in neurofibroma development. CNFs are thought to arise from skin-derived precursor cells. Both benign neurofibromas and MPNSTs are composed of SCs, degranulating mast cells, macrophages, fibroblasts, and extracellular matrix. Elucidation of the intrinsic and extrinsic factors within the peripheral nerve microenvironment that play essential roles in neurofibroma development have provided therapeutic targets for treatment of tumors. In addition, several recent studies have highlighted the potential of targeting the immune system for NF1 treatment.
NF1−/+ mast cells play a key role in both tumorigenesis and neoangiogenesis by producing matrix metalloproteinases, heparin, and a range of growth factors (Fig. 3). Within NF1-associated tumors, mast cells differentiate, proliferate, and secrete cytokines in response to stem cell factor (SCF), a secreted ligand from NF1−/− SCs, which signals through the c-KIT receptor tyrosine kinase [96]. A study using MPNST xenografts suggests PLX3397, a c-KIT inhibitor, is superior to imatinib in the treatment of MPNSTs [97]. In addition to inhibiting c-KIT signaling, PLX3397 also inhibits CSF1 receptor activity to prevent macrophage recruitment to tumors. Further evidence of macrophage involvement in neurofibroma formation comes from Nf1 mouse studies showing that injury signals recruit macrophages to relieve the normally tumor-suppressive environment of adult peripheral nerve [98]. Further clues from the tumor microenvironment for possible therapeutic strategies come from experiments on the formation of optic gliomas (OPGs) in Nf1-mutant mice [99]. OPGs contain microglial cells expressing the chemokine receptor CX3CR1; genetic reduction of Cx3cr1 expression in Nf1 optic glioma mice has been found to delay OPG formation.
Targeted immunotherapy options for NF1 tumors will require identifying their immunologic profiles. SCs and macrophages in neurofibromas show distinct differences in their inflammatory gene signatures compared to cells in the normal peripheral nervous system [100]. Further, network analysis predicted decreased type-I interferon signaling as playing a central role in neurofibromas. Treatment of a GEMM neurofibroma model with polyethylene glycolyated (PEGylated) type-I interferon-α2b reduced the expression of many cytokines overexpressed in NF tumors.
Immune checkpoint inhibitors targeting the programmed cell death 1 (PD-1) receptor or its ligand (PD-L1) have been shown to be effective in treating patients with a variety of cancers [101]. Clinical response rates correlate with PD-L1 expression on tumor cells, as well as the presence of tumor-infiltrating lymphocytes (TILs). Recently, immunohistochemistry has revealed the expression of PD-L1 as well as the presence of CD68-, CD3-, or CD8-positive TILs in PNF and CNF specimens [102], suggesting adaptive resistance to cell- mediated immunity may play a major role in the tumor immune microenvironment of NF1-associated tumors. Moreover, expression of PD-L1 and the presence of TILs suggest that NF1-deficient tumors may be responsive to immunotherapy with immune checkpoint inhibitors.
An increase in plasma levels of transforming growth factor-beta (TGF-β) in NF1 patients suggests that the role of TGF-β signaling in MPNST pathogenesis might be worth investigating [103]. While revealing that both neurofibromas and MPNSTS show potential targets for T-cell-based immunotherapies, a recent study also showed considerable tumor heterogeneity in immune profiles, even within an individual [104]. This could pose a significant clinical challenge to using immunotherapies for NF1, which are likely to require a highly individualized, patient immunology-based, and tumor profile-based strategy.
5.10. Therapeutic targets for peripheral neuropathy in NF1
Peripheral neuropathy in NF1 constitutes a potentially severe clinical complication and is associated with increased morbidity and mortality [101]. Recently rodent models of NF1 have provided potential clues to the mechanism underlying pain in NF1 [102, 103]. Loss of neurofibromin results in functional remodeling of peripheral nociceptors characterized by enhancement of interactions of the tetrodotoxin-sensitive (TTX-S) Na+ voltage-gated sodium channel (NaV1.7) and the collapsin response mediator protein 2 (CRMP2). Inhibition of CRMP2 phosphorylation with (S)-lacosamide is sufficient to normalize channel current densities observed with NF1 loss, suggesting CRMP2 could be a key target for therapeutic intervention for NF1-associated pain.
5.11. Therapeutic targets for NF1 cardiovascular disease
NF1 predisposes individuals to an increased risk for premature and severe arterial stenosis. Arterial lesions associated with NF1 are characterized by smooth muscle cell (SMC) hyperplasia, leukocyte infiltration and arterial remodeling leading to vaso-occlusion and tissue ischemia [1]. Similar to NF1 patients, Nf1+/− mice develop a marked arterial stenosis and have increased circulating CCR2+ proinflammatory monocytes [108]. Monocyte chemotactic protein-1 (MCP-1) has been shown to be a potent chemokine for CCR2 receptor activation and is critical for monocyte infiltration into the arterial wall and neointima formation in this GEMM [109]. Administration of a CCR2 antagonist (INCB3284) significantly reduced Nf1+/− neointima formation, suggesting it may be a viable therapeutic target for NF1 arterial stenosis [109].
5.12. Therapeutic targets for skeletal abnormalities in NF1
Recalcitrant bone healing following fracture (pseudarthrosis) is one of the most problematic skeletal complications associated with NF1. Multiple studies have shown reduced osteogenic potential of NF1-deficient osteoprogenitors due to increased RAS-ERK signaling [110]. The MEK inhibitor PD0325901 in combination with recombinant human bone morphogenetic protein (rhBMP-2) has been shown to increase bone formation in a GEMM of NF1 pseudarthrosis [111]. Further murine studies have demonstrated a role for aberrant c-Jun N-terminal Kinase (JNK), a non-canonical RAS effector pathway [112], TGF-β1 [113] and fibroblast growth factor receptor 1 (FGFR1) signaling [114] in NF1 skeletal defects, suggesting further potential therapeutic targets for NF1 osseous defects. Two recent reports have also highlighted the importance of elevated β-catenin levels at fracture sites in Nf1 mice [115, 116]. The5increased fibrous tissue at fracture sites was rescued by local treatment with a WNT antagonist, Dickkopf-1 (Dkk1) or by conditional β-catenin gene inactivation, suggesting pharmacological modulation of β-catenin could be used to treat pseudarthrosis in NF1 patients.
5.13. Therapeutic targets for NF1 cognitive dysfunction
Cognitive manifestations of NF1 are the result of neurofibromin deficiency on both brain development (aberrant myelination) and brain function [117]. Defects in hippocampal spatial learning in Nf1+/− mice are due to enhanced RAS activity, leading to enhanced GABA-mediated inhibitory neurotransmission and subsequent impaired long-term potentiation (LTP) [118]. In addition, some defects observed in NF1-deficient neurons are dependent on cyclic AMP signaling [119].
Lovastatin, a HMG-CoA-reductase inhibitor, has so far been unsuccessful in improving learning in children with NF1, despite promising preclinical test in Nf1 mice [120]. Studies showing that defective dopaminergic function is a contributing factor underlying impaired spatial learning and memory in Nf1 mice [121, 122] have led to clinical trials of a dopamine–norepinephrine reuptake inhibitor, methylphenidate, originally developed for ADHD. The interneuron-specific attenuation of hyperpolarization-activated cyclic nucleotide-gated (HCN) current has been suggested as a cause for increased neuronal inhibition [123]. The HCN channel agonist, lamotrigine (a sodium channel blocker) was able to rescue the electrophysiological and cognitive deficits in two Nf1 mouse models and is currently in clinical trials. Studies in Drosophila and mice have implicated the neuronal-specific RTK, Anaplastic Lymphoma Kinase (ALK), as an upstream activator of RAS signaling in neurons [124, 125, 126]. Altering ALK activity in these animal models partially ameliorates NF1 cognitive deficits, suggesting it represents a potential therapeutic target.
6. Conclusions
NF1 is a complex multi-system disorder affecting many different tissues. While benign and malignant tumors are often considered to be the main characteristics of NF1, other symptoms including cognitive problems, skeletal and vascular abnormalities contribute to the morbidity of the disease. Neurofibromin controls RAS signaling in many cell types and its deficiency affects a variety of molecular and cellular processes. Rather than seeking a panacea for the diverse manifestations of the disease, therapeutic strategies will require an understanding of the specific defects caused by loss of NF1 in defined cell types underlying a particular symptom. Elucidation of the canonical and non-canonical effector pathways downstream of RAS activation and their ultimate cell-specific consequences has identified promising therapeutic targets for this chronic disease.
7. Expert opinion
Despite the NF1 gene having been identified in 1990, a multitude of factors have hampered research into the disease, for which there is still no effective treatment. In the laboratory, the technical limitations of working with such a large gene (the genomic locus spans >350 kb) have hindered experiments using transfection of full-length NF1 cDNAs (>9kb). In the clinic, the multisystem nature of NF1, coupled with the extreme variability of complications between individuals within and between families has also been challenging. Environmental factors, age, as well as genetic modifiers all play a role in disease development. Even though the underlying cause of NF1 is monogenic, the heterogeneity of both germline and somatic NF1 mutations adds an additional level of complexity in attempting to understand the disease. However, improved molecular diagnosis of patients has revealed several genotype-phenotype correlations, which may provide new clues into the pathogenesis of NF1 and potentially reveal critical regions of neurofibromin outside of its RAS-GAP domain.
One of the biggest challenges for devising therapies for NF1 will be considering treatments of both the acute and chronic aspects of the disorder. While short-term aggressive therapies for MPNSTs may be tolerated, the use of anti-tumor agents are of limited value to the NF population for prolonged administration in order to achieve tumor shrinkage or stabilization. Furthermore, many of the non-tumor symptoms will require long-term treatment to improve the quality of life of the individual, while safety issues are of particular concern in treating infants with NF1.
The cellular pathways affected in NF1 tumors and their downstream effects continue to be elucidated. A great deal of evidence indicates that multiple pathways function cooperatively in tumor formation. Many of the therapeutic strategies currently being tested for NF1 involve inhibiting the RAS/MEK and AKT/mTORC1 pathways. Several studies highlight combinatorial approaches as likely to be more effective than single agents, as well as delaying or preventing the emergence of preexisting drug-resistant clones to extend the duration of response.
Neurofibromas usually arise during puberty, with their growth being influenced by hormones and increase in number with age. Understanding the mechanism that underlies the transition of NF1−/+ SCs into tumors may provide potential therapeutic interventions. For example, recently, EGFR-STAT3 signaling has been shown to be involved in the self-renewal of SC precursors (SCPs), which result in PNF initiation [127], raising the possibility that it might be possible to devise a pre-emptive strategy to inhibit PNF formation. The recent development of three-dimensional (3D) cell culture models of PNFs may facilitate pre-clinical identification of potential targeted therapeutics for these tumors [128].
MPNSTs represent one of the more serious complications in NF1 and do not respond to standard chemotherapy or radiation therapy. The most effective treatment of MPNSTs appears to be early diagnosis and surgery; discovery of biomarkers that can be used for the early detection of MPNSTs could greatly aid treatment of these tumors. Synthetic lethal screens using the latest technologies (e.g., CRISPR-based libraries) may reveal specific vulnerabilities of NF1-deficient tumors that could be exploited for therapy. While GEMMs offer systems for validation of potential tar- gets and therapeutics, the establishment of orthoxenograft mouse models of patient-derived MPNSTs offers a chance for personalized therapy and the ability to perform preclinical drug testing [129].
The recent advances in therapeutic genome editing and targeted immunotherapy offer personalized approaches for treating NF1. CRISPR/Cas9 or gene editing technology may potentially be used to alter the genome of NF1 patients, to provide a long-term solution. However, while these approaches have yet to be implemented in the clinic, they may be impractical in overcoming neurofibromin deficiency in all cells and tissues within an individual, especially in patients with de novo NF1 mutations. Targeted immunotherapy has dramatically changed cancer treatment recently. Efforts to identify the immunologic profiles of NF1 tumors may provide a therapeutic option for using this personalized approach on neurofibromas and MPNSTs.
The continuing development of novel cell and animal disease models will greatly aid both the understanding of basic NF1 biology as well as providing tools for screening novel therapeutics. We anticipate that studies using induced pluripotent stem cell (iPSC) will help both to elucidate the pathways and cell-specific defects caused by NF1 loss and the subsequent discovery of novel therapeutic interventions [130]. Not only do iPSCs allow the derivation of the different cell types involved in NF1 (e.g. neurons, SCs, melanocytes), this approach also provides an opportunity to develop patient-specific cell models of NF1 (B. Korf, pers. comm). The continued identification of biomarkers associated with NF1 deficiency in specific cell types should be useful in both devising effective treatments, as well as for assessing whether therapeutic interventions are successful. Preclinical testing of potential therapeutic agents for NF1 has been aided by the availability of well-characterized mouse models of PNFs, MPNSTs JMML, OPG and cognitive dysfunction. It is anticipated that the ongoing development of a porcine NF1 model (CTF) may be more accurate for testing new therapeutic targets.
The establishment of the multiple-institutional Neurofibromatosis Preclinical Consortium (NFPC) and Neurofibromatosis Clinical Trials Consortium (NFCTC, http://www.uab.edu/nfconsortium) has accelerated therapeutic trials for both the benign and malignant features of NF1 [131, 132] (Table 1). However, it is not certain whether treatment with these drugs will change the natural history of tumors, therefore long-term patient follow-up is warranted. The availability of emerging NF1 genotype-phenotype correlations, development of sophisticated animal models accurately reflecting human pathology and NF1 patient-associated iPSC reagents will continue to improve our knowledge of the function of neurofibromin, the cellular pathways it regulates and identify novel biomarkers (Fig. 5). The synthesis, high-throughput screening and application of small molecule libraries using these disease models will expand the repertoire of targeted agents. Genome-guided therapeutics including gene editing may offer new options for the application of precision medicine for NF1. Furthermore, establishing a preclinical framework culminating in actionable drug targets, marked collaborations between academia, hospitals and pharmacological industries to implement the clinical drug pipeline will offer promising opportunities for imminent NF1 treatment.
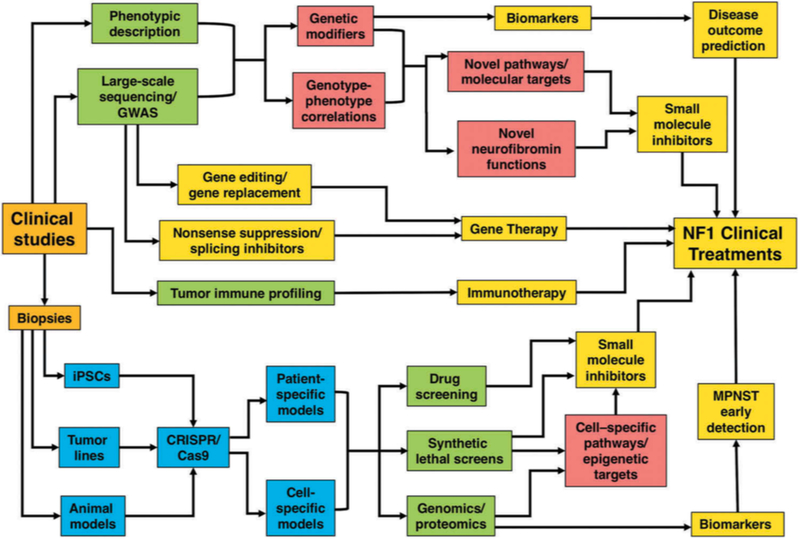
Figure 5. Network of approaches leading to discovery and testing of new therapeutic targets in NF1.
1) Clinical examination of patients combined with molecular analyses is beginning to reveal NF1 genotype-phenotype correlations - findings that may define novel functions of neurofibromin and identify new therapeutic targets. In addition, genome-wide association studies (GWAS) offer a platform to identify genetic modifiers, which will facilitate the identification of novel targets and biomarkers. Understanding the effect of environmental factors and hormones on NF1 disease progression may also reveal novel treatments. 2) Patient-derived biopsies will aid in the generation of: a) induced pluripotent stem cells (iPSCs) to allow development of patient and cell-specific models and give insights for new targets and biomarkers for different NF1 clinical manifestations; b) tumor-specific cell lines and animal models accurately reflecting the human disease that will permit improved screening of small molecule inhibitors; c) patient-derived cell line models will facilitate cellular pathway analysis (in particular RAS pathway and its upstream and downstream effectors (including receptor tyrosine kinases and micro RNAs) identification of therapeutic targets and biomarkers, and will also allow the testing of novel drugs including small molecule inhibitors prior to their use in clinical trials; d) synthetic lethal screening (using CRISPR libraries) could be exploited to devise therapies to selectively kill NF1-deficient tumors; e) immune profiling leading to immunotherapy and generation of novel biomarkers for NF1-associated tumors. 3) Gene therapy approaches focus on antisense oligonucleotides (ASOs) and nonsense suppression, whereas potential correction of mutations via gene editing offers a possibility of restoring endogenous NF1 gene function, thereby providing a long-term solution for NF1 patients. Highlighted boxes: clinical studies/biopsies (orange), genetic analysis and screening (green), disease models (blue), new insights into basic NF1 biology (red), potential therapeutic approaches and clinical treatments (yellow).
Acknowledgments
Figures created using elements from Servier Medical Art according to a Creative Commons Attribution 3.0 Unported License. The authors thank Dr. Andre Bernards for critical reading of the manuscript and for helpful discussions and Brittany Leger for help with figures.
Funding
This work was funded by the National Institute of Health (NIH R21 NS096402-01A1), the Department of Defense (W81XWH-16-1-0220), the Neurofibromatosis Therapeutic Acceleration Program (NTAP), the Kanter family and Ian Owen funds.
References
Papers or patents of particular interest should be identified using one or two asterisk symbols (• = of interest, •• = of considerable interest), and annotated with a brief sentence explaining why the reference is considered to be of interest.
- 1.Upadhyaya M, Cooper DN. “Molecular and Cellular Biology of Neurofibromatosis Type 1”. 2012:Eds: Upadhyaya M and Cooper DN Springer Verlag. [Google Scholar]
- 2.Upadhyaya M Neurofibromatosis type 1: diagnosis and recent advances. Expert Opin Med Diagn. 2010. July;4(4):307–22. doi: 10.1517/17530059.2010.494660. [DOI] [PubMed] [Google Scholar]
- 3.Evans DG, Baser ME, McGaughran J, et al. Malignant peripheral nerve sheath tumours in neurofibromatosis 1. J Med Genet. 2002. May;39(5):311–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farid M, Demicco EG, Garcia R, et al. Malignant peripheral nerve sheath tumors. Oncologist. 2014. February;19(2):193–201. doi: 10.1634/theoncologist.2013-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutmann DH, Ferner RE, Listernick RH, et al. Neurofibromatosis type 1. Nat Rev Dis Primers. 2017. February 23;3:17004. doi: 10.1038/nrdp.2017.4. [DOI] [PubMed] [Google Scholar]
- 6.Tidyman WE, Rauen KA. Pathogenetics of the RASopathies. Hum Mol Genet. 2016. October 1;25(R2):R123–R132. doi: ddw191 [pii]10.1093/hmg/ddw191.** Excellent review focusing on pathogenetic mechanisms associated with RASopathies.
- 7.Mautner VF, Asuagbor FA, Dombi E, et al. Assessment of benign tumor burden by whole-body MRI in patients with neurofibromatosis 1. Neuro Oncol. 2008. August;10(4):593–8. doi: 15228517–2008-011 [pii] 10.1215/15228517-2008-011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miettinen MM, Antonescu CR, Fletcher CDM, et al. Histopathologic evaluation of atypical neurofibromatous tumors and their transformation into malignant peripheral nerve sheath tumor in patients with neurofibromatosis 1-a consensus overview. Hum Pathol. 2017. September;67:1–10. doi: 10.1016/j.humpath.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beert E, Brems H, Daniels B, et al. Atypical neurofibromas in neurofibromatosis type 1 are premalignant tumors. Genes Chromosomes Cancer. 2011. December;50(12):1021–32. doi: 10.1002/gcc.20921.**This is the first study providing evidence that benign-atypical neurofibromas can develop into MPNSTs
- 10.Acosta MT, Gioia GA, Silva AJ. Neurofibromatosis type 1: new insights into neurocognitive issues. Curr Neurol Neurosci Rep. 2006. March;6(2):136–43. [DOI] [PubMed] [Google Scholar]
- 11.Mautner VF, Kluwe L, Thakker SD, et al. Treatment of ADHD in neurofibromatosis type 1. Dev Med Child Neurol. 2002. March;44(3):164–70. [DOI] [PubMed] [Google Scholar]
- 12.Thomas PK, King RH, Chiang TR, et al. Neurofibromatous neuropathy. Muscle Nerve. 1990. February;13(2):93–101. doi: 10.1002/mus.880130202. [DOI] [PubMed] [Google Scholar]
- 13.Brems H, Legius E. Legius syndrome, an Update. Molecular pathology of mutations in SPRED1. Keio J Med. 2013;62(4):107–12. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Romero MT, Parkin P, Lara-Corrales I. Mosaic Neurofibromatosis Type 1: A Systematic Review. Pediatr Dermatol. 2016. Jan-Feb;33(1):9–17. doi: 10.1111/pde.12673. [DOI] [PubMed] [Google Scholar]
- 15.Messiaen LM, Callens T, Mortier G, et al. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat. 2000;15(6):541–55. doi: . [DOI] [PubMed] [Google Scholar]
- 16.Messiaen LM, Wimmer K. NF1 Mutational Spectrum In: Neurofibromatoses Edited by Kaufmann D: Karger; 2008:63–77. [Google Scholar]
- 17.Legius E, Dierick H, Wu R, et al. TP53 mutations are frequent in malignant NF1 tumors. Genes Chromosomes Cancer. 1994. August;10(4):250–5. [DOI] [PubMed] [Google Scholar]
- 18.De Raedt T, Beert E, Pasmant E, et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature. 2014. October 9;514(7521):247–51. doi: 10.1038/nature13561.** This is the first study to demonstrate that SUZ12 is a tumour suppressor in NF1 tumors. SUZ12 inactivation triggers an epigenetic switch that sensitises these tumors to bromodomain inhibitors.
- 19.Lee W, Teckie S, Wiesner T, et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet. 2014. November;46(11):1227–32. doi: 10.1038/ng.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Philpott C, Tovell H, Frayling IM, et al. The NF1 somatic mutational landscape in sporadic human cancers. Hum Genomics. 2017. June 21;11(1):13. doi: 10.1186/s40246-017-0109-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasmant E, Sabbagh A, Spurlock G, et al. NF1 microdeletions in neurofibromatosis type 1: from genotype to phenotype. Hum Mutat. 2010. June;31(6):E1506–18. doi: 10.1002/humu.21271. [DOI] [PubMed] [Google Scholar]
- 22.Zhang M, Wang Y, Jones S, et al. Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nat Genet. 2014. November;46(11):1170–2. doi: 10.1038/ng.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinna V, Lanari V, Daniele P, et al. p.Arg1809Cys substitution in neurofibromin is associated with a distinctive NF1 phenotype without neurofibromas. Eur J Hum Genet. 2015. August;23(8):1068–71. doi: ejhg2014243 [pii]10.1038/ejhg.2014.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Upadhyaya M, Huson SM, Davies M, et al. An absence of cutaneous neurofibromas associated with a 3-bp inframe deletion in exon 17 of the NF1 gene (c.2970–2972 delAAT): evidence of a clinically significant NF1 genotype-phenotype correlation. Am J Hum Genet. 2007. January;80(1):140–51. doi: S0002–9297(07)60928–8 [pii] 10.1086/510781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rojnueangnit K, Xie J, Gomes A, et al. High Incidence of Noonan Syndrome Features Including Short Stature and Pulmonic Stenosis in Patients carrying NF1 Missense Mutations Affecting p.Arg1809: Genotype-Phenotype Correlation. Hum Mutat. 2015. November;36(11):1052–63. doi: 10.1002/humu.22832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koczkowska M, Chen Y, Callens T, et al. Genotype-Phenotype Correlation in NF1: Evidence for a More Severe Phenotype Associated with Missense Mutations Affecting NF1 Codons 844–848. Am J Hum Genet. 2018. January 4;102(1):69–87. doi: S0002–9297(17)30490–1 [pii] 10.1016/j.ajhg.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Easton DF, Ponder MA, Huson SM, et al. An analysis of variation in expression of neurofibromatosis (NF) type 1 (NF1): evidence for modifying genes. Am J Hum Genet. 1993. August;53(2):305–13.* This is the first study to provide evidence of modifying genes for NF1
- 28.Rieley MB, Stevenson DA, Viskochil DH, et al. Variable expression of neurofibromatosis 1 in monozygotic twins. Am J Med Genet A. 2011. March;155A(3):478–85. doi: 10.1002/ajmg.a.33851. [DOI] [PubMed] [Google Scholar]
- 29.Sabbagh A, Pasmant E, Laurendeau I, et al. Unravelling the genetic basis of variable clinical expression in neurofibromatosis 1. Hum Mol Genet. 2009. August 1;18(15):2768–78. doi: ddp212 [pii]10.1093/hmg/ddp212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin GA, Viskochil D, Bollag G, et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990. November 16;63(4):843–9. [DOI] [PubMed] [Google Scholar]
- 31.Simanshu DK, Nissley DV, McCormick F. RAS Proteins and Their Regulators in Human Disease. Cell. 2017. June 29;170(1):17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohba Y, Mochizuki N, Yamashita S, et al. Regulatory proteins of R-Ras, TC21/R-Ras2, and M-Ras/R-Ras3. J Biol Chem. 2000. June 30;275(26):20020–6. doi: 10.1074/jbc.M000981200. [DOI] [PubMed] [Google Scholar]
- 33.Johannessen CM, Reczek EE, James MF, et al. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci U S A. 2005. June 14;102(24):8573–8. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shilyansky C, Lee YS, Silva AJ. Molecular and cellular mechanisms of learning disabilities: a focus on NF1. Annu Rev Neurosci. 2010;33:221–43. doi: 10.1146/annurev-neuro-060909-153215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunzendorfer-Matt T, Mercado EL, Maly K, et al. The neurofibromin recruitment factor Spred1 binds to the GAP related domain without affecting Ras inactivation. Proc Natl Acad Sci U S A. 2016. July 5;113(27):7497–502. doi: 10.1073/pnas.1607298113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keeling KM, Xue X, Gunn G, et al. Therapeutics based on stop codon readthrough. Annu Rev Genomics Hum Genet. 2014;15:371–94. doi: 10.1146/annurev-genom-091212-153527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee HL, Dougherty JP. Pharmaceutical therapies to recode non- sense mutations in inherited diseases. Pharmacol Ther. 2012. November;136(2):227–266. [DOI] [PubMed] [Google Scholar]
- 38.Pros E, Fernandez-Rodriguez J, Canet B, et al. Antisense therapeutics for neurofibromatosis type 1 caused by deep intronic mutations. Hum Mutat. 2009. March;30(3):454–62. doi: 10.1002/humu.20933. [DOI] [PubMed] [Google Scholar]
- 39.Nelson CE, Gersbach CA. Engineering Delivery Vehicles for Genome Editing. Annu Rev Chem Biomol Eng. 2016. June 7;7:637–62. doi: 10.1146/annurev-chembioeng-080615-034711. [DOI] [PubMed] [Google Scholar]
- 40.Liang XH, Sun H, Shen W, et al. Antisense oligonucleotides targeting translation inhibitory elements in 5’ UTRs can selectively increase protein levels. Nucleic Acids Res. 2016. September 19;45(16):9528–9546. doi: 3984484 [pii] 10.1093/nar/gkx632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gori JL, Hsu PD, Maeder ML, et al. Delivery and Specificity of CRISPR-Cas9 Genome Editing Technologies for Human Gene Therapy. Hum Gene Ther. 2015. July;26(7):443–51. doi: 10.1089/hum.2015.074. [DOI] [PubMed] [Google Scholar]
- 42.Hanemann CO, Blakeley JO, Nunes FP, et al. Current status and recommendations for biomarkers and biobanking in neurofibromatosis. Neurology. 2016. August 16;87(7 Suppl 1):S40–8. doi: 10.1212/WNL.0000000000002932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones RE, Grimstead JW, Sedani A, et al. Telomere erosion in NF1 tumorigenesis. Oncotarget. 2017. June 20;8(25):40132–40139. doi: 10.18632/oncotarget.16981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mantripragada KK, Caley M, Stephens P, et al. Telomerase activity is a biomarker for high grade malignant peripheral nerve sheath tumors in neurofibromatosis type 1 individuals. Genes Chromosomes Cancer. 2008. March;47(3):238–46. doi: 10.1002/gcc.20525. [DOI] [PubMed] [Google Scholar]
- 45.Klochkov SG, Neganova ME, Yarla NS, et al. Implications of farnesyltransferase and its inhibitors as a promising strategy for cancer therapy. Semin Cancer Biol. 2017. October 31. doi: 10.1016/j.semcancer.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 46.Baines AT, Xu D, Der CJ. Inhibition of Ras for cancer treatment: the search continues. Future Med Chem. 2011. October;3(14):1787–808. doi: 10.4155/fmc.11.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bakker AC, La Rosa S, Sherman LS, et al. Neurofibromatosis as a gateway to better treatment for a variety of malignancies. Prog Neurobiol. 2017. May;152:149–165. doi: 10.1016/j.pneurobio.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 48.Ostrem JM, Shokat KM. Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nat Rev Drug Discov. 2016. November;15(11):771–785. doi: 10.1038/nrd.2016.139. [DOI] [PubMed] [Google Scholar]
- 49.Holderfield M Efforts to Develop KRAS Inhibitors. Cold Spring Harb Perspect Med. 2017. November 3. doi: cshperspect.a031864 [pii] 10.1101/cshperspect.a031864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Athuluri-Divakar SK, Vasquez-Del Carpio R, Dutta K, et al. A Small Molecule RAS-Mimetic Disrupts RAS Association with Effector Proteins to Block Signaling. Cell. 2016. April 21;165(3):643–55. doi: 10.1016/j.cell.2016.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dombi E, Baldwin A, Marcus LJ, et al. Activity of Selumetinib in Neurofibromatosis Type 1-Related Plexiform Neurofibromas. N Engl J Med. 2016. December 29;375(26):2550–2560. doi: 10.1056/NEJMoa1605943.** This is the first large study that reveals that NF1 children with inoperable plexiform neurofibromas benefited from long-term treatment of selumetinib without toxic effects.
- 52.Jessen WJ, Miller SJ, Jousma E, et al. MEK inhibition exhibits efficacy in human and mouse neurofibromatosis tumors. J Clin Invest. 2013. January;123(1):340–7. doi: 10.1172/JCI60578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jousma E, Rizvi TA, Wu J, et al. Preclinical assessments of the MEK inhibitor PD-0325901 in a mouse model of Neurofibromatosis type 1. Pediatr Blood Cancer. 2015. October;62(10):1709–16. doi: 10.1002/pbc.25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang T, Krisman K, Theobald EH, et al. Sustained MEK inhibition abrogates myeloproliferative disease in Nf1 mutant mice. J Clin Invest. 2013. January;123(1):335–9. doi: 10.1172/JCI63193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lake D, Correa SA, Muller J. Negative feedback regulation of the ERK1/2 MAPK pathway. Cell Mol Life Sci. 2016. December;73(23):4397–4413. doi: 10.1007/s00018-016-2297-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nissan MH, Rosen N, Solit DB. ERK pathway inhibitors: how low should we go? Cancer Discov. 2013. July;3(7):719–21. doi: 10.1158/2159-8290.CD-13-0245. [DOI] [PubMed] [Google Scholar]
- 57.Johannessen CM, Johnson BW, Williams SM, et al. TORC1 is essential for NF1-associated malignancies. Curr Biol. 2008. January 8;18(1):56–62. doi: 10.1016/j.cub.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 58.Hegedus B, Banerjee D, Yeh TH, et al. Preclinical cancer therapy in a mouse model of neurofibromatosis-1 optic glioma. Cancer Res. 2008. March 1;68(5):1520–8. doi: 10.1158/0008-5472.CAN-07-5916. [DOI] [PubMed] [Google Scholar]
- 59.Watson AL, Anderson LK, Greeley AD, et al. Co-targeting the MAPK and PI3K/AKT/mTOR pathways in two genetically engineered mouse models of schwann cell tumors reduces tumor grade and multiplicity. Oncotarget. 2014. March 30;5(6):1502–14. doi: 10.18632/oncotarget.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malone CF, Fromm JA, Maertens O, et al. Defining key signaling nodes and therapeutic biomarkers in NF1-mutant cancers. Cancer Discov. 2014. September;4(9):1062–73. doi: 10.1158/2159-8290.CD-14-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Massacesi C, Di Tomaso E, Urban P, et al. PI3K inhibitors as new cancer therapeutics: implications for clinical trial design. Onco Targets Ther. 2016;9:203–10. doi: 10.2147/OTT.S89967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Upadhyaya M, Spurlock G, Thomas L, et al. Microarray-based copy number analysis of neurofibromatosis type-1 (NF1)-associated malignant peripheral nerve sheath tumors reveals a role for Rho-GTPase pathway genes in NF1 tumorigenesis. Hum Mutat. 2012. April;33(4):763–76. doi: 10.1002/humu.22044. [DOI] [PubMed] [Google Scholar]
- 63.Yang FC, Ingram DA, Chen S, et al. Nf1-dependent tumors require a microenvironment containing Nf1+/−- and c-kit-dependent bone marrow. Cell. 2008. October 31;135(3):437–48. doi: 10.1016/j.cell.2008.08.041.** This is the first study which reveals that mast cells increase neoplastic Schwann cell growth through KIT ligand, suggesting the usage of imatinib mesylate for the treatment of plexiform neurofibromas.
- 64.Demestre M, Herzberg J, Holtkamp N, et al. Imatinib mesylate (Glivec) inhibits Schwann cell viability and reduces the size of human plexiform neurofibroma in a xenograft model. J Neurooncol. 2010. May;98(1):11–9. doi: 10.1007/s11060-009-0049-4. [DOI] [PubMed] [Google Scholar]
- 65.Robertson KA, Nalepa G, Yang FC, et al. Imatinib mesylate for plexiform neurofibromas in patients with neurofibromatosis type 1: a phase 2 trial. Lancet Oncol. 2012. December;13(12):1218–24. doi: 10.1016/S1470-2045(12)70414-X.** This is the first report to demonstrate that Imatinib mesylate could be used to treat plexiform neurofibromas in NF1 patients. Following the treatment the tumor volume reduced by 20% or more.
- 66.Gesundheit B, Parkin P, Greenberg M, et al. The role of angiogenesis in the transformation of plexiform neurofibroma into malignant peripheral nerve sheath tumors in children with neurofibromatosis type 1. J Pediatr Hematol Oncol. 2010. October;32(7):548–53. doi: 10.1097/MPH.0b013e3181e887c7. [DOI] [PubMed] [Google Scholar]
- 67.Rad E, Dodd K, Thomas L, et al. STAT3 and HIF1alpha Signaling Drives Oncogenic Cellular Phenotypes in Malignant Peripheral Nerve Sheath Tumors. Mol Cancer Res. 2015. July;13(7):1149–60. doi: 10.1158/1541-7786.MCR-14-0182. [DOI] [PubMed] [Google Scholar]
- 68.Luscan A, Shackleford G, Masliah-Planchon J, et al. The activation of the WNT signaling pathway is a Hallmark in neurofibromatosis type 1 tumorigenesis. Clin Cancer Res. 2013. January 15;20(2):358–71. doi: 1078–0432.CCR-13–0780 [pii] 10.1158/1078-0432.CCR-13-0780.* This report suggests implication of the Wnt pathway in NF1-associated tumorigenesis.
- 69.Mo W, Chen J, Patel A, et al. CXCR4/CXCL12 mediate autocrine cell- cycle progression in NF1-associated malignant peripheral nerve sheath tumors. Cell. 2013. February 28;152(5):1077–90. doi: 10.1016/j.cell.2013.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu J, Keng VW, Patmore DM, et al. Insertional Mutagenesis Identifies a STAT3/Arid1b/beta-catenin Pathway Driving Neurofibroma Initiation. Cell Rep. 2016. March 1;14(8):1979–90. doi: S2211–1247(16)30067–5 [pii] 10.1016/j.celrep.2016.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reilly KM, Kim A, Blakely J, et al. Neurofibromatosis Type 1-Associated MPNST State of the Science: Outlining a Research Agenda for the Future. J Natl Cancer Inst. 2017. August 1;109(8). doi: 10.1093/jnci/djx124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim A, Pratilas CA. The promise of signal transduction in genetically driven sarcomas of the nerve. Exp Neurol. 2018. January;299(Pt B):317–325. doi: 10.1016/j.expneurol.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 73.De Raedt T, Walton Z, Yecies JL, et al. Exploiting cancer cell vulnerabilities to develop a combination therapy for ras-driven tumors. Cancer Cell. 2011. September 13;20(3):400–13. doi: 10.1016/j.ccr.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malone CF, Emerson C, Ingraham R, et al. mTOR and HDAC Inhibitors Converge on the TXNIP/Thioredoxin Pathway to Cause Catastrophic Oxidative Stress and Regression of RAS-Driven Tumors. Cancer Discov. 2017. December;7(12):1450–1463. doi: 10.1158/2159-8290.CD-17-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patel AJ, Liao CP, Chen Z, et al. BET bromodomain inhibition triggers apoptosis of NF1-associated malignant peripheral nerve sheath tumors through Bim induction. Cell Rep. 2014. January 16;6(1):81–92. doi: 10.1016/j.celrep.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Varin J, Poulain L, Hivelin M, et al. Dual mTORC1/2 inhibition induces anti-proliferative effect in NF1-associated plexiform neurofibroma and malignant peripheral nerve sheath tumor cells. Oncotarget. 2016. June 14;7(24):35753–35767. doi: 7099 [pii] 10.18632/oncotarget.7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Patel AV, Eaves D, Jessen WJ, et al. Ras-driven transcriptome analysis identifies aurora kinase A as a potential malignant peripheral nerve sheath tumor therapeutic target. Clin Cancer Res. 2012. September 15;18(18):5020–30. doi: 10.1158/1078-0432.CCR-12-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Patel AV, Chaney KE, Choi K, et al. An ShRNA Screen Identifies MEIS1 as a Driver of Malignant Peripheral Nerve Sheath Tumors . EBioMedicine. 2016. July;9:110–119. doi: 10.1016/j.ebiom.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Radu M, Semenova G, Kosoff R, et al. PAK signalling during the development and progression of cancer. Nat Rev Cancer. 2014. January;14(1):13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Semenova G, Stepanova DS, Dubyk C, et al. Targeting group I p21-activated kinases to control malignant peripheral nerve sheath tumor growth and metastasis. Oncogene. 2017. September 21;36(38):5421–5431. doi: 10.1038/onc.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fischer-Huchzermeyer S, Dombrowski A, Hagel C, et al. The Cellular Retinoic Acid Binding Protein 2 Promotes Survival of Malignant Peripheral Nerve Sheath Tumor Cells. Am J Pathol. 2017. July;187(7):1623–1632. doi: 10.1016/j.ajpath.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 82.Fischer-Huchzermeyer S, Dombrowski A, Wilke G, et al. MEK inhibitors enhance therapeutic response towards ATRA in NF1 associated malignant peripheral nerve sheath tumors (MPNST) in-vitro. PLoS One. 2017;12(11):e0187700. doi: 10.1371/journal.pone.0187700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kobayashi D, Hirayama M, Komohara Y, et al. Translationally controlled tumor protein is a novel biological target for neurofibromatosis type 1-associated tumors. J Biol Chem. 2014. September 19;289(38):26314–26. doi: 10.1074/jbc.M114.568253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Varelas X The Hippo pathway effectors TAZ and YAP in devel- opment, homeostasis and disease. Development. 2014. April;141 (8):1614–1626. [DOI] [PubMed] [Google Scholar]
- 85.Kim YH, Ohta T, Oh JE, et al. TP53, MSH4, and LATS1 germline mutations in a family with clustering of nervous system tumors. Am J Pathol. 2014. September;184(9):2374–2381. [DOI] [PubMed] [Google Scholar]
- 86.Oh JE, Ohta T, Satomi K, et al. Alterations in the NF2/LATS1/LATS2/ YAP pathway in schwannomas. J Neuropathol Exp Neurol. 2015. October;74(10):952–959. [DOI] [PubMed] [Google Scholar]
- 87.Wu LMN, Deng Y, Wang J, et al. Programming of Schwann Cells by Lats1/2-TAZ/YAP Signaling Drives Malignant Peripheral Nerve Sheath Tumorigenesis. Cancer Cell. 2018. February 12;33(2):292–308 e7. doi: 10.1016/j.ccell.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chau V, Lim SK, Mo W, et al. Preclinical therapeutic efficacy of a novel pharmacologic inducer of apoptosis in malignant peripheral nerve sheath tumors. Cancer Res. 2014. January 15;74(2):586–97. doi: 10.1158/0008-5472.CAN-13-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oblinger JL, Burns SS, Akhmametyeva EM, et al. Components of the eIF4F complex are potential therapeutic targets for malignant peripheral nerve sheath tumors and vestibular schwannomas. Neuro Oncol. 2016. September;18(9):1265–77. doi: 10.1093/neuonc/now032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90..Lee MJ, Cho JH, Galas DJ, et al. The systems biology of neurofibromatosis type 1--critical roles for microRNA. Exp Neurol. 2012. June;235(2):464–8. doi: 10.1016/j.expneurol.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 91.Subramanian S, Thayanithy V, West RB, et al. Genome-wide transcriptome analyses reveal p53 inactivation mediated loss of miR-34a expression in malignant peripheral nerve sheath tumours. J Pathol. 2010. January;220(1):58–70. doi: 10.1002/path.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gong M, Ma J, Li M, et al. MicroRNA-204 critically regulates carcinogenesis in malignant peripheral nerve sheath tumors. Neuro Oncol. 2012. August;14(8):1007–17. doi: 10.1093/neuonc/nos124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Presneau N, Eskandarpour M, Shemais T, et al. MicroRNA profiling of peripheral nerve sheath tumours identifies miR-29c as a tumour suppressor gene involved in tumour progression. Br J Cancer. 2013. March 5;108(4):964–72. doi: 10.1038/bjc.2012.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Masliah-Planchon J, Pasmant E, Luscan A, et al. MicroRNAome profiling in benign and malignant neurofibromatosis type 1-associated nerve sheath tumors: evidences of PTEN pathway alterations in early NF1 tumorigenesis. BMC Genomics. 2013. July 13;14:473. doi: 10.1186/1471-2164-14-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Staser K, Yang FC, Clapp DW. Pathogenesis of plexiform neurofibroma: tumor-stromal/hematopoietic interactions in tumor progression. Annu Rev Pathol. 2012;7:469–95. doi: 10.1146/annurev-pathol-011811-132441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Staser K, Yang FC, Clapp DW. Mast cells and the neurofibroma microenvironment. Blood. 2010. July 15;116(2):157–64. doi: 10.1182/blood-2009-09-242875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Patwardhan PP, Surriga O, Beckman MJ, et al. Sustained inhibition of receptor tyrosine kinases and macrophage depletion by PLX3397 and rapamycin as a potential new approach for the treatment of MPNSTs. Clin Cancer Res. 2014. June 15;20(12):3146–58. doi: 10.1158/1078-0432.CCR-13-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ribeiro S, Napoli I, White IJ, et al. Injury signals cooperate with Nf1 loss to relieve the tumor-suppressive environment of adult peripheral nerve. Cell Rep. 2013. October 17;5(1):126–36. doi: 10.1016/j.celrep.2013.08.033. [DOI] [PubMed] [Google Scholar]
- 99.Pong WW, Higer SB, Gianino SM, et al. Reduced microglial CX3CR1 expression delays neurofibromatosis-1 glioma formation. Ann Neurol. 2013. February;73(2):303–8. doi: 10.1002/ana.23813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Choi K, Komurov K, Fletcher JS, et al. An inflammatory gene signature distinguishes neurofibroma Schwann cells and macrophages from cells in the normal peripheral nervous system. Sci Rep. 2017. March 3;7:43315. doi: 10.1038/srep43315.* This report describes inflammatory gene signatures that distinguishes neurofibroma Schwann cells and macrophages from cells in normal peripheral nervous system in mice and opens the door for many therapeutic targets
- 101.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018. March 23;359(6382):1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang S, Liechty B, Patel S, et al. Programmed death ligand 1 expression and tumor infiltrating lymphocytes in neurofibromatosis type 1 and 2 associated tumors. J Neurooncol. 2018. February 9. doi: 10.1007/s11060-018-2788-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Torres KC, Lima G, Simoes ESAC, et al. Immune markers in the RASopathy neurofibromatosis type 1. J Neuroimmunol. 2016. June 15;295–296:122–9. doi: 10.1016/j.jneuroim.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 104.Haworth KB, Arnold MA, Pierson CR, et al. Immune profiling of NF1-associated tumors reveals histologic subtype distinctions and heterogeneity: implications for immunotherapy. Oncotarget. 2017. October 10;8(47):82037–82048. doi: 10.18632/oncotarget.18301.* This is the first study providing immune profiling of NF1-associated tumors although highlights that tumor heterogeneity might pose a significant challenge to this therapeutic approach.
- 105.Schulz A, Grafe P, Hagel C, et al. Neuropathies in the setting of Neurofibromatosis tumor syndromes: Complexities and opportunities. Exp Neurol. 2018. January;299(Pt B):334–344. doi: 10.1016/j.expneurol.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 106.Moutal A, Wang Y, Yang X, et al. Dissecting the role of the CRMP2-neurofibromin complex on pain behaviors. Pain. 2017. November;158(11):2203–2221. doi: 10.1097/j.pain.0000000000001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Moutal A, Yang X, Li W, et al. CRISPR/Cas9 editing of Nf1 gene identifies CRMP2 as a therapeutic target in neurofibromatosis type 1-related pain that is reversed by (S)-Lacosamide. Pain. 2017. December;158(12):2301–2319. doi: 10.1097/j.pain.0000000000001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stansfield BK, Bessler WK, Mali R, et al. Heterozygous inactivation of the Nf1 gene in myeloid cells enhances neointima formation via a rosuvastatin-sensitive cellular pathway. Hum Mol Genet. 2013. March 1;22(5):977–88. doi: dds502 [pii] 10.1093/hmg/dds502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bessler WK, Kim G, Hudson FZ, et al. Nf1+/− monocytes/macrophages induce neointima formation via CCR2 activation. Hum Mol Genet. 2016. March 15;25(6):1129–39. doi: ddv635 [pii] 10.1093/hmg/ddv635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stevenson DA, Elefteriou F. Molecular Basis of Bone Abnormalities in NF1 in “Molecular and Cellular Biology of Neurofibromatosis Type 1” 2012:Eds: Upadhyaya M and Cooper DN Springer Verlag. .* A clear and comprehensive review of clinical skeletal phenotype of NF1, the molecular and cellular findings of bone in NF1 and the correlative animal models.
- 111.El-Hoss J, Cheng T, Carpenter EC, et al. A Combination of rhBMP-2 (Recombinant Human Bone Morphogenetic Protein-2) and MEK (MAP Kinase/ERK Kinase) Inhibitor PD0325901 Increases Bone Formation in a Murine Model of Neurofibromatosis Type I Pseudarthrosis. J Bone Joint Surg Am. 2014. July 16;96(14):e117. doi: 10.2106/JBJS.M.00862. [DOI] [PubMed] [Google Scholar]
- 112.Sullivan K, El-Hoss J, Little DG, et al. JNK inhibitors increase osteogenesis in Nf1-deficient cells. Bone. 2011. December;49(6):1311–6. doi: 10.1016/j.bone.2011.09.043. [DOI] [PubMed] [Google Scholar]
- 113.Rhodes SD, Wu X, He Y, et al. Hyperactive transforming growth factor-beta1 signaling potentiates skeletal defects in a neurofibromatosis type 1 mouse model. J Bone Miner Res. 2013. December;28(12):2476–89. doi: 10.1002/jbmr.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Karolak MR, Yang X, Elefteriou F. FGFR1 signaling in hypertrophic chondrocytes is attenuated by the Ras-GAP neurofibromin during endochondral bone formation. Hum Mol Genet. 2015. May 1;24(9):2552–64. doi: 10.1093/hmg/ddv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ghadakzadeh S, Kannu P, Whetstone H, et al. beta-Catenin modulation in neurofibromatosis type 1 bone repair: therapeutic implications. FASEB J. 2016. September;30(9):3227–37. doi: 10.1096/fj.201500190RR. [DOI] [PubMed] [Google Scholar]
- 116.Baht GS, Nadesan P, Silkstone D, et al. Pharmacologically targeting beta-catenin for NF1 associated deficiencies in fracture repair. Bone. 2017. May;98:31–36. doi: 10.1016/j.bone.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 117.Gutmann DH, Parada LF, Silva AJ, et al. Neurofibromatosis type 1: modeling CNS dysfunction. J Neurosci. 2012. October 10;32(41):14087–93. doi: 32/41/14087 [pii] 10.1523/JNEUROSCI.3242-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cui Y, Costa RM, Murphy GG, et al. Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell. 2008. October 31;135(3):549–60. doi: 10.1016/j.cell.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brown JA, Gianino SM, Gutmann DH. Defective cAMP generation underlies the sensitivity of CNS neurons to neurofibromatosis-1 heterozygosity. J Neurosci. 2010. April 21;30(16):5579–89. doi: 30/16/5579 [pii] 10.1523/JNEUROSCI.3994-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Payne JM, Barton B, Ullrich NJ, et al. Randomized placebo-controlled study of lovastatin in children with neurofibromatosis type 1. Neurology. 2016. December 13;87(24):2575–2584. doi: 10.1212/WNL.0000000000003435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brown JA, Emnett RJ, White CR, et al. Reduced striatal dopamine underlies the attention system dysfunction in neurofibromatosis-1 mutant mice. Hum Mol Genet. 2010. November 15;19(22):4515–28. doi: 10.1093/hmg/ddq382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Diggs-Andrews KA, Tokuda K, Izumi Y, et al. Dopamine deficiency underlies learning deficits in neurofibromatosis-1 mice. Ann Neurol. 2013. February;73(2):309–15. doi: 10.1002/ana.23793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Omrani A, van der Vaart T, Mientjes E, et al. HCN channels are a novel therapeutic target for cognitive dysfunction in Neurofibromatosis type 1. Mol Psychiatry. 2015. November;20(11):1311–21. doi: 10.1038/mp.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gouzi JY, Moressis A, Walker JA, et al. The receptor tyrosine kinase Alk controls neurofibromin functions in Drosophila growth and learning. PLoS Genet. 2011. September;7(9):e1002281. doi: 10.1371/journal.pgen.1002281PGENETICS-D-11-00724 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Walker JA, Gouzi JY, Long JB, et al. Genetic and functional studies implicate synaptic overgrowth and ring gland cAMP/PKA signaling defects in the Drosophila melanogaster neurofibromatosis-1 growth deficiency. PLoS Genet. 2013. November;9(11):e1003958. doi: 10.1371/journal.pgen.1003958PGENETICS-D-12-03054 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Weiss JB, Weber S, Marzulla T, et al. Pharmacological inhibition of Anaplastic Lymphoma Kinase rescues spatial memory impairments in Neurofibromatosis 1 mutant mice. Behav Brain Res. 2017. August 14;332:337–342. doi: S0166–4328(17)30814–8 [pii] 10.1016/j.bbr.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 127.Wu J, Liu W, Williams JP, et al. EGFR-Stat3 signalling in nerve glial cells modifies neurofibroma initiation. Oncogene. 2017. March 23;36(12):1669–1677. doi: 10.1038/onc.2016.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kraniak JM, Chalasani A, Wallace MR, et al. Development of 3D culture models of plexiform neurofibroma and initial application for phenotypic characterization and drug screening. Exp Neurol. 2018. January;299(Pt B):289–298. doi: 10.1016/j.expneurol.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Castellsague J, Gel B, Fernandez-Rodriguez J, et al. Comprehensive establishment and characterization of orthoxenograft mouse models of malignant peripheral nerve sheath tumors for personalized medicine. EMBO Mol Med. 2015. May;7(5):608–27. doi: 10.15252/emmm.201404430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wegscheid ML, Anastasaki C, Gutmann DH. Human stem cell modeling in neurofibromatosis type 1 (NF1). Exp Neurol. 2018. January;299(Pt B):270–280. doi: 10.1016/j.expneurol.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gutmann DH, Blakeley JO, Korf BR, et al. Optimizing biologically targeted clinical trials for neurofibromatosis. Expert Opin Investig Drugs. 2013. April;22(4):443–62. doi: 10.1517/13543784.2013.772979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Maertens O, McCurrach ME, Braun BS, et al. A Collaborative Model for Accelerating the Discovery and Translation of Cancer Therapies. Cancer Res. 2017. November 1;77(21):5706–5711. doi: 10.1158/0008-5472.CAN-17-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Weiss B, Widemann BC, Wolters P, et al. Sirolimus for progressive neurofibromatosis type 1-associated plexiform neurofibromas: a neurofibromatosis Clinical Trials Consortium phase II study. Neuro Oncol. 2015. April;17(4):596–603. doi: 10.1093/neuonc/nou235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Babovic-Vuksanovic D, Ballman K, Michels V, et al. Phase II trial of pirfenidone in adults with neurofibromatosis type 1. Neurology. 2006. November 28;67(10):1860–2. doi: 10.1212/01.wnl.0000243231.12248.67. [DOI] [PubMed] [Google Scholar]
- 135.Widemann BC, Babovic-Vuksanovic D, Dombi E, et al. Phase II trial of pirfenidone in children and young adults with neurofibromatosis type 1 and progressive plexiform neurofibromas. Pediatr Blood Cancer. 2014. September;61(9):1598–602. doi: 10.1002/pbc.25041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kim A, Dombi E, Tepas K, et al. Phase I trial and pharmacokinetic study of sorafenib in children with neurofibromatosis type I and plexiform neurofibromas. Pediatr Blood Cancer. 2013. March;60(3):396–401. doi: 10.1002/pbc.24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Widemann BC, Dombi E, Gillespie A, et al. Phase 2 randomized, flexible crossover, double-blinded, placebo-controlled trial of the farnesyltransferase inhibitor tipifarnib in children and young adults with neurofibromatosis type 1 and progressive plexiform neurofibromas. Neuro Oncol. 2014. May;16(5):707–18. doi: 10.1093/neuonc/nou004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Albritton KH, Rankin C, Coffin CM, et al. Phase II study of erlotinib in metastatic or unresectable malignant peripheral nerve sheath tumors (MPNST). Journal of Clinical Oncology. 2006;24(18_suppl):9518–9518. doi: 10.1200/jco.2006.24.18_suppl.9518. [DOI] [Google Scholar]
- 139.Chugh R, Wathen JK, Maki RG, et al. Phase II Multicenter Trial of Imatinib in 10 Histologic Subtypes of Sarcoma Using a Bayesian Hierarchical Statistical Model. Journal of Clinical Oncology. 2009;27(19):3148–3153. doi: 10.1200/jco.2008.20.5054. [DOI] [PubMed] [Google Scholar]
- 140.Maki RG, D’Adamo DR, Keohan ML, et al. Phase II study of sorafenib in patients with metastatic or recurrent sarcomas. J Clin Oncol. 2009. July 1;27(19):3133–40. doi: 10.1200/JCO.2008.20.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schuetze SM, Wathen JK, Lucas DR, et al. SARC009: Phase 2 study of dasatinib in patients with previously treated, high-grade, advanced sarcoma. Cancer. 2016. March 15;122(6):868–74. doi: 10.1002/cncr.29858. [DOI] [PubMed] [Google Scholar]
- 142.Dickson MA, Mahoney MR, Tap WD, et al. Phase II study of MLN8237 (Alisertib) in advanced/metastatic sarcoma. Ann Oncol. 2016. October;27(10):1855–60. doi: 10.1093/annonc/mdw281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Widemann BC, Meyer CF, Cote GM, et al. SARC016: Phase II study of everolimus in combination with bevacizumab in sporadic and neurofibromatosis type 1 (NF1) related refractory malignant peripheral nerve sheath tumors (MPNST). Journal of Clinical Oncology. 2016;34(15_suppl):11053–11053. doi: 10.1200/JCO.2016.34.15_suppl.11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Karajannis MA, Legault G, Fisher MJ, et al. Phase II study of sorafenib in children with recurrent or progressive low-grade astrocytomas. Neuro Oncol. 2014. October;16(10):1408–16. doi: 10.1093/neuonc/nou059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ater JL, Xia C, Mazewski CM, et al. Nonrandomized comparison of neurofibromatosis type 1 and non-neurofibromatosis type 1 children who received carboplatin and vincristine for progressive low-grade glioma: A report from the Children’s Oncology Group. Cancer. 2016. June 15;122(12):1928–36. doi: 10.1002/cncr.29987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lion-Francois L, Gueyffier F, Mercier C, et al. The effect of methylphenidate on neurofibromatosis type 1: a randomised, double-blind, placebo-controlled, crossover trial. Orphanet J Rare Dis. 2014. September 10;9:142. doi: 10.1186/s13023-014-0142-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.van der Vaart T, Rietman AB, Plasschaert E, et al. Behavioral and cognitive outcomes for clinical trials in children with neurofibromatosis type 1. Neurology. 2016. January 12;86(2):154–60. doi: 10.1212/WNL.0000000000002118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.van der Vaart T, Plasschaert E, Rietman AB, et al. Simvastatin for cognitive deficits and behavioural problems in patients with neurofibromatosis type 1 (NF1-SIMCODA): a randomised, placebo-controlled trial. Lancet Neurol. 2013. November;12(11):1076–83. doi: 10.1016/S1474-4422(13)70227-8. [DOI] [PubMed] [Google Scholar]
- 149.Stivaros S, Garg S, Tziraki M, et al. Randomised controlled trial of simvastatin treatment for autism in young children with neurofibromatosis type 1 (SANTA). Mol Autism. 2018;9:12. doi: 10.1186/s13229-018-0190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]