Summary
Clostridia are obligate anaerobic bacteria that can produce solvents such as acetone, butanol and ethanol. Alcohol dehydrogenases (ADHs) play a key role in solvent production; however, their regulatory mechanisms remain largely unknown. In this study, we characterized the regulatory mechanisms of two ADH‐encoding genes in C. beijerinckii. SigL (sigma factor σ54) was found to be required for transcription of adhA1 and adhA2 genes. Moreover, a novel transcriptional activator AdhR was identified, which binds to the σ54 promoter and activates σ54‐dependent transcription of adhA1 and adhA2. Clostridia beijerinckii mutants deficient in SigL or AdhR showed severely impaired butanol and ethanol production as well as altered acetone and butyrate synthesis. Overexpression of SigL resulted in significantly improved solvent production by C. beijerinckii when butyrate was added to cultures. Our results reveal SigL as a novel engineering target for improving solvent production by C. beijerinckii and other solvent‐producing clostridia. Moreover, this study gains an insight into regulation of alcohol metabolism in diverse clostridia.
A novel regulatory system of alcohol synthesis in Clostridia beijerinckii was characterized. This study provides a new insight into regulation of ADH‐encoding genes in diverse Clostridium species and a novel engineering target to improve solvent production in solvent‐producing clostridia.
![]()
Introduction
The clostridial solvent production, also known as the acetone–butanol–ethanol (ABE) fermentation, was one of the largest industrial fermentation processes early in the 20th century (Durre, 2007). Due to the growing concerns on energy security and climate change, biological production of butanol has been receiving renewed interest during the last 10 years (Lee et al., 2008; Papoutsakis, 2008; Cheng et al., 2019). This is because butanol possesses superior fuel properties compared with ethanol (Durre, 2007; Lee et al., 2008). Clostridia, which are Gram‐positive obligate anaerobes, can produce fuels and chemicals through fermentation of a variety of carbon sources including sugars, cellulose, and CO2 or CO (Durre, 2007; Tracy et al., 2012; Ren et al., 2016). Among solventogenic clostridia, Clostridium beijerinckii is one of the best‐studied species (Lee et al., 2008; Ezeji et al., 2010). Random mutagenesis has been used to enhance its solvent production (Green, 2011). Attempts have also been made to improve the strain performance based on genetic engineering strategies (Charubin et al., 2018; Cheng et al., 2019).
Alcohol dehydrogenases (ADHs) play a key role in solvent production in clostridia, which catalyze the interconversion between aldehydes and ketones and their corresponding alcohols (Reid and Fewson, 1994; Radianingtyas and Wright, 2003). A few global transcriptional regulators including Spo0A, CcpA, AbrB and Rex have been found to regulate ADH‐encoding genes in Clostridium acetobutylicum (Yang et al., 2018). However, little is known about the regulatory mechanisms of ADH‐encoding genes in other clostridia such as C. beijerinckii. Among the 20 ADH‐encoding genes in C. beijerinckii, two genes (Cbei_2181 and Cbei_1722) encoding iron‐containing/activated ADHs were strongly induced at the onset of solvent production, suggesting that they are responsible for synthesis of butanol and ethanol (Wang, et al., 2012). Unlike in C. acetobutylicum, lack of the redox‐sensing regulator Rex did not affect the fermentation product formation in C. beijerinckii, suggesting that this species may have another mechanism for modulating alcohol production Zhang et al., 2014). Recently, sigma factor σ54 was found to play a central role in carbon metabolism in C. beijerinckii (Hocq et al., 2019), but the exact regulatory mechanism remains to be elucidated.
σ54 is unique in that it shares no detectable homology with any of the other known sigma factors and binds to conserved −12 and −24 promoter elements (Buck and Cannon, 1992; Barrios et al., 1999). The σ54‐dependent transcription absolutely requires the presence of an activator that couples the energy generated from ATP hydrolysis to the isomerization of the RNA polymerase‐σ54 closed complex (Schumacher et al., 2004; Bush and Dixon, 2012). These activators, also called enhancer‐binding proteins (EBPs), bind to upstream activator sequences (UAS) located upstream of the promoter. In a previous study, we have identified putative EBPs and reconstructed σ54 regulons in diverse Clostridium species by using comparative genomic approaches (Nie et al., 2019). The reconstructed σ54 regulons contain the genes associated with butyrate and alcohol synthesis. A large number of ADH‐encoding genes were predicted to be regulated by σ54 (SigL) and individual EBPs (Nie et al., 2019).
In this study, we verified the putative σ54 promoter elements upstream of ADH‐encoding genes in the genomes of Clostridium spp. by using in vitro binding assays. We found that SigL is required for transcription of the adhA1 (Cbei_2181) and adhA2 (Cbei_1722) genes in C. beijerinckii. Then, a novel EBP, namely AdhR, was experimentally characterized as an activator of σ54‐dependent transcription of adhA1 and adhA2. Furthermore, we studied the effects of disruption of SigL‐ or AdhR‐encoding genes on fermentation product formation. Our results indicated that SigL and AdhR control alcohol synthesis in C. beijerinckii and overexpression of SigL can improve the solvent production.
Results
SigL binds to the −12 and −24 promoter elements in vitro
To validate the predicted regulation of clostridial ADH‐encoding genes by σ54 (Fig. 1A), electrophoretic mobility shift assays (EMSAs) were performed using the recombinant SigL protein from C. beijerinckii, which was overexpressed in E. coli with the N‐terminal His6 tag and purified with a nickel‐chelating affinity column. The DNA fragments from the promoter regions of adhA1 and adhA2 in C. beijerinckii, which contain the putative −12 and −24 promoter elements, were tested in EMSAs. We observed that SigL protein binds to the DNA fragments in a concentration‐dependent manner (Fig. 1B and C). The DNA fragments were completely shifted with 200 nM SigL. In contrast, no binding was observed for the mutated fragments with substitutions of the strictly conserved GC and GG dinucleotides at −12 and −24 regions, respectively, even at 500 nM SigL protein (Fig. 1B and C). Thus, SigL binds to the −12 and −24 promoter elements of adhA1 and adhA2 in C. beijerinckii.
Figure 1.
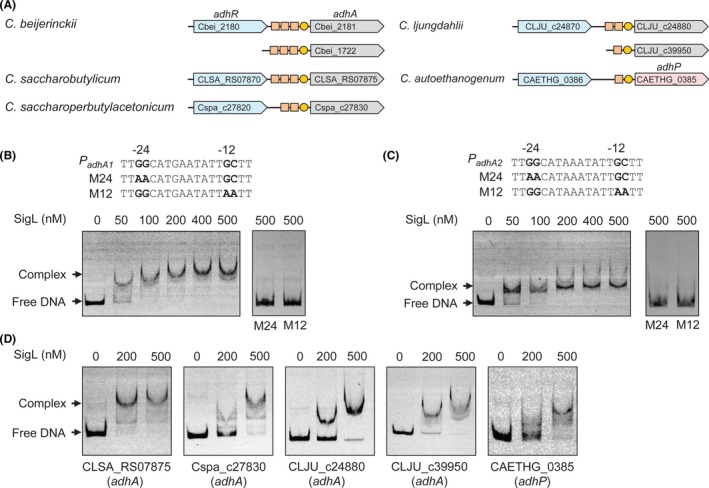
SigL binds to −12 and −24 promoter elements upstream of ADH‐encoding genes.
A. Genomic context of ADH‐encoding genes in Clostridium species. Genes are shown by arrows. Predicted binding sites of SigL and AdhR are shown by yellow circles and squares respectively.
B, C. EMSAs with purified SigL protein and DNA fragments from the promoter region of adhA1 (B) and adhA2 (C). Binding of SigL to the promoter fragments with mutations at −24 (M24) and −12 (M12) elements was also shown.
D. EMSAs with purified SigL protein and DNA fragments from the promoter regions of ADH‐encoding genes in Clostridium species other than C. beijerinckii. The DNA fragments contain the putative −12 and −24 elements.
Electrophoretic mobility shift assays were also performed to assess the predicted SigL‐binding sites upstream of the ADH‐encoding genes in other clostridia. Conserved −12 and −24 elements were identified in the promoter region of 17 adhA genes encoding iron‐containing/activated ADHs and of 20 adhP genes encoding zinc‐dependent ADHs in our previous study (Nie et al., 2019). Among them, five DNA fragments were amplified from the promoter regions of adhA or adhP genes in Clostridium saccharobutylicum, Clostridium saccharoperbutylacetonicum, Clostridium ljungdahlii and Clostridium autoethanogenum (Fig. 1A). These DNA fragments were tested for binding of C. beijerinckii SigL protein that is well conserved in clostridia. A shift in the presence of purified SigL was observed for all the five fragments (Fig. 1D), suggesting that regulation of the ADH‐encoding genes by SigL is conserved in diverse Clostridium species.
SigL is required for transcription of adhA1 and adhA2 genes in C. beijerinckii
To examine whether SigL regulates expression of adhA1 and adhA2 genes, the sigL gene (Cbei_0595) in C. beijerinckii was disrupted by inserting an intron (Fig. S1). We compared the gene expression between the sigL‐inactivated mutant (sigL mutant) and the wild type by using quantitative real‐time PCR (qRT‐PCR). The transcript levels of both adhA1 and adhA2 were decreased by more than 90% in the sigL mutant compared with the wild type (Fig. 2A). Complementation of the sigL mutant by using a plasmid construct constitutively expressing sigL restored the transcript levels of adhA1 and adhA2. We constructed C. beijerinckii strains expressing lacZ under the control of adhA1 or adhA2 promoters. The sigL mutant, compared with the wild type, exhibited markedly lower levels of expression from adhA1 and adhA2 promoters, as determined by β‐galactosidase assays (Fig. 2B). Moreover, mutations of the −12 and −24 elements for SigL binding decreased the activities of adhA1 and adhA2 promoters by 72−95% (Fig. 2C). These results indicate that SigL is required for transcription of adhA1 and adhA2 genes in C. beijerinckii.
Figure 2.
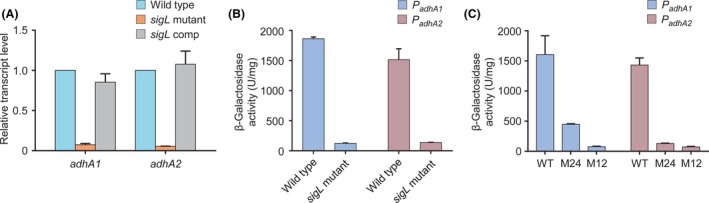
SigL is required for transcription of adhA1 and adhA2 genes.
A. Effect of sigL inactivation on transcript levels of adhA1 and adhA2. The transcript levels of each gene in the sigL‐inactivated mutant (sigL mutant) and sigL‐complemented strain (sigL comp) were normalized to the gene expression in the wild type.
B. Effect of sigL inactivation on expression from adhA1 and adhA2 promoters. The activities of adhA1 and adhA2 promoters were determined using β‐galactosidase assays.
C. Effect of mutations of SigL‐binding sites on adhA1 and adhA2 promoter activities. See Fig. 1 for M24 and M12. Error bars represent the standard deviation of three biological replicates.
AdhR activates σ54‐dependent transcription of adhA1 and adhA2
In a previous study, we made the hypothesis that adhA1 and adhA2 genes are regulated by a novel EBP, namely AdhR, in C. beijerinckii (Nie et al., 2019). To test it, we investigated the effect of inactivation of adhR (Cbei_2180) on target gene expression by using qRT‐PCR and β‐galactosidase assays. We observed that the transcript levels of adhA1 and adhA2 were decreased by 90% in the adhR mutant compared with the wild type (Fig. 3A). Consistently, compared with the wild type, the adhR mutant showed substantially lower levels of expression from adhA1 and adhA2 promoters (Fig. 3B). Complementation of the adhR mutant by using a plasmid expressing adhR increased the expression of adhA1 and adhA2 (Fig. 3A). Thus, AdhR is a positive regulator of adhA1 and adhA2 genes. AdhR contains a central AAA+ (ATPase associated with various cellular activities) domain that is highly homologous (> 50% sequence identity) to the σ54‐interacting domain of transcriptional activators NtrC and NifA (Martinez‐Argudo et al., 2004; De Carlo et al., 2006). We found that a small histone‐like protein HU (encoded by Cbei_0091), which shares sequence homology with the HBsu protein from B. subtilis (Micka and Marahiel, 1992), was also involved in transcription of adhA1 and adhA2 (Fig. 3C). The HU protein may facilitate DNA looping to enable the interaction between σ54 and AdhR (Hoover et al., 1990).
Figure 3.
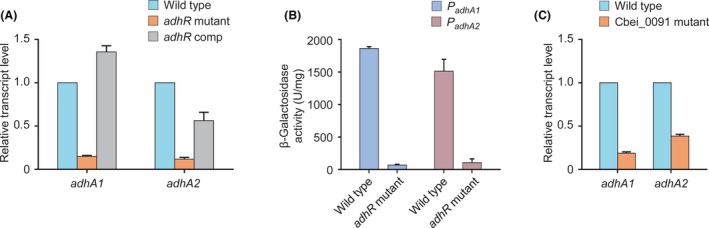
Effect of adhR inactivation on transcription of adhA1 and adhA2 genes.
A. Effect of adhR inactivation on transcript levels of adhA1 and adhA2. The transcript levels of each gene in the adhR‐inactivated mutant (adhR mutant) and adhR‐complemented strain (adhR comp) were normalized to the gene expression in the wild type.
B. Effect of adhR inactivation on expression from promoters of adhA1 and adhA2.
C. Effect of inactivation of the gene encoding a small heterodimeric protein HU (Cbei_0091) on transcript levels of adhA1 and adhA2. The transcript levels of each gene were normalized to the gene expression in the wild‐type strain. Error bars represent the standard deviation of three biological replicates.
The C‐terminal domain of AdhR protein is a helix–turn–helix (HTH) DNA‐binding domain (DBD). Three AdhR‐binding sites (i.e. UAS sites) upstream of the σ54 promoter elements was predicted for both adhA1 and adhA2 genes in our previous study (Nie, et al., 2019). To test whether these UAS sites are involved in expression of adhA1 and adhA2, mutations were introduced into individual UAS (Fig. 4A). We observed that disruption of any one of the three UAS sites severely impaired the activities of both adhA1 and adhA2 promoters and the triple mutation resulted in < 2% of the promoter activities (Fig. 4B). Thus, all the three UAS sites contribute to transcriptional activation of adhA1 and adhA2. To confirm the direct binding of AdhR to the UAS sites, EMSAs were performed using purified recombinant DBD of AdhR protein (AdhR‐DBD) from C. beijerinckii and the DNA fragments containing all three UAS sites. For both adhA1 and adhA2 promoter fragments, shifted bands were visible in the presence of increasing amounts of AdhR‐DBD protein (Fig. 4C). Therefore, the above results indicate that AdhR binds to the three UAS sites upstream of the promoter and activates the transcription of adhA1 and adhA2 genes in C. beijerinckii.
Figure 4.
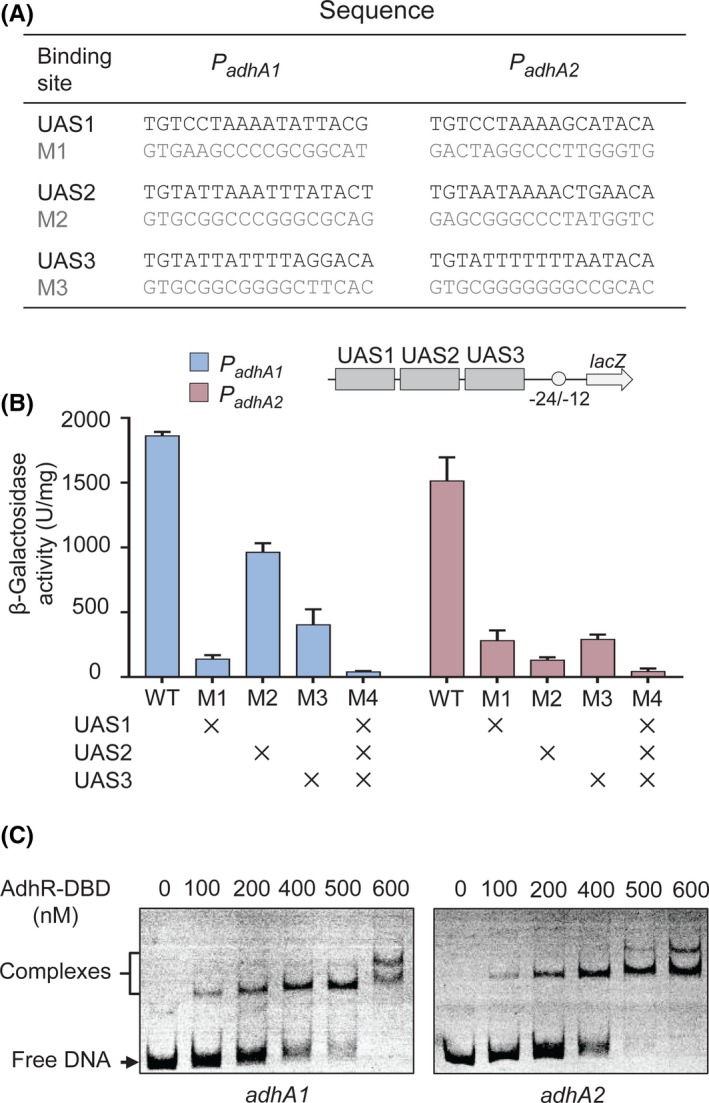
AdhR activates adhA1 and adhA2 promoters by binding to three UAS sites.
A. Mutations introduced into each of the three UAS sites (M1 − M3) in the adhA1 and adhA2 promoters. The mutated sequences are shown below the wild‐type sequences.
B. Effect of UAS mutations on activities of adhA1 and adhA2 promoters. The mutations are indicated by ‘×’. Error bars represent the standard deviation of three biological replicates.
C. EMSAs with purified C. beijerinckii AdhR‐DBD and DNA fragment from the promoter region of adhA1 and adhA2.
SigL and AdhR control solvent synthesis
To elucidate the role of σ54 and AdhR in regulation of central metabolism in C. beijerinckii, we compared the cell growth and fermentation products between wild type and adhR and sigL mutants. Inactivation of adhR or sigL severely impaired the growth of the resulting strains on glucose (Fig. 5). Compared with the wild type, ethanol, butanol and acetone production by both sigL and adhR mutants decreased by 74%, 85% and 70% respectively (Fig. 5). Both mutants accumulated a high level (~ 3 g l−1) of butyric acid (Fig. 5). The production of acetic acid by the mutants was not changed significantly compared with other fermentation products. These results indicate that σ54 and AdhR not only control the butanol and ethanol production by directly regulating adhA1 and adhA2 genes but also influence the synthesis of acetone and butyric acid.
Figure 5.
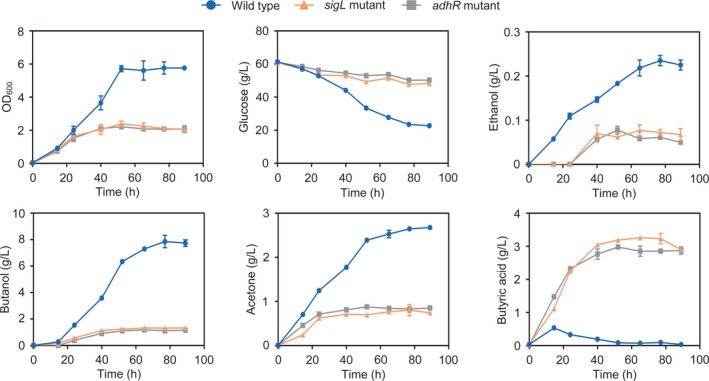
Cell growth and fermentation product formation in batch cultures of C. beijerinckii wild‐type strain and adhR and sigL mutants. The OD600, concentrations of glucose, ethanol, butanol, acetone and butyric acid in the medium were measured during the cultivation. Error bars represent the standard deviation of three biological replicates.
Overexpression of SigL can improve solvent production by C. beijerinckii
To investigate the effect of overexpression of SigL and AdhR, we constructed C. beijerinckii strains, in which sigL or adhR genes from C. beijerinckii were expressed in a plasmid by the promoter of thiolase (thl). The strain carrying an empty‐vector plasmid was used as a control. Then, we compared the fermentation performances of the SigL‐ or AdhR‐overexpressing strains and the control strain in batch cultures. During the exponential growth phase, the SigL‐overexpressing strain exhibited slightly increased rates of glucose consumption and ethanol production compared to the control strain (Fig. 6). Although butanol production was almost unchanged by SigL overexpression, acetone production was greatly enhanced throughout the cultivation, leading to a 37% increase in the titre of acetone (Fig. 6). However, overexpression of AdhR resulted in decreases in cell growth and solvent production (Fig. 6). Interestingly, we observed that SigL overexpression led to drastically reduced synthesis and reassimilation of butyric acid (Fig. 6). This result suggests that the availability of butyryl coenzyme A (butyryl‐CoA) is limited in the SigL‐overexpressing strain (Tummala et al., 2003; Sillers et al., 2009).
Figure 6.
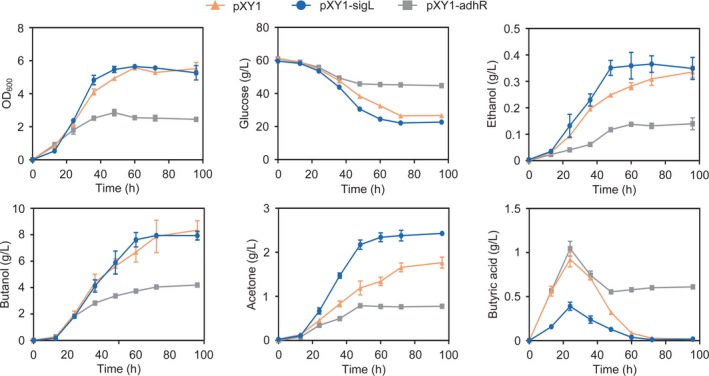
Cell growth and fermentation product formation in batch cultures of C. beijerinckii SigL‐ or AdhR‐overexpressing strains. The strain carrying an empty‐vector plasmid (pXY1) was used as a control. The OD600, concentrations of glucose, ethanol, butanol, acetone and butyric acid in the medium were measured during the cultivation. Error bars represent the standard deviation of three biological replicates.
To study whether an increased availability of butyryl‐CoA can improve the butanol production by the SigL‐overexpressing strain, we added 3.5 g l−1 of butyrate to the culture medium when the butyrate concentration started to decline. Exogenously added butyrate was quickly consumed by the SigL‐overexpressing strain, leading to a marked increase in butanol production from 7.9 to 11.25 g l−1 (Fig. 7). Compared with the control strain, the SigL‐overexpressing strain showed significantly increased rates of cell growth and glucose consumption after butyrate was added (Fig. 7). Production of butanol, ethanol and acetone was increased by 50%, 96% and 93%, respectively, by SigL overexpression (Fig. 7). The titre of total solvents was increased 1.6‐fold from 10.7 to 17.6 g l−1 in the SigL‐overexpressing strain compared to that in the control strain. By adding butyrate at the beginning of batch culture, we also observed enhanced solvent production by the SigL‐overexpressing strain compared with the control strain. Therefore, overexpression of SigL can significantly improve solvent production by C. beijerinckii.
Figure 7.
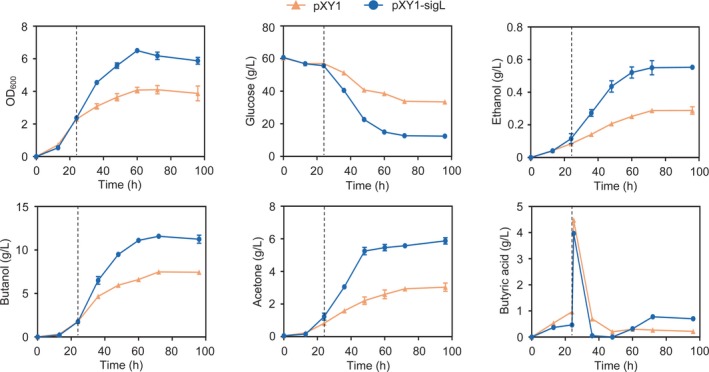
Product profiles of C. beijerinckii SigL‐overexpressing strain fermentation with addition of butyrate. The strain carrying an empty‐vector plasmid (pXY1) was used as a control. The vertical dashed line indicates the time at which butyrate was added to both the control strain and SigL‐overexpressing strain cultures. The OD600, concentrations of glucose, ethanol, butanol, acetone and butyric acid in the medium were measured during the cultivation. Error bars represent the standard deviation of three biological replicates.
Discussion
In this study, SigL (σ54) was identified as a master regulator of solvent synthesis in C. beijerinckii. We found that SigL, which binds to −12 and −24 promoter elements, is required for transcription of adhA1 and adhA2 in C. beijerinckii (Fig. 8). Moreover, we identified AdhR as an activator of σ54‐dependent transcription of adhA1 and adhA2. AdhR binds to three UAS sites upstream of the promoter. C. beijerinckii mutants deficient in SigL or AdhR showed severely impaired butanol and ethanol synthesis. We propose that AdhR interacts with σ54 via DNA looping facilitated by HU protein and activates the σ54‐dependent transcription of adhA1 and adhA2, leading to ADH‐catalyzed conversion of aldehydes to alcohols (Fig. 8). Based on the understanding of regulation of alcohol synthesis, we overexpressed SigL in C. beijerinckii, which resulted in significantly improved solvent production.
Figure 8.
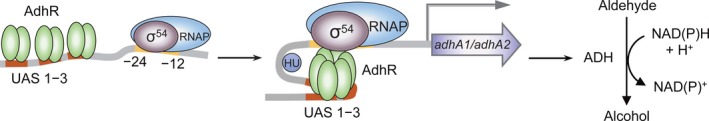
Schematic of the proposed mechanism for regulation of adhA1 and adhA2 genes by σ54 (SigL) and AdhR. σ54 directs the RNA polymerase holoenzyme to bind at the −12 and −24 promoter elements, and AdhR binds to three UAS sites upstream of the promoter. AdhR interacts with σ54 via DNA looping facilitated by HU protein and activates the σ54‐dependent transcription of adhA1 and adhA2 encoding ADHs, which catalyzes the conversion of aldehydes to alcohols.
Genetic engineering has been applied to enhance solvent production by C. beijerinckii; however, most of the attempts did not result in a desired solvent producer (Lu et al., 2017; Wen et al., 2017). Although a few transcriptional factors including Spo0A, CcpA, AbrB and Rex have been found to regulate solvent production in C. acetobutylicum (Yang et al., 2018), little is known about the regulatory mechanisms of solvent production in other clostridia. Our finding, which indicates that alcohol synthesis is controlled by σ54, proposes a novel engineering target for improving solvent production by C. beijerinckii and other solvent‐producing clostridia. As expected, SigL overexpression resulted in increased ethanol synthesis by directly upregulating adhA1 and adhA2 in C. beijerinckii. Butanol production was also significantly enhanced by SigL overexpression when butyryl‐CoA availability was increased by adding butyrate to cultures. The SigL‐overexpressing strain also exhibited a remarkable increase in acetone production compared to the control strain. A previous report has shown that overexpression of the gene for alcohol synthesis led to upregulation of the genes involved in acetone synthesis (ctfA, ctfB and adc) in C. acetobutylicum (Tummala et al., 2003). Thus, we speculate that SigL may indirectly modulate acetone synthesis via upregulation of adhA1 and adhA2. In addition, the increased glucose consumption rate of the SigL‐overexpressing strain may be explained by enhancement of glycolytic activity by decreased NADH/NAD+ ratio due to an increase in alcohol production (Liang et al., 2013). The solvent production by the SigL‐overexpressing strain could be further improved by engineering cellular metabolism for the increased availability of butyryl‐CoA as well as optimizing fermentation and product recovery processes (e.g. using gas stripping for solvent recovery) (Ezeji et al., 2003; Hou et al., 2013).
AdhR is a novel activator of σ54‐dependent transcription. The C‐terminal domain of AdhR directs its binding to UAS sites, and the central AAA+ domain is responsible for interaction with σ54. In addition, AdhR contains an N‐terminal GAF domain (named for cyclic GMP‐specific and stimulated phosphodiesterases, Anabaena adenylate cyclases and E. coli FhlA) and a PAS (Per, ARNT and Sim) domain, which are assumed to have a role in signal perception, and modulates the activity of AdhR (Aravind and Ponting, 1997; Taylor and Zhulin, 1999; Henry and Crosson, 2011). We speculate that transcriptional activation of adhA1 and adhA2 by AdhR is controlled by these sensory modules rather than AdhR abundances. Instead, the presence of a large amount of inactive AdhR protein may be a burden to cells. This may explain why overexpression of AdhR resulted in impaired cell growth and did not enhance solvent production. The studies on the mechanism of transcriptional activation by AdhR, including the signals perceived by the N‐terminal regulatory domains and how these domains control the activity of the central AAA+ domain, are now under way. A deep insight into the mechanism for AdhR regulation may allow designing sophisticated strategies of AdhR protein engineering to improve solvent production by C. beijerinckii.
A previous report has shown that adhA1 and adhA2 are members of Rex regulon in C. beijerinckii (Zhang et al., 2014). However, inactivation of rex had only a modest effect on expression of adhA1 and adhA2 and did not change the fermentation product profile in C. beijerinckii. Our study indicates that SigL and AdhR are required for transcription of adhA1 and adhA2. Compared with the wild type, ethanol production and butanol production were drastically reduced in sigL and adhR mutants. This may lead to an increase in intracellular NADH/NAD+ ratio and thus depression of Rex target operons including those involved in butyrate synthesis (crt‐bcd‐etfBA‐hbd and ptb‐buk operons). Consistently, we observed markedly increased butyrate synthesis in both sigL and adhR mutants, which can consume large amounts of NADH. The regulatory system composed of SigL and AdhR and the Rex regulator may sense different signals and act cooperatively to maintain redox homeostasis in C. beijerinckii.
This study provided experimental evidence that SigL binds upstream of adhA genes in other solvent‐producing species including C. saccharobutylicum and C. saccharoperbutylacetonicum, suggesting that SigL may also regulate solvent synthesis in these species. We also observed binding of SigL to the promoter regions of ADH‐encoding genes in gas‐fermenting species including C. ljungdahlii and C. autoethanogenum. Of these adhA and adhP genes, CAETHG_0386 is involved in 2,3‐butanediol synthesis (Kopke et al., 2011), and CLJU_c24880 and CLJU_c39950 are involved in butanol degradation (Tan et al., 2014). Thus, SigL may play a role in regulation of alcohol metabolism in various Clostridium species.
Experimental procedures
Strains and culture conditions
The bacterial strains and plasmids used in this study were summarized in Table S1. Clostridium beijerinckii NCIMB 8052 strains were grown anaerobically in clostridial growth medium (CGM) (Wiesenborn et al., 1989). Erythromycin (30 μg ml−1) or spectinomycin (350 μg ml−1) was added when needed. A single colony was inoculated into a test tube containing 5 ml CGM, anaerobically cultured at 37°C for 12 h. Cells were transferred to 50 ml fresh CGM and grown anaerobically to mid‐exponential growth phase (optical density at 600 nm [OD600] of ~ 0.6). Then, 2.5 ml of the culture aliquot was transferred to a flask with 60 ml of P2 minimal medium, which contains (per litre) 0.5 g KH2PO4, 0.5 g K2HPO4, 0.01 g NaCl, 0.2 g MgSO4 · 7H2O, 0.01 g MnSO4 · H2O, 0.01 g FeSO4 · 7H2O, 1 mg p‐aminobenzoic acid, 1 mg vitamin B1, 0.01 mg biotin, 2.2 g CH3COONH4 and 60 g glucose (Baer et al., 1987). The culture was grown anaerobically at 37°C, and samples were taken from the flasks for measurements of OD600 and cellular metabolites, RNA isolation and β‐galactosidase assay.
Mutant construction
Gene disruption in C. beijerinckii was performed by using group II intron‐based targetron technology as described previously (Shao et al., 2007). Briefly, a 350 bp fragment for retargeting an intron to insert within the sigL (Cbei_0595), adhR (Cbei_2180) or Cbei_0091 genes was generated by one‐step assembly PCR using the primers shown in Table S2 based on the protocol of TargeTronTM gene knockout system (Sigma, Sigma‐Aldrich, Darmstadt, Germany). The PCR product was ligated to a targetron vector pWJ1 (Xiao et al., 2011) using One Step Seamless Assembly Super Kit (Paisiwen Co., Ltd, Shanghai, China), yielding the plasmids pWJ1‐sigL, pWJ1‐adhR and pWJ1‐Cbei0091 (Table S1). Each plasmid was electroporated into C. beijerinckii. The transformants were selected on CGM plate supplemented with erythromycin. The resulting mutant with an intron insertion in the respective gene was confirmed by PCR.
For complementation of the gene inactivation, the sigL and adhR genes were PCR‐amplified from the C. beijerinckii NCIMB 8052 genomic DNA and cloned into the pXY1 vector under the control of the constitutive P thl promoter (Tummala et al., 1999). The obtained plasmids pXY1‐sigL and pXY1‐adhR were electroporated into the sigL‐inactivated mutant and the adhR‐inactivated mutant respectively (Table S1). For overexpression of sigL or adhR genes, plasmid pXY1‐sigL or pXY1‐adhR was electroporated into wild‐type C. beijerinckii (Table S1).
RNA isolation and real‐time PCR analysis
Total RNA was isolated from C. beijerinckii cells that were harvested at OD600 of ~ 2.0. RNA isolation, and real‐time PCR analysis was performed as described previously (Zhang et al., 2014).
β‐Galactosidase assay
For lacZ reporter experiments, plasmids containing either a wild‐type or a mutated promoter DNA of adhA genes were constructed (Table S1). A 380 bp fragment of adhA1 (Cbei_2181) promoter and a 475 bp fragment of adhA2 (Cbei_1722) promoter, both of which contain the putative ‐12 and ‐24 elements and three UAS sites, were PCR‐amplified using C. beijerinckii genome as template and the primers shown in Table S2. The mutated promoter DNAs were synthesized by GenScript. The wild‐type or mutated promoter DNA was cloned into pIMPI‐lacZ plasmid to control lacZ expression (Feustel et al., 2004). Clostridium beijerinckii strains harbouring either the adhA1 or adhA2 promoters upstream of the lacZ reporter were grown in P2 minimal medium and harvested at OD600 of about 2.0 by centrifugation. The crude cell extract was prepared, and the β‐galactosidase activity was determined by measuring the absorbance at 420 nm at 60°C (Tummala et al., 1999). One unit is defined as the amount of the enzyme that catalyzes the formation of 1 nmol of o‐nitrophenol (extinction coefficient, 0.0045 μM−1 cm−1) per min.
Protein purification
The sigL gene and the DNA coding for truncated AdhR (AdhR‐DBD) were PCR‐amplified from C. beijerinckii genome using the primers shown in Table S2. The PCR fragments were cloned into the expression vector pET28a (Table S1). The resulting plasmids pET28a‐sigL and pET28a‐adhR‐DBD (Table S1) were sequenced to exclude unwanted mutations and used to produce the N‐terminal hexahistidine‐tagged SigL and AdhR‐DBD respectively. Recombinant protein overexpression and purification were performed as described previously (Nie et al., 2016).
Electrophoretic mobility shift assay
A 200 bp DNA fragment containing the putative −24 and −12 elements in the promoter region of adhA1 or adhA2 genes was PCR‐amplified from C. beijerinckii genome using the primers shown in Table S2. The DNA fragments containing the putative −24 and −12 elements upstream of CLSA_RS07875 gene from C. saccharobutylicum, Cspa_c27830 gene from C. saccharoperbutylacetonicum, CLJU_c24880 and CLJU_c39950 genes from C. ljungdahlii and CAETHG_0385 gene from C. autoethanogenum were chemically synthesized by GenScript. The DNA fragment containing the three UAS sites in the promoter region of adhA1 or adhA2 genes was PCR‐amplified from C. beijerinckii genome using the primers shown in Table S2. Both forward and reverse primers were Cy5 fluorescence labelled at the 5′‐end. Mobility shift assays were performed as described previously (Zhang et al., 2014).
Metabolite analysis
For analysis of extracellular metabolites, culture samples were centrifuged for 10 min at 4°C and 15 000 g to remove the cells. Acetone, butanol and ethanol were detected by gas chromatography as described previously (Liu et al., 2012). Glucose concentration in culture supernatant was detected by high‐pressure liquid chromatography using an Agilent model 1260 instrument equipped with a Sugar‐PakTM I column (Waters, Milford, MA, USA) and a refractive index detector (Agilent Technologies, Santa Clara, CA, USA).
Statistical analysis
Unless noted otherwise, data are presented as the mean ± SD of n independent experiments.
Conflict of interests
None declared.
Supporting information
Fig. S1. Construction and validation of gene inactivation mutants of C. beijerinckii. The coding region of sigL (Cbei_0595) or adhR (Cbei_2180) genes was inserted with an intron. The resulting mutants were confirmed by PCR.
Table S1. Strains and plasmids used in this study.
Table S2. Oligonucleotides used in this study.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31630003, 31900044 and 31921006), National Key R&D Program of China (2016YFC1303303) and Chinese Academy of Sciences (XDB27020201).
Microbial Biotechnology (2020) 13(2), 328–338
Funding Information
This work was supported by the National Natural Science Foundation of China (31630003, 31900044 and 31921006), National Key R&D Program of China (2016YFC1303303) and Chinese Academy of Sciences (XDB27020201).
References
- Aravind, L. , and Ponting, C.P. (1997) The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci 22: 458–459. [DOI] [PubMed] [Google Scholar]
- Baer, S.H. , Blaschek, H.P. , and Smith, T.L. (1987) Effect of butanol challenge and temperature on lipid composition and membrane fluidity of butanol‐tolerant Clostridium acetobutylicum . Appl Environ Microbiol 53: 2854–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios, H. , Valderrama, B. , and Morett, E. (1999) Compilation and analysis of sigma(54)‐dependent promoter sequences. Nucleic Acids Res 27: 4305–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, M. , and Cannon, W. (1992) Specific binding of the transcription factor sigma‐54 to promoter DNA. Nature 358: 422–422. [DOI] [PubMed] [Google Scholar]
- Bush, M. , and Dixon, R. (2012) The role of bacterial enhancer binding proteins as specialized activators of sigma54‐dependent transcription. MMBR 76: 497–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charubin, K. , Bennett, R.K. , Fast, A.G. , and Papoutsakis, E. T. (2018) Engineering Clostridium organisms as microbial cell‐factories: challenges & opportunities. Metab Eng 50: 173–191. [DOI] [PubMed] [Google Scholar]
- Cheng, C. , Bao, T. , and Yang, S.T. (2019) Engineering Clostridium for improved solvent production: recent progress and perspective. Appl Microbiol Biotechnol 103: 5549–5566. [DOI] [PubMed] [Google Scholar]
- De Carlo, S. , Chen, B. , Hoover, T.R. , Kondrashkina, E. , Nogales, E. , and Nixon, B.T. (2006) The structural basis for regulated assembly and function of the transcriptional activator NtrC. Genes Dev 20: 1485–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durre, P. (2007) Biobutanol: an attractive biofuel. Biotechnol J 2: 1525–1534. [DOI] [PubMed] [Google Scholar]
- Ezeji, T.C. , Qureshi, N. , and Blaschek, H.P. (2003) Production of acetone, butanol and ethanol by Clostridium beijerinckii BA101 and in situ recovery by gas stripping. World J Microb Biot 19: 595–603. [Google Scholar]
- Ezeji, T. , Milne, C. , Price, N.D. , and Blaschek, H.P. (2010) Achievements and perspectives to overcome the poor solvent resistance in acetone and butanol‐producing microorganisms. Appl Microbiol Biotechnol 85: 1697–1712. [DOI] [PubMed] [Google Scholar]
- Feustel, L. , Nakotte, S. , and Durre, P. (2004) Characterization and development of two reporter gene systems for Clostridium acetobutylicum . Appl Environ Microbiol 70: 798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, E.M. (2011) Fermentative production of butanol–the industrial perspective. Curr Opin Biotechnol 22: 337–343. [DOI] [PubMed] [Google Scholar]
- Henry, J.T. , and Crosson, S. (2011) Ligand‐binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol 65: 261–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocq, R. , Bouilloux‐Lafont, M. , Lopes Ferreira, N. , and Wasels, F. (2019) sigma(54) (sigma(L)) plays a central role in carbon metabolism in the industrially relevant Clostridium beijerinckii . Sci Rep 9: 7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover, T.R. , Santero, E. , Porter, S. , and Kustu, S. (1990) The integration host factor stimulates interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell 63: 11–22. [DOI] [PubMed] [Google Scholar]
- Hou, X.H. , Peng, W.F. , Xiong, L. , Huang, C. , Chen, X.F. , Chen, X.D. , and Zhang, W.G. (2013) Engineering Clostridium acetobutylicum for alcohol production. J Biotechnol 166: 25–33. [DOI] [PubMed] [Google Scholar]
- Kopke, M. , Mihalcea, C. , Liew, F. , Tizard, J.H. , Ali, M.S. , Conolly, J.J. , et al. (2011) 2,3‐butanediol production by acetogenic bacteria, an alternative route to chemical synthesis, using industrial waste gas. Appl Environ Microbiol 77: 5467–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.Y. , Park, J.H. , Jang, S.H. , Nielsen, L.K. , Kim, J. , and Jung, K.S. (2008) Fermentative butanol production by Clostridia. Biotechnol Bioeng 101: 209–228. [DOI] [PubMed] [Google Scholar]
- Liang, L.Y. , Liu, R.M. , Chen, X. , Ren, X.Y. , Ma, J.F. , Chen, K.Q. , et al. (2013) Effects of overexpression of NAPRTase, NAMNAT, and NAD synthetase in the NAD(H) biosynthetic pathways on the NAD(H) pool, NADH/NAD(+) ratio, and succinic acid production with different carbon sources by metabolically engineered Escherichia coli . Biochem Eng J 81: 90–96. [Google Scholar]
- Liu, L. , Zhang, L. , Tang, W. , Gu, Y. , Hua, Q. , Yang, S. , et al. (2012) Phosphoketolase pathway for xylose catabolism in Clostridium acetobutylicum revealed by 13C metabolic flux analysis. J Bacteriol 194: 5413–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C. , Yu, L. , Varghese, S. , Yu, M. , and Yang, S.T. (2017) Enhanced robustness in acetone‐butanol‐ethanol fermentation with engineered Clostridium beijerinckii overexpressing adhE2 and ctfAB. Bioresour Technol 243: 1000–1008. [DOI] [PubMed] [Google Scholar]
- Martinez‐Argudo, I. , Little, R. , Shearer, N. , Johnson, P. , and Dixon, R. (2004) The NifL‐NifA System: a multidomain transcriptional regulatory complex that integrates environmental signals. J Bacteriol 186: 601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micka, B. , and Marahiel, M.A. (1992) The DNA‐binding protein hbsu is essential for normal growth and development in Bacillus‐Subtilis . Biochimie 74: 641–650. [DOI] [PubMed] [Google Scholar]
- Nie, X. , Yang, B. , Zhang, L. , Gu, Y. , Yang, S. , Jiang, W. , and Yang, C. (2016) PTS regulation domain‐containing transcriptional activator CelR and sigma factor sigma‐54 control cellobiose utilization in Clostridium acetobutylicum . Mol Microbiol 100: 289–302. [DOI] [PubMed] [Google Scholar]
- Nie, X. , Dong, W. , and Yang, C. (2019) Genomic reconstruction of sigma(54) regulons in Clostridiales. BMC Genom 20: 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoutsakis, E. T. (2008) Engineering solventogenic clostridia. Curr Opin Biotechnol 19: 420–429. [DOI] [PubMed] [Google Scholar]
- Radianingtyas, H. , and Wright, P.C. (2003) Alcohol dehydrogenases from thermophilic and hyperthermophilic archaea and bacteria. FEMS Microbiol Rev 27: 593–616. [DOI] [PubMed] [Google Scholar]
- Reid, M.F. , and Fewson, C.A. (1994) Molecular characterization of microbial alcohol dehydrogenases. Crit Rev Microbiol 20: 13–56. [DOI] [PubMed] [Google Scholar]
- Ren, C. , Wen, Z. , Xu, Y. , Jiang, W. , and Gu, Y. (2016) Clostridia: a flexible microbial platform for the production of alcohols. Curr Opin Chem Biol 35: 65–72. [DOI] [PubMed] [Google Scholar]
- Schumacher, J. , Zhang, X. , Jones, S. , Bordes, P. , and Buck, M. (2004) ATP‐dependent transcriptional activation by bacterial PspF AAA+protein. J Mol Biol 338: 863–875. [DOI] [PubMed] [Google Scholar]
- Shao, L. , Hu, S. , Yang, Y. , Gu, Y. , Chen, J. , Yang, Y. , et al. (2007) Targeted gene disruption by use of a group II intron (targetron) vector in Clostridium acetobutylicum . Cell Res. 17: 963–965. [DOI] [PubMed] [Google Scholar]
- Sillers, R. , Al‐Hinai, M.A. , and Papoutsakis, E.T. (2009) Aldehyde‐alcohol dehydrogenase and/or thiolase overexpression coupled with CoA transferase downregulation lead to higher alcohol titers and selectivity in Clostridium acetobutylicum fermentations. Biotechnol Bioeng 102: 38–49. [DOI] [PubMed] [Google Scholar]
- Tan, Y. , Liu, J. , Liu, Z. , and Li, F. (2014) Characterization of two novel butanol dehydrogenases involved in butanol degradation in syngas‐utilizing bacterium Clostridium ljungdahlii DSM 13528. J Basic Microbiol 54: 996–1004. [DOI] [PubMed] [Google Scholar]
- Taylor, B.L. , and Zhulin, I.B. (1999) PAS domains: internal sensors of oxygen, redox potential, and light. MMBR 63: 479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy, B.P. , Jones, S.W. , Fast, A.G. , Indurthi, D.C. , and Papoutsakis, E.T. (2012) Clostridia: the importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr Opin Biotechnol 23: 364–381. [DOI] [PubMed] [Google Scholar]
- Tummala, S.B. , Welker, N.E. , and Papoutsakis, E.T. (1999) Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 65: 3793–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummala, S.B. , Junne, S.G. , and Papoutsakis, E.T. (2003) Antisense RNA downregulation of coenzyme A transferase combined with alcohol‐aldehyde dehydrogenase overexpression leads to predominantly alcohologenic Clostridium acetobutylicum fermentations. J Bacteriol 185: 3644–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Li, X. , Mao, Y. , and Blaschek, H.P. (2012) Genome‐wide dynamic transcriptional profiling in Clostridium beijerinckii NCIMB 8052 using single‐nucleotide resolution RNA‐Seq. BMC Genom 13: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, Z. , Minton, N.P. , Zhang, Y. , Li, Q. , Liu, J. , Jiang, Y. , and Yang, S. (2017) Enhanced solvent production by metabolic engineering of a twin‐clostridial consortium. Metab Eng 39: 38–48. [DOI] [PubMed] [Google Scholar]
- Wiesenborn, D.P. , Rudolph, F.B. , and Papoutsakis, E.T. (1989) Coenzyme A transferase from Clostridium acetobutylicum ATCC 824 and its role in the uptake of acids. Appl Environ Microbiol 55: 323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, H. , Gu, Y. , Ning, Y. , Yang, Y. , Mitchell, W.J. , Jiang, W. , and Yang, S. (2011) Confirmation and elimination of xylose metabolism bottlenecks in glucose phosphoenolpyruvate‐dependent phosphotransferase system‐deficient Clostridium acetobutylicum for simultaneous utilization of glucose, xylose, and arabinose. Appl Environ Microbiol 77: 7886–7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Nie, X. , Jiang, Y. , Yang, C. , Gu, Y. , and Jiang, W. (2018) Metabolic regulation in solventogenic clostridia: regulators, mechanisms and engineering. Biotechnol Adv 36: 905–914. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Nie, X. , Ravcheev, D.A. , Rodionov, D.A. , Sheng, J. , Gu, Y. , et al. (2014) Redox‐responsive repressor Rex modulates alcohol production and oxidative stress tolerance in Clostridium acetobutylicum . J Bacteriol 196: 3949–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Construction and validation of gene inactivation mutants of C. beijerinckii. The coding region of sigL (Cbei_0595) or adhR (Cbei_2180) genes was inserted with an intron. The resulting mutants were confirmed by PCR.
Table S1. Strains and plasmids used in this study.
Table S2. Oligonucleotides used in this study.


