Summary
Multi‐omics can informally be described as the combined use of high‐throughput techniques allowing the characterization of complete microbial communities by the sequencing/identification of total pools of biomolecules including DNA, proteins or metabolites. These techniques have allowed an unprecedented level of knowledge on complex microbial ecosystems, which is having key implications in land and marine ecology, industrial biotechnology or biomedicine. Multi‐omics have recently been applied to artistic or archaeological objects, with the goal of either contributing to shedding light on the original context of the pieces and/or to inform conservation approaches. In this minireview, we discuss the application of ‐omic techniques to the study of prehistoric artworks and ancient man‐made objects in three main technical blocks: metagenomics, proteomics and metabolomics. In particular, we will focus on how proteomics and metabolomics can provide paradigm‐breaking results by unambiguously identifying peptides associated with a given, palaeo‐cultural context; and we will discuss how metagenomics can be central for the identification of the microbial keyplayers on artworks surfaces, whose conservation can then be approached by a range of techniques, including using selected microorganisms as ‘probiotics’ because of their direct or indirect effect in the stabilization and preservation of valuable art objects.
We discuss the application of ‐omic techniques to the study of prehistoric artworks and ancient man‐made objects in three main technical blocks: metagenomics, proteomics and metabolomics.
![]()
Biomolecules and archaeology: from targeted analysis to high‐throughput, untargeted approaches
Archaeology is a discipline under constant evolution, in which numerous methodologies are typically combined in order to get more knowledge about past civilizations, including their lifestyles, their relationships with the environment, their traditions and their social organization. Very often, the same combination of techniques and procedures are applied with the purpose of preserving the cultural heritage, which is a great challenge for our own society. Archaeology is thus a deeply interdisciplinary field. The choice of the technique or techniques to be applied in archaeological research depends on the goal of the study, which can focus on the chronological determination for some event in the past (David et al., 2013; Sauvet, 2015; Bonneau et al., 2017), identifying the identity of biological traces (Kamp et al., 1999; Kamp, 2001; Crown et al., 2009) or analyzing the composition of a particular object (Reiche et al., 2011; Brandt et al., 2014). Several factors such as the type of material under study (i.e. rock art, man‐made objects and hearths), the requirements of the technique (i.e. invasive vs non‐invasive) and the versatility of the generated data strongly determine the experimental design and the methodology chosen for archaeological research.
In particular, physico‐chemical analysis are the main group of methodologies enabling the investigation of the origin of archaeological objects and the raw materials used for their construction, the description of the elaboration processes of human‐made objects or artworks, and the follow‐up of conservation and restoration efforts. Three main types of physico‐chemical techniques have traditionally been applied in this field (Neff, 1993): (i) thermal analysis, (ii) mineralogical‐crystallographic analysis and (iii) techniques for the identification and quantification of chemical elements or compounds. The latter group includes, among others, a range of physico‐chemical techniques targeting specific biomolecules such as DNA, proteins and other metabolites.
Even though the vast majority of biomolecular techniques are invasive and require the destruction of the sample, they have greatly expanded among the archaeological community in the last decades due to the valuable information they are able to produce. In origin, these techniques were designed to target particular biomolecules with a high specificity. For instance, specific PCR (Polymerase Chain Reaction) protocols were developed to analyze hypervariable regions of human DNA (Harder et al., 2012; Olasz et al., 2019), and a range of ELISA (Enzyme‐Linked ImmunoSorbent Assay) protocols were designed for the detection of particularly relevant proteins such as albumin (Cattaneo et al., 1994), animal keratin (Wang et al., 2018) or silk fibroin (Liu et al., 2015) in ancient textiles. However, the development of the so‐called omic techniques (genomics, proteomics and metabolomics), able to perform a complete analysis of the total pool of DNA (genome), proteins (proteome) and other metabolites (metabolomics) in a single experiment (Jansson and Baker, 2016), has dramatically changed the nature of the biomolecular studies applied in the field of archaeology. The two main advantages of ‐omic technologies for archaeological research are their untargeted nature and the high‐throughput generation of data, since they are able to simultaneously yield information on a broad repertoire of biomolecules present in a sample. Hence, with the same amount of sample used in traditional biomolecular analysis to obtain information on a given molecule, omic technologies are able to generate an exhaustive dataset on thousands of molecules. In the following sections, the application of proteomic, metabolomic and metagenomic techniques in archaeological research (especially in the study of ancient art and objects) is briefly reviewed.
Application of proteomic and metabolomic techniques to the study of artwork and ancient man‐made objects
Multi‐omics have recently arisen as strong, alternative approaches for chronologic and conservation purposes of artistic and/or prehistoric objects. The main goal of proteomics on ancient art paintings has been the identification of the proteinaceous binders used for the preparation of the pigments. The two main approaches for this type of studies consist of (i) untargeted LC–MS/MS (liquid chromatography coupled to tandem mass spectrometry), aimed at identifying a broad representation of the peptides present in the sample; and (ii) MRM (multiple reaction monitoring), targeting particular peptides which are known markers for a range of putative binders. This is the case of casein (an obvious protein marker for milk), ovoalbumin (egg) or vitellogenin (yolk) peptides. These techniques have been applied to the analysis of ancient protein binders in a range of prehistoric art elements. The oldest sample analyzed by proteomics is an ochre‐painted stone flake from a 49 000‐year‐old layer of Sibudu (South Africa), in which casein from a wild bovid was detected as the main protein binder (Villa et al., 2015). Other sound cases include the proteomic analysis of the first historical overpaintings of the lost Giant Buddhas of Bāmiyān (Afghanistan), where a mixture of milk from different mammals was pointed out as the most likely binder (Lluveras‐Tenorio et al., 2017), and a set of rock art paintings from the Mediterranean basin (Valltorta, Spain), in which bovid casein traces were also detected (Roldán et al., 2018). The latter case is an example of how proteomic techniques can shed light not only on the composition of the binders, but also on art dating, since the detection of milk from wild or farmed animals has direct implications in the putative lifestyle of the authors of the art elements (i.e. early stockbreeding communities). Indeed, it has to be stressed that the Levantine rock art lacks a consensus dating and it is estimated to span over a culturally crucial period between, approximately 8000 and 3000 years BC. This period saw a transition from hunter‐gatherer economic systems to a Neolithic society (http://whc.unesco.org/en/list/874). The paintings include animals that could be used as cattle, and the identification of milk on the paintings would thus further support this conclusion (Roldán et al., 2018).
Out of the art framework, proteomics has been also essential to unveil the composition of food remains found in ancient ceramic vessels, yielding valuable evidence of the lifestyle and traditions of prehistoric societies. A range of dietary foodstuffs, including dairy, cereal grains, legumes and non‐dairy animal proteins were detected in ceramic sherds from the early farming site of Çatalhöyük, in central Anatolia (Hendy et al., 2018). Animal milk proteins were also detected in the ancient Subeixi region, the furthest eastern location of prehistoric milking in the Old World (Hong et al., 2012), and evidence for the early exploitation of aquatic resources was obtained in pottery from inland prehistorical localities in Brandenburg (Germany), where processed fish caviar meal traces were detected through proteomics (Shevchenko et al., 2018).
Finally, the proteomic analysis of other ancient objects such as wooden staffs (Rao et al., 2015), baskets (Solazzo et al., 2016) and textiles (Gong et al., 2016) has enabled the detection of bovine and fish tissues used as glues or coatings and the early use of silk for textile elaboration. Albeit controversial, the direct proteomic analysis of archaeological soils has been proposed as a way to detect ancient hairs, feedstock remains and other traces during the sampling of archaeological sites (Oonk et al., 2012).
On the other hand, different biomolecules such as lipids and secondary metabolites have been analyzed through metabolomics, by using GC–MS (gas chromatography coupled to mass spectrometry) to investigate the composition of art elements and prehistoric objects beyond their protein binders. This is the case of paint microsamples from different artworks (a painted hull of a shipwreck, an easel painting and a painted Etrurian sarcophagus), where linseed oil, beeswax and Pinaceae resin were identified as part of the paint mixture (Andreotti et al., 1978). However, the most informative studies based on GC–MS shed light on the contents of ancient pottery or other archaeological elements. The oldest objects analyzed with this approach are indeed pottery elements from Xianrendong Cave (China), dated about 20 000–19 000 years BP, in which the lipid composition (rich in markers dominated by medium‐ and long‐chain saturated and monounsaturated fatty acids and isoprenoid fatty acids) suggested the use and storage of high‐trophic‐level aquatic food (Craig et al., 2013). Similar results were later obtained in a study comprising samples from a wide chronological spectrum (9000‐years sequence) from the Jōmon site of Torihama, in Western Japan (Lucquin et al., 2016). The first direct evidence for the exploitation of palm fruit in antiquity and the use of pottery vessels in its processing was obtained by means of the same technique on samples from Qasr Ibrim (Egypt) (Copley et al., 2001).
GC–MS lipid analysis has also contributed to archaeological evidences in the analysis of different sites within hearths. Despite the degradation suffered by lipids as a consequence of fire or other catastrophic events, the information obtained from key fatty acid ratios has helped to distinguish between plant and animal lipids, and in some cases can even help to identify specific plant and animal taxa (Kedrowski et al., 2009).
The particular case of metagenomics: conservation and art probiotics
The microbial composition of ancient, artificial objects or artworks has been studied with some detail with worldwide examples, although many of those were limited because of the use of only culture‐dependent techniques. For example, a collection of actinomycetes strains from mural Egyptian paintings has been reported (Elhagrassy, 2018). These isolates proved able to modify the colour of the original pigments while, paradoxically, were also able to biocontrol potential microbial degraders (Elhagrassy, 2018). A painting in Herculanum, Italy, was also studied through culture techniques that allowed the identification of heterotrophic bacteria and fungi (Pepe et al., 2011). Culture‐depending techniques can yield a moderately diverse set of microorganisms. Indeed, the isolation of novel species from ancient artworks is not rare, and examples include the Myroides new species isolated from prehistoric paintings in Bulgaria (Tomova et al., 2013); several Rubrobacter strains from deteriorated ancient monuments (Laiz et al., 2009); and a new Paenibacillus and several new Arthrobacter species from a Roman tomb in Spain (Heyrman et al., 2005; Smerda et al., 2006). That said, Next Generation Sequencing (NGS) techniques can obviously yield a much broader view of the actual biological diversity of the microbial communities associated with man‐made ancient objects.
Duan et al. (2017) reported a culture‐independent Illumina MiSeq sequencing characterization of the microbial composition of the Maijishan Grottoes in a cave in China, a World Cultural Heritage site listed by UNESCO, by sampling paintings carried out during the Northern Zhou Dynasty (557‐581AD). A rich bacterial diversity including the bacterial groups Actinobacteria, Acidobacteria, Bacteroidetes, Cyanobacteria, Chloroflexi, Firmicutes, Proteobacteria and Verrucomicrobia was unveiled, while fungi exhibited a lower diversity, consisting of Ascomycota, Basidiomycota and Chytridiomycota. Some of these microorganisms, particularly the Pseudonocardia genus, have been reported to be associated with biodeterioration of art pieces or other culturally relevant objects, such as Etruscan paintings in Tuscany suffering from a heavy microbial colonization (Diaz‐Herraiz et al., 2013). In Nara, Japan, Sugiyama et al. (2017) used a combination of culture‐dependent and culture‐independent techniques to study some of the 1300‐year‐old polychrome mural paintings of the historical sites of Takamatsuzuka and Kitora Tumulus, which resulted in the identification of a new bacterial species of the Stenotrophomonas genus (Handa et al., 2016). Interestingly, severe outbreaks of microbial contaminants were reported, involving genera such as Doratomyces sp. and Streptomyces sp. Such outbreaks even required dramatic corrective actions such as UV light treatments, with limited success in controlling some of the microbial keyplayers in degradation (Sugiyama et al., 2017) .
One of the first culture‐independent studies performed on rock paintings describe an important pool of novel species in Altamira, Spain (Schabereiter‐Gurtner et al., 2002). The authors concluded that the high number of clones that were most closely related to environmental 16S rDNA clones suggests a wide set of unknown bacteria in the cave. In this same location, other researchers further reported that the metabolically active microbial community, dominated by proteobacteria, is only a small fraction of the total one, which implies that microorganisms with undetectable activity are paradoxically a potential risk for the conservation of ancient artworks since environmental variations can trigger their flourishing (Portillo et al., 2008).
The current development of NGS techniques applied to art conservation implies that an accurate knowledge of the microbial composition of paintings, buildings or sculptures is accessible with relatively fast and easy techniques. Beyond taxonomic characterization and the eventual description of new taxa, a major goal of metagenomics is the artificial modification of the microbiome, often with the objective of improving the pieces’ conservation. For example, a recent report describing the microbiome of a 17th century baroque easel demonstrated the potential of Bacillus spp. to control fungal populations associated with poor conservation and deterioration of the pictural layer (Caselli et al., 2018). The use of microorganisms in restoration of painting and other artworks, often defined as ‘biocleaning’ has been shown to be less hazardous than traditional, mechanical or chemical techniques, as well as to be non‐toxic for both restorers and the environment. One example of a bacterium used in biocleaning is Pseudomonas stutzeri strain 5190, which can use a wide range of carbon and nitrogen sources and is very tolerant to several stressing abiotic factors found on the painting surface (Bosch‐Roig et al., 2016). Paradoxically, these strategies imply the use of culture techniques, which are still imperative to carry out in controlled environments the confrontation tests between target and ‘probiotic’ microorganisms. This is, in our view, a very good example of both the power and limitations of ‐omic techniques, as we have previously highlighted (Vilanova and Porcar, 2016): such techniques are a priceless source of biological information, but yet require cultivation when it comes to translate massive genomic information into practical (i.e. conservation) actions.
Beware of degradation and modern contaminants: current limitations of ‐omic studies in archaeological research
Art and archaeological objects are always precious materials that are central to contribute to the understanding of our cultural and biological history. Multi‐omics (Fig. 1) have arisen as a new range of techniques that have proven helpful to either indirectly calculating the chronology or the cultural context of the pieces, as well as to provide relevant information on potential microbial degraders and stabilizers of the objects (typically, but not restricted to, paintings). That said, there are important limitations of the assays linked to the degradation of biomolecules in time and the contamination of the samples with modern biomolecules.
Figure 1.
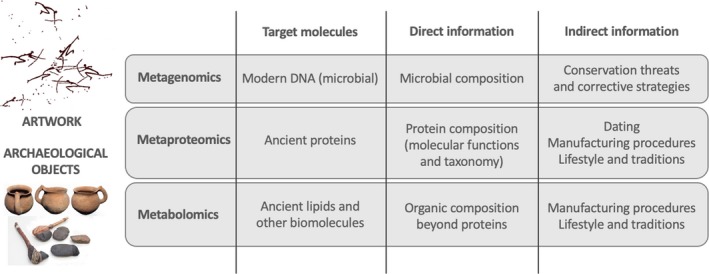
Summary of ‐omic approaches applied to the study of artwork and archaeological objects and expected evidences depending on the biomolecules under study.
On the degradation of biomolecules, it is well known that DNA can survive up to hundreds of thousands of years under certain circumstances, but in practical circumstances and under already human‐contaminated and/or warm climates, the determination of the palaeo‐metagenomic composition of a piece may be impossible. Therefore, and in normal conditions, a metagenomic DNA analysis will provide insights on the actual microbial community on the object, not the original one. By contrast, both proteins and lipids are less sensitive to degradation (Evershed et al., 1990; Cappellini et al., 2012), and therefore are suitable markers for determining the ancestral composition of an object/sample. Nevertheless, it has to be noted that microbial degradation can play a significant role in lipid degradation in archaeological samples (Malainey et al., 1999; Evershed et al., 2002).
On the other side, and especially in the case of proteomic studies, contamination of human origin is almost always present in the samples and can be due to a poor sub‐recent preservation frame (i.e. prehistoric paintings vandalized during the last years/decades) and/or to inadequate handling of the samples (objects handled without gloves, for example). That is why specific strategies for the filtering of modern contaminants have been set in place, being the analysis of glutamine deamidation rates (Wilson et al., 2012; Solazzo et al., 2014) the most reliable method to distinguish ancient peptides (with high deamidation levels) from contemporary ones (showing a low rate of deamidation). However, a range of factors different from age (i.e. preservation conditions, protein primary sequence and tridimensional structure) are known to influence glutamine deamidation (Schroeter and Cleland, 2016), so the direct correlation between deamidation rates and protein antiquity cannot be taken for granted in all cases. This suggests the need of combining such analytical methodologies with a robust sampling strategy, including a range of control samples, which may help to detect modern contaminant proteins.
Provided that suitable controls are set, multi‐omics will certainly consolidate as powerful tools to contribute to shed light on our past as well as to preserve modern and ancient work pieces. We anticipate that the current progress in proteomic analysis in the frame of human palaeontology will be linked to a parallel explosion of protein‐ and maybe metalobite‐based studies on man‐made objects, which may translate into a revolutionary biologization of the way we approach our past.
Authors’ contribution
Both authors conceived, wrote, read and approved the final manuscript.
Conflict of interest
None declared.
Acknowledgements
We thank our colleagues from ICMUV (Universitat de València) for introducing us in the exciting field of archaeology. MP is indebted to Radiohead for continuous musical support during the preparation of this manuscript.
Microbial Biotechnology (2020) 13(2), 435–441
Funding information
No funding information provided.
Contributor Information
Cristina Vilanova, Email: cristina@darwinbioprospecting.com.
Manuel Porcar, Email: manuel.porcar@uv.es.
References
- Andreotti, A. , Bonaduce, I. , Perla Colombini, M. , nae, G. , Gautier, l. , Modugno, F. and Ribechini, E. (1978) Cennini, C. Il Libro dell'Arte XIV sec. Anal Chem 4: 4490. [DOI] [PubMed] [Google Scholar]
- Bonneau, A. , Pearce, D. , Mitchell, P. , Staff, R. , Arthur, C. , Mallen, L. , et al (2017) The earliest directly dated rock paintings from southern Africa: new AMS radiocarbon dates. Antiquity 91: 322–333. [Google Scholar]
- Bosch‐Roig, P. , Decorosi, F. , Giovannetti, L. , Ranalli, G. , and Viti, C. (2016) Connecting phenome to genome in Pseudomonas stutzeri 5190: an artwork biocleaning bacterium. Res Microbiol 167: 757–765. [DOI] [PubMed] [Google Scholar]
- Brandt, L.Ø. , Schmidt, A.L. , Mannering, U. , Sarret, M. , Kelstrup, C.D. , Olsen, J.V. , and Cappellini, E. (2014) Species identification of archaeological skin objects from Danish bogs: comparison between mass spectrometry-based peptide sequencing and microscopy-based methods. PLoS One 9: e106875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellini, E. , Jensen, L.J. , Szklarczyk, D. , Ginolhac, A. , da Fonseca, R.A.R. , Stafford, T.W. , et al (2012) Proteomic Analysis of a Pleistocene Mammoth Femur Reveals More than One Hundred Ancient Bone Proteins. J Proteome Res 11: 917–926. [DOI] [PubMed] [Google Scholar]
- Caselli, E. , Pancaldi, S. , Baldisserotto, C. , Petrucci, F. , Impallaria, A. , Volpe, L. , et al (2018) Characterization of biodegradation in a 17th century easel painting and potential for a biological approach. PLoS ONE 13: e0207630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo, C. , Gelsthorpe, K. , Sokol, R.J. , and Phillips, P. (1994) Immunological Detection of Albumin in Ancient Human Cremations using ELISA and Monoclonal Antibodies. J Archaeol Sci 21: 565–571. [Google Scholar]
- Copley, M.S. , Rose, P.J. , Clapham, A. , Edwards, D.N. , Horton, M.C. and Evershed, R.P. (2001) Detection of palm fruit lipids in archaeological pottery from Qasr Ibrim, Egyptian Nubia. [DOI] [PMC free article] [PubMed]
- Craig, O.E. , Saul, H. , Lucquin, A. , Nishida, Y. , Taché, K. , Clarke, L. , et al (2013) Earliest evidence for the use of pottery. Nature 496: 351–354. [DOI] [PubMed] [Google Scholar]
- Crown, P.L. , Hurst, W.J. , and Smith, B.D. (2009) Evidence of cacao use in the Prehispanic American Southwest. Proc Natl Acad Sci USA 106: 2110–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, B. , Geneste, J.‐M. , Petchey, F. , Delannoy, J.‐J. , Barker, B. , and Eccleston, M. (2013) How old are Australia's pictographs? A review of rock art dating. Journal of Archaeological Science 40: 3–10. [Google Scholar]
- Diaz‐Herraiz, M. , Jurado, V. , Cuezva, S. , Laiz, L. , Pallecchi, P. , Tiano, P. , et al (2013) The actinobacterial colonization of Etruscan paintings. Sci Rep 3: 1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Y. , Wu, F. , Wang, W. , He, D. , Gu, J.‐D. , Feng, H. , et al (2017) The microbial community characteristics of ancient painted sculptures in Maijishan Grottoes. China. PLoS One 12: e0179718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhagrassy, A.F. (2018) Isolation and characterization of actinomycetes from Mural paintings of Snu‐ Sert‐Ankh tomb, their antimicrobial activity, and their biodeterioration. Microbiol Res 216: 47–55. [DOI] [PubMed] [Google Scholar]
- Evershed, R.P. , Heron, C. , and Goad, L.J. (1990) Analysis of organic residues of archaeological origin by high‐temperature gas chromatography and gas chromatography‐mass spectrometry. Analyst 115: 1339. [Google Scholar]
- Evershed, R.P. , Dudd, S.N. , Copley, M.S. , Berstan, R. , Stott, A.W. , Mottram, H. , et al (2002) Chemistry of archaeological animal fats. Acc Chem Res 35: 660–668. [DOI] [PubMed] [Google Scholar]
- Gong, Y. , Li, L. , Gong, D. , Yin, H. , and Zhang, J. (2016) Biomolecular Evidence of Silk from 8,500 Years Ago. PLoS ONE 11: e0168042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa, Y. , Tazato, N. , Nagatsuka, Y. , Koide, T. , Kigawa, R. , Sano, C. , and Sugiyama, J. (2016) Stenotrophomonas tumulicola sp. nov., a major contaminant of the stone chamber interior in the Takamatsuzuka Tumulus. Int J Syst Evol Microbiol 66: 1119–1124. [DOI] [PubMed] [Google Scholar]
- Harder, M. , Renneberg, R. , Meyer, P. , Krause-Kyora, B. , and von Wurmb-Schwark, N. (2012) STRtyping of ancient skeletal remains: which multiplex-PCR kit is the best? Croat Med J 53: 416–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendy, J. , Colonese, A.C. , Franz, I. , Fernandes, R. , Fischer, R. , Orton, D. , et al (2018) Ancient proteins from ceramic vessels at Çatalhöyük West reveal the hidden cuisine of early farmers. Nat Commun 9: 4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyrman, J. , Verbeeren, J. , Schumann, P. , Swings, J. , and De Vos, P. (2005) Six novel Arthrobacter species isolated from deteriorated mural paintings. Int J Syst Evol Microbiol 55: 1457–1464. [DOI] [PubMed] [Google Scholar]
- Hong, C. , Jiang, H. , Lü, E. , Wu, Y. , Guo, L. , Xie, Y. , et al (2012) Identification of Milk Component in Ancient Food Residue by Proteomics. PLoS ONE 7: e37053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson, J.K. , and Baker, E.S. (2016) A multi-omic future for microbiome studies. Nat Microbiol 1: 16049. [DOI] [PubMed] [Google Scholar]
- Kamp, K.A. (2001) Prehistoric Children Working and Playing: A Southwestern Case Study in Learning Ceramics. J Anthropol Res 57: 427–450. [Google Scholar]
- Kamp, K.A. , Timmerman, N. , Lind, G. , Graybill, J. , and Natowsky, I. (1999) Discovering Childhood: Using Fingerprints to Find Children in the Archaeological Record. Am Antiq 64: 309–315. [Google Scholar]
- Kedrowski, B.L. , Crass, B.A. , Behm, J.A. , Luetke, J.C. , Nichols, A.L. , Moreck, A.M. , and Holmes, C.E. (2009) GC/MS analysis of fatty acids from ancient hearth residues at the swan point archaeological site. Archaeometry 51: 110–122. [Google Scholar]
- Laiz, L. , Miller, A.Z. , Jurado, V. , Akatova, E. , Sanchez‐Moral, S. , Gonzalez, J.M. , et al (2009) Isolation of five Rubrobacter strains from biodeteriorated monuments. Naturwissenschaften 96: 71–79. [DOI] [PubMed] [Google Scholar]
- Liu, M. , Xie, J. , Zheng, H. , Zhou, Y. , Wang, B. , and Hu, Z. (2015) Identification of Ancient Silk Using an Enzyme-linked Immunosorbent Assay and Immuno-fluorescence Microscopy. Anal Sci 31: 1317 –1323. [DOI] [PubMed] [Google Scholar]
- Lluveras‐Tenorio, A. , Vinciguerra, R. , Galano, E. , Blaensdorf, C. , Emmerling, E. , Perla Colombini, M. , et al (2017) GC/MS and proteomics to unravel the painting history of the lost Giant Buddhas of Bāmiyān (Afghanistan). PLoS ONE 12: e0172990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucquin, A. , Gibbs, K. , Uchiyama, J. , Saul, H. , Ajimoto, M. , Eley, Y. , et al (2016) Ancient lipids document continuity in the use of early hunter‐gatherer pottery through 9,000 years of Japanese prehistory. Proc. Natl. Acad. Sci. U. S. A. 113: 3991–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malainey, M.E. , Przybylski, R. , and Sherriff, B.L. (1999) The Effects of Thermal and Oxidative Degradation on the Fatty Acid Composition of Food Plants and Animals of Western Canada: Implications for the Identification of Archaeological Vessel Residues. J Archaeol Sci 26: 95–103. [Google Scholar]
- Neff, H. (1993) Theory, Sampling, and Analytical Techniques in the Archaeological Study of Prehistoric Ceramics. Am Antiq 58: 23–44. [Google Scholar]
- Olasz, J. , Seidenberg, V. , Hummel, S. , Szentirmay, Z. , Szabados, G. , Melegh, B. , and Kásler, M. (2019) DNA profiling of Hungarian King Béla III and other skeletal remains originating from the Royal Basilica of Székesfehérvár. Archaeol Anthropol Sci 11: 1345–1357. [Google Scholar]
- Oonk, S. , Cappellini, E. , and Collins, M.J. (2012) Soil proteomics: an assessment of its potential for archaeological site interpretation. Org Geochem 50: 57–67. [Google Scholar]
- Pepe, O. , Palomba, S. , Sannino, L. , Blaiotta, G. , Ventorino, V. , Moschetti, G. , and Villani, F. (2011) Characterization in the archaeological excavation site of heterotrophic bacteria and fungi of deteriorated wall painting of Herculaneum in Italy. J Environ Biol 32: 241–250. [PubMed] [Google Scholar]
- Portillo, M.C. , Gonzalez, J.M. , and Saiz‐Jimenez, C. (2008) Metabolically active microbial communities of yellow and grey colonizations on the walls of Altamira Cave, Spain. J Appl Microbiol 104: 681–691. [DOI] [PubMed] [Google Scholar]
- Rao, H. , Yang, Y. , Abuduresule, I. , Li, W. , Hu, X. , and Wang, C. (2015) Proteomic identification of adhesive on a bone sculpture‐inlaid wooden artifact from the Xiaohe Cemetery, Xinjiang, China. J Archaeol Sci 53: 148–155. [Google Scholar]
- Reiche, I. , Müller, K. , Staude, A. , Goebbels, J. , and Riesemeier, H. (2011) Synchrotron radiation and laboratory micro X-ray computed tomography—useful tools for the material identification of prehistoric objects made of ivory, bone or antler. J Anal At Spectrom 26: 1802. [Google Scholar]
- Roldán, C. , Murcia‐Mascarós, S. , López‐Montalvo, E. , Vilanova, C. , and Porcar, M. (2018) Proteomic and metagenomic insights into prehistoric Spanish Levantine Rock Art. Sci Rep 8: 10011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvet, G. (2015) In Search of Lost Time. Dating Methods for Prehistoric Art: the Example of Aurignacian Sites. Palethnologie 7. [Google Scholar]
- Schabereiter‐Gurtner, C. , Saiz‐Jimenez, C. , Piñar, G. , Lubitz, W. , and Rölleke, S. (2002) Altamira cave Paleolithic paintings harbor partly unknown bacterial communities. FEMS Microbiol Lett 211: 7–11. [DOI] [PubMed] [Google Scholar]
- Schroeter, E.R. , and Cleland, T.P. (2016) Glutamine deamidation: an indicator of antiquity, or preservational quality? Rapid Commun Mass Spectrom 30: 251–255. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A. , Schuhmann, A. , Thomas, H. , and Wetzel, G. (2018) Fine Endmesolithic fish caviar meal discovered by proteomics in foodcrusts from archaeological site Friesack 4 (Brandenburg, Germany). PLoS ONE 13: e0206483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerda, J. , Sedlácek, I. , Pácová, Z. , Krejcí, E. , and Havel, L. (2006) Paenibacillus sepulcri sp. nov., isolated from biodeteriorated mural paintings in the Servilia tomb. Int J Syst Evol Microbiol 56: 2341–2344. [DOI] [PubMed] [Google Scholar]
- Solazzo, C. , Wilson, J. , Dyer, J.M. , Clerens, S. , Plowman, J.E. , von Holstein, I. , et al (2014) Modeling Deamidation in Sheep α‐Keratin Peptides and Application to Archeological Wool Textiles. Anal Chem 86: 567–575. [DOI] [PubMed] [Google Scholar]
- Solazzo, C. , Courel, B. , Connan, J. , van Dongen, B.E. , Barden, H. , Penkman, K. , et al (2016) Identification of the earliest collagen‐ and plant‐based coatings from Neolithic artefacts (Nahal Hemar cave, Israel). Sci Rep 6: 31053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama, J. , Kiyuna, T. , Nishijima, M. , An, K.‐D. , Nagatsuka, Y. , Tazato, N. , et al (2017) Polyphasic insights into the microbiomes of the Takamatsuzuka Tumulus and Kitora Tumulus. J Gen Appl Microbiol 63: 63–113. [DOI] [PubMed] [Google Scholar]
- Tomova, A. , Tomova, I. , Vasileva‐Tonkova, E. , Lazarkevich, I. , Stoilova‐Disheva, M. , Lyutskanova, D. , and Kambourova, M. (2013) Myroides guanonis sp. nov., isolated from prehistoric paintings. Int J Syst Evol Microbiol 63: 4266–4270. [DOI] [PubMed] [Google Scholar]
- Vilanova, C. , and Porcar, M. (2016) Are multi‐omics enough? Nat. Microbiol. 1: 16101. [DOI] [PubMed] [Google Scholar]
- Villa, P. , Pollarolo, L. , Degano, I. , Birolo, L. , Pasero, M. , Biagioni, C. , et al (2015) A Milk and Ochre Paint Mixture Used 49,000 Years Ago at Sibudu. South Africa. PLoS One 10: e0131273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B. , Gu, J. , Chen, B. , Xu, C. , Zheng, H. , Peng, Z. , et al (2018) Development of an Enzyme-Linked Immunosorbent Assay and Gold-Labelled Immunochromatographic Strip Assay for the Detection of Ancient Wool. J Anal Methods Chem 2018: 2641624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J. , van Doorn, N.L. , and Collins, M.J. (2012) Assessing the extent of bone degradation using glutamine deamidation in collagen. Anal Chem 84: 9041–9048. [DOI] [PubMed] [Google Scholar]


