Abstract
Ganoderma lucidum is an important medicinal mushroom in traditional Chinese medicine. However, the lack of adequate genetic tools has hindered molecular genetic research in and the genetic modification of this species. Here, we report that the presence of an intron is necessary for the efficient expression of the heterologous phosphinothricin‐resistance and green fluorescent protein genes in G. lucidum. Moreover, we improved the CRISPR/Cas9‐mediated gene disruption frequency in G. lucidum by adding an intron upstream of the Cas9 gene. Our results showed that the disruption frequency of the orotidine 5’‐monophosphate decarboxylase gene (ura3) in transformants containing the glyceraldehyde‐3‐phosphate dehydrogenase gene intron in the Cas9 plasmid is 14–18 in 107 protoplasts, which is 10.6 times higher than that in transformants without any intron sequence. Furthermore, genomic fragment deletions in the ura3 and GL17624 genes were achieved via a dual sgRNA‐directed CRISPR/Cas9 system in G. lucidum. We achieved a ura3 deletion frequency of 36.7% in G. lucidum. The developed method provides a powerful platform to generate gene deletion mutants and will facilitate functional genomic studies in G. lucidum.
An improved CRISPR/Cas9 system that contained the intron from the glyceraldehyde‐3‐phosphate dehydrogenase gene (gpd) for gene disruption in G. lucidum is described. An effective platform for target gene deletion was first established using a dual sgRNA‐directed CRISPR/Cas9 system in G. lucidum.
Introduction
Medicinal mushrooms are rich sources of pharmacologically active compounds (Chaturvedi et al., 2018). Ganoderma lucidum, a traditional medicinal mushroom, has been used to improve health and longevity in China for several millennia (Hsu and Cheng, 2018). These mushrooms exhibit several therapeutic activities, such as antitumor, immunomodulatory and antihypertensive activities (Russell and Paterson, 2006). Due to its wide range of pharmacological activities, Ganoderma has drawn widespread interest in recent years. The genomes of Ganoderma strains have recently been sequenced (Chen et al., 2012; Kües et al., 2015); however, functional characterization of the genes of interest is time‐consuming due to the lack of adequate molecular genetic tools.
Genetic transformation systems, including Agrobacterium tumefacien‐mediated and polyethylene glycol (PEG)‐mediated transformations, have been developed in G. lucidum (Shi et al., 2012; Xu et al., 2012; Xu and Zhong, 2015) and have been successfully used to enhance the production of bioactive compounds, such as ganoderic acids and polysaccharides (Zhou et al., 2014; Li et al., 2016; Zhang et al., 2017a, 2017b; Fei et al., 2019; Xu et al., 2019). Gene silencing systems have also been used for the partial suppression of genes of interest in G. lucidum (Mu et al., 2012; Liu et al., 2018; Zhu et al., 2019). So far, however, targeted gene deletion has not been established in Ganoderma, although it would be a powerful tool for G. lucidum research in the post‐genomic era.
Generally, homologous recombination (HR)‐mediated gene deletion occurs at a low frequency, because DNA integration mostly occurs ectopically through non‐homologous end joining (NHEJ) in filamentous fungi, animals and plants (Kück and Hoff, 2010). Although the frequency of HR is higher in NHEJ‐deficient filamentous fungi such as Neurospora, Coprinopsis cinerea and Pleurotus ostreatus (Ninomiya et al., 2004; Nakazawa et al., 2011; Salame et al., 2012), such strains may show growth defects, genomic instability and a low regeneration rate of protoplasts (de Jong et al., 2010; Zhang et al., 2011). Therefore, the development of novel molecular tools is required for efficient gene deletion in filamentous fungi.
The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR‐associated protein 9 (Cas9) system has been successfully applied to genome editing in animals, plants and microbes (Arazoe et al., 2015; Bortesi et al., 2016; Li et al., 2019; Schultz et al., 2019). Recently, the CRISPR/Cas9 system has been used for the disruption of genes in higher fungi such as Coprinopsis cinerea, Cordyceps militaris and Schizophyllum commune (Sugano et al., 2017; Chen et al., 2018; Vonk et al., 2019). Qin et al. reported the disruption of the ura3 gene of G. lucidum and G. lingzhi, which revealed the potential of the CRISPR/Cas9 system for disrupting the genes of Ganoderma (Qin et al., 2017). However, the disruption of the target gene must be highly efficient to facilitate genome editing approaches in Ganoderma. NHEJ‐mediated gene disruption using the CRISPR/Cas9 system typically produces small insertions and deletions in the target genes. This method usually lacks the ability to disrupt the function of regulatory sequences or elements in the non‐coding genome, whereas gene deletion is an effective alternative for achieving these aims (Cai et al., 2018). Recently, dual sgRNA‐directed gene deletion using the CRISPR/Cas9 system has also been demonstrated in some species, such as humans, rabbits, Caenorhabditis elegans and Arabidopsis (Chen et al., 2014; Zheng et al., 2014; Song et al., 2016; Durr et al., 2018). However, there has been no report of dual sgRNA‐directed deletion of target genes in Ganoderma spp.
In this study, we show that the presence of an intron is necessary for the efficient expression of the heterologous phosphinothricin‐resistance gene (bar) and the green fluorescent protein gene (gfp) in G. lucidum. We describe an improved CRISPR/Cas9 system that contains an intron of the glyceraldehyde‐3‐phosphate dehydrogenase gene (gpd) for gene disruption in G. lucidum. Moreover, we demonstrate that the dual sgRNA‐directed CRISPR/Cas9 system is an efficient tool for gene deletion in G. lucidum. This technology provides a useful platform in basic and applied research for gene deletion in medicinal mushrooms.
Results
Expression of the heterologous genes bar and gfp in G. lucidum
To efficiently express the Cas9 gene, we first evaluated the effects of codon optimization and intron addition on the expression of heterologous genes in G. lucidum. Currently, only the carboxin‐resistance gene cbx and hygromycin B‐resistance gene hph have proven useful as selection markers for the genetic manipulation of G. lucidum. In order to explore more genetic selection markers, the expression of bar, which encodes the phosphinothricin‐resistance gene, was investigated in G. lucidum. Plasmids pJW‐EXP‐bar, pJW‐EXP‐opbar and pJW‐EXP‐intron‐opbar (Fig. 1), which carry the bar gene (opbar), the codon‐optimized bar gene, and the 5′ intron of G. lucidum gpd with opbar, were constructed and transformed into protoplasts of G. lucidum, respectively. After the genetic transformation of G. lucidum, protoplasts with pJW‐EXP‐bar and pJW‐EXP‐opbar were unable to grow on the phosphinothricin‐containing CYM plate. However, four carboxin‐ and phosphinothricin‐resistant colonies were obtained with pJW‐EXP‐intron‐opbar (Fig. 1). These results indicated that the extra intron at the 5′ end of the phosphinothricin‐resistance gene is required for its efficient expression in G. lucidum. To confirm these results, we also constructed plasmid pJW‐EXP‐gfp, pJW‐EXP‐opgfp and pJW‐EXP‐intron‐opgfp (Fig. 2A) and transformed them into the protoplast of G. lucidum. Transgenes from carboxin‐resistant colonies were selected by PCR and subsequently microscopically screened for GFP expression. No fluorescence was detected in any of the carboxin‐resistant pJW‐EXP‐gfp and pJW‐EXP‐opgfp transformants (Fig. 2B and 2), despite confirmation of the presence of gfp and opgfp by PCR (data not shown). In contrast, all pJW‐EXP‐intron‐opgfp transformants exhibited green fluorescence in the G. lucidum mycelia (Fig. 2D). GFP expression was shown to be stable across five rounds of subculturing (data not shown). These results confirmed that a 5′ intron was essential for GFP expression in G. lucidum.
Figure 1.
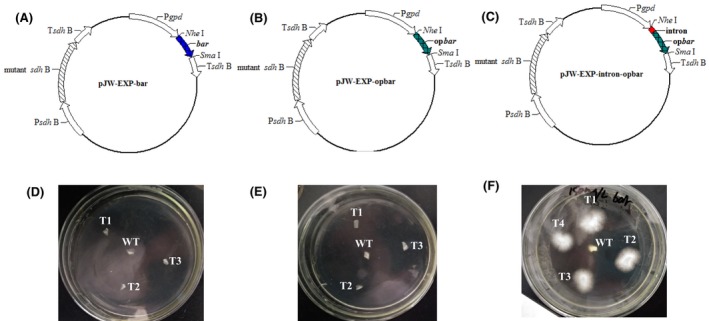
Selection of carboxin‐ and phosphinothricin‐resistant transformants on a selective CYM plate. Plasmid pJW‐EXP‐bar (A), pJW‐EXP‐opbar (B) and pJW‐EXP‐intron‐opbar (C). Selection of carboxin‐resistant pJW‐EXP‐bar transformants (D), pJW‐EXP‐opbar transformants (E) and pJW‐EXP‐intron‐opbar transformants (F) on a selective CYM plate with 150 mg l−1 phosphinothricin.
Figure 2.
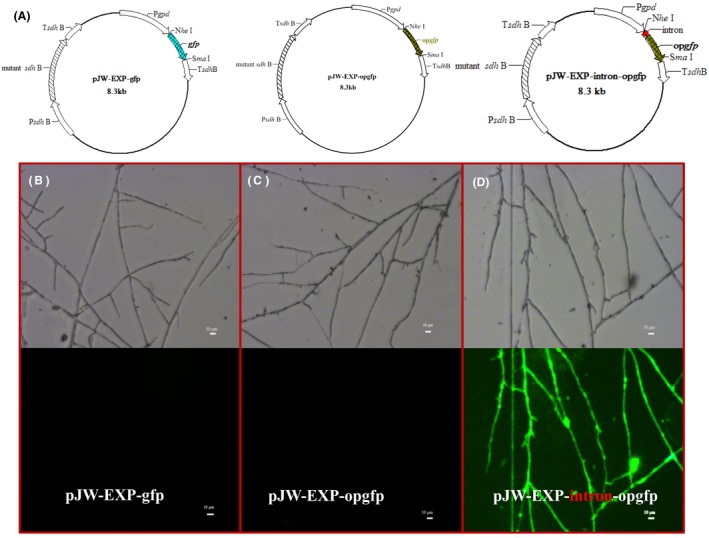
Expression of GFP in G. lucidum transformants. Plasmids pJW‐EXP‐bar, pJW‐EXP‐opbar and pJW‐EXP‐intron‐opbar (A). Phase‐contrast and fluorescence image of mycelium containing the plasmids pJW‐EXP‐bar (B), pJW‐EXP‐opbar (C) and pJW‐EXP‐intron‐opbar (D).
Disruption of ura3 in the engineered G. lucidum pJW‐EXP‐intron‐opcas9
To disrupt ura3 in G. lucidum, plasmids GL‐opcas9 and pJW‐EXP‐intron‐opcas9 (Fig. 3A and 3) were first constructed and transformed into the protoplasts of G. lucidum, separately. After screening on selective CYM plates with 2 mg/L carboxin, and confirmation via genomic PCR analysis and sequencing, we obtained the stable transformants GL‐opcas9 and pJW‐EXP‐intron‐opcas9 for gene disruption. No morphological difference was observed between the transformants GL‐opcas9 and pJW‐EXP‐intron‐opcas9, and the growth rates of both transformants were similar (Fig. S1). In vitro‐transcribed sgRNA1 (100 µg) targeting ura3 (Fig. 3C) was delivered into transformants GL‐opcas9 and pJW‐EXP‐intron‐opcas9, and 1‐3 and 28‐36 transformants resistant to 5‐fluoroorotic acid (FOA) were obtained per 2 × 107 protoplasts, respectively (Fig. 3). The average disruption efficiency of ura3 in pJW‐EXP‐intron‐opcas9 was 16 in 107 protoplasts, which is 10.6 times higher than that in GL‐opcas9 (1.5 in 107 protoplasts). The obtained ura3 disruption frequency in GL‐opcas9 was comparable to that (0.2–1.78 in 107 protoplasts) reported by Qin et al. (2017). Thirty 5‐FOA‐resistant colonies from the transformant pJW‐EXP‐intron‐opcas9 were selected, and transformation was confirmed using genome PCR with primers ura3‐F/ura3‐R and sequence analysis. These results are summarized in Table 1 and Data S3. The sequences of ura3 of the wild‐type strain (WT) and four randomly selected 5‐FOA‐resistant colonies are shown in Fig. S2. The replacement, deletion and insertion sites were located 3 bp upstream of the protospacer‐adjacent motif (PAM) sequence, which indicated that DNA double‐strand breaks (DSBs) were directed by CRISPR/Cas9 and repaired via NHEJ. When sgRNA2 (100 µg) targeting ura3 (Data S4) was delivered into pJW‐EXP‐intron‐opcas9, 28 transformants were obtained in 5‐FOA‐containing minimal medium (MM) plates. In Ashbya gossypii, different editing efficiency between the three ADE2 sgRNA‐dDNAs was also found (Jimenez et al., 2019). The observed difference in the disruption efficiency of ura3 may be due to the fact that the Cas9‐generated alleles display sequence‐dependent bias (Allen et al., 2019). These results are summarized in Data S4. The selected transformants contain deletions and insertions close to the PAM sequence of ura3. In contrast, no transformants were observed in the 5‐FOA‐containing MM plates without sgRNA1 or sgRNA2 from the control transformation of transformant pJW‐EXP‐intron‐opcas9. Our results demonstrated that both sgRNA1 and sgRNA2 are effective in disrupting ura3 in the engineered G. lucidum pJW‐EXP‐intron‐opcas9.
Figure 3.
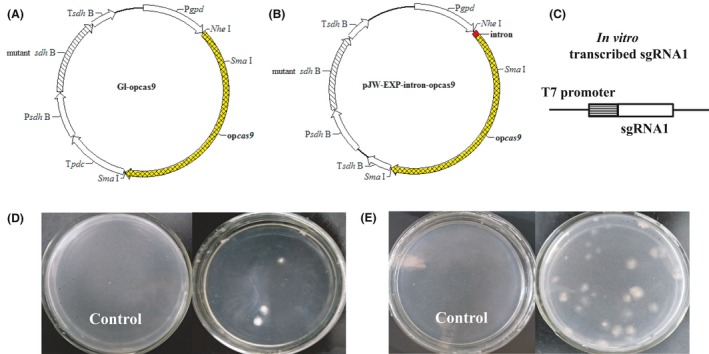
Selection of 5‐FOA‐resistant mutants on a selective MM plate. The design of plasmids GL‐opcas9 (A), pJW‐EXP‐intron‐opcas9 (B) and in vitro‐transcribed sgRNA1 (C). Selection of 5‐FOA‐resistant mutants after sgRNA1 targeting ura3 was delivered into the transformants GL‐opcas9 (D) and pJW‐EXP‐intron‐opcas9 (E) on a selective MM plate.
Table 1.
The mutation efficiency of insertion, deletion and replacement of ura3.
| Mutant of ura3 | Number of mutants | Efficiency (%) |
|---|---|---|
| Disruption | 30 | 100 |
| Insertion | 25 | 83.3 |
| Deletion | 4 | 13.3 |
| Replacement | 1 | 3.3 |
sgRNA1 targeting ura3 was delivered into protoplasts of G. lucidum transformant pJW‐EXP‐intron‐opcas9. Thirty individual 5‐FOA‐resistant mutants were assessed for insertion, deletion and replacement efficiency using the PCR and TA‐cloning sequence assay.
Dual sgRNA‐directed deletion of ura3 in the engineered G. lucidum pJW‐EXP‐intron‐opcas9
To perform the targeted deletion of ura3 in G. lucidum using the CRISPR/Cas9 system, in vitro‐transcribed sgRNA1 and sgRNA2 targeting ura3 were mixed and transformed into protoplasts of the engineered G. lucidum pJW‐EXP‐intron‐opcas9 at a concentration of 100 µg (Fig. 4A). Figure 4B shows that numerous colonies appeared on the selective MM plate containing 5‐FOA after transformation. The transformants that were chosen continued to grow on the selective medium after three rounds of growth on a non‐selective medium. We characterized those transformants by genome PCR (Fig. 4C) and sequence analysis (Fig. 4D). The desired fragment deletion of ura3 was found in transformants 4 and 5, demonstrating that the sequence between sgRNA1 and sgRNA2 can be deleted by CRISPR/Cas9‐mediated cleavage. The mutation frequency of the CRISPR/Cas9 system for each sgRNA was also determined by PCR and TA‐cloning. Primers ura3‐F and ura3‐R were used to detect the mutation of sgRNA1 and sgRNA2. A total of 30, 28 and 49 transformants from each genetic transformation were selected and then confirmed by sequence analysis. These results are summarized in Table 2 and Data S5. The average mutation frequency of ura3 for sgRNA1 and sgRNA2 was 28.5% and 34.6%, respectively. The average frequency for dual sgRNA‐directed deletion (simultaneous mutation of two sites) of ura3 was 36.7% in the engineered G. lucidum pJW‐EXP‐intron‐opcas9. These results illustrated that the CRISPR/Cas9 system is an effective tool for gene deletion in G. lucidum.
Figure 4.
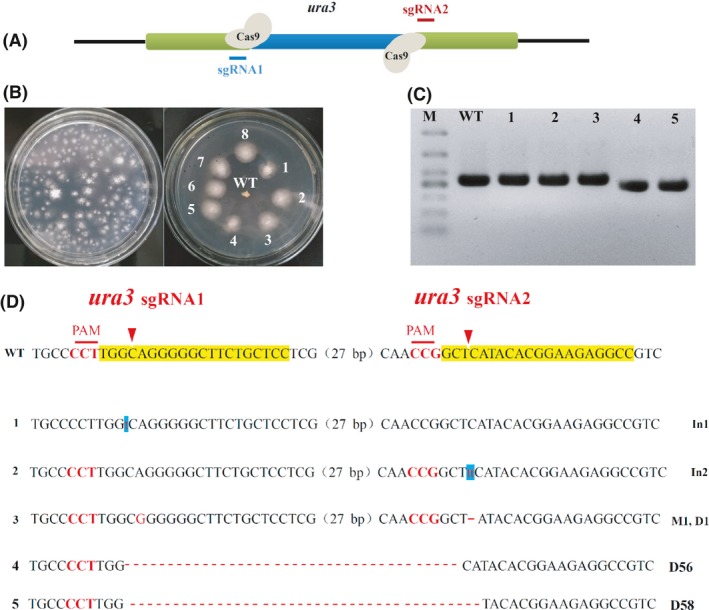
Dual sgRNA‐directed deletion of ura3 in G. lucidum.
A. Schematic representation of ura3 of G. lucidum and sequences targeted by Cas9.
B. Screening and re‐selection of 5‐FOA‐resistant mutants after sgRNA1 and sgRNA2 targeting ura3 were delivered into transformant pJW‐EXP‐intron‐opCas9.
C. Determination of the deletions of ura3 in G. lucidum transformants by PCR.
D. TA‐cloning of the target sites for each sgRNA in selected transformants. The sgRNA‐guiding sequences are highlighted in yellow. WT, the wild‐type strain; 1, 2, 3, 4, 5, G. lucidum transformants; M, DL500 DNA marker; In1 and In2, 1bp insertion and 2 bp insertion; M1, 1 bp replacement; D1, D56 and D58, 1 bp deletion, 56 bp deletion and 58 bp deletion, respectively.
Table 2.
The gene deletion frequencies of ura3 in the pJW‐EXP‐intron‐opcas9 strain by dual sgRNA.
| Target site | Number of mutants | Frequency (%) |
|---|---|---|
| sgRNA1 | 14 ± 2 | 28.5 ± 4 |
| sgRNA2 | 17 ± 1 | 34.6 ± 2 |
| sgRNA1 and sgRNA2 | 18 ± 3 | 36.7 ± 6 |
Forty‐nine individual 5‐FOA‐resistant mutants were assessed using the PCR and TA‐cloning sequence assay.
Targeted deletion of the GL17624 gene in G. lucidum pJW‐EXP‐intron‐opcas9
To investigate whether dual sgRNA‐directed gene deletion is applicable to other genomic loci in G. lucidum, the GL 17624 gene, which encodes a DNA‐binding protein, was targeted. We designed sgRNA1‐GL17624 and sgRNA2‐GL17624 (Data S3), which have targets 185 bp and 2311 bp downstream of the start codon of the GL 17624 gene, respectively. The two sgRNAs and the plasmid pJW‐EXP‐intron‐opbar were mixed and transformed into protoplasts of the engineered G. lucidum pJW‐EXP‐intron‐opcas9. Fifteen phosphinothricin‐resistant colonies were obtained in the selective CYM plate after 14 days of culture. The WT and all transformants were subjected to genome PCR analysis using the primers GL17624‐F and GL17624‐R (Table S1). The sequences of the PCR products are shown in Figure 5 and Data S6. The desired fragment deletion of the GL17624 gene was detected in 2 out of 15 transformants (13.3%). The PCR product showed a clear band for GL17624 (2469 bp) in the WT, but only an approximately 340 bp band was observed in the gene‐deleted transformants. The sequence analysis confirmed the expected deletion of the GL17624 gene in the obtained transformants. These results further validated that the use of the dual sgRNA‐directed CRISPR/Cas9 system is a promising approach for the deletion of target genes in G. lucidum.
Figure 5.
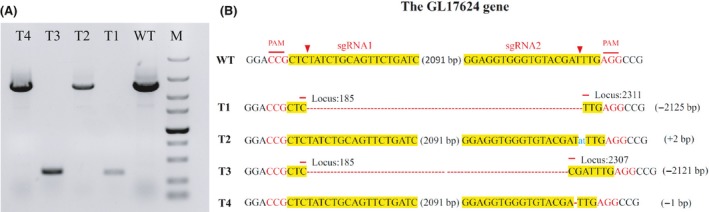
Dual sgRNA‐directed deletion of the GL 17624 gene in G. lucidum.
A. Determination of the deletions of the GL 17624 gene in the G. lucidum transformants by PCR.
B. TA‐cloning of the deletions of the GL 17624 gene in selected transformants. The sgRNA‐guiding sequences are highlighted in yellow. WT, the wild‐type strain; T, G. lucidum transformants; M, DL5000 DNA Marker
Discussion
Although the CRISPR/Cas9 system has been used in the higher fungus Ganoderma for gene disruption (Qin et al., 2017), its low disruption frequency hampered its application to genome engineering. We improved the frequency of gene disruption in G. lucidum by the introduction of an intron to the codon‐optimized Cas9. Furthermore, target gene deletion was achieved by employing the dual sgRNA‐directed CRISPR/Cas9 system in G. lucidum (Figure 6).
Figure 6.
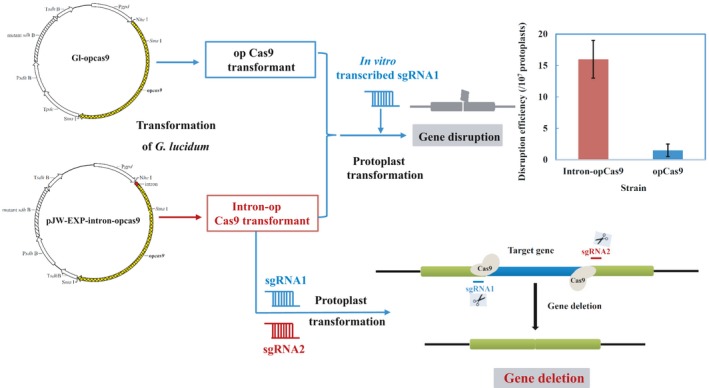
An effective platform for disruption and deletion of target genes in G. lucidum.
Phosphinothricin‐resistant and green fluorescent transformants were generated by adopting intron addition and codon optimization in G. lucidum. The results suggested that a 5′ intron was necessary for the efficient expression of heterologous genes in G. lucidum. Other heterologous genes may also be successfully expressed in G. lucidum by introducing an intron. This report agrees with previous reports on Clitopilus passeckerianus, Phanerochaete chrysosporium and Agaricus bisporus. The addition of an intron at the 5′ end of the phleomycin‐resistance gene was required for its efficient expression in C. passeckerianus (Kilaru et al., 2009). The presence of a 5′ intron significantly affected the expression level of the enhanced GFP (EGFP) and GFP in P. chrysosporium and A. bisporus (Ma et al., 2001; Burns et al., 2005). An intron introduced before heterologous genes may increase the level of mRNA accumulation, thus upregulating protein expression in basidiomycete mushrooms. Previously, Mass et al. reported that the addition of the first intron of the maize Shrunken‐1 gene to constructs containing prokaryotic reporter genes dramatically increased the expression level of chloramphenicol transacetylase marker gene in monocotyledonous plants (Maas et al., 1991). In Schizophyllum commune, the addition of an intron was sufficient to increase the mRNA accumulation of SC3 hydrophobin cDNA to a level similar to that of the genomic SC3 gene (Lugones et al., 1999). Until now, only the carboxin and hygromycin resistance cassettes can be used in G. lucidum as a selectable marker. Now, the modified phosphinothricin‐resistance cassette is also available and is just as efficient for transformant selection. The expression of GFP will facilitate the determination of gene expression and protein localization in G. lucidum.
The gene disruption frequency significantly increased in the transformants when they contained the Cas9 gene with the gpd 5′ intron of G. lucidum compared with those in the transformant without the intron. Similar observations were also reported in potato and rice, in which insertion of a 5′‐untranslated region (UTR) upstream of the Cas9 gene elevated the mutation frequency of the target gene (Table 3). A previous study found that a large amount of transfected Cas9‐encoding mRNA increased gene mutation frequencies in zebrafish (Hwang et al., 2013). In the liverwort Marchantia polymorpha L., the mutation frequency of the ARF1 locus was increased by the strong promoter MpEPpro, thereby driving Cas9 gene expression (Sugano et al., 2014). A positive correlation between the Cas9 expression level and CRISPR/Cas9‐mediated gene mutation frequency has also been reported in rice (Mikami et al., 2015a, 2015b). The addition of an intron led to an increase in the transcription levels of the Cas9 gene (Fig. S3), resulting in an increased gene disruption frequency in G. lucidum. The engineered strain pJW‐EXP‐intron‐opcas9 is useful for efficient gene mutagenesis using the CRISPR/Cas9 system in G. lucidum.
Table 3.
The effect of the insertion of a 5′‐UTR or intron upstream of the Cas9 gene on mutation frequencies.
The ura3 and GL17624 gene fragments were successfully deleted in G. lucidum by applying the Cas9/dual sgRNA technology. To the best of our knowledge, this is the first description of target gene deletion in non‐model mushrooms using the dual sgRNA system. Our results suggest that this method is efficient for gene deletion in G. lucidum. The dual sgRNA system has also been used to delete target DNA fragments in rabbit and indica rice (Song et al., 2016; Wang et al., 2017). The application of dual sgRNAs would result in the simultaneous cleavage of the target gene by Cas9, leading to the elimination of the intervening fragment and ligation of both ends by DNA repair (Essletzbichler et al., 2014). Our developed tool, based on dual sgRNA, is suitable for the deletion of target genes in basidiomycete mushrooms.
Experimental procedures
Strains and culture conditions
Ganoderma lucidum strain 260125 (a monokaryotic strain derived from the dikaryotic strain CGMCC 5.26) was used for CRISPR/Cas9 gene deletion experiments. This strain was maintained on potato dextrose agar slants. Escherichia coli strain DH5α was used in the construction of recombinant plasmids. G. lucidum cells were cultured in CYM plates (20 g l−1 glucose, 10 g l−1 maltose, 0.6 m mannitol, 2 g l−1 yeast extract, 2 g l−1 tryptone, 0.5 g l−1 MgSO4, 4.6 g l−1 KH2PO4 and 10 g l−1 agar) as described by Xu et al. (2012).
Plasmid construction and in vitro transcription
All plasmids used were derived from the plasmid pJW‐EXP as a backbone. The pJW‐EXP was generated from the plasmid pMD19‐T by inserting the G. lucidum gpd promoter, the iron‐sulphur protein subunit of succinate dehydrogenase gene terminator, and the carboxin‐resistance gene as a selection marker (Yu et al., 2014). The bar gene was acquired from the pBARGPE1 plasmid (University of Kansas Medical Center, Kansas City, KS, USA) using primers bar‐NheI‐F and bar‐SmaI‐R (Supplementary data, Table S1). This PCR fragment was digested with NheI and SmaI and ligated into NheI‐ and SmaI‐digested pJW‐EXP plasmids to yield pJW‐EXP‐bar. The plasmid pJW‐EXP‐opbar is similar to pJW‐EXP‐bar, except that bar was replaced by the codon‐optimized bar (opbar, Data S1), which was obtained from Shanghai Sangon Ltd., Crop. (Shanghai, China). The opbar was amplified using the primers opbar‐NheI‐F and opbar‐SmaI‐R (Supplementary data, Table S1). The pJW‐EXP plasmid was modified to produce pJW‐EXP‐intron‐opbar plasmid (5′ intron). It contained the G. lucidum gpd promoter sequence followed by the fragment intron (Data S2), which includes, from 5′ to 3′, gpd’s exon 1 (6 bp), intron 1 (67 bp), the 5′ end of exon 2 (3 bp) (Fei et al., 2006), the opbar coding sequence, and the succinate dehydrogenase gene terminator Tsdh B (Yu et al., 2014). The fragment intron was cloned from G. lucidum genomic DNA by the primers gpd‐NheI‐Intron‐F and intron‐R (Supplementary data, Table S1). The opbar was cloned using the primers intron‐opbar‐F and ter‐SmaI‐opbar‐R (Supplementary data, Table S1). The fragment intron and the opbar were ligated into NheI‐ and SmaI‐digested pJW‐EXP plasmid using the ClonExpress MultiS one Step Cloning Kit (Vazyme, Nanjing, China) to generate the pJW‐EXP‐intron‐opbar plasmid.
The gfp and NheI‐Intron‐codon‐optimized GFP (opgfp)‐SmaI genes were ordered from Shanghai Sangon Ltd., Crop. (Shanghai, China). Gfp and opgfp were amplified from the synthetic sequences using the primers gfp‐NheI‐F/gfp‐SmaI‐R and opgfp‐NheI‐F/opgfp‐SmaI‐R (Supplementary data, Table S1), respectively. The fragments NheI‐gfp‐SmaI, NheI‐opgfp‐SmaI and NheI‐Intron‐opgfp‐SmaI were ligated into the NheI‐ and SmaI‐digested pJW‐EXP plasmid to produce the pJW‐EXP‐gfp, pJW‐EXP‐opgfp and pJW‐EXP‐intron‐opgfp plasmids, respectively.
The plasmid GL‐opcas9 was provided by Jian‐Jiang Zhong Laboratory. The plasmid pJW‐EXP‐intron‐opcas9 is similar to pJW‐EXP‐intron‐opbar, except that the opbar was replaced by the opcas9. The opcas9 was amplified from plasmid GL‐opcas9 using the primers intron‐opcas9‐F and ter‐opcas9‐SmaI‐R (Supplementary data, Table S1). The fragment intron and opcas9 were ligated into the NheI‐ and SmaI‐digested pJW‐EXP plasmid to produce the pJW‐EXP‐intron‐opcas9 plasmid using the ClonExpress MultiS one Step Cloning Kit (Vazyme, Nanjing, China).
The four sgRNA cassettes (Data S3), including two ura3 and two GL26016 gene targeting sequences, and an sgRNA sequence were driven by a T7 promoter. The T7 promoter and four sgRNA cassettes were synthesized by Shanghai Sangon Ltd., Corp. (Shanghai, China). These sgRNA cassettes were transcribed in vitro using the HiScribe™ T7 High Yield RNA Synthesis Kit (NEB, Beijing, China) and purified using the RNA Clean & Concentrator™‐25 Kit (Zymo Research, Beijing, China).
Genetic transformation of G. lucidum
The preparation of G. lucidum protoplasts and PEG‐mediated genetic transformation were conducted as previously described (Xu et al., 2012, 2015; Yu et al., 2014). Transformants were screened using carboxin (2 mg l−1) or phosphinothricin (150 mg l−1) on a CYM regeneration plate. For the protoplast transformation of sgRNA cassettes, the generated RNA (10 µg) was transformed into the opcas9 or the intron‐opCas9 transformant using the PEG‐mediated transformation procedure.
Microscopic analysis for GFP expression
GFP expression was detected by microscopic screening with an excitation filter of 450‐490 nm, a dichroic filter of 510 nm and an emission filter of 515 nm as previously described (Ford et al., 2016). Images were captured with a Nikon Coolpix 900 camera. Samples for microscopy were mycelia grown in a CYM plate for five days.
Screening of ura3 disruption or deletion mutants
Following the PEG‐mediated transformation of sgRNA cassettes targeting ura3, transformants were screened on an MM selection plate (20 g/L glucose, 2 g/L L‐asparagine, 0.6 m mannitol, 0.5 g l−1 MgSO4, 0.46 g l−1 KH2PO4, 1 g l−1 K2HPO4, 0.125 mg l−1 vitamin B1 and 10 g l−1 agar), including 400 mg l−1 5‐FOA and 100 mg l−1 uridine (Sangon, Shanghai, China). The genomic DNA of the transformants was extracted using the Wizard Genomic DNA purification Kit (Promega, Beijing, China) for PCR amplification of ura3 using the primers ura3‐F and ura3‐R (Supplementary data, Table S1). Gene disruption or deletion was confirmed using Sanger sequencing.
Screening of the GL 17624 gene deletion mutants
The GL17624 gene of G. lucidum was acquired by genomic PCR using the primers GL17624‐F and GL17624‐R. Following the genetic transformation of protoplasts of the intron‐opCas9 strain with the pJW‐EXP‐intron‐opbar plasmid and the transcribed sgRNAs targeting the GL 17624 gene, transformants were picked from a CYM selection plate containing 150 mg l−1 phosphinothricin. The GL 17624 gene was amplified from the genomic DNA of transformants and the WT and sequenced to confirm the gene deletion.
Conflicts of interest
None declared.
Supporting information
Data S1. Sequences
Data S2. Sequences
Data S3. Sequences
Data S4. Sequences
Table S1. Primers used in the study
Fig. S1. Mycelia growth of the transformants GL‐opcas9 and pJW‐EXP‐intron‐opcas9 in CYM plates
Fig. S2. CRISPR/Cas9‐directed mutation of ura3 in G. lucidum. Alignments of ura3 mutants (M1, D1, In1, and In37) from 5‐FOA‐resistant colonies in sgRNA1 transformants. WT is the wild‐type strain ura3 from G. lucidum. The sgRNA1‐guiding sequence is highlighted in yellow. Replacements, deletions and insertions were found near the PAM sequence.
Fig. S3. Transcriptional levels of Cas9 gene in the GL‐opcas9 and pJW‐EXP‐intron‐opcas9 strains. Expression of Cas9 gene from the GL‐opcas9 strain is defined as 1.0, and expression levels in the pJW‐EXP‐intron‐opcas9 strain are displayed as fold increases over the reference sample. The following primers were used: Cas9‐f, 5’‐GAGGTCGCCTACCACGAGAAGT‐3’ and Cas9‐r, 5’ ‐TGGACGAGCTGGATGAAGAGC‐3’. * indicates statistical significance (P < 0.05) compared to the GL‐opcas9 strain (control).
Acknowledgement
This work was supported by the National Natural Science Foundation of China (Nos. 81860668 and 21566016) and Yunnan Applied Basic Research Project (2018FB065). We thank Professor Jian‐Jiang Zhong and Dr. Han Xiao (Shanghai Jiao Tong University) for providing the plasmid GL‐opcas9.
Microbial Biotechnology (2020) 13(2), 386–396
Funding information
This work was supported by the National Natural Science Foundation of China (Nos. 81860668 and 21566016) and Yunnan Applied Basic Research Project (2018FB065).
References
- Allen, F. , Crepaldi, L. , Alsinet, C. , Strong, A.J. , Kleshchevnikov, V. , De Angeli, P. , et al (2019) Predicting the mutations generated by repair of Cas9‐induced double‐strand breaks. Nat Biotechnol 37: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arazoe, T. , Miyoshi, K. , Yamato, T. , Ogawa, T. , Ohsato, S. , Arie, T. , et al (2015) Tailor‐made CRISPR/Cas system for highly efficient targeted gene replacement in the rice blast fungus. Biotechnol Bioeng 112: 2543–2549. [DOI] [PubMed] [Google Scholar]
- Bortesi, L. , Zhu, C. , Zischewski, J. , Perez, L. , Bassie, L. , Nadi, R. , et al (2016) Patterns of CRISPR/Cas9 activity in plants, animals and microbes. Plant Biotechnol J 14: 2203–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, C. , Gregory, K.E. , Kirby, M. , Cheung, M.K. , Riquelme, M. , Elliott, T.J. , et al (2005) Efficient GFP expression in the mushrooms Agaricus bisporus and Coprinus cinereus requires introns. Fungal Genet Biol 42: 191–199. [DOI] [PubMed] [Google Scholar]
- Cai, Y. , Chen, L. , Sun, S. , Wu, C. , Yao, W. , Jiang, B. , et al (2018) CRISPR/Cas9‐mediated deletion of large genomic fragments in soybean. Int J Mol Sci 19: 3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi, V.K. , Agarwal, S. , Gupta, K.K. , Ramteke, P.W. , and Singh, M.P. (2018) Medicinal mushroom: boon for therapeutic applications. 3. Biotech 8: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Xu, J. , Liu, C. , Zhu, Y. , Nelson, D.R. , Zhou, S. , et al (2012) Genome sequence of the model medicinal mushroom Ganoderma lucidum . Nat Commun 3: 913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Xu, F. , Zhu, C. , Ji, J. , Zhou, X. , Feng, X. , and Guang, S. (2014) Dual sgRNA‐directed gene knockout using CRISPR/Cas9 technology in Caenorhabditis elegans . Sci Rep 4: 7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B.X. , Wei, T. , Ye, Z.W. , Yun, F. , Kang, L.Z. , Tang, H.B. , et al (2018) Efficient CRISPR‐Cas9 gene disruption system in edible‐medicinal mushroom Cordyceps militaris . Front Microbiol 9: 1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr, J. , Papareddy, R. , Nakajima, K. , and Gutierrez‐Marcos, J. (2018) Highly efficient heritable targeted deletions of gene clusters and non‐coding regulatory regions in Arabidopsis using CRISPR/Cas9. Sci Rep 8: 4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essletzbichler, P. , Konopka, T. , Santoro, F. , Chen, D. , Gapp, B.V. , Kralovics, R. , et al (2014) Megabase‐scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res 24: 2059–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, X. , Zhao, M.W. , and Li, Y.X. (2006) Cloning and sequence analysis of a glyceraldehyde‐3‐phosphate dehydrogenase gene from Ganoderma lucidum . J Microbiol 44: 515–522. [PubMed] [Google Scholar]
- Fei, Y. , Li, N. , Zhang, D.H. , and Xu, J.W. (2019) Increased production of ganoderic acids by overexpression of homologous farnesyl diphosphate synthase and kinetic modeling of ganoderic acid production in Ganoderma lucidum . Microb Cell Fact 18: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, K.L. , Baumgartner, K. , Henricot, B. , Bailey, A.M. , and Foster, G.D. (2016) A native promoter and inclusion of an intron is necessary for efficient expression of GFP or mRFP in Armillaria mellea . Sci Rep 6: 29226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, K.D. , and Cheng, K.C. (2018) From nutraceutical to clinical trial: frontiers in Ganoderma development . Appl Microbiol Biotechnol 102: 9037–9051. [DOI] [PubMed] [Google Scholar]
- Hwang, W.Y. , Fu, Y. , Reyon, D. , Maeder, M.L. , Tsai, S.Q. , Sander, J.D. , et al (2013) Efficient genome editing in zebrafish using a CRISPR‐Cas system. Nat Biotechnol 31: 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez, A. , Munoz‐Fernandez, G. , Ledesma‐Amaro, R. , Buey, R.M. , and Revuelta, J.L. (2019) One‐vector CRISPR/Cas9 genome engineering of the industrial fungus Ashbya gossypii . Microb Biotechnol 12: 1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, J.F. , Ohm, R.A. , de Bekker, C. , Wosten, H.A.B. , and Lugones, L.G. (2010) Inactivation of ku80 in the mushroom‐forming fungus Schizophyllum commune increases the relative incidence of homologous recombination. FEMS Microbiol Lett 310: 91–95. [DOI] [PubMed] [Google Scholar]
- Kilaru, S. , Collins, C.M. , Hartley, A.J. , Bailey, A.M. , and Foster, G.D. (2009) Establishing molecular tools for genetic manipulation of the pleuromutilin‐producing fungus Clitopilus passeckerianus . Appl Environ Microbiol 75: 7196–7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück, U. , and Hoff, B. (2010) New tools for the genetic manipulation of filamentous fungi. Appl Microbiol Biotechnol 86: 51–62. [DOI] [PubMed] [Google Scholar]
- Kües, U. , Nelson, D.R. , Liu, C. , Yu, G.J. , Zhang, J. , Li, J. , et al (2015) Genome analysis of medicinal Ganoderma spp. with plant‐pathogenic and saprotrophic life‐styles. Phytochemistry 114: 18–37. [DOI] [PubMed] [Google Scholar]
- Kusano, H. , Ohnuma, M. , Mutsuro‐Aoki, H. , Asahi, T. , Ichinosawa, D. , Onodera, H. , et al (2018) Establishment of a modified CRISPR/Cas9 system with increased mutagenesis frequency using the translational enhancer dMac3 and multiple guide RNAs in potato. Sci Rep 8: 4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.J. , Zhang, D.H. , Yue, T.H. , Jiang, L.X. , Yu, X. , Zhao, P. , et al (2016) Improved polysaccharide production in a submerged culture of Ganoderma lucidum by the heterologous expression of Vitreoscilla hemoglobin gene. J Biotechnol 217: 132–137. [DOI] [PubMed] [Google Scholar]
- Li, P.S. , Fu, X.F. , Zhang, L. , and Li, S.Z. (2019) CRISPR/Cas‐based screening of a gene activation library in Saccharomyces cerevisiae identifies a crucial role of OLE1 in thermotolerance. Microb Biotechnol 12: 1154–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, R. , Shi, L. , Zhu, T. , Yang, T. , Ren, A. , Zhu, J. , and Zhao, M.W. (2018) Cross talk between nitric oxide and calcium‐calmodulin regulates ganoderic acid biosynthesis in Ganoderma lucidum under heat stress. Appl Environ Microbiol 84: e00043‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugones, L.G. , Scholtmeijer, K. , Klootwijk, R. , and Wessels, J.G.H. (1999) Introns are necessary for mRNA accumulation in Schizophyllum commune . Mol Microbiol 32: 681–689. [DOI] [PubMed] [Google Scholar]
- Ma, B. , Mayfield, M. B. , and Gold, M.H. (2001) The green fluorescent protein gene functions as a reporter of gene expression in Phanerochaete chrysosporium . Appl Environ Microbiol 67: 948–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas, C. , Laufs, J. , Grant, S. , Korfhage, C. , and Werr, W. (1991) The combination of a novel stimulatory element in the first exon of the maize Shrunken‐1 gene with the following intron 1 enhances reporter gene expression up to 1000‐fold. Plant Mol Biol 16: 199–207. [DOI] [PubMed] [Google Scholar]
- Mikami, M. , Toki, S. , and Endo, M. (2015a) Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol Biol 88: 561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami, M. , Toki, S. , and Endo, M. (2015b) Parameters affecting frequency of CRISPR/Cas9 mediated targeted mutagenesis in rice. Plant Cell Rep 34: 1807–1815. [DOI] [PubMed] [Google Scholar]
- Mu, D. , Shi, L. , Ren, A. , Li, M. , Wu, F. , Jiang, A. , and Zhao, M.W. (2012) The development and application of a multiple gene co‐silencing system using endogenous URA3 as a reporter gene in Ganoderma lucidum . PLoS ONE 7: e43737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa, T. , Ando, Y. , Kitaaki, K. , Nakahori, K. , and Kamada, T. (2011) Efficient gene targeting in ∆ Cc.ku70 or ∆Cc.lig4 mutants of the agaricomycete Coprinopsis cinerea . Fungal Genet Biol 48: 939–946. [DOI] [PubMed] [Google Scholar]
- Ninomiya, Y. , Suzuki, K. , Ishii, C. , and Inoue, H. (2004) Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end‐joining. P Natl Acad Sci USA 101: 12248–12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, H. , Xiao, H. , Zou, G. , Zhou, Z. , and Zhong, J.J. (2017) CRISPR‐Cas9 assisted gene disruption in the higher fungus Ganoderma species . Process Biochem 56: 57–61. [Google Scholar]
- Russell, R. , and Paterson, M. (2006) Ganoderma‐A therapeutic fungal biofactory. Phytochemistry 67: 1985–2001. [DOI] [PubMed] [Google Scholar]
- Salame, T.M. , Knop, D. , Tal, D. , Levinson, D. , Yarden, O. , and Hadar, Y. (2012) Predominance of a versatile‐peroxidase‐encoding gene, mnp4, as demonstrated by gene replacement via a gene targeting system for Pleurotus ostreatus . Appl Environ Microbiol 78: 5341–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, J.C. , Cao, M. , and Zhao, H. (2019) Development of a CRISPR/Cas9 system for high efficiency multiplexed gene deletion in Rhodosporidium toruloides . Biotechnol Bioeng 116: 2103–2109. [DOI] [PubMed] [Google Scholar]
- Shi, L. , Fang, X. , Li, M. , Mu, D. , Ren, A. , Tan, Q. , and Zhao, M.W. (2012) Development of a simple and efficient transformation system for the basidiomycetous medicinal fungus Ganoderma lucidum . World J Microb Biotehnol 28: 283–291. [DOI] [PubMed] [Google Scholar]
- Song, Y. , Yuan, L. , Wang, Y. , Chen, M. , Deng, J. , Lv, Q. , et al (2016) Efficient dual sgRNA‐directed large gene deletion in rabbit with CRISPR/Cas9 system. Cell Mol Life Sci 73: 2959–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano, S.S. , Shirakawa, M. , Takagi, J. , Matsuda, Y. , Shimada, T. , Hara‐Nishimura, I. , and Kohchi, T. (2014) CRISPR/Cas9‐mediated targeted mutagenesis in the Liverwort Marchantia polymorpha L. Plant Cell Physiol 55: 475–481. [DOI] [PubMed] [Google Scholar]
- Sugano, S.S. , Suzuki, H. , Shimokita, E. , Chiba, H. , Noji, S. , Osakabe, Y. , and Osakabe, K. (2017) Genome editing in the mushroom‐forming basidiomycete Coprinopsis cinerea, optimized by a high‐throughput transformation system. Sci Rep 7: 1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonk, P.J. , Escobar, N. , Wosten, H.A.B. , Lugones, L.G. , and Ohm, R.A. (2019) High‐throughput targeted gene deletion in the model mushroom Schizophyllum commune using pre‐assembled Cas9 ribonucleoproteins. Sci Rep 9: 7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Geng, L. , Yuan, M. , Wei, J. , Jin, C. , Li, M. , et al (2017) Deletion of a target gene in Indica rice via CRISPR/Cas9. Plant Cell Rep 36: 1333–1343. [DOI] [PubMed] [Google Scholar]
- Xu, J.W. , and Zhong, J.J. (2015) Genetic engineering of Ganoderma lucidum for the efficient production of ganoderic acids. Bioengineered 6: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J.W. , Xu, Y.N. , and Zhong, J.J. (2012) Enhancement of ganoderic acid accumulation by overexpression of an N‐terminally truncated 3‐hydroxy‐3‐methylglutaryl coenzyme A reductase gene in the basidiomycete Ganoderma lucidum . Appl Environ Microbiol 78: 7968–7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J.W. , Ji, S.L. , Li, H.J. , Zhou, J.S. , Duan, Y.Q. , Dang, L.Z. , and Mo, M.H. (2015) Increased polysaccharide production and biosynthetic gene expressions in a submerged culture of Ganoderma lucidum by the overexpression of the homologous alpha‐phosphoglucomutase gene. Bioproc Biosys Eng 38: 399–405. [DOI] [PubMed] [Google Scholar]
- Xu, J.W. , Yue, T.H. , Yu, X. , Zhao, P. , Li, T. , and Li, N. (2019) Enhanced production of individual ganoderic acids by integrating Vitreoscilla haemoglobin expression and calcium ion induction in liquid static cultures of Ganoderma lingzhi . Microb Biotechnol 12: 1180–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Ji, S.L. , He, Y.L. , Ren, M.F. , and Xu, J.W. (2014) Development of an expression plasmid and its use in genetic manipulation of Lingzhi or Reishi medicinal mushroom, Ganoderma lucidum (higher basidiomycetes). Int J Med Mushrooms 16: 161–168. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Mao, Z. , Xue, W. , Li, Y. , Tang, G. , Wang, A. , et al (2011) Ku80 gene is related to non‐homologous end‐joining and genome stability in Aspergillus niger . Curr Microbiol 62: 1342–1346. [DOI] [PubMed] [Google Scholar]
- Zhang, D.H. , Jiang, L.X. , Li, N. , Yu, X. , Zhao, P. , Li, T. , and Xu, J.W. (2017a) Overexpression of the squalene epoxidase gene alone and in combination with the 3‐hydroxy‐3‐methylglutaryl coenzyme A gene increases ganoderic acid production in Ganoderma lingzhi . J Agr Food Chem 65: 4683–4690. [DOI] [PubMed] [Google Scholar]
- Zhang, D.H. , Li, N. , Yu, X. , Zhao, P. , Li, T. , and Xu, J.W. (2017b) Overexpression of the homologous lanosterol synthase gene in ganoderic acid biosynthesis in Ganoderma lingzhi . Phytochemistry 134: 46–53. [DOI] [PubMed] [Google Scholar]
- Zheng, Q. , Cai, X. , Tan, M.H. , Schaffert, S. , Arnold, C.P. , Gong, X. , et al (2014) Precise gene deletion and replacement using the CRISPR/Cas9 system in human cells. Biotechniques 57: 115–124. [DOI] [PubMed] [Google Scholar]
- Zhou, J.S. , Ji, S.L. , Ren, M.F. , He, Y.L. , Jing, X.R. , and Xu, J.W. (2014) Enhanced accumulation of individual ganoderic acids in a submerged culture of Ganoderma lucidum by the overexpression of squalene synthase gene. Biochem Eng J 90: 178–183. [Google Scholar]
- Zhu, J. , Sun, Z. , Shi, D. , Song, S. , Lian, L. , Shi, L. , et al (2019) Dual functions of AreA, a GATA transcription factor, on influencing ganoderic acid biosynthesis in Ganoderma lucidum . Environ Microbiol 21: 4166–4179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Sequences
Data S2. Sequences
Data S3. Sequences
Data S4. Sequences
Table S1. Primers used in the study
Fig. S1. Mycelia growth of the transformants GL‐opcas9 and pJW‐EXP‐intron‐opcas9 in CYM plates
Fig. S2. CRISPR/Cas9‐directed mutation of ura3 in G. lucidum. Alignments of ura3 mutants (M1, D1, In1, and In37) from 5‐FOA‐resistant colonies in sgRNA1 transformants. WT is the wild‐type strain ura3 from G. lucidum. The sgRNA1‐guiding sequence is highlighted in yellow. Replacements, deletions and insertions were found near the PAM sequence.
Fig. S3. Transcriptional levels of Cas9 gene in the GL‐opcas9 and pJW‐EXP‐intron‐opcas9 strains. Expression of Cas9 gene from the GL‐opcas9 strain is defined as 1.0, and expression levels in the pJW‐EXP‐intron‐opcas9 strain are displayed as fold increases over the reference sample. The following primers were used: Cas9‐f, 5’‐GAGGTCGCCTACCACGAGAAGT‐3’ and Cas9‐r, 5’ ‐TGGACGAGCTGGATGAAGAGC‐3’. * indicates statistical significance (P < 0.05) compared to the GL‐opcas9 strain (control).


