Summary
Xylella fastidiosa is a xylem‐limited bacterial pathogen causing devastating diseases in many economically important crops. Calcium (Ca) is a major inorganic nutrient in xylem sap that influences virulence‐related traits of this pathogen, including biofilm formation and twitching motility. This study aimed to adapt a microfluidic system, which mimics the natural habitat of X. fastidiosa, for whole transcriptome analysis under flow conditions. A microfluidic chamber with two parallel channels was used, and RNA isolated from cells grown inside the system was analysed by RNA‐Seq. Ca transcriptionally regulated the machinery of type IV pili and other genes related to pathogenicity and host adaptation. Results were compared to our previous RNA‐Seq study in biofilm cells in batch cultures (Parker et al., 2016, Environ Microbiol 18, 1620). Ca‐regulated genes in both studies belonged to similar functional categories, but the number and tendencies (up‐/downregulation) of regulated genes were different. Recombination‐related genes were upregulated by Ca, and we proved experimentally that 2 mM Ca enhances natural transformation frequency. Taken together, our results suggest that the regulatory role of Ca in X. fastidiosa acts differently during growth in flow or batch conditions, and this can correlate to the different phases of growth (planktonic and biofilm) during the infection process.
Whole transcriptome analysis was conducted on cells of the bacterial plant pathogen Xylella fastidiosa grown inside microfluidic chambers. Comparison of cells grown in synthetic media with or without supplemental calcium (Ca) revealed that Ca regulates movement and other virulence factors of this pathogen under flow conditions.

Introduction
Plant xylem, the system that conveys water and dissolved minerals from the roots to the photosynthetic organs, can serve as a growth niche for microbes including bacterial pathogens. When xylem‐residing bacterial pathogens enter the water‐conducting xylem vessels through natural openings, wounds or helped by xylem‐feeding insects, they proliferate. Inside the xylem, they secrete cell wall‐degrading enzymes and obstruct the transportation of water and minerals, which leads to disease of the host plant and eventually death (Yadeta and Thomma, 2013). Since these bacterial pathogens live deep in the interior of plants and can infect a broad range of host plants; cultural and chemical methods to control these pathogens are generally ineffective (Yadeta and Thomma, 2013). Xylella fastidiosa, a xylem‐limited bacterial pathogen, is the causative agent of devastating plant diseases in many economically important crops in the Americas, Europe and Asia, including Pierce’s disease on grape, citrus variegated chlorosis, olive quick decline syndrome and others (Almeida and Nunney, 2015; Sicard et al., 2018). Xylella fastidiosa is one of the two plant bacterial pathogens to date that has been reported to be naturally competent (Kung and Almeida, 2011). This ability was hypothesized to confer this pathogen with adaptive advantages (Kandel et al., 2016; Sicard et al., 2018; Potnis et al., 2019).
Calcium is an important inorganic compound in xylem sap, which has multiple functions for both plant host and bacterial pathogens (De La Fuente et al., 2014). The role of Ca in plants has been extensively studied, which not only involves the stabilization of cell wall structures but also acts as a versatile messenger that mediates a sophisticated signalling network (Demarty et al., 1984; Dodd et al., 2010). The role of Ca in bacteria is less studied; however, recent indirect evidence suggests multiple regulatory functions of Ca in bacteria (Dominguez, 2004; Dominguez et al., 2015). Calcium regulates a variety of bacterial behaviours including biofilm formation (Sarkisova et al., 2005; Rinaudi et al., 2006), motility (Gode‐Potratz et al., 2010; Guragain et al., 2013), secretion activity (DeBord et al., 2003; Dasgupta et al., 2006), spore germination (Wang et al., 2008) and quorum sensing (Werthén and Lundgren, 2001). A series of studies conducted by our group has shown that Ca influences key virulence‐related traits of X. fastidiosa. An increase of external Ca concentration leads to higher surface adhesion force, twitching speed and more biofilm formation by X. fastidiosa (Cruz et al., 2012). Calcium‐enhanced twitching motility is associated with the interaction between Ca and the Ca‐binding protein PilY1 of the type IV pili (TFP) (Cruz et al., 2014). Whole transcriptome analysis of X. fastidiosa biofilm cells in batch culture at different Ca concentration showed that genes involved in attachment and biofilm formation maintained a higher expression level in Ca‐supplemented media; in contrast, these gene expression levels were gradually decreased in non‐supplemented media, which indicates Ca promotes continued biofilm development in X. fastidiosa (Parker et al., 2016). Unravelling the molecular basis of the effect of Ca on virulence‐related traits of X. fastidiosa is essential for understanding the biology of X. fastidiosa in plant xylem.
Xylem‐residing bacterial pathogens grown in the xylem vessels are under continuous flow conditions. However, the majority of in vitro studies of xylem‐residing bacterial pathogens have been performed in batch cultures. X. fastidiosa only survives in two natural habitats, the plant xylem vessels and the feeding canal of insects, both considered flow channels for xylem fluid (Chatterjee et al., 2008). Microfluidic chambers (MC), an artificial device mimicking the natural environment of plant xylem vessels and insect foreguts, provide a continuous flow condition for growth of xylem‐residing bacterial pathogens (Meng et al., 2005; De La Fuente et al., 2007; Bahar et al., 2010). Bacterial cells grown in MC are monitored by time‐lapse microscopy, which provides important spatial and temporal information for understanding physiological processes of bacteria that we are unable to observe directly inside the host in vivo. Previously, this MC system has been used to study virulence‐related traits of X. fastidiosa (De La Fuente et al., 2007; Cruz et al., 2012; Navarrete and De La Fuente, 2014; Chen et al., 2017). In this present study, RNA‐Seq was conducted on X. fastidiosa Temecula1, the causal agent of Pierce’s disease on grape, grown in MC comparing 2 mM CaCl2‐supplemented and non‐supplemented media. Results revealed regulation of genes by Ca that are responsible for enhanced twitching motility and are associated with pathogenicity and adaptation to the host. Comparison of these RNA‐Seq data with our previous assessment in biofilm cells in batch culture (Parker et al., 2016) suggests that the regulation by Ca in X. fastidiosa is different between cells in biofilm and under flow conditions. In addition, phenotypic assessment revealed that Ca enhanced natural transformation of X. fastidiosa. These findings support that the regulatory role of Ca in X. fastidiosa acts differently depending on the stage of the infection process.
Results and discussion
Adaptation of a microfluidics system for Xylella fastidiosa whole transcriptome analysis
Both natural habitats of X. fastidiosa, that is insect foregut and plant xylem vessels, are under continuous flow conditions. Our MC dual parallel channel design (Fig. 1A, B) not only mimic these natural habitats but also offers the possibility to compare simultaneously bacterial behaviour under two different treatments. To confirm a previously observed phenotype (Cruz et al., 2012), twitching motility of X. fastidiosa Temecula1 in PD2 and 2Ca was quantified by tracking cells upstream movement. Cell twitching speed in 2Ca was significantly higher (P < 0.001) (0.65 ± 0.3 μm min−1) than that in PD2 medium (0.38 ± 0.2 μm min−1) (Fig. S1). Xylella fastidiosa Temecula1 cells were harvested from the MC system 6 days post‐inoculation, when biofilm started to form (Fig. 1C). At this time point, the majority of Temecula1 cells in the channel were motile, and enough Temecula1 cells could be collected from the channel with the help of DNA/RNA Shield. Initially, Temecula1 cells were harvested by increasing media flow rate, which could not remove all cells from the channel, resulting in a low cell concentration (data not shown). RNA isolated from those samples were in low quantity and quality and were not suitable for RNA‐Seq analysis. When media was replaced by DNA/RNA Shield for harvest, all cells can be collected in a short time (Fig. 1C and Video S1), and more importantly, composition of mRNA from cells in the RNA protection buffer was not modified during the harvesting process and was protected for further processing.
Figure 1.
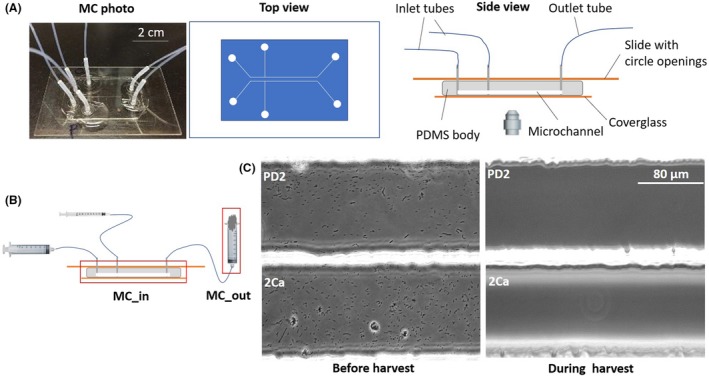
Preparation of X. fastidiosa cultures in microfluidic chamber (MC) for whole transcriptome analysis.
A. MC with dual parallel channel design. Left picture shows an assembled MC, middle diagram shows top view, and right diagram shows side view of the MC.
B. Side view of the MC during X. fastidiosa growth experiments. 5 ml glass syringes filled with media were connected to medium inlets, 1 ml plastic syringes filled with X. fastidiosa suspension were connected to bacterium inlets, and 10 ml plastic syringes sealed with cotton balls were connected to outlets. During cell harvest, 1 ml plastic syringe filled with RNA/DNA Shield buffer replaced the 5 ml glass syringes, and the 10 ml plastic syringes were replaced by 1.5 ml microcentrifuge tubes.
C. Micrographs of the two parallel channels showing the growth of X. fastidiosa and harvest of cells from channels.
In total, nine independent MC experiments were conducted, each consisting of two treatments (PD2 and 2Ca); therefore,18 RNA samples were obtained. Although X. fastidiosa cells collected from each microchannel of MC were in a limited amount, RNA isolated from these samples fulfilled the requirements of quantity and quality for RNA sequencing analysis on Illumina HiSeq platform. Three groups of RNA samples with the best quality (Table S1) were used for whole transcriptome sequencing. Six RNA sequencing libraries were constructed and each library generated more than 9.7 million reads (sequencing reads available from NCBI BioProject ID PRJNA556203). After adapter and quality trimming and ribosome RNA filtering, 44–73% clean reads remained. The average length of post‐trim reads ranged from 73 to 89 bp, and 92–96% of post‐trim reads aligned to the X. fastidiosa Temecula1 genome (Data Set S1). Mapped reads were used to determine transcript boundaries and normalized expression for all protein‐coding genes (Data Set S2). These results suggest our MC system is suitable for whole transcriptome analysis of X. fastidiosa. Recently, microfluidic systems have been demonstrated to be suitable diagnostic devices measuring RNA expression of bacteria (by microarray) in human blood (Gandi et al., 2015). High‐throughput microfluidics were used to assess the genome of a single bacterial cell (Pamp et al., 2012) and transcriptome of a single mammalian cell (Tang et al., 2009; Streets et al., 2014). These studies suggest the advantage for using a microfluidic system to discover novel finding in genetics. The present study is the first time where MC and RNA‐Seq were combined to analyse the whole transcriptome of bacterial cells, which is a suitable system for novel discoveries in bacteriology.
Functional classification of differentially expressed genes (DEGs)
A total of 122 protein‐coding genes were differentially expressed (q < 0.05, FC ≥ 1.5) when comparing 2Ca treatment to control. These DEGs represent 5 % of the annotated genes of X. fastidiosa Temecula1. Among the 122 genes, 85 genes were upregulated, and 37 genes were downregulated (Fig. 2 and Data Set S3). To reveal the putative function of the DEGs, they were annotated and classified using Gene ontology (GO) analysis. The 225 GO terms were annotated and classified into three main categories: 82 biological processes (BP), 93 molecular functions (MF) and 50 cellular components (CC) (Fig. S2). Most of the DEGs were mainly related to cellular and metabolic process in BP, binding and catalytic activity in MF, and component of the membrane in CC. According to the GO classification of the DEGs, 56 genes could not be annotated with any GO term and were annotated as unknown function genes. In addition, all DEGs were searched against the KEGG database using a pathway mapping tool to obtain an overview of the metabolic pathways and other functional systems of X. fastidiosa Temecula1 regulated by external Ca supplementation. A total of 30 genes were mapped to different pathways involved in a series of compound metabolism and biogenesis, two‐component system, homologous recombination, ABC transporters and bacterial secretion system (Data Set S4).
Figure 2.
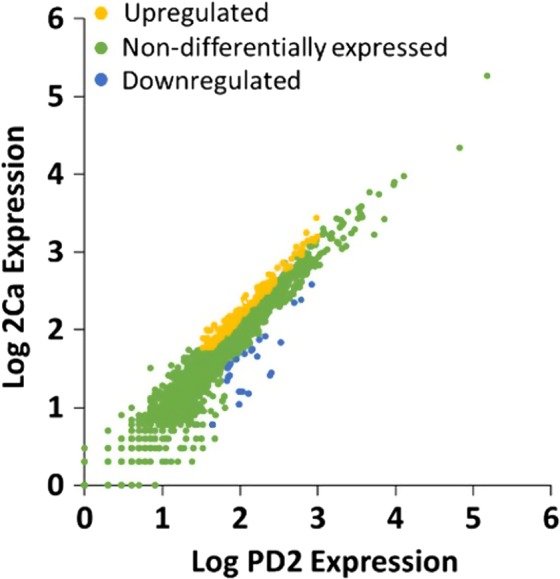
Scatter plots of log gene expression from X. fastidiosa cells cultured in MC (2Ca vs. PD2) as computed from the RNA‐Seq data. Genes significantly differentially expressed in 2Ca/PD2 (1.5‐fold change; q < 0.05) are indicated in yellow (upregulation) or blue (downregulation). Non‐differentially expressed genes are indicated in green.
According to the GO classification and KEGG analysis of potential function of DEGs, Ca may be involved in many aspects of X. fastidiosa including nutrient metabolism, stimulus response, signal transduction and cellular component biosynthesis. Around 18% of DEGs encode enzymes with functions in metabolism of sugars, nucleic acids, and amino acids, and biosynthesis of secondary metabolites and antibiotics. Interestingly, some of these enzymes are required for the glycolysis pathway (Data Set S4), indicating Ca could regulate glycolysis of X. fastidiosa. Former studies in eukaryotes have shown that glycolysis is regulated by Ca via its effect on activities of glycolytic enzyme (Schönekess et al., 1995; Ørtenblad et al., 2009; Nichols et al., 2017), suggesting this role of Ca may be conserved in prokaryotes and eukaryotes. Our results also suggest that Ca is an important signal messenger in this bacterium since many DEGs were associated with signal binding, transport and transcriptional regulation. Similar results were observed in other transcriptome analysis of bacterial responses to Ca (Oomes et al., 2009; Gode‐Potratz et al., 2010; Parker et al., 2016; Zhu et al., 2017).
RT‐qPCR was used to validate the RNA‐Seq differential gene expression results. Expression levels of six selected genes related to pili, surface proteins and virulence were assessed using the same RNA samples that were used in the RNA‐Seq analysis. Pearson’s correlation coefficient for RT‐qPCR and RNA‐Seq results was 0. 941 (P = 0.005, Fig. S3), which indicate a positive correlation and significant similarity between these two experiments.
Differential expression of genes associated with twitching motility in 2Ca compared to PD2
Twitching motility is an important virulence‐related trait of X. fastidiosa (Mattick, 2002; Chatterjee et al., 2008; Chen et al., 2017). This ability is mediated by extension and retraction of TFP that contributes to upstream movement of X. fastidiosa, which is very important for this pathogen to colonize the plant xylem system (Meng et al., 2005; De La Fuente et al., 2007). TFP is complex surface appendages, at least 30 genes in X. fastidiosa are involved in biogenesis and function of TFP (Wang et al., 2012). Our data have shown that nine genes involved in TFP assembly and functional regulation were differentially expressed between 2Ca and PD2 treatments (Fig. 3, Table 1). Genes encoding PilN, PilO and PilP for TFP pili assembly subcomplex were upregulated. These proteins, together with PilQ and PilM, constitute the secretion complex of the pilin protein that is the basic unit for the extracellular part of TFP (Goosens et al., 2017). Mutation of this complex leads to absence of TFP (Li et al., 2007). Three components of the chemosensory system, pilI, pilG and pilH were upregulated as well. This chemosensory system is known to play a role in pili formation, extension and retraction (Bertrand et al., 2010; Cursino et al., 2011). pilR, encoding the response regulator of the two‐component system (TCS) PilS‐PilR, was upregulated by 2Ca. PilS‐PilR TCS plays an important regulatory role in TFP biosynthesis (Hobbs et al., 1993; Kilmury and Burrows, 2016). It has been reported that the regulator PilR can bind to an enhancer sequence associated with the promoter of the pilin gene in P. aeruginosa (Jin et al., 1994). Additionally, two of the three predicted pilA homologues in X. fastidiosa Temecula1, PD1077 and PD1924 (pilA1), were downregulated. The pilA gene encodes pilin, which is the basic component of the TFP filaments (Mattick, 2002). Proteins encoded by the three pilA homologue genes, PD1077, PD1924 (pilA1) and PD1926 (pilA2), contain conserved protein domains related to PilA. Comparison of amino acid sequences of these three proteins indicated that PilA1 has 65% identity with PilA2, but PD1077 has 45% identity with PilA1 and 36% identity with PilA2. The transcriptome data show that expression of PD1077 was similar to pilA1 but much lower than pilA2. The function of PD1077 has not been characterized yet; however, pilA1 and pilA2 were studied by our group, and we demonstrated that pilA1 affects the number and position of TFP, while pilA2 is needed for twitching movement (Kandel et al., 2018). This suggests that PilA1 may be a regulator protein rather than the major pilin of TFP, but this needs to be further studied. 2Ca suppressed pilA1 but did not affect pilA2, which could contribute to a more effective twitching motility of X. fastidiosa. According to these results, we hypothesize that Ca transcriptionally regulated TFP assembly and function to enhance twitching motility of X. fastidiosa.
Figure 3.
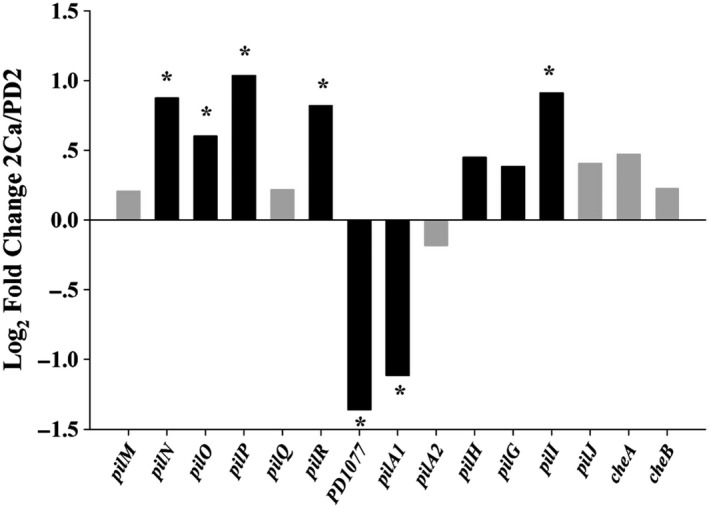
Expression changes of selected type IV pili genes in X. fastidiosa cultured in microfluidic chamber. Each bar shows the log2 fold change of gene expression in 2Ca versus PD2 as computed from the RNA‐Seq data. Black bars indicate q < 0.05. Asterisk indicates fold changes > 1.5.
Table 1.
Selected genes transcriptionally regulated by Ca in MC‐RNA‐Seq and Flask‐RNA‐Seq.
| Functional group | Gene name | Locus tag | Product | MC | Flask |
|---|---|---|---|---|---|
| Twitching motility | pilG* | PD0845 | Pili chemotaxis CheY homolog PilG | Up | – |
| pilI | PD0846 | Pili chemotaxis CheW homolog Pil | Up | – | |
| ‐ | PD1077 | Fimbrial protein | Down | – | |
| pilH* | PD1632 | Regulatory protein pilH family | Up | Down | |
| pilP | PD1692 | Fimbrial assembly protein | Up | – | |
| pilO | PD1693 | Fimbrial assembly membrane protein | Up | – | |
| pilN | PD1694 | Fimbrial assembly membrane protein | Up | – | |
| pilA1 | PD1924 | Fimbrial protein | Down | Down | |
| pilA2 | PD1926 | Fimbrial protein | ‐ | Down | |
| pilR | PD1928 | Regulatory protein | Up | – | |
| Adhesins and EPS biosynthesis | xanA | PD0213 | Phosphoglucomutase | Down | – |
| xadA | PD0731 | Outer membrane protein XadA | – | Up | |
| hsf | PD0744 | Surface protein | Up | Up | |
| pspA | PD0986 | Hemagglutinin‐like secreted protein | – | Up | |
| gumM | PD1387 | GumM protein | – | – | |
| gumJ | PD1389 | GumJ protein | – | Up | |
| gumH | PD1391 | GumH protein | – | Up | |
| gumF | PD1392 | GumF protein | – | Up | |
| hxfA | PD2118 | Hemagglutinin‐like secreted protein | – | Up | |
| Regulatory function | – | PD1087 | XRE family transcriptional regulator | Down | Down |
| gacA | PD1984 | Transcriptional regulator | Down | ‐ | |
| Phage‐related proteins | – | PD0377 | Phage‐related protein | Down | Up |
| – | PD0906 | Phage‐related protein | Up | Down | |
| – | PD0916 | Phage‐related protein | Up | Down | |
| – | PD0929 | Phage‐related protein | Up | Down | |
| – | PD0932 | Phage‐related protein | Up | Down | |
| – | PD0936 | Phage‐related protein | Up | Down | |
| – | PD0943 | Phage‐related protein | Up | Down | |
| Colicin V‐like bacteriocins | cvaC | PD0215 | colicin V precursor | Up | Up |
| Recombination and competence | recF | PD0003 | Recombination protein F | Up | – |
| recJ | PD0402 | ssDNA‐specific exonuclease RecJ | – | Up | |
| recD | PD1651 | exodeoxyribonuclease V subunit alpha | – | Up | |
| comA | PD0358 | DNA uptake protein | – | Up | |
| comM | PD0464 | Competence‐like protein | Down | – | |
| comB | PD1558 | DNA transport competence protein | Down | – | |
| ruvC | PD0886 | Holliday junction resolvase | Up | Down | |
| ung | PD2049 | Uracil‐DNA glycosylase | Up | – | |
| Uncharacterized Proteins | PD0556 | hypothetical protein | Up | Down | |
| PD1111 | hypothetical protein | Down | Up |
–, No changes; Down, downregulated gene expression; Flask, Flask‐RNA‐Seq (Parker et al., 2016); MC, MC‐RNA‐Seq (this study); and Up, upregulated gene expression.
Locus tag in bold text indicates genes overlapped between MC and Flask.
Asterisk indicates q value of gene is < 0.05, but fold change is not > 1.5.
Results in this study have shown that more than ten genes encoding response and transcriptional regulators were regulated by 2Ca in X. fastidiosa (Data Set S3). A key transcriptional regulator downregulated by 2Ca is gacA, which is a global activator in many plant‐associated gram‐negative bacteria (Heeb and Haas, 2001). The phenotype and transcriptome of X. fastidiosa gacA mutant were previously studied, concluding that GacA in X. fastidiosa contributes to cell adhesion and surface attachment (Shi et al., 2009). In P. aeruginosa, GacA is a key component in the Gac/Rsm pathway which mediates Ca signaling to regulate motility and biofilm formation of this pathogen (Broder et al., 2017). Since our previous results (Cruz et al., 2012) showed that Ca regulates attachment and motility of X. fastidiosa, we speculate that gacA may be part of the regulatory cascade involving Ca.
Comparison of gene expression profiles of X. fastidiosa in microfluidic chamber and batch cultures
The whole transcriptome of X. fastidiosa biofilm cells grown in flasks with PD2 and 4Ca (PD2 supplemented 4 mM CaCl2) for 72 h was previously studied by our group using RNA‐Seq (Parker et al., 2016), which is referred here as ‘Flask‐RNA‐Seq’. For comparisons, results from this present study are referred as ‘MC‐RNA‐Seq’. Here, the two RNA‐Seq experiments were compared (Fig. 4, Table 1). Even considering that many parameters between these two experiments were different, including culture conditions (MC and Flasks), cell physiological state (all cells and biofilm cells) and Ca concentrations (2 and 4 mM); we believe that comparison of these two studies has significance for understanding the molecular basis of the role of Ca during the X. fastidiosa infection process.
Figure 4.
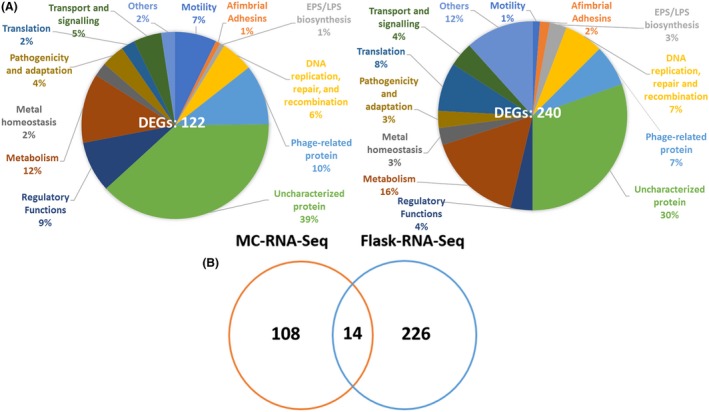
Comparison of the effect of Ca on gene expression profiles of X. fastidiosa grown in microfluidic chamber (MC‐RNA‐Seq) and batch cultures (Flask‐RNA‐Seq, Parker et al., 2016).
A. Pie charts for distribution of differentially expressed genes (DEGs) in different functional categories between MC‐RNA‐Seq and Flask‐RNA‐Seq.
B. Venn diagram showing number of DEGs overlapping between the two RNA‐Seq experiments.
The overall distribution of DEGs based on functional annotation in both RNA‐Seq studies in X. fastidiosa was similar, which confirmed the regulatory function of Ca in X. fastidiosa. Around 40% DEGs were classified as unknown function genes and phage‐related genes (Fig. 4A), suggesting the Ca‐regulated genetic network in X. fastidiosa remains elusive. According to this distribution, the percentage of genes belonging to the motility functional group in MC‐RNA‐Seq was much higher than that in Flask‐RNA‐Seq. The majority of DEGs belonging to motility functional group in MC‐RNA‐Seq were upregulated by Ca, while genes belonging to this group in Flask‐RNA‐Seq were downregulated (Table 1). However, in Flask‐RNA‐Seq, the percentage of DEGs belonging to afimbrial adhesins and exopolysaccharides (EPS) biosynthesis functional groups was higher than that in MC‐RNA‐Seq (Fig. 4A). Adhesion and EPS production are two critical aspects for biofilm formation in many bacteria (Marques et al., 2002; Castiblanco and Sundin, 2015). Genes belonging to these two groups in Flask‐RNA‐Seq were upregulated, but genes belonging to EPS biosynthesis functional groups in MC‐RNA‐Seq were downregulated (Table 1). These results corresponded with the physiological states that were used in these two studies: MC included both planktonic and biofilm cells, but most cells were actively moving against the flow (Fig. 1C and Video S2); while cells in flasks were collected exclusively from biofilms (Parker et al., 2016).
Only 14 DEGs overlapped between MC‐RNA‐Seq and Flask‐RNA‐Seq (Fig. 4B, Table 1), and only 4 of the 14 genes regulated by Ca showed the same tendency (viz., upregulated or downregulated in both cases), including PD1087, pilA1, cvaC and hsf (Table 1). PD1087 and pilA1 were downregulated by Ca in both RNA‐Seq experiments. PD1087 encodes an XRE family transcriptional regulator and is located upstream of multiple phage‐related gene operons. Regulators of the XRE family are widely distributed and regulate functions such as oxidant tolerance and virulence in Streptococcus suis (Hu et al., 2018); repression of a defective prophage in Bacillus subtilis (Wood et al., 1990; McDonnell and McConnell, 1994) and resistance of antibiotics in Lactococcus lactis (McAuliffe et al., 2001 ). However, the role of PD1087 in X. fastidiosa is unclear and needs further characterization. pilA1, as mentioned above, is associated with the number and location of TFP. Deletion of pilA1 led to an increase in biofilm formation by X. fastidiosa but did not affect its twitching motility (Kandel et al., 2018). This protein seems to be an important component for both stages of X. fastidiosa growth, planktonic and biofilm, and this can explain the same regulatory tendency in both transcriptome studies discussed here.
The cvaC and hsf were upregulated by Ca in both experiments. cvaC is predicted to encode a colicin V‐like bacteriocin, an antimicrobial peptide first reported in E. coli (Waters and Crosa, 1991). Genes required for production and secretion of colicin V‐like bacteriocin were found among the genome of X. fastidiosa, including cvaC (PD0215), cvaA (PD0496), cvaB (PD0499) and cvpA (PD0852). Here, we found that in addition to cvaC, cvpA was also upregulated by 2Ca. In host plant xylem systems, X. fastidiosa is not the only bacterium to reside in that environment, previous studies showed that there is an endophytic bacterial community, and the communities are different between healthy and X. fastidiosa infected hosts (Araújo et al., 2002). Xylella fastidiosa should have some arsenals to compete with other endophytic bacteria, enabling the colonization of xylem vessels. According to previous transcriptome analysis of X. fastidiosa under other xylem‐related environmental contexts, including basic medium supplemented with the inorganic nutrient iron (Fe) (Zaini et al., 2008), organic nutrient glucose (Pashalidis et al., 2005) and grape sap (Ciraulo et al., 2010), cvaC was upregulated in all of these xylem‐related conditions, suggesting that this gene may play an important role in the environmental adaptation of X. fastidiosa. The hsf gene encodes a surface protein similar to Hsf (Haemophilus surface fibril), and the C‐terminal region of this protein shares characteristics with autotransporter proteins (de Souza et al., 2003). Homologues of this protein in other bacteria are important for cell adhesion and attachment to the host cell (de Souza et al., 2003; Singh et al., 2015). The hsf gene expression level in recently isolated X. fastidiosa 9a5c was significantly higher than that in 9a5c after many passages in axenic culture (de Souza et al., 2003), suggesting hsf could be a pathogenicity‐related factor in X. fastidiosa. Interestingly, expression of hsf was also upregulated in grape sap‐supplemented media (Ciraulo et al., 2010) but suppressed by Fe (Zaini et al., 2008), and did not change in basic medium supplemented with glucose (Pashalidis et al., 2005). Accordingly, modification of hsf gene expression is associated with increased concentrations of mineral nutrients Ca and Fe and grape sap; therefore, we speculate that hsf encodes a type of adhesin that may be involved in ion bridge‐mediated adherence. The function of hsf in X. fastidiosa still needs further studies to be elucidated.
Another interesting finding among these 14 overlapped DEGs was that the regulation of 7 phage‐related genes by Ca that is exactly opposite between MC and flasks (Table 1). PD0906, PD0916, PD0926, PD0932, PD0936 and PD0943 were upregulated in MC and downregulated in flasks. These genes are located in a putative genomic island comprising PD0906‐PD0943 (Parker et al., 2016). 21 out of the 38 genes in this island do not have homologues in the closely related avirulent strain X. fastidiosa EB92‐1 (Zhang et al., 2011; Parker et al., 2016). These findings suggest that the role of these phage‐related genes may be associated with twitching motility, biofilm formation and virulence of X. fastidiosa, but to confirm this, further investigation is needed.
Effect of 2 mM external Ca on natural competence of Xylella fastidiosa
Xylella fastidiosa is naturally competent, with the ability to take environmental DNA fragments and integrate them into its genome via homologous recombination (Lorenz and Wackernagel, 1994; Kung and Almeida, 2011); this was hypothesized to impact adaption of this pathogen to new hosts and cause disease emergence (Almeida and Nunney, 2015; Kandel et al., 2016; Sicard et al., 2018; Potnis et al., 2019). The differential gene expression data in this study showed that genes associated with homologous recombination, including recF, ruvD and ung, were upregulated in 2Ca (Table 1). Interestingly, in Flask‐RNA‐Seq, genes recJ, recD and comA involved in recombination and competence were also upregulated in 4Ca (Table 1) (Parker et al., 2016). Moreover, twitching motility of X. fastidiosa, which has been reported to be important for natural transformation (Kandel et al., 2017), was enhanced in 2Ca. Taking these results into consideration, we hypothesized that 2 mM external Ca contributes to natural competence of X. fastidiosa.
To determine whether natural competence of X. fastidiosa is influenced by Ca, X. fastidiosa Temecula1 was incubated with plasmid pAX1.Cm or pAX1.Km and cultured in MC or on plates with and without amendment of 2 mM CaCl2 (2Ca vs. PD3), and the frequency of acquisition of antibiotic resistance marker by X. fastidiosa Temecula1 was assessed (Fig. 5). Under MC conditions, using pAX1.Cm as donor DNA, the recombination frequency (recombinant/total cells) in PD3 and 2Ca was 1.42 × 10−3 and 1.50 × 10−3 respectively. For MC_in fraction (X. fastidiosa cells collected from inside the microchannels of MC, Fig. 1B), no significant differences (P = 0.287) between treatments were found. However, for MC_out fraction (X. fastidiosa cells collected from the outlet containers of MC, Fig. 1B), the recombination frequency in PD3 and 2Ca was significantly different (P = 0.026) with 1.74 × 10−4 and 1.70 × 10−3 respectively. We also observed that the recombination frequency in PD3 for MC_in fraction was higher than that for MC_out fraction, similar to that in previous studies (Kandel et al., 2016), which confirmed flow conditions in MC contributes to natural competence of X. fastidiosa. Under agar plate conditions, using pAX1.Km plasmid as donor DNA, the recombination frequency on 2Ca plates (7.7 × 10−5) was significantly higher (P < 0.001) than that on PD3 plates (1.5 × 10−5). Similarly, using pAX1.Cm as donor DNA, the recombination frequency in 2Ca treatment and control (1.22 × 10−4 and 4.80 × 10−5) was significant different (P = 0.014). These results suggest that 2 mM external Ca increased natural recombination frequency in X. fastidiosa, confirming our hypotheses.
Figure 5.
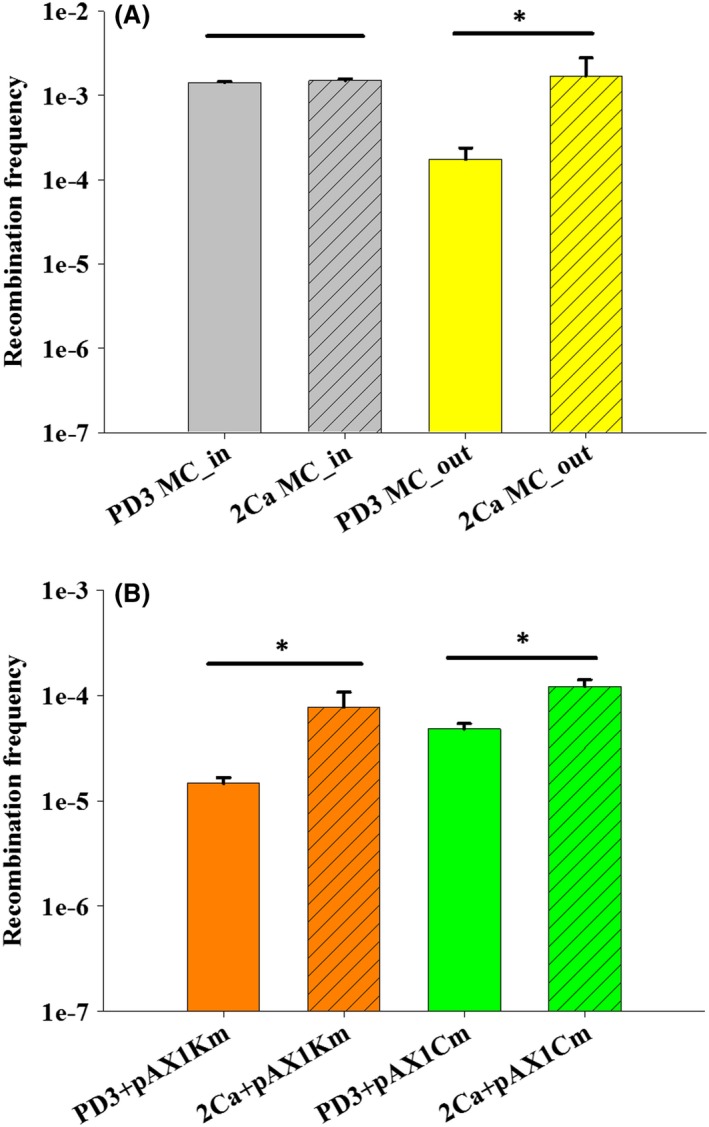
Effect of 2 mM external Ca on natural recombination in X. fastidiosa Temecula1.
A. X. fastidiosa was grown in microfluidic chambers. Plasmid pAX1.Cm was used as donor DNA.
B. X. fastidiosa was grown on agar media plates. Plasmids pAX1.Km and pAX1.Cm were used as donor DNA. For each growth condition, three independent experiments were conducted. Data represent means (A; MC: n = 6, B, plates: n = 9), and error bars represent standard errors form different experiments. Asterisk indicates significant difference as analysed by t‐test (P < 0.05).
Ca has been reported to be necessary for natural competence in some bacteria: 1 mM Ca2+ was critical for competence induction in Streptococcus pneumoniae (Trombe, 1993); 1–2 mM Ca2+ was enough to induce Escherichia coli to natural competence, which has been originally thought to be incapable of this process (Baur et al., 1996); and in Azotobater vinelandii, 0.5 and 1 mM Ca2+ was required to optimally trigger natural competence (Page and Doran, 1981). Similarly, contribution of Ca to natural competence has been reported in other bacteria. In the human pathogen Acinetobacter baumannii, a naturally competent strain, its natural transformation frequency was increased upon the addition of Ca (Traglia et al., 2016). In another naturally competent pathogen Aggregatibacter actinomycetemcomitans, 1 mM external Ca enhanced its cell aggregation and natural competence (Hisano et al., 2014), and the author of that study postulated Ca‐promoted biofilm may increase the ability of the bacteria to uptake environmental DNA (Hisano et al., 2014). This may be one explanation for the finding that Ca increased natural recombination frequency in MC_out fraction but not in MC_in fraction. Ca enhanced natural competence is associated with Ca‐promoted biofilm (accumulation of biofilm in MC_out fraction is more than that in MC_in fraction). Our transcriptome data support this concept. In MC‐RNA‐Seq, 2Ca can increase expression of twitching motility and recombination‐related genes; however, competence‐related genes comM and comE were downregulated (Table 1). For batch cultures (Flask‐RNA‐Seq), where biofilms cells were sampled, 4Ca increased expression of competence‐related gene comA (Table 1) (Parker et al., 2016).
Conclusions
Although it is important to study the whole transcriptome of bacteria by RNA‐Seq in planta, as it was done in a few cases (Ailloud et al., 2016; Lee et al., 2017; Nobori et al., 2018), this is technically challenging with X. fastidiosa. The main limitation is the small amount and uneven distribution of bacteria found in planta (e.g. 105 to 107 CFU g−1 for X. fastidiosa vs. 108 to 109 CFU g−1 for Ralstonia sp.) (Hill and Purcell, 1995; Jacobs et al., 2012). Another limitation is that woody plants are hosts for X. fastidiosa, therefore, making it harder to extract the relatively small amount of bacteria inside xylem vessels. However, our MC provides a uniform, precisely controlled growth environment close to the natural conditions for X. fastidiosa. Through time‐lapse microscopy, MC also provides critical spatial and temporal information regarding physiological processes of bacteria that we are unable to observe directly in planta. Therefore, we feel that our contribution of our system combining MC with RNA‐Seq is a valid approximation to understand gene regulation under flow conditions mimicking the natural habitats of these bacteria. In susceptible grapes, the host of X. fastidiosa Temecula1, Ca concentration in its xylem sap is approximately 2–3 mM (Cobine et al., 2013), the same concentration we used in this present study. We conclude that under flow conditions Ca transcriptionally regulates the machinery of TFP to enhance twitching motility and other key genes involved in pathogenicity and adaptation of X. fastidiosa to the host environment. The transcriptome profile described in this study represents that of X. fastidiosa during the initial stages of colonization when they were either recently injected into the host xylem vessels by insect vectors, or they broke free from mature biofilms, and they are moving in the xylem. Through comparisons with our previous transcriptome study with mature biofilms (Parker et al., 2016), we conclude that Ca influences gene expression differently at different growth stages. Nevertheless, we found a few common regulation patterns between both studies; in particular, we found that Ca can modulate expression of genes related to natural competence and increase natural competence of X. fastidiosa. Collectively, this study suggests the Ca concentration in susceptible grape contributes for pathogenicity, natural competence and host adaptation of X. fastidiosa Temecula1.
Experimental procedures
Bacterial strains and culture conditions
Xylella fastidiosa subsp. fastidiosa type strain Temecula1 was used throughout this study. Temecula1 was cultured on PW (Davis et al., 1980) or PD3 (Davis et al., 1981a) plates at 28°C. Cell suspensions of Temecula1 used for MC experiments were prepared as follows: Temecula1 was recovered from glycerol stocks on PW plates and grown for 7 days. Temecula1 cultures were streaked to new PW plates, and seven‐day‐old Temecula1 subcultures were scraped from PW plates, suspended in PD2 (Davis et al., 1981b) or PD2 supplemented with 2 mM CaCl2 (2Ca) liquid media and diluted to an OD600 of 1.0.
Xylella fastidiosa cultures in microfluidic chambers for transcriptome analysis
The MC with dual parallel channel design (Fig. 1A) used in this study was fabricated as previously described (De La Fuente et al., 2007). The major portion of MC consisted of a moulded polydimethylsiloxane (PDMS) body with two parallel microchannels on a surface that was sealed by a cover glass (Fig. 1A). The dimensions of these microchannels were 80 µm wide, 50 µm deep and 3.7 cm long. For each microchannel, there are two inlets for introduction of liquid medium and bacteria suspension, and an outlet for collection of fluid flow.
The Temecula1 suspension was used to fill a pair of 1 ml plastic syringes (Becton Dickinson & Company, Franklin Lakes, NJ, USA) that were, respectively, connected to the bacterial inlet of each microchannel (Fig. 1A). Another pair of 5 ml glass syringes (Hamilton Company, Reno, NV) was filled with PD2 liquid medium (‘PD2’ treatment) or 2Ca (‘2Ca’ treatment) and connected to the media inlet of each microchannel (Fig. 1B). Media and Temecula1 suspension were injected into microchannels by two programmable dual channel syringe pumps (Pico Plus; Harvard Apparatus, Holliston, MA, USA). Fluid flow was collected in two sealed containers connected to the outlets (Fig. 1B). To allow Temecula1 cells to attach to the inside of the microchannels, the flow rates of media and suspension were 1 µl min‐1 and 0.6 µl min‐1 respectively. After 1 h, cell suspension injection was stopped, and the flow rate of media was adjusted to 0.25 µl min‐1, which is close to average xylem sap flow rate inside grapevines (Andersen and Brodbeck, 1989; Greenspan et al., 1996). MC was monitored under a Nikon Eclipse Ti inverted microscope (Nikon, Melville, NY, USA), using Nomarski differential interference contrast optics and phase contrast. Time‐lapse images were acquired every 30 s, using a Nikon DS‐Q1 digital camera (Nikon) controlled by NIS‐Elements Advanced Research 3.01 (Nikon). After 6 days, in order to harvest X. fastidiosa cells inside microchannels, 1.5 ml Eppendorf tubes replaced the collection containers. Media was changed to DNA/RNA ShieldTM (Zymo Research, Irvine, CA, USA), used to preserve nucleic acids and help remove X. fastidiosa cells attached to surfaces inside the MC. Flow rate during cell removal using DNA/RNA ShieldTM was adjusted to 20 µl min‐1. Finally, each X. fastidiosa sample in 500 µl DNA/RNA Shield was stored at −80°C. Samples from three independent MC experiments were used for sequencing analysis.
Library construction and sequencing analysis
RNA preparation, cDNA library construction and sequencing were performed as previously described (Parker et al., 2016). Briefly, total RNA was extracted and purified using Quick‐RNATM MiniPrep and RNA Clean and ConcentrationTM (Zymo Research). Purity, concentration and integrity of RNA samples were measured by using a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA samples with a RNA:DNA ratio > 15 and RNA integrity number (RIN) > 7 were treated with Ribo‐ZeroTM rRNA Removal Kit (Gram‐Negative Bacteria) (Epicentre, Madison, WI, USA) to deplete ribosomal RNA. Libraries were prepared with the TruSeq Stranded mRNA Sample Prep Kit (Illumina, Inc., San Diego, CA, USA), and paired‐end sequencing was performed on an Illumina HiSeqTM 1500 system at the Auburn University Genomics and Sequencing Laboratory.
RNA‐Seq data processing and analysis
Adapter and quality trimming of the raw sequencing data was conducted using trimmomatic version 0.33 (Bolger et al., 2014). Reads aligning to Ribosomal RNA were filtered using geneious version 10.0 (Biomatters Ltd., Newark, NJ, USA) to reduce background noise. Gene expression analysis was conducted as previously described using rockhopper version 2.03 (McClure et al., 2013; Parker et al., 2016). In this study, q‐value < 0.05 and fold change (FC) ≥ 1.5 were considered as the threshold values for selecting DEGs. GO annotation of DEGs was performed using blast2go 5.0. DEGs were searched against the Kyoto Encyclopedia of Gene and Genomes (KEGG) database using KEGG mapper.
Quantitative PCR (qPCR)
The remaining total RNA from the three MC experiments was adjusted to 1 ng µl‐1 and reverse‐transcribed to cDNA for qPCR analysis using the qScriptTM cDNA Supermix (Quanta Biosciences, Beverly, MA, USA). TaqMan qPCR primer and probe (labelled with 5’ 6‐carboxyfluorescein [FAM] and 3’ Black Hole 588 Quencher‐1 [BHQ‐1]) sets were used from previous publications or designed using PrimerQuest (Integrated DNA Technologies, Coralville, IA, USA). The primer/probe sets targeted 7 genes (2 upregulated, 3 downregulated, 1 no differentially expressed and 1 internal control; Table S2). qPCR was conducted on a CFX96TM Real‐Time System (Bio‐Rad Laboratories, Hercules, CA, USA) in 20 µl reactions contains: 1 µl cDNA, 1 × PerfeCTa MultiPlex qPCR SupperMix Low‐ROX (Quanta Biosciences), 0.4 µM forward and reverse primers and 0.2 µM TaqMan probe. The following cycling parameters were used 95°C for 1 min, followed by 40 cycles of 95°C for 15 sand 60°C for 1 min. Fold change in expression of target genes was calculated by the 2−∆∆ C T method (Livak and Schmittgen, 2001). Transcripts of the nuoA gene were used as internal control to perform normalization (Cruz et al., 2014).
Effect of calcium on natural competence of X . fastidiosa
Plasmids pAX1.Km and pAX1.Cm from a previous study (Matsumoto et al., 2009) were used as donor DNA for natural transformation of X. fastidiosa Temecula1. Experiments were performed under two growth conditions: solid agar plates (plates) and MC. PD3 medium (PD3) was used as control, and PD3 supplemented with 2 mM CaCl2 (2Ca) was used as treatment. Natural competence assays under different growth conditions were conducted as previously described (Kandel et al., 2016). Briefly, for agar plate conditions 10 µl of X. fastidiosa Temecula1 cell suspension at OD600 of 0.25 was spotted on top of PD3 or 2Ca plates, and 1 µg of plasmid (pAX1.Km/ pAX1.Cm) in a 10 µl volume was added to the spots. Following incubation at 28°C for ~ 3 days, spots were suspended in 500 µl PD3 and serial dilutions were plated on PW plates with or without the respective antibiotics: 30 µg ml‐1 kanamycin (Km) and 10 µg ml‐1 chloramphenicol (Cm). After 10–14 days of incubation at 28°C, CFUs on all plates were enumerated for recombinants and total viable cells. Recombination frequency was calculated as a ratio of the number of recombinants to total viable cells (recombinant/total cells) (Kandel et al., 2016). MC recombination experiments were performed as described before (Kandel et al., 2016). PD3/2Ca media with 1 µg ml‐1 pAX1.Cm plasmid flowed through the microchannel with a rate of 0.25 µl ml‐1. Cell suspensions of OD600 of 0.25 were inoculated into the microchannel. After 7 days, the fraction of cells collected in the outlet containers (MC_out, Fig. 1B) was harvested; then, the fraction inside the microchannel (MC_in, Fig. 1B) was detached and harvested. Serial dilution, plating, CFU counts and recombination frequency calculations were performed following standard procedures (Kandel et al., 2016). For agar plates condition, three independent experiments were performed, and three replications were included in each experiment (n = 9). For MCs condition, two independent experiments were performed with six replicates in total (n = 6). Recombination frequency data were analysed by Students’ t‐test (P < 0.05) using r Project 3.4.3 for Windows.
Conflict of interest
None declared.
Supporting information
Fig. S1. Twitching speed assessments of X. fastidiosa Temecula1 in microfluidic chambers comparing PD2 and 2Ca treatments. Different letters on top of the bars indicate significant differences (P < 0.05) according to Student’s t test. Data presented was obtained from six independent experiments. Error bars correspond to SE of the mean (n = 18).
Fig. S2. Gene Ontology classification of the differentially expressed genes in X. fastidiosa microfluidic transcriptome study. Gene Ontology (GO) term assignment to the 122 DEGs in this study were summarized into three main GO categories (biological process, cellular component, molecular function) and 28 sub‐categories (Level 2).
Fig. S3. RNA‐Seq confirmation using qPCR. Log2 fold change of qPCR data is plotted against that of RNA‐Seq data for 6 genes. Equation of the line and correlation coefficients are presented.
Video S1. Harvest of X. fastidiosa Temecula1 cells in microfluidic chambers. DNA/RNA shield buffer constantly flows from left to right in both channels. All Temecula1 cells grown in the channel were removed quickly.
Video S2. X. fastidiosa Temecula1 cells grown in microfluidic chambers. PD2 and 2Ca media constantly flows from left to right in both channels.
Data Set S1. RNA‐Seq read and assembly statistics for the 6 X. fastidiosa samples.
Data Set S2. RNA‐Seq data analysis for all X. fastidiosa protein‐coding genes. Includes transcript boundaries, normalized expression levels and statistical significance of differential gene expression for 2Ca and PD2 treatments.
Data Set S3. Functional categorization of X. fastidiosa protein‐coding genes differentially expressed in 2Ca/PD2. The fold changes are shown for these genes.
Data Set S4. Mapping of X. fastidiosa protein‐coding genes differentially expressed in 2Ca/PD2 to the KEGG pathways.
Table S1. Summary of purified total RNA samples.
Table S2. Primers and probesa used in qPCR analysis.
Acknowledgements
We would like to thank Lee Zhang at Auburn University Genomics and Sequencing Laboratory for sequencing the RNA‐Seq samples. We also appreciate comments from Jennifer Parker, Jeffrey Coleman, Tonia Schwartz, Neha Potnis and Cova Arias that helped our data analysis. This work was funded by NIFA (National Institute of Food and Agriculture) grant 2015‐67014‐23085, CDFA (California Department of Food and Agriculture) and the Alabama Agricultural Experiment Station (AAES‐HATCH).
Microbial Biotechnology (2020) 13(2), 548–561
Funding information
This work was funded by NIFA (National Institute of Food and Agriculture) grant 2015‐67014‐23085, CDFA (California Department of Food and Agriculture) and the Alabama Agricultural Experiment Station (AAES‐HATCH).
References
- Ailloud, F. , Lowe, T.M. , Robène, I. , Cruveiller, S. , Allen, C. , and Prior, P. (2016) In planta comparative transcriptomics of host‐adapted strains of Ralstonia solanacearum . PeerJ 4: e1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida, R.P. , and Nunney, L. (2015) How do plant diseases caused by Xylella fastidiosa emerge? Plant Dis 99: 1457–1467. [DOI] [PubMed] [Google Scholar]
- Andersen, P.C. , and Brodbeck, B.V. (1989) Diurnal and temporal changes in the chemical profile of xylem exudate from Vitis rotundifolia . Physiol Plant 75: 63–70. [Google Scholar]
- Araújo, W.L. , Marcon, J. , Maccheroni, W. , van Elsas, J. D. , van Vuurde, J.W. , and Azevedo, J.L. (2002) Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl Environ Microbiol 68: 4906–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar, O. , De La Fuente, L. , and Burdman, S. (2010) Assessing adhesion, biofilm formation and motility of Acidovorax citrulli using microfluidic flow chambers. FEMS Microbiol Lett 312: 33–39. [DOI] [PubMed] [Google Scholar]
- Baur, B. , Hanselmann, K. , Schlimme, W. , and Jenni, B. (1996) Genetic transformation in freshwater: Escherichia coli is able to develop natural competence. Appl Environ Microbiol 62: 3673–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand, J.J. , West, J.T. , and Engel, J.N. (2010) Genetic analysis of the regulation of type IV pilus function by the Chp chemosensory system of Pseudomonas aeruginosa . J Bacteriol 192: 994–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A.M. , Lohse, M. , and Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder, U.N. , Jaeger, T. , and Jenal, U. (2017) LadS is a calcium‐responsive kinase that induces acute‐to‐chronic virulence switch in Pseudomonas aeruginosa . Nat Microbiol 2: 16184. [DOI] [PubMed] [Google Scholar]
- Castiblanco, L.F. , and Sundin, G.W. (2015) New insights on molecular regulation of biofilm formation in plant‐associated bacteria. J Integr Plant Biol 58: 362–372. [DOI] [PubMed] [Google Scholar]
- Chatterjee, S. , Almeida, R.P. , and Lindow, S. (2008) Living in two worlds: the plant and insect lifestyles of Xylella fastidiosa . Annu Rev Phytopathol 46: 243–271. [DOI] [PubMed] [Google Scholar]
- Chen, H. , Kandel, P.P. , Cruz, L.F. , Cobine, P.A. , and De La Fuente, L. (2017) The major outer membrane protein MopB is required for twitching movement and affects biofilm formation and virulence in two Xylella fastidiosa strains. Mol Plant‐Microbe Interact 30: 896–905. [DOI] [PubMed] [Google Scholar]
- Ciraulo, M.B. , Santos, D.S. , Rodrigues, A.C.d.F.O. , Oliveira, M.V.d. , Rodrigues, T. , Oliveira, R.C.d. , and Nunes, L.R. (2010) Transcriptome analysis of the phytobacterium Xylella fastidiosa growing under xylem‐based chemical conditions. J BioMed Res 2010: 18 10.1155/2010/781365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobine, P.A. , Cruz, L.F. , Navarrete, F. , Duncan, D. , Tygart, M. , and De La Fuente, L. (2013) Xylella fastidiosa differentially accumulates mineral elements in biofilm and planktonic cells. PLoS ONE 8: e54936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz, L.F. , Cobine, P.A. , and De La Fuente, L. (2012) Calcium increases Xylella fastidiosa surface attachment, biofilm formation, and twitching motility. Appl Environ Microbiol 78: 1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz, L.F. , Parker, J.K. , Cobine, P.A. , and De La Fuente, L. (2014) Calcium‐enhanced twitching motility in Xylella fastidiosa is linked to a single PilY1 homolog. Appl Environ Microbiol 80: 7176–7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cursino, L. , Galvani, C.D. , Athinuwat, D. , Zaini, P.A. , Li, Y. , De La Fuente, L. , et al (2011) Identification of an operon, Pil‐Chp, that controls twitching motility and virulence in Xylella fastidiosa . Mol Plant‐Microbe Interact 24: 1198–1206. [DOI] [PubMed] [Google Scholar]
- Dasgupta, N. , Ashare, A. , Hunninghake, G.W. , and Yahr, T.L. (2006) Transcriptional induction of the Pseudomonas aeruginosa type III secretion system by low Ca2+ and host cell contact proceeds through two distinct signaling pathways. Infect Immun 74: 3334–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, M. , Purcell, A. , and Thomson, S. (1980) Isolation media for the Pierce's disease bacterium. Phytopathology 70: 425–429. [Google Scholar]
- Davis, M. , Whitcomb, R. , and Gillaspie, A. Jr . (1981a) Fastidious bacteria of plant vascular tissue and invertebrates (including so called rickettsia‐like bacteria) In The prokaryotes: a handbook on habitats, isolation, and identification of bacteria. Starr M. P., Stolp H. G., Trüper H. G., Balows A., and Schlegel H. G. (eds). New York: Springer‐Verlag, pp. 2172–2188. [Google Scholar]
- Davis, M.J. , French, W.J. , and Schaad, N.W. (1981b) Axenic culture of the bacteria associated with phony disease of peach and plum leaf scald. Curr Microbiol 6: 309–314. [Google Scholar]
- De La Fuente, L. , Montanes, E. , Meng, Y. , Li, Y. , Burr, T. J. , Hoch, H. , and Wu, M. (2007) Assessing adhesion forces of type I and type IV pili of Xylella fastidiosa bacteria by use of a microfluidic flow chamber. Appl Environ Microbiol 73: 2690–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fuente, L. , Navarrete, F. , Oliver, J.E. , Cruz, L. , and Cobine, P.A. (2014) The influence of metal elements on virulence in plant pathogenic bacteria In Virulence mechanisms of plant pathogenic bacteria. [Google Scholar]
- DeBord, K.L. , Galanopoulos, N.S. , and Schneewind, O. (2003) The ttsA gene is required for low‐calcium‐induced type III secretion of Yop proteins and virulence of Yersinia enterocolitica W22703. J Bacteriol 185: 3499–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarty, M. , Morvan, C. , and Thellier, M. (1984) Calcium and the cell wall. Plant Cell Environ 7: 441–448. [Google Scholar]
- Dodd, A.N. , Kudla, J. , and Sanders, D. (2010) The language of calcium signaling. Annu Rev Plant Biol 61: 593–620. [DOI] [PubMed] [Google Scholar]
- Dominguez, D.C. (2004) Calcium signalling in bacteria. Mol Microbiol 54: 291–297. [DOI] [PubMed] [Google Scholar]
- Dominguez, D.C. , Guragain, M. , and Patrauchan, M. (2015) Calcium binding proteins and calcium signaling in prokaryotes. Cell Calcium 57: 151–165. [DOI] [PubMed] [Google Scholar]
- Gandi, S.K. , Watson, D. , Kersaudy‐Kerhoas, M. , Desmulliez, M.P. , Bachmann, T. , and Bridle, H. (2015) Impact of microfluidic processing on bacterial ribonucleic acid expression. Biomicrofluidics 9: 031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gode‐Potratz, C.J. , Chodur, D.M. , and McCarter, L.L. (2010) Calcium and iron regulate swarming and type III secretion in Vibrio parahaemolyticus . J Bacteriol 192: 6025–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosens, V.J. , Busch, A. , Georgiadou, M. , Castagnini, M. , Forest, K.T. , Waksman, G. , and Pelicic, V. (2017) Reconstitution of a minimal machinery capable of assembling periplasmic type IV pili. Proc Natl Acad Sci USA 114: E4978–E4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan, M.D. , Schultz, H.R. , and Matthews, M.A. (1996) Field evaluation of water transport in grape berries during water deficits. Physiol Plant 97: 55–62. [Google Scholar]
- Guragain, M. , Lenaburg, D.L. , Moore, F.S. , Reutlinger, I. , and Patrauchan, M.A. (2013) Calcium homeostasis in Pseudomonas aeruginosa requires multiple transporters and modulates swarming motility. Cell Calcium 54: 350–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeb, S. , and Haas, D. (2001) Regulatory roles of the GacS/GacA two‐component system in plant‐associated and other gram‐negative bacteria. Mol Plant‐Microbe Interact 14: 1351–1363. [DOI] [PubMed] [Google Scholar]
- Hill, B. , and Purcell, A. (1995) Multiplication and movement of Xylella fastidiosa within grapevine and four other plants. Phytopathology 85: 1368–1372. [Google Scholar]
- Hisano, K. , Fujise, O. , Miura, M. , Hamachi, T. , Matsuzaki, E. , and Nishimura, F. (2014) The pga gene cluster in Aggregatibacter actinomycetemcomitans is necessary for the development of natural competence in Ca2+‐promoted biofilms. Mol Oral Microbiol 29: 79–89. [DOI] [PubMed] [Google Scholar]
- Hobbs, M. , Collie, E.S.R. , Free, P.D. , Livingston, S.P. , and Mattick, J.S. (1993) PilS and PilR, a two‐component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa . Mol Microbiol 7: 669–682. [DOI] [PubMed] [Google Scholar]
- Hu, Y. , Hu, Q. , Wei, R. , Li, R. , Zhao, D. , Ge, M. , et al (2018) The XRE family transcriptional regulator SrtR in Streptococcus suis is involved in oxidant resistance and virulence. Front Cell Infect Mi 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, J.M. , Babujee, L. , Meng, F. , Milling, A. , and Allen, C. (2012) The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato. MBio 3: e00114–00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, S. , Ishimoto, K.S. , and Lory, S. (1994) PilR, a transcriptional regulator of piliation in Pseudomonas aeruginosa, binds to a cis‐acting sequence upstream of the pilin gene promoter. Mol Microbiol 14: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Kandel, P.P. , Lopez, S.M. , Almeida, R.P. , and De La Fuente, L. (2016) Natural competence of Xylella fastidiosa occurs at a high frequency inside microfluidic chambers mimicking the bacterium's natural habitats. Appl Environ Microbiol 82: 5269–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel, P.P. , Almeida, R.P.P. , Cobine, P.A. , and De La Fuente, L. (2017) Natural competence rates are variable among Xylella fastidiosa strains and homologous recombination occurs in vitro between subspecies fastidiosa and multiplex . Mol Plant‐Microbe Interact 30: 589–600. [DOI] [PubMed] [Google Scholar]
- Kandel, P.P. , Chen, H. , and De La Fuente, L. (2018) A short protocol for gene knockout and complementation in Xylella fastidiosa shows that one of the type IV pilin paralogs (PD1926) is needed for twitching while another (PD1924) affects pilus number and location. Appl Environ Microbiol 84: e01167–01118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmury, S.L. , and Burrows, L.L. (2016) Type IV pilins regulate their own expression via direct intramembrane interactions with the sensor kinase PilS. Proc Natl Acad Sci 113: 6017–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung, S.H. , and Almeida, R.P. (2011) Natural competence and recombination in the plant pathogen Xylella fastidiosa . Appl Environ Microbiol 77: 5278–5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.E. , Gupta, R. , Jayaramaiah, R.H. , Lee, S.H. , Wang, Y. , Park, S.‐R. , and Kim, S.T. (2017) Global transcriptome profiling of Xanthomonas oryzae pv. oryzae under in planta growth and in vitro culture conditions. Plant Pathol J 33: 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Hao, G. , Galvani, C.D. , Meng, Y. , De La Fuente, L. , Hoch, H. , and Burr, T.J. (2007) Type I and type IV pili of Xylella fastidiosa affect twitching motility, biofilm formation and cell–cell aggregation. Microbiology 153: 719–726. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. , and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lorenz, M.G. , and Wackernagel, W. (1994) Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Mol Biol Rev 58: 563–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques, L. , Ceri, H. , Manfio, G. , Reid, D. , and Olson, M. (2002) Characterization of biofilm formation by Xylella fastidiosa in vitro. Plant Dis 86: 633–638. [DOI] [PubMed] [Google Scholar]
- Matsumoto, A. , Young, G.M. , and Igo, M.M. (2009) Chromosome‐based genetic complementation system for Xylella fastidiosa . Appl Environ Microbiol 75: 1679–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick, J.S. (2002) Type IV pili and twitching motility. Annu Rev Microbiol 56: 289–314. [DOI] [PubMed] [Google Scholar]
- McAuliffe, O. , O'Keeffe, T. , Hill, C. , and Ross, R.P., (2001) Regulation of immunity to the two‐component lantibiotic, lacticin 3147, by the transcriptional repressor LtnR. Mol Microbiol 39: 982–993. [DOI] [PubMed] [Google Scholar]
- McClure, R. , Balasubramanian, D. , Sun, Y. , Bobrovskyy, M. , Sumby, P. , Genco, C.A. , et al (2013) Computational analysis of bacterial RNA‐Seq data. Nucleic Acids Res 41: e140–e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell, G.E. , and McConnell, D.J. (1994) Overproduction, isolation, and DNA‐binding characteristics of Xre, the repressor protein from the Bacillus subtilis defective prophage PBSX. J Bacteriol 176: 5831–5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Y. , Li, Y. , Galvani, C.D. , Hao, G. , Turner, J.N. , Burr, T.J. , and Hoch, H. (2005) Upstream migration of Xylella fastidiosa via pilus‐driven twitching motility. J Bacteriol 187: 5560–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarrete, F. , and De La Fuente, L. (2014) Response of Xylella fastidiosa to zinc: decreased culturability, increased exopolysaccharide production, and formation of resilient biofilms under flow conditions. Appl Environ Microbiol 80: 1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols, M. , Elustondo, P.A. , Warford, J. , Thirumaran, A. , Pavlov, E.V. , and Robertson, G.S. (2017) Global ablation of the mitochondrial calcium uniporter increases glycolysis in cortical neurons subjected to energetic stressors. J Cerebr Blood F Met 37: 3027–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobori, T. , Velásquez, A.C. , Wu, J. , Kvitko, B.H. , Kremer, J.M. , Wang, Y. , et al (2018) Transcriptome landscape of a bacterial pathogen under plant immunity. Proc Natl Acad Sci USA 115: E3055–E3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oomes, S. , Jonker, M. , Wittink, F. , Hehenkamp, J. , Breit, T. , and Brul, S. (2009) The effect of calcium on the transcriptome of sporulating B. subtilis cells. Int J Food Microbiol 133: 234–242. [DOI] [PubMed] [Google Scholar]
- Ørtenblad, N. , Macdonald, W.A. , and Sahlin, K. (2009) Glycolysis in contracting rat skeletal muscle is controlled by factors related to energy state. Biochem J 420: 161–168. [DOI] [PubMed] [Google Scholar]
- Page, W.J. , and Doran, J.L. (1981) Recovery of competence in calcium‐limited Azotobacter vinelandii . J Bacteriol 146: 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamp, S.J. , Harrington, E.D. , Quake, S.R. , Relman, D.A. , and Blainey, P.C. (2012) Single‐cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB). Genome Res 22: 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.K. , Chen, H. , McCarty, S.E. , Liu, L.Y. , and De La Fuente, L. (2016) Calcium transcriptionally regulates the biofilm machinery of Xylella fastidiosa to promote continued biofilm development in batch cultures. Environ Microbiol 18: 1620–1634. [DOI] [PubMed] [Google Scholar]
- Pashalidis, S. , Moreira, L.M. , Zaini, P.A. , Campanharo, J.C. , Alves, L.M. , Ciapina, L.P. , et al (2005) Whole‐genome expression profiling of Xylella fastidiosa in response to growth on glucose. OMICS 9: 77–90. [DOI] [PubMed] [Google Scholar]
- Potnis, N. , Kandel, P.P. , Merfa, M.V. , Retchless, A.C. , Parker, J.K. , Stenger, D.C. , et al (2019) Patterns of inter‐ and intra‐subspecific homologous recombination inform eco‐evolutionary dynamics of Xylella fastidiosa . ISME J 13: 2319–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaudi, L. , Fujishige, N.A. , Hirsch, A.M. , Banchio, E. , Zorreguieta, A. , and Giordano, W. (2006) Effects of nutritional and environmental conditions on Sinorhizobium meliloti biofilm formation. Res Microbiol 157: 867–875. [DOI] [PubMed] [Google Scholar]
- Sarkisova, S. , Patrauchan, M.A. , Berglund, D. , Nivens, D.E. , and Franklin, M.J. (2005) Calcium‐induced virulence factors associated with the extracellular matrix of mucoid Pseudomonas aeruginosa biofilms. J Bacteriol 187: 4327–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönekess, B. , Brindley, P.G. , and Lopaschuk, G.O. (1995) Calcium regulation of glycolysis, glucose oxidation and fatty acid oxidation in the aerobic and ischemic heart. Can J Physiol Pharmacol 73: 1632–1640. [DOI] [PubMed] [Google Scholar]
- Shi, X.Y. , Dumenyo, C.K. , Hernandez‐Martinez, R. , Azad, H. , and Cooksey, D.A. (2009) Characterization of regulatory pathways in Xylella fastidiosa: genes and phenotypes controlled by gacA . Appl Environ Microbiol 75: 2275–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard, A. , Zeilinger, A.R. , Vanhove, M. , Schartel, T.E. , Beal, D.J. , Daugherty, M.P. , and Almeida, R.P. (2018) Xylella fastidiosa: Insights into an emerging plant pathogen. Annu Rev Phytopathol 56: 181–202. [DOI] [PubMed] [Google Scholar]
- Singh, B. , Al Jubair, T. , Mörgelin, M. , Sundin, A. , Linse, S. , Nilsson, U.J. , and Riesbeck, K. (2015) Haemophilus influenzae surface fibril (Hsf) is a unique twisted hairpin‐like trimeric autotransporter. Int J Med Microbiol 305: 27–37. [DOI] [PubMed] [Google Scholar]
- de Souza, A.A. , Takita, M.A. , Coletta‐Filho, H.D. , Caldana, C. , Goldman, G.H. , Yanai, G.M. , et al (2003) Analysis of gene expression in two growth states of Xylella fastidiosa and its relationship with pathogenicity. Mol Plant‐Microbe Interact 16: 867–875. [DOI] [PubMed] [Google Scholar]
- Streets, A.M. , Zhang, X. , Cao, C. , Pang, Y. , Wu, X. , Xiong, L. , et al (2014) Microfluidic single‐cell whole‐transcriptome sequencing. Proc Natl Acad Sci USA 111: 7048–7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, F. , Barbacioru, C. , Wang, Y. , Nordman, E. , Lee, C. , Xu, N. , et al (2009) mRNA‐Seq whole‐transcriptome analysis of a single cell. Nat Methods 6: 377. [DOI] [PubMed] [Google Scholar]
- Traglia, G.M. , Quinn, B. , Schramm, S.T. , Soler‐Bistue, A. , and Ramirez, M.S. (2016) Serum albumin and Ca2+ are natural competence inducers in the human pathogen Acinetobacter baumannii . Antimicrob Agents Chemother 60: 4920–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombe, M.‐C. (1993) Characterization of a calcium porter of Streptococcus pneumoniae involved in calcium regulation of growth and competence. Microbiology 139: 433–439. [DOI] [PubMed] [Google Scholar]
- Wang, S.‐L. , Fan, K.‐Q. , Yang, X. , Lin, Z.‐X. , Xu, X.‐P. , and Yang, K.‐Q. (2008) CabC, an EF‐hand calcium‐binding protein, is involved in Ca2+‐mediated regulation of spore germination and aerial hypha formation in Streptomyces coelicolor . J Bacteriol 190: 4061–4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N. , Li, J.‐L. , and Lindow, S.E. (2012) RpfF‐dependent regulon of Xylella fastidiosa . Phytopathology 102: 1045–1053. [DOI] [PubMed] [Google Scholar]
- Waters, V.L. , and Crosa, J.H. (1991) Colicin V virulence plasmids. Microbiol Mol Biol Rev 55: 437–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werthén, M. , and Lundgren, T. (2001) Intracellular Ca2+ mobilization and kinase activity during acylated homoserine lactone‐dependent quorum sensing in Serratia liquefaciens . J Biol Chem 276: 6468–6472. [DOI] [PubMed] [Google Scholar]
- Wood, H.E. , Devine, K.M. , and McConnell, D.J. (1990) Characterisation of a repressor gene (xre) and a temperature‐sensitive allele from the Bacillus subtilis prophage, PBSX. Gene 96: 83–88. [DOI] [PubMed] [Google Scholar]
- Yadeta, K. , and Thomma, B. (2013) The xylem as battleground for plant hosts and vascular wilt pathogens. Front Plant Sci 4: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaini, P.A. , Fogaça, A.C. , Lupo, F.G. , Nakaya, H.I. , Vêncio, R.Z. , and da Silva, A.M. (2008) The iron stimulon of Xylella fastidiosa includes genes for type IV pilus and colicin V‐like bacteriocins. J Bacteriol 190: 2368–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. , Flores‐Cruz, Z. , Kumar, D. , Chakrabarty, P. , Hopkins, D.L. , and Gabriel, D.W. (2011) The Xylella fastidiosa biocontrol strain EB92‐1 genome is very similar and syntenic to Pierce's disease strains. J Bacteriol 193: 5576–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Ma, N. , Jin, W. , Wu, S. , and Sun, C. (2017) Genomic and transcriptomic insights into calcium carbonate biomineralization by marine actinobacterium Brevibacterium linens BS258. Front Microbiol 8: 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Twitching speed assessments of X. fastidiosa Temecula1 in microfluidic chambers comparing PD2 and 2Ca treatments. Different letters on top of the bars indicate significant differences (P < 0.05) according to Student’s t test. Data presented was obtained from six independent experiments. Error bars correspond to SE of the mean (n = 18).
Fig. S2. Gene Ontology classification of the differentially expressed genes in X. fastidiosa microfluidic transcriptome study. Gene Ontology (GO) term assignment to the 122 DEGs in this study were summarized into three main GO categories (biological process, cellular component, molecular function) and 28 sub‐categories (Level 2).
Fig. S3. RNA‐Seq confirmation using qPCR. Log2 fold change of qPCR data is plotted against that of RNA‐Seq data for 6 genes. Equation of the line and correlation coefficients are presented.
Video S1. Harvest of X. fastidiosa Temecula1 cells in microfluidic chambers. DNA/RNA shield buffer constantly flows from left to right in both channels. All Temecula1 cells grown in the channel were removed quickly.
Video S2. X. fastidiosa Temecula1 cells grown in microfluidic chambers. PD2 and 2Ca media constantly flows from left to right in both channels.
Data Set S1. RNA‐Seq read and assembly statistics for the 6 X. fastidiosa samples.
Data Set S2. RNA‐Seq data analysis for all X. fastidiosa protein‐coding genes. Includes transcript boundaries, normalized expression levels and statistical significance of differential gene expression for 2Ca and PD2 treatments.
Data Set S3. Functional categorization of X. fastidiosa protein‐coding genes differentially expressed in 2Ca/PD2. The fold changes are shown for these genes.
Data Set S4. Mapping of X. fastidiosa protein‐coding genes differentially expressed in 2Ca/PD2 to the KEGG pathways.
Table S1. Summary of purified total RNA samples.
Table S2. Primers and probesa used in qPCR analysis.


