Summary
Its features as a microbial and eukaryotic organism have turned Komagataella phaffii (Pichia pastoris) into an emerging cell factory for recombinant protein production (RPP). As a key step of the bioprocess development, this work aimed to demonstrate the importance of tailor designing the cultivation strategy according to the production kinetics of the cell factory. For this purpose, K. phaffii clones constitutively expressing (P GAP) Candida rugosa lipase 1 (Crl1) with different gene dosage were used as models in continuous and fed‐batch cultures. Production parameters were much greater with a multicopy clone (MCC) than with the single‐copy clone (SCC). Regarding production kinetics, the specific product generation rate (q P ) increased linearly with increasing specific growth rate (µ) in SCC; by contrast, q P exhibited saturation in MCC. A transcriptional analysis in chemostat cultures suggested the presence of eventual post‐transcriptional bottlenecks in MCC. After the strain characterization, in order to fulfil overall development of the bioprocess, the performance of both clones was also evaluated in fed‐batch mode. Strikingly, different optimal strategies were determined for both models due to the different production kinetic patterns observed as a trade‐off for product titre, yields and productivity. The combined effect of gene dosage and adequate µ enables rational process development with a view to optimize K. phaffii RPP bioprocesses.
The combined effect of strain and bioprocess engineering should be considered during a rational process development towards bioprocess optimization. The synergy between gene dosage and specific growth rate have been evaluated for the Komagataella phaffii (Pichia pastoris) recombinant protein production (RPP) of an industrial lipase in chemostat and fed‐batch cultures. Different optimal strategies were designed from a trade‐off for product titer, yields and productivity for two clone models with different gene dosage and production kinetics, from which further transcription analysis supported potential bottlenecks after transcription.
Introduction
Komagataella phaffii, formerly known as Pichia pastoris, is a widely used yeast for recombinant protein production (RPP), for both biopharmaceuticals and an increasing number of industrial enzymes of interest (Puxbaum et al., 2015; Burgard et al., 2017). This host has major advantages such as a wide range of genetic modification tools including genome editing toolkits are available (e.g. the CRISPR/Cas9 system); its ability to grow to a high cell density in defined media, to perform eukaryotic post‐translational modifications and to release target products extracellularly. These features in combination make K. phaffii a promising cell factory for industrial biotechnology (Potvin et al., 2012; Vogl and Glieder, 2013; Weninger et al., 2018).
The increasing demand for recombinant proteins has generated a multibillion‐dollar market over the last few decades (Highsmith, 2015; Dewan, 2017). Therefore, important efforts are being dedicated to increase bioprocess efficiency and profitability. Two widely reviewed complementary approaches are currently being developed to reach these goals, namely strain engineering (Zahrl et al., 2017; Juturu and Wu, 2018; Vogl et al., 2018a) and bioprocess optimization (Theron et al., 2018; Yang and Zhang, 2018).
Using efficient promoters is essential to ensure efficient recombinant protein expression in this context. The methanol‐inducible alcohol oxidase promoter (PAOX1) has been widely used in K. phaffii bioprocesses by virtue of allowing a strong and tight regulation for the recombinant expression in the presence of methanol (Barrigón et al., 2013; Ponte et al., 2016; Vogl et al., 2016). However, PAOX1‐driven expression bioprocesses are subject to constraints derived from the use of methanol as inducer. Thus, using methanol at the industrial scale requires adopting safety measures that raise production costs and is subject to operational problems arising from heavy high heat production and oxygen demand (Prielhofer et al., 2013), cell metabolic burdens (Hartner and Glieder, 2006), culture cell lysis and potential subsequent proteolysis of the target product (Mattanovich et al., 2009).
Alternative promoters avoiding the use of methanol have recently been explored (Liang et al., 2013; Prielhofer et al., 2013; Shen et al., 2016; Vogl et al., 2018b; Robert et al., 2019). The glyceraldehyde‐3‐phosphate dehydrogenase GAP promoter (PGAP), which is involved in a key step of the glycolysis pathway, was the first to emerge as a benchmark for efficient protein expression on various carbon sources. Thus, by avoiding all methanol‐related drawbacks, PGAP‐based bioprocesses present relevant advantages for large‐scale production (Zhang et al., 2009; Ahmad et al., 2014; Çalık et al., 2015).
Some authors have found copy number integration of the expression cassette in the genome, also called gene dosage, to play a central role in specific productivity (Schwarzhans et al., 2016a; Vogl et al., 2018a). Using large numbers of gene copies results in increased productivity in some cases (Nordén et al., 2011; Prielhofer et al., 2013; Zhu et al., 2014) but has the opposite effect in others (Zhu et al., 2009; Liu et al., 2014; Cámara et al., 2016).In fact, as claimed, integrating several expression cassettes in the genome may have adverse effects owing to the physiological limitations in the transcriptional capacity of gene AOX1, which is governed by its transcriptional factors (Cámara et al., 2017).
Production kinetics, the relationship between specific production rate (q P) and specific growth rate (μ), is considered a key factor to be considered in the bioprocess development. It reflects the equilibrium between the various steps until the product is secreted, as a balance of the different processes involved during the protein synthesis, folding and secretion. This relationship is crucial to bioprocess development and optimization (Potvin et al., 2012; Looser et al., 2015; Çalik et al., 2015). Thus, Garcia‐Ortega et al. (2016) and Maurer et al. (2006) characterized PGAP‐based strains producing an antibody fragment and obtained robust results with them in chemostat systems; so, they found q P to increase eight times with increasing μ. Rebnegger et al. (2014) examined the response of this expression system producing human serum albumin (HSA) at different specific growth rates at transcriptomic level and observed marked upregulation of genes involved in translation. However, genes of the glycolytic pathways such as TDH3, which is the endogenous gene regulated by PGAP, were unregulated or weakly regulated, therefore suggesting that effect in the translational machinery played a major role in by causing q P to increase with increasing μ. The results obtained with some fed‐batch cultures are also consistent with synergism in these two variables (Zhao et al., 2008; Garcia‐Ortega et al., 2013). Therefore, because gene dosage is expected to affect production rates, one should assume that it may considerably influence production kinetics in assessing its effects.
Candida rugosa lipase (Crl) is one of the most promising lipase enzymes for biocatalytic applications (Ken Ugo et al., 2017). At least seven genes of C. rugosa lipases (CRL1‐CRL7) have been identified; also, all except CRL6 and CRL7 have been identified and sequenced (Ferrer et al., 2001). Crl1, which accounts for about 80% of all lipase present in commercial powders, is the most widely studied (Sánchez et al., 1999) Because of the difficulty involved in isolating the pure isoenzyme from the native microorganism, it has been alternatively obtained from K. phaffii cell factory (Valero, 2018).
For the present work, the isoenzyme Candida rugosa lipase 1 (Crl1) was selected as model protein to elucidate the differences in the rational design of optimal bioprocess strategies for two clones of K. phaffii with different gene dosage, as example of clone variability in terms of protein production. For this purpose, an accurate characterization of physiological parameters and the production kinetics was firstly performed on chemostat cultures. The obtained results allowed the selection of the optimal bioprocess strategy to maximize the RPP, in order to be applied further in fed‐batch cultivations, which is currently considered as the most used operational mode for industrial RPP (García‐Ortega et al., 2019). As a major outcome, this contribution discusses the influence of gene dosage linked to production kinetics on the determination of the optimal operating strategies with a view to maximizing bioprocess production rates and yields.
Results and discussion
Strain construction and gene dosage
In order to get two clones with contrasting production performances, several recombinant clones with different gene dosage of CRL1 were obtained by transforming different amounts of plasmid in which the CRL1 expression cassette is placed under PGAP regulation. According to the transformation method used, these cassettes are expected to be integrated by homologous recombination into the native PGAP locus. However, constructing producer strains from K. phaffii may result in non‐homologous end‐joining recombination and/or multiple insertion of the gene expression cassette, which usually expands the spectrum of clonal variability (Schwarzhans et al., 2016b; Jiao et al., 2018; Vogl et al., 2018a).
Later, transformed clones were screened in order to identify the best producer clone, which was a clone that integrated five copies of the expression cassette. Thus, it was selected for further studies in which it was compared with a clone with a single copy of gene of interest. The determination of gene dosage was performed by ddPCR. This method allows to determine the exact number of expression cassettes that were integrated in multicopy clone (MCC), five copies, and to confirm the presence of only one copy of CRL1 gene in the single‐copy clone (SCC).
Strain characterization in chemostat cultures
Cell growth
The impact of gene dosage on clone production kinetics was assessed with two sets of chemostat cultures grown at different dilution rates (D). The specific growth rates spanned the range of 0.025–0.15 h−1. The carbon and electron balances were verified and closure found to exceed 95% prior to reconciliation. Fig. 1A and B show the variation of the main physiological variables at different D in both strains.
Figure 1.
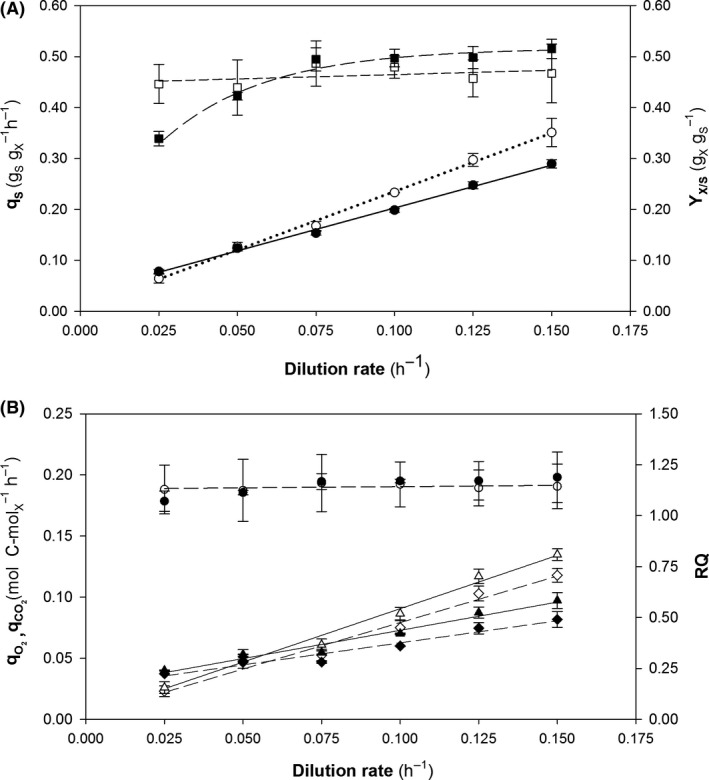
Main physiological parameters for continuous cultures of multicopy (black symbols) and single‐copy clones (white symbols).
A. (●, ○) specific glucose uptake rate (q S); (■, □) overall biomass‐to‐substrate yield Y X/S.
B. (♦, ◊) specific oxygen uptake rate (); (▲, Δ) specific carbon dioxide production rate (); and (●,○) respiratory quotient (RQ). Error bars represent SE of the mean values.
As can be seen in Fig. 1A, the specific substrate uptake rate (q S) increased linearly with increasing μ. Interestingly, there were no significant differences in q S between clones, which suggests that this rate was unaffected by gene dosage. On the other hand, the overall biomass substrate yield (Y X/S) remained fairly constant with a value of about 0.5 gX gS –1, which is consistent with most reported values (Garcia‐Ortega et al., 2013; Çalik et al., 2015; Adelantado et al., 2017). However, MCC exhibited a slight decrease in Y X/S at the lowest μ values (0.05 and 0.025 h‐1) possibly due to requirement of more energy for maintenance (m s) than the single‐copy strain (0.029 vs. 0.0023 gS gX –1 h–1). This factor strongly influenced Y X/S, which is consistent with some previous studies where biomass production decreased with decreasing μ (Rebnegger et al., 2016).
The lower values observed in maintenance coefficient could be expected since K. phaffii has been described as a robust system in terms of present lower maintenance requirements over other alternative platforms such as E. coli (Zhu et al., 2019). However, in this work, a relevant difference on m S has been described between SCC and MCC, specifically the difference is about one order of magnitude (0.0023 and 0.029 respectively). This notable change could be related to the RPP. It exerts a strong effect on metabolic fluxes that often leads to an increase in the maintenance requirements (Carnicer et al., 2012; Moser et al., 2017).
Accordingly, when comparing MCC respect to SCC in relative terms, the cell maintenance requirements consume a higher proportion of the overall energy resources obtained from carbon source uptake.
In Fig. 1B is shown how the specific O2 uptake () and CO2 production () rates increased linearly with μ. Slight differences between both strains were observed at high μ. However, the proportion between these two specific rates are constant, and consequently, the respiratory quotient (RQ) was always about 1.15.
Similarly to q S, and showed also to be strongly coupled with μ, therefore, fit into a linear equation pattern (Herbert, Pirt, Luedeking‐Piret), which describes how is distributed a determined bioprocess parameter (specific rate) for cell growth and maintenance.
Regarding the maintenance coefficient, which is represented by the intercept, for both specific rates of each clone, a rather similar value was obtained. Like q S (Fig. 1A), MCC maintenance coefficient was slightly higher, suggesting that this difference is produced due to the gene dosage effect. Thus, considering that five functional CRL1 copies are integrated on MCC, it exerts a relevant demand of resources in comparison with SCC.
On the other hand, if it is compared and trend for SCC and MCC, both display similar values across the μ until 0.10 h‐1. Nevertheless, at higher μ’s, a relatively slight decrease is detected for MCC in front of SCC. It has to be considered that RPP consumes energetic resources that drain precursors from the central carbon metabolism to sustain the productivity, which probably results in a readjustment of metabolism, being more inefficient (Peña et al., 2018). For the MCC, it can be hypothesized that at low μ, Crl1 synthesis does not produce any significant metabolic readjustment; therefore, not significant changes are observed on gas‐related specific rates. However, at higher μ this readjustment is shown as a reduction in the oxygen consumption and carbon dioxide production rates.
Target protein production
The specific product generation rate (q P) and the overall product‐to‐biomass yield (Y P/X) were evaluated as main key production parameters. Both are shown in Fig. 2.
Figure 2.
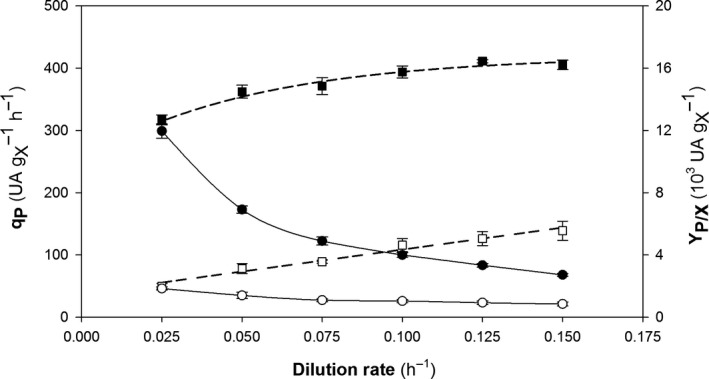
Production parameters for chemostat cultivation at different dilution rates of the multicopy (black symbols) and single‐copy clones (white symbols). (■, □) Specific product generation rate (q P); (●, ○) overall product‐to‐biomass yield coefficient (Y P/X). Error bars represent SE of the mean values.
Most of the studies involving PGAP have shown q P to increase linearly with increasing µ (Khasa et al., 2007; Rebnegger et al., 2014; Garcia‐Ortega et al., 2016). In the present work, similar behaviour was observed for the SCC. In the previous studies, heterologous protein expression was strongly coupled with cell growth. With the methanol‐inducible promoter, P AOX1, q P may not increase with increase in µ and substrate inhibition may arise as a result (Cos et al., 2006; Ahmad et al., 2014; Schwarzhans et al., 2017; Ponte et al., 2018).
As expected, q P was greater with MCC than with SCC (roughly 3–6 times). Some studies have shown protein production to be correlated with gene dosage and hence suggest that the number of gene copies influences q P up an optimum number above which the synergistic effect usually is lost (Schwarzhans et al., 2016a; Betancur et al., 2017; Dagar and Khasa, 2018; Vogl et al., 2018a).
Unlike SCC, the variation of q P for MCC with µ was not linear. In fact, a saturation effect was observed at µ > 0.10 h–1, from which the q P reaches a rather constant value at about 390 UA·gX −1 h−1. As a result, the proportional difference in q P between the two clones decreased with increasing µ (from roughly six times in the low µ range to only about three times in the high µ range).
During the chemostat cultures, the stability of the strains has been demonstrated by confirming the gene copy number of the expression cassette genome integration by ddPCR. Other works also have evaluated strain stability for K. phaffii strains in long‐run chemostats describing a high genetic stability of the recombinant strains (Cankorur‐Cetinkaya et al., 2018).
Looser et al. (2015) described that the strains expressing heterologous proteins under PGAP normally show a linear pattern of production kinetics. Therefore, the optimum µ value for production purposes must be close to μ max. However, in this work q P for MCC results exhibited saturation at specific growth rates lower than μ max, so identifying the optimum µ value should not be straightforward, which was also suggested by other authors (Maurer et al., 2006; Buchetics et al., 2011).
As shown in Fig. 2, the variation of Y P/X with µ differed between SC and MC clones. While the variation pattern for Y P/X in SCC was quite constant irrespective of D, Y P/X in MCC exhibited a substantial decrease in the higher D range. Thus, the highest values of Y P/X were obtained for low specific growth rates (0.025 and 0.05 h–1).
Protein production kinetics reflects the equilibrium between the various steps involved in the synthesis, folding and secretion of proteins, which are influenced by a variety of physiological factors. Based on the results for MCC, one may hypothesize that some of the processes involved in protein production are subject to a bottleneck in the high µ range that leads to the saturation effect observed in the production kinetic profile. This result is consistent with previous reports where the expression system seemingly saturated during transcription, protein synthesis, post‐translational modifications or even secretion (Puxbaum et al., 2015; Cámara et al., 2017). Further research is therefore required to improve existing knowledge about the events. In this work, we focused the efforts on the transcriptional analysis of key genes.
Transcriptional analysis
As suggested in the previous section, further research into subjects such as transcriptional analysis was deemed necessary to throw further light onto the differences in q P variation patterns between SC and MC clones in chemostat cultures. For this purpose, relative transcript levels in relevant target genes were determined under all the culture conditions studied in chemostat cultures, using qPCR as described in the experimental procedures section.
Figure 3 compares the correlation of q P with the relative transcript levels of the genes CRL1 and TDH3. Initially, TDH3 transcription increased linearly with increasing D in both clones regardless the CRL1 gene dosage. This result can be ascribed to the TDH3 product corresponding to a critical node in the glucose uptake pathway, which is closely associated with biomass growth (Nocon et al., 2014; Çalik et al., 2015). Consequently, with high specific biomass growth rates, high fluxes through the glycolytic pathway can only be maintained by increasing production in TDH3. These results would be in contrast with those microarrays results presented by Rebnegger et al. (2014) in which the TDH3 transcription appeared to be unregulated with the dilution rate. Remarkably, it is considered that qPCR is thought to provide increased quantitative resolution in transcriptional analyses of genes that present slight differences among samples (Morey et al., 2006). Finally, on constancy of dilution rate (D), TDH3 relative transcript levels were essentially identical for the two strains, which suggest that the levels were influenced by D but not by CRL1 gene dosage.
Figure 3.
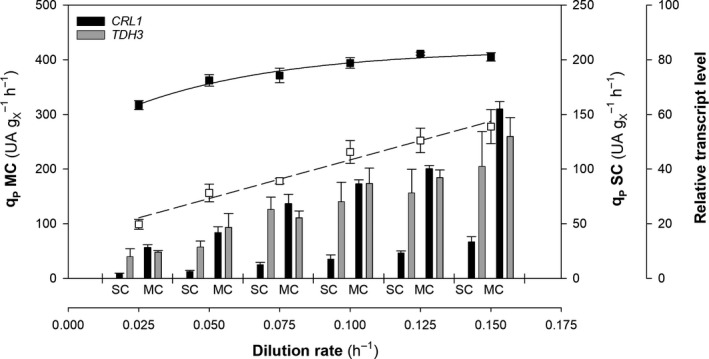
Specific product generation rate, q P, of the single‐copy (□) and multicopy clones (■); and relative transcript level of the clones for CRL1 (black bars) and TDH3 (grey bars) at each dilution rate. Error bars represent SE of the mean values.
Consistent with the results of the enzymatic activity analysis, CRL1 gene expression was initially greater in MC than in SC clones. In SCC, CRL1 relative transcript levels increased linearly with increasing D, which suggests coupling of µ, CRL1 relative transcript levels and q P. These results indicate that PGAP regulates genes TDH3 and CRL1 similarly and hence that the effects are strongly related with cell growth. This was not the case with MCC, however. Thus, although CRL1 gene expression was seemingly not affected by D, q P clearly saturated at high D levels. This may have resulted from CRL1 expression coupling with biomass growth and the total amount of product secreted being limited. Therefore, there might be a bottleneck after transcription precluding conversion of all CRL1 transcripts into proper folded and secreted Crl1.
In Fig. 4, in which the transcriptional regulation of TDH3 and CRL1 for different µ is compared by means of plotting the quotient of the relative transcript levels between the MCC and the SCC, can be observed the above‐mentioned trends. Interestingly, while no effect was observed for the TDH3 quotient, the positive effect of CRL1 gene dosage in terms of transcription is confirmed for all the culture conditions tested. According to these results, therefore, no limitation in the transcriptional machinery for CRL1 can be stated. Nevertheless, and despite the CRL1 quotient of transcript levels remains constant regardless the different cultures conditions at different D, there is a significant decrease in the q P quotient when increasing µ. This would support the hypothesis that the relevant increases achieved in terms of transcript levels due to the higher gene dosage of the target gene cannot be therefore converted into functional protein of interest.
Figure 4.
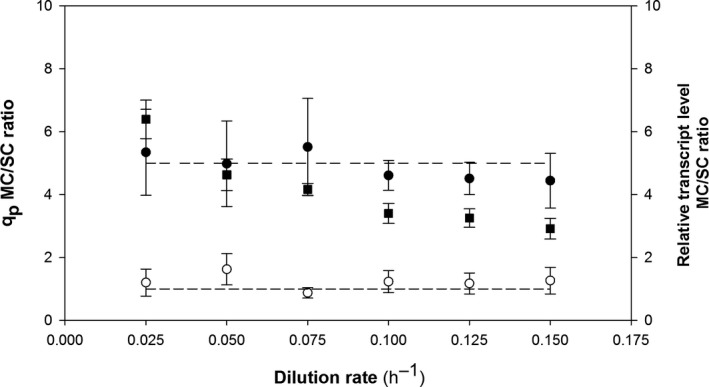
Product and relative transcript‐level ratios between multicopy and single‐copy clones at different dilution rates in chemostat cultures. (■) Ratio of specific product generation rates (q P); (●) ratio of relative transcript level for CRL1; and (○) ratio of relative transcript level for TDH3. Error bars represent SE of the mean values.
Strikingly, the gene dosage effect observed in this PGAP‐driven example of protein expression differs markedly from that reported by Cámara et al. (2017) for PAOX1‐driven Rhizopus oryzae lipase (Rol) recombinant expression. The increased number of cassettes in Rol attenuated transcription of methanol metabolism and decreased methanol consumption, cell growth and recombinant protein production as a result.
Because K. phaffii may be limited in terms of protein folding, glycosylation and secretion, reduced product yields and productivity could be expected. These limitations were widely discussed by Puxbaum et al. (2015) and are related to ER‐associated degradation (ERAD), ER‐Golgi trafficking in the secretory pathway and unfolded protein response (UPR). There have been some attempts at circumventing these constraints and enhancing production of various recombinant proteins. For instance, some authors have used increased gene dosages of target genes involved in product folding and secretion such as HAC1, PDI1 and/or KAR2 (Bankefa et al., 2018; Guan et al., 2016; Liu et al., 2014; Yang et al., 2016). Other authors (Barrero et.al., 2018) have engineered the secretion signal in order to improve translocation into the endoplasmic reticulum (ER).
No limitation in TDH3 or CRL1 gene transcription was observed here. With MCC, however, CRL1 relative transcript levels increased linearly with increasing D, but the transcripts could not be converted into functional secreted Crl1 owing to potential post‐transcriptional constraints. In order to shed further light on the presumed bottlenecks, the relative expression of other target genes including PGK, KAR2 and HAC1 was examined. No limitation in chemostat cultures was observed at any D value, so we can conclude that the specific growth rate had no regulatory effect on the expression of these genes. Overall, this result reveals that, although no transcriptional limitation was identified – and hence CRL1 gene transcription in MCC increased linearly with D – this trend was inconsistent with that in the amount of product formed and hence suggests the presence of post‐transcriptional constraints.
Fed‐batch cultures
In addition to the chemostat cultures, the performance of SC and MC clones was also examined in fed‐batch cultures, which is the operational mode typically used for industrial RPP. All fed‐batch fermentations were conducted according to a carbon‐limited pre‐programmed exponential feeding profile in order to maintain a constant µ throughout the cultivation period. The ending criteria selected for these cultures were to reach a final biomass concentration about 100 g l–1 to compare the experiments with a similar biomass concentration and always below maximal working volume. Thus, the culture is stopped before the bioprocess may be limited due to some biological and physical restrictions, such as heat and mass transfer, and lack of homogeneity, keeping the pseudo‐stationary state in the system. μ spanned the range of 0.05–0.15 h–1 and included an additional condition of 0.025 h–1 for MCC.
Figure 5 presents the time‐course of total lipolytic activity in the fed‐batch cultures with SC and MC clones. Although the figure only shows the results for the fed phase, it should be noted that lipolytic activity at the end of the batch phase was roughly 3.5 times higher with MC clone. Since biomass grew at the highest possible µ during the batch phase, these results seemingly confirm the effect of gene dosage on high µ cultures suggested in the previous section.
Figure 5.
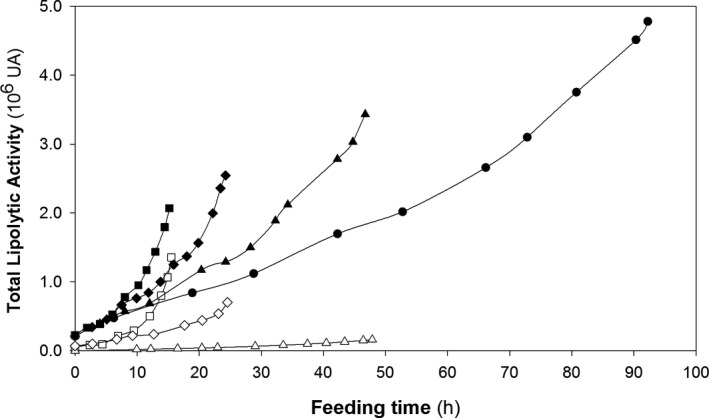
Variation of the total lipolytic activity in fed‐batch cultures at different specific growth rates (µ) with the multicopy (black symbols) and single‐copy clones (white symbols). Specific growth rates: (●) 0.025 h–1, (▲) 0.05 h–1, (♦) 0.10 h–1, (■) 0.15 h–1, (Δ) 0.05 h–1, (◊) 0.10 h–1 and (□) 0.15 h–1.
As detailed in Table 1, lipolytic activity peaked at 372 UA ml–1 at the highest µ value with SCC and at 1009 UA ml–1 at low µ (0.05 h–1) with MCC.
Table 1.
Main production parameters obtained with the single‐copy and multicopy clones at different specific growth rates (µ) in fed‐batch cultures. For the comparing rows, cultivation at the lowest µ of the single‐copy clone is taken as reference.
| Single‐copy clone | Multicopy clone | ||||||
|---|---|---|---|---|---|---|---|
|
Nominal µ (h−1) |
0.050 | 0.100 | 0.150 | 0.025 | 0.050 | 0.100 | 0.150 |
|
Experimental µ (h−1) |
0.048 | 0.088 | 0.133 | 0.023 | 0.051 | 0.087 | 0.147 |
|
Product titre (UA ml−1) |
180 | 195 | 372 | 1414 | 1009 | 827 | 600 |
|
Product titre increase (%) |
− | +8% | +107% | +685% | +460% | +359% | +233% |
|
q P (UA gX −1 h−1) |
77 | 149 | 292 | 374 | 550 | 697 | 777 |
|
q P increase (%) |
− | +92% | +277% | +383% | +611% | +800% | +903% |
|
Q P (103 UA l−1 h−1) |
3.3 | 6.3 | 22.9 | 14.6 | 20.1 | 31.1 | 34.9 |
|
Q P increase (%) |
− | +87% | +586% | +337% | +502% | +831% | +943% |
|
Y P/S (103 UA gS −1) |
0.75 | 0.71 | 0.94 | 7.33 | 5.54 | 3.72 | 2.61 |
|
Y P/S increase (%) |
− | −5% | +26% | +880% | +640% | +398% | +248% |
|
Y P/X (103 UA gX −1) |
1.61 | 1.70 | 2.19 | 16.1 | 10.9 | 8.02 | 5.28 |
|
Y P/X increase (%) |
‐ | +6% | +36% | +902% | +577% | +399% | +229% |
µ, specific growth rate; q P , specific product generation rate; Q P, volumetric productivity; Y P/S, overall product‐to‐substrate yield; Y P/X, overall product‐to‐biomass yield.
Based on these results for MCC, an additional fermentation run at 0.025 h–1 was used to confirm that Crl1 production would peak at the lowest μ level. As hypothesized, lipolytic activity (1414 UA ml–1) was highest under those conditions and four times greater than the highest value for SCC. A systematic comparison based on the main production parameters obtained at the different operating conditions followed with the two producer clones is presented in Table 1.
In Fig. 6, the mean values of q P and Y P/X for the fed‐batch cultures at different µ values are depicted. As expected for consistency with the chemostat results, q P increased linearly with increasing µ in SCC but exhibited saturation in MCC. The Y P/X variation pattern was similar to that for the chemostat cultures; thus, Y P/X decreased exponentially with increasing µ in MCC, but remained fairly constant in SCC.
Figure 6.
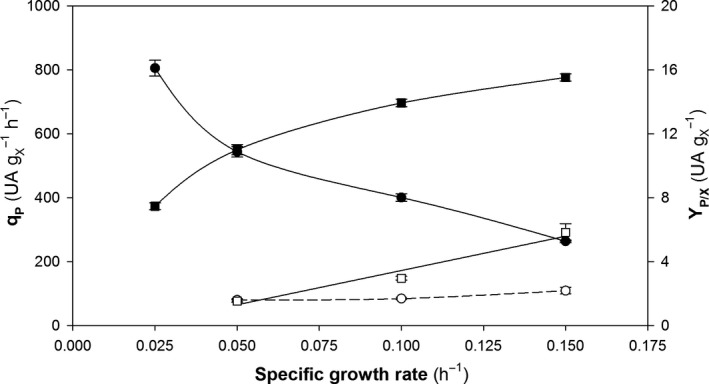
Production parameters for fed‐batch cultures at different specific growth rates of the multicopy (black symbols) and single‐copy clones (white symbols). (■,) Specific product generation rate (q P); (●, ○) overall product‐to‐biomass yield coefficient (Y P/X). Error bars show SE from regression analysis.
As in the chemostat cultures, q P was higher with MCC than with SCC, and their ratio (3–6) was dependent on the µ applied. Although the overall patterns for the two operational modes were similar, q P and Y P/X were substantially higher for the fed‐batch cultures.
Fed‐batch cultivation usually ends when biomass production reaches a critical concentration limit imposed by biological and physical restrictions. As a result, growth‐associated biomass in fermentation processes is an unavoidable by‐product and requires controlling the biomass growth in order not to exceed the limiting concentration. Thus, the lower μ is, the longer a bioprocess can be extended to increase product titres until the termination criterion is fulfilled.
In Table 1 are gathered the main production parameters obtained in fed‐batch cultures for the single‐copy and multicopy clones. Product titre, yields and productivities are detailed as key performance indexes along the different specific growth rates applied. Some aspects can be assessed for discussion, mainly relating to the synergistic effect of µ and q P, and how these two specific rates affect the bioprocess efficiency.
Regarding the production kinetics, that is, how affects μ on q P, it was demonstrated that the kinetic pattern is markedly different for the MCC and SCC. The MCC exhibited a 30% q P decrease when μ decreased from 0.15 to 0.05 h–1, by effect of its rather flat production kinetic curve at the highest μ, as shown in Fig. 6 and Table 1. In a similar comparison for the SCC, q P decreased by 70 % from 0.15 to 0.05 h–1. It is due to the more marked coupling of q P to the cell growth.
As expected, productivities (Q P) varied on µ similarly to that observed for q P in MCC and SCC. Consequently, the rational selection of µ to optimize the bioprocess performance would require a trade‐off.
In terms of parameters focused on production and yield, the SCC at 0.15 h–1 reached product titres 107 % higher compared with the obtained at 0.05 h–1. Accordingly, Y P/S and Y P/X increased a 26% and 36% respectively. Therefore, high µ should be clearly recommended for production kinetic patterns similarly to SCC.
On the contrary, for the MCC, product titre reached a 68 % increase when μ decreased from 0.15 to 0.05 h–1. Strikingly, titre was even better at 0.025 h–1, equivalent to a 140 % increase compared with 0.15 h–1, which lead to important increases in terms of Y P/S and Y P/X (180% and 204% respectively).
Rational‐based bioprocess design
The results previously exposed support the idea that combined effect of µ and gene dosage on PGAP‐based K. phaffii clones expressing CRL1 can be used to design optimal operating strategies.
In this way, the results obtained from chemostat cultures reveal that q P increases linearly with increasing specific growth rate in SCC but exhibits a non‐linear pattern suggestive of saturation of the production kinetics in MCC. Interestingly, q P is considerably higher with MCC than with SCC (3–6 times depending on the particular µ value). These results confirm that gene dosage impacts directly on the production kinetic profile, both in the levels of titre achieved as well as the profile pattern. From the transcriptional analysis performed, one can hypothesize the presence of a potential bottleneck after transcription.
The continuous cultures were used as tool to the design of optimized fed‐batch strategies, since this operational mode is the most widely operational mode used for industrial RPP. Therefore, a rational‐based bioprocess design can be carried out considering the effect of gene dosage and μ on the bioprocess efficiency. Although trends were similar as those observed in the chemostat cultures, there were some differences, especially with MCC. Saturation was more marked than in the chemostat tests, and a plateau was reached in the higher µ range.
When the data for the main production parameters in Table 1 are jointly considered, the different trends in production kinetics and yields can be useful to identify the best operational strategies by characterizing producer clones. Usually, when µ has a strong impact on specific production rates and yields (e.g. with SCC), a high µ level should be used to increase q P and Y P/X. On the other hand, if the impact of µ is weak (e.g. with MCC), low‐to‐moderate µ values can help to extend the bioprocess time until the limit imposed by a critical amount of biomass is reached. In this case, these differences have been caused by the different gene dosage.
Summing up, it can be stated as main outcome of this contribution that the production kinetics of the cell factory and specific growth rate must be jointly considered to tailor operating strategies to production clones for maximizing expression in K. phaffii. This work presents how a comprehensively elucidation of the gene dosage effect on production kinetics towards an overall optimization of the RPP bioprocesses can be afforded. Hence, properly understanding these features and their correlation provides a more rational, robust knowledge of the behaviour of cell factories and allows bioprocesses to be engineered for greater efficiency.
Experimental procedures
Strains
Two recombinant clones of K. phaffii X‐33 from Invitrogen (Carlsbad, CA, USA) expressing CRL1 regulated by the GAP promoter were obtained and used. A chimeric vector assembled with the restriction–ligation method was constructed by using commercial pGAPZαA plasmid from Invitrogen and a codon‐optimized synthetic CRL1 coding sequence from GeneScript (Piscataway, NJ, USA). Different amounts of plasmid were transformed by electroporation in order to obtain clones with different numbers of integrated expression cassettes. However, only the clones containing 1 or 5 gene copies were used. Both clones can secrete Crl1 to the extracellular medium through the Saccharomyces cerevisiae α‐mating factor signal sequence.
Gene copy number
Droplet digital PCR (ddPCR) was used to determine the number of cassette integrations in both clones, using a slightly modified version of the method of Cámara et al. (2016). Actin gene was used as housekeeping agent, but the primer sequence was that proposed by Landes et.al. (2016). The specific primers used are shown in Table S1.
Transcriptional analysis
Transcriptional analyses were only done on chemostat cultures, where steady‐state conditions ensured homogeneity in cell population.
Total RNA extraction
Samples of 1 ml were withdrawn under different chemostat conditions and centrifuged at 4 ºC at maximum speed for 2 min. The resulting pellets were resuspended in 1 ml of TRIzolTM (Waltham, MA, USA) and 200‐mg glass beads. Total RNA was extracted as per the manufacturer’s instructions. Then, RNA integrity and concentration were checked by agarose electrophoresis and Nanodrop analysis (Thermo ScientificTM, Waltham, MA, USA) respectively.
cDNA synthesis and transcriptional levels
cDNA was synthetized by using the iScript™ cDNA Synthesis Kit from Bio‐Rad (Hercules, CA, USA) according to the manufacturer’s instructions. A set of primers was designed to amplify cDNA for the target genes by qPCR. The set comprised CRL1 (heterologous product); TDH3, which is the gene natively expressed under P GAP control (glycolytic); and PGK gene, phosphoglycerate kinase (glycolytic). Additional genes involved in the unfolded protein response (UPR) and the secretory mechanisms such as KAR2 and HAC1 were also studied. Transcription was assessed by qPCR amplification. For maximum accuracy, mixes were made by the robot EpMotion® (Eppendorf, Germany). SYBR™ Select Master Mix was used as polymerase mix and a QuantStudio 12K Flex Real‐Timer from Thermo ScientificTM for amplification cycles and data acquisition.
The qPCR programme was implemented as prescribed by the manufacturer but using a primer annealing temperature of 57.4°C. Relative transcript level was determined by using the MTH1 glucose‐responsive transcriptional factors, which code for a negative regulator of the glucose‐sensing signal transduction pathway, as housekeeping agents. Rebnegger et al. (2014) previously found the specific growth rate to be uninfluential.
Cultivation methods
Inoculum cultures for the bioreactor tests were prepared according to Garcia‐Ortega et al. (2013).
Chemostat cultivation
Chemostat cultures of the two strains were prepared in duplicate as described elsewhere (Garcia‐Ortega et al., 2016). Different specific growth rates from 0.025 to 0.15 h–1 were evaluated. For every dilution rate, the continuous cultures were carried out for at least five residence times. In order to ensure that steady state was achieved, several samples were taken and analysed since three residence times during three consecutive residence times up to confirming the stability of the studied parameters.
Fed‐batch cultivation
Both strains were also cultivated in the fed‐batch mode, at different specific growth rates from 0.025 to 0.15 h–1. This strategy is based on a carbon‐limited feeding profile, keeping a constant µ during the culture reaching a pseudo‐stationary state. The process is described in detail elsewhere (Garcia‐Ortega et al., 2013).
Analytical methods
Determination of biomass as dry cell weight (DCW)
Biomass concentrations were measured in triplicate in terms of DCW as described elsewhere (Cos et al., 2005). The relative standard deviation (RSD) was about 3%.
Quantification of the carbon source and by‐products
The concentrations of the different carbon sources used in the batch (glycerol) and fed‐batch tests (glucose), and of potential fermentation by‐products (arabitol or ethanol), were all determined by HPLC. The column and procedure used for this purpose are described elsewhere (Garcia‐Ortega et al., 2017). RSD was always < 1%.
Off‐gas analyses
BlueInOne Cell gas analysers were used to monitor the cultures exhaust gas (BlueSens, Herten, Germany). CO2 and O2 mole fractions were measured on‐line with provision for off‐gas pressure and humidity. The data thus obtained were used to estimate the oxygen uptake rate (OUR), carbon dioxide evolution rate (CER), specific rates ( and ) and the respiratory quotient (RQ). RSD was < 5% in all cases.
Lipolytic activity assay
Crl1 activity was determined by using a modified version of an existing enzymatic assay based on p‐nitrophenyl butyrate (pNPB) (Chang et al., 2006). The reaction buffer consisted of 1 mM pNPB, 50 mM phosphate buffer at pH 7 and 4 (v/v)% acetone. A volume of 980 µl of buffer was mixed with 20 µl of sample. The absorbance at 348 nm was measured on‐line at 30°C for 2 min on a Specord 200 Plus instrument from Analytic Jena (Jena, Germany). One unit of activity was defined as the amount of enzyme needed to release 1 mmol of p‐nitrophenol per minute under assay conditions. RSD was < 1%.
Process parameters
Mass balance and stoichiometric equations
All equations derived from the mass balances used to calculate yields and rates in the chemostat (Garcia‐Ortega et al., 2016) and fed‐batch cultures (Ponte et al., 2016) were described elsewhere.
Data consistency and reconciliation
Measurement consistency was checked by using the standard test with carbon and electron balances as constraints. Both on‐line and off‐line measurements allowed five key specific rates in the black‐box process model to be calculated, namely biomass generation (µ), glucose uptake (qS), product generation (qP), oxygen uptake () and carbon dioxide production (). The methodology used is described in detail in a previous paper (Ponte et al., 2016).
Conflict of interest
None declared.
Supporting information
Table S1. Primer pairs used for gene dosage and transcript‐level determination by means of ddPCR and qPCR respectively.
Acknowledgements
This work was funded by MINECO and FEDER under Project CTQ2016‐74959‐R. The authors’ group is member of 2017‐SGR‐1462 and the Reference Network in Biotechnology (XRB) of Generalitat de Catalunya. M.A. Nieto‐Taype acknowledges award by the National Council of Science, Technology and Technological Innovation (CONCYTEC) through its executing unit, the National Fund for Scientific, Technological and Technological Innovation Development (FONDECYT) and J. Garrigós‐Martínez, a PIF scholarship from the Universitat Autònoma de Barcelona.
Microbial Biotechnology (2020) 13(2), 315–327
Funding information
This work was funded by MINECO and FEDER under Project CTQ2016‐74959‐R.
References
- Adelantado, N. , Tarazona, P. , Grillitsch, K. , García‐Ortega, X. , Monforte, S. , Valero, F. , et al. (2017) The effect of hypoxia on the lipidome of recombinant Pichia pastoris . Microb Cell Fact 16: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad, M. , Hirz, M. , Pichler, H. , and Schwab, H. (2014) Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol 98: 5301–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankefa, O.E. , Wang, M. , Zhu, T. , and Li, Y. (2018) Hac1p homologues from higher eukaryotes can improve the secretion of heterologous proteins in the yeast Pichia pastoris . Biotechnol Lett 40: 1149–1156. [DOI] [PubMed] [Google Scholar]
- Barrero, J.J. , Casler, J.C. , Valero, F. , Ferrer, P. , and Glick, B.S. (2018) An improved secretion signal enhances the secretion of model proteins from Pichia pastoris . Microb Cell Fact 17: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrigón, J.M. , Montesinos, J.L. , and Valero, F. (2013) Searching the best operational strategies for Rhizopus oryzae lipase production in Pichia pastoris Mut+ phenotype: Methanol limited or methanol non‐limited fed‐batch cultures? Biochem Eng J 75: 47–54. [Google Scholar]
- Betancur, M.O. , Reis, V.C.B. , Nicola, A.M. , De Marco, J.L. , de Moraes, L.M.P. , and Torres, F.A.G. (2017) Multicopy plasmid integration in Komagataella phaffii mediated by a defective auxotrophic marker. Microb Cell Fact 16: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchetics, M. , Dragosits, M. , Maurer, M. , Rebnegger, C. , Porro, D. , Sauer, M. , et al. (2011) Reverse engineering of protein secretion by uncoupling of cell cycle phases from growth. Biotechnol Bioeng 108: 2403–2412. [DOI] [PubMed] [Google Scholar]
- Burgard, J. , Valli, M. , Graf, A.B. , Gasser, B. , and Mattanovich, D. (2017) Biomarkers allow detection of nutrient limitations and respective supplementation for elimination in Pichia pastoris fed‐batch cultures. Microb Cell Fact 16: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çalık, P. , Ata, Ö. , Güneş, H. , Massahi, A. , Boy, E. , Keskin, A. , et al. (2015) Recombinant protein production in Pichia pastoris under glyceraldehyde‐3‐phosphate dehydrogenase promoter: From carbon source metabolism to bioreactor operation parameters. Biochem Eng J 95: 20–36. [Google Scholar]
- Cámara, E. , Albiol, J. , and Ferrer, P. (2016) Droplet digital PCR‐aided screening and characterization of Pichia pastoris multiple gene copy strains. Biotechnol Bioeng 113: 1542–1551. [DOI] [PubMed] [Google Scholar]
- Cámara, E. , Landes, N. , Albiol, J. , Gasser, B. , Mattanovich, D. , and Ferrer, P. (2017) Increased dosage of AOX1 promoter‐regulated expression cassettes leads to transcription attenuation of the methanol metabolism in Pichia pastoris . Sci Rep 7: 44302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cankorur‐Cetinkaya, A. , Narraidoo, N. , Kasavi, C. , Slater, N.K.H. , Archer, D.B. , and Oliver, S.G. (2018) Process development for the continuous production of heterologous proteins by the industrial yeast, Komagataella phaffii . Biotechnol Bioeng 115: 2962–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnicer, M. , Ten Pierick, A. , Van Dam, J. , Heijnen, J.J. , Albiol, J. , Van Gulik, W. , and Ferrer, P. (2012) Quantitative metabolomics analysis of amino acid metabolism in recombinant Pichia pastoris under different oxygen availability conditions. Microb Cell Fact 11: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S.W. , Lee, G.C. , and Shaw, J.F. (2006) Codon optimization of Candida rugosa lip1 gene for improving expression in Pichia pastoris and biochemical characterization of the purified recombinant LIP1 lipase. J Agric Food Chem 54: 815–822. [DOI] [PubMed] [Google Scholar]
- Cos, O. , Serrano, A. , Montesinos, J.L. , Ferrer, P. , Cregg, J.M. , and Valero, F. (2005) Combined effect of the methanol utilization (Mut) phenotype and gene dosage on recombinant protein production in Pichia pastoris fed‐batch cultures. J Biotechnol 117: 321–335. [DOI] [PubMed] [Google Scholar]
- Cos, O. , Ramón, R. , Montesinos, J.L. , and Valero, F. (2006) Operational strategies, monitoring and control of heterologous protein production in the methylotrophic yeast Pichia pastoris under different promoters: a review. Microb Cell Fact 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagar, V.K. , and Khasa, Y.P. (2018) Combined effect of gene dosage and process optimization strategies on high‐level production of recombinant human interleukin‐3 (hIL‐3) in Pichia pastoris fed‐batch culture. Int J Biol Macromol 108: 999–1009. [DOI] [PubMed] [Google Scholar]
- Dewan, S. S. (2017) Global markets for enzymes in industrial applications. BCC Res (BIO030J).
- Ferrer, P. , Montesinos, J.L. , Valero, F. , and Solà, C. (2001) Production of native and recombinant lipases by Candida rugosa: a review. Appl Biochem Biotechnol 95: 221–256. [DOI] [PubMed] [Google Scholar]
- Garcia‐Ortega, X. , Ferrer, P. , Montesinos, J.L. , and Valero, F. (2013) Fed‐batch operational strategies for recombinant Fab production with Pichia pastoris using the constitutive GAP promoter. Biochem Eng J 79: 172–181. [Google Scholar]
- Garcia‐Ortega, X. , Adelantado, N. , Ferrer, P. , Montesinos, J.L. , and Valero, F. (2016) A step forward to improve recombinant protein production in Pichia pastoris: from specific growth rate effect on protein secretion to carbon‐starving conditions as advanced strategy. Process Biochem 51: 681–691. [Google Scholar]
- Garcia‐Ortega, X. , Valero, F. , and Montesinos‐Seguí, J.L. (2017) Physiological state as transferable operating criterion to improve recombinant protein production in Pichia pastoris through oxygen limitation. J Chem Technol Biotechnol 92: 2573–2582. [Google Scholar]
- García‐Ortega, X. , Cámara, E. , Ferrer, P. , Albiol, J. , Montesinos‐Seguí, J.L. , and Valero, F. (2019) Rational development of bioprocess engineering strategies for recombinant protein production in Pichia pastoris (Komagataella phaffii) using the methanol‐free GAP promoter. Where do we stand? N Biotechnol 53: 24–34. [DOI] [PubMed] [Google Scholar]
- Guan, B. , Chen, F. , Su, S. , Duan, Z. , Chen, Y. , Li, H. , and Jin, J. (2016) Effects of co‐overexpression of secretion helper factors on the secretion of a HSA fusion protein (IL2‐HSA) in Pichia pastoris . Yeast 33: 587–600. [DOI] [PubMed] [Google Scholar]
- Hartner, F.S. , and Glieder, A. (2006) Regulation of methanol utilisation pathway genes in yeasts. Microb Cell Fact 5: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highsmith, J. (2015) Biological Therapeutic Drugs: Technologies and Global Markets. BCC Res. ( BIO079C).
- Jiao, L. , Zhou, Q. , Su, Z. , Xu, L. , and Yan, Y. (2018) High‐level extracellular production of Rhizopus oryzae lipase in Pichia pastoris via a strategy combining optimization of gene‐copy number with co‐expression of ERAD‐related proteins. Protein Expr Purif 147: 1–12. [DOI] [PubMed] [Google Scholar]
- Juturu, V. , and Wu, J.C. (2018) Heterologous protein expression in Pichia pastoris : latest research progress and applications. ChemBioChem 19: 7–21. [DOI] [PubMed] [Google Scholar]
- Ken Ugo, A. , Vivian Amara, A. , CN, I. , and Kenechuwku, U., (2017) Microbial lipases: a prospect for biotechnological industrial catalysis for green products: a review. Ferment Technol 6: 2. [Google Scholar]
- Khasa, Y.P. , Khushoo, A. , Srivastava, L. , and Mukherjee, K.J. (2007) Kinetic studies of constitutive human granulocyte‐macrophage colony stimulating factor (hGM‐CSF) expression in continuous culture of Pichia pastoris . Biotechnol Lett 29: 1903–1908. [DOI] [PubMed] [Google Scholar]
- Landes, N. , Gasser, B. , Vorauer‐Uhl, K. , Lhota, G. , Mattanovich, D. , and Maurer, M. (2016) The vitamin‐sensitive promoter PTHI11 enables pre‐defined autonomous induction of recombinant protein production in Pichia pastoris . Biotechnol Bioeng 113: 2633–2643. [DOI] [PubMed] [Google Scholar]
- Liang, S. , Zou, C. , Lin, Y. , Zhang, X. , and Ye, Y. (2013) Identification and characterization of PGCW14: a novel, strong constitutive promoter of Pichia pastoris . Biotechnol Lett 35: 1865–1871. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Qin, Y. , Huang, Y. , Chen, Y. , Cong, P. , and He, Z. (2014) Direct evaluation of the effect of gene dosage on secretion of protein from yeast Pichia pastoris by expressing EGFP. J Microbiol Biotechnol 24: 144–151. [DOI] [PubMed] [Google Scholar]
- Looser, V. , Bruhlmann, B. , Bumbak, F. , Stenger, C. , Costa, M. , Camattari, A. , et al. (2015) Cultivation strategies to enhance productivity of Pichia pastoris: a review. Biotechnol Adv 33: 1177–1193. [DOI] [PubMed] [Google Scholar]
- Mattanovich, D. , Graf, A. , Stadlmann, J. , Dragosits, M. , Redl, A. , Maurer, M. , et al. (2009) Genome, secretome and glucose transport highlight unique features of the protein production host Pichia pastoris . Microb Cell Fact 8: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer, M. , Kühleitner, M. , Gasser, B. , and Mattanovich, D. (2006) Versatile modeling and optimization of fed batch processes for the production of secreted heterologous proteins with Pichia pastoris . Microb Cell Fact 5: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey, J.S. , Ryan, J.C. , and Dolah, F.M. (2006) Microarray validation: factors influencing correlation between oligonucleotide microarrays and real‐time PCR. Biol Proced Online 8: 175–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser, J.W. , Prielhofer, R. , Gerner, S.M. , Graf, A.B. , Wilson, I.B.H. , Mattanovich, D. , and Dragosits, M. (2017) Implications of evolutionary engineering for growth and recombinant protein production in methanol‐based growth media in the yeast Pichia pastoris . Microb Cell Fact 16: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocon, J. , Steiger, M.G. , Pfeffer, M. , Sohn, S.B. , Kim, T.Y. , Maurer, M. , et al. (2014) Model based engineering of Pichia pastoris central metabolism enhances recombinant protein production. Metab Eng 24: 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordén, K. , Agemark, M. , Danielson, J.Å.H. , Alexandersson, E. , Kjellbom, P. , and Johanson, U. (2011) Increasing gene dosage greatly enhances recombinant expression of aquaporins in Pichia pastoris . BMC Biotechnol 11: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña, D.A. , Gasser, B. , Zanghellini, J. , Steiger, M.G. , and Mattanovich, D. (2018) Metabolic engineering of Pichia pastoris . Metab Eng 50: 2–15. [DOI] [PubMed] [Google Scholar]
- Ponte, X. , Montesinos‐Seguí, J.L. , and Valero, F. (2016) Bioprocess efficiency in Rhizopus oryzae lipase production by Pichia pastoris under the control of PAOX1 is oxygen tension dependent. Process Biochem 51: 1954–1963. [Google Scholar]
- Ponte, X. , Barrigón, J.M. , Maurer, M. , Mattanovich, D. , Valero, F. , and Montesinos‐Seguí, J.L. (2018) Towards optimal substrate feeding for heterologous protein production in Pichia pastoris (Komagataella spp) fed‐batch processes under PAOX1 control: a modeling aided approach. J Chem Technol Biotechnol 93: 3208–3218. [Google Scholar]
- Potvin, G. , Ahmad, A. , and Zhang, Z. (2012) Bioprocess engineering aspects of heterologous protein production in Pichia pastoris: a review. Biochem Eng J 64: 91–105. [Google Scholar]
- Prielhofer, R. , Maurer, M. , Klein, J. , Wenger, J. , Kiziak, C. , Gasser, B. , and Mattanovich, D. (2013) Induction without methanol: novel regulated promoters enable high‐level expression in Pichia pastoris . Microb Cell Fact 12: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puxbaum, V. , Mattanovich, D. , and Gasser, B. (2015) Quo vadis? The challenges of recombinant protein folding and secretion in Pichia pastoris . Appl Microbiol Biotechnol 99: 2925–38. [DOI] [PubMed] [Google Scholar]
- Rebnegger, C. , Graf, A. B. , Valli, M. , Steiger, M.G. , Gasser, B. , Maurer, M. , and Mattanovich, D. (2014) In Pichia pastoris, growth rate regulates protein synthesis and secretion, mating and stress response. Biotechnol J 9: 511–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebnegger, C. , Vos, T. , Graf, A.B. , Valli, M. , Pronk, J.T. , Daran‐Lapujade, P. , and Mattanovich, D. (2016) Pichia pastoris exhibits high viability and a low maintenance energy requirement at near‐zero specific growth rates. Appl Environ Microbiol 82: 4570–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, J.M. , Garcia‐Ortega, X. , Montesinos‐Seguí, J.L. , Freire, D.M.G. , and Valero, F. (2019) Continuous operation, a realistic alternative to fed‐batch fermentation for the production of recombinant lipase B from Candida antarctica under the constitutive promoter PGK in Pichia pastoris . Biochem Eng J 147: 39–47. [Google Scholar]
- Sánchez, A. , Ferrer, P. , Serrano, A. , Pernas, M. A. , Valero, F. , Rúa, M.L. , et al. (1999) Characterization of the lipase and esterase multiple forms in an enzyme preparation from a Candida rugosa pilot‐plant scale fed‐batch fermentation. Enzyme Microb Technol 25: 214–223. [Google Scholar]
- Schwarzhans, J.P. , Wibberg, D. , Winkler, A. , Luttermann, T. , Kalinowski, J. , and Friehs, K. (2016a) Integration event induced changes in recombinant protein productivity in Pichia pastoris discovered by whole genome sequencing and derived vector optimization. Microb Cell Fact 15: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzhans, J.P. , Wibberg, D. , Winkler, A. , Luttermann, T. , Kalinowski, J. , and Friehs, K. (2016b) Non‐canonical integration events in Pichia pastoris encountered during standard transformation analysed with genome sequencing. Sci Rep 6: 38952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzhans, J.P. , Luttermann, T. , Geier, M. , Kalinowski, J. , and Friehs, K. (2017) Towards systems metabolic engineering in Pichia pastoris . Biotechnol Adv 35: 681–710. [DOI] [PubMed] [Google Scholar]
- Shen, W. , Xue, Y. , Liu, Y. , Kong, C. , Wang, X. , Huang, M. , et al. (2016) A novel methanol‐free Pichia pastoris system for recombinant protein expression. Microb Cell Fact 15: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theron, C.W. , Berrios, J. , Delvigne, F. , and Fickers, P. (2018) Integrating metabolic modeling and Komagataella (Pichia) pastoris . Appl Microbiol Biotechnol 102: 63–80. [DOI] [PubMed] [Google Scholar]
- Valero, F. (2018) Recent advances in Pichia pastoris as host for heterologous expression system for lipases: a review In: Methods in Molecular Biology. New York, NY: Humana Press, pp. 205–216. [DOI] [PubMed] [Google Scholar]
- Vogl, T. , and Glieder, A. (2013) Regulation of Pichia pastoris promoters and its consequences for protein production. N Biotechnol 30: 385–404. [DOI] [PubMed] [Google Scholar]
- Vogl, T. , Sturmberger, L. , Kickenweiz, T. , Wasmayer, R. , Schmid, C. , Hatzl, A.M. , et al. (2016) A toolbox of diverse promoters related to methanol utilization: functionally verified parts for heterologous pathway expression in Pichia pastoris . ACS Synth Biol 5: 172–186. [DOI] [PubMed] [Google Scholar]
- Vogl, T. , Gebbie, L. , Palfreyman, R.W. , and Speight, R. (2018a) Effect of plasmid design and type of integration event on recombinant protein expression in Pichia pastoris . Appl Environ Microbiol 84: e02712‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl, T. , Sturmberger, L. , Fauland, P.C. , Hyden, P. , Fischer, J.E. , Schmid, C. , et al. (2018b) Methanol independent induction in Pichia pastoris by simple derepressed overexpression of single transcription factors. Biotechnol Bioeng 115: 1037–1050. [DOI] [PubMed] [Google Scholar]
- Weninger, A. , Fischer, J.E. , Raschmanová, H. , Kniely, C. , Vogl, T. , and Glieder, A. (2018) Expanding the CRISPR/Cas9 toolkit for Pichia pastoris with efficient donor integration and alternative resistance markers. J Cell Biochem 119: 3183–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. , and Zhang, Z. (2018) Engineering strategies for enhanced production of protein and bio‐products in Pichia pastoris: a review. Biotechnol Adv 36: 182–195. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Lu, Z. , Chen, J. , Chu, P. , Cheng, Q. , Liu, J. , et al. (2016) Effect of cooperation of chaperones and gene dosage on the expression of porcine PGLYRP‐1 in Pichia pastoris . Appl Microbiol Biotechnol 100: 5453–5465. [DOI] [PubMed] [Google Scholar]
- Zahrl, R.J. , Peña, D.A. , Mattanovich, D. , and Gasser, B. (2017) Systems biotechnology for protein production in Pichia pastoris . FEMS Yeast Res 17: 7. [DOI] [PubMed] [Google Scholar]
- Zhang, A.L. , Luo, J.X. , Zhang, T.Y. , Pan, Y.W. , Tan, Y.H. , Fu, C.Y. , et al. (2009) Recent advances on the GAP promoter derived expression system of Pichia pastoris . Mol Biol Rep 36: 1611–1619. [DOI] [PubMed] [Google Scholar]
- Zhao, W. , Wang, J. , Deng, R. , and Wang, X. (2008) Scale‐up fermentation of recombinant Candida rugosa lipase expressed in Pichia pastoris using the GAP promoter. J Ind Microbiol Biotechnol 35: 189–195. [DOI] [PubMed] [Google Scholar]
- Zhu, T. , Guo, M. , Tang, Z. , Zhang, M. , Zhuang, Y. , Chu, J. , and Zhang, S. (2009) Efficient generation of multi‐copy strains for optimizing secretory expression of porcine insulin precursor in yeast Pichia pastoris . J Appl Microbiol 107: 954–63. [DOI] [PubMed] [Google Scholar]
- Zhu, T. , Sun, H. , Li, P. , Xue, Y. , Li, Y. , and Ma, Y. (2014) Constitutive expression of alkaline β‐mannanase in recombinant Pichia pastoris . Process Biochem 49: 2025–2029. [Google Scholar]
- Zhu, T. , Sun, H. , Wang, M. , and Li, Y. (2019) Pichia pastoris as a versatile cell factory for the production of industrial enzymes and chemicals: current status and future perspectives. Biotechnol J 14: 1800694. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer pairs used for gene dosage and transcript‐level determination by means of ddPCR and qPCR respectively.


