Abstract
Little is known about the infections of double-stranded DNA (dsDNA) viruses in fungi. Here, we use a paleovirological method to systematically identify the footprints of past dsDNA virus infections within the fungal genomes. We uncover two distinct groups of endogenous nucleocytoplasmic large DNA viruses (NCLDVs) in at least seven fungal phyla (accounting for about a third of known fungal phyla), revealing an unprecedented diversity of dsDNA viruses in fungi. Interestingly, one fungal dsDNA virus lineage infecting six fungal phyla is closely related to the giant virus Pithovirus, suggesting giant virus relatives might widely infect fungi. Co-speciation analyses indicate fungal NCLDVs mainly evolved through cross-species transmission. Taken together, our findings provide novel insights into the diversity and evolution of NCLDVs in fungi.
Keywords: paleovirology, fungi, endogenous viral elements, nucleocytoplasmic large DNA viruses, phylogenetics
1. Introduction
As a paradigm, viruses have long been thought to be smaller than cellular organisms. The discovery of Acanthamoeba polyphaga mimivirus, a giant virus with a double-stranded DNA linear genome of ∼1.2 Mb, challenged this ‘well-established’ concept (La Scola 2003; Raoult et al. 2004). Since then, many giant viruses have been identified in eukaryotes (primarily amoebae) and diverse environments across the globe (Koonin 2009; Koonin and Yutin 2010; Philippe et al. 2013; Maumus et al. 2014). Phylogenetic analyses suggest that giant viruses fall within the diversity of nucleocytoplasmic large DNA viruses (NCLDVs), a group of double-stranded DNA (dsDNA) viruses (Colson et al. 2012; Koonin and Yutin 2018; Guglielmini et al. 2019).
Despite their various gene repertoires and genome sizes, NCLDVs form a monophyletic group. Currently, NCLDVs comprise at least seven families (Iridoviridae, Ascoviridae, Asfarviridae, Phycodnaviridae, Poxviridae, Mimiviridae, and Marseilleviridae) and some unclassified viruses (Koonin and Yutin 2018). NCLDVs possess highly diverse genomes ranging from ∼100 kb (iridoviruses) to > 2.5 Mb (pandoraviruses) (Philippe et al. 2013; Legendre et al. 2018). Some giant viruses can even encode protein translation components, further blurring the boundary between viruses and cellular world (Arslan et al. 2011). NCLDVs infect a remarkably wide range of eukaryotes, from protists to green algae and animals (Delaroque and Boland 2008; Koonin and Yutin 2010; Colson et al. 2012; Gallot-Lavallée and Blanc 2017; Koonin and Yutin 2018). However, only few NCLDVs have been described in land plants and fungi (Maumus et al. 2014; Gallot-Lavallée and Blanc 2017).
To date, double-stranded RNA (dsRNA) viruses (e.g. Quadriviridae, Megabirnaviridae, Partitiviridae, Reoviridae, and Totiviridae), positive-sense single-stranded RNA [(+)ssRNA] viruses (e.g. Alphaflexiviridae, Gammaflexiviridae, and Hypoviridae), negative-sense single-stranded RNA [(-)ssRNA] viruses (e.g. Bunyaviridae), and single-stranded DNA (ssDNA) viruses (e.g. Genomoviridae) have been known to infect fungi (Ghabrial et al. 2015; Krupovic et al. 2016; Roossinck 2019). Among them, dsRNA viruses are most commonly observed in fungi (Ghabrial et al. 2015; Roossinck 2019).
Virus can adventitiously integrate into host germline genomes and become vertically inherited to next generation, forming the so called endogenous viral elements (EVEs). Notably, the replication of phaeoviruses requires integration into the host genomes using integrase encoded by their genomes (Delaroque et al. 1999; Meints et al. 2008). EVEs record past viral infections and thus represent ‘molecular fossils’ for studying the deep history and ancient ecology of viruses (Patel, Emerman, and Malik 2011; Feschotte and Gilbert 2012). EVEs have facilitated the rise of an emerging field, Paleovirology. With the recent development of next generation sequencing (NGS), hundreds of eukaryote genomes have been sequenced, providing a rich resource for uncovering the footprints of past viral infections. Recently, NCLDV-like sequences have been identified in three fungus species (Gallot-Lavallée and Blanc 2017). However, much remains unknown about the distribution and evolution of dsDNA viruses in fungi (Colson et al. 2013).
In this study, we used a paleovirological approach to systematically identify endogenous dsDNA virus elements in the genomes of fungi. We performed phylogenetic analyses to investigate the relationship between the newly identified viral sequences in fungal genomes and NCLDVs. We also performed co-speciation analyses to explore the evolutionary mode of fungal viruses.
2. Results and discussion
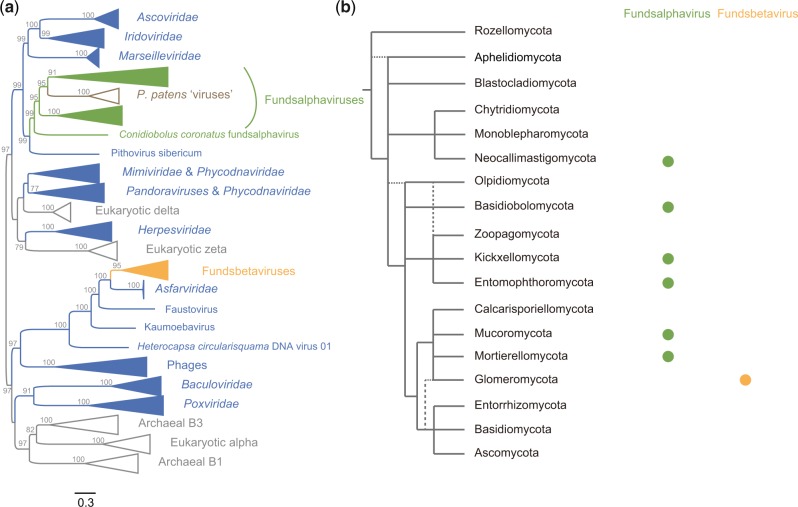
To explore the diversity of dsDNA viruses in fungi, we used a similarity search and phylogenetic analysis combined approach to screen the presence of NCLDV-like elements in 1,006 fungal genomes (see Methods). We identified a total of 87 NCLDV-like sequences within 15 fungal genomes (Supplementary Table S1). Phylogenetic analysis based on the family B DNA polymerase (DNAP), a core gene conserved among NCLDVs and cellular organisms, shows that the fungus NCLDV-like sequences identified here cluster into two distinct lineages, designated as fungus dsDNA alpha virus (fundsalphavirus) and fungus dsDNA beta virus (fundsbetavirus) (Fig. 1a and Supplementary Fig. S1). Premature stop codons and frameshift mutations exist in some DNAP sequences, suggesting these viruses are endogenous viral elements and are not generated by laboratory contamination. Interestingly, fundsalphaviruses appear to be closely related to Pithovirus sibericum, a giant virus isolated from Siberian permafrost (Fig. 1a and Supplementary Fig. S5) (Legendre et al. 2014). Surprisingly, endogenous fundsalphaviruses were identified in six fungus phyla (accounting for a third of known fungal phyla) (Fig. 1b) (Tedersoo et al. 2018). Endogenous fundsbetaviruses were only identified within Rhizophagus irregularis. Phylogenetic analysis shows that fundsbetaviruses are closely related with African swine fever virus (the Asfarviridae family). Taken together, our results suggest NCLDVs might infect/have infected a wide range of fungi.
Figure 1.
Phylogeny and host distribution of fungus NCLDV-like sequences. (a) Phylogenetic tree of fundsalphaviruses and fundsbetaviruses based on DNAP. Fundsalphaviruses, fundsbetaviruses, other dsDNA viruses, and P. patens ‘virus’ were labeled in green, orange, blue, and brown, respectively. This unrooted tree was reconstructed using a maximum likelihood method. Ultrafast bootstrap (UFBoot) values (>75) were shown on selected nodes. (b) Host distribution of fundsalphaviruses and fundsbetaviruses. The fungi phylogeny at the level of phylum was inferred from Tedersoo et al. (2018). The presence of fundsalphaviruses and fundsbetaviruses in fungus phylum was labeled with green solid circle and orange solid circle, respectively.
To further characterize fundsalphaviruses and fundsbetaviruses, we investigated the gene contents flanking DNAP. We found that at least six genes might be shared among fundsalphaviruses, namely D5 helicase-primase (D5), D6/D11-like SNF2 helicase (DEXDc-HELICc), S-adenosylmethionine-dependent methyltransferase (SAM), DNA-dependent RNA polymerase (RNAP) subunit 2 and 5 (RPB2 and RPB5), and RNAP subunit 1 (N-terminal [RP1n] and C-terminal [RP1c]) (Supplementary Fig. S2). In some cases, retrotransposon-related sequences were found near the endogenous fundsalphavirus elements, suggesting retrotransposons might potentially play a role in the integration and proliferation of fundsalphaviruses (Supplementary Fig. S2; Aswad and Katzourakis 2017). The variation in gene contents among different endogenous fundsalphavirus elements may reflect the variation of fundsalphaviruses infecting different fungi and/or be due to the long-term evolution after the integrations of ancestral viruses. Phylogenetic analyses based on these six genes show that fundsalphaviruses consistently cluster together with NCLDVs (Supplementary Figs S3–S8). In particular, our phylogenetic analyses of RNAP1 and RNAP2 clearly distinguish NCLDVs from cellular species, which is consistent with a previous study by Guglielmini et al. (2019), and show that fundsalphaviruses cluster together with NCLDVs (Supplementary Figs S4 and S6). But fundsalphaviruses do not share similar gene orders with other NCLDVs (Supplementary Fig. S2). Taken together, these results indicate that fundsalphaviruses might be derived from a distinct NCLDV family.
Moreover, we performed similarity search of major capsid protein (MCP) and A32 packaging ATPase, proteins involved in virion morphogenesis, against fungal genomes. The evolutionary history of MCP and A32 proteins are complex with sequences from prokaryotes (perhaps phages) dispersed across the trees, precluding meaningful evolutionary interpretation and classification (Supplementary Figs S9 and S10). However, we did find that some significant hits from fungal genomes cluster together with NCLDVs, further confirming the presence of dsDNA viruses in fungal genomes (Supplementary Figs S9 and S10).
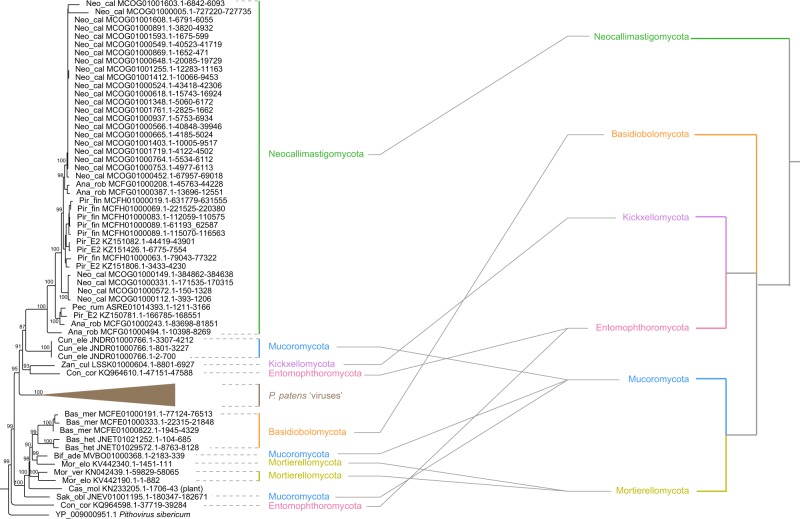
Endogenous fundsalphavirus elements were identified in fourteen fungus species that belong to six fungal phyla (Fig. 1b). One might argue that they may arise through an ancestral integration or horizontal gene transfer event in the most recent common ancestor of these fungi. To test this possibility, we performed co-speciation analyses using an event-based approach at the level of host phylum. There is no significant congruence between host and fundsalphavirus phylogenies (Fig. 2; Table 1), revealing that these NCLDV-like sequences within fungal genomes may not derive from an ancestral integration event. Moreover, we did not find shared host genes among endogenous fundsalphaviruses of different fungi (Supplementary Fig. S2). These results suggest these endogenous fundsalphaviruses might arise through multiple integration events, and fundsalphaviruses evolved in fungi mainly via cross-species transmission.
Figure 2.
Evolutionary mode of fundsalphaviruses. The fundsalphavirus tree (left) is compared with its fungus host tree (right) at the level of phylum. Gray lines connect the fundsalphaviruses to their corresponding host phyla. Fungus name abbreviations: Neo_cal, Neocallimastix californiae; Pir_fin, Piromyces finnis; Ana_rob, Anaeromyces robustus; Pir_ E2, Piromyces sp. E2; Pec_rum, Pecoramyces ruminatium; Bas_mer, Basidiobolus meristosporus; Bas_het, Basidiobolus heterosporus; Cun_ele, Cunninghamella elegans; Bif_ade, Bifiguratus adelaidae; Sak_obl, Saksenaea oblongispora; Con_cor, Conidiobolus coronatus; Mor_elo, Mortierella elongate; Mor_ver, Mortierella verticillata; Zan_cul, Zancudomyces culisetae.
Table 1.
Number of events experienced by fundsalphaviruses
| Event costsa | Total cost | Co-speciationb | Duplicationb | Duplication and host switchb | Lossb | Failure to divergeb | P-valuec | P-valued |
|---|---|---|---|---|---|---|---|---|
| −1, 0, 0, 0, 0 | −4 | 4-4 | 0-2 | 3-5 | 4-8 | 0-0 | 0.85 | 0.89 |
| 0, 1, 1, 2, 0 | 7 | 2-2 | 1-1 | 6-6 | 0-0 | 0-0 | 0.818 | 0.874 |
Event costs are for co-speciation, duplication, duplication and host switch, loss, and failure to diverge, respectively.
Number of events is expressed as ranges that result in the same cost.
Random tip mapping method with sample size of 500.
Random parasite tree method with sample size of 500.
Interestingly, the NCLDV-like sequences previously identified in the genome of the moss Physcomitrella patens (Filée 2014; Maumus et al. 2014) fall within the diversity of fundsalphaviruses (Fig. 1a), raising the possibility that land plant NCLDVs might originate through cross-species transmission from fungi. Indeed, many plant viruses are closely related to fungal viruses (Roossinck 2019).
Much remains unknown about the distribution and evolution of dsDNA viruses in fungi. In this study, we found two lineages of dsDNA viruses infecting fungi, revealing a potentially unprecedented diversity of dsDNA virus families in fungi and raising the possibility that other dsDNA viruses might be still circulating in fungi. Further work is needed to characterize the diversity of dsDNA viruses in fungi. Our findings come with two caveats: (1) the diversity of dsDNA viruses might be underestimated in fungi, because we only used DNAP as a probe to screen dsDNA virus insertions; (2) the fungus NCLDV sequences might be generated by laboratory contamination or sequencing error. However, many fungus NCLDV sequences are in long genomic contigs. Fundsalphaviruses have been identified in as many as fourteen species across six phyla. There are premature stop codons and frameshift mutations in fungus NCLDV sequences. All these lines make the possibility of contamination highly unlikely (Naccache et al. 2013; Kjartansdóttir et al. 2015). Nevertheless, our findings reveal dsDNA viruses, especially giant virus relatives, could infect a previously neglected major eukaryotic kingdom, Fungi.
3. Methods and materials
3.1 Identification of NCLDV-like sequences in fungal genomes
We screened a total of 1,006 whole genome shotgun (WGS) sequences of fungi for the NCLDV-like sequences through a similarity search and phylogenetic analysis combined approach. First, we used the tBLASTn algorithm to search against the fungal genomes with an e-cut-off value of 10−5 and DNAP of representative dsDNA viruses as queries. DNAP has been used widely to identify and classify NCLDVs due to strong sequence conservation, clear evolutionary history, and few horizontal gene transfer (Monier, Claverie, and Ogata 2008; Colson et al. 2013). Next, the significant hits together with DNAP sequences of representative NCLDVs, herpesviruses, baculoviruses, eukaryotes, and archaea were aligned using MAFFT (Koonin and Yutin 2010; Katoh and Standley 2013) (accession numbers are available in Supplementary Fig. S1). Initial phylogenetic analyses were performed through an approximate maximum likelihood method implemented in FastTree (Price, Dehal, and Arkin 2010). The fungal sequences that group with NCLDVs were extracted for further studies. Moreover, we performed similarity searches using MCP and A32 protein as queries with e-cut-off value of 0.01. Phylogenetic analyses were also performed through an approximate maximum likelihood method implemented in FastTree (Price, Dehal, and Arkin 2010).
To explore the gene contents of fundsalphaviruses and fundsbetaviruses, we extracted the sequences containing DNAP within their genomes, covering eight flanking protein domains for each side of DNAP. The conserved domains were detected through the BLASTx algorithm against non-redundant protein database in National Center for Biotechnology Information (NCBI) website, and then confirmed by the Conserved Domain search.
3.2 Phylogenetic analyses
To explore the phylogenetic relationship among fungal NCLDV-like sequences, other dsDNA viruses, bacteria, archaea, and eukaryotes, we performed phylogenetic analyses of seven proteins (DNAP, SAM, RPB2, RPB5, RPB1, D5 helicase-primase, and D6/D11-like SNF2 helicase). We used the BLASTp algorithm to search against non-redundant protein database with an e-cut-off value of 0.01 and the fungal virus sequences as queries and selected reference sequences of eukaryotes, bacteria, and archaea. Sequences of representative eukaryotes, bacteria, and archaea used by Guglielmini et al. (2019) were also included in our dataset. Protein sequences were aligned using MAFFT with the L-INS-i strategy, followed by manual edition (Katoh and Standley 2013). The ambiguous regions within the alignments were removed using trimAl (Capella-Gutiérrez, Silla-Martínez, and Gabaldón 2009). The phylogenetic analyses were performed using a maximum likelihood method implemented in IQ-TREE (Nguyen et al. 2015). The best-fit model for each alignment was selected by using the ModelFinder algorithm in IQ-TREE (Kalyaanamoorthy et al. 2017). The branch support values were estimated by using the ultrafast bootstrap (UFBoot) approach with 1,000 replicates (Hoang et al. 2018).
3.3 Co-speciation analyses
To investigate the evolutionary mode of fundsalphaviruses, we detected fundsalphavirus-host co-speciation signal at the level of fungus phylum. Co-speciation analyses were performed by an event-based approach implemented in Jane that compares topologies between host and viral phylogenies (Conow et al. 2010). Briefly, five evolutionary events (co-speciation, duplication, duplication and host switch, loss, and failure to diverge) were assigned with a cost. The number of each event can be calculated by combining five events and finding the best solution with the minimum total cost. We conducted analyses for two event cost schemes (co-speciation–duplication–duplication and host switch-loss-failure to diverge), that is −1-0-0-0-0 (Ronquist 1997; Aiewsakun and Katzourakis 2017) and 0-1-1-2-0 (Charleston 1998; Conow et al. 2010). The congruence between fundsalphavirus and host trees were assessed by statistical tests with two methods, random-tip-mapping method and random-parasite-tree method, with a sample size of 500. The minimum costs generated by random samples were then statistically compared to the original cost. If the original cost was significantly different from random costs, there is co-speciation signal between host-virus phylogeny.
Supplementary Material
Acknowledgements
This work was supported by National Natural Science Foundation of China (31922001 and 31701091) and Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.
Conflict of interest: None declared.
References
- Aiewsakun P., Katzourakis A. (2017) ‘Marine Origin of Retroviruses in the Early Palaeozoic Era’, Nature Communications, 8: 13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arslan D., et al. (2011) ‘Distant Mimivirus Relative with a Larger Genome Highlights the Fundamental Features of Megaviridae’, Proceedings of the National Academy of Sciences United States of America, 8: 17486–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aswad A., Katzourakis A. (2017) ‘A Novel Viral Lineage Distantly Related to Herpesviruses Discovered within Fish Genome Sequence Data’, Virus Evolution, 3: vex016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T. (2009) ‘trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses’, Bioinformatics (Oxford, England), 25: 1972–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charleston M. A. (1998) ‘Jungles: A New Solution to the Host/Parasite Phylogeny Reconciliation Problem’, Mathematical Biosciences, 149: 191–223. [DOI] [PubMed] [Google Scholar]
- Colson P., et al. (2013) ‘Megavirales’, a Proposed New Order for Eukaryotic Nucleocytoplasmic Large DNA Viruses’, Archives of Virology, 158: 2517–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., et al. (2012) ‘Reclassification of Giant Viruses Composing a Fourth Domain of Life in the New Order Megavirales’, Intervirology, 55: 321–32. [DOI] [PubMed] [Google Scholar]
- Conow C., et al. (2010) ‘Jane: A New Tool for the Cophylogeny Reconstruction Problem’, Algorithms for Molecular Biology, 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaroque N., et al. (1999) ‘Persistent Virus Integration into the Genome of Its Algal Host, Ectocarpus Siliculosus (Phaeophyceae)’, Journal of General Virology, 80: 1367–70. [DOI] [PubMed] [Google Scholar]
- Delaroque N., Boland W. (2008) ‘The Genome of the Brown Alga Ectocarpus Siliculosus Contains a Series of Viral DNA Pieces, Suggesting an Ancient Association with Large dsDNA Viruses’, BMC Evolutionary Biology, 8: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C., Gilbert C. (2012) ‘Endogenous Viruses: Insights into Viral Evolution and Impact on Host Biology’, Nature Reviews Genetics, 13: 283–96. [DOI] [PubMed] [Google Scholar]
- Filée J. (2014) ‘Multiple Occurrences of Giant Virus Core Genes Acquired by Eukaryotic Genomes: The Visible Part of the Iceberg?’, Virology, 466-467: 53–9. [DOI] [PubMed] [Google Scholar]
- Gallot-Lavallée L., Blanc G. (2017) ‘A Glimpse of Nucleo-cytoplasmic Large DNA Virus Biodiversity through the Eukaryotic Genomics Window’, Viruses, 9: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial S. A., et al. (2015) ‘50-plus Years of Fungal Viruses’, Virology, 479-480: 356–68. [DOI] [PubMed] [Google Scholar]
- Guglielmini J., et al. (2019) ‘Diversification of Giant and Large Eukaryotic dsDNA Viruses Predated the Origin of Modern Eukaryotes’, Proceedings of the National Academy of Sciences United States of America, 116: 19585–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang D. T., et al. (2018) ‘UFBoot2: Improving the Ultrafast Bootstrap Approximation’, Molecular Biology and Evolution, 35: 518–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S., et al. (2017) ‘ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates’, Nature Methods, 14: 587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013) ‘MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability’, Molecular Biology and Evolution, 30: 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjartansdóttir K. R., et al. (2015) ‘Traces of ATCV-1 Associated with Laboratory Component Contamination’, Proceedings of the National Academy of Sciences United States of America, 112: E925–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Yutin N. (2018) ‘Multiple evolutionary origins of giant viruses’, F1000Research, 7: F1000. [DOI] [PMC free article] [PubMed]
- Koonin E. V., Yutin N. (2010) ‘Origin and Evolution of Eukaryotic Large Nucleo-cytoplasmic DNA Viruses’, Intervirology, 53: 284–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V. (2009) ‘Evolution of Genome Architecture’, The International Journal of Biochemistry & Cell Biology, 41: 298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., et al. (2016) ‘Genomoviridae: A New Family of Widespread Single-Stranded DNA Viruses’, Archives of Virology, 161: 2633–43. [DOI] [PubMed] [Google Scholar]
- La Scola B., et al. (2003) ‘A Giant Virus in Amoebae’, Science, 299: 2033. [DOI] [PubMed] [Google Scholar]
- Legendre M., et al. (2018) ‘Diversity and Evolution of the Emerging Pandoraviridae Family’, Nature Communications, 9: 2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M., et al. (2014) ‘Thirty-Thousand-Year-Old Distant Relative of Giant Icosahedral DNA Viruses with a Pandoravirus Morphology’, Proceedings of the National Academy of Sciences United States of America, 111: 4274–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maumus F., et al. (2014) ‘Plant Genomes Enclose Footprints of past Infections by Giant Virus Relatives’, Nature Communications, 5: 4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meints R. H., et al. (2008) ‘Identification of Two Virus Integration Sites in the Brown Alga Feldmannia Chromosome’, Journal of Virology, 82: 1407–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier A., Claverie J., Ogata H. (2008) ‘Taxonomic Distribution of Large DNA Viruses in the Sea’, Genome Biology, 9: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naccache S. N., et al. (2013) ‘The Perils of Pathogen Discovery: Origin of a Novel Parvovirus-like Hybrid Genome Traced to Nucleic Acid Extraction Spin Columns’, Journal of Virology, 87: 11966–11977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. T., et al. (2015) ‘IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum Likelihood Phylogenies’, Molecular Biology and Evolution, 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M. R., Emerman M., Malik H. S. (2011) ‘Paleovirology—Ghosts and Gifts of Viruses Past’, Current Opinion in Virology, 1: 304–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe N., et al. (2013) ‘Pandoraviruses: Amoeba Viruses with Genomes up to 2.5 Mb Reaching That of Parasitic Eukaryotes’, Science, 341: 281–286. [DOI] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2010) ‘FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments’, PLoS One, 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D., et al. (2004) ‘The 1.2-Megabase Genome Sequence of Mimivirus’, Science, 306: 1344–1350. [DOI] [PubMed] [Google Scholar]
- Ronquist F. (1997) ‘Phylogenetic Approaches in Coevolution and Biogeography’, Zoologica Scripta, 26: 313–322. [Google Scholar]
- Roossinck M. J. (2019) ‘Evolutionary and Ecological Links between Plant and Fungal Viruses’, New Phytologist, 221: 86–92. [DOI] [PubMed] [Google Scholar]
- Tedersoo L., et al. (2018) ‘High-Level Classification of the Fungi and a Tool for Evolutionary Ecological Analyses’, Fungal Diversity, 90: 135–159. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.