Summary
Genomic conflicts may play a central role in the evolution of reproductive barriers. Theory predicts that early-onset hybrid inviability may stem from conflict between parents for resource allocation to offspring. Here we describe M. decorus; a group of cryptic species within the M. guttatus species complex that are largely reproductively isolated by hybrid seed inviability (HSI). HSI between M. guttatus and M. decorus is common and strong, but populations of M. decorus vary in the magnitude and directionality of HSI with M. guttatus. Patterns of HSI between M. guttatus and M. decorus, as well as within M. decorus conform to the predictions of parental conflict: firstly, reciprocal F1s exhibit size differences and parent-of-origin specific endosperm defects, secondly the extent of asymmetry between reciprocal F1 seed size is correlated with asymmetry in HSI, and lastly, inferred differences in the extent of conflict predict the extent of HSI between populations. We also find that HSI is rapidly evolving, as populations that exhibit the most HSI are each others’ closest relative. Lastly, while all populations appear largely outcrossing, we find that the differences in the inferred strength of conflict scale positively with π, suggesting that demographic or life history factors other than transitions to self-fertilization may influence the rate of parental conflict driven evolution. Overall, these patterns suggest the rapid evolution of parent-of-origin specific resource allocation alleles coincident with HSI within and between M. guttatus and M. decorus. Parental conflict may therefore be an important evolutionary driver of reproductive isolation.
Keywords: Speciation, Endosperm, BDMIs, arms race, cryptic species
Graphical Abstract
eTOC
Genomic conflict may play a central role in speciation. Coughlan et al. find that parental conflict may play a role in the evolution of hybrid seed inviability between Mimulus guttatus and a newly discovered species complex M. decorus. Differences in conflict between these lineages may stem from demographic or life history differences.
Introduction
The origin and maintenance of species ultimately depends on the formation of reproductive barriers between populations. Research to date has highlighted the role of ecological and sexual selection in the formation of many pre-zygotic and extrinsic post-zygotic barriers (e.g. [1–4]). However, relatively less is known about the evolutionary forces driving the formation of intrinsic barriers.
The formation of early-onset hybrid inviability is a common and powerful intrinsic reproductive barrier in nature [5,6], especially in placental mammals (e.g. [7–10]) and seed plants (e.g. [11–16]). One hypothesis to explain the evolution of early-onset hybrid inviability is that of parental conflict [17–19]. Parental conflict can arise in non-monogamous systems because maternal and paternal optima for resource allocation to offspring differ [17–19]. Since maternity is guaranteed, selection should favor equivalent resource allocation from mothers to children. However, in non-monogamous systems, selection should favor the evolution of paternally derived alleles that increase resource allocation to offspring [17–19]. Therefore, a co-evolutionary arms race for resource-acquiring paternal alleles and resource-repressive maternal alleles can evolve. Reproductive isolation can manifest if populations have evolved at different rates and/or with different genetic responses to conflict, resulting in a mismatch between maternal and paternal alleles in hybrids. While there is some evidence for parental conflict dynamics within species [e.g. 20–22], the role of parental conflict in the evolution of reproductive isolation requires further study.
Parental conflict poses an enticing hypothesis for the evolution of early-onset hybrid inviability, because early-onset hybrid inviability is common in systems that partition resources after fertilization (e.g. seed plants and placental mammals; [23,24]) relative to systems that do not (e.g. birds and frogs; [25–27]). It is also consistent with the observation that many examples of early-onset hybrid inviability are asymmetric [e.g. 7,13], and arise from issues of excessive or limited growth, consistent with the idea that these incompatibilities involve parent-of-origin resource allocation alleles (i.e. alleles whose effects are mediated by whether they are maternally or paternally inherited; [7,13,14]). Yet, hybrid inviability may also evolve as a byproduct of other selective pressures (e.g. ecological differences), through neutral processes (e.g. gene duplication; [28]), or other sources of conflict (e.g. TE/repressor dynamics [29,30]). As most studies of early-onset hybrid inviability are based on a single population pair, [e.g. 7,10, 12,13], we lack a comparative framework for understanding whether parental conflict can shape early-onset hybrid inviability, and what factors affect the magnitude and direction of inviability.
In seed plants, parental conflict is thought to manifest in the endosperm- a nutritive tissue that is essential for proper embryo development [31]. Like the mammalian placenta, endosperm represents a direct nutritive connection between developing offspring and mother. Unlike placenta, endosperm is a triploid tissue that is comprised of 2:1 maternal:paternal genetic composition. In inter-ploidy crosses-where hybrid seed inviability (HSI) has been best studied- departures from this 2:1 ratio often result in irregular endosperm growth, and seed death, even if the embryo itself is viable ([11, 32; ‘triploid block’- 31, 33]. In Arabidopsis inter-ploidy crosses, improper dosage of normally imprinted genes in the endosperm cause endosperm irregularities and ultimately, inviability [12, 34]. Because of the shared phenotypic defects in HSI between intra-diploid and inter-ploidy crosses [e.g. 35], many researchers use ‘effective ploidy’ or ‘Endosperm Balance Number’ (EBN) as a way to quantify the strength of conflict of a given species relative to another, regardless of actual ploidy [36]. While EBNs can be used to describe differences in both inter-ploidy and intra-diploid crosses, the underlying genetic mechanisms between these two types of crosses may differ. While inter-ploidy HSI can be caused by differences in dosage of maternal versus paternal alleles as a result of whole genome duplication, intra-diploid HSI must be caused by either sequence change or duplication of specific genes. Whether parental conflict is responsible for genic changes that contribute to HSI among diploids remains relatively unexplored [but see 37].
The strength of reproductive isolation often varies between populations [38–45]. Leveraging this natural variation can help researchers to test the potential evolutionary drivers responsible for different reproductive barriers. If parental conflict drives the evolution of HSI, we would predict that reciprocal F1 seeds should show differences in size, indicating parent-of-origin effects on growth. Secondly, if these parent-of-origin effects on growth cause reproductive isolation, then the degree of asymmetry in reciprocal F1 size should correlate with inviability. Lastly, differences in the strength of conflict between populations (e.g. differences in EBN between populations) should predict the degree of reproductive isolation in subsequent crosses.
We use the Mimulus guttatus species complex as a system to study the evolution of HSI. The Mimulus guttatus species complex is a diverse group of wildflowers that are well known for adaptations to extreme ecological conditions [e.g. 46–49], mating system evolution [50–53], and life history variation [54–57], and is a model system for ecology, evolution, and genetics [58,59]. The group consists of both annual and perennial species, and while many annual species have been well studied [e.g. 14, 48, 51, 60–62], studies of perennials in this group have been largely limited to morphological anecdotes [63, 64]. Thus we lack a basic understanding of the diversity of species in one of the most well studied, non-crop plant systems [65].
Here we combine a common garden experiment, population genomics, and quantification of reproductive isolation to reveal the presence of a cryptic species complex- M. decorus- nested within the M. guttatus species complex. All lineages of M. decorus exhibit a largely outcrossing morphology, therefore increasing the chance of multiple paternity and conditions for parental conflict to evolve. Using both crossing and developmental surveys, we find that reproductive isolation between these cryptic species is largely conferred via HSI. We leverage diversity in the extent and direction of HSI in this group to assess the role of parental conflict in the evolution of HSI.
Results
M. decorus is a phenotypically cryptic, but genetically distinct group of species
In order to assess the phenotypic and genetic divergence between perennial members of the M. guttatus species complex we combined previously published genomic datasets with new re-sequencing data and performed a common garden experiment. While several perennial morphological variants have been described based on subtle phenotypic differences [63,64], it remains unknown whether these morphological variants comprise reproductively isolated species or are simply localized morphological oddities. We find that one such morphological variant- M. decorus- is genetically unique from other perennial variants in the M. guttatus species complex, despite minimal phenotypic divergence (Figure 1; also see [66]). In contrast, populations of annual, inland perennial, and coastal perennial M. guttatus show minimal genetic separation despite substantial phenotypic differentiation among life histories types (Figure 1, Figure S1; also see [67]). Mimulus decorus can further be categorized into three genetic clusters: a northern diploid that occur in the central Cascade Mountains of Oregon, a southern diploid group from the southern Cascades, and several tetraploids that occur primarily in the northern end of the range (Figure 1, Figure S1).
Figure 1: Phenotypic and genetic description of focal members of the M. guttatus species complex.

(A) Geographic sampling of genetic samples from M. guttatus and M. decorus. Inland versus coastal perennials were not distinguished in the previous study and are thus both indicated by darker pink points in this panel. (B) Legend of the colors used to denote each species (C) NJ tree constructed using 4-fold degenerate sites and rooted using M. dentilobus. (D) PCA of 15 morphological characteristics for perennials within the M. guttatus species complex, as well as perennial M. tilingii and annual M. guttatus. Trait codes as follows: FT-days to first flower, FN- node of first flower, ST-stem thickness, CW- corolla width, TL- tube length, CL- corolla length, LL- leaf length, LW- leaf width, IL1- first internode length, IL2- second internode length, NS- number of stolons, SL- stolon length, SW- stolon width, SBL- side branch length, NSB- number of side branches. (E) PCA of re-sequencing data for focal members of the M. guttatus species complex. In both PCAs the percent of total variance explained by that PC is indicated in parentheses. Further PCA and NJ analyses are given in Figure S1.
Neighbor-joining trees also place M. decorus as a monophyletic group that is is sister to M. guttatus, with the more divergent M. tilingii as an outgroup (Figure 1; Figure S1). Divergence between M. decorus and M. guttatus is similar to that of the recently diverged M. guttatus and M. nasutus (genome-wide dxy using 4-fold degenerate sites between M. guttatus and M. decorus is 5.7% and 5.8% for the northern and southern clades of M. decorus, respectively, and 5.8% between M. guttatus and M. nasutus; Table S3; [52]). In contrast, divergence between more distantly related species M. guttatus and M. tilingii is 6.5% (Table S3; [15]) Using the same approach as [52], we estimate the split between M. guttatus and M. decorus to be approximately 230 kya (i.e. t = πguttxdec - πdec/2μ, where μ is estimated to be 1.5 × 10−8. Therefore, t = 0.007/2μ = ~233 ky). Northern and southern clades of M. decorus may thus represent a relatively recent split from M. guttatus, and/or have experienced substantial introgression with M. guttatus post secondary contact. In order to determine if this population genetic structuring is a consequence of reproductive isolation between these taxa, we next assessed crossing barriers between M. guttatus and M. decorus by completing a range-wide crossing survey.
Hybrid seed inviability is a significant, but variable reproductive barrier between M. decorus and M. guttatus
There is significant HSI between M. guttatus and each genetic clade of M. decorus (Figure 2; RI is 0.53, 0.39, and 0.38 for M. guttatus and northern, southern, and tetraploid M. decorus, respectively). However, we find no evidence for a reduction in seed set in interspecific crosses relative to intraspecific crosses (Figure S3). Thus, while there is limited reproductive isolation conferred by pollen pistil interactions, HSI imposes a substantial reproductive barrier.
Figure 2: Geographic distribution of hybrid seed inviability between M. decorus and M. guttatus.

(A) Sampling locales of 19 populations of M. decorus that were crossed to four M. guttatus populations reciprocally. Points are colored as in Figure 1, but points that are lighter in hue and outlined in grey are populations that exhibit limited reproductive isolation with M. guttatus. (B) Crossing patterns for four populations of M. decorus, representing the four main crossing phenotypes observed (from top left to bottom right): 2 populations of tetraploid M. decorus that show minimal HSI with M. guttatus, most tetraploid populations of M. decorus exhibits an asymmetric barrier with M. guttatus, wherein seeds are largely inviable when M. guttatus is the maternal donor, northern M. decorus exhibit strong, asymmetric inviability when M. decorus is the maternal parent, southern M. decorus exhibit intermediate -strong asymmetric inviability when M. guttatus is the maternal donor. For all crosses, maternal parent is listed first. C) representative viable hybrid seeds, D) representative inviable hybrid seeds. Figure S2 depicts population-level patterns of HSI for each population of M. decorus and M. guttatus. Figure S3 depicts patterns of HSI, as well as total seed set for inter- and intraspecific crosses for each genetic clade of M. decorus.
We find substantial variation in HSI throughout the range of M. decorus. Northern, southern, and tetraploid clades of M. decorus differed significantly from each other in the degree and directionality of HSI with M. guttatus (clade effect: χ2=6.69, df=2, p=0.035; as well as a clade × cross type interaction: χ2=135.56, df=6, p<0.0001; Figure S2 & S3). Northern M. decorus populations tend to produce inviable seeds when they are the maternal donors in crosses with M. guttatus (Figure 2; Figure S3), while southern M. decorus populations produce more inviable seeds when they are the paternal donor in crosses with M. guttatus (Figure 2; Figure S3). Tetraploids exhibit stronger HSI when they are the paternal donor, although we note two populations with limited HSI (i.e. Figure 2, Figure S3). In contrast, populations of M. guttatus did not drastically differ in their average HSI when crossed with M. decorus (Figure S2).
Patterns of hybrid seed inviability show parent of origin effects on growth and development
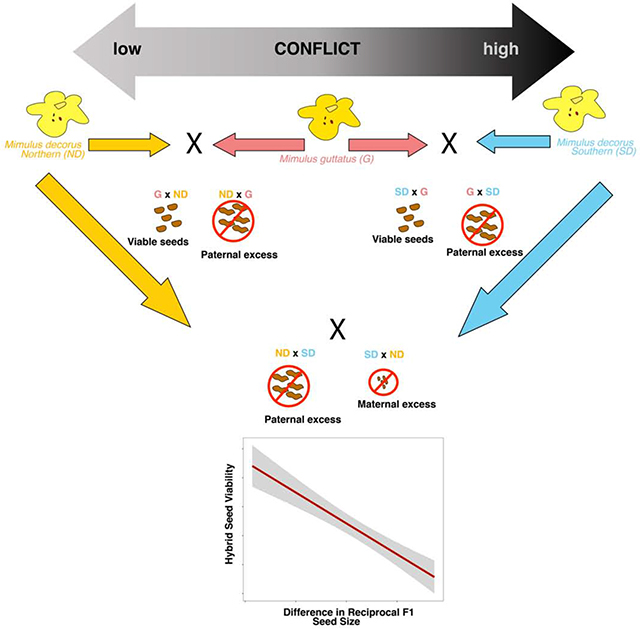
We sought to leverage the diversity of HSI in this system to test for a role of parental conflict in the evolution of HSI. If parental conflict was driving HSI, we would first predict that reciprocal F1 seeds should show differences in size, indicating parent-of-origin effects on growth. Secondly if these parent-of-origin effects on growth cause reproductive isolation, then the degree of asymmetry in reciprocal F1s size should correlate with inviability.
To determine if HSI is associated with parent-of-origin effects on growth, we measured reciprocal F1 seed size and completed a survey of developing hybrid and pure-species seeds for intra-diploid crosses only. HSI was always paired with significant differences in reciprocal F1 hybrid seed size (Figure 3). Northern M. decorus produced F1s with larger seeds when they were the maternal donor, while southern M. decorus produced larger F1 seeds when they were the paternal donor in crosses with M. guttatus (Figure 3; northern clade crosses: χ2==296, df=3, p<0.0001; southern clade crosses: χ2==298, df=3, p<0.0001). The level of asymmetry in seed size was correlated with the level of asymmetry in viability (Figure 3; r2=−0.69, df=22, p<0.0001), such that the reciprocal F1 seed that is larger tended to be the seed that was inviable. Thus, HSI is strongly associated with parent-of-origin effects on size; a key prediction of parental conflict.
Figure 3: A test for conflict using reciprocal F1 seed sizes and symmetries.

(A) Seed sizes for reciprocal F1s and parents for northern M. decorus (yellow) and southern M. decorus (blue) when crossed to M. guttatus (pink). Letters above denote significantly different groups, wherein comparable groups are denoted by color. (B) Seed viability symmetry between reciprocal F1s as a function of seed size symmetry between reciprocal F1s. Color of the points denote the line of M. guttatus that was used in the cross. The black line indicates the linear regression line of best fit, and grey denotes the 95% confidence intervals of the linear regression. (C) Parent of origin effects on endosperm proliferation between independent incidences of hybrid seed inviability at 8 Days After Pollination. G= M. guttatus, D= M. decorus. Maternal parent is listed first. Tissues are labeled in panel (a): em=embryo, en=endosperm, sc=seed coat. Arrows denote the location of the developing embryo; red stars indicate that these seeds will eventually become inviable. Full developmental surveys between M. guttatus and each clade of M. decorus are available in Figure S5.
To determine if reciprocal F1 size differences were associated with inappropriate development of endosperm we completed developmental surveys of reciprocal F1 seeds from M. guttatus crossed to both a focal northern M. decorus accession (IMP) and a focal southern M. decorus accession (Odell Creek). Reciprocal F1s between each set of crosses show substantial and parallel parent-of-origin effects on endosperm growth based on seed fate (i.e. whether the seed will remain viable or not; Figure 3; Figure S5). Hybrid seeds that remain viable (e.g. M. guttatus × northern M. decorus and southern M. decorus × M. guttatus, with the maternal parent listed first) display a precocious development, with small endosperm cells that are quickly degraded by the embryo (Figure 3; Figure S5), and is almost entirely gone by by 14 Days After Pollination (DAP; Figure S5). In contrast, hybrid seeds that will eventually become inviable (i.e. northern M. decorus × M. guttatus and M. guttatus × southern M. decorus, with the maternal parent listed first) exhibit a chaotic endosperm growth, producing fewer, but larger and more diffuse endosperm cells (Figure 3; Figure S5). Embryos in this direction of the cross remain small, relative to both parents and the reciprocal hybrid, and are usually degraded by 14 DAP (Figure 3; Figure S5). While the endosperm exhibits parent-of-origin specific growth defects, additional incompatibilities in the embryo cannot be ruled out, as embryo rescues from all crosses (including intraspecific crosses) were generally unsuccessful. Thus, HSI in this system is associated with parent-of-origin specific growth defects of the endosperm that are replicated across independent incidences of HSI, in line with parental conflict.
Our last prediction is that differences in the strength of conflict between populations (e.g. differences in EBN between populations) should predict the degree of reproductive isolation in subsequent crosses. In order to test this prediction, we must first infer EBNs for M. guttatus and each clade of M. decorus. Other researchers have used mating system and/or ploidy to infer EBNs, however the species we describe here are diploid and exhibit highly outcrossing morphologies (Figure 5). We therefore use a similar approach to the originators of the concept of EBNs, wherein the EBNs of focal species are inferred from crosses to a common tester line. Predictions can then be made about subsequent crosses between focal species based on these inferred EBNs [32,36]. Using M. guttatus as our common tester, we predict that northern populations of M. decorus likely have a lower EBN (and therefore have experienced a history of weaker conflict) than M. guttatus, as they are unable to prevent M. guttatus paternal alleles from exploiting maternal resources and inducing growth. In contrast, southern populations of M. decorus likely have a higher EBN (and therefore have experienced a history of stronger conflict) than M. guttatus, as this cross exhibits a paternal excess phenotype when southern M. decorus is the paternal donor. We next sought to determine whether these designations of EBNs were predictive of reproductive isolation in subsequent crosses using untested intra-diploid and inter-ploidy crosses.
Figure 5: Potential correlates of EBN variation in M. decorus and M. guttatus.

(A) No difference in the average stigma-anther separation between different clades of M. decorus (p=0.29), but (B) significant variation in genome-wide π between northern and southern clades of M. decorus, as well as between northern M. decorus and M. guttatus. Error bars in panel (B) represent standard error. G= M. guttatus, N= northern M. decorus, and S= southern M. decorus. Complete pairwise measurements of Fst, Dxy, and π for all species are listed in Table S3.
First, we crossed all diploid accessions of M. decorus to two focal populations; a northern, weaker conflict/ lower EBN M. decorus population (IMP) and a southern, stronger conflict/ high EBN M. decorus population (Odell Creek). Secondly, we crossed all tetraploid accessions to each of these focal diploid populations. Given our predicted differences in conflict between diploid clades of M. decorus, we can form two distinct predictions: firstly, diploid populations of M. decorus that exhibit the largest difference in inferred EBN should exhibit the most HSI, and HSI should be accompanied by reciprocal F1 seed size differences (wherein seeds are larger when lower EBN species are the maternal parents). Secondly, tetraploid populations of M. decorus should exhibit strong reproductive isolation with low EBN northern M. decorus populations (with accompanying reciprocal F1 seed size differences, wherein seeds are larger when northern M. decorus is the maternal parent), while high EBN southern M. decorus populations should exhibit little or no HSI when crossed to tetraploid M. decorus (and minimal seed size differences between reciprocal F1s). We also crossed all four populations of M. guttatus used above in all possible combinations to determine if alleles that contribute to HSI were naturally segregating throughout M. guttatus.
In line with our predictions, we find nearly complete reproductive isolation between northern and southern clades of M. decorus (Figure 4; IMP crossed to southern clade: χ2=12513, df=3, p<0.0001; Odell Creek crossed to northern clade: χ2==2321.6, df=3, p<0.0001), but no reproductive isolation within groups (Figure S4). In addition, northern M. decorus show complete reproductive isolation via HSI with tetraploid M. decorus (Figure 4; χ2=4107, df=3, p<0.0001), but southern M. decorus show no HSI with tetraploid M. decorus (χ2==1.69, df=3, p=0.6). We note that geographically and phenotypically distinct populations of M. guttatus do not exhibit any signs of HSI (F=0.772, df=1, p=0.39; Figure S4).
Figure 4: Assessing whether inferred EBNs are predictive of HSI in intra-diploid and inter-ploidy crosses.
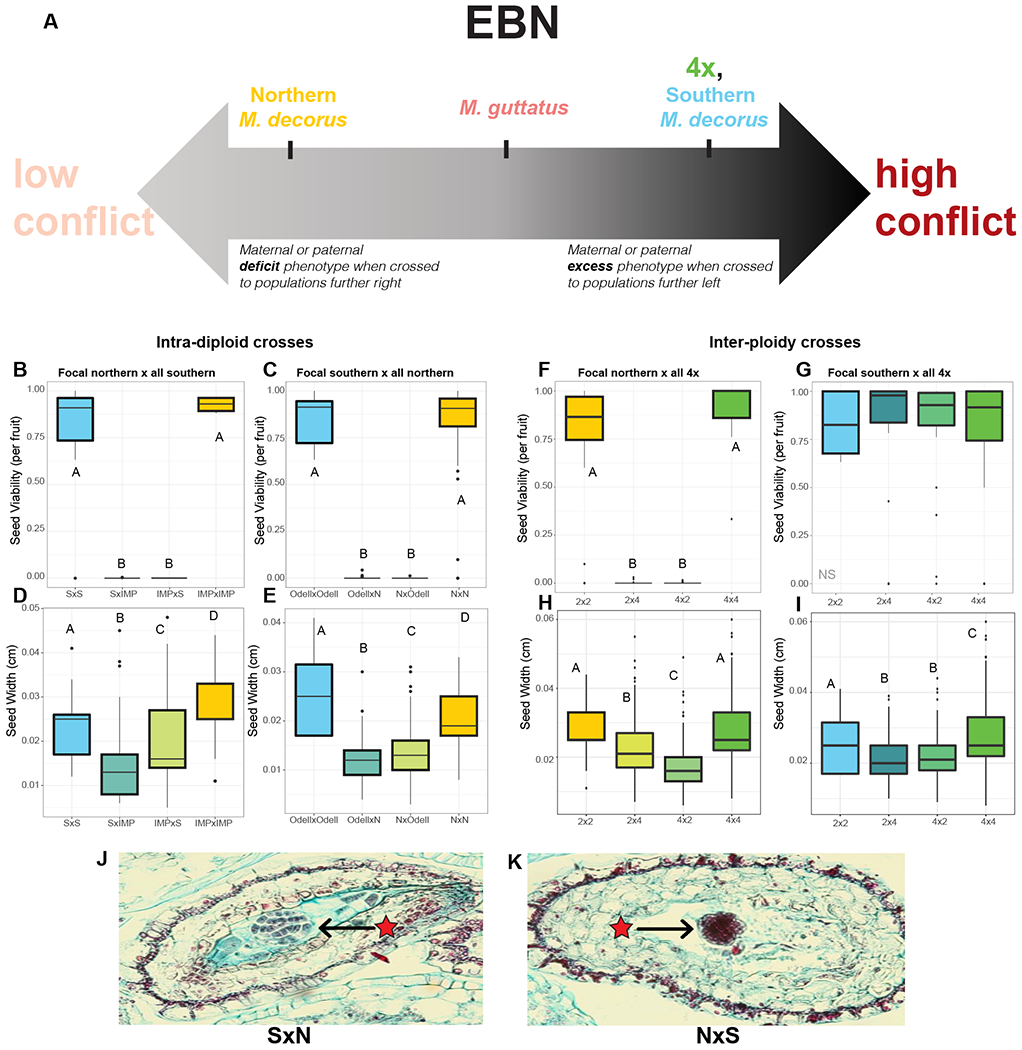
(A) A carton representation of the variance in EBN (i.e. our inference of the historical level of conflict), where further to the right denotes higher EBN/more conflict.
Proportion viable seed between (B) the focal northern (e.g. IMP, low conflict) and all southern (high conflict) populations (C) the focal southern (e.g. Odell Creek, high conflict) and all northern (low conflict) populations. Seed sizes for crosses between (D) the focal northern and all southern populations, (E) the focal southern and all northern populations. Crosses between all tetraploid accessions and (F) the focal northern accession and (G) the focal southern accession. Seed sizes for crosses between all tetraploids and (H) the focal northern accession and (I) the focal southern accession. Crosses are denoted with the maternal parent first, S= Southern M. decorus, N= Northern M. decorus. Letters denote significantly different groups. (J,K) Developing seeds at 8 DAP for crosses between southern and northern clades of M. decorus (Odell Creek and IMP, respectively). (J) Odell Creek x IMP, (K) IMP x Odell Creek. Maternal parent is listed first. Crosses within clades, as well as within M. guttatus crosses are shown in Figure S4.
In all crosses with strong HSI, reciprocal F1s vary in seed size in the direction predicted by parental conflict (Figure 4; IMP crossed to southern clade: χ2=79, df=3, p<0.0001; Odell Creek crossed to northern clade: χ2=301, df=3, p<0.0001, northern clade M. decorus crossed to tetraploids: ; χ2=349, df=3, p<0.0001). That is, when higher EBN taxa served as paternal donors F1 seeds were larger, while F1 seeds were smaller when higher EBN taxa severed as the maternal donor. These differences in reciprocal F1 seed size did not occur in crosses within clades (Figure S4), nor in crosses between taxa with more evenly matched EBNs (e.g. southern M. decorus and tetraploid M. decorus: p=0.7).
In addition, developmental surveys of crosses between southern and northern clades of M. decorus exhibited substantial defects in endosperm development in the directions predicted by parental conflict. These defects were more extreme than in crosses between M. guttatus and either clade of M. decorus (Figure 3, Figure 4). When northern M. decorus (i.e. IMP) was the maternal parent, hybrid seeds produced large and diffuse endosperm cells, while when southern M. decorus (i.e. Odell Creek) was the maternal parent, hybrid seeds exhibited precociously developing endosperm cells, but hardly any endosperm tissue. Crosses in both directions often had a relatively healthy globular stage embryo at 8 DAP.
Hybrid seed inviability is rapidly evolving within the M. guttatus species complex, and inferred levels of conflict are positively associated with nucleotide diversity
Despite being each others’ closest extant relative, northern and southern clades of M. decorus have the most extreme differences in inferred EBN and also the most reproductive isolation via HSI than either do to M. guttatus. Given that the northern and southern clades form a monophyletic group that diverged from M. guttatus roughly 230 kya (Figure 1), HSI appears to be rapidly evolving in this group. The rapid evolution of alleles associated with parent-of-origin effects on growth is consistent a parental conflict driven arms race of maternal and paternal resource acquisition alleles.
The rapid evolution of EBN and HSI in this group suggests that these species vary in some key factor influencing the amount of variance in paternity. Transitions from outcrossing to a self-fertilization are most commonly used to explain differences in EBN between populations [21, 37, 68]. While all species presented here have highly outcrossing morphologies (Figure 5), it is possible that lineages of M. decorus vary in other aspects of their demography in ways that would influence the amount of variance in paternity that plants experience in nature. For example, these populations may vary in the rate of clonal reproduction via stolon production relative to sexual reproduction, the levels of bi-parental inbreeding brought on by small population sizes, or historical differences in population sizes during the last glacial maxima. We tested whether variation in EBN was associated with genome-wide diversity, as a proxy for effective population size. Using a 4-fold degenerate sites across the genome, we find that inferred EBNs scale positively with genome-wide π, wherein the northern clade of M. decorus is the least diverse group, and southern M. decorus is slightly more diverse than M. guttatus (Figure 5; Table S3). Therefore, differences in inferred conflict between these populations may stem from differences in demographic or life history factors other than transitions from outcrossing to self-fertilization.
Discussion
Here we use a combination of population genomics, common garden experiments, and crossing surveys to describe a cryptic species group- M. decorus- nested within the M. guttatus species complex. We find that M. decorus and M. guttatus are largely reproductively isolated via HSI, but the magnitude of reproductive isolation and the direction of the cross that results in inviable hybrid seeds varies throughout the range of M. decorus. We leverage variation in HSI between M. guttatus and different genetic clades of diploid M. decorus to assess whether patterns of HSI can be explained by parental conflict. We find that all three predictions of parental conflict are met in this group, namely: reciprocal F1s show differences in size, the strength of asymmetry in HSI is correlated with asymmetric growth defects between reciprocal F1s, and that inferred EBNs can predict the outcome of crosses. In addition, HSI appears to be rapidly evolving in this group, as the two species that exhibit the most HSI are each others’ closest relative. Lastly, the inferred strength of conflict for each clade of M. decorus and M. guttatus scale positively with the level of within-species π, suggesting that demographic or life history factors other than transitions from outcrossing to self-fertilization may affect the speed at which parental conflict driven arms races may evolve.
Phenotypic and genetic diversity within perennials of the M. guttatus species complex
Despite morphological similarity, M. decorus is genetically unique from and exhibits strong reproductive isolation with M. guttatus. Work in the M. guttatus species complex has focused on the diversity of annual forms [e.g. 48, 51, 61, 69–72], and differences between annuals and perennials [e.g. 55, 57, 73]. Relatively less work has explored diversity within perennials of the complex. Here we find substantial cryptic species diversity within perennials of one of the most well studied, non-crop, plant model systems. Not only do we find that M. decorus is a genetically distinct and reproductively isolated species from M. guttatus, but it likely represents three reproductively isolated taxa: a diploid northern clade that produces inviable seeds when it is the maternal donor in crosses with M. guttatus, a diploid southern clade that produces inviable seeds when it is the paternal donor in crossed with M. guttatus, and at least one tetraploid taxon. This highlights a potential difference between annuals and perennials in the M. guttatus species complex: annual species generally exhibit both ecological and phenotypic divergence, but very few exhibit substantial post-zygotic reproductive isolation with M. guttatus. In contrast, M. decorus is phenotypically and ecologically very similar to perennial M. guttatus, but exhibits relatively strong post-zygotic reproductive isolation.
HSI is a common and substantial reproductive barrier between M. guttatus and M. decorus. This contrasts with other, more geographically restricted intrinsic post-zygotic barriers in this complex [e.g. 38, 40, 43, 74, 75]. Because of its commonality, strength, and the fact that it stops gene flow in the first generation of hybridization, HSI may represent an important barrier in nature. Mimulus decorus exhibits striking variation throughout the range in the extent and direction of HSI, which allows us to dissect, in part, the evolutionary drivers of HSI.
Patterns of hybrid seed inviability conform to the predictions of parental conflict
We tested three predictions of the role of parental conflict in HSI: Firstly, reciprocal F1s will show differences in size, secondly, size differences between F1s will correlate with the degree of reproductive isolation, and lastly, differences in inferred levels of parental conflict are predictive of reproductive isolation in subsequent crosses. We find support for each of these predictions in patterns of HSI between M. guttatus and diploid accessions of the morphological variant M. decorus, highlighting the potential role of parental conflict in HSI in this group.
Evidence for parental conflict in HSI between diploid species pairs has been found in some systems (e.g. Capsella [13, 37]; Arabidopsis [35]; wild tomato [16]; peromyscus mice [7]; and dwarf hamsters [10]). In both Capsella and Mimulus, which show asymmetric inviability, a paternal-excess phenotype appears more lethal [13], which is consistent with inter-ploidy crosses (reviewed in [24]). In systems with symmetric HSI (e.g. in Arabidopsis lyrata and A. arenosa, [35]; wild tomato species, [16]; and the northern x southern M. decorus crosses here), parent-of-origin effects on growth have been shown, although both directions of the cross are often smaller than either parent, perhaps suggesting that in cases of stronger HSI, endosperm defects are more substantial earlier in development, and thus seeds are aborted earlier. In many of these systems, inviability is caused by malformation of endosperm, rather than an incompatibility manifesting in the embryo [13, 35, 37], although in both wild tomatoes and Mimulus, HSI tends to produce both malformed endosperm and also early aborting embryos [e.g. 14, 16], which may be related to differences in endosperm developmental programs between Brassica and Solanum/Mimulus.
While that patterns of HSI in the Mimulus guttatus species complex conform to the predictions of parental conflict, it is possible that evolutionary explanations other than parental conflict could explain HSI. It has been hypothesized that mismatches in small RNAs carried by male and female gametes may result in de-repression of TEs in developing seeds, as imprinted genes are often associated with TEs [29, 76]. However, unlike parental conflict, the TE hypothesis does not predict asymmetries in growth between reciprocal F1s, as we have found here. In addition, the TE and parental conflict hypotheses are not mutually exclusive; TEs may provide the proximate, molecular mechanism by which genes are imprinted during endosperm development, while parental conflict ultimately drives the evolution of imprinted resource allocation alleles [31]. A molecular genetic dissection of this incompatibility will be needed to fully understand the proximate and ultimate causes of HSI.
Hybrid seed inviability is rapidly evolving
HSI between northern and southern clades of M. decorus highlights the rapidity at which HSI has evolved in this group. These two diploid species are morphologically almost indistinguishable, inhabit similar habitats, and are each others’ closest relative. Despite this, HSI between northern and southern M. decorus is stronger than HSI between either clade of M. decorus and M. guttatus. While parental conflict would predict rapid evolution of maternal and paternal resource allocation alleles [17,19], the variance in the levels of conflict between northern and southern clades of M. decorus is curious. Much work on the rate of evolution of HSI due to parental conflict focuses on differences between outcrossing and highly selfing species, or between species of differing ploidies [i.e. 11, 13, 24, 35, 37, 68, 77]. Both the southern and northern clades of M. decorus are diploid, and exhibit highly outcrossing floral morphologies (i.e. large corollas and significant stigma/anther separation; Figure 5). One possibility is that differences in conflict may be due to differences in effective population size between M. guttatus and each clade of M. decorus. This could be caused by a number of factors, including variance in the rates of clonal reproduction via stolon production, bi-parental inbreeding, or historical population sizes in glacial refugia. Like transitions to selfing, each of these factors can affect the effective number of fathers and therefore the variance in paternity in natural populations, ultimately changing the strength of selection due to parental conflict. Patterns of nucleotide diversity support this hypothesis; northern M. decorus is substantially less genetically diverse than either M. guttatus or southern M. decorus, while southern M. decorus is slightly more genetically diverse than M. guttatus. Observationally, we also note that populations of northern M. decorus tend to be much smaller than populations of southern M. decorus. However, more detailed demographic and population genomic analyses are needed to investigate the demographic and life history differences that are ultimately responsible for differences in conflict between perennial members of this complex.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for materials, resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jenn Coughlan (jcoug@email.unc.edu). Seeds from field collections and experimental crosses are available by request to the Lead Contact.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The Mimulus guttatus species complex
The Mimulus guttatus species complex is a morphologically, ecologically, and genetically diverse group [58,59]. The species complex is home to several known annual species, as well as many perennial morphological variants. In fact, M. guttatus itself comprises several morphological variants, including two inland perennials (M. guttatus sensu stricto, and M. decorus), and a coastal perennial (M. grandis), that are phenotypically differentiated by subtle distinctions in corolla length, leaf shape, and the types, densities and locations of trichomes [63,64]. We refer to these as inland M. guttatus, M. decorus, and coastal M. guttatus, respectively. Mimulus tilingii is the most divergent perennial species, and represents its own species complex [15, 63, 64]. Little is known about the phenotypic, genetic relationships and reproductive barriers between morphologically described perennials of the M. guttatus species complex.
Plant rearing and common garden conditions
For the phenotypic survey and crossing survey described below, we grew plants in a common garden in the Duke Greenhouses under long day conditions (18h days, 21C days/18C nights). Between 1–5 maternal families with 5 replicates/family for each of six populations of annual M. guttatus, seven populations of coastal perennial M. guttatus, 16 populations of inland perennial M. guttatus, 18 populations of M. decorus and seven populations of M. tilingii were grown. Seeds were first cold stratified for one week on moist Fafard 4P soil then transferred to the greenhouses. Germinants were transplanted on the day of germination and phenotyped on the day of first flower.
METHOD DETAILS
Phenotypic survey
To assess the degree of phenotypic differentiation among members of the M. guttatus species complex, we use data from the common garden experiment described in [66] Briefly, on the day of first flower, we measured 15 morphological traits (days from germination to first flower, node of first flower, total corolla length, corolla tube length, corolla width, stem thickness, leaf length and width, internode length between the cotyledons and first true leaf, internode length between the first and second pair of true leaves, the number of stolons, length of the longest stolon, width of the longest stolon, number of side branches, length of the longest side branch). We also assessed stigma-anther separation on individuals from each maternal family of M. decorus.
Population genetics survey
To determine the genetic relationship among perennials of the M. guttatus species complex, we leveraged previously published whole genome re-sequencing [78] and GBS data [67] with new re-sequence data for several members of the M. guttatus species complex (outlined in Table S1). In total, we include genomes from 222 accessions: 199 M. guttatus (113 perennials, 74 annuals, and 12 individuals of unknown or intermediate life history), two additional perennial species, including 15 M. decorus and two M. tilingii accessions, and five annual M. nasutus accessions. We also included a single accession of M. dentilobus as the outgroup. For the whole-genome re-sequencing data, all genomic DNA was extracted from young bud or leaf tissue, and 150bp, paired-end read Illumina libraries were made by either the Duke Sequencing facility or by using Illumina Nextera DNA kit. For all sequencing, samples were multiplexed by adding individual barcodes during library construction and pooling samples with equal molar amounts. For the libraries constructed using Illumina Nextera DNA kits, up to 24 samples were pooled and run on an Illumina HiSeq 2500 Rapid-Run platform [as in 78], while for the libraries made by the Duke Sequencing Facility, 11 libraries were pooled, then sequenced on an Illumina HiSeq 4000 platform. Samples had an average of 15x coverage (range: 3.3x-51.5x Table S1).
Determining genome size and ploidy
We performed a combination of flow cytometry and chromosome squashes to assess ploidy variation within M. decorus. We surveyed 1-2 individuals per population of M. decorus to determine total genomic content using flow cytometry [outlined in 79]. Briefly, we chopped freshly collected, young leaf tissue in Cystain UV Precise P buffer for each individual, as well as an internal standard, Arabidopsis thaliana (2n=2x, 2C=0.431pg; [80]). Samples in buffer were filtered through a 40 μm, then 20 μm pore-sized nylon mesh. Shortly before analysis, we added 0.1mg/ml of 4′,6-diamidino-2-phenylindole (DAPI). Nuclei stained for at least ten minutes before being run on a Becton Dickinson LSRII flow cytometer. Total DNA content was calculated by the following equation:
Total genomic content ranged from 2C= 0.61- 3.06 pg (Figure S3). To confirm that larger genomic content corresponded with changes in ploidy, we performed meiotic chromosomal squashes on select populations that spanned the range of genome size (0.67pg, 1.06pg, 1.22 pg, 1.43 pg, 2 pg, and 2.96 pg). From these squashes, all individuals with genome sizes <1.22pg were found to be diploid (2n=2x=14), while all individuals with genomes between 1.43-2.96pg were found to be tetraploid (2n=4x=28). As meiotic chromosomal squashes were not possible for all accessions due to constraints of both labor and plant material, we inferred ploidy for the remaining samples based on these ranges, wherein individuals with a genome size of 1.2pg or less were classified as diploid, and those with a genome size of 1.43 or higher were classified as tetraploid. Two accessions (HJA and Kink Creek) exhibited genome sizes that were intermediate to our cutoffs (1.25pg and 1.41pg, respectively). However, these accessions tightly clustered with northern diploids in our genomic PCA and are in close geographic proximity to all other northern diploids. In contrast, almost all of the tetraploids sampled were from more northerly latitudes and formed a separate genetic cluster in a PCA. We therefore treat these two accessions as northern diploids. We note that our results do not qualitatively change whether these two samples are retained or omitted from analyses.
Crossing survey between M. guttatus and M. decorus
To assess hybrid seed inviability between perennials in this group we crossed 19 populations of M. decorus to an average of 4 populations of M. guttatus reciprocally, as well as within population crosses, with an average of 13 replicates per cross (ranging from 2-37 replicate crosses, totaling 1020 fruits; Table S1 & S2). The 19 populations of M. decorus represented all three genetic clades, including: five northern M. decorus populations, two southern M. decorus populations, and twelve tetraploid populations. The four populations of M. guttatus that were used maximize the phenotypic and genetic diversity of the species and include one annual, two inland perennials, and one coastal perennial population. Due to the sheer number of crosses, individuals were used for multiple crosses and acted as both seed and pollen parents. For each fruit, we calculated the proportion of viable seeds, assessing viability based on visible malformations. We confirmed inviability by performing germination assays (100 seeds plated across 5 replicate plates of 0.6% agar for each cross. Plates were cold stratified for one week, then transferred to warm, long days in the Duke growth chambers. Final germination was recorded after three weeks). Results based on germination assays and morphologically assessed HSI agree qualitatively (r2= 0.66, df=137, p<0.0001; Figure S4). We therefore focus on morphologically determined HSI (results based on germination proclivity are presented in the supplement). We calculated reproductive isolation [2]
and the symmetry of reproductive isolation:
Where ‘G’ and ‘D’ refer to M. guttatus and M. decorus, respectively. These values are bounded between −1 and 1. Values closer to −1 indicate an asymmetry where seeds fail when M. decorus is the maternal donor and values closer to 1 indicate an asymmetry where seeds fail when M. guttatus is the maternal donor, and values close to 0 indicate little asymmetry (but do not denote the severity of HSI).
Assessing the role of parental conflict in hybrid seed inviability
Each population of M. guttatus exhibited a consistent crossing phenotype when crossed to multiple populations of M. deorus (i.e. Figure S3), but we found substantial variation between populations of M. decorus in the magnitude and the direction of HSI (Figure S3). While some of the variation in HSI between populations of M. decorus and M. guttatus can be attributed to variation in ploidy within M. decorus, we also find that the two genetic clades of M. decorus exhibit oppositely asymmetric HSI with M. guttatus. That is to say, HSI manifests when northern M. decorus are the maternal contributors in crosses to M. guttatus, while HSI manifests when southern M. decorus are the paternal contributor in crosses to M. guttatus.
If HSI is largely driven by parental conflict, we would predict that hybrid seeds should display parent-of-origin effects on growth and development, resulting in a size difference between reciprocal F1s. Specifically, populations that have stronger conflict/ higher EBNs should produce hybrid seeds which are larger when they are the paternal donor and smaller when they are the maternal donor when crossed to a population with weaker conflict/ lower EBNs. Secondly, we predict that the magnitude of asymmetry in growth defects between reciprocal F1s should correspond to the magnitude of HSI. Lastly, we predict that inferences of the presumed level of conflict that each species has experienced (e.g. EBN) should predict of the outcomes of untested crosses.
Assessing parent-of-origin effects on resource allocation to offspring
We measured parent of origin effects on resource allocation to offspring using two approaches. Firstly, we used photographs to measure total seed width for all hybrid and parental crosses in ImageJ [81]. Secondly, we sought to assess development of hybrid and parental seeds using crosses between M. guttatus and a focal population from each of northern and southern M. decorus. To do this, we crossed plants, as in above, but collected fruits at 4,6,8,10 and 14 Days After Pollination (DAP). Fruits were collected and immediately placed in FAA fixative (10 Ethanol: 1 Glacial acetic acid: 2 Formalin: 7 H20). Fruits were processed in a similar manner to [82]. In brief, after at least 24 hours in fixative, fruits were gradually dehydrated in a Tert-Butyl alcohol (TBA) dehydration series, then mounted in paraffin wax with ~5% gum elemi resin. Paraffin mounted specimens were then sliced to 8 micron ribbons and mounted onto slides. We performed a staining series using Safranin-O and Fast-Green, which stain for nucleic acids and carbohydrates, respectively. We visualized and photographed seeds from a single fruit for each cross type and time point combination.
Assessing the correlation between the magnitude of asymmetry in growth defects between reciprocal F1s and HSI
Using the measurements of seed width from above, we simply correlated the degree of symmetry of HSI and the degree of symmetry in reciprocal F1 seed size to determine if the magnitude of growth defect differences were related to the extent of HSI.
Assessing whether our designations of conflict predict HSI in subsequent crosses
While other studies have used proxies for the extent of differences in EBN/inferred conflict between species (e.g. mating system or ploidy; e.g. [37]), the species we describe here exhibit a highly outcrossing morphology (for example, they do not differ in anther-stigma separation; Figure 5), and potential drivers of variation in the level of conflict that different species experience are unknown. We therefore use the patterns of HSI, reciprocal F1 seed sizes, and hybrid seed development between M. guttatus and each clade of diploid M. decorus to infer EBN/the extent of conflict that different species have experienced. This mirrors earlier work on EBNs, wherein focal species were crossed to a common test line to infer EBN, then specific predictions were formed based on the inferred EBNs and tested using subsequent crosses between lines with inferred EBNs [i.e. 32. 36]. In our case, if parental conflict drives HSI, then we infer that northern M. decorus must have weaker conflict than M. guttatus, as hybrid seeds between the two species are larger and display excessive endosperm development when northern M. decorus is the maternal parent, but smaller and exhibit more precocious endosperm development when northern M. decorus is the paternal parent. In contrast, we infer that southern M. decorus has stronger conflict than M. guttatus, because when M. guttatus and southern M. decorus are crossed, seeds display a paternal-excess phenotype when southern M. decorus is the paternal genotype, and a maternal-excess phenotype when M. guttatus is the paternal parent. We can then use these inferences to make predictions about subsequent, untested crosses, if HSI is driven by patterns of conflict.
Parental conflict theory predicts that populations that show the most extreme levels of conflict should display the strongest reproductive isolation, and this reproductive isolation should be accompanied by differences in reciprocal F1 seed size. To test this, we performed two additional sets of crosses. Firstly, we leveraged the diversity of inferred EBNs between diploid species by crossing accessions from all populations of the northern and southern clades of diploid M. decorus to a focal accession from each clade (IMP from the northern clade; Odell Creek from the southern clade). As these clades are presumed to exhibit the most extreme difference in parental conflict, we would predict that they should exhibit the most extreme HSI, and that patterns of HSI should be accompanied by differences in reciprocal F1 seed sizes, wherein F1 seeds are larger when northern M. decorus is the maternal parent. Secondly, we leveraged diversity in inferred EBNs caused by differences in ploidy by crossing the focal accessions from the northern and southern clades of M. decorus from above to all populations of tetraploid M. decorus. If parental conflict is driving patterns of HSI in this group, we would predict that crosses between tetraploids and low-conflict northern M. decorus should exhibit more extreme HSI than crosses between northern M. decorus and M. guttatus, and reciprocal F1 seeds should exhibit differences in growth and development. In contrast, southern M. decorus should exhibit much less, if any, HSI when crossed to tetraploid M. decorus as these two clades exhibit a smaller difference in presumed conflict. Accordingly, reciprocal F1 seeds should exhibit minimal differences in growth and development. We also crossed all four populations of M. guttatus used above in all possible combinations to determine if alleles that contribute to HSI were naturally segregating throughout M. guttatus. Crosses were processed as above to score average seed viability based on morphology, average germination rate, and average size.
QUANTIFICATION AND STATISTICAL ANALYSIS
Phenotypic analyses
To determine how species varied in multivariate trait space, we completed a PCA of all traits, with traits scaled to have unit variance. PCs 1 and 2 accounted for 47.84% and 19.58% trait variance, respectively, and were the only PCs to explain a significant amount of variance using a broken stick model.
Genotypic analyses
Genome Re-sequencing: Processing of Files
For population genomic analyses, we used only the whole-genome sequences. For these samples, we trimmed adapter and low-quality sequences using Trim Galore! [83], then aligned the trimmed sequences to the M. guttatus Version 2.0 hard masked reference genome using BWA mem [84; https://phytozome.jgi.doe.gov/]. We cleaned, sorted and marked duplicate reads using Picard tools [85], then used HaplotypeCaller in GATK to call SNPs for each genome separately, and finally performed GenotypeGVCFs in GATK with all samples and both variant and invariant sites [86]. We then filtered the resultant VCF file to remove INDELs, and keep sites with a minimum quality score of 30 and a depth of coverage of 5x per individual using VCFtools [87]
For a PCA we combined the whole genome re-sequence data with previously published GBS dataset from [67]. For these samples, we processed the raw reads from [67] similarly, but split individuals by barcode, discarded low quality reads, and trimmed barcodes using STACKs before alignment [88].
Population Genomic Analyses
To estimate diversity and divergence among taxa, we used only diploid, whole-genome re-sequenced accessions. We first filtered the VCF file to retain only 4-fold degenerate sites, remove INDELs and retain sites with a quality score =/>30 and a minimum read depth of 5x. This left 1,201,466 sites total (including both variant and invariant sites). We then estimated within-species diversity and between species divergence and differentiation by calculating pairwise nucleotide diversity (π), dxy, and Fst in non-overlapping 500 kb windows using custom Python scripts courtesy of Simon Martin (available: https://github.com/simonhmartin/genomics_general). Both variant and invariant sites were used to calculate π and dxy, which were estimated by dividing the total number of pairwise differences by the total number of genotyped sites for within and between-species, respectively. Estimates of divergence and differentiation were completed for each pairwise comparison of the 5 species (20 comparison), while nucleotide diversity was calculated for each species separately. Genome-wide averages were then calculated.
Phylogenetic Analyses
We next constructed Neighbor-Joining (NJ) trees to infer relationships among diploid taxa in our sample. We further filtered the VCF to retain only sites with <5% missing data, leaving 25,765 SNPs for analyses. We then constructed a consensus NJ tree using the ape package in R [89] We also calculated 56 NJ trees for 500 SNP windows, with a step size of 100 SNPs to assess the level of discordance throughout the genome (Figure S2), and plotted a densitree using the phangorn package in R [90]. All trees were rooted the outgroup M. dentilobus.
Principal Components Analyses
To summarize genetic relationships among diploid accessions we performed a PCA using ANGSD [91]. ANGSD uses .bam files to calculate a genotype likelihood at each site, and is therefore able to integrate genotype uncertainty into various analyses, which is useful for low-coverage re-sequencing or GBS data. For our PCA, we filtered our dataset to keep sites that contained a minimum mapping quality score of 30 or above, a base quality score of 20 or above, and contained genotypic information for at least ~85% of accessions (i.e. 203/238), which leaves a total of 25,405 sites. We find that PCs 1-6 explained a significant proportion of the variation using a broken stick model, with PCs 1 and 2 explaining 33.1% and 25.7% of the variance, respectively. PCs 3, 4, 5 and 6 accounted for 19.0, 16.3, 10.1, and 5.7% of the variance, respectively (Figure S1).
For comparison, we also performed an additional PCA that included only diploid samples, to determine if the addition of tetraploid samples biased our PCA. The results from the two analyses qualitatively agree, and the relationships among species in PC space did not change by the addition of tetraploid individuals (Figure 1; Figure S1).
Analyses of crossing data
To determine the extent of HSI, the extent of pollen/pistil interactions, and reciprocal F1 seed size differences between M. guttatus and M. decorus, as well as within M. decorus, we performed series of a linear mixed models using the lme4 and car packages in the statistical interface R [92–94]. To determine the extent of HSI and pollen/pistil interactions, we used the proportion of viable seeds per fruit and the total number of seeds produced as the dependent variable, respectively. Cross type, genetic clade, and their interaction were treated as fixed effect predictor variables, while population was treated as a random effect, and the maternal and paternal individual used in each cross were treated as random effects nested within population. We find a significant interaction effect between cross type and genetic clade for the proportion of viable seeds, and so also performed these linear mixed models on each clade separately, again treating population as a random effect and the maternal and paternal individual used as random effects nested within population. In all cases where cross type was significant, we performed a post hoc pairwise T-test with Holm correction for multiple testing to determine which crosses differed significantly.
To determine the extent of size differences between reciprocal F1s, we also performed a linear mixed model on seed widths with cross type as a fixed effect and population as a random effect. As seeds were pooled in order to photograph them, the exact maternal and paternal individual were unknown for each particular seed, and therefore could not be included in the model. These analyses were done for each genetic clade separately.
To assess differences in stigma-anther separation, we first averaged the distance between the stigma and the upper and lower sets of anthers. We then performed a linear mixed model on the average stigma-anther separation with genetic clade as a fixed effect and maternal family as a random effect.
DATA AND CODE AVAILABILITY
Phenotypic data collected from a common garden experiment for members of the M. guttatus species complex are available on dryad (https://doi.org/10.5061/dryad.881j0ds), as are the raw data for cross compatibility between M. guttatus and M. decorus, as well as within M. decorus (https://doi.org/10.5061/dryad.cz8w9ghzr). Re-sequencing data is available on the NCBI Short Reads Archive (SRA), under the BioProject ID PRJNA574603. Accession numbers for each sample are listed in Table S1.
Supplementary Material
KEY RESOURCES TABLE
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Chemicals | ||
| Cystain UV Precise P buffer | Sysmex | Catalogue number: 05-5002 |
| DAPI (4′,6-diamidino-2-phenylindole) | ThermoFisher Scientific | Catalogue number: D1306 |
| Safranin-O | Sigma-Aldrich | Catalogue number: S2255-25G |
| Fast-Green | Sigma-Aldrich | Catalogue number: F7252-5G |
| Critical commercial assays | ||
| Genejet extraction kit | ThermoFisher Scientific | Catalogue number: K0691 |
| Nextra library kit | Illumina | Catalogue number: FC-131-1024 |
| Deposited data | ||
| Phenotypic data from common garden study | This paper; [66] | https://doi.org/10.5061/dryad.881j0ds |
| Raw crossing survey data | This paper | https://doi.org/10.5061/dryad.cz8w9ghzr |
| Re-sequence genomes | This paper; [78] | NCBI SRA (https://www.ncbi.nlm.nih.gov/sra), BioProject ID PRJNA574603 |
| Experimental models | ||
| Mimulus seeds | Seeds collected by JMC, or generously given to JMC by former members of the Willis lab | N/A |
| Software and algorithms | ||
| TrimGalore! | [83] | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| BWA | [84] | http://bio-bwa.sourceforge.net/ |
| GATK | [86] | https://software.broadinstitute.org/gatk/ |
| VCFTools | [87] | http://vcftools.sourceforge.net/ |
| ANGSD | [91] | http://www.popgen.dk/angsd/index.php/ANGSD |
| Genome Tools | Python scripts provided by Simon Martin | http://github.com/simonhmartin/genomics_general |
| R: ape | [89] | N/A |
| R: phangorn | [90] | N/A |
| R: lme4 | [93] | N/A |
| R: car | [94] | N/A |
| Stacks | [88] | http://catchenlab.life.illinois.edu/stacks/ |
Highlights.
Mimulus decorus is a cryptic species complex within the M. guttatus species complex
M. decorus is reproductive isolated from M. guttatus by hybrid seed inviability
Patterns of hybrid seed inviability conform to the predictions of parental conflict
Differences in conflict between species scale with genome-wide diversity
Acknowledgements
We thank Shivam Dave for help with seed counting, and Madison Zamora for assistance with field work. Miguel Flores provided valuable expertise and help with flow cytometry, and chromosomal squashes were only possible with the patient help of Michael Windham. We thank Yaniv Brandvain and Josh Puzey for access to the whole genome re-sequencing data for several accessions of M. guttatus. We are deeply grateful to members of the Willis and Matute lab, particularly Daniel Matute, who provided useful comments on this manuscript. We also thank those who provided insightful comments during the 2019 Gordon Speciation Conferences in Ventura, CA. The manuscript was substantially improved by the thoughtful comments of 5 anonymous reviewers. This project was funded from NSF grants IOS-1558113 and IOS-1354688 to JHW, a DDIG (DEB- 1501758), American Society of Naturalists’ Student research award and Society for the Study of Evolution student research award to JMC. JMC was also funded through the Duke Graduate School via the Myra and William Waldo Boone fellowship, the Duke Biology Department, and NIGMS grant R01GM121750 to Daniel Matute. Duke BioCore provided support for MWB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no conflict of interest
References
- 1.Bradshaw H, & Schemske DW (2003). Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature, 44(3), 138–147. [DOI] [PubMed] [Google Scholar]
- 2.Ramsey J, Bradshaw HD, & Schemske DW (2003). Components of reproductive isolation between the monkeyflowers Mimulus lewisii and M. cardinalis (Phrymaceae). Evolution, 57(7), 1520–1534. [DOI] [PubMed] [Google Scholar]
- 3.Nosil P, Vines TH, & Funk DJ (2005). Reproductive isolation casued by natural selection against immigrants from divergent habitats. Evolution, 59, 705–719. [PubMed] [Google Scholar]
- 4.Arnegard ME, McGee MD, Matthews B, Marchinko KB, Conte GL, Kabir S, ... Schluter D (2014). Genetics of ecological divergence during speciation. Nature, 511, 307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tiffin P, Olson MS, & Moyle LC (2001). Asymmetrical crossing barriers in angiosperms. Proceedings. Biological Sciences / The Royal Society, 268(1469), 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowry DB, Modliszewski JL, Wright KM, Wu CA, & Willis JH (2008). The strength and genetic basis of reproductive isolating barriers in flowering plants. Philosophical Transactions of the Royal Society B, 363(1506), 3009–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vrana PB, Fossella J. a., Matteson P, del Rio T, O’Neill MJ, & Tilghman SM (2000). Genetic and epigenetic incompatibilities underlie hybrid dysgenesis in Peromyscus. Nature Genetics, 25(1), 120–124. [DOI] [PubMed] [Google Scholar]
- 8.Zechner U, Shi W, Hemberger M, Himmelbauer H, Otto S, Orth a., … Fundele R (2004). Divergent genetic and epigenetic post-zygotic isolation mechanisms in Mus and Peromyscus. Journal of Evolutionary Biology, 17(2), 453–460. [DOI] [PubMed] [Google Scholar]
- 9.Brekke Th.D.. , & Good JM (2014). Parent-of-origin growth effects and the evolution of hybrid inviability in dwarf hamsters. Evolution, 68(11), 3134–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brekke TD, Henry L. a., & Good JM (2016). Genomic imprinting, disrupted placental expression, and speciation. Evolution, 3(Haig 1996), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott RJ, Spielman M, Bailey J, & Dickinson HG (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development (Cambridge, England), 125(October 2014), 3329–3341. [DOI] [PubMed] [Google Scholar]
- 12.Wolff P, Jiang H, Wang G, Santos-González J, & Köhler C (2015). Paternally expressed imprinted genes establish postzygotic hybridization barriers in Arabidopsis thaliana. ELife, 4, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rebernig C. a., Lafon-Placette C, Hatorangan MR, Slotte T, & Köhler C (2015). Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm. PLOS Genetics, 11(6), e1005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oneal E, Willis JH, & Franks RG (2016). Disruption of endosperm development is a major cause of hybrid seed inviability between Mimulus guttatus and Mimulus nudatus. New Phytologist, 210, 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garner AG, Kenney AM, Fishman L, & Sweigart AL (2016). Genetic loci with parent of origin effects cause hybrid seed lethality between Mimulus species. New Phytologist, 211, 319–331. [DOI] [PubMed] [Google Scholar]
- 16.Roth M, Florez-Rueda AM, Griesser S, Paris M, & Städler T (2017). Incidence and developmental timing of endosperm failure in postzygotic isolation between wild tomato lineages. Annals of Botany, (January), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trivers RL (1974). Parent-Offspring Conflict. American Zoology, 14(1), 249–264. [Google Scholar]
- 18.Charnov EL, & Hutchinson GE (1979). Population Biology Simultaneous hermaphroditism and sexual selection, 76(5), 2480–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haig D, & Mark Westoby. (1989). Parent-specific gene expression and the triploid endosperm. American Naturalist, 134(1), 147–155. [Google Scholar]
- 20.Willi Y (2013). The Battle of the Sexes over Seed Size: Support for Both Kinship Genomic Imprinting and Interlocus Contest Evolution. The American Naturalist, 181(6), 787–798. [DOI] [PubMed] [Google Scholar]
- 21.Raunsgard A, Opedal ØH, Ekrem RK, Wright J, Bolstad GH, Armbruster WS, & Pélabon C (2018). Intersexual conflict over seed size is stronger in more outcrossed populations of a mixed-mating plant. Proceedings of the National Academy of Sciences, 201810979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cailleau A, Grimanelli D, Blanchet E, Cheptou P-O, & Lenormand T (2018). Dividing a Maternal Pie among Half-Sibs: Genetic Conflicts and the Control of Resource Allocation to Seeds in Maize. The American Naturalist, 192(5), 000–000. [DOI] [PubMed] [Google Scholar]
- 23.Vrana PB (2007). Genomic Imprinting As a Mechanism of Reproductive Isolation in Mammals, 88(1), 5–23. [Google Scholar]
- 24.Lafon-Placette C, & Köhler C (2016). Endosperm-based postzygotic hybridization barriers: Developmental mechanisms and evolutionary drivers. Molecular Ecology, 2620–2629. [DOI] [PubMed] [Google Scholar]
- 25.Wilson a. C., Maxson LR, & Sarich VM (1974). Two types of molecular evolution: evidence from studies of interspecific hybridization. Proceedings of the National Academy of Sciences of the United States of America, 71(7), 2843–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prager EM, & Wilson a. C. (1975). Slow evolutionary loss of the potential for interspecific hybridization in birds: a manifestation of slow regulatory evolution. Proceedings of the National Academy of Sciences, 72(1), 200–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fitzpatrick BM (2004). Rates of evolution of hybrid inviability in birds and mammals. Evolution, 58(8), 1865–1870. [DOI] [PubMed] [Google Scholar]
- 28.Lynch M, & Force AG (2000). The Origin of Interspecific Genomic Incompatibility via Gene Duplication. The American Naturalist, 156(6), 590–605. [DOI] [PubMed] [Google Scholar]
- 29.Martienssen RA (2010). Heterochromatin, small RNA and post-fertilization dysgenesis in allopolyploid and interploid hybrids of Arabidopsis. New Phytologist, 186(1), 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castillo DM, & Moyle LC (2012). Evolutionary Implications of Mechanistic Models of TE-Mediated Hybrid Incompatibility. International Journal of Evolutionary Biology, 2012, 698198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Köhler C, Mittelsten Scheid O, & Erilova A (2010). The impact of the triploid block on the origin and evolution of polyploid plants. Trends in Genetics, 26(3), 142–148. [DOI] [PubMed] [Google Scholar]
- 32.Lin BY (1984). Ploidy barrier to endosperm development in maize. Genetics, 107(1), 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Comai L (2005). The advantages and disadvantages of being polyploid. Nature Reviews, 6(11), 836–846. [DOI] [PubMed] [Google Scholar]
- 34.Kradolfer D, Wolff P, Jiang H, Siretskiy A, & Köhler C (2013). An imprinted gene underlies postzygotic reproductive isolation in arabidopsis thaliana. Developmental Cell, 26(5), 525–535. [DOI] [PubMed] [Google Scholar]
- 35.Lafon-Placette C, Johannessen IM, Hornslien KS, Ali MF, Bjerkan KN, Bramsiepe J, … Köhler C (2017). Endosperm-based hybridization barriers explain the pattern of gene flow between Arabidopsis lyrata and Arabidopsis arenosa in Central Europe. Proceedings of the National Academy of Sciences, 201615123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnston S. a., den Nijs TPM, Peloquin SJ, & Hanneman RE (1980). The significance of genic balance to endosperm development in interspecific crosses. Theoretical and Applied Genetics, 57(1), 5–9. [DOI] [PubMed] [Google Scholar]
- 37.Lafon-Placette C, Hatorangan MR, Steige KA, Cornille A, Lascoux M, Slotte T, & Köhler C (2018). Paternally expressed imprinted genes associate with hybridization barriers in Capsella. Nature Plants. [DOI] [PubMed] [Google Scholar]
- 38.Sweigart AL, Mason AR, & Willis JH (2007). Natural variation for a hybrid incompatibility between two species of mimulus. Evolution, 61(1), 141–151. [DOI] [PubMed] [Google Scholar]
- 39.Bomblies K, & Weigel D (2007). Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nature Reviews. Genetics, 8(5), 382–393. [DOI] [PubMed] [Google Scholar]
- 40.Case AL, & Willis JH (2008). Hybrid male sterility in Mimulus (Phrymaceae) isassociated with a geographically restricted mitochondrial rearrangement. Evolution, 62(5), 1026–1039. [DOI] [PubMed] [Google Scholar]
- 41.].Martin NH, & Willis JH (2010). Geographical variation in postzygotic isolation and its genetic basis within and between two Mimulus species. Philosophical Transactions of the Royal Society B: Biological Sciences, 365(1555), 3261–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sicard A, Kappel C, Josephs EB, Lee YW, Marona C, Stinchcombe JR, … Lenhard M (2015). Divergent sorting of a balanced ancestral polymorphism underlies the establishment of gene-flow barriers in Capsella. Nature Communications, 6, 7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Case AL, Finseth FR, Barr CM, & Fishman L (2016). Selfish evolution of cytonuclear hybrid incompatibility in Mimulus. Proceedings of the Royal Society B, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barnard-Kubow KB, & Galloway LF (2017). Variation in reproductive isolation across a species range. Ecology and Evolution, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bracewell RR, Bentz BJ, Sullivan BT, & Good JM (2017). Rapid neo-sex chromosome evolution and incipient speciation in a major forest pest. Nature Communications, 8(1), 1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lowry DB, Hall MC, Salt DE, & Willis JH (2009). Genetic and physiological basis of adaptive salt tolerance divergence between coastal and inland Mimulus guttatus. The New Phytologist, 183(3), 776–788. [DOI] [PubMed] [Google Scholar]
- 47.Wright KM, Lloyd D, Lowry DB, Macnair MR, & Willis JH (2013). Indirect evolution of hybrid lethality due to linkage with selected locus in Mimulus guttatus. PLoS Biology, 11(2), e1001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferris KG, & Willis JH (2018). Differential Adaptation to a Harsh Granite Outcrop Habitat between Sympatric. Evolution, 72(6), 1225–1241. [DOI] [PubMed] [Google Scholar]
- 49.Selby JP, & Willis JH (2018). Major QTL controls adaptation to serpentine soils in Mimulus guttatus, (August), 5073–5087. [DOI] [PubMed] [Google Scholar]
- 50.Fishman L, Kelly AJ, & Willis JH (2002). Minor Quantitative Trait Loci Underlie Floral Traits Associated with Mating System Divergence in Mimulus. Evolution, 56(11), 2138–2155. [DOI] [PubMed] [Google Scholar]
- 51.Martin NH, & Willis JH (2007). Ecological divergence associated with mating system causes nearly complete reproductive isolation between sympatric Mimulus species. Evolution; International Journal of Organic Evolution, 61(1), 68–82. [DOI] [PubMed] [Google Scholar]
- 52.Brandvain Y, Kenney AM, Flagel LE, Coop G, & Sweigart AL (2014). Speciation and Introgression between Mimulus nasutus and Mimulus guttatus. PLoS Genetics, 10(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fishman L, & Kelly JK (2015). Centromere-associated meiotic drive and female fitness variation in Mimulus. Evolution, 69, 1208–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hall MC, & Willis JH (2006). Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution, 60(12), 2466–2477. [PubMed] [Google Scholar]
- 55.Lowry DB, & Willis JH (2010). A Widespread Chromosomal Inversion Polymorphism Contributes to a Major Life-History Transition, Local Adaptation , and Reproductive Isolation. PLoS Biology, 8(9), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friedman J, Twyford AD, Willis JH, & Blackman BK (2015). The extent and genetic basis of phenotypic divergence in life history traits in Mimulus guttatus. Molecular Ecology, 24(1), 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peterson ML, Kay KM, & Angert AL (2016). The scale of local adaptation in Mimulus guttatus : comparing life history races , ecotypes , and populations. New Phytologist, 211, 345–356. [DOI] [PubMed] [Google Scholar]
- 58.Wu CA, Lowry DB, Cooley a M., Wright KM, Lee YW, & Willis JH (2008). Mimulus is an emerging model system for the integration of ecological and genomic studies. Heredity, 100(2), 220–230. [DOI] [PubMed] [Google Scholar]
- 59.Twyford AD, Streisfeld MA, Lowry DB, & Friedman J (2015). Genomic studies on the nature of species: Adaptation and speciation in Mimulus. Molecular Ecology, 24(11), 2601–2609. [DOI] [PubMed] [Google Scholar]
- 60.Sweigart AL, Fishman L, & Willis JH (2006). A simple genetic incompatibility causes hybrid male sterility in mimulus. Genetics, 172(4), 2465–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peterson ML, Rice KJ, & Sexton JP (2013). Niche partitioning between close relatives suggests trade-offs between adaptation to local environments and competition. Ecology and Evolution, 3(3), 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toll K, & Willis JH (2019). Hybrid inviability and differential submergence tolerance drive habitat segregation between two congeneric monkey fl owers, 99(12), 2776–2786. [DOI] [PubMed] [Google Scholar]
- 63.Grant AL (1924). A monograph of the genus Mimulus. Annals of the Missouri Botanical Garden, 11(2), 99–388. [Google Scholar]
- 64.Nesom GL (2012). Taxonomy of erythranthe sect. simiolus (phrymaceae) in the usa and mexico. Phytoneuron, (1978). [Google Scholar]
- 65.Lowry DB, Sobel JM, Angert AL, Ashman T, Baker RL, Blackman BK, Brandvain Y, Byers KJRP, Cooley AM, Coughlan JM, Dudash MR, Fenster CB, Ferris KG, Fishman L, Friedman J, Grossenbacher DL, Holeski LM, Ivey CT, Kay KM, Koelling VA, Kooyers NJ, Murren CJ, Muir CD, Nelson TC, Peterson ML, Puzey JR, Rotter MC, Seemann JR, Sexton JP, Sheth SN, Streisfeld MA, Sweigart AL, Twyford AD, Vallejo-Marin M, Willis JH, Wu CA, Yuan Y-W. The case for the continued use of the genus name Mimulus for all monkeyflowers. Taxon in press. [Google Scholar]
- 66.Coughlan JM, & Willis JH (2018). Dissecting the role of a large chromosomal inversion in life history divergence throughout the Mimulus guttatus species complex. Molecular Ecology, 1–15. [DOI] [PubMed] [Google Scholar]
- 67.Twyford AD, & Friedman J (2015). Adaptive divergence in the monkey flower Mimulus guttatus is maintained by a chromosomal inversion. Evolution, 69(9), 1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brandvain Y, & Haig D (2005). Divergent mating systems and parental conflict as a barrier to hybridization in flowering plants. The American Naturalist, 166(3), 330–338. [DOI] [PubMed] [Google Scholar]
- 69.Gardner M, & Macnair M (2000). Factors affecting the co-existence of the serpentine endemic Mimulus nudatus Curran and it presumed progenitor, Mimulus guttatus Fischer ex DC. Biological Journal of the Linnean Society, 69, 443–459. [Google Scholar]
- 70.Habecker NM (2012). Ecogeographical and intrinsic postzygotic isolation between Mimulus glaucescens and Mimulus guttatus (Masters thesis). Retrieved from California State University Chico repository. [Google Scholar]
- 71.Kenney AM, Sweigart AL, & Kenney AM (2016). Reproductive isolation and introgression between sympatric Mimulus species. Molecular Ecology, 25, 2499–2517. [DOI] [PubMed] [Google Scholar]
- 72.Ferris KG, Sexton JP, & Willis JH (2014). Speciation on a local geographic scale: the evolution of a rare rock outcrop specialist in Mimulus. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 369(1648), 20140001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hall MC, & Willis JH (2005). Transmission ratio distortion in intraspecific hybrids of Mimulus guttatus: implications for genomic divergence. Genetics, 170(1), 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zuellig MP, & Sweigart AL (2018)a. A two-locus hybrid incompatibility is widespread, polymorphic, and active in natural populations of Mimulus. Evolution, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zuellig MP, & Sweigart AL (2018)b. Gene duplicates cause hybrid lethality between sympatric species of Mimulus. PLoS, 14(4), e1007130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gehring M, Bubb KL, & Henikoff S (2009). Extensive Demethylation of Repetitive. Science, 324, 1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pennington PD, Costa LM, Gutierrez-Marcos JF, Greenland AJ, & Dickinson HG (2008). When genomes collide: Aberrant seed development following maize interploidy crosses. Annals of Botany, 101(6), 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Puzey JR, Willis JH, & Kelly JK (2017). Population structure and local selection yield high genomic variation in Mimulus guttatus. Molecular Ecology, 26, 519–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Galbraith DW, Lambert GM, Macas J, & Dolezel J (2001). Analysis of nuclear DNA content and ploidy in higher plants. Current Protocols in Cytometry, Chapter 7, Unit 7.6. [DOI] [PubMed] [Google Scholar]
- 80.Schmuths H, Meister A, Horres R, & Bachmann K (2004). Genome size variation among accessions of Arabidopsis thaliana. Annals of Botany, 93(3), 317–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schneider C. a., Rasband WS, & Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nature Methods, 9(7), 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.White RA, & Turner MD (2012). The Anatomy and Occurrence of Foliar Nectaries in Cyathea (Cyatheaceae ). American Fern Journal, 102(2), 91–113. [Google Scholar]
- 83.Kreuger F Trim Galore! 0.3. 7 In. [Google Scholar]
- 84.Li H, & Durbin R (2009). Fast and accurate short read alignment with BurrowsWheeler transform. Bioinformatics, 25(14), 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25(16), 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kerntsky A, … DePristo MA (2010). The Genome Analysis Toolkit: A MapReduce framework for analysing next-generation DNA sequencing data. Genome Research, 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, Depristo MA, … Vcf T (2011). The variant call format and VCFtools, 27(15), 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Catchen J, Hohenlohe P. a., Bassham S, Amores A, & Cresko W. a. (2013). Stacks: An analysis tool set for population genomics. Molecular Ecology, 22(11), 3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paradis E & Schliep K 2018. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35: 526–528. [DOI] [PubMed] [Google Scholar]
- 90.Schliep KP 2011. phangorn: phylogenetic analysis in R. Bioinformatics, 27(4) 592–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Korneliussen TS, Albrechtsen A, & Nielsen R (2014). Open Access ANGSD©: Analysis of Next Generation Sequencing Data, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.R Core Team (2017). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: URL https://www.R-project.org/. [Google Scholar]
- 93.Bates D, Maechler M, Bolker B, Walker S (2015). Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software, 67(1), 1–48. [Google Scholar]
- 94.Fox J & Weisberg S (2011). An {R} Companion to Applied Regression, Second Edition Thousand Oaks CA: Sage. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Phenotypic data collected from a common garden experiment for members of the M. guttatus species complex are available on dryad (https://doi.org/10.5061/dryad.881j0ds), as are the raw data for cross compatibility between M. guttatus and M. decorus, as well as within M. decorus (https://doi.org/10.5061/dryad.cz8w9ghzr). Re-sequencing data is available on the NCBI Short Reads Archive (SRA), under the BioProject ID PRJNA574603. Accession numbers for each sample are listed in Table S1.


