Although philosophers might argue about it, anyone who owns a dog will agree that non-primate animals can think. Cats are a bit more questionable due to their screwy logic but still they show evidence of thought. As one proceeds downward in size to smaller animals, the ability to think, as judged by the ability to make decisions and learn from experience, continues to be seen. This raises the question, how many cells does an organism need in order to think? How about just one? It sounds crazy to talk about a cell thinking, but cells make decisions all the time. Examples include the decision to enter the cell cycle or to undergo apoptosis. Even bacteriophages make decisions to undergo lysis or lysogeny. These examples illustrate binary decision to either take a specific irrevocable action, or not. Can cells go beyond such simple Hamlet-like decisions? A new study in this issue of Current Biology from Dexter et al, working with the ciliate Stentor suggests this is indeed likely [1]
Herbert Spencer Jennings (1868–1947), along with a number of his contemporaries [2–5], observed the behaviors of a variety of single-celled organisms, and concluded their behavior showed evidence of complex decision making processes in which a cell could choose between many different possible actions, and in which the decisions made depended on past experience. In subsequent decades, researchers carried out more careful quantitative studies of single cell behavior. For example, Wood showed that a single cell can display a primitive form of learning [6]. Wood’s experiments built on earlier observations by Jennings [2] on Stentor, a giant ciliate over a millimeter long [7]. Stentor cells are cone-shaped sessile organisms that, when mechanically stimulated by a predator, rapidly contract into a ball for safety. This contraction is energetically expensive and so the cell only “wants” to contact if it is facing a novel threat. Consequently, the cell “learns” to ignore repeated mechanical stimulation, thus representing an example of habituation, the most fundamental form of learning [8]. Other reported examples of learning by Stentor, such as the ability to learn how to escape from glass capillary tubes, were harder to reproduce or interpret [9,10].
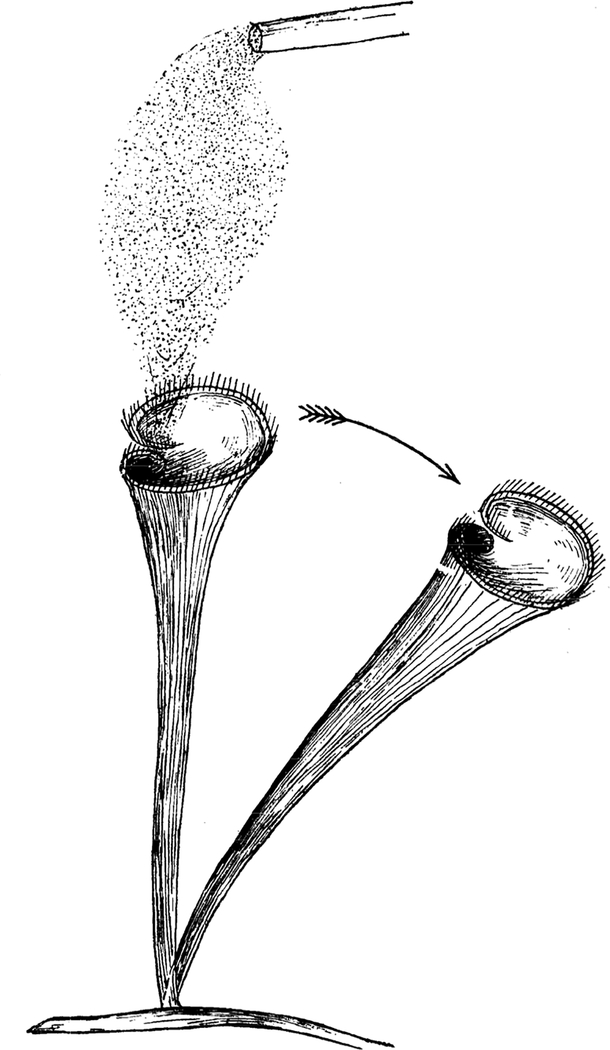
One of the most provocative early reports on Stentor was a study from Jennings in which he exposed Stentor cells to an unpleasant chemical stimulus (carmine dye). Jennings found that each cell executed a stereotyped sequence of distinct behaviors in response to the stimulus [3]. First, the cell would try to bend out of the way (Figure 1). If this bending failed to stop the noxious stimulus, the cell would reverse the direction of its ciliary beating. If the stimulus still continued, the cell would eventually contract. Sometimes, after many stimuli, the cell would detach from the substrate and swim off, apparently looking for a safer environment to resettle. Jennings noted that these behaviors typically occurred in the same sequence, with contraction only happening after the cell had already spent some time on the first two behaviors. Intriguingly, if the stimulus was applied again and again, the cell would start skipping the first two behaviors and go straight to contraction [2].
Figure 1.
Avoidance behavior of Stentor roeselli. Image shows a Stentor cell being exposed to a cloud of carmine dye particles. As indicated by the arrow, the cell bends away from the cloud of dye. Alternative avoidance behaviors include reversing the direction of ciliary beating, contraction, and swimming away from the stimulus. Image taken from [3].
The interpretation of Jennings’ experiments was that the cell could “decide” which behavior to do, and that it could “learn” which behaviors worked and which didn’t. These two features of selecting among multiple behaviors and altering the choices based on past experience are the very essence of decision making, and so if a cell can really do these things, it suggests a thought-like or computational process is at work. Unfortunately, researchers following Jennings failed to reproduce the phenomenon [11]. Instead of a complex series of responses, they reported that the Stentor cells sometimes contracted but mostly just swam away. However, it turns out that they weren’t using the same species of Stentor. Jennings studied Stentor roeselli, a transparent Stentor that builds a mucus tube to live in. But the later researchers, who claimed they couldn’t reproduce Jennings’ findings, used Stentor coeruleus, a blue Stentor that does not built a tube. When faced with a noxious stimulus, S. coeruleus tends to detach from the substrate and swim away, a fact that Jennings himself had reported. S. roeselli, in contrast, stands its ground and tries to evade the stimulus by bending, ciliary reversals, and contraction. It is thus hardly surprising that studies of S. coeruleus failed to show the sequence of behaviors that Jennings had reported.
In this issue of Current Biology, Dexter et al. [1] revisit Jennings’ classical experiments, this time using the same species (S. roeselli) that Jennings used. By delivering precise bursts of plastic beads (which the cell doesn’t like) they found that S. roeselli indeed shows the three behaviors reported by Jennings. Importantly, they confirmed, using rigorous statistical testing, that the first two avoidance behaviors (bending and ciliary reversal) tend to precede contractions, just as Jennings reported. The new paper also analyzed the detachment response and found that every time the cell contracts, there is a constant probability of detaching. Remarkably, the probability of detaching appears to be exactly 50%. In abstract terms, the cell is flipping a fair coin to make the decision. The one major claim of Jennings that wasn’t directly addressed is the idea that repeated stimulation will cause the cell to give up on the earlier responses and switch over entirely to contraction. Although not analyzed statistically, this trend is in fact visible in the data found within the paper. For example, considering just the experiments in which a cell was exposed to two rounds of stimulation, and looking at the six examples in which one of the two responses was ciliary reversal and the other was contraction, in 5/6 cases the response sequence to the first stimulus began with a reversal and the response sequence to the second stimulus began with a contraction. Because the new paper carefully reports the actual results for each cell examined, the data is there to be mined for interesting trends like this. In any case the evidence that a cell executes a series of different responses in a roughly specific order is quite convincing from the reported data, confirming the key claim that Jennings made 100 years ago.
What sort of molecular machinery might underlie these complex behaviors? In a provocative book entitled “Wetware: a computer in every living cell”, Dennis Bray has argued that biochemical pathways can act like logic circuits or neural networks [12], suggesting that we might be able to understand cell behavior by drawing analogies to robotics or digital electronics. It is clear that signaling proteins and gene expression networks can act like digital logic gates [13,14], combining two or more input signals to produce an output signal. Such combinatorial logic represents one of the two main flavors of digital electronics. The other flavor is sequential logic, in which the system switches between a series of distinct states in response to a sequence of inputs, and these states determine different outputs. Simple examples are traffic lights that sequence between three different colors, or a counter that counts down from ten before launching a rocket. Sequential logic is implemented using circuit elements known as flip-flops that can be switched between two different states and that retain their state until reset back to the original state. A series of flip-flops connected one after another can generate a sequence of states in an orderly progression. The ability of Stentor roeselli to switch between a series of behaviors in a defined order suggest that cells must have some form of sequential logic, raising the question question of how a flip-flop might be implemented in a living cell. In fact one answer is already known – protein phosphorylation. Given a protein that is regulated by the presence of absence of a phosphate, one could view it as a flip-flop that is set by phosphorylation and reset by dephosphorylation. More complicated artificial sequential systems have been implemented in cells, for example using protein-DNA interactions [15]. The challenge going forward is to figure out what molecules are used to implement the sequential controller that determines the different behaviors of S. roeselli.
Is sequential logic limited to Stentor? The fast, dramatic behaviors of Stentor make it much easier to investigate the logic underlying these behaviors compared to other cell types, since the behaviors can be unambiguously detected and quantified. In the long run, though, such experiments will need to be adapted to more general cell types, requiring more subtle ways to classify cell behavioral states based on, for example, motility, morphology, or biochemical output. Someday it may become quite natural to think of cells as robots or computers, in which future behavior can be predicted from past experience using abstract models of state transitions [16], just as has now been done for Stentor.
References
- 1.Dexter JP, Prabakaran S, Gunawardena J 2019. A complex hierarchy of avoidance behaviours in a single-cell eukaryote. Current Biology 29, 4323–4329. [DOI] [PubMed] [Google Scholar]
- 2.Jennings HS. 1906. Behavior of the Lower Organisms, Columbia University Press, New York, NY, USA. [Google Scholar]
- 3.Jennings HS. 1902. Studies on reactions to stimuli in unicellular organisms. IX. On the behavior of fixed infusoria (Stentor and Vorticella) with special reference to the modifiability of protozoan reactions. Am. J. Physiol 8, 23–60. [Google Scholar]
- 4.Gibbs D, Dellinger OP. 1908. The daily life of Amoeba proteus. Am. J. Psych 19 232–241. [Google Scholar]
- 5.Kepner WA and Taliaferro WH. 1913. Reactions of Amoeba proteus to food. Biol. Bull 24, 411–428. [Google Scholar]
- 6.Wood DC. 1970. Parametric studies of the response decrement produced by mechanical stimuli in the protozoan, Stentor coeruleus. J. Neurobiol 1, 345–60. [DOI] [PubMed] [Google Scholar]
- 7.Tartar V 1961. The Biology of Stentor, Pergammon Press, Oxford, UK. [Google Scholar]
- 8.Thompson RF, Spencer WA. 1966. Habituation: a model phenomenon for the study of neuronal substrates of behavior. Psych. Rev 73, 16–43. [DOI] [PubMed] [Google Scholar]
- 9.Bennet DA, Francis D. 1972. Learning in Stentor. J. Protozool 19, 484–487. [Google Scholar]
- 10.Hinkle DJ, Wood DC. 1994. Is tube-escape learning by protozoa associative learning? Behavioral Neurosci 108, 94–90. [DOI] [PubMed] [Google Scholar]
- 11.Reynierse JH, Walsh GL. 1967. Behavior modification in the protozoan Stentor re-examined. Psychol. Rec 17, 161–5. [Google Scholar]
- 12.Bray D 2009. Wetware: A Computer in Every living Cell, Yale University Press, New Haven, CT, USA. [Google Scholar]
- 13.Dueber JE, Yeh BJ, Chak K, Lim WA. 2003. Reprogramming control of an allosteric signalling switch through molecular recombination. Science 301, 1904–8. [DOI] [PubMed] [Google Scholar]
- 14.Mayo AE, Setty Y, Shavit S, Zaslaver A, Alon U. 2006. Plasticity of the cis-regulatory input function of a gene. PLoS Biol 4, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bar-Ziv R, Tlusty T, Libchaber A. 2002. Protein-DNA computation by stochastic assembly cascade. Proc Natl Acad Sci U S A 99, 11589–11592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Condon A, Kirchner H, Lariviere D, Marshall WF, Noireaux V, Tlusty T, Fourmentin E 2018. Will biologists become computer scientists? EMBO Reports pii: e46628 [DOI] [PMC free article] [PubMed] [Google Scholar]