Abstract
Many species of bacteria can manufacture materials on a finer scale than those that are synthetically made. These products are often produced within intracellular compartments that bear many hallmarks of eukaryotic organelles. One unique and elegant group of organisms is at the forefront of studies into the mechanisms of organelle formation and biomineralization. Magnetotactic bacteria (MTB) produce organelles called magnetosomes that contain nanocrystals of magnetic material, and understanding the molecular mechanisms behind magnetosome formation and biomineralization is a rich area of study. In this Review, we focus on the genetics behind the formation of magnetosomes and biomineralization. We cover the history of genetic discoveries in MTB and key insights that have been found in recent years and provide a perspective on the future of genetic studies in MTB.
Author summary
Open any biology textbook and you are likely to learn that bacteria—unlike the cells of plants, animals, and other eukaryotes—do not contain organelles to compartmentalize and facilitate cellular functions. However, over the past several decades, many different bacterial organelles have been discovered. In this Review, we highlight magnetotactic bacteria (MTB), which are a group of organisms capable of producing organelles called magnetosomes where nano-sized crystals of magnetic material are synthesized and housed. In order to understand how and why MTB form magnetosomes, it is important to study the genes involved. Here, we lay out the history of genetic studies in MTB and more recent discoveries about which genes are involved at each step in the process of magnetosome formation and discuss where the field is headed.
Introduction
Bacteria, according to the canonical definition, do not have subcellular compartments for organization or specialized functions. Yet microbiologists are becoming increasingly aware that many bacteria do have organelles, some of which are capable of manufacturing biomaterials with specialized functions [1]. MTB present a particularly elegant example of the biological behaviors that are mediated by intracellular compartments [2]. MTB are a group of bacteria spanning multiple phyla that can be found in aquatic environments all over the world [3–5], where they inhabit low oxygen environments, and are most often found at the oxic–anoxic interface in the water column or sediments [6]. MTB are characterized by their ability to form organelles called magnetosomes—lipid-bounded compartments in which biomineralization of magnetic crystals of magnetite (Fe3O4) and/or greigite (Fe3S4) occurs [3]. Magnetosomes align in one or multiple chains along the cell, creating a magnetic dipole that allows MTB to passively align along Earth’s magnetic field lines [7]. This is thought to help MTB perform more efficient chemotaxis and aerotaxis in the water column as their swimming behavior is restricted to one dimension instead of a three dimensional run and tumble search: a process called magneto-aerotaxis [6]. The regulated process of biomineralization has made MTB an attractive area of study in basic and applied biology, geochemistry, and physics alike. A greater understanding of the molecular processes needed to form magnetosomes will enhance studies in each of these fields, as knowledge of what is occurring on a molecular scale can lend greater precision to technical applications and provide systems-level information for studying the large-scale impacts of MTB.
The ecological impact of MTB is one rising area of research in the field. In recent years, our understanding of the diversity of MTB has expanded greatly. In the process of biomineralization of magnetite or greigite, MTB take up large amounts of dissolved iron from the surrounding environment and sequester it in magnetosomes as iron crystals. As such, the role that MTB play in iron cycling in both freshwater bodies and the ocean is potentially quite large [5,8]. Conservative estimates from Amor and colleagues indicate that estuarine and oceanic MTB may take up anywhere from approximately 1% to 50% of dissolved iron inputs (approximately 9 × 108 kg per year) in their environments [8]. Having a greater understanding of the iron regulation strategies encoded within the genomes of MTB, as well as how iron is taken up and distributed to magnetosomes, is important for a more accurate picture of the role of MTB in their aquatic environments.
In this same vein, fossils of magnetosomes can help us understand the environmental conditions that were present when MTB originated and the evolution of life on Earth. Magnetofossils—currently dating back to approximately 1.9 Ga—may reflect changes that occurred in sediments and the water column and have the potential to serve as an indicator of redox and oxygen levels in ancient environments [9]. Based on phylogenetic analyses, the origins of biomineralization may have occurred much earlier, in the mid-Archaean (approximately 3 Ga), when the ability to biomineralize may have provided an advantage in coping with reactive oxygen species, avoiding harmful UV radiation, and/or navigating ferrous-iron gradients [10,11]. Understanding the genetic factors behind the formation of magnetosomes that are common across modern MTB can provide insight into the conditions and processes that were present when the first MTB originated [12]. Genetic analysis may also uncover unknown functions of magnetosomes and can hint at the conditions that were needed to produce ancient magnetosomes.
Additionally, the use of MTB in various biotechnological applications is promising. Magnetosomes are currently being developed for use as magnetic resonance imaging (MRI) contrast agents, drug delivery systems, hyperthermic and photothermic treatments for cancer, bioremediation of heavy metals, and other nanotechnologies [13–15]. In order to efficiently produce the large numbers of magnetosomes required in these applications, it is critical to understand how magnetosomes are produced.
Taking up iron from the environment for biomineralization, producing phospholipid membranes of a specific size, and aligning magnetosomes in a chain is a complex, tightly controlled process—one that is interesting in itself but also provides more general insights into the precise formation of organelles. MTB encode the genes necessary for these processes in magnetosome gene clusters (MGCs) [16,17]. The MGCs in the most well-studied model organisms—Magnetospirillum magneticum AMB-1, M. gryphiswaldense MSR-1, and Desulfovibrio magneticus RS-1 (Fig 1)—are structured as magnetosome gene islands (MAIs). In both AMB-1 and RS-1, the MAI is defined by repeat regions on either side of the large chromosomal region [18,19]. Across related species, there is a large amount of genetic homology in the MAI [20]. The functions of many of the genes within MGCs have been investigated, but much remains unknown in each of the model organisms, and there is even more to be discovered about other species and phyla of MTB.
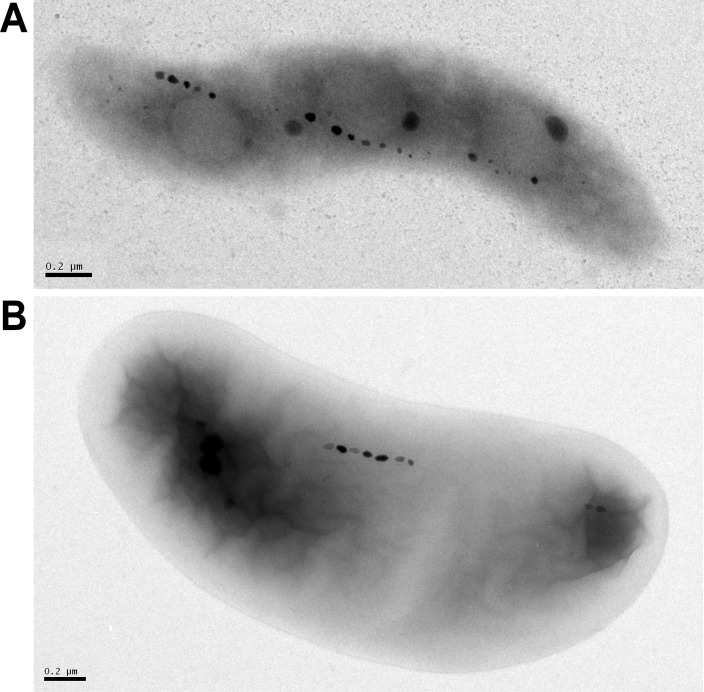
Fig 1. The MTB model systems.
(A) TEM image of wild-type AMB-1 cell. (B) TEM image of wild-type RS-1 cell, scale bar 200 nm. Reprinted with permission from Rahn-Lee and colleagues. [21]. TEM, transmission electron microscopy.
In this Review, we lay out the history of landmark genetic discoveries in revealing important insights into the process of magnetosome and biomineral formation by MTB. We also dive into more nuanced views of genetics that have come out in recent years in step with advances in genetic techniques. Finally, we will address the next directions that the field is taking to learn more about the genetics of MTB.
Genetics in Magnetospirilla
The discovery of MTB [7] fueled broad interest in understanding and exploiting the process of organelle formation and biomineralization. Development of genetic systems greatly accelerated the discovery of the molecular basis of magnetosome formation at a refined level. M. magnetotacticum MS-1 was the first magnetotactic bacterium to be isolated in pure culture [22]. However, MS-1 did not become a major model system for genetics since conditions that support colony growth on solid media have not been identified. Subsequently, AMB-1 and MSR-1 were successfully cultured and established as model organisms in the lab, making it possible to manipulate and investigate their genomes [23–25].
Early studies of MTB genetics
The first genetic studies in MTB involved transposon mutagenesis coupled with magnetic selection and transmission electron microscopy (TEM). Matsunaga and colleagues (1992) performed mutagenesis in AMB-1 with the Tn5 transposon [23]. They identified several genomic fragments involved in magnetosome synthesis by picking out mutant cultures that no longer responded to a magnet under a light microscope. After confirming the mutants were deficient in magnetosome formation with electron microscopy, they used restriction mapping to narrow down the location of each insertion site in the genome. One of these mutants carries a transposon insertion in magA, a gene encoding a cation efflux pump proposed to function in iron transport [26]. Importantly, this study also demonstrated that it was possible to transfer plasmid DNA to AMB-1 using conjugation.
Large strides were made in understanding MTB genomes in the early 2000s. Wahyudi and colleagues (2001) also isolated Tn5 transposon mutants in AMB-1 and found colonies with defects in biomineralization by looking at colony color, which is thought to be an indicator of how much magnetite has accumulated in the cell [27]. They concluded that at least 10, and up to 60, genes could be involved in magnetosome formation. The publication of the genome sequences of MS-1 and Magnetococcus marinus MC-1 also opened the door to genome-level studies [28,29]. Grünberg and colleagues (2001) compared protein sequences isolated from MSR-1 magnetosomes to those in the MS-1 and MC-1 genomes and found two gene clusters containing genes (mamA, mamB, mamC, and mamD) we now know to be critical for magnetosome formation [30].
The key genomic region needed for magnetosome formation, the MAI, was discovered when spontaneous nonmagnetic mutants of MSR-1 were isolated from a wild-type population of cells [17]. It was characterized as a 130-kb region containing multiple insertion sequence (IS) elements [18]. The AMB-1 gene island was described as a 98-kb region flanked by two 1.1-kb repeat sequences [16]. A study of transcription of MAI genes indicated that while magnetosome genes are organized in operons, they are constitutively expressed [31]. Identifying the MAI narrowed down the pool of genes to investigate and provided a foundation for more targeted genetic studies.
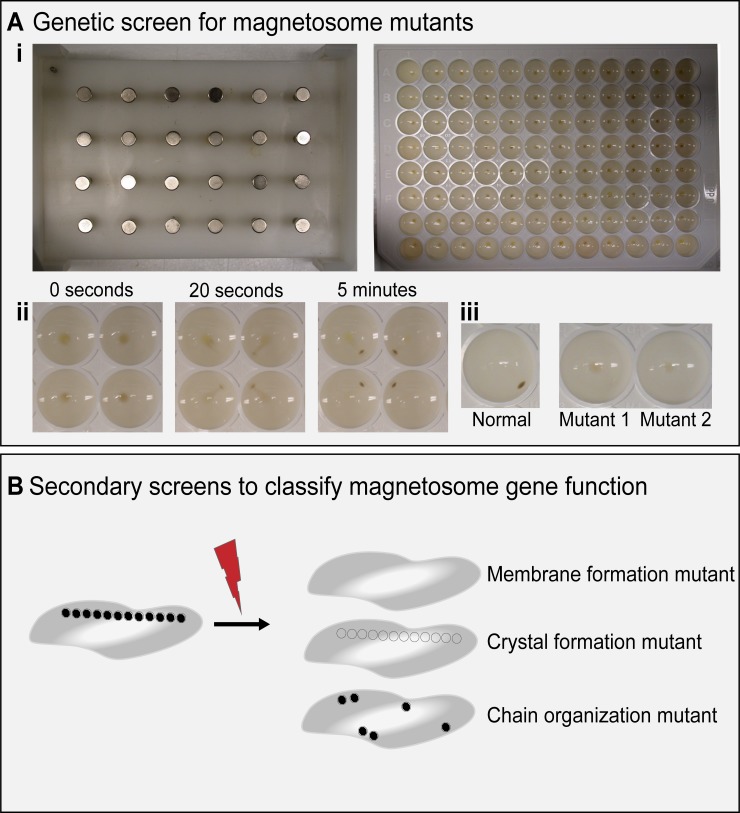
In addition to defining the MAI, the establishment of MSR-1 and AMB-1 as model systems allowed for more detailed molecular and genetic analyses [32,33]. A transposon mutagenesis screen by Komeili and colleagues (2004) used a magnetic selection to enrich for nonmagnetic mutants [34]. Colonies were then grown in 96-well plates and screened for magnetic response using a 24-pin magnetic plate (Fig 2A). In this study, transposon insertions within the mamAB gene cluster of the MAI resulted in nonmagnetic mutants. This work proved to be a great complement to proteomic studies that had found the same MAI-encoded proteins associated with magnetosomes [35].
Fig 2. Magnetic screening technique.
(A) (i) 24-pin magnetic plate (left) and 96-well plate of AMB-1 cells (right) used in Komeili and colleagues. [34]. (ii) Movement of AMB-1 cells on magnetic plate at 0 seconds, 20 seconds, and 5 minutes. (iii) Phenotype of normal magnetic cells (left) and two representative, nonmagnetic mutants (right). (B) Diagram of secondary screens to classify magnetosome mutants.
Transposon mutagenesis studies proved to be a key turning point in using genetics to understand the process of biomineralization in MTB. However, much like many other genetic studies, their interpretation and broader utility were complicated by several confounding factors. First, due to homologous recombination between repeated sequences or potential action of transposases, the MAI is unstable and can be lost spontaneously, an event that is more likely to occur under stress conditions [18]. If transposition occurs in a bacterium that has lost its MAI or if the MAI is lost following transposition, otherwise neutral events may appear linked to changes in the magnetic phenotype. Screening potential mutants for the presence of the MAI proved essential in isolating mutations in the mamAB region [34].
Second, many magnetosome genes contain functional paralogs that play redundant roles. In AMB-1, three genes (mamQ, mamR, and, mamB) from the mamAB operon are perfectly duplicated in another segment of the MAI [36]. Additionally, the AMB-1 genome contains a magnetosome gene islet, a region outside of the MAI, which includes several homologs of mamAB genes [37,38]. As a result, the absence of a distinct phenotype when any of the duplicated genes are deleted individually does not rule out the possibility that they play a role in magnetosome formation. Thus, in many cases, multiple genes must be deleted to understand the function of a specific gene and its interactions with other genes. Additionally, complementing deletions becomes critical for the evaluation of gene function.
Third, magnetosome genes are often organized as operons and transposon insertions result in the polar loss of expression for all downstream genes. Thus, it is difficult to link a specific phenotype to the loss of one single gene. Finally, by necessity, these studies used the magnetic phenotype as a quick screening method to find relevant mutants. The secondary screens of transposon mutants were important for establishing which step in the magnetosome formation process was affected by a particular mutation and allowed for assigning more specific functions to genes (Fig 2B). For example, a nonmagnetic mutant might make magnetosome membranes but not form crystals, indicating the interrupted gene was likely involved in crystal formation. Or a nonmagnetic mutant might not make membranes at all, suggesting the site of transposon insertion is a key part of magnetosome membrane formation.
Dissecting Magnetospirillum genomes
Obtaining the full genome sequences of the primary model organisms (MSR-1 and AMB-1) propelled genetic investigations of MTB forward [33,39]. Previous studies provided limited functional detail about any particular gene, at times involved looking at large deletions of multiple genes, and had complicating factors, as mentioned above. Deleting individual genes was necessary for a more complete picture of magnetosome formation.
Murat and colleagues (2010) used previously developed methodology to thoroughly dissect the MAI in AMB-1 by creating targeted deletions of genes and operons [36]. They began with the observation that the loss of the MAI results in complete absence of both magnetosome membranes and magnetic particles. Using a double recombination method for generating nonpolar deletions, they first made mutants lacking larger subsections of the MAI [34,36]. Next, they focused on the regions that showed dramatic phenotypes such as small particles or complete loss of the magnetosome membrane. Finally, they deleted individual genes within these flagged regions and used a suite of secondary screens to assign specific functions to the genes. Various electron microscopy techniques were used to visualize the magnetosome membrane as well as the size, morphology, and subcellular arrangement of magnetic particles. Green fluorescent protein (GFP) fusions to model magnetosome proteins were used to monitor protein localization.
Through multiple layers of analysis, Murat and colleagues described the possible functions of many of the key magnetosome-formation genes in AMB-1, like mamE, mamN, mamM, mamO, mamI, mamL, mamQ, and mamB. Lohße and colleagues (2011, 2014) dissected the MAI in MSR-1 [40,41] and found similar results, except that mamI and mamN were not essential for magnetosome formation in MSR-1. This discrepancy might be due to the particular growth conditions used obscuring more subtle differences between the two species. It may also reflect broader divergence between the two organisms. In a landmark study, heterologous expression of the mamAB and mms6 operons, plus mamGFDC and mamXYZ, was found to be sufficient to produce magnetosomes in the nonmagnetic α-Proteobacterium Rhodospirillum rubrum [42], highlighting both the importance of these operons in magnetosome formation and the minimal gene set needed to make magnetosomes under laboratory conditions.
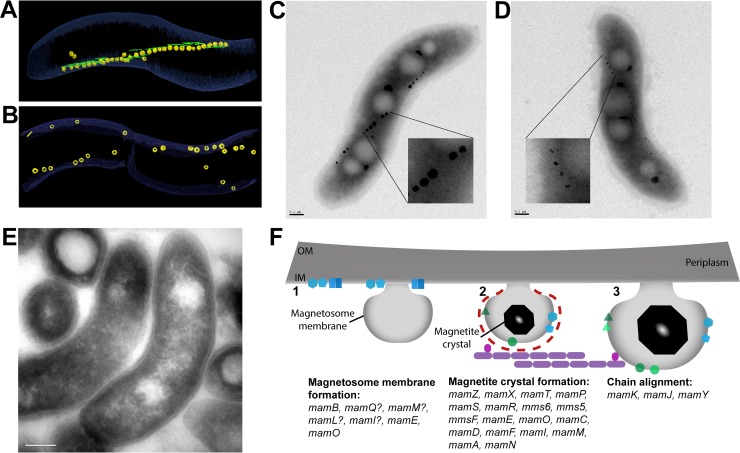
These studies of the AMB-1 and MSR-1 islands provided a broad overview of the functions of genes in the MAI. Further studies dove into investigating the functions of individual genes in magnetosome formation and their mechanisms of action. The general framework that magnetosome genes are important for either membrane formation (Fig 3E) [43–45] or biomineralization (Fig 3C and 3D) [46–53] holds true. A third category of genes that are involved in chain organization has become an important area of study in recent years [38,54–59] (Fig 3A and 3B). At the center of the chain-arrangement process is an actin-like protein called MamK. Dynamic polymerization behavior of MamK is required for the integrity and proper segregation of the chain during cell division [54–58,60,61]. The proteins MamJ and MamY are also key components of chain formation. MamJ acts as a link between MamK filaments and magnetosomes [54]. MamY is a cytoplasmic membrane protein that works to align the magnetosome chain along the cell’s motility axis, which likely improves the efficiency of magnetotaxis [59]. To add nuance to the broad categories, more specific functions of genes involved in each stage of the process are being discovered as techniques and tools improve. More detailed summaries of the genes and proteins responsible for magnetosome formation can be found in previously published Review articles (Fig 3F) [62–64].
Fig 3. AMB-1 and MSR-1 strains with defects in magnetosome formation.
(A) Wild-type, AMB-1–cell image taken from segmentation of an electron cryotomogram. MamK filaments (green) run parallel to magnetosomes (yellow). (B) Electron cryotomogram image of a ΔmamK AMB-1 cell that shows disorganized magnetosomes. Images provided by Komeili. (C) TEM image of a wild-type, AMB-1 cell. Scale bar is 0.2 μm. Close-up of magnetosomes is magnified 6×. (D) TEM image of a ΔmamT AMB-1 cell showing small, misshapen magnetosomes. Scale bar is 0.2 μm. Close-up of magnetosomes is magnified 6×. Image provided by McCausland and colleagues. (E) TEM image of a cryosection of ΔmamL AMB-1 cells showing that magnetosome membranes are absent. Scale bar is 0.2 μm. Image provided by Komeili. (F) Diagram of the stepwise process of magnetosome formation and the proteins involved from membrane invagination (1), to crystal nucleation (2), and to membrane growth and formation of a mature magnetic crystal (3). Genes that have been found to be involved at each step are listed. AMB-1, Magnetospirillum magneticum AMB-1; IM, inner membrane; OM, outermembrane; MSR-1, Magnetospirillum gryphiswaldense MSR-1; TEM, transmission electron microscopy.
In addition to the limits of techniques used in genetic analysis, the growth conditions of MTB are an important factor to consider when examining the phenotype of particular genes. Genes both inside and outside the MAI have been found to have important roles in biomineralization but only under certain conditions. For example, within the MAI ΔmamX, ΔmamZ, ΔmamH, and ΔftsZm strains only show defects in biomineralization when MSR-1 is grown with ammonium in place of nitrate, indicating that the use of oxygen instead of nitrate as the terminal electron acceptor is detrimental to magnetosome formation [65–67]. Genes outside the island, like the nap operon encoding nitrate-reductase genes [68] and the cytochrome c oxidase cbb3 [69], are also key in the biomineralization process, again highlighting the importance of redox processes in magnetosome formation [70]. It is possible that these metabolic pathways generate an overall redox balance within the cell that is compatible with biomineralization, which requires both ferric and ferrous iron to be present. Alternatively, they may participate directly in generating a redox-balanced iron pool that will allow for magnetite biomineralization.
Another key environmental factor to consider in magnetosome formation is the availability of iron. Unsurprisingly, genes involved in regulating the uptake of iron have been connected to biomineralization. MSR-1 contains a homolog of the ferric uptake regulator (fur) gene common among bacteria [71]. This fur-like gene affects magnetosome size and number—potentially due to reduced incorporation of iron into magnetite and an increase in cytoplasmic iron concentrations—as well as transcription levels of several key MAI genes, including the mamGFDC and mms6 operons [72]. Deletion of feoB1—which is involved in transport of ferrous iron into the cell—in MSR-1 resulted in fewer and smaller magnetosomes, as well as decreased uptake of iron [73]. In AMB-1, the feoAB operon showed increased levels of transcription under iron-rich conditions and was associated with an increase in intracellular iron levels, suggesting that it is a component of iron uptake in AMB-1 [74]. Ferric iron transporters were down-regulated in the same high-iron conditions. While the feoAB1 operon is within the MAI, it is not a magnetosome-associated protein and thus its regulation of magnetosome formation is indirect. MS-1 also expresses feoB, indicating that iron uptake systems are conserved across species of MTB [75]. The iron response regulator IrrB was shown to be important for magnetosome formation in MSR-1, and deletion affected the transcription of several genes involved in the regulation of iron uptake [76]. These studies taken together indicate that iron availability is an important factor to consider in magnetosome formation and genetic regulation.
Genetics and genomics of diverse MTB
As described above, work in the model organisms MSR-1 and AMB-1 has identified many of the genes that are important for the formation and positioning of magnetosomes. However, MSR-1 and AMB-1 are both α-Proteobacteria species and MTB are an incredibly diverse group of organisms with strains found in several classes of Proteobacteria, as well as Nitrospirae and the candidate division OP3 [3]. Little is known about the formation of magnetosomes in nonmodel organisms. The genetic studies in MSR-1 and AMB-1 have been used as jumping off points for the newer model system D. magneticus RS-1, as well as uncultured species of MTB like those from other classes of Proteobacteria and the phyla Nitrospirae [20]. Additionally, improved sequencing technologies have allowed the field to study the phylogeny of MTB using more relevant magnetosome genes instead of standard housekeeping genes.
The MAI in D. magneticus RS-1
The establishment of the δ-Proteobacterium RS-1 as a model system has opened up the field to studying the genetic diversity of magnetosome formation. In the phylogeny of Proteobacteria, the δ-Proteobacteria class is deeply branching, relative to α-Proteobacteria. As such, research into δ-Proteobacteria can provide valuable insight into the origins of MTB. Through the study of RS-1, it has been found that, in addition to a core set of genes required across all MTB, different types of MTB have distinct genes for magnetosome formation. Presumably the genes evolved to adapt to the diverse lifestyles of each organism.
Comparative genome analysis of a variety of δ-Proteobacteria revealed that many of the genes required for magnetosome formation in α-Proteobacteria are shared by the δ-Proteobacteria, though some of the genes in the mamAB operon and the entire mamGFDC operon are missing [19]. It was discovered in the same study that the δ-Proteobacteria and Nitrospirae MTB have a separate set of class-specific genes termed the mad genes, which are likely involved in the production of bullet-shaped magnetite crystals. Additionally, Nitrospirae MTB have another set of genes, the man genes, that may be involved in the processes of magnetosome formation and/or chain arrangement that are particular to Nitrospirae [77]. The magnetosome gene sets that appear in different phyla provide a convincing link between genetic differences and the clear phenotypic differences seen across MTB. Model organisms from the δ-Proteobacteria and Nitrospirae are necessary to study the functions of mad and man genes.
In 1993, RS-1 was discovered as a sulfate-reducing MTB [78] and later identified as a Desulfovibrio species [79]. RS-1 is an obligate anaerobe that synthesizes bullet-shaped magnetite crystals, as opposed to the cubo-octahedral crystals produced by the α-Proteobacteria MTB species. While RS-1 was successfully cultured in the lab, attempts to delete individual genes were unsuccessful at first. Rahn-Lee and colleagues got around this problem with a classic forward genetic screen using random chemical and UV mutagenesis followed by whole-genome sequencing of nonmagnetic mutants of RS-1 [21]. Both mam genes and mad genes were found to be important in biomineralization, as were several novel MAI genes, including the ion transport genes tauE and kup. Genetic tools for deleting specific genes in RS-1 were only recently developed. Using suicide vectors for targeted gene deletion—as is commonly done in other bacterial systems—does not work in RS-1 due to low transconjugation and recombination rates. Grant and colleagues developed a strategy using replicative plasmids that carry positive and negative selectable markers to replace the gene of interest with an antibiotic-resistance gene [80].
The study of magnetosome formation in RS-1 also identified a novel organelle consisting of iron–phosphorous granules surrounded by a membrane [81]. Byrne and colleagues showed that these granules are separate organelles and not precursors to the formation of magnetite with a pulse-chase experiment using different stable isotopes of iron. This conclusion was further solidified by the finding that the deletion of the entire MAI of RS-1 had no impact on the formation of the iron-rich granules [21]. The genetics of these novel bacterial organelles is a rich area for future investigation.
The MAI in nonmodel and uncultured MTB
The genetic differences that have already been found between MSR-1, AMB-1, and RS-1 highlight the need to study diverse MTB species in order to more fully understand magnetosome formation and function(s). The rapid improvements in sequencing technologies in recent years—in addition to the elegant method for isolating MTB using an external magnetic field—have made it possible to study uncultured organisms in greater detail [82]. The genomes of many uncultured MTB have been sequenced and analyzed in detail [19,28,42,77,83–85], revealing that MTB belong to a wide variety of bacterial phyla. Metagenomic analyses are also contributing greatly to our knowledge of the diversity of MTB and their evolutionary history [10,20].
In the realm of nonmodel organisms, greigite-producing strains provide an interesting case to study the evolution of MTB. Work by DeLong and colleagues analyzing 16S rRNA gene sequences suggested that greigite-producing strains and magnetite-producing strains evolved separately [86]. However, a later analysis by Abreu and colleagues found that the greigite-producing strain Candidatus Magnetoglobus multicellularis has some of the mam genes that magnetite-producing strains require to produce magnetosomes, suggesting a monophyletic origin for MTB [87]. Lefèvre and colleagues (2011) discovered the δ-Proteobacteria Desulfamplus magnetovallimortis BW-1 and found that it is capable of producing both magnetite and greigite [85]. Interestingly, the BW-1 genome has mam genes in two separate MGCs. Proteins encoded in one cluster are closely related to proteins found in magnetite-producing species, while those in the second cluster are more closely related to the proteins encoded in the MGCs of greigite producers. The simplest hypothesis emerging from these genomic insights is that each cluster is responsible for producing a chemically distinct, magnetic mineral. Lefèvre and colleagues used the unique MGCs of BW-1 to examine phylogenetic differences between magnetite-producing and greigite-producing strains [19]. Genes required for producing magnetite-containing magnetosomes are clustered together, as are those required for producing greigite-containing magnetosomes, suggesting that there are separate sets of genes (and proteins) involved in forming each type of crystal. However, the mad genes, which are needed to form bullet-shaped magnetosomes, are present in both clusters. It is still unclear if mad genes were lost during the evolution of magnetite-producing strains that do not form bullet-shaped crystals or if they were acquired separately by δ-Proteobacteria and Nitrospirae strains of MTB. Analysis of the α-Proteobacteria PR-1 also indicated that evolution of MTB likely involved both vertical inheritance and horizontal gene transfer (HGT) or duplication events [84].
Looking further into the origins of MTB, Lefèvre and colleagues (2013) compared phylogenies of several α-, δ-, and γ-Proteobacteria and one Nitrospirae MTB species. They constructed phylogenetic trees using either 16S rRNA gene sequences and housekeeping genes or common Mam proteins [88]. They found that both trees showed a similar pattern of divergence, leading to the conclusion that all modern day Proteobacteria and Nitrospirae had a magnetotactic common ancestor, though they did not rule out the possibility of an ancient HGT event. Two recent studies from Lin and colleagues analyzed metagenomic data to gain insight into the origins of MTB [10,20]. The first study analyzed the genomes of multiple magnetotactic, Nitrospirae strains and found that the gene content and order in the MGCs were conserved across the Nitrospirae, indicating a common origin. In the second study, a wide variety of MTB genomes were analyzed using core magnetosome proteins and the phylogenetic trees showed MTB clustering together. The authors concluded, like Lefèvre and colleagues, that HGT of magnetosome genes were likely rare events. The simplest conclusion based on these studies is that all MTB originated from a common ancestor. In fact, using commonly accepted molecular clocks, it can be estimated that the original MTB—and presumably the first instance of magnetosome formation—appeared approximately 3.2 billion years ago [10]. An additional implication of this work is that at some point in the past the last common ancestor of the Proteobacteria, Nitrospirae, and Omnitrophica phyla had the genes necessary for formation of magnetic particles. The origins of magnetotactic Latescibacteria and Planctomycetes are less clear. These phyla could have emerged from the last common ancestor of the magnetotactic Proteobacteria, Nitrospirae, and Omnitrophica or acquired the genes through HGT. Subsequently, most descendants of these founding members lost the magnetosome genes leaving behind the handful of modern-day MTB. The environmental conditions and changes that initially favored the evolution and expansion of magnetosome-formation genes and later selected against them in the majority of bacteria remain to be elucidated. Perhaps, genetic studies of other model MTB are needed to understand the potential contributions of group-specific genes (such as mad and man genes) to the evolution and phenotypic diversification of magnetosomes.
The study of uncultured MTB has also been aided through the analysis of gene function in model MTB. Take, for example, the MAI genes mamE and mamO, which are critical for biomineralization in both AMB-1 and MSR-1 [36,89]. Both gene products are predicted serine proteases, and initial genetic studies concerning their functions concluded that this was indeed the case [90]. However, further biochemical and structural studies revealed that the active site of MamO is not functional and that it is in reality a metal-binding protein that controls biomineralization and regulates the proteolytic activity of MamE [91,92]. Phylogenetic analyses showed that, similar to AMB-1, all proteobacterial MTB encode an active and inactive protease in their MGCs [91]. The active protease is ancestral to all MTB and has been diversified through vertical descent. However, the inactive protease has arisen multiple times in MTB through duplications of the active protease or acquisition via HGT. These insights were only possible through a combination of genetic, genomic, and biochemical studies. They highlight the critical analyses needed in studies in which duplication events and diversification of function of similar proteins can blur the accuracy of phylogenetic studies. They also show that the study of a protein in one species of MTB may not clarify the function of a homologous protein in another related organism.
Outlook
Huge strides have been made in understanding the genetics behind magnetosome formation. While the minimum set of genes required to generate magnetosomes is known, the specific roles of many of these factors remain unknown. Additionally, there are several genes within the MAI that when deleted have subtle or not obvious phenotypes. There are also presumably many genes outside the island that have key, though indirect, roles in magnetosome formation.
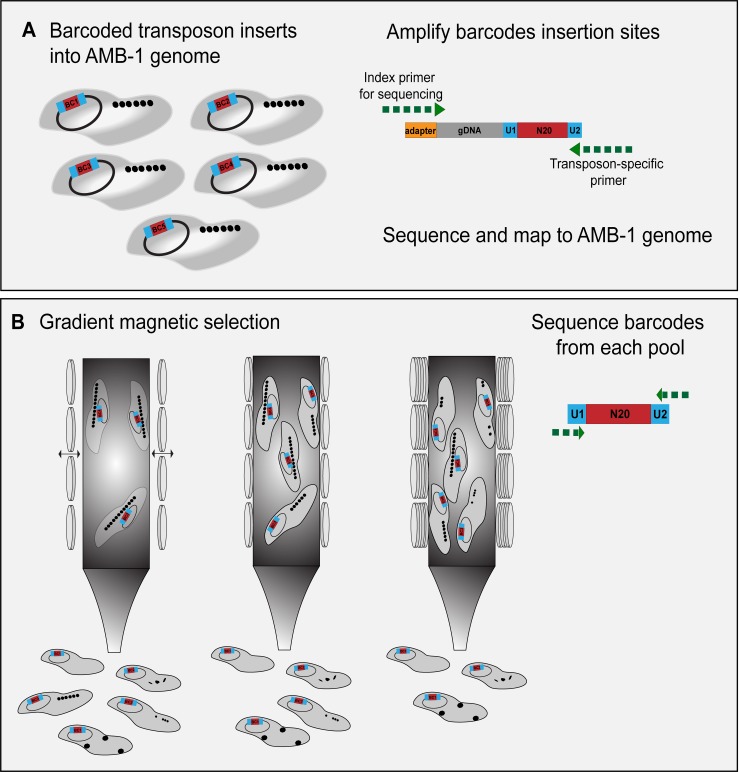
Genetic screens provide a high-throughput strategy for uncovering novel genes. Transposon mutagenesis, in particular, has been used multiple times to study the genomes of AMB-1 and MSR-1. In the future, we envision several improvements that can make transposon mutagenesis an even more useful method for genetic investigation of MTB. The latest techniques in transposon mutagenesis involve pooling tens to hundreds of thousands of labeled mutants with a method called random barcoded transposon-site sequencing (RB-TnSeq) [93] (Fig 4A). This strategy allows for saturated coverage of a bacterial genome and averaged impact of gene loss across multiple mutants. As such, polar effects and individual off-target effects are minimized during phenotyping. However, genes identified in transposon-mutagenesis screens still require phenotypic validation with gene deletions. The development of more advanced gene-editing technologies, like CRISPR, will be just as valuable for the study of MTB as it has been for other organisms [80,94]. For example, CRISPR interference (CRISPRi) can be used to knockdown multiple genes at a time more readily than traditional methods or to study essential genes by tuning their expression [95,96]. Additionally, stepping aside from transposon mutagenesis and screening instead for point mutants that have conditional phenotypes, act as dominant alleles, or suppress known mutant phenotypes can help to expand our understanding of the genetic networks that participate in magnetosome formation.
Fig 4. Barcoded transposon mutagenesis and a potential magnetic screen.
(A) Diagram of RB-TnSeq in AMB-1. Each transposon insertion carries a unique 20-nucleotide sequence that acts as a barcode. The mutated strains are pooled, and then the barcodes are mapped to their insertion site in the genome. (B) Diagram of magnetic selection with the TnSeq library using different magnetic strengths to select for a range of mutant phenotypes. From left to right, as magnetic strength applied to the column increases, strains with weaker magnetic responses will be able to stick to the column, yielding a gradient of magnetic phenotypes to analyze. AMB-1, Magnetospirillum magneticum AMB-1; BC1, barcode inserted into AMB-1 genome; gDNA, genomic DNA; N20, unique 20-nucleotide sequence; RB-TnSeq, random barcoded transposon-site sequencing; U1, universal polymerase chain reaction priming site.
The methods for screening after the initial mutagenesis can also be refined. Screens thus far have mostly relied on a simple binary identification of magnetic versus nonmagnetic cells or have used colony color as a proxy for magnetite formation. The ability to identify mutants on a spectrum of magnetic responses would be highly informative for understanding the process of magnetosome formation (Fig 4B). It may be possible to use microfluidics for this approach [97,98]. Methods that simply allow for the capture of more magnetic mutants are also key to saturate screens and identify potential negative regulators of magnetosome formation.
Screening methods that allow for the identification of genes with more subtle phenotypes on a spectrum will open the field to studying both genes in the MAI previously thought to have little to no effect on magnetosome formation or genes outside the MAI that are key in magnetosome formation under specific growth conditions. Multiple genes outside the MAI—primarily involved in metabolism—have already been connected to magnetosome formation. For example, the nitrate-reductase genes of the nap operon are important for magnetosome formation in MSR-1, even when oxygen is available as the terminal electron acceptor [68]. And the metabolic regulator crp has also been tied to magnetosome formation [99]. Different species of MTB migrate to a variety of preferred oxygen concentrations (all under 25 μM) using one of three patterns of magneto-aerotaxis [100]. The genetic mechanisms behind aerotactic behavior have begun to be investigated. For example, Popp and colleagues showed that the chemotaxis operon cheOp1 was necessary for the aerotactic response in MSR-1 [101].
The insights into metabolism and magnetosome formation are naturally connected to the increasing interest in the field in studying the ecological role of MTB in their natural environments [102–104]. How MTB interact with and adapt to changing conditions will also be an interesting problem from the perspective of geneticists and cell biologists. Most studies on MTB have been done under tightly controlled laboratory conditions, but in nature, MTB encounter changes in pH, temperature, oxygen gradients, and nutrient levels. A Review by Moisescu and colleagues summarizes the effects of these changes on magnetosome formation [105]. In addition, the study of environmental conditions may help us understand how extremophile MTB evolved or retained the ability to form magnetosomes in conditions that are not viable for most MTB species [106].
Conclusion
The set of genes needed for magnetosome formation has been clearly determined across multiple organisms, and many of their functions have been investigated. However, it is clear that even in the most well-studied MTB, like AMB-1 and MSR-1, the roles of genes that work in tandem with other factors, that participate in multiple aspects of magnetosome formation, or that are only required conditionally have yet to be fully understood. Going forward, more nuanced study of genes involved in magnetosome formation will be key to expanding our knowledge of MTB for basic cell biology, ecology, and biotechnology applications. Additionally, the study of diverse MTB using both targeted genetic analyses and whole-genome studies will potentially clarify the functions of many genes, while also adding layers to our picture of MTB.
Funding Statement
AK was supported by a grant from the National Institutes of Health (R35GM127114). The funder had no role in the decision to publish or the preparation of the manuscript.
References
- 1.Grant CR, Wan J, Komeili A. Organelle Formation in Bacteria and Archaea. Annu Rev Cell Dev Biol. 2018. October 6;34(1):217–38. [DOI] [PubMed] [Google Scholar]
- 2.Rahn-Lee L, Komeili A. The magnetosome model: insights into the mechanisms of bacterial biomineralization. Front Microbiol. 2013;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefèvre CT, Bazylinski DA. Ecology, Diversity, and Evolution of Magnetotactic Bacteria. Microbiol Mol Biol Rev MMBR. 2013. September;77(3):497–526. 10.1128/MMBR.00021-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin W, Bazylinski DA, Xiao T, Wu L-F, Pan Y. Life with compass: diversity and biogeography of magnetotactic bacteria. Environ Microbiol. 2014;16(9):2646–58. 10.1111/1462-2920.12313 [DOI] [PubMed] [Google Scholar]
- 5.Lin W, Pan Y, Bazylinski DA. Diversity and ecology of and biomineralization by magnetotactic bacteria. Environ Microbiol Rep. 2017;9(4):345–56. 10.1111/1758-2229.12550 [DOI] [PubMed] [Google Scholar]
- 6.Frankel RB, Bazylinski DA, Johnson MS, Taylor BL. Magneto-aerotaxis in marine coccoid bacteria. Biophys J. 1997. August;73(2):994–1000. 10.1016/S0006-3495(97)78132-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blakemore R. Magnetotactic bacteria. Science. 1975. October 24;190(4212):377–9. 10.1126/science.170679 [DOI] [PubMed] [Google Scholar]
- 8.Amor M, Tharaud M, Gélabert A, Komeili A. Single-cell determination of iron content in magnetotactic bacteria: implications for the iron biogeochemical cycle. Environ Microbiol. 2019. 10.1111/1462-2920.14708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopp RE, Kirschvink JL. The identification and biogeochemical interpretation of fossil magnetotactic bacteria. Earth-Sci Rev. 2008. January 1;86(1):42–61. [Google Scholar]
- 10.Lin W, Paterson GA, Zhu Q, Wang Y, Kopylova E, Li Y, et al. Origin of microbial biomineralization and magnetotaxis during the Archean. Proc Natl Acad Sci U S A. 2017. 28;114(9):2171–6. 10.1073/pnas.1614654114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin W, Kirschvink JL, Paterson GA, Bazylinski DA, Pan Y. On the origin of microbial magnetoreception. Natl Sci Rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jimenez-Lopez C, Romanek CS, Bazylinski DA. Magnetite as a prokaryotic biomarker: A review. J Geophys Res Biogeosciences. 2010. June;115(G2):n/a-n/a. [Google Scholar]
- 13.Yan L, Da H, Zhang S, López VM, Wang W. Bacterial magnetosome and its potential application. Microbiol Res. 2017. October 1;203:19–28. 10.1016/j.micres.2017.06.005 [DOI] [PubMed] [Google Scholar]
- 14.Alphandéry E. Applications of magnetosomes synthesized by magnetotactic bacteria in medicine. Front Bioeng Biotechnol. 2014;2:5 10.3389/fbioe.2014.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vargas G, Cypriano J, Correa T, Leão P, Bazylinski DA, Abreu F. Applications of Magnetotactic Bacteria, Magnetosomes and Magnetosome Crystals in Biotechnology and Nanotechnology: Mini-Review. Mol Basel Switz. 2018. September 24;23(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuda Y, Okamura Y, Takeyama H, Matsunaga T. Dynamic analysis of a genomic island in Magnetospirillum sp. strain AMB-1 reveals how magnetosome synthesis developed. FEBS Lett. 2006. February 6;580(3):801–12. 10.1016/j.febslet.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 17.Schübbe S, Kube M, Scheffel A, Wawer C, Heyen U, Meyerdierks A, et al. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J Bacteriol. 2003. October;185(19):5779–90. 10.1128/JB.185.19.5779-5790.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ullrich S, Kube M, Schübbe S, Reinhardt R, Schüler D. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J Bacteriol. 2005. November;187(21):7176–84. 10.1128/JB.187.21.7176-7184.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lefèvre CT, Trubitsyn D, Abreu F, Kolinko S, Jogler C, de Almeida LGP, et al. Comparative genomic analysis of magnetotactic bacteria from the Deltaproteobacteria provides new insights into magnetite and greigite magnetosome genes required for magnetotaxis. Environ Microbiol. 2013. October;15(10):2712–35. 10.1111/1462-2920.12128 [DOI] [PubMed] [Google Scholar]
- 20.Lin W, Zhang W, Zhao X, Roberts AP, Paterson GA, Bazylinski DA, et al. Genomic expansion of magnetotactic bacteria reveals an early common origin of magnetotaxis with lineage-specific evolution. ISME J. 2018. June;12(6):1508 10.1038/s41396-018-0098-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahn-Lee L, Byrne ME, Zhang M, Sage DL, Glenn DR, Milbourne T, et al. A Genetic Strategy for Probing the Functional Diversity of Magnetosome Formation. PLoS Genet. 2015. January 8;11(1):e1004811 10.1371/journal.pgen.1004811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blakemore RP, Maratea D, Wolfe RS. Isolation and pure culture of a freshwater magnetic spirillum in chemically defined medium. J Bacteriol. 1979. November;140(2):720–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsunaga T, Nakamura C, Burgess JG, Sode K. Gene transfer in magnetic bacteria: transposon mutagenesis and cloning of genomic DNA fragments required for magnetosome synthesis. J Bacteriol. 1992. May;174(9):2748–53. 10.1128/jb.174.9.2748-2753.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsunaga T, Sakaguchi T, Tadakoro F. Magnetite formation by a magnetic bacterium capable of growing aerobically. Appl Microbiol Biotechnol. 1991. August 1;35(5):651–5. [Google Scholar]
- 25.Schleifer KH, Schüler D, Spring S, Weizenegger M, Amann R, Ludwig W, et al. The Genus Magnetospirillum gen. nov. Description of Magnetospirillum gryphiswaldense sp. nov. and Transfer of Aquaspirillum magnetotacticum to Magnetospirillum magnetotacticum comb. nov. Syst Appl Microbiol. 1991. October 1;14(4):379–85. [Google Scholar]
- 26.Nakamura C, Burgess JG, Sode K, Matsunaga T. An Iron-regulated Gene, magA, Encoding an Iron Transport Protein of Magnetospirillum sp. Strain AMB-1. J Biol Chem. 1995. November 24;270(47):28392–6. 10.1074/jbc.270.47.28392 [DOI] [PubMed] [Google Scholar]
- 27.Wahyudi AT, Takeyama H, Matsunaga T. Isolation of Magnetospirillum magneticum AMB-1 mutants defective in bacterial magnetic particle synthesis by transposon mutagenesis. Appl Biochem Biotechnol. 2001;91–93:147–54. 10.1385/abab:91-93:1-9:147 [DOI] [PubMed] [Google Scholar]
- 28.Schübbe S, Williams TJ, Xie G, Kiss HE, Brettin TS, Martinez D, et al. Complete Genome Sequence of the Chemolithoautotrophic Marine Magnetotactic Coccus Strain MC-1. Appl Env Microbiol. 2009. July 15;75(14):4835–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smalley MD, Marinov GK, Bertani LE, DeSalvo G. Genome Sequence of Magnetospirillum magnetotacticum Strain MS-1. Genome Announc. 2015. April 2;3(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grünberg K, Wawer C, Tebo BM, Schüler D. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl Environ Microbiol. 2001. October;67(10):4573–82. 10.1128/AEM.67.10.4573-4582.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schübbe S, Würdemann C, Peplies J, Heyen U, Wawer C, Glöckner FO, et al. Transcriptional organization and regulation of magnetosome operons in Magnetospirillum gryphiswaldense. Appl Environ Microbiol. 2006. September;72(9):5757–65. 10.1128/AEM.00201-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultheiss D, Schüler D. Development of a genetic system for Magnetospirillum gryphiswaldense. Arch Microbiol. 2003. February;179(2):89–94. 10.1007/s00203-002-0498-z [DOI] [PubMed] [Google Scholar]
- 33.Matsunaga T, Okamura Y, Fukuda Y, Wahyudi AT, Murase Y, Takeyama H. Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. strain AMB-1. DNA Res Int J Rapid Publ Rep Genes Genomes. 2005;12(3):157–66. [DOI] [PubMed] [Google Scholar]
- 34.Komeili A, Vali H, Beveridge TJ, Newman DK. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc Natl Acad Sci U S A. 2004. March 16;101(11):3839–44. 10.1073/pnas.0400391101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grünberg K, Müller E-C, Otto A, Reszka R, Linder D, Kube M, et al. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl Environ Microbiol. 2004. February;70(2):1040–50. 10.1128/AEM.70.2.1040-1050.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murat D, Quinlan A, Vali H, Komeili A. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc Natl Acad Sci U S A. 2010. March 23;107(12):5593–8. 10.1073/pnas.0914439107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rioux J-B, Philippe N, Pereira S, Pignol D, Wu L-F, Ginet N. A second actin-like MamK protein in Magnetospirillum magneticum AMB-1 encoded outside the genomic magnetosome island. PLoS ONE. 2010. February 10;5(2):e9151 10.1371/journal.pone.0009151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abreu N, Mannoubi S, Ozyamak E, Pignol D, Ginet N, Komeili A. Interplay between two bacterial actin homologs, MamK and MamK-Like, is required for the alignment of magnetosome organelles in Magnetospirillum magneticum AMB-1. J Bacteriol. 2014. September;196(17):3111–21. 10.1128/JB.01674-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Wang Q, Zhang W, Wang Y, Li L, Wen T, et al. Complete Genome Sequence of Magnetospirillum gryphiswaldense MSR-1. Genome Announc. 2014. March 13;2(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohße A, Borg S, Raschdorf O, Kolinko I, Tompa É, Pósfai M, et al. Genetic Dissection of the mamAB and mms6 Operons Reveals a Gene Set Essential for Magnetosome Biogenesis in Magnetospirillum gryphiswaldense. J Bacteriol. 2014. July;196(14):2658–69. 10.1128/JB.01716-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lohße A, Ullrich S, Katzmann E, Borg S, Wanner G, Richter M, et al. Functional Analysis of the Magnetosome Island in Magnetospirillum gryphiswaldense: The mamAB Operon Is Sufficient for Magnetite Biomineralization. PLoS ONE [Internet]. 2011. October 17;6(10). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3197154/. [cited 2019 Feb 7]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolinko I, Lohße A, Borg S, Raschdorf O, Jogler C, Tu Q, et al. Biosynthesis of magnetic nanostructures in a foreign organism by transfer of bacterial magnetosome gene clusters. Nat Nanotechnol. 2014. March;9(3):193–7. 10.1038/nnano.2014.13 [DOI] [PubMed] [Google Scholar]
- 43.Cornejo E, Subramanian P, Li Z, Jensen GJ, Komeili A. Dynamic Remodeling of the Magnetosome Membrane Is Triggered by the Initiation of Biomineralization. mBio. 2016. March 2;7(1):e01898–15. 10.1128/mBio.01898-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raschdorf O, Forstner Y, Kolinko I, Uebe R, Plitzko JM, Schüler D. Genetic and Ultrastructural Analysis Reveals the Key Players and Initial Steps of Bacterial Magnetosome Membrane Biogenesis. PLoS Genet. 2016. June 10;12(6):e1006101 10.1371/journal.pgen.1006101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uebe R, Junge K, Henn V, Poxleitner G, Katzmann E, Plitzko JM, et al. The cation diffusion facilitator proteins MamB and MamM of Magnetospirillum gryphiswaldense have distinct and complex functions, and are involved in magnetite biomineralization and magnetosome membrane assembly. Mol Microbiol. 2011. November;82(4):818–35. 10.1111/j.1365-2958.2011.07863.x [DOI] [PubMed] [Google Scholar]
- 46.Murat D, Falahati V, Bertinetti L, Csencsits R, Körnig A, Downing K, et al. The magnetosome membrane protein, MmsF, is a major regulator of magnetite biomineralization in Magnetospirillum magneticum AMB-1. Mol Microbiol. 2012. August;85(4):684–99. 10.1111/j.1365-2958.2012.08132.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siponen MI, Legrand P, Widdrat M, Jones SR, Zhang W-J, Chang MCY, et al. Structural insight into magnetochrome-mediated magnetite biomineralization. Nature. 2013. October 31;502(7473):681–4. 10.1038/nature12573 [DOI] [PubMed] [Google Scholar]
- 48.Rawlings AE, Bramble JP, Walker R, Bain J, Galloway JM, Staniland SS. Self-assembled MmsF proteinosomes control magnetite nanoparticle formation in vitro. Proc Natl Acad Sci. 2014. November 11;111(45):16094–9. 10.1073/pnas.1409256111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kashyap S, Woehl TJ, Liu X, Mallapragada SK, Prozorov T. Nucleation of Iron Oxide Nanoparticles Mediated by Mms6 Protein in Situ. ACS Nano. 2014. September 23;8(9):9097–106. 10.1021/nn502551y [DOI] [PubMed] [Google Scholar]
- 50.Amemiya Y, Arakaki A, Staniland SS, Tanaka T, Matsunaga T. Controlled formation of magnetite crystal by partial oxidation of ferrous hydroxide in the presence of recombinant magnetotactic bacterial protein Mms6. Biomaterials. 2007. December;28(35):5381–9. 10.1016/j.biomaterials.2007.07.051 [DOI] [PubMed] [Google Scholar]
- 51.Yamagishi A, Tanaka M, Lenders JJM, Thiesbrummel J, Sommerdijk NAJM, Matsunaga T, et al. Control of magnetite nanocrystal morphology in magnetotactic bacteria by regulation of mms7 gene expression. In: Scientific reports. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arakaki A, Yamagishi A, Fukuyo A, Tanaka M, Matsunaga T. Co-ordinated functions of Mms proteins define the surface structure of cubo-octahedral magnetite crystals in magnetotactic bacteria. Mol Microbiol. 2014. August;93(3):554–67. 10.1111/mmi.12683 [DOI] [PubMed] [Google Scholar]
- 53.Jones SR, Wilson TD, Brown ME, Rahn-Lee L, Yu Y, Fredriksen LL, et al. Genetic and biochemical investigations of the role of MamP in redox control of iron biomineralization in Magnetospirillum magneticum. Proc Natl Acad Sci U S A. 2015. March 31;112(13):3904–9. 10.1073/pnas.1417614112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schüler D. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature. 2006. March;440(7080):110–4. 10.1038/nature04382 [DOI] [PubMed] [Google Scholar]
- 55.Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006. January 13;311(5758):242–5. 10.1126/science.1123231 [DOI] [PubMed] [Google Scholar]
- 56.Draper O, Byrne ME, Li Z, Keyhani S, Barrozo JC, Jensen G, et al. MamK, a bacterial actin, forms dynamic filaments in vivo that are regulated by the acidic proteins MamJ and LimJ. Mol Microbiol. 2011. October;82(2):342–54. 10.1111/j.1365-2958.2011.07815.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toro-Nahuelpan M, Müller FD, Klumpp S, Plitzko JM, Bramkamp M, Schüler D. Segregation of prokaryotic magnetosomes organelles is driven by treadmilling of a dynamic actin-like MamK filament. BMC Biol. 2016. 12;14(1):88 10.1186/s12915-016-0290-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ozyamak E, Kollman J, Agard DA, Komeili A. The bacterial actin MamK: in vitro assembly behavior and filament architecture. J Biol Chem. 2013. February 8;288(6):4265–77. 10.1074/jbc.M112.417030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toro-Nahuelpan M, Giacomelli G, Raschdorf O, Borg S, Plitzko JM, Bramkamp M, et al. MamY is a membrane-bound protein that aligns magnetosomes and the motility axis of helical magnetotactic bacteria. Nat Microbiol. 2019. July 29; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katzmann E, Scheffel A, Gruska M, Plitzko JM, Schüler D. Loss of the actin-like protein MamK has pleiotropic effects on magnetosome formation and chain assembly in Magnetospirillum gryphiswaldense. Mol Microbiol. 2010;77(1):208–24. 10.1111/j.1365-2958.2010.07202.x [DOI] [PubMed] [Google Scholar]
- 61.Taoka A, Kiyokawa A, Uesugi C, Kikuchi Y, Oestreicher Z, Morii K, et al. Tethered Magnets Are the Key to Magnetotaxis: Direct Observations of Magnetospirillum magneticum AMB-1 Show that MamK Distributes Magnetosome Organelles Equally to Daughter Cells. mBio. 2017. 08;8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uebe R, Schüler D. Magnetosome biogenesis in magnetotactic bacteria. Nat Rev Microbiol. 2016. 13;14(10):621–37. 10.1038/nrmicro.2016.99 [DOI] [PubMed] [Google Scholar]
- 63.Barber-Zucker S, Zarivach R. A Look into the Biochemistry of Magnetosome Biosynthesis in Magnetotactic Bacteria. ACS Chem Biol. 2017. 20;12(1):13–22. 10.1021/acschembio.6b01000 [DOI] [PubMed] [Google Scholar]
- 64.Schüler D. Genetics and cell biology of magnetosome formation in magnetotactic bacteria. FEMS Microbiol Rev. 2008;32(4):654–72. 10.1111/j.1574-6976.2008.00116.x [DOI] [PubMed] [Google Scholar]
- 65.Raschdorf O, Müller FD, Pósfai M, Plitzko JM, Schüler D. The magnetosome proteins MamX, MamZ and MamH are involved in redox control of magnetite biomineralization in Magnetospirillum gryphiswaldense. Mol Microbiol. 2013. September;89(5):872–86. 10.1111/mmi.12317 [DOI] [PubMed] [Google Scholar]
- 66.Müller FD, Raschdorf O, Nudelman H, Messerer M, Katzmann E, Plitzko JM, et al. The FtsZ-Like Protein FtsZm of Magnetospirillum gryphiswaldense Likely Interacts with Its Generic Homolog and Is Required for Biomineralization under Nitrate Deprivation. J Bacteriol. 2014. February 1;196(3):650–9. 10.1128/JB.00804-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ding Y, Li J, Liu J, Yang J, Jiang W, Tian J, et al. Deletion of the ftsZ-Like Gene Results in the Production of Superparamagnetic Magnetite Magnetosomes in Magnetospirillum gryphiswaldense. J Bacteriol. 2010. February;192(4):1097–105. 10.1128/JB.01292-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Y, Katzmann E, Borg S, Schüler D. The Periplasmic Nitrate Reductase Nap Is Required for Anaerobic Growth and Involved in Redox Control of Magnetite Biomineralization in Magnetospirillum gryphiswaldense. J Bacteriol. 2012. September 15;194(18):4847–56. 10.1128/JB.00903-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, Raschdorf O, Silva KT, Schüler D. The Terminal Oxidase cbb3 Functions in Redox Control of Magnetite Biomineralization in Magnetospirillum gryphiswaldense. J Bacteriol. 2014. July;196(14):2552–62. 10.1128/JB.01652-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Y, Sabaty M, Borg S, Silva KT, Pignol D, Schüler D. The oxygen sensor MgFnr controls magnetite biomineralization by regulation of denitrification in Magnetospirillum gryphiswaldense. BMC Microbiol. 2014. June 10;14(1):153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fillat MF. The FUR (ferric uptake regulator) superfamily: Diversity and versatility of key transcriptional regulators. Arch Biochem Biophys. 2014. March 15;546:41–52. 10.1016/j.abb.2014.01.029 [DOI] [PubMed] [Google Scholar]
- 72.Uebe R, Voigt B, Schweder T, Albrecht D, Katzmann E, Lang C, et al. Deletion of a fur-like gene affects iron homeostasis and magnetosome formation in Magnetospirillum gryphiswaldense. J Bacteriol. 2010. August;192(16):4192–204. 10.1128/JB.00319-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rong C, Huang Y, Zhang W, Jiang W, Li Y, Li J. Ferrous iron transport protein B gene (feoB1) plays an accessory role in magnetosome formation in Magnetospirillum gryphiswaldense strain MSR-1. Res Microbiol. 2008. October;159(7–8):530–6. 10.1016/j.resmic.2008.06.005 [DOI] [PubMed] [Google Scholar]
- 74.Suzuki T, Okamura Y, Calugay RJ, Takeyama H, Matsunaga T. Global Gene Expression Analysis of Iron-Inducible Genes in Magnetospirillum magneticum AMB-1. J Bacteriol. 2006. March;188(6):2275–9. 10.1128/JB.188.6.2275-2279.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taoka A, Umeyama C, Fukumori Y. Identification of iron transporters expressed in the magnetotactic bacterium Magnetospirillum magnetotacticum. Curr Microbiol. 2009. February;58(2):177–81. 10.1007/s00284-008-9305-7 [DOI] [PubMed] [Google Scholar]
- 76.Wang Q, Wang M, Wang X, Guan G, Li Y, Peng Y, et al. Iron Response Regulator Protein IrrB in Magnetospirillum gryphiswaldense MSR-1 Helps Control the Iron/Oxygen Balance, Oxidative Stress Tolerance, and Magnetosome Formation. Appl Environ Microbiol. 2015. December;81(23):8044–53. 10.1128/AEM.02585-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin W, Deng A, Wang Z, Li Y, Wen T, Wu L-F, et al. Genomic insights into the uncultured genus “Candidatus Magnetobacterium” in the phylum Nitrospirae. ISME J. 2014. December;8(12):2463–77. 10.1038/ismej.2014.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sakaguchi T, Burgess JG, Matsunaga T. Magnetite formation by a sulphate-reducing bacterium. Nature. 1993. September;365(6441):47. [Google Scholar]
- 79.Sakaguchi T, Arakaki A, Matsunaga T. Desulfovibrio magneticus sp. nov., a novel sulfate-reducing bacterium that produces intracellular single-domain-sized magnetite particles. Int J Syst Evol Microbiol. 2002. January;52(1):215–21. [DOI] [PubMed] [Google Scholar]
- 80.Grant CR, Rahn-Lee L, LeGault KN, Komeili A. Genome Editing Method for the Anaerobic Magnetotactic Bacterium Desulfovibrio magneticus RS-1. Appl Env Microbiol. 2018. November 15;84(22):e01724–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Byrne ME, Ball DA, Guerquin-Kern J-L, Rouiller I, Wu T-D, Downing KH, et al. Desulfovibrio magneticus RS-1 contains an iron- and phosphorus-rich organelle distinct from its bullet-shaped magnetosomes. Proc Natl Acad Sci. 2010. July 6;107(27):12263–8. 10.1073/pnas.1001290107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kolinko S, Richter M, Glöckner F-O, Brachmann A, Schüler D. Single-cell genomics of uncultivated deep-branching magnetotactic bacteria reveals a conserved set of magnetosome genes. Environ Microbiol. 2016. January;18(1):21–37. 10.1111/1462-2920.12907 [DOI] [PubMed] [Google Scholar]
- 83.Descamps ECT, Monteil CL, Menguy N, Ginet N, Pignol D, Bazylinski DA, et al. Desulfamplus magnetovallimortis gen. nov., sp. nov., a magnetotactic bacterium from a brackish desert spring able to biomineralize greigite and magnetite, that represents a novel lineage in the Desulfobacteraceae. Syst Appl Microbiol. 2017. July;40(5):280–9. 10.1016/j.syapm.2017.05.001 [DOI] [PubMed] [Google Scholar]
- 84.Monteil CL, Perrière G, Menguy N, Ginet N, Alonso B, Waisbord N, et al. Genomic study of a novel magnetotactic Alphaproteobacteria uncovers the multiple ancestry of magnetotaxis. Environ Microbiol. 2018. December;20(12):4415–30. 10.1111/1462-2920.14364 [DOI] [PubMed] [Google Scholar]
- 85.Lefèvre CT, Menguy N, Abreu F, Lins U, Pósfai M, Prozorov T, et al. A cultured greigite-producing magnetotactic bacterium in a novel group of sulfate-reducing bacteria. Science. 2011. December 23;334(6063):1720–3. 10.1126/science.1212596 [DOI] [PubMed] [Google Scholar]
- 86.DeLong EF, Frankel RB, Bazylinski DA. Multiple Evolutionary Origins of Magnetotaxis in Bacteria. Science. 1993;259(5096):803–6. 10.1126/science.259.5096.803 [DOI] [PubMed] [Google Scholar]
- 87.Abreu F, Cantão ME, Nicolás MF, Barcellos FG, Morillo V, Almeida LG, et al. Common ancestry of iron oxide- and iron-sulfide-based biomineralization in magnetotactic bacteria. ISME J. 2011. October;5(10):1634–40. 10.1038/ismej.2011.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lefèvre CT, Trubitsyn D, Abreu F, Kolinko S, Almeida LGP de, Vasconcelos ATR de, et al. Monophyletic origin of magnetotaxis and the first magnetosomes. Environ Microbiol. 2013;15(8):2267–74. 10.1111/1462-2920.12097 [DOI] [PubMed] [Google Scholar]
- 89.Yang W, Li R, Peng T, Zhang Y, Jiang W, Li Y, et al. mamO and mamE genes are essential for magnetosome crystal biomineralization in Magnetospirillum gryphiswaldense MSR-1. Res Microbiol. 2010. October;161(8):701–5. 10.1016/j.resmic.2010.07.002 [DOI] [PubMed] [Google Scholar]
- 90.Quinlan A, Murat D, Vali H, Komeili A. The HtrA/DegP family protease MamE is a bifunctional protein with roles in magnetosome protein localization and magnetite biomineralization. Mol Microbiol. 2011. May;80(4):1075–87. 10.1111/j.1365-2958.2011.07631.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hershey DM, Ren X, Melnyk RA, Browne PJ, Ozyamak E, Jones SR, et al. MamO Is a Repurposed Serine Protease that Promotes Magnetite Biomineralization through Direct Transition Metal Binding in Magnetotactic Bacteria. PLoS Biol. 2016. March;14(3):e1002402 10.1371/journal.pbio.1002402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hershey DM, Browne PJ, Iavarone AT, Teyra J, Lee EH, Sidhu SS, et al. Magnetite Biomineralization in Magnetospirillum magneticum Is Regulated by a Switch-like Behavior in the HtrA Protease MamE. J Biol Chem. 2016. 19;291(34):17941–52. 10.1074/jbc.M116.731000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wetmore KM, Price MN, Waters RJ, Lamson JS, He J, Hoover CA, et al. Rapid Quantification of Mutant Fitness in Diverse Bacteria by Sequencing Randomly Bar-Coded Transposons. mBio. 2015. July 1;6(3):e00306–15. 10.1128/mBio.00306-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen H, Zhang S-D, Chen L, Cai Y, Zhang W-J, Song T, et al. Efficient Genome Editing of Magnetospirillum magneticum AMB-1 by CRISPR-Cas9 System for Analyzing Magnetotactic Behavior. Front Microbiol. 2018. July 17;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Peters JM, Colavin A, Shi H, Czarny TL, Larson MH, Wong S, et al. A Comprehensive, CRISPR-based Functional Analysis of Essential Genes in Bacteria. Cell. 2016. June 2;165(6):1493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peters JM, Koo B-M, Patino R, Heussler GE, Hearne CC, Qu J, et al. Enabling genetic analysis of diverse bacteria with Mobile-CRISPRi. Nat Microbiol. 2019. February;4(2):244–50. 10.1038/s41564-018-0327-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tay A, Pfeiffer D, Rowe K, Tannenbaum A, Popp F, Strangeway R, et al. High-Throughput Microfluidic Sorting of Live Magnetotactic Bacteria. Appl Env Microbiol. 2018. September 1;84(17):e01308–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tay A, McCausland H, Komeili A, Carlo DD. Nano and Microtechnologies for the Study of Magnetotactic Bacteria. Adv Funct Mater. 2019;29(38):1904178. [Google Scholar]
- 99.Wen T, Guo F, Zhang Y, Tian J, Li Y, Li J, et al. A novel role for Crp in controlling magnetosome biosynthesis in Magnetospirillum gryphiswaldense MSR-1. Sci Rep. 2016. February 16;6:21156 10.1038/srep21156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lefèvre CT, Bennet M, Landau L, Vach P, Pignol D, Bazylinski DA, et al. Diversity of magneto-aerotactic behaviors and oxygen sensing mechanisms in cultured magnetotactic bacteria. Biophys J. 2014. July 15;107(2):527–38. 10.1016/j.bpj.2014.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Popp F, Armitage JP, Schüler D. Polarity of bacterial magnetotaxis is controlled by aerotaxis through a common sensory pathway. Nat Commun. 2014. November 14;5:5398 10.1038/ncomms6398 [DOI] [PubMed] [Google Scholar]
- 102.Simmons SL, Sievert SM, Frankel RB, Bazylinski DA, Edwards KJ. Spatiotemporal distribution of marine magnetotactic bacteria in a seasonally stratified coastal salt pond. Appl Environ Microbiol. 2004. October;70(10):6230–9. 10.1128/AEM.70.10.6230-6239.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Flies CB, Jonkers HM, de Beer D, Bosselmann K, Böttcher ME, Schüler D. Diversity and vertical distribution of magnetotactic bacteria along chemical gradients in freshwater microcosms. FEMS Microbiol Ecol. 2005. April 1;52(2):185–95. 10.1016/j.femsec.2004.11.006 [DOI] [PubMed] [Google Scholar]
- 104.Liu J, Zhang W, Du H, Leng X, Li J-H, Pan H, et al. Seasonal changes in the vertical distribution of two types of multicellular magnetotactic prokaryotes in the sediment of Lake Yuehu, China. Environ Microbiol Rep. 2018;10(4):475–84. 10.1111/1758-2229.12652 [DOI] [PubMed] [Google Scholar]
- 105.Moisescu C, Ardelean II, Benning LG. The effect and role of environmental conditions on magnetosome synthesis. Front Microbiol. 2014. February 11;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bazylinski DA, Lefèvre CT. Magnetotactic bacteria from extreme environments. Life Basel Switz. 2013. March 26;3(2):295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]