Abstract
Prediction and early detection of kidney damage induced by nonsteroidal anti-inflammatories (NSAIDs) would provide the best chances of maximizing the anti-inflammatory effects while minimizing the risk of kidney damage. Unfortunately, biomarkers for detecting NSAID-induced kidney damage in cats remain to be discovered. To identify potential urinary biomarkers for monitoring NSAID-based treatments, we applied an untargeted metabolomics approach to urine collected from cats treated repeatedly with meloxicam or saline for up to 17 days. Applying multivariate analysis, this study identified a panel of seven metabolites that discriminate meloxicam treated from saline treated cats. Combining artificial intelligence machine learning algorithms and an independent testing urinary metabolome data set from cats with meloxicam-induced kidney damage, a panel of metabolites was identified and validated. The panel of metabolites including tryptophan, tyrosine, taurine, threonic acid, pseudouridine, xylitol and lyxitol, successfully distinguish meloxicam-treated and saline-treated cats with up to 75–100% sensitivity and specificity. This panel of urinary metabolites may prove a useful and non-invasive diagnostic tool for monitoring potential NSAID induced kidney injury in feline patients and may act as the framework for identifying urine biomarkers of NSAID induced injury in other species.
1. Introduction
Non-steroidal anti-inflammatory drugs (NSAIDs) are massively used in both veterinary and human patients for treating pain and inflammation with approximately 111 million prescriptions dispensed each year in the U.S alone [1].
The main goal of the pharmacological control of pain and inflammation is to prevent suffering, avoid further tissue damage, and promote the recovery of normal tissue structure and organ function(s). Nevertheless, one of the greatest challenges in therapeutics is the effective and safe long-term use of NSAIDs for conditions requiring sustained anti-inflammatory or pain relief because the repeated administration of NSAIDS can result in gastrointestinal and kidney damage [2], [3], [4].
Early stages of kidney injury are often subclinical. If kidney damage following acute kidney injury (AKI) is not appropriately repaired with normal renal parenchyma, this can lead to chronic kidney disease (CKD) [5]. NSAID-induced kidney damage severely limits the possibility to provide optimal treatment for a variety of conditions, including: degenerative joint disease, cancer, etc., all of which require long-term anti-inflammatory therapy.
A potential contributing factor to the development of kidney disease is delayed detection due to the lack of non-invasive and sensitive diagnostic tests. The most commonly utilized clinical diagnostic tests for detection and monitoring of AKI and CKD include serum biochemical values including blood urea nitrogen (BUN), creatinine and urine concentrating ability. However, these tests have severe limitations which hamper early detection and diagnosis of kidney damage [6, 7]. Unfortunately, serum creatinine and BUN are late markers of kidney filtration and function, with limited specificity and impaired capacity to accurately reflect the timing and severity of injury.
The European Medicines Agency and the US Food and Drug Administration have encouraged further development of eight biomarkers of drug-induced-kidney-injury in early clinical trials (EMEA/FDA guideline on detection of drug-induced nephrotoxicity. 2009). To date, however, no specific biomarkers have been identified to detect early kidney damage induced by NSAIDs. A single-biomarker-to-single-disease approach is highly unlikely to capture the multifactorial nature of a dynamic progressive process such as kidney disease; whereas a panel of biomarkers would increase the likelihood of detecting early subclinical kidney damage.
Metabolomics can be applied to many body fluids, such as urine and blood, to identify alterations in concentrations of 0.8–1.2 kDa molecules. Altered metabolites act as a chemical fingerprint and can then potentially be utilized as biomarkers for detecting or monitoring biological processes [8, 9]. Urine is commonly used to study diseases of the renal system because it offers advantages owing to its minimal invasiveness and non–volume-limited nature. In addition, urine can be used for measuring time-related metabolic alterations—a valuable feature for the study of progression and prognostication of acute and chronic diseases [10]. Urine metabolomics has been successfully applied to understand the mechanism(s) underlying drug toxicity and discover novel biomarkers for detecting kidney disease [11].
The repeated administration of NSAIDs in cats is known to damage kidneys [12]. Therefore we hypothesize that the repeated administration of meloxicam alters the urine metabolome. We addressed this hypothesis by comparing the urine metabolome in meloxicam- and saline treated cats. Considering that the metabolite profile is analogous to a chemical fingerprint left behind by NSAID-induced alterations in cellular processes another objective of this study is to identify small molecular weight substances in urine that could be further studied as putative biomarkers for monitoring the administration of NSAIDs. Identification of putative biomarkers for monitoring the administration of NSAIDs was accomplished by applying multivariate analysis and artificial intelligence machine learning algorithms.
Artificial intelligence is a rapidly expanding specialty within the field of computer science. Current uses for artificial intelligence include prediction of Amazon shopping preferences, search engine recommendations, the training of Google self-driving cars and prediction of patient outcomes from medical ailments such as myocardial infarction [13]. Machine learning is a subbranch of artificial intelligence in which patterns and statistical models are extracted from large complex data sets to train algorithms to identify potentially complex patterns and interactions within large data sets (e.g. metabolomics data) [14, 15]. These learned patterns can be applied to test data sets of interest to make outcome predictions. In the present study, we trained machine learning algorithms on an initial urine metabolomics training data set to make data driven identification of putative urinary biomarkers for monitoring cats treated repeatedly with an anti-inflammatory dose of meloxicam. Then, the panel of metabolites was validated using an independent testing urinary metabolome data set from cats with meloxicam-induced kidney damage.
2. Materials and methods
The Washington State University Institutional Animal Care and Use Committee approved all study procedures before their initiation (ASAF#04915 and #04662). All methods were carried out in accordance with the relevant guidelines and regulations.
2.1 Study population and inclusion criteria
For the biomarker discovery experiment, a training data set of 12 female clinically healthy intact adult (1–1.5 years old) purpose bred cats (2.5–3.8 kg) were obtained from a USDA-licensed commercial breeder (Nutrition and pet Care Center UC DAVIS, Davis, USA).
For the biomarker validation experiment, a testing data set of 8 female clinically healthy intact adult (1–2 years old) cats (2.8–3.7 kg) were obtained from a USDA-licensed commercial breeder (Liberty Research, Inc., Waverly, NY).
The enrollment criteria for both training and testing data set cats were no signs of illness or infection and no history of drug treatment within 30 days prior to initiation of the study. Each cat enrolled in the study was given a baseline complete history, physical examination, and laboratory evaluation. All cats had normal complete blood count, serum chemistry, and urinalysis. All cats were vaccinated for rabies prior to transfer to Washington State University.
2.1.1 Animal management and monitoring
Cats from both groups were acclimated to the new housing environment at least 10 days before beginning the study. Each training data set cat was housed separately in cages 49” wide, 37” tall and 38” deep. Cages were set up in the room in a way compatible cats could see one another. Each testing data set cat was housed separately in a 4’ X 8’ partitioned room. A see-through barrier partitioned the room so the cats could see each other. Bedding, cat trees, and additional enrichment, including toys, were provided to all cats.
For both groups of cats, the room was temperature (21–23°C), humidity (25–35%) and 12h light/dark cycle controlled. The cats had free access to drinking water and food (Purina Cat Chow Indoor Formula) throughout the study. Fresh water and food were changed every day. Cages/rooms were cleaned, and litter boxes changed every day. Cats were examined at least twice daily during the entire study to rule out possible health problems. Body weight and condition scores were assessed every 2 days during the duration of the study.
Following the acclimation period, vascular access ports (VAP, petite size) (Le Port Companion Port, Norfolk Vet, Skokie, IL, USA) were implanted subcutaneously in the training data set of cats (n = 12) at least 7 days before starting the administration of the treatments following standard procedures as recommended by the manufacturer. VAPs were implanted aseptically in the jugular vein. The aspiration port was implanted subcutaneously in the dorsal aspect of the neck cranial to the interscapular space. The VAPs were maintained with heparinized saline solution (100 I.U./mL, 0.7 mL each time) following the manufacturer recommendations.
A long-term indwelling jugular catheter (Mila International, Inc) was placed in each testing data set of cats at least 1 day before starting the study. The jugular catheters were maintained with 0.5 mL of heparinized saline solution (10 IU/mL, 2–4 times a day).
2.1.2 Controlled randomized experimental design
Following the VAPs implantation, the training data cats were randomly allocated to two groups; a saline control group (n = 6) and a meloxicam treatment group (n = 6). For the testing data set cats, following the acclimation period, cats were allocated randomly to control (n = 4) and meloxicam (n = 4) groups. Randomization of the treatments for both the training and testing set was done using simple randomization in R studio. At the end of the treatment phase, three cats from each training data set group were euthanized within 24h after the last treatment. The remaining cats in the training data set group (n = 3 per group) were monitored for up to 16 days after the last administration of treatments and were euthanized within 24h after the last monitoring day. All testing data set cats (n = 8) were euthanized within 24h of the last treatment. An intravenous overdose of pentobarbital (Beuthanasia-D, Intervet/Merck Animal Health, Giralda Farms, Madison, NJ) was used for euthanasia of all cats.
2.1.3 Treatment administrations
Prior to initiation of the biomarker discovery experiment, training data set cats (n = 12) were treated subcutaneously with 0.1 mL/kg body weight of saline every 24h on days -3, -2 and -1 (6 p.m. +/- 1h). Beginning on day zero, cats in the meloxicam group were treated subcutaneously with meloxicam at a dosage of 0.3 mg meloxicam/kg body weight (equivalent to 0.1 mL/kg) (Metacam® injectable, Boehringer Ingelheim Vetmedica, inc) every 24h for 31 days. Cats in the control group received 0.1 mL/kg body weight of saline subcutaneously every 24h. The duration of these treatments was defined based on preliminary experiment information.
For the biomarker validation experiment, testing data set cats in the control group were treated subcutaneously with a volume of saline at 0.1 ml/kg of body weight (equivalent volume) every 24 h (7 a.m. +/- 2 h) for 17 days. Testing data set cats were treated with 0.3 mg meloxicam/kg body weight (Metacam® Injectable, Boehringer Ingelheim Vetmedica, Inc) every 24 h (7 a.m. +/- 1 h) for 17 days.
2.1.4 Blood, plasma and urine collection
Blood and urine samples were collected on days -3, -1, 4, 9, 13, 17, 23, 26, 31, 34, 40, and 47 for the training data set of cats and on days -1, 2, 13 and 17 for the testing data set of cats. For both sets of cats, food was withheld 8h prior to collection of blood and urine samples. Blood and urine samples were collected during the afternoon (7 p.m. +/- 1h). Blood samples were collected into 3.2% sodium citrate tubes and kept in ice until centrifugation. Plasma samples were collected by centrifugation (1800 x g during 8 min) within one hour upon blood sample collection. Urine samples were collected by ultrasound-guided cystocentesis following standard procedures. Upon urine collection, approximately 100 μL of urine was used to estimate the urinary specific gravity. The remaining volume of the urine sample was maintained in ice and was centrifuged at 1800 x g for 8 min within one hour upon collection. The supernatant was aliquoted and stored at -80°C until metabolomic analysis. Urine metabolomic analysis was done within 6 months of sample collection. One cat in the treatment group of the testing data set of cats (M4) was euthanized early due to declining health and samples were collected on days 4, 9, 11 for this cat. The urine samples collected for cat M4 on days 9 and 11 were included in the metabolomics statistical analysis for days 13 and 17, respectively.
2.1.5 Creatinine quantification and urine specific gravity (USG) determination
For both the training and testing data set urine samples, plasma creatinine concentrations were measured using the QuantiChrom TM Creatinine Assay Kit (BioAssay Systems, Hayward, CA). Urinary specific gravity was determined at room temperature within 2h upon sample collection using a refractometry.
2.1.6 Kidney histopathology
Immediately following euthanasia, cross sections of the left and right kidney, encompassing the capsule, cortex, and medulla, from each animal from both the training and testing data sets were trimmed to a maximum thickness of 0.5 cm and fixed in 10% formaldehyde. Paraffin-embedded kidney specimens were cut at a 5 μm thickness and stained with either hematoxylin and eosin (H&E), Masson’s trichrome, or periodic acid-Schiff methenamine silver (PAMS) stains. Each of the three stains was performed on each kidney sample. Histologic changes of the renal tissues were scored by a blinded pathologist using a modified semiquantitative scoring system [16]. A representative section of at least one kidney from each testing data set animal was scored. Traits related to tubular-interstitial disease, including cortical and medullary tubular damage, basement membrane integrity, interstitial fibrosis, inflammation and if present, the distribution of the inflammation were evaluated counting 50 cortical and medullary tubules from at least 10 different fields for each animal. A semiquantitative scoring system was used as follows: 0, no disease; 1, 1–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100% of tissue affected. Higher scores represent more severe histologic changes.
2.2 Urine metabolomics
2.2.1 Metabolite determination
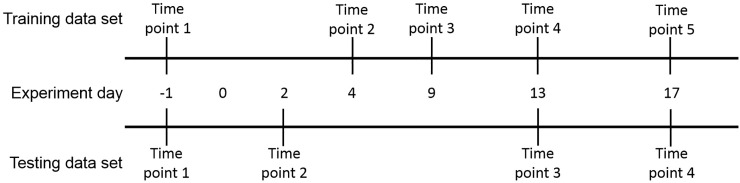
Metabolomics analyses for the training and testing data sets were performed at the West Coast Metabolomics Center (Davis, CA), a National Institute of Health Regional resource core. For the training data set, metabolomics was performed on urine samples collected from meloxicam- and saline-treated cats day -1 (time point 1) and on days 4, 9, 13, and 17 (time points 2–5, respectively). For the testing data set, metabolomics was performed on urine samples from control and treated cats collected on day -1 (time point 1) and on days 2, 13, and 17 (time points 2–4, respectively). A graphic representation of the sampling time line is provided in Fig 1. The methods used to determine the urine feline metabolome was fully described elsewhere [17, 18] and described briefly as follows.
Fig 1. Sampling schedule for metabolite determination in the training and testing groups.
Day -1 represents the pretreatment time point. For both the training and testing data sets, starting on day 0 cats in the treatment groups for each set received 0.3 mg/kg meloxicam every 24h till the end of the study. Cats in the control groups received 0.1 mL/kg body weight of saline every 24h until the studies end.
2.2.2 Metabolite identification
The metabolomics data acquisition for feline plasma and urine samples was performed at the West Coast Metabolomics Center (Davis, California), a National Institute of Health regional resource laboratory on May 2017 for the testing data set samples and March 2016 for the training data set samples. An in-depth description of the methods utilized have been previously published and are summarized below ([17], [19], [20]).
2.2.2.1 Extraction, clean-up and derivatization. A 30 μL aliquot of urine sample was added to a 1.0 mL cooled extraction solution composed of acetonitrile and isopropanol (JT Baker Center Valley, PA, USA) and 18.2 MΩ water (MilliQ) at a 3:3:2 v/v/v ratio in a 1.5 mL polypropylene tube. Before adding the extraction mixture to the study samples, oxygen was removed by sparging nitrogen for 5 minutes. Internal standards with isotope-labeled metabolite surrogates were added to the study samples. The study samples were mixed by vortexing for 10 s and then placed in a chilled (4°C) shaker for 5 mins. Samples were then centrifuged at 14,000 rcf for 2 minutes to pellet insoluble material. A 450 μL aliquot of the supernatant was removed, placed in a fresh 1.5 mL polypropylene tube and evaporated using a cold trap concentrator. The dried samples were re-suspended with 450 μL of 50% acetonitrile and centrifuged at 14,000 rcf for 2 minutes. The supernatant was transferred to a new tube and desiccated using a cold trap concentrator. The dried samples were derivatized by adding 10 μL of 40 mg/mL of methoxyamine hydrochloride (Aldrich, St. Louis, MO, USA) and shaken at maximum speed at room temp 30°C for 1.5 h. A 10 μl aliquot of fatty acid methyl esters (FAME, recipe provided in Fiehn 2017) was added to 1 mL of N-methyl-N-(trimethylsilyl)-trifluoroactamide (MSTFA: Aldrich, St. Louis, MO, USA) and vortexed for 10 s. Finally, 91 μL of the MSTFA + FAME mixture was added to each sample at shaken at 37°C at maximum speed for 30 minutes. The final mixture was transferred to glass vials containing micro-inserts and immediately sealed.
A reagent and derivatization blank was used per batch of 50 sample extractions as negative control. A reagent blank uses all reagents, but does not use extraction tubes or extraction solvents.
2.2.2.2 Chromatography and mass spectrometry data acquisition. Samples were separated using an RTX-5Sil MS analytical column made of 0.25 μm thick 95% dimethyl/5%diphenylpolysiloxane film which is 30 m long with a 0.25 mm internal diameter (Restek corporation, Bellefonte, PA USA). This analytical column is surrounded and protected by a 10 m long guard column. The mobile phase consisted of pure helium with a flow-rate of 1 mL/min and a column temperature of 50–330°C. The sample injection volume was 0.5 μL at 50°C ramped to 250°C by 12°C/s. The oven was held at 50°C for 1 minute and then ramped to 330°C and held for 5 minutes. The ramp speed for the oven is 20°C/min. An Agilent 6890 gas chromatograph with a Leco Pegasus IV gas chromatography, time-of-flight mass spectrophotometer (GC-TOF-MS) controlled with Leco ChromaTOF software version 2.32 were used for metabolite data acquisition. The mass spectrophotometer parameters included unit mass resolution at 17 spectra/s from 80–500 Da at -70 eV ionization energy and 1800 V detector voltage with a 230°C transfer line and a 250°C ion source.
2.2.2.3 Spectra data processing and metabolite identification. Raw data files were preprocessed using ChromaTOF version 2.32 with the following settings: no smoothing, 3 s peak width, baseline subtraction, automatic spectral deconvolution and peak detection at signal/noise levels of 5:1 throughout the chromatogram. Spectra which failed to meet at least one of these parameters were considered not identified for purposes of this study. All spectra identification was done using the BinBase database.
The BinBase algorithm (rtx5) used the following settings: validity of chromatogram (10^7 counts s -1), unbiased retention index marker detection (MS similarity>800, validity of intensity range for high m/z marker ions), retention index calculation by 5th order polynomial regression. Spectra were cut to 5% base peak abundance and matched to database entries from most to least abundant spectra using the following matching filters: Parameters used to identify spectra included retention index window ± 2000 U (around ± 2 s retention time deviation), unique ion and apex mass validation, mass spectrum similarity and a final isomer filter. Peak height was reported as quantification using the unique ion as default. A quantification report table was produced for all database entries that are positively detected in more than 10% of the samples of a study design class for unidentified metabolites. A subsequent post-processing module was employed to automatically replace missing values. Replaced values were labeled as ‘low confidence’ by color coding, and for each metabolite, the number of high-confidence peak detections was recorded as well as the ratio of the average height of replaced values to high-confidence peak detections. These ratios and numbers were used for manual curation of automatic report data sets to data sets released for submission. Peak heights (mz value) at the specific retention index were reported. Peak heights instead of peak areas because peak heights are more precise for low abundant metabolites than peak areas, due to the larger influence of baseline determinations on areas compared to peak heights.
2.2.3 Data processing
All metabolomics data analyses carried out in this study were performed using Metaboanalyst 4.0, an open source R-based program specifically designed for metabolomics [21]. The raw data was filtered using standard deviation to identify and remove variables which were non-informative to downstream modeling [22]. Additionally, the raw data was normalized through log (base 2) generalized transformation to the metabolite peak intensities to stabilize the variance and autoscaled by mean-centering and dividing by the standard deviation of each variable [23], [24].
2.2.4 Identification of putative biomarkers using a training metabolome data set
In order to minimize the likelihood of capturing irrelevant metabolites, we applied in tandem three statistical approaches to the training data set including partial least squares discriminant analysis (PLS-DA) variable importance of projection scores (VIP), random forest (RF) mean decrease in accuracy (MDA) values and, univariate receiver operating curve (ROC) area-under the curve (AUC) analysis to the complete set of identified metabolites, for each time point separately.
A combination of four selection criteria were used to identify metabolites as potential biomarkers in the training data set for discriminating meloxicam-treated from saline-treated cats. These criteria include metabolites with an i) VIP >1, ii) MDA > 0.4%, iii) AUC >0.85, in at least one treatment time point and iv) only metabolites which were identified at each treatment time point [25], [26], [27]. Metabolites which met these inclusion criteria were considered putative biomarkers and selected for additional analyses in the testing data set.
2.2.5 Supervised dimension reduction
PLS-DA was applied to each time point, 1 through 5, individually to compare meloxicam treated cats against the control groups for supervised dimension reduction and initial identification of phenotypically significant urine metabolites of acute feline kidney injury. VIP values were used as one method of identifying biomarkers with predictive value in separating meloxicam treated cats from control cats. Metabolites with VIP scores ≤1 at each time point were considered irrelevant to the prediction and excluded from analysis [25], [28]. The predictive ability of the model for each time point was internally validated based on leave-one-out cross validation using the Q2 diagnostic statistic [29].
2.2.6 Random forest classification
The supervised machine learning random forest algorithm was applied to cluster cats based on their phenotype (meloxicam and saline treatments) and classify metabolites according with their contributions to classification accuracy. The parameters used for the analysis included 500 trees, 7 predictors and randomness. Each tree attempts to identify metabolites which contribute the most information to discriminate treatment from controls. Those metabolites with >0.4% MDA in the random forest analysis during at least one treatment time point were considered for inclusion in the model [26].
2.2.7 Receiver operating characteristic curve analysis
Univariate ROC curve analysis was used to identify molecules that may represent candidate biomarkers for detecting changes in urine induced by the repeated administration of meloxicam. We initially used this approach by assessing the complete set of identified urine metabolites between the saline and meloxicam group at each time point. The area under the ROC curve (AUC) value was used as a measure of the prognostic accuracy of individual metabolites. An AUC value of 1.0 indicates a perfect test due to absence of overlap of the test data from the control and diseased states, whereas an AUC value of 0.5 shows the test is no better than random chance and therefore has no diagnostic or prognostic value. We only considered metabolites with an AUC >0.85 for inclusion in the model [27].
2.2.8 Validation of putative biomarkers using a separate testing metabolome data set
The putative biomarker model was trained on training data set cats and then applied to a separate testing urine metabolite data set generated from an independent experiment of cats (n = 8).The select set of biomarkers was utilized in the test data set to perform multivariate ROC curve analysis with PLS-DA and RF machine learning, separately, as the classification and feature ranking method performed. Sensitivity and specificity, ROC curves, Monte Carlo cross-validation confusion matrices, predictive accuracies and mean importance were calculated for each testing data set time point to validate the value of the metabolite model derived from the training data set. [30], [31].
3. Results
3.1 Training metabolome data set
All animals included in this study were clinically healthy according to physical examinations, cellular blood counts and blood chemistry panels prior to administration of treatments. Once treatments were started, cats in the control group remained healthy for the duration of the study. One cat in the control group vomited once on day 4 after saline administration. In the meloxicam treated group, the cats’ body-weights and condition scores were relatively stable, with the exception of one cat (M5) whose body weight was reduced by ~ 7%, likely due to a decrease in food intake. During the period of sample collection (0–17 days of treatments), 5 out of 6 cats in the meloxicam group vomited 2 to 15 times but no more than once a day. However, their food intake, body weight, and condition scores were consistent with pre-treatment values. One cat in the training experiment, meloxicam-treated group failed to develop clinical pathologic signs of AKI kidney damage (changes in serum creatinine concentrations and USG) during the study and lacked histopathologic lesions consistent with kidney injury. This cat had comparable plasma concentrations of meloxicam as compared to other cats within the meloxicam treatment group (unpublished data). Due to the lack of clinical kidney disease, this single cat was censored from the biomarker discovery analysis.
3.2 Training data set creatinine and USG
Serum creatinine concentrations initially rose in the meloxicam treated group on day 9 and stayed markedly elevated for the remainder of the study, consistent with a decreased glomerular filtration rate (S1 Fig). Similarly, the USG began to drop on day 9 within the meloxicam treated group, consistent with perturbations in concentrating ability secondary to renal tubular damage (S2 Fig).
3.3 Urine metabolome of training data set
By applying an untargeted metabolomics approach, we were able to detect 318 low molecular-weight (LMW) substances in the urine. Of these, 114 have been identified (Table 1), while 204 remain unknown (when spectra or the retention index is not matched to library entries).
Table 1. List of 114 low molecular-weight molecules identified in feline urine in the training data set using an untargeted metabolomics approach.
| 3-(3-hydroxyphenyl)propionic acid | glycerol | phenaceturic acid |
| 3-(4-hydroxyphenyl)propionic acid | glycerol-3-galactoside | phenol |
| 3,4-dihydroxycinnamic acid | glycine | phosphate |
| 3,4-dihydroxyhydrocinnamic acid NIST | glycocyamine | pimelic acid |
| 3,4-dihydroxyphenylacetic acid | glycolic acid | pinitol |
| 3-aminoisobutyric acid | hexadecane | propane-1,3-diol NIST |
| 3-hydroxy-3-methylglutaric acid | hexitol | pseudo uridine |
| 3-phosphoglycerate | hexuronic acid | putrescine |
| 4-hydroxybenzoate | hippuric acid | pyruvic acid |
| 4-hydroxybutyric acid | hydroxylamine | quinic acid |
| 4-hydroxycinnamic acid | indole-3-acetate | raffinose |
| 4-hydroxyhippuric acid NIST | indoxyl sulfate | ribitol |
| 4-hydroxyphenylacetic acid | inosine | ribonic acid |
| 5-hydroxy-3-indoleacetic acid | isocitric acid | ribose |
| aconitic acid | isohexonic acid | saccharic acid |
| adenosine | isomaltose | serine |
| alanine | isoribose | sorbitol |
| allantoic acid | isothreonic acid | stearic acid |
| alpha-ketoglutarate | kynurenic acid | succinic acid |
| benzoic acid | lactic acid | sucrose |
| benzylalcohol | lysine | sulfuric acid |
| beta-alanine | lyxitol | tagatose |
| beta-gentiobiose | lyxose | taurine |
| catechol | malic acid | threitol |
| citramalic acid | mannose | threonic acid |
| citric acid | mucic acid | trehalose |
| citrulline | myo-inositol | tryptophan |
| conduritol-beta-expoxide | myristic acid | tyrosine |
| creatinine | N-acetylaspartic acid | tyrosol |
| deoxypentitol | n-acetyl-d-hexosamine | urea |
| erythritol | N-acetylglutamate | uric acid |
| ferulic acid | N-acetylmannosamine | uridine |
| fructose | ornithine | valine |
| fucose | oxalic acid | vanillic acid |
| galactinol | oxoproline | xylitol |
| galactonic acid | palmitic acid | xylonic acid |
| gluconic acid | pelargonic acid | xylose |
| glyceric acid | pentitol | xylulose NIST |
3.4 Identification of putative biomarkers using a training metabolome data set
3.4.1 Dimension reduction and cluster analysis
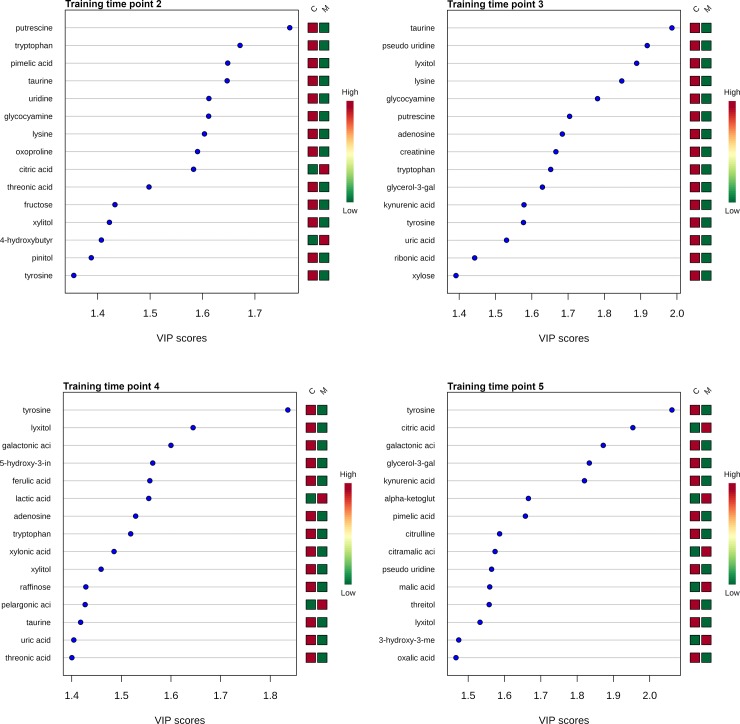
PLS-DA was efficient at discriminating between groups as these supervised multivariate techniques take group identity (i.e. treatment vs control) into account during dimension reduction analysis. The Q2 statistics of performance, an estimate of the predictive ability of the discriminant analysis model, for time points 1 through 5 were 0.03, 0.55, 0.52, 0.52, and 0.33, respectively. These Q2 values support that there was not overfitting of the data, except for at time point 1 (prior to meloxicam administrations). However, this is expected for time point 1 as PLS-DA will attempt to force a distinction utilizing group identity, even prior to treatment (overfitting). The top 15 VIP results for each time point of the PLS-DA are presented in Fig 2. VIP scores summarize the influence of an individual metabolite on the overall PLS-DA model (S1 Table). VIP scores ≥ 1 indicate metabolites which contribute significantly to the model.
Fig 2. Variable importance in projection (VIP) scores for the top 15 metabolites.
VIP scores of top 15 urine metabolites used to differentiate meloxicam-treated (n = 5) and saline-treated cats (n = 6). VIP scores are derived from PLS-DA analysis performed at each time point, 1–5. VIP scores ≥ 1.0 were considered significant when selecting metabolites for the final model. The column to the right of each figure display variations in metabolite peak intensities.
3.4.2 Random forest classification
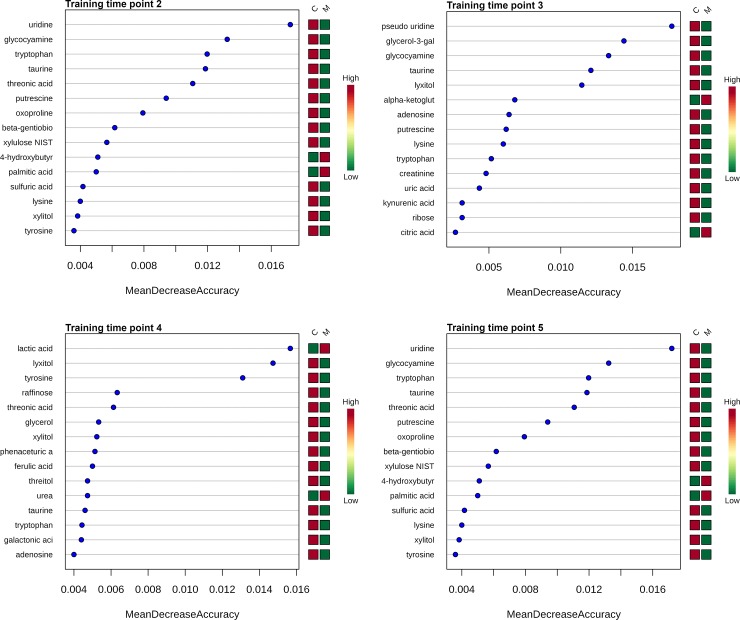
Metabolites for time points 2–5 were ranked using machine learning algorithms by contributions to the performance of the model as measured by the MDA value (Fig 3). A higher MDA value indicate increasing importance of a metabolite in predicting phenotype group (control vs. meloxicam). There are 15, 14, 10 and 8 metabolites which met this inclusion criteria at time points, 2, 3, 4 and 5, respectively during the RF classification step (S2 Table).
Fig 3. Mean decrease in accuracy (MDA) of top 15 urine metabolites.
MDA of the top 15 urine metabolites for the discrimination of meloxicam-treated (n = 5) and saline-controls (n = 6) for each time points (2–5) after the administration of meloxicam derived from random forest analysis. The higher the MDA value the more important a metabolite is to the performance of the model. Urine metabolites with a mean decrease in accuracy ≥ 0.004 in at least one post-treatment time point (2–5) were considered for inclusion in the predictive model.
3.4.3 Receiver operating characteristic curve analysis
Univariate ROC AUC values were calculated for all metabolites. AUC values represent the accuracy of an individual metabolite in correctly separating treatment from control animals. Prior to administration of treatments (time 1), 7 metabolites had an AUC > 0.85 including citrulline, alpha-ketoglutarate, tyrosol, mucic acid, citric acid, mannose and 3-hydroxy-3-methylglutaric acid. These seven metabolites were excluded from the training data set time points 2–5 for all analyses performed on the training data set i.e PLS-DA and RF analyses (S3 Table).
3.4.4 Training data set putative biomarkers
Metabolites present at all treatment time points which met the inclusion criteria of AUC ROC > 0.85, VIP score > 1.0 and an MDA > 0.004 in at least one time point include tyrosine, taurine, tryptophan, pseudouridine, lyxitol, xylitol and threonic acid (Table 2). These seven metabolites were selected for inclusion in the model as potential biomarker candidates. To evaluate the performance of the proposed seven metabolite model, multivariate ROC analysis was performed on each treatment time point, 2–5, and results were verified by the Monte Carlo cross-validation method. The sensitivity and specificity of the seven metabolite model based on either PLS-DA or RF as the final classifier are listed in Table 3 when applied to the training data set.
Table 2. List of putative urine biomarkers derived from the training data set.
The selected seven urine metabolites were the only compounds in this study which met the following inclusion criteria: MDA ≥ 0.004, VIP score > 1.0, ROC AUC > 0.85 and in at least one treatment time-point.
| Time 2 | MDA | VIP score | ROC AUC |
| Tryptophan | 1.20% | 1.7 | 1.0 |
| Taurine | 1.19% | 1.6 | 1.0 |
| Threonic acid | 1.11% | 1.5 | 1.0 |
| Tyrosine | 0.36% | 1.4 | 0.93 |
| Lyxitol | 0.23% | 1.4 | 0.93 |
| Xylitol | 0.38% | 1.4 | 0.93 |
| Pseudouridine | 0.35% | 1.3 | 0.9 |
| Time 3 | MDA | VIP score | ROC AUC |
| Tryptophan | 0.52% | 1.6 | 0.9 |
| Taurine | 1.21% | 2.0 | 1.0 |
| Threonic acid | 0.00% | 1.3 | 0.9 |
| Tyrosine | 0.17% | 1.6 | 0.93 |
| Lyxitol | 1.15% | 1.9 | 1.0 |
| Xylitol | 0.16% | 1.2 | 0.97 |
| Pseudouridine | 1.77% | 1.9 | 1.0 |
| Time 4 | MDA | VIP score | ROC AUC |
| Tryptophan | 0.44% | 1.5 | 0.97 |
| Taurine | 0.46% | 1.4 | 0.93 |
| Threonic acid | 0.61% | 1.4 | 0.97 |
| Tyrosine | 1.31% | 1.8 | 1.0 |
| Lyxitol | 1.47% | 1.6 | 1.0 |
| Xylitol | 0.52% | 1.5 | 0.93 |
| Pseudouridine | 0.31% | 1.2 | 0.93 |
| Time 5 | MDA | VIP score | ROC AUC |
| Tryptophan | -0.07% | 1.2 | 0.9 |
| Taurine | 0.40% | 1.5 | 0.93 |
| Threonic acid | -0.23% | 1.3 | 0.87 |
| Tyrosine | 2.85% | 2.1 | 1.0 |
| Lyxitol | 0.33% | 1.5 | 0.9 |
| Xylitol | 0.08% | 1.4 | 0.87 |
| Pseudouridine | 0.58% | 1.6 | 0.9 |
Table 3. Binary classification for the 7 metabolite model.
These values were derived from multivariate exploratory ROC analysis with PLS-DA or RF as the final classification methods. The 7 metabolite model is composed of tyrosine, tryptophan, threonic acid, pseudouridine, taurine, xylitol, and lyxitol. Sens, sensitivity; Spec, specificity. When both training and testing time points are listed, the model is trained using data from training time point and used to predict the testing time point classifier (i.e treated vs control).
| Classification Method | Training Time Point | Testing Time Point | True pos. | False neg. | True neg. | False pos. | Sens. | Spec. |
|---|---|---|---|---|---|---|---|---|
| PLS-DA | 1 | NA | 3 | 2 | 3 | 3 | 60% | 50% |
| PLS-DA | 2 | NA | 5 | 0 | 6 | 0 | 100% | 100% |
| PLS-DA | 3 | NA | 5 | 0 | 6 | 0 | 100% | 100% |
| PLS-DA | 4 | NA | 5 | 0 | 6 | 0 | 100% | 100% |
| PLS-DA | 5 | NA | 5 | 0 | 5 | 1 | 100% | 83.33% |
| RF | 1 | NA | 2 | 3 | 2 | 4 | 40% | 66.67% |
| RF | 2 | NA | 4 | 1 | 6 | 0 | 80% | 100% |
| RF | 3 | NA | 5 | 0 | 5 | 1 | 100% | 83.33% |
| RF | 4 | NA | 5 | 0 | 6 | 0 | 100% | 100% |
| RF | 5 | NA | 5 | 0 | 5 | 1 | 100% | 83.33% |
| PLS-DA | 2 | 2 | 4 | 0 | 4 | 0 | 100% | 100% |
| PLS-DA | 4 | 3 | 0 | 4 | 3 | 1 | 0% | 75% |
| PLS-DA | 5 | 4 | 4 | 0 | 2 | 2 | 100% | 50% |
| RF | 2 | 2 | 3 | 1 | 3 | 1 | 75% | 75% |
| RF | 4 | 3 | 0 | 4 | 2 | 2 | 0% | 50% |
| RF | 5 | 4 | 3 | 1 | 3 | 1 | 75% | 75% |
3.5 Testing metabolome data set
All animals were clinically healthy according to physical examinations, cellular blood counts and blood chemistry panels before the administration of the treatments. Once treatments were started, cats in the control group remained healthy for the duration of the study. One cat in the control group vomited once on day 4 after the first drug administration. In the meloxicam treated group, the cats’ body-weights and condition scores were relatively stable.
3.5.1 Kidney histopathology for testing data set
Following 17 days of consecutive subcutaneous meloxicam treatment every 24h, renal histologic lesions within the treatment group (n = 4) included multifocal cortical tubular dilation, proximal tubular epithelial degeneration and necrosis, degeneration and swelling of medullary tubular epithelial cells, heterogeneous cortical peri-degenerate tubular inflammatory infiltrate composed of peri-degenerate tubular aggregates of lymphocytes, plasma cells macrophages with fewer neutrophils and rare eosinophil. In contrast, cats within the saline control group (n = 4) had no to scattered tubular degeneration and minimal interstitial inflammatory infiltrate. These findings are consistent with acute kidney injury predominantly focused on the renal tubules. Representative photomicrographs, histologic scores, and aggregates scores for each cat are available in the supplementary data (S4 and S5 Figs, S4 Table).
3.5.2 Testing data set creatinine concentrations and USG
The serum creatinine and USG values followed a similar clinical progression as observed in the training data set. Serum creatinine concentrations rose starting on day 7 and stayed elevated for the remainder of the study (17 days) consistent with decreased glomerular filtration rate (S6 Fig). Similarly, USG values did not drop below the cut-off of 1.035 until day 11 within the meloxicam treated group, consistent with renal tubular concentrating ability (S7 Fig).
3.5.3 Validation of putative biomarkers using testing metabolome data set
The seven metabolite model was applied to the testing data set obtained from a separate meloxicam treatment experiment to confirm the value of these biomarkers in identifying cats treated with meloxicam.
A model using multivariate ROC analysis with either PLS-DA or RF as the classifier was fitted using the 7 putative biomarkers in the training data set and subsequently employed to predict the classification of unlabeled treated and control cats in the testing data set. The sensitivity and specificity for the seven metabolite complete model when applied to the test data set at each time point is reported in Table 3.
4. Discussion
In this study, we utilized an untargeted metabolomics approach to identify urine LMWs that could be further studied as biomarker candidates for monitoring during the administration of NSAIDs in cats. The subcutaneous administration of meloxicam rapidly and consistently altered the urine metabolome of cats with marked changes as early as 2 days (training data set time point 2) following the administration of meloxicam.
One of the earliest and strongest metabolic changes induced by meloxicam was a decrease in the intensity of most of the urinary metabolites (S5 Table). The exact mechanism for a decrease in the urine metabolites is unclear. Humans with chronic kidney disease also had decrease of several metabolites in urine [32]. However, our findings contrast with some earlier studies done in rodents reporting increased amino acid excretion upon dosing with cisplatin, aminoglycosides, and other nephrotoxins [33], [34], [35], [36], [37]. Under nephrotoxic conditions, amino acid excretion increases because of impaired reabsorption by the renal tubules, increased cellular turnover, or increased permeability of the glomerular membranes [38]. In our study, there was a decrease in the USG that was evident in most of the training data set cats after 13 days of treatment. A decrease in the USG is likely a result of alteration of the urine concentration capacity of kidneys. Moreover a decrease in the secretion or increase of reabsorption of metabolites could also explain a decrease in the intensity of some urinary metabolites. Interspecies, and experimental model differences and meloxicam-specific effects may be important factors dictating the studies dissimilarities. Another study suggests that the NSAID indomethacin increases solute reabsorption at the medullary segment of the thick ascending limb of the loop of Henle [39]. Whether meloxicam has similar effects as indomethacin in the kidney remains to be determined but it could also explain, at least partially, a lower urinary intensity of some metabolites.
Piroxicam (a drug that belongs to the same family of drugs as meloxicam) and other NSAIDs have been reported to inhibit organic anion transporters (hOATs) and human organic cation transporters (hOCTs). The exact mechanism of solutes’ renal excretion in cats is unknown but it is possible that at least some of the metabolites reported in this study are excreted by transport-mediated mechanisms which could have been altered by meloxicam [40].
A group of 7 LMWs, including taurine, tryptophan, tyrosine, lyxitol, pseudouridine, xylitol and threoinic acid were consistently able to discern meloxicam-treated from saline-treated cats in the training data set. The alterations in peak intensity of these metabolites in urine may be the result of kidney damage caused by the repeated administration of meloxicam. Urine samples can be considered a liquid biopsy of the kidneys [10], thus some of the changes in the urine metabolome might reflect effects of meloxicam on this organ. Interesting, within the training data set all these LMWs changed in urine earlier (at least 4 days after the first treatment) than creatinine (~10 days after the first treatment) in plasma, suggesting that these molecules could be used as urinary biomarkers for early detection of meloxicam-induced kidney damage. However, the metabolite changes in urine observed in this study cannot be solely associated with kidney damage as NSAIDs have effects in a myriad of body cells and organs which could also be reflected in the urine metabolome [4], [41], [42].
Notably, taurine, tryptophan, tyrosine, and pseudouridine were identified as potential biomarker candidates of kidney disease in other species [43], [44], [45], [46], [47], [48]. Several of the putative biomarkers proposed here have previously been shown to play a biologically significant role both within normal renal homeostasis and disease states. For example, taurine, a β-amino acid, is involved in regulation of renal blood flow [49, 50], apoptosis of renal tubular epithelial cells [51], [52] and protection against renal injury [53], [54]. Tyrosine, a non-essential amino acid, is produced through metabolism of phenylalanine by renal tubular epithelial cells. As renal function decreases following injury, there is decreased production of tyrosine.
ROC analysis results suggest that a cats’ phenotype can be discriminated based on seven metabolites identified in urine metabolome across multiple time points, with an average sensitivity of 95–100% and specificity of 91.7–95.8% in the training data set, depending on the classification algorithm utilized. Even though it is necessary to assess the robustness of the selected biomarkers through validation of the findings using independent data sets [55, 56], biological validation of putative biomarkers is rarely found in scientific biomarker discovery publications but necessary for increasing the value of the experimental findings. Distinctively, this study tested prospectively sensitivity and specificity of the 7 metabolites (from the training data set) on a separate metabolome data set (testing dataset) generated in an independent feline study. When the 7 metabolites are applied to a separate test data set this resulted in an average of 66–75%% sensitivity and 50–66% specificity across multiple time points. If testing time point 3 is excluded from the analysis, this increased the sensitivity and specificity to 75–100% and 75% respectively. These results using an independent data set support the relevance of the seven metabolite biomarkers for dichotomizing meloxicam-treated from saline-treated cats. When interpreting the calculated binary classification values it should be cautioned that these values are limited by the small sample size of this study. Additional studies in a larger population is required to confirm the diagnostic potential of the metabolites identified in this study.
This study discovered LMWs that are altered by the repeated administration of meloxicam and could be used to detect or predict early adverse effects in cats. However, the seven metabolites identified in this study should not be considered biomarkers yet because this study has several limitations. First, this pilot exploratory study has been performed in a relatively small and highly homogenous population of intact female domestic cats maintained under experimentally controlled conditions. Many extrinsic factors have been shown to affect the urinary metabolome including the gastrointestinal microbiota [57], [58], obesity/diet [59], [60], age [61], exercise [62], diurnal variation [63], gender [61], and environmental chemicals [64]. The elegance of the study reported here lies in the fact that we tested the metabolite selection using a separate data set generated during an independent study that included cats from a different breeder source, hosting conditions, and sampling times which increases the validity of this study’s findings. However, the data presented in this study should be considered for hypothesis generation. The validity of the metabolites found in this research should be assessed in studies including both genders, different ages, diets and other environmental conditions. Even though the sample sizes were enough to detect differences in the urine metabolome a larger study will increase the robustness and external validity of the present study’s findings. It is important to remark that this study provides, untargeted and nonbiased information regarding urinary metabolites, as the hypothesis is not based on pre-selection of potential candidates to be investigated. Nevertheless, currently, there is no single-instrument platform that can cover all urine metabolites (>3000) [65] therefore the LMWs evaluated in this study are likely to be a fraction of the actual urine metabolome because this study was limited to one analytical platform [18]. It is expected that expanding the metabolome analytical platforms will help to discover other metabolites that could serve as biomarkers for monitoring cats treated with NSAIDs.
5. Conclusions
This is the first study reporting changes in the urinary metabolome of cats with meloxicam-induced kidney disease. Using a machine leaning technique, this study identified a novel panel of 7 metabolites that may help to identify early non-symptomatic effects of the repeated administration of meloxicam. This panel of metabolites could be useful for drug development and clinical trials involving NSAIDs or monitoring clinical patients. However, the clinical value and robustness of this panel of metabolites need to be assessed in future experiments with a larger target population of animals.
Supporting information
The dashed line indicates a USG of 1.035. Urine samples were collected on days -3, 0, 4, 9, 13, 17, 23, 26, 31, 34, 40 and 47.
(DOCX)
Cats were treated with saline (n = 6) or meloxicam (n = 5) at 0.3 mg/kg every 24 hr for up to 47 days. The dashed line indicates a USG of 1.035. Urine samples were collected on days -3, 0, 4, 9, 13, 17, 23, 26, 31, 34, 40 and 47.
(DOCX)
Each data point represents a urine sample from an individual cat. Axis values are normalized principal component scores. Shaded backgrounds define the 95% confidence interval for each group. The sampling time point is denoted in the upper left of each plot.
(DOCX)
A) Kidney, cortical, control, testing data set. Normal cortical tubular components including proximal convoluted tubule with intracytoplasmic lipid vacuoles, thick ascending limb of the nephron and a glomerulus, hematoxylin and eosin (H&E, 200x). B) Kidney, cortical, meloxicam treated, testing data set. Renal tubular dilation, tubular epithelial cell degeneration and interstitial mixed inflammation (H&E, 200x). C) Kidney, cortical, meloxicam treated, testing data set. Tubular degeneration characterized by tubular epithelial cells with granular, faintly eosinophilic and vacuolated cytoplasm with renal tubular epithelial necrosis (H&E, 400x). D) Kidney, medullary, meloxicam treated. Nodular accumulation of lymphocytes, neutrophils and macrophages admixed with necrotic cellular debris within the medullary interstitium (H&E, 400x). E) Kidney, cortical, control, testing data set. Diffusely intact and histologically unremarkable renal tubular basement membranes (PAMS, 200x). F) Kidney, cortical, meloxicam treated, testing data set. Multifocal fraying and loss of basement membrane integrity which correlates to regions of interstitial inflammation and tubular necrosis in the H&E replicate sections (PAMS, 200x).
(DOCX)
The bold center line represents the median score, the boxes above and below the median line are the first and third quartile. The whiskers represent the maximum and minimum scores for each group. Utilizing the non-parametric Mann-Whiney test, there is a significant difference in renal histologic scores between the meloxicam treated group (M) and the saline treated control cats (C) (p = 0.006766).
(DOCX)
Creatinine values are displayed in percent change from baseline samples. Serum samples were collected on days -1, 1, 11, 14 and 17.
(DOCX)
Cats were treated with saline (n = 4) or meloxicam (n = 4) at 0.3 mg/kg every 24 hr for 17 days. The dashed line indicates a USG of 1.035. Urine samples were collected on days -1, 2, 11, 14 and 17.
(DOCX)
The list of identified metabolites is derived from urine collected from control cats treated with saline (n = 6) or meloxicam (n = 5) at 0.3 mg/kg every 24 hr for up to 17 days (time point 5). Metabolites with VIP scores > 1 were considered for inclusion in the model. Blank values indicate the metabolite was not detected at that time point or was removed during the data filtering step.
(DOCX)
The list of identified metabolites is derived from urine collected from control cats treated with saline (n = 6) or meloxicam (n = 5) at 0.3 mg/kg every 24 hr for up to 17 days (time point 5). Metabolites with MDA scores > 0.0004 were considered for inclusion in the model. Blank values indicate the metabolite was not detected at that time point or was removed during the data filtering step.
(DOCX)
The list of identified metabolites is derived from urine collected from control cats treated with saline (n = 6) or meloxicam (n = 5) at 0.3 mg/kg every 24 hr for up to 17 days (time point 5). Metabolites with AUC scores > 0.85 were considered for inclusion in the model. Metabolites with AUC scores > 0.85 at time point 1 were excluded from the model. Blank values indicate the metabolite was not detected at that time point or was removed during the data filtering step.
(DOCX)
Tubule damage and inflammation were assessed on hematoxylin and eosin (H&E) stained sections and basement membrane integrity were assessed on periodic acid-Schiff methenamine silver (PAMS) stained sections. Cortical and corticomedullary tubules were assessed for degeneration, necrosis, regeneration, dilation, attenuation and hypertrophy. Any subtype of inflammatory cell within the interstitium (i.e. neutrophil, lymphocyte, macrophage, eosinophil, etc.) were counted when determining extent of inflammation. The maximum score per category is 4 with a maximum possible score of 16.
(DOCX)
The list of identified metabolites is derived from training data set urine collected from control cats treated with saline (n = 6) or meloxicam (n = 5) at 0.3 mg/kg every 24 hrs for up to 17 days (time point 5). Values highlighted in red indicate an increase in average intensity (> 1.2 fold) and values highlighted in yellow represent a decrease in average intensity (< 0.8) when comparing the meloxicam treated group vs. the saline treated control group.
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
N.F.V: The Autzen Foundation and the John and Ruth Ottmar Memorial Endowment intramural funds from the College of Veterinary Medicine at Washington State University, USA funded this research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Laine L. Approaches to nonsteroidal anti-inflammatory drug use in the high-risk patient. Gastroenterology. 2001;120(3):594–606. . [DOI] [PubMed] [Google Scholar]
- 2.Harirforoosh S, Jamali F. Renal adverse effects of nonsteroidal anti-inflammatory drugs. Expert Opin Drug Saf. 2009;8(6):669–81. 10.1517/14740330903311023 . [DOI] [PubMed] [Google Scholar]
- 3.Hörl WH. Nonsteroidal Anti-Inflammatory Drugs and the Kidney. Pharmaceuticals (Basel). 2010;3(7):2291–321. Epub 2010/07/21. 10.3390/ph3072291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sostres C, Gargallo CJ, Lanas A. Nonsteroidal anti-inflammatory drugs and upper and lower gastrointestinal mucosal damage. Arthritis Res Ther. 2013;15 Suppl 3:S3. Epub 2013/07/24. 10.1186/ar4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chawla LS, Eggers PW, Star RA, Kimmel PL. Acute kidney injury and chronic kidney disease as interconnected syndromes. N Engl J Med. 2014;371(1):58–66. 10.1056/NEJMra1214243 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bargnoux AS, Beaufils O, Oguike M, Lopasso A, Dupuy AM, Sebbane M, et al. Point-of-care creatinine testing in patients receiving contrast-enhanced computed tomography scan. Clin Chim Acta. 2018;478:111–3. Epub 2017/12/18. 10.1016/j.cca.2017.12.025 . [DOI] [PubMed] [Google Scholar]
- 7.Rácz O, Lepej J, Fodor B, Lepejová K, Jarčuška P, Kováčová A, et al. Pitfalls in the Measurements and Assesment of Glomerular Filtration Rate and How to Escape them. EJIFCC. 2012;23(2):33–40. Epub 2012/07/18. [PMC free article] [PubMed] [Google Scholar]
- 8.Kosmides AK, Kamisoglu K, Calvano SE, Corbett SA, Androulakis IP. Metabolomic fingerprinting: challenges and opportunities. Crit Rev Biomed Eng. 2013;41(3):205–21. 10.1615/critrevbiomedeng.2013007736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134(5):714–7. 10.1016/j.cell.2008.08.026 . [DOI] [PubMed] [Google Scholar]
- 10.Chan EC, Pasikanti KK, Nicholson JK. Global urinary metabolic profiling procedures using gas chromatography-mass spectrometry. Nat Protoc. 2011;6(10):1483–99. Epub 2011/09/08. 10.1038/nprot.2011.375 . [DOI] [PubMed] [Google Scholar]
- 11.Silva RE, Baldim JL, Chagas-Paula DA, Soares MG, Lago JHG, Gonçalves RV, et al. Predictive metabolomic signatures of end-stage renal disease: A multivariate analysis of population-based data. Biochimie. 2018;152:14–30. Epub 2018/06/15. 10.1016/j.biochi.2018.06.009 . [DOI] [PubMed] [Google Scholar]
- 12.Ross L. Acute kidney injury in dogs and cats. Vet Clin North Am Small Anim Pract. 2011;41(1):1–14. Epub 2010/12/04. 10.1016/j.cvsm.2010.09.003 . [DOI] [PubMed] [Google Scholar]
- 13.Krittanawong C, Zhang H, Wang Z, Aydar M, Kitai T. Artificial Intelligence in Precision Cardiovascular Medicine. J Am Coll Cardiol. 2017;69(21):2657–64. 10.1016/j.jacc.2017.03.571 . [DOI] [PubMed] [Google Scholar]
- 14.Alakwaa FM, Chaudhary K, Garmire LX. Deep Learning Accurately Predicts Estrogen Receptor Status in Breast Cancer Metabolomics Data. J Proteome Res. 2018;17(1):337–47. Epub 2017/11/27. 10.1021/acs.jproteome.7b00595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuperlovic-Culf M. Machine Learning Methods for Analysis of Metabolic Data and Metabolic Pathway Modeling. Metabolites. 2018;8(1). Epub 2018/01/11. 10.3390/metabo8010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLeland SM, Cianciolo RE, Duncan CG, Quimby JM. A comparison of biochemical and histopathologic staging in cats with chronic kidney disease. Vet Pathol. 2015;52(3):524–34. Epub 2014/12/16. 10.1177/0300985814561095 . [DOI] [PubMed] [Google Scholar]
- 17.Fiehn O, Kind T. Metabolite profiling in blood plasma. Methods Mol Biol. 2007;358:3–17. 10.1007/978-1-59745-244-1_1 . [DOI] [PubMed] [Google Scholar]
- 18.Rivera-Vélez SM, Villarino NF. Feline urine metabolomic signature: characterization of low-molecular-weight substances in urine from domestic cats. J Feline Med Surg. 2018;20(2):155–63. Epub 2017/04/03. 10.1177/1098612X17701010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiehn O. Metabolomics by Gas Chromatography-Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr Protoc Mol Biol. 2016;114:30.4.1–.4.2. Epub 2016/04/01. 10.1002/0471142727.mb3004s114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivera-Velez SM, Broughton-Neiswanger LE, Suarez M, Piñeyro P, Navas J, Chen S, et al. Repeated administration of the NSAID meloxicam alters the plasma and urine lipidome. Sci Rep. 2019;9(1):4303 Epub 2019/03/13. 10.1038/s41598-019-40686-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46(W1):W486–W94. 10.1093/nar/gky310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hackstadt AJ, Hess AM. Filtering for increased power for microarray data analysis. BMC Bioinformatics. 2009;10:11 Epub 2009/01/08. 10.1186/1471-2105-10-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enot DP, Lin W, Beckmann M, Parker D, Overy DP, Draper J. Preprocessing, classification modeling and feature selection using flow injection electrospray mass spectrometry metabolite fingerprint data. Nat Protoc. 2008;3(3):446–70. 10.1038/nprot.2007.511 . [DOI] [PubMed] [Google Scholar]
- 24.van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142 Epub 2006/06/08. 10.1186/1471-2164-7-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chong I-G, Jun C-H. Performance of some variable selection methods when multicollinearity is present. Chemometrics and Intelligent Laboratory Systems. 2005;78(1–2):103–12. [Google Scholar]
- 26.Chen T, Cao Y, Zhang Y, Liu J, Bao Y, Wang C, et al. Random forest in clinical metabolomics for phenotypic discrimination and biomarker selection. Evid Based Complement Alternat Med. 2013;2013:298183 Epub 2013/02/02. 10.1155/2013/298183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia J, Broadhurst DI, Wilson M, Wishart DS. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics. 2013;9(2):280–99. Epub 2012/12/04. 10.1007/s11306-012-0482-9 ; PubMed Central PMCID: PMC3608878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jansson J, Willing B, Lucio M, Fekete A, Dicksved J, Halfvarson J, et al. Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS One. 2009;4(7):e6386 Epub 2009/07/28. 10.1371/journal.pone.0006386 ; PubMed Central PMCID: PMC2713417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szymańska E, Saccenti E, Smilde AK, Westerhuis JA. Double-check: validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics. 2012;8(Suppl 1):3–16. Epub 2011/07/08. 10.1007/s11306-011-0330-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagana Gowda GA, Raftery D. Biomarker Discovery and Translation in Metabolomics. Curr Metabolomics. 2013;1(3):227–40. 10.2174/2213235X113019990005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor JM, Ankerst DP, Andridge RR. Validation of biomarker-based risk prediction models. Clin Cancer Res. 2008;14(19):5977–83. 10.1158/1078-0432.CCR-07-4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rysz J, Gluba-Brzózka A, Franczyk B, Jabłonowski Z, Ciałkowska-Rysz A. Novel Biomarkers in the Diagnosis of Chronic Kidney Disease and the Prediction of Its Outcome. Int J Mol Sci. 2017;18(8). Epub 2017/08/04. 10.3390/ijms18081702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghiculescu RA, Kubler PA. Aminoglycoside-associated Fanconi syndrome. Am J Kidney Dis. 2006;48(6):e89–93. 10.1053/j.ajkd.2006.08.009 . [DOI] [PubMed] [Google Scholar]
- 34.Kim YK, Byun HS, Kim YH, Woo JS, Lee SH. Effect of cisplatin on renal function in rabbits: mechanism of reduced glucose reabsorption. Toxicol Appl Pharmacol. 1995;130(1):19–26. 10.1006/taap.1995.1003 . [DOI] [PubMed] [Google Scholar]
- 35.Macpherson NA, Moscarello MA, Goldberg DM. Aminoaciduria is an earlier index of renal tubular damage than conventional renal disease markers in the gentamicin-rat model of acute renal failure. Clin Invest Med. 1991;14(2):101–10. . [PubMed] [Google Scholar]
- 36.Niemann CU, Serkova NJ. Biochemical mechanisms of nephrotoxicity: application for metabolomics. Expert Opin Drug Metab Toxicol. 2007;3(4):527–44. 10.1517/17425225.3.4.527 . [DOI] [PubMed] [Google Scholar]
- 37.Portilla D, Li S, Nagothu KK, Megyesi J, Kaissling B, Schnackenberg L, et al. Metabolomic study of cisplatin-induced nephrotoxicity. Kidney Int. 2006;69(12):2194–204. Epub 2006/05/03. 10.1038/sj.ki.5000433 . [DOI] [PubMed] [Google Scholar]
- 38.van de Poll MC, Soeters PB, Deutz NE, Fearon KC, Dejong CH. Renal metabolism of amino acids: its role in interorgan amino acid exchange. Am J Clin Nutr. 2004;79(2):185–97. 10.1093/ajcn/79.2.185 . [DOI] [PubMed] [Google Scholar]
- 39.Kaojarern S, Chennavasin P, Anderson S, Brater DC. Nephron site of effect of nonsteroidal anti-inflammatory drugs on solute excretion in humans. Am J Physiol. 1983;244(2):F134–9. 10.1152/ajprenal.1983.244.2.F134 . [DOI] [PubMed] [Google Scholar]
- 40.Khamdang S, Takeda M, Noshiro R, Narikawa S, Enomoto A, Anzai N, et al. Interactions of human organic anion transporters and human organic cation transporters with nonsteroidal anti-inflammatory drugs. J Pharmacol Exp Ther. 2002;303(2):534–9. 10.1124/jpet.102.037580 . [DOI] [PubMed] [Google Scholar]
- 41.Matsui H, Shimokawa O, Kaneko T, Nagano Y, Rai K, Hyodo I. The pathophysiology of non-steroidal anti-inflammatory drug (NSAID)-induced mucosal injuries in stomach and small intestine. J Clin Biochem Nutr. 2011;48(2):107–11. Epub 2011/02/26. 10.3164/jcbn.10-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morita I. Distinct functions of COX-1 and COX-2. Prostaglandins Other Lipid Mediat. 2002;68–69:165–75. 10.1016/s0090-6980(02)00029-1 . [DOI] [PubMed] [Google Scholar]
- 43.Chesney RW, Han X, Patters AB. Taurine and the renal system. J Biomed Sci. 2010;17 Suppl 1:S4 Epub 2010/08/24. 10.1186/1423-0127-17-S1-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niewczas MA, Sirich TL, Mathew AV, Skupien J, Mohney RP, Warram JH, et al. Uremic solutes and risk of end-stage renal disease in type 2 diabetes: metabolomic study. Kidney Int. 2014;85(5):1214–24. Epub 2014/01/15. 10.1038/ki.2013.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boudonck KJ, Mitchell MW, Német L, Keresztes L, Nyska A, Shinar D, et al. Discovery of metabolomics biomarkers for early detection of nephrotoxicity. Toxicol Pathol. 2009;37(3):280–92. 10.1177/0192623309332992 . [DOI] [PubMed] [Google Scholar]
- 46.Hocher B, Adamski J. Metabolomics for clinical use and research in chronic kidney disease. Nat Rev Nephrol. 2017;13(5):269–84. Epub 2017/03/06. 10.1038/nrneph.2017.30 . [DOI] [PubMed] [Google Scholar]
- 47.Sharma K, Karl B, Mathew AV, Gangoiti JA, Wassel CL, Saito R, et al. Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J Am Soc Nephrol. 2013;24(11):1901–12. Epub 2013/10/10. 10.1681/ASN.2013020126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei Q, Xiao X, Fogle P, Dong Z. Changes in metabolic profiles during acute kidney injury and recovery following ischemia/reperfusion. PLoS One. 2014;9(9):e106647 Epub 2014/09/05. 10.1371/journal.pone.0106647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hagar HH, El Etter E, Arafa M. Taurine attenuates hypertension and renal dysfunction induced by cyclosporine A in rats. Clin Exp Pharmacol Physiol. 2006;33(3):189–96. 10.1111/j.1440-1681.2006.04345.x . [DOI] [PubMed] [Google Scholar]
- 50.Hu J, Xu X, Yang J, Wu G, Sun C, Lv Q. Antihypertensive effect of taurine in rat. Adv Exp Med Biol. 2009;643:75–84. 10.1007/978-0-387-75681-3_8 . [DOI] [PubMed] [Google Scholar]
- 51.Das J, Sil PC. Taurine ameliorates alloxan-induced diabetic renal injury, oxidative stress-related signaling pathways and apoptosis in rats. Amino Acids. 2012;43(4):1509–23. 10.1007/s00726-012-1225-y . [DOI] [PubMed] [Google Scholar]
- 52.Verzola D, Bertolotto MB, Villaggio B, Ottonello L, Dallegri F, Frumento G, et al. Taurine prevents apoptosis induced by high ambient glucose in human tubule renal cells. J Investig Med. 2002;50(6):443–51. 10.1136/jim-50-06-04 . [DOI] [PubMed] [Google Scholar]
- 53.Kim YS, Sung SH, Tang Y, Choi EJ, Choi YJ, Hwang YJ, et al. Protective Effect of Taurine on Mice with Doxorubicin-induced Acute Kidney Injury. Adv Exp Med Biol. 2017;975 Pt 2:1191–201. 10.1007/978-94-024-1079-2_95 . [DOI] [PubMed] [Google Scholar]
- 54.Erdem A, Gündoğan NU, Usubütün A, Kilinç K, Erdem SR, Kara A, et al. The protective effect of taurine against gentamicin-induced acute tubular necrosis in rats. Nephrol Dial Transplant. 2000;15(8):1175–82. 10.1093/ndt/15.8.1175 . [DOI] [PubMed] [Google Scholar]
- 55.Drucker E, Krapfenbauer K. Pitfalls and limitations in translation from biomarker discovery to clinical utility in predictive and personalised medicine. EPMA J. 2013;4(1):7 Epub 2013/02/25. 10.1186/1878-5085-4-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srivastava S, Kramer BS. Validation: a critical step in bringing biomarkers to clinical fruition. Ann Epidemiol. 2018;28(2):135–8. Epub 2017/11/09. 10.1016/j.annepidem.2017.10.013 . [DOI] [PubMed] [Google Scholar]
- 57.Marcobal A, Kashyap PC, Nelson TA, Aronov PA, Donia MS, Spormann A, et al. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013;7(10):1933–43. Epub 2013/06/06. 10.1038/ismej.2013.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vernocchi P, Del Chierico F, Putignani L. Gut Microbiota Profiling: Metabolomics Based Approach to Unravel Compounds Affecting Human Health. Front Microbiol. 2016;7:1144 Epub 2016/07/26. 10.3389/fmicb.2016.01144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Campbell R, Tasevska N, Jackson KG, Sagi-Kiss V, di Paolo N, Mindell JS, et al. Association between urinary biomarkers of total sugars intake and measures of obesity in a cross-sectional study. PLoS One. 2017;12(7):e0179508 Epub 2017/07/19. 10.1371/journal.pone.0179508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Söder J, Hagman R, Dicksved J, Lindåse S, Malmlöf K, Agback P, et al. The urine metabolome differs between lean and overweight Labrador Retriever dogs during a feed-challenge. PLoS One. 2017;12(6):e0180086 Epub 2017/06/29. 10.1371/journal.pone.0180086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thévenot EA, Roux A, Xu Y, Ezan E, Junot C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J Proteome Res. 2015;14(8):3322–35. Epub 2015/07/02. 10.1021/acs.jproteome.5b00354 . [DOI] [PubMed] [Google Scholar]
- 62.Kohler M, Schänzer W, Thevis M. Effects of exercise on the urinary proteome. Adv Exp Med Biol. 2015;845:121–31. 10.1007/978-94-017-9523-4_12 . [DOI] [PubMed] [Google Scholar]
- 63.Giskeødegård GF, Davies SK, Revell VL, Keun H, Skene DJ. Diurnal rhythms in the human urine metabolome during sleep and total sleep deprivation. Sci Rep. 2015;5:14843 Epub 2015/10/09. 10.1038/srep14843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Z, Wolff MS, Moline J. Analysis of environmental biomarkers in urine using an electrochemical detector. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;819(1):155–9. 10.1016/j.jchromb.2005.02.005 . [DOI] [PubMed] [Google Scholar]
- 65.Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, et al. The human urine metabolome. PLoS One. 2013;8(9):e73076 Epub 2013/09/04. 10.1371/journal.pone.0073076 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The dashed line indicates a USG of 1.035. Urine samples were collected on days -3, 0, 4, 9, 13, 17, 23, 26, 31, 34, 40 and 47.
(DOCX)
Cats were treated with saline (n = 6) or meloxicam (n = 5) at 0.3 mg/kg every 24 hr for up to 47 days. The dashed line indicates a USG of 1.035. Urine samples were collected on days -3, 0, 4, 9, 13, 17, 23, 26, 31, 34, 40 and 47.
(DOCX)
Each data point represents a urine sample from an individual cat. Axis values are normalized principal component scores. Shaded backgrounds define the 95% confidence interval for each group. The sampling time point is denoted in the upper left of each plot.
(DOCX)
A) Kidney, cortical, control, testing data set. Normal cortical tubular components including proximal convoluted tubule with intracytoplasmic lipid vacuoles, thick ascending limb of the nephron and a glomerulus, hematoxylin and eosin (H&E, 200x). B) Kidney, cortical, meloxicam treated, testing data set. Renal tubular dilation, tubular epithelial cell degeneration and interstitial mixed inflammation (H&E, 200x). C) Kidney, cortical, meloxicam treated, testing data set. Tubular degeneration characterized by tubular epithelial cells with granular, faintly eosinophilic and vacuolated cytoplasm with renal tubular epithelial necrosis (H&E, 400x). D) Kidney, medullary, meloxicam treated. Nodular accumulation of lymphocytes, neutrophils and macrophages admixed with necrotic cellular debris within the medullary interstitium (H&E, 400x). E) Kidney, cortical, control, testing data set. Diffusely intact and histologically unremarkable renal tubular basement membranes (PAMS, 200x). F) Kidney, cortical, meloxicam treated, testing data set. Multifocal fraying and loss of basement membrane integrity which correlates to regions of interstitial inflammation and tubular necrosis in the H&E replicate sections (PAMS, 200x).
(DOCX)
The bold center line represents the median score, the boxes above and below the median line are the first and third quartile. The whiskers represent the maximum and minimum scores for each group. Utilizing the non-parametric Mann-Whiney test, there is a significant difference in renal histologic scores between the meloxicam treated group (M) and the saline treated control cats (C) (p = 0.006766).
(DOCX)
Creatinine values are displayed in percent change from baseline samples. Serum samples were collected on days -1, 1, 11, 14 and 17.
(DOCX)
Cats were treated with saline (n = 4) or meloxicam (n = 4) at 0.3 mg/kg every 24 hr for 17 days. The dashed line indicates a USG of 1.035. Urine samples were collected on days -1, 2, 11, 14 and 17.
(DOCX)
The list of identified metabolites is derived from urine collected from control cats treated with saline (n = 6) or meloxicam (n = 5) at 0.3 mg/kg every 24 hr for up to 17 days (time point 5). Metabolites with VIP scores > 1 were considered for inclusion in the model. Blank values indicate the metabolite was not detected at that time point or was removed during the data filtering step.
(DOCX)
The list of identified metabolites is derived from urine collected from control cats treated with saline (n = 6) or meloxicam (n = 5) at 0.3 mg/kg every 24 hr for up to 17 days (time point 5). Metabolites with MDA scores > 0.0004 were considered for inclusion in the model. Blank values indicate the metabolite was not detected at that time point or was removed during the data filtering step.
(DOCX)
The list of identified metabolites is derived from urine collected from control cats treated with saline (n = 6) or meloxicam (n = 5) at 0.3 mg/kg every 24 hr for up to 17 days (time point 5). Metabolites with AUC scores > 0.85 were considered for inclusion in the model. Metabolites with AUC scores > 0.85 at time point 1 were excluded from the model. Blank values indicate the metabolite was not detected at that time point or was removed during the data filtering step.
(DOCX)
Tubule damage and inflammation were assessed on hematoxylin and eosin (H&E) stained sections and basement membrane integrity were assessed on periodic acid-Schiff methenamine silver (PAMS) stained sections. Cortical and corticomedullary tubules were assessed for degeneration, necrosis, regeneration, dilation, attenuation and hypertrophy. Any subtype of inflammatory cell within the interstitium (i.e. neutrophil, lymphocyte, macrophage, eosinophil, etc.) were counted when determining extent of inflammation. The maximum score per category is 4 with a maximum possible score of 16.
(DOCX)
The list of identified metabolites is derived from training data set urine collected from control cats treated with saline (n = 6) or meloxicam (n = 5) at 0.3 mg/kg every 24 hrs for up to 17 days (time point 5). Values highlighted in red indicate an increase in average intensity (> 1.2 fold) and values highlighted in yellow represent a decrease in average intensity (< 0.8) when comparing the meloxicam treated group vs. the saline treated control group.
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.