Abstract
Adenocarcinoma is the most common type of non-small cell lung cancer. Some causative genomic alterations in epidermal growth factor receptor (EGFR), including deletions in exon 19 (E19 dels) and a point mutation in E21, are known to have favourable prognoses due to sensitivity to tyrosine kinase inhibitors; however, the prognoses of other uncommon mutations are unclear. This study analysed the clinical significance of EGFR mutation types in lung adenocarcinoma. We retrospectively reviewed 1,020 subjects (mean age: 66.8 years, female: 41.7%) who were diagnosed with advanced lung adenocarcinoma, had EGFR mutation data, and did not undergo surgery from five medical institutes between 2010 and 2016. Subjects were classified according to EGFR mutation status, particularly for exon-specific mutations. EGFR positivity was defined as the presence of mutation and EGFR negativity was defined as wild-type EGFR. EGFR positivity was 38.0%, with the incidence of mutations in E18, E19, E20, and E21 was 3.6%, 51.0%, 3.4%, and 42.0%, respectively. The EGFR positive group survived significantly longer than the negative group (p<0.001), and there was a significant difference in survival among the four EGFR mutation sites (p = 0.003); E19 dels were the only significant factor that lowered mortality (HR: 0.678, p = 0.002), while an E21 mutation was the prognostic factor associated with the most increased mortality (HR: 1.365, p = 0.015). Amongst EGFR positive subjects, the proportion of E19 dels in TKI-responders was significantly higher and that of E21 mutations significantly lower, compared with non-responders. In TKI treatment, mutations in E18 and E20 were not worse factors than the E21 L858R mutation. In conclusion, the presence of EGFR mutations in advanced lung adenocarcinoma can predict a good prognosis; E19 dels prospect to have a better prognosis than other mutations, while an E21 mutation is expected to increase mortality.
Introduction
Non-small cell lung cancer (NSCLC), particularly in advanced stages, has a very poor prognosis, and conventional systemic chemotherapy only results in an increase of less than one year for overall survival (OS) with a high possibility for toxicity [1–3]. Mutations in epidermal growth factor receptor (EGFR) lead to increased downstream signalling, which promotes cell proliferation, differentiation, and growth [4]. Tyrosine kinase inhibitors (TKIs) that block EGFR-derived signal transduction show excellent efficacy in many patients with EGFR mutations [5–8]. According to the National Comprehensive Cancer Network guidelines, TKIs are currently recommended as first-line treatment for advanced EGFR-mutant NSCLC [9].
EGFR mutations are commonly detected in adenocarcinoma, with higher rates amongst Asians (38.8%–64.0%) than amongst Caucasians (4.9%–17.4%) [10–14]. Almost 90% of all EGFR mutations are deletions in exon 19 (E19 dels) or a leucine to arginine substitution (L858R) in E21, which are generally referred to as “common mutations” [15]. Clinical trials have demonstrated efficacy of TKIs for advanced EGFR-mutant NSCLC patients with these common mutations; however, only a small number (n) of patients with other EGFR mutations were enrolled [5, 6, 16]. Uncommon mutations, including E18, E20 and other complex mutations are relatively rare in NSCLC patients, with a prevalence ranging from 10%–18% [5, 6, 17, 18]. Although some studies have reported sensitivities to TKIs according to EGFR mutation types [19–22], these studies have only focused on differences between the common mutation types. The response to TKIs of NSCLC patients with uncommon mutations, including E18 and E20, and their prognoses has not been fully investigated and previous studies have found conflicting results. In recent studies, uncommon mutations were associated with poorer prognoses compared with common mutations [23–25]. Additionally, there are differences in prognosis among the uncommon mutations; specific uncommon mutations, including G719X in E18, have a good prognosis and were associated with improved TKI responses [26]. However, there have been limited studies comparing the prognoses of common and uncommon EGFR mutations in real-world clinical settings. The purpose of this study was to investigate outcomes of advanced lung adenocarcinomas with regard to EGFR mutation status and TKI treatment responses.
Methods
Study population
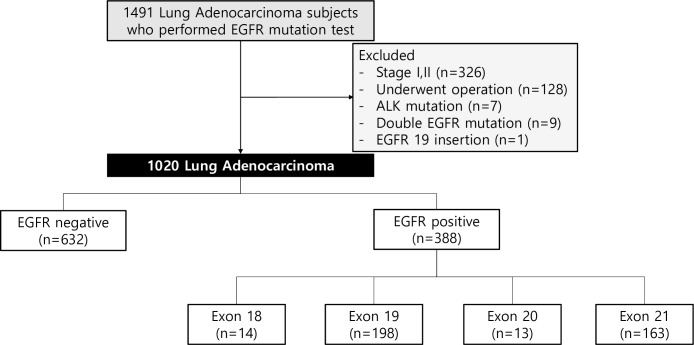
Between January 2010 and December 2016, 1491 lung adenocarcinoma subjects who had EGFR sequencing data from five secondary or tertiary medical institutes were screened for our study (Fig 1) (doi.org/10.17504/protocols.io.bahwib7e). Among them, 471 were excluded due to non-advanced stage (n = 326) and surgery (n = 128). Because the frequencies of EGFR duplicate exonal mutations (n = 9), ALK mutation (n = 7) and E19 insertion (n = 1) were relatively low compared with those of other EGFR mutations, these were excluded in this analysis due to concerns of confounding variables in the analysis; thus, 1,020 subjects were included in our study. All subjects were initially classified into EGFR positive and EGFR negative groups. EGFR positivity was defined as the presence of EGFR mutation or insertion/deletion and EGFR negativity was defined as wild-type EGFR. The EGFR positive group was divided according to mutation sites of E18, E19, E20, and E21. Also, EGFR positive lung adenocarcinoma subjects who received more than four TKI cycles were defined as TKI-responders and separately analysed. Informed consent was waived due to the retrospective study design, and the study was approved by the Institutional Review Board of all participating institutes (Ewha Womans University: EUMC 2018-04-043, Hallym University: HKS 2018-04-009, Inha University: 2018-03-017-001, Korean University: 2018GR0013, and Soonchunhyang University: 2018-05-010).
Fig 1. Patient enrolment.
EGFR, epidermal growth factor receptor; ALK, anaplastic lymphoma kinase.
Study design
Demographics and clinical information of subjects at time of diagnosis were obtained through medical records. The following variables were collected: age, sex, low body mass index (BMI <18.5 kg/m2), smoking history, presence of EGFR mutation, stage according to 7th TNM, status of TKI treatment, other treatments (chemotherapy, radiotherapy), OS, forced expiratory volume in 1 second (FEV1), and forced vital capacity (FVC) at diagnosis. Variables for Charlson Comorbidity Index (CCI) were also collected to evaluate baseline comorbidities. OS was defined as the time from diagnosis to death or last follow-up and was evaluated for all subjects.
Tissue preparation and DNA extraction
When tumour tissues were obtained from biopsy, the tissues were fixed with paraffin embedding and stored in the form of cytology slides. When lung cancer was diagnosed in this state, DNA extraction and amplification from tissue proceeded as follows. Tumour cells were scraped with a 26-gauge needle. DNA extraction buffer solution (50 mM Tris buffer, pH 8.3, 1 mM EDTA, pH 8.0, 5% Tween-20, and 100 μg/mL proteinase K) with 10% resin (20–50 μL) was added to the scraped cells. After incubation at 56°C for at least 1 hour, each tube was heated to 100°C for 20 minutes (min) followed by centrifugation at 12,000 rpm for 10 min at 4°C to pellet the debris [27].
EGFR mutation testing
EGFR mutation analysis was conducted in two ways according to the facilities and timing of each institution. Between 2010 and 2013, mutation testing was performed using direct sequencing by ISU ABXIS Co Ltd (Seoul, Korea), an independent commercial laboratory. From 2013 to 2016, the peptide nucleic acid (PNA)-mediated PCR clamping method (PNAClamp™ EGFR Mutation Detection Kit, Panagene, Daejeon, Korea) was used to identify EGFR mutation according to the manufacturer’s instructions in the department of pathology at each institute. Complete data analysis and quality control according to each department’s own specific protocols were performed. The target somatic mutations included E19 dels, E21 L858R mutation, E18 G719X mutation, E20 S768I mutation, E20 insertions, E20 T790M mutation, and E21 L861Q mutation. The subtypes of detected mutations are described in S1 Table.
Direct sequencing. The mutational analyses of EGFR were performed by directional sequencing using polymerase chain reaction (PCR) fragments amplified from genomic DNA. The DNA was amplified with standard PCR technique using each exon-specific primer. PCR products were electrophoresed on 2% agarose gels and were purified with a QIAquick PCR purification kit (QIAGEN, Hilden, Germany). Bidirectional sequencing was performed using the BigDye Terminator v 1.1 kit (Applied Biosystems, Foster City, CA, USA) on an ABI 3130xl DNA analyser (Applied Biosystems) [28].
PNA clamping method. The principle of the technology is that PNA inhibits amplification of wild-type DNA by hybridizing wild-type sequences, and therefore mutant DNA is dominantly amplified. The amplified mutant type DNA is detected by intercalating dye. PNAClamp analysis was performed using the PNAClamp™ EGFR Mutation Detection Kit (Panagene) following the manual provided by the manufacturer. Briefly, 7 μl of DNA template, 3 μl of each PNA mix, and 10 μl of 2X premix are mixed for a single amplification reaction. The amplification of the mixture (20 μl) was performed in a CFX96 real-time PCR instrument (Bio-Rad, Richmond, CA, USA) with the following thermal program: a pre-incubation at 94°C for 5 min and 40 cycles of amplification (94°C for 30 seconds (s); 70°C for 30 s; 63°C for 30 s; and 72°C for 30 s). Detection of signal of intercalating dye was measured at every step at 63°C. Ct value for the reaction (Sample Ct value) was determined based on the fluorescence values measured at every step at 63°C. If a mutation occurs in a specific codon site, it is not hybridized and amplified so that the Ct value is low. Assessment of the result was determined according to the delta (Δ) Ct value, which was calculated according to each kit manual. ΔCt is the standard Ct value minus the Ct value that was obtained from an unknown sample. The presence of each codon mutation is confirmed by the unique value of ΔCt [29].
Statistical analysis
All continuous variables were presented as mean ± standard deviation (SD) and categorical variables were presented as number (%). Among EGFR mutation types, continuous variables were analysed by one-way analysis of variance (ANOVA) test or Kruskal-Wallis test and categorical variables were analysed by chi-square test or Fisher’s exact test, when appropriate. Student t-test or Mann-Whitney test were used for comparison of continuous variables between two groups. Bonferroni post hoc test was applied with a p value < 0.01 in variables that showed significant differences in ANOVA for the comparison of five institutes. OS was estimated by using the Kaplan-Meier methods, and difference between groups was assessed by log-rank test. A Cox proportional hazard regression model was used to identify independent factors of OS in subjects with EGFR mutations, presenting hazard ratio (HR) with 95% confidential interval (CI). P-value < 0.05 (two-tailed) was considered statistically significant. All statistical analyses were calculated using SPSS version 24.0 (IBM Corporation, Armonk, NY, USA).
Results
Baseline characteristics between EGFR positive and negative subjects
The study cohort included 1,020 subjects; the mean age was 66.8 years, 41.7% were female, 58.7% were never-smokers, and 388 (38.0%) had EGFR mutations (Table 1). The median follow-up period was 13.3 months, and 779 (76.4%) patients died during follow-up. Significant differences by institutes were found in age, smoking status, stage, pulmonary functional parameters, CCI, treatment patterns, and OS (S2 Table). In post hoc analysis of mean OS, there was no statistical difference between any two groups when the p value was cut off at 0.01. Also in univariate and multivariate analysis, OS was not different among institutes (S3 Table).
Table 1. Baseline demographics in EGFR positive and negative lung adenocarcinoma subjects.
| EGFR positive | EGFR negative | Total | p-value | |
|---|---|---|---|---|
| Number(n) (%) | 388 (38.0)1 | 632 (62.0) | 1,020 | |
| Age | 66.2 ± 11.5 | 67.2 ± 11.4 | 66.8 ± 11.42 | 0.190 |
| Sex | <0.001 | |||
| Male | 154 (39.7) | 441 (69.8) | 595 (58.3) | |
| Female | 234 (60.3) | 191 (30.2) | 425 (41.7) | |
| Low BMI (<18.5 kg/m2) | 28 (7.2) | 63 (10.0) | 91 (8.9) | 0.134 |
| Smoking status (n = 1,017)3 | <0.001 | |||
| Ever smoker | 97/387 (25.1) | 323/630 (51.3) | 420/1017 (41.3) | |
| Never smoker | 290/387 (74.9) | 307/630 (48.7) | 597/1017 (58.7) | |
| Smoking amount in smoker, pack-years | 27.9 ± 18.2 | 37.1 ± 20.5 | 35.0 ± 20.3 | <0.001 |
| Stage (n = 1012)3 | 0.001 | |||
| III | 43/386 (11.1) | 121/626 (19.3) | 164/1012 (16.2) | |
| IV | 343/386 (88.9) | 505/626 (80.7) | 848/1012 (83.8) | |
| Treatment | ||||
| Chemotherapy | 321 (82.7) | 442 (69.9) | 763 (74.8) | <0.001 |
| TKI | 284 (73.2) | 131 (20.7) | 415 (40.7) | <0.001 |
| Radiation therapy | 106 (27.3) | 166 (26.3) | 272 (26.7) | 0.712 |
| FEV1 (n = 706)3 | 79.2 ± 19.3 | 76.0 ± 20.3 | 77.2 ± 20.0 | 0.041 |
| FVC (n = 706)3 | 77.6 ± 17.7 | 78.0 ± 17.3 | 77.9 ± 17.4 | 0.745 |
| CCI | 5.6 ± 2.2 | 5.9 ± 2.2 | 5.8 ± 2.2 | 0.044 |
| Median overall survival, month (95% CI) | 22.8 (20.5–25.0) | 10.0 (8.9–11.1) | 13.8 (12.2–15.5) | <0.001 |
1Number (%);
2Mean+/- SD; BMI, body mass index;
3Differences in total number are due to missing values. TKI, tyrosine kinase inhibitor; FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; CCI, Charlson comorbidity index; CI, confidence interval.
Subjects with EGFR positive lung adenocarcinoma showed higher rates of female sex, never smokers, and TNM stage IV. They also received more conventional chemotherapy and TKIs compared with EGFR negative subjects. Baseline FEV1 was better in EGFR positive subjects than in EGFR negative subjects (Table 1).
Baseline characteristics of EGFR positive subjects
Among the 388 EGFR positive subjects, E19 dels (51.0%) were most common, followed by E21 mutations (42.0%). Mutations in E18 and E20 were identified in 3.6% and 3.4% of EGFR positive cases, respectively (Table 2). There were significant differences among the four EGFR mutation sites in age (67.0 [E18] vs. 64.1 [E19] vs. 66.4 [E20] vs. 68.8 [E21] years, p = 0.002) and rates of never-smokers (71.4% vs. 74.7% vs. 38.5% vs. 78.4%, p = 0.016). Other baseline characteristics were not different among the four groups.
Table 2. Baseline demographics according to mutation types among EGFR positive lung adenocarcinoma subjects.
| Exon 18 | Exon 19 | Exon 20 | Exon 21 | p-value | |
|---|---|---|---|---|---|
| Number(n) (%) | 14 (3.6)1 | 198 (51.0) | 13 (3.4) | 163 (42.0) | |
| Age | 67.0 ± 10.12 | 64.1 ± 11.9 | 66.4 ± 9.2 | 68.8 ± 10.7 | 0.002 |
| Sex | 0.102 | ||||
| Male | 7 (50.0) | 79 (39.9) | 9 (69.2) | 59 (36.2) | |
| Female | 7 (50.0) | 119 (60.1) | 4 (30.8) | 104 (63.8) | |
| Low BMI (<18.5 kg/m2) | 0 | 17 (8.6) | 0 | 11 (6.7) | 0.439 |
| Smoking status (n = 387)3 | 0.016 | ||||
| Ever smoker | 4/14 (28.6) | 50/198 (25.3) | 8/13 (61.5) | 35/162 (21.6) | |
| Never smoker | 10/14 (71.4) | 148/198 (74.7) | 5/13 (38.5) | 127/162 (78.4) | |
| Smoking amount (PY) in smoker | 38.5 ± 19.9 | 27.8 ± 16.9 | 33.2 ± 29.6 | 25.7 ± 16.6 | 0.475 |
| Stage (n = 386)3 | 0.941 | ||||
| III | 1/13 (7.7) | 22/197 (11.2) | 2/13 (15.4) | 18/163 (11.0) | |
| IV | 12/13 (92.3) | 175/197 (88.8) | 11/13 (84.6) | 145/163 (89.0) | |
| Treatment | |||||
| Chemotherapy | 11 (78.6) | 157 (79.3) | 12 (92.3) | 141 (86.5) | 0.234 |
| TKI | 9 (64.3) | 140 (70.7) | 8 (61.5) | 127 (77.9) | 0.268 |
| Radiation therapy | 4 (28.6) | 56 (28.3) | 4 (30.8) | 42 (25.8) | 0.944 |
| FEV1 (n = 269)3 | 77.5 ± 25.8 | 79.0 ± 17.8 | 80.1 ± 16.4 | 79.6 ± 21.1 | 0.982 |
| FVC (n = 269)3 | 79.9 ± 23.5 | 76.9 ± 16.8 | 83.0 ± 18.6 | 77.9 ± 18.3 | 0.763 |
| CCI | 5.7 ± 2.2 | 5.5 ± 2.2 | 4.8 ± 1.6 | 5.7 ± 2.2 | 0.440 |
| Median overall survival, month (95% CI) | 17.2 (10.5–24.0) | 29.9 (22.4–37.4) | 19.1 (8.3–30.0) | 20.6 (16.9–24.3) | 0.003 |
1 Number (%);
2Mean+/- SD;
3Differences in total number are due to missing values. BMI, body mass index; PY, pack-years; TKI, tyrosine kinase inhibitor; FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; CCI, Charlson comorbidity index; CI, confidence interval.
Of the 284 subjects who received TKI, 250 (88.0%) were TKI-responders. TKI-responders were younger; the TKI- responder group had a significantly higher proportion of E19 dels and a significantly lower rate of E21 mutations than the TKI non-responder (Table 3).
Table 3. Comparison of baseline characteristics between TKI-responders and non-responders in EGFR positive lung adenocarcinoma.
| Responder1 | Non-responder | Total | p-value | |
|---|---|---|---|---|
| Number(n) (%) | 250 (88.0)2 | 34 (12.0) | 284 | |
| Age | 66.5 ± 10.73 | 72.0 ± 10.8 | 67.2 ± 10.8 | 0.006 |
| Sex | 0.653 | |||
| Male | 93 (37.2) | 14 (41.2) | 107 (37.7) | |
| Female | 157 (62.8) | 20 (58.8) | 177 (62.3) | |
| Low BMI (<18.5 kg/m2) | 20 (8.0) | 3 (8.8) | 23 (8.1) | 0.745 |
| Smoking status (n = 283)4 | 0.924 | |||
| Ever smoker | 64/249 (25.7) | 9/34 (26.5) | 73/283 (25.7) | |
| Never smoker | 185/249 (74.3) | 25/34 (73.5) | 210/283 (74.2) | |
| Smoking amount (PY) in smoker | 28.1 ± 19.1 | 30.3 ± 21.4 | 28.3 ± 19.2 | 0.765 |
| Stage | 0.054 | |||
| III | 25 (10.0) | 0 (0.0) | 25 (8.8) | |
| IV | 225 (90.0) | 34 (100.0) | 259 (91.2) | |
| FEV1 (n = 208)4 | 79.7 ± 19.9 | 77.5 ± 19.7 | 79.4 ± 19.8 | 0.609 |
| FVC (n = 208)4 | 78.4 ± 17.8 | 78.4 ± 16.2 | 78.4 ± 17.6 | 1.000 |
| CCI | 5.6 ± 2.1 | 6.2 ± 2.4 | 5.7 ± 2.2 | 0.138 |
| Mutation type | ||||
| E18 | 9 (3.6) | 0 | 9 (3.2) | 0.606 |
| E19 | 129 (51.6) | 11 (32.4) | 140 (49.3) | 0.035 |
| E20 | 6 (2.4) | 2 (5.9) | 8 (2.8) | 0.246 |
| E21 | 106 (42.4) | 21 (61.8) | 127 (44.7) | 0.033 |
| Median overall survival, month (95% CI) | 28.2 (23.6–32.8) | 11.2 (NO-24.0) | 25.1 (20.9–29.4) | <0.001 |
1TKI responder was defined as a patient who received more than 4 cycles of TKI;
2Number (%);
3Mean+/- SD;
4Differences in total number are due to missing values. TKI, tyrosine kinase inhibitor; BMI, body mass index; PY, pack-years; FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; CCI, Charlson comorbidity index; E, Exon; NO, not obtained; CI, confidence interval.
Comparison of survival curves
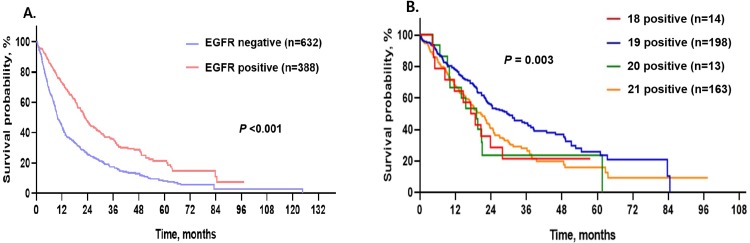
Amongst all subjects, the median OS was 13.8 months (95% CI: 12.2–15.5 months). EGFR positive subjects had better median OS (22.8 months, 95% CI: 20.5–25.0 months) than EGFR negative subjects (10.0 months, 95% CI: 8.9–11.1 months, p < 0.001; Table 1 and Fig 2A).
Fig 2. Survival curves in subjects with advanced lung adenocarcinoma.
A. Comparison of survival curves between EGFR positive and negative lung adenocarcinoma. B. Comparison of survival curves among mutation types in EGFR positive lung adenocarcinoma.
Within EGFR positive subjects, there were also significant differences in survival curves among the four EGFR mutation sites (Table 2; p = 0.003). Subjects with E19 dels had the best median OS (29.9 months, 95% CI: 22.4–37.4 months), while E18 mutations were associated with the worst median OS (17.2 months, 95% CI: 10.5–24.0 months). The median OS of subjects with E20 and E21 mutations were 19.1 months (95% CI: 8.3–30.0 months) and 20.6 months (95% CI: 16.9–24.3 months), respectively (Table 2 and Fig 2B).
Comparison of survival curves in subjects with TKI treatment
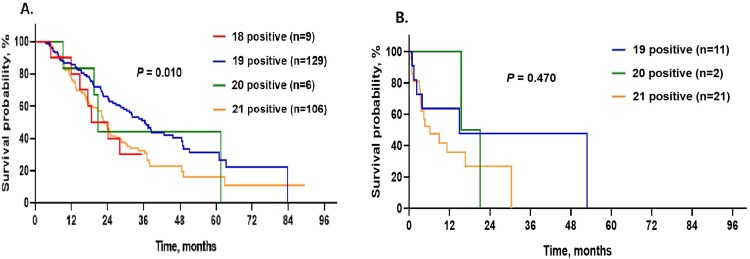
The median OS of TKI-responders was 28.2 months (95% CI: 23.6–32.8 months), while that of TKI non-responders was 11.2 months (95% CI: not obtained–24.0 months) (Table 3). Amongst TKI-responders, subjects with E19 dels had significantly longer median OS (36.4 months, 95% CI: 30.5–42.3 months, p = 0.010) than subjects with E18 (18.6 months, 95% CI: 14.7–22.5 months), E20 (20.8 months, 95% CI: 18.2–23.3 months), and E21 (22.9 months, 95% CI: 19.6–26.2 months) mutations (Fig 3A). There were no significant differences in OS between TKI non-responders with regard to mutation site (p = 0.470; Fig 3B). Amongst all subjects who were treated with TKI, there were no significant differences in median OS between subjects with E18 (18.6 months, 95% CI: 14.7–22.5 months) and E21 (22.0 months, 95% CI, 17.6–26.4 months, p = 0.818) mutations, nor between subjects with E20 (20.8 months, 95% CI, 18.9–22.6 months) and E21 (22.0 months, 95% CI, 17.6–26.4 months, p = 0.927) mutations.
Fig 3. Survival curves in subjects with EGFR positive lung adenocarcinoma according to TKI response.
A. Comparison of survival curves among EGFR mutation types in TKI-responders. B. Comparison of survival curves among EGFR mutation types in TKI non-responders.
Risk factors for mortality in EGFR positive subjects
E19 dels and an E21 mutation were inversely associated with mortality compared with EGFR negative subjects, while E18 and E20 mutations did not show statistically significant associations with mortality (Table 4). In EGFR positive subjects, E19 dels, an E21 mutation, age, low BMI, smoking status, stage, CCI, and use of TKI were associated with mortality in univariate analysis. In a multivariate analysis model including E19 dels, age (HR: 1.029, 95% CI: 1.017–1.041), low BMI (HR: 1,729, 95% CI: 1.108–2.700), stage IV (HR: 2.246, 95% CI: 1.426–3.536), use of TKI (HR: 0.533, 95% CI: 0.405–0.702) as well as E19 dels (HR: 0.678, 95% CI: 0.527–0.872) were significant factors for mortality. Conversely, a multivariate analysis model including an E21 mutation revealed that age (HR: 1.030, 95% CI: 1.018–1.042), low BMI (HR, 1,658, 95% CI: 1.065–2.582), stage IV (HR:2.296, 95% CI: 1.458–3.615), use of TKI (HR: 0.527, 95% CI: 0.400–0.695) as well as an E21 mutation (HR: 1,365, 95% CI: 1.063–1.754) were significant factors for mortality.
Table 4. Univariate and multivariate Cox proportional hazard analysis for mortality in EGFR positive lung adenocarcinoma subjects.
| Univariate | Multivariate model-1 | Multivariate model-2 | ||||
|---|---|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Mutated exon1 | ||||||
| 18 | 1.398 (0.763–2.559) | 0.278 | ||||
| 19 | 0.638 (0.498–0.818) | <0.001 | 0.678 (0.527–0.872) | 0.002 | ||
| 20 | 1.648 (0.899–3.022) | 0.106 | ||||
| 21 | 1.408 (1.099–1.804) | 0.007 | 1.365 (1.063–1.754) | 0.015 | ||
| Age | 1.025 (1.013–1.037) | <0.001 | 1.029 (1.017–1.041) | <0.001 | 1.030 (1.018–1.042) | <0.001 |
| Female | 0.837 (0.651–1.078) | 0.168 | ||||
| Low BMI (<18.5 kg/m2) | 1.591 (1.025–2.468) | 0.038 | 1.729 (1.108–2.700) | 0.016 | 1.658 (1.065–2.582) | 0.025 |
| Never smoker | 1.086 (0.811–1.456) | 0.579 | ||||
| Stage IV | 1.916 (1.225–2.999) | 0.004 | 2.246 (1.426–3.536) | <0.001 | 2.296 (1.458–3.615) | <0.001 |
| CCI | 1.110 (1.050–1.173) | <0.001 | ||||
| Chemotherapy | 0.758 (0.550–1.043) | 0.089 | ||||
| TKI | 0.674 (0.515–0.883) | 0.004 | 0.533 (0.405–0.702) | <0.001 | 0.527 (0.400–0.695) | <0.001 |
| Radiation therapy | 1.120 (0.856–1.464) | 0.408 | ||||
1Mutations on a specific exon were compared with mutations on the rest of exons. BMI, body mass index; CCI, Charlson comorbidity index; TKI, tyrosine kinase inhibitor.
Comparison between E19 dels and E18/20/21 mutations in EGFR positive subjects
We also compared E19 dels to E18/20/21 mutations in terms of baseline demographics and a survival curve in EGFR positive subjects. Subjects with E19 dels were younger than subjects with other mutations (64.1 ± 11.9 [E19 dels] vs. 68.5 ± 10.5 [E18/20/21 mutations] years; p < 0.001). In addition, subjects with E19 dels had better median OS (29.9 months, 95% CI: 22.4–37.4 months) than subjects with E18/20/21 mutations (19.2 months, 95% CI: 16.1–22.4 months, p < 0.001) (S4 Table and S1 Fig).
Discussion
This study first investigated clinical and prognostic features of advanced adenocarcinoma according to EGFR mutation status in Korean patients, particularly for specific EGFR mutation types (mutations in E18, E19, E20, and E21). Similar studies have previously examined common and uncommon EGFR mutations with limited prognostic comparisons for NSCLC [23, 25, 30]. Our study identified that E19 dels were a predictor for good prognosis in EGFR positive lung cancer, similar to previous results [31–35]; conversely, an E21 mutation significantly increased mortality. These results were maintained when we analysed only TKI-responders. Mutations in E18 and E20 were not worse factors than an E21 mutation in lung adenocarcinoma.
In this study, EGFR-mutant lung adenocarcinoma patients had better survival rate than patients with wild-type EGFR. It is well known that TKI treatment leads to a dramatically improved prognosis in patients with EGFR positive lung cancer [5–8]. In our cohort, the EGFR positive group had a higher prevalence of stage IV lung cancer, but a significantly better prognosis, which was probably related to higher rates of TKI treatment. This result is supported by the fact that 73.2% of EGFR positive patients received TKI treatment in our study. Additionally, never-smoker lung cancer, especially of adenocarcinoma histology, was highly associated with female sex of East Asian ethnicity and higher prevalence of genomic alterations [36], which was in accordance with our findings.
Our study demonstrated significant differences in survival curves amongst the four EGFR mutation sites, with a good prognosis in patients with E19 dels and relatively adverse prognosis in patients with an E21 mutation. The fact that TKI is more beneficial in patients with E19 dels than in those with an E21 mutation with regard to OS and progression-free survival (PFS) has already been reported in a number of studies [31–34]. In a meta-analysis by Zhang et al. [34], six clinical trials documented that patients with E19 dels were associated with a better prognosis than patients with an E21 L858R mutation (HR of E19 dels to E21 L858R mutation: 0.59, 95% CI: 0.38–0.92, p = 0.019). These findings were in line with our results.
According to our study, mutations in E18 and E20 did not significantly influence prognosis compared with previous studies on the prognosis of uncommon mutations [22, 24, 37]. Due to the low incidence of E18 and E20 mutations, limited studies are available on the prognosis of patients with uncommon EGFR mutations. Sutiman et al. [37] found significant differences in PFS (p < 0.001) between patients with different exon mutations, with the best median PFS among E19 dels (n = 202, 10.9 months) and the worst among E20 mutations (n = 9, 1.2 months), which serve as independent predictors for PFS (HR: 16.0, 95% CI: 6.88–37.1, p < 0.001). However, the authors also reported no significant differences in OS curves among EGFR mutation types. Beau-Faller et al. [22] found that OS of patients with E18 mutations (n = 18, median: 22 months) was better than that of patients with E20 mutations (n = 25, median: 9.5 month) in patients with TKI treatment. Chiu et al. [24] reported that patients with uncommon mutations, including G719X (n = 78), L861Q (n = 57), and S768I (n = 7), benefited from TKI treatment, but with lower efficacy than those with common mutations (median OS: 24 [uncommon] vs. 29.7 [common] months, p = 0.005). The study by Yang et al. [21] that was based on afatinib phase 2 and 3 clinical trials revealed that patients with a point mutation or duplications in E18–21 had better PFS and OS than those with E20 insertions or T790M mutations. This discordance might be attributable to a small sample size or different definitions of the “uncommon group.” Thus, a single prognostic evaluation under the umbrella of “uncommon mutation” to represent several exonal variations cannot be justified because prognosis is different for each type. Krawczyk et al. [25] found that among 180 NSCLC patients who were treated with 1st or 2nd line TKIs, patients with uncommon mutations (n = 13) had shorter PFS (median: 5 [uncommon] vs. 10 [common] months, p = 0.005), but similar median OS (26 vs. 22 months, p = 0.251) compared with patients with common mutations of E19 dels and a L858R point mutation. Considering the different TKI sensitivity between patients with E19 dels and an E21 mutation, it might be inappropriate to simply divide into common and uncommon mutations when evaluating OS. In our study, the prognosis of uncommon mutations in adenocarcinoma treated with TKIs was worse than that of E19 dels and similar to that of an E21 mutation. When an uncommon EGFR mutation is detected in lung adenocarcinoma, TKI treatment should be actively considered. Comparison of E19 dels to E18/20/21 mutations showed younger age and better OS in patients with E19 dels. The prognostic differences in EGFR mutations may be due to functional differences between deletions and other kinds of mutations. The functional loss due to exonal deletion in cancer cells will be a good prognostic factor, compared with other types of mutation, but further research is needed at this point.
This study had several limitations. First, although statistically corrected for the effects of variables, bias may exist from two kinds of EGFR test methods and a long observational study. Second, survival differences among institutes existed, but Bonferroni post hoc test showed no significance, and the differences of OS by institutes were influenced by age, sex, BMI, stage, CCI, and use of TKI. Third, early in the study, mutations other than EGFR were not routinely checked, so the rate of ALK mutations was relatively low. Fourth, the number of cases with E18 or E20 mutations was relatively small and can create bias in the analysis. Despite these limitations, the strength of this paper is that the prognosis according to EGFR exon mutational status was targeted to lung adenocarcinoma, a representative type for EGFR mutations, and exon locations of EGFR mutations that included specific types were focused to help assess clinical response to TKIs and their prognosis.
Conclusions
We found that EGFR positive advanced adenocarcinoma patients had better OS compared with EGFR negative due to response to TKI treatment. In lung adenocarcinoma with EGFR positivity, E19 dels predicts a good prognosis, while an E21 mutation is expected to increase mortality. In TKI treatment, mutations in E18 and E20 were not worse factors than the E21 L858R mutation, unlike previous reports. Further large-scale prospective studies will be needed to verify these results.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(TIF)
(XLSX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
A part of this study conducted by JS Ryu was supported by Korea Health Industry Development Institute (Korea Health Technology R&D Project HI15C0554). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346(2):92–8. Epub 2002/01/11. 10.1056/NEJMoa011954 [DOI] [PubMed] [Google Scholar]
- 2.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, et al. Paclitaxel–Carboplatin Alone or with Bevacizumab for Non–Small-Cell Lung Cancer. N Engl J Med. 2006;355(24):2542–50. 10.1056/NEJMoa061884 [DOI] [PubMed] [Google Scholar]
- 3.Marino P, Pampallona S, Preatoni A, Cantoni A, Invernizzi F. Chemotherapy vs supportive care in advanced non-small-cell lung cancer. Results of a meta-analysis of the literature. Chest. 1994;106(3):861–5. Epub 1994/09/01. 10.1378/chest.106.3.861 [DOI] [PubMed] [Google Scholar]
- 4.Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003;284(1):31–53. Epub 2003/03/22. 10.1016/s0014-4827(02)00098-8 [DOI] [PubMed] [Google Scholar]
- 5.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–57. Epub 2009/08/21. 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 6.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380–8. Epub 2010/06/25. 10.1056/NEJMoa0909530 [DOI] [PubMed] [Google Scholar]
- 7.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. The Lancet Oncol. 2012;13(3):239–46. Epub 2012/01/31. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 8.Tamura K, Okamoto I, Kashii T, Negoro S, Hirashima T, Kudoh S, et al. Multicentre prospective phase II trial of gefitinib for advanced non-small cell lung cancer with epidermal growth factor receptor mutations: results of the West Japan Thoracic Oncology Group trial (WJTOG0403). Br J Cancer. 2008;98(5):907–14. Epub 2008/02/20. 10.1038/sj.bjc.6604249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Comprenhensive Cancer Network (2017). Non-small cell lung cancer guidelines. https://wwwtri-kobeorg/nccn/guideline/lung/english/non_smallpdf 2017;v17.
- 10.Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res. 2015;5(9):2892–911. PMID: PMC4633915 [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y-L, Yuan J-Q, Wang K-F, Fu X-H, Han X-R, Threapleton D, et al. The prevalence of EGFR mutation in patients with non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget. 2016;7(48):78985–93. 10.18632/oncotarget.12587 PMID: PMC5346692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gahr S, Stoehr R, Geissinger E, Ficker JH, Brueckl WM, Gschwendtner A, et al. EGFR mutational status in a large series of Caucasian European NSCLC patients: data from daily practice. Br J Cancer. 2013;109(7):1821–8. 10.1038/bjc.2013.511 PMID: PMC3790166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boch C, Kollmeier J, Roth A, Stephan-Falkenau S, Misch D, Gruning W, et al. The frequency of EGFR and KRAS mutations in non-small cell lung cancer (NSCLC): routine screening data for central Europe from a cohort study. BMJ open. 2013;3(4). Epub 2013/04/06. 10.1136/bmjopen-2013-002560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y, Au JS, Thongprasert S, Srinivasan S, Tsai CM, Khoa MT, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol. 2014;9(2):154–62. Epub 2014/01/15. 10.1097/JTO.0000000000000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li AR, Chitale D, Riely GJ, Pao W, Miller VA, Zakowski MF, et al. EGFR mutations in lung adenocarcinomas: clinical testing experience and relationship to EGFR gene copy number and immunohistochemical expression J Mol Diagn. 2008;10(3):242–8. Epub 2008/04/12. 10.2353/jmoldx.2008.070178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sequist LV, Yang JC, Yamamoto N, O'Byrne K, Hirsh V, Mok T, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31(27):3327–34. Epub 2013/07/03. 10.1200/JCO.2012.44.2806 [DOI] [PubMed] [Google Scholar]
- 17.De Pas T, Toffalorio F, Manzotti M, Fumagalli C, Spitaleri G, Catania C, et al. Activity of epidermal growth factor receptor-tyrosine kinase inhibitors in patients with non-small cell lung cancer harboring rare epidermal growth factor receptor mutations J Thorac Oncol. 2011;6(11):1895–901. Epub 2011/08/16. 10.1097/JTO.0b013e318227e8c6 [DOI] [PubMed] [Google Scholar]
- 18.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Final overall survival results from a randomised, phase III study of erlotinib versus chemotherapy as first-line treatment of EGFR mutation-positive advanced non-small-cell lung cancer (OPTIMAL, CTONG-0802). Ann Oncol. 2015;26(9):1877–83. Epub 2015/07/05. 10.1093/annonc/mdv276 [DOI] [PubMed] [Google Scholar]
- 19.Lee CK, Wu YL, Ding PN, Lord SJ, Inoue A, Zhou C, et al. Impact of Specific Epidermal Growth Factor Receptor (EGFR) Mutations and Clinical Characteristics on Outcomes After Treatment With EGFR Tyrosine Kinase Inhibitors Versus Chemotherapy in EGFR-Mutant Lung Cancer: A Meta-Analysis. J Clin Oncol. 2015;33(17):1958–65. Epub 2015/04/22. 10.1200/JCO.2014.58.1736 [DOI] [PubMed] [Google Scholar]
- 20.Riely GJ, Pao W, Pham D, Li AR, Rizvi N, Venkatraman ES, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib Clin Cancer Res. 2006;12(3 Pt 1):839–44. Epub 2006/02/10. 10.1158/1078-0432.Ccr-05-1846 [DOI] [PubMed] [Google Scholar]
- 21.Yang JC, Sequist LV, Geater SL, Tsai CM, Mok TS, Schuler M, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. The Lancet Oncol. 2015;16(7):830–8. Epub 2015/06/09. 10.1016/S1470-2045(15)00026-1 [DOI] [PubMed] [Google Scholar]
- 22.Beau-Faller M, Prim N, Ruppert AM, Nanni-Metellus I, Lacave R, Lacroix L, et al. Rare EGFR exon 18 and exon 20 mutations in non-small-cell lung cancer on 10 117 patients: a multicentre observational study by the French ERMETIC-IFCT network. Ann Oncol. 2014;25(1):126–31. Epub 2013/11/29. 10.1093/annonc/mdt418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arrieta O, Cardona AF, Corrales L, Campos-Parra AD, Sanchez-Reyes R, Amieva-Rivera E, et al. The impact of common and rare EGFR mutations in response to EGFR tyrosine kinase inhibitors and platinum-based chemotherapy in patients with non-small cell lung cancer. Lung cancer. 2015;87(2):169–75. Epub 2015/01/07. 10.1016/j.lungcan.2014.12.009 [DOI] [PubMed] [Google Scholar]
- 24.Chiu CH, Yang CT, Shih JY, Huang MS, Su WC, Lai RS, et al. Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Treatment Response in Advanced Lung Adenocarcinomas with G719X/L861Q/S768I Mutations. J Thorac Oncol. 2015;10(5):793–9. Epub 2015/02/11. 10.1097/JTO.0000000000000504 [DOI] [PubMed] [Google Scholar]
- 25.Krawczyk P, Kowalski DM, Ramlau R, Kalinka-Warzocha E, Winiarczyk K, Stencel K, et al. Comparison of the effectiveness of erlotinib, gefitinib, and afatinib for treatment of non-small cell lung cancer in patients with common and rare EGFR gene mutations. Oncol Lett. 2017;13(6):4433–44. Epub 2017/06/11. 10.3892/ol.2017.5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Arcangelo M, Hirsch FR. Clinical and comparative utility of afatinib in non-small cell lung cancer. Biologics. 2014;8:183–92. 10.2147/BTT.S40567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang TS. Molecular biological techniques in cytopathologic diagnosis. Korean J Pathol 2009, 43(5):387–92. 10.4132/KoreanJPathol.2009.43.5.387 [DOI] [Google Scholar]
- 28.Choi YL, Sun JM, Cho J, Rampal S, Han J, Parasuraman B, et al. EGFR mutation testing in patients with advanced non-small cell lung cancer: a comprehensive evaluation of real-world practice in an East Asian tertiary hospital. PLoS One. 2013;8(2):e56011 Epub 2013/03/08. 10.1371/journal.pone.0056011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee KY, Kim HJ, Kim SJ, Yoo GH, Kim WD, Oh SY, et al. PNA-mediated PCR clamping for the detection of EGFR mutations in non-small cell lung cancer. Tuberc Respir Dis. 2010;69(4):271–8. 10.4046/trd.2010.69.4.271 [DOI] [Google Scholar]
- 30.Rossi S, D'Argento E, Basso M, Strippoli A, Dadduzio V, Cerchiaro E, et al. Different EGFR Gene Mutations in Exon 18, 19 and 21 as Prognostic and Predictive Markers in NSCLC: A Single Institution Analysis. Mol Diagn Ther. 2016;20(1):55–63. Epub 2015/12/10. 10.1007/s40291-015-0176-x [DOI] [PubMed] [Google Scholar]
- 31.Choi YW, Jeon SY, Jeong GS, Lee HW, Jeong SH, Kang SY, et al. EGFR Exon 19 Deletion is Associated With Favorable Overall Survival After First-line Gefitinib Therapy in Advanced Non-Small Cell Lung Cancer Patients. Am J Clin Oncol. 2018;41(4):385–90. Epub 2016/03/12. 10.1097/COC.0000000000000282 [DOI] [PubMed] [Google Scholar]
- 32.Wang H, Huang J, Yu X, Han S, Yan X, Sun S, et al. Different efficacy of EGFR tyrosine kinase inhibitors and prognosis in patients with subtypes of EGFR-mutated advanced non-small cell lung cancer: a meta-analysis. J Cancer Res Clin Oncol. 2014;140(11):1901–9. Epub 2014/06/09. 10.1007/s00432-014-1709-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackman DM, Yeap BY, Sequist LV, Lindeman N, Holmes AJ, Joshi VA, et al. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12(13):3908–14. Epub 2006/07/05. 10.1158/1078-0432.CCR-06-0462 [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Sheng J, Kang S, Fang W, Yan Y, Hu Z, et al. Patients with exon 19 deletion were associated with longer progression-free survival compared to those with L858R mutation after first-line EGFR-TKIs for advanced non-small cell lung cancer: a meta-analysis. PLoS One. 2014;9(9):e107161 Epub 2014/09/16. 10.1371/journal.pone.0107161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang H, Zhu M, Li Y, Li Q. Association between EGFR exon 19 or exon 21 mutations and survival rates after first-line EGFR-TKI treatment in patients with non-small cell lung cancer. Mol Clin Oncol. 2019;11(3):301–8. Epub 2019/08/07. 10.3892/mco.2019.1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito S, Espinoza-Mercado F, Liu H, Sata N, Cui X, Soukiasian HJ. Current status of research and treatment for non-small cell lung cancer in never-smoking females. Cancer Biol Ther. 2017; 18(6):359–368. Epub 2017/05/11. 10.1080/15384047.2017.1323580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutiman N, Tan SW, Tan EH, Lim WT, Kanesvaran R, Ng QS, et al. EGFR Mutation Subtypes Influence Survival Outcomes following First-Line Gefitinib Therapy in Advanced Asian NSCLC Patients. J Thorac Oncol. 2017;12(3):529–38. Epub 2016/12/03. 10.1016/j.jtho.2016.11.2225 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.