Abstract
Fritillariae cirrhosae bulbus is a famous type of traditional Chinese medicine used for cough relief and eliminating phlegm. The medicine originates from dried bulbs of five species and one variety of Fritillaria. Recently, immature bulbs from other congeneric species, such as F. ussuriensis, have been sold as adulterants of Fritillariae cirrhosae bulbus in medicine markets owing to the high price and limited availability of the genuine medicine. However, it is difficult to accurately identify the bulbs from different original species of Fritillariae cirrhosae bulbus and its adulterants based on traditional methods, although such medicines have different prices and treatment efficacies. The present study adopted DNA barcoding to identify these different species and compared the discriminatory power of super, universal, and specific barcodes in Fritillaria. The results revealed that the super-barcode had strong discriminatory power (87.5%). Among universal barcodes, matK provided the best species resolution (87.5%), followed by ITS (62.5%), rbcL (62.5%), and trnH-psbA (25%). The combination of these four universal barcodes provided the highest discriminatory power (87.5%), which was equivalent to that of the super-barcode. Two plastid genes, ycf1 and psbM-psbD, had much better discriminatory power (both 87.5%) than did other plastid barcodes, and were suggested as potential specific barcodes for identifying Fritillaria species. Phylogenetic analyses indicated that F. cirrhosa was not a “good” species that was composed of multiple lineages, which might have affected the evaluation of the discriminatory ability. This study revealed that the complete plastid genome, as super barcode, was an efficient and reliable tool for identifying the original species of Fritillariae cirrhosae bulbus and its adulterants.
Introduction
Fritillaria L. is one of the most important genera in the family Liliaceae, and it includes approximately 140 species of perennial herbaceous plants worldwide [1,2]. Almost all known species in this genus grow in temperate regions of the Northern Hemisphere [3]. There are 24 species in China, most of which have medicinal value and are used for cough relief and eliminating phlegm, such as F. cirrhosa D. Don, F. ussuriensis Maxim., F. walujewii Regel, F. thunbergii Miq., and so on [4–6]. According to the Chinese Pharmacopoeia (2015), there are five traditional Chinese medicines that originate from the dried bulbs of Fritillaria species. Among them, Fritillariae cirrhosae bulbus (also known as “Chuanbeimu” in traditional Chinese medicine) has been regarded the best throughout Chinese history. The original species used to make this type of medicine include five species and one variety, namely F. cirrhosa, F. przewalskii Maxim., F. unibracteata Hsiao et K.C. Hsia, F. delavayi Franch., F. taipaiensis P.Y. Li, and F. unibracteata var. wabuensis (S.Y. Tang et S.C. Yue) Z.D. Liu., S. Wang et S.C. Chen [7]. Recently, the market price of Fritillariae cirrhosae bulbus has been increasing sharply due to its limited output from wild habitats, and thus immature bulbs of other Fritillaria species (e.g., F. ussuriensis, F. thunbergii, F. pallidiflora Schrenk, etc.) have been sold in medicinal markets imitating the original Fritillariae cirrhosae bulbus. Genuine Fritillariae cirrhosae bulbus originating from bulbs with different botanical origins are difficult to identify based on the morphological characteristics and traditional methods. The bulbs from each original species obviously differ in price and might also differ in their medicinal treatment efficacy, which could seriously affect their health benefits to consumers [8–11]. Thus, it is extremely important to be able to accurately discriminate between Fritillariae cirrhosae bulbus and its adulterants, as well as among the different original species used to make these medicines.
DNA barcoding is a technology used for species identification that employs a short and standardized DNA region [12–15]. It has been proven to be an effective tool for rapid and accurate species discrimination [16–19]. Based on a large number of studies, three plastid loci (trnH-psbA, matK, and rbcL) and one ribosomal DNA spacer region (the internal transcribed spacer or ITS), or their combinations, were proposed as universal barcodes for plants that could discriminate most plant species [20–22]. However, for taxonomically complicated groups, such as Fritillaria, the use of these universal barcodes and their combinations has achieved lower success rates in species discrimination, except matK (this barcode had low success rate of PCR and Sanger sequencing in Fritillaria), due to insufficient sequence variation [23, 24].
The complete plastid genome was suggested as a super-barcode for identification of plant species, especially for taxonomically difficult taxa such as the genera Citrus, Oncidium, and Gossypium [25–27]. This genome has been also used successfully to explore phylogenetic relationships among plant taxa [28–30]. Typically, the plastid genome in angiosperms is circular, ranges in size from 72 to 217 kb, and has a quadripartite structure composed of a large single copy region (LSC), small single copy region (SSC), and a pair of inverted repeats (IRs) [31–34]. In contrast with the nuclear and mitochondrial genomes, the plastid genome is largely conserved among taxa in terms of its gene content, organization, and structure. Moreover, plastid genomes are maternally inherited and therefore appropriate for use in genetic engineering due to lack of cross-recombination [35,36]. These characteristics of the plastid genome render it suitable for species discrimination within complex plant taxa [37,38]. The results of recent studies also supported the strong discriminatory power of this super-barcode when used in species identification [39,40]. For the genus Fritillaria, complete plastid genomes were previously employed to evaluate phylogenetic relationships among some species and produced results that were supported by high bootstrap values [41–43]. However, those previous studies generally sequenced only one individual per species, and thus they could not effectively compare the intra- and inter-specific genetic distances in Fritillaria species. This leads to the question of whether species of Fritillaria, especially the original plants used to make Fritillariae cirrhosae bulbus and its adulterants, could be correctly identified at the species level by analyses of the complete plastid genome.
In the present study, we used the complete plastid genome sequence as a super-barcode to: identify the botanical origins of Fritillariae cirrhosae bulbus and its adulterants; compare the discriminatory power of the super-barcode with that of universal DNA barcodes and their combinations; and scan the highly variable gene regions as potential specific barcodes for species identification of Fritillariae cirrhosae bulbus. The present study provided abundant information for further development of super-barcodes and broadened the horizon over which other rapid and accurate molecular techniques for species identification in Fritillaria can be explored.
Materials and methods
Material sampling
A total of 32 individuals of Fritillaria representing the five original species of Fritillariae cirrhosae bulbus and three species of its adulterants, as well as an individual of F. anhuiensis S.C. Chen & S.F. Yin as an outgroup, were used in tree-building analysis in this study (Fig 1, S1 Table). Among these species, individuals of the five original species that produce the genuine medicine were collected from wild habitats in Southwest China, while those of the adulterants were collected from cultivated populations in provinces of Zhejiang, Jilin, and Xinjiang, China. None of the species from the genus Fritillaria are listed as the national protected plants in China (Information System of Chinese Rare and Endangered Plants, http://www.iplant.cn/rep/) and therefore their collection is allowed for scientific research. Fresh leaves were sampled from healthy, mature individuals in the field or cultivation bases and then dried by using silica gel. Meanwhile, 3–5 individuals with flowers were dug up and preserved as voucher specimens. Subsequently, geographic information (latitude, longitude, altitude, etc.) for the sampling locations was obtained by a Global Positioning System receiver (GPS; Garmin, Olathe, KS, USA). All voucher specimens of Fritillaria were identified and then deposited at the Herbarium of Medicinal Plants and Crude Drugs of the College of Pharmacy and Chemistry, Dali University, Dali, China (S1 Table).
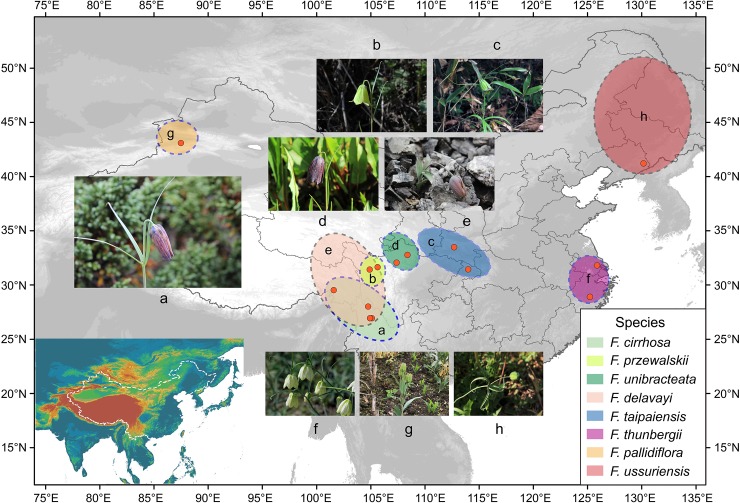
Fig 1. Distribution of the five original species of Fritillariae cirrhosae bulbus and three species of its adulterants.
The distributional range of each species is drawn based on the records by Luo et al. [8,9] and herbarium specimens (http://www.cvh.ac.cn/). The a-f refer to these species and their distribution. Photos of the eight Fritillaria species studied are also added: a. F. cirrhosa; b. F. przewalskii; c. F. taipaiensis; d. F. unibracteata; e. F. delavayi; f. F. thunbergii; g. F. pallidiflora; h. F. ussuriensis.
DNA extraction, sequencing, and assembly
Total genomic DNA was extracted from about 100 mg of dried leaf material using a modified CTAB method [44,45]. The DNA content was checked by electrophoresis on 1.2% agarose gels, and its concentration was determined using a SmartSpecTM Plus Spectrophotometer (Bio-Rad, Hercules, CA, USA). DNA extracts were then fragmented for the construction of 300 bp short-insert paired-end (PE) libraries and sequenced on an Illumina HiSeq 2000/2500 sequencer at the Beijing Genomics Institute (BGI, Shenzhen, China).
The raw data were filtered using Trimmomatic v.0.32 [46] with default settings. Paired-end reads in the clean data were then filtered and assembled into contigs using GetOrganelle.py [47] with Fritillaria cirrhosa (accession number: KF769143) as reference [48], calling the bowtie2 v., Blastn v., and SPAdes v.3.10 [49]. The de novo assembly graphs were visualized and edited using Bandage Linux dynamic v.8.0 [50], and then a whole or nearly whole circular plastid genome (plastome) was generated.
Annotation and sequence submission
The plastomes were annotated by aligning them to the complete plastid genome sequence available in NCBI using MAFFT [48,51] with default parameters, which was coupled with manual adjustment using Geneious v.11.1.4 [52]. Circular genome visualization was generated with OGDRAW v.1.2 [53]. Furthermore, the ITS sequences were sequenced using Illumina sequencing technology and extracted from the raw data. The annotated plastid genomes of the nine Fritillaria species and their ITS sequences were submitted to the NCBI under the accession numbers MH588404-MH588436 for ITS sequences and those listed in Table 1 for the complete plastid genomes.
Table 1. Summary of complete plastid genomes obtained for the five original species of Fritillariae cirrhosae bulbus and three species of its adulterants, as well as the outgroup (F. anhuiensis).
| Species | Code | Total length (bp) | Large single copy (LSC, bp) | Small single copy (SSC, bp) | Inverted repeat (IR, bp) | GC% | Number of genes | Accession number |
|---|---|---|---|---|---|---|---|---|
| F. cirrhosa | BM 1–1 | 151,546 | 81,402 | 17,542 | 26,301 | 37.0% | 115 | MH593342 |
| BM 1–2 | 151,546 | 81,402 | 17,542 | 26,301 | 37.0% | 115 | MH593343 | |
| BM 2–1 | 151,998 | 81,755 | 17,545 | 26,349 | 37.0% | 115 | MH244906 | |
| BM 2–2 | 151,605 | 81,467 | 17,534 | 26,302 | 37.0% | 115 | MH593344 | |
| BM 3–1 | 152,035 | 81,794 | 17,541 | 26,350 | 36.9% | 115 | MH593345 | |
| BM 3–2 | 152,035 | 81,794 | 17,541 | 26,350 | 36.9% | 115 | MH593346 | |
| F. przewalskii | BM 6–1 | 151,983 | 81,744 | 17,539 | 26,350 | 36.9% | 115 | MH244908 |
| BM 6–2 | 152,054 | 81,816 | 17,538 | 26,350 | 36.9% | 115 | MH593347 | |
| BM 7–1 | 151,955 | 81,715 | 17,540 | 26,350 | 37.0% | 115 | MH593348 | |
| BM 7–2 | 151,960 | 81,722 | 17,538 | 26,350 | 37.0% | 115 | MH593349 | |
| F. unibracteata | BM 8–1 | 151,058 | 81,339 | 17,539 | 26,090 | 37.0% | 115 | MH244909 |
| BM 8–2 | 151,057 | 81,338 | 17,539 | 26,090 | 37.0% | 115 | MH593350 | |
| BM 9–1 | 151,012 | 81,295 | 17,537 | 26,090 | 37.0% | 115 | MH593351 | |
| BM 9–2 | 151,078 | 81,398 | 17,538 | 26,071 | 37.0% | 115 | MH593352 | |
| F. delavayi | BM 10–1 | 151,853 | 81,602 | 17,513 | 26,369 | 37.0% | 115 | MH593353 |
| BM 10–2 | 151,854 | 81,603 | 17,513 | 26,369 | 37.0% | 115 | MH593354 | |
| BM 10–3 | 151,854 | 81,603 | 17,513 | 26,369 | 37.0% | 115 | MH593355 | |
| F. taipaiensis | BM 11–1 | 151,707 | 81,451 | 17,552 | 26,352 | 37.0% | 115 | MH244910 |
| BM 11–2 | 151,518 | 81,268 | 17,546 | 26,352 | 37.0% | 115 | MH593356 | |
| BM 12–1 | 151,741 | 81,478 | 17,561 | 26,351 | 37.0% | 115 | MH593357 | |
| BM 12–2 | 151,741 | 81,478 | 17,561 | 26,351 | 37.0% | 115 | MH593358 | |
| BM 12–3 | 151,741 | 81,478 | 17,561 | 26,351 | 37.0% | 115 | MH593359 | |
| F. thunbergii | BM 16–1 | 152,160 | 81,895 | 17,565 | 26,350 | 37.0% | 115 | MH244914 |
| BM 16–2 | 152,160 | 81,895 | 17,565 | 26,350 | 37.0% | 115 | MH593360 | |
| BM 17–1 | 152,160 | 81,895 | 17,565 | 26,350 | 37.0% | 115 | MH593361 | |
| BM 17–2 | 152,160 | 81,895 | 17,565 | 26,350 | 37.0% | 115 | MH593362 | |
| F. pallidiflora | BM 23–1 | 152,073 | 81,779 | 17,514 | 26,390 | 37.0% | 115 | MH593364 |
| BM 23–2 | 152,067 | 81,763 | 17,528 | 26,388 | 37.0% | 115 | MH593365 | |
| BM 23–3 | 152,073 | 81,780 | 17,513 | 26,390 | 37.0% | 115 | MH593366 | |
| F. ussuriensis | BM 26–1 | 151,571 | 81,773 | 17,126 | 26,336 | 36.9% | 115 | MH593367 |
| BM 26–2 | 151,523 | 81,741 | 17,122 | 26,330 | 37.0% | 115 | MH593368 | |
| BM 26–3 | 151,552 | 81,764 | 17,124 | 26,332 | 37.0% | 115 | MH593369 | |
| F. anhuiensis | BM 20–2 | 152,119 | 81,817 | 17,560 | 26,371 | 37.0% | 115 | MH593363 |
Variable site analysis
After using MAFFT v.7.129 to align the plastid genome sequences, BioEdit software was used to adjust the alignment manually [51,54]. A sliding window analysis was conducted to determine the nucleotide variability (Pi) in the whole plastid genome using DnaSP v.6.11 [55]. The step size was set to 200 bp, with a 600 bp window length. Moreover, the DnaSP software was used to identify and quantify the insertions/deletions (indels), mutations, and nucleotide variability (Pi) in all aligned datasets. The p-distances, GC content, variable sites, and parsimony informative sites in the genomes were identified and analyzed by the software MEGA v.7.0.26 [56].
Species discrimination of universal, super, and specific barcodes
To evaluate the success rates of species discrimination with each barcode, we used a tree-building method to analyze 14 datasets for each of the single regions examined and their combinations. All single regions, including universal DNA barcodes and highly variable loci, were extracted from the complete plastid genome sequences, except the ITS/ITS2 region. These datasets were aligned with MAFFT [51] and used to build neighbor-joining trees (NJ) based on p-distances in the software MEGA [56]. The plastome of F. anhuiensis (accession number: MH593363) was used as the outgroup in these tree-building analyses. Species were regarded as being successfully discriminated if all the individuals of a given species formed a monophyletic group [57].
Results
Plastid genome organization of Fritillaria
The 32 complete plastid genomes, consisting of a circular double-stranded DNA, ranged from 151,518 bp in F. taipaiensis (accession number: MH593356) to 152,073 bp in F. pallidiflora (accession number: MH593366). The genomes possessed a typical quadripartite structure, which comprised a pair of IRs (26,090–26,390 bp), LSC (81,295–82,085 bp), and SSC (17,122–17,565 bp) regions (Table 1, S1 Fig). Overall GC content of the complete plastid genomes was 36.9%-37.0% (Table 1). Moreover, a total of 115 genes were found, namely 78 protein coding genes, 30 tRNA genes, and 4 rRNA genes, as well as 3 pseudogenes (Table 2, S1 Fig). The protein coding genes present in the plastid genome of Fritillaria included 9 genes for large ribosomal proteins, 12 genes for small ribosomal proteins, 5 genes for photosystem I, 15 genes for photosystem II, and 6 genes for ATP synthase (Table 2, S1 Fig).
Table 2. Genes included in Fritillaria plastid genomes.
| Category for gene | Group of genes | Name of genes |
|---|---|---|
| Self-replication | Large subunit of ribosome | rpl2a*, rpl14, rpl16*, rpl20, rpl22, rpl23a, rpl32, rpl33, rpl36 |
| Small subunit of ribosome | rps2, rps3, rps4, rps7a, rps8, rps11, rps12a*, rps14, rps15, rps16*, rps18, rps19 | |
| DNA dependent RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 | |
| rRNA gene | rrn4.5a, rrn5a, rrn16a, rrn23a | |
| tRNA gene | trnK-UUU*, trnI-GAUI*, trnA-UGCa*, trnG-GCC*, trnV-UAC*, trnL-UAA*, trnS-UGA, trnS-GCU, trnS-GGA, trnY-GUA, trnC-GCA, trnL-CAAa, trnL-UAG, trnH-GUGa, trnD-GUC, trnfM-CAU, trnW-CCA, trnP-UGG, trnI-CAUa, trnR-ACGa, trnI-CAUa, trnE-UUC, trnT-UGU, trnF-GAA, trnQ-UUG, trnR-UCU, trnT-GGU, trnM-CAU, trnV-GACa, trnN-GUUa, trnN-GUUa, trnV-GACa, trnG-UCC | |
| Gene for photosynthesis | Subunits of photosystem Ⅰ | psaA, psaB, psaC, psaI, psaJ |
| Subunits of photosystem Ⅱ | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of NADH-dehydrogenase | ndhA*, ndhBI*, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Subunits of cytochrome b/f complex | petA, petB*, petD*, petG, petL, petN | |
| Subunit for ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | |
| Large subunit of rubisco | rbcL | |
| Other genes | Translational initiation factor | infA |
| Maturase | matK | |
| Protease | clpP* | |
| Envelope membrane protein | cemA | |
| Subunit of Acetyl-carboxylase | accD | |
| C-type cytochrome synthesis gene | ccsA | |
| Open reading frames (ORF,ycf) | ycf1, ycf2a, ycf3*, ycf4, ycf15a, ycf68a |
The lowercase letter a in superscript after gene names indicates genes located in IR regions. Asterisks indicate intron-containing genes.
Highly variable regions in plastid genome
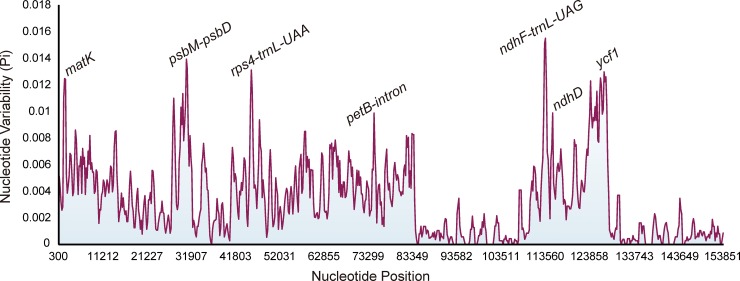
Seven highly variable regions from the plastid genomes, namely three intergenic regions (psbM-psdD, rps4-trnL-UAA, and ndhF-trnL-UAG), three gene regions (matK, ndhD, and ycf1), and one intron region (petB-intron), were selected as potential specific barcodes for use in species identification in Fritillaria (Fig 2). Among these regions, ycf1 was the longest, followed by psbM-psbD, while petB-intron region was the shortest (Table 3). Furthermore, all highly divergent fragments were found in the LSC and SSC regions, whereas none were present in the IR regions.
Fig 2. Sliding window analysis of 32 Fritillaria plastid genomes (window length: 600 bp, step size: 200 bp).
X-axis: position of the midpoint of a window; Y-axis: nucleotide diversity of each window.
Table 3. Analysis of the variability in different fragments and combination of fragments.
| No. sites | No. variable sites | No. parsimony information sites | No. mutations | No. InDels | Intraspecific distance | Interspecific distance | Nucleotide diversity (Pi) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Range | Mean (± SE) | Range | Mean (± SE) | |||||||
| Genome | 154,404 | 2,667 | 2,436 | 2,449 | 4,273 | 0–0.0021 | 0.0005±0.0001 | 0.0007–0.0078 | 0.0037±0.0001 | 0.00337 |
| HKLI | 3,890 | 180 | 132 | 170 | 44 | 0–0.0052 | 0.0012±0.0002 | 0.0008–0.0191 | 0.0082±0.0006 | 0.00784 |
| HKL | 3,255 | 76 | 74 | 71 | 19 | 0–0.0034 | 0.0006±0.0002 | 0.0003–0.0117 | 0.0051±0.0001 | 0.00466 |
| ITS2 | 239 | 52 | 22 | 55 | 9 | 0–0.0807 | 0.0094±0.0027 | 0–0.1211 | 0.0326±0.0013 | 0.03304 |
| ITS | 635 | 104 | 58 | 99 | 25 | 0–0.0338 | 0.0044±0.0011 | 0–0.0709 | 0.0235±0.0008 | 0.02560 |
| trnH-psbA | 253 | 17 | 17 | 16 | 19 | 0–0.0043 | 0.0010±0.0003 | 0–0.0598 | 0.0147±0.0009 | 0.01288 |
| matK | 1539 | 37 | 37 | 37 | 0 | 0–0.0039 | 0.0007±0.0002 | 0.0006–0.0123 | 0.0059±0.0001 | 0.00537 |
| rbcL | 1464 | 21 | 19 | 21 | 0 | 0–0.0027 | 0.0005±0.0001 | 0–0.0075 | 0.0033±0.0001 | 0.00295 |
| ndhF-trnL-UAG | 1,381 | 60 | 57 | 52 | 77 | 0–0.0052 | 0.0008±0.0002 | 0–0.0321 | 0.0112±0.0011 | 0.00994 |
| psbM-psdD | 3608 | 163 | 150 | 130 | 582 | 0–0.0058 | 0.0014±0.0003 | 0.0020–0.0202 | 0.0101±0.0002 | 0.00907 |
| rps4-trnL-UAA | 1,323 | 56 | 53 | 49 | 138 | 0–0.0061 | 0.0016±0.0003 | 0–0.0238 | 0.0098±0.0003 | 0.00892 |
| ycf1 | 5,565 | 214 | 203 | 223 | 63 | 0–0.0044 | 0.0010±0.0002 | 0.0016–0.0240 | 0.0092±0.0003 | 0.00836 |
| petB-intron | 855 | 30 | 29 | 28 | 17 | 0–0.0050 | 0.0012±0.0002 | 0–0.0200 | 0.0078±0.0002 | 0.00703 |
| ndhD | 1,509 | 39 | 36 | 41 | 6 | 0–0.0020 | 0.0006±0.0001 | 0–0.0160 | 0.0056±0.0002 | 0.00502 |
HKL and HKLI represent a combination of three fragments of the plastid genome and a combination of four fragments of universal barcodes, respectively (H: trnH-psbA; K: matK; L: rbcL; I: ITS)
Variability of the barcodes and their combinations
Among the regions and their combinations examined, the complete plastid genome clearly had the highest number of variable sites, parsimony-informative sites, and mutation sites (Table 3). Among the universal DNA barcodes, the ITS2 region was the most mutation-rich (Pi: 0.03304), followed by the ITS (0.02560), trnH-psbA (0.01288), and matK regions (0.00537), whereas the rbcL region (0.00295) was highly conserved (Table 3). Furthermore, among the highly variable regions, ndhF-trnL-UAG showed the highest variability (0.00994), and it was followed by psbM-psdD (0.00907), rps4-trnL-UAA (0.00892), ycf1 (0.00836), petB-intron (0.00703), and ndhD (0.00502).
DNA barcoding gap assessment
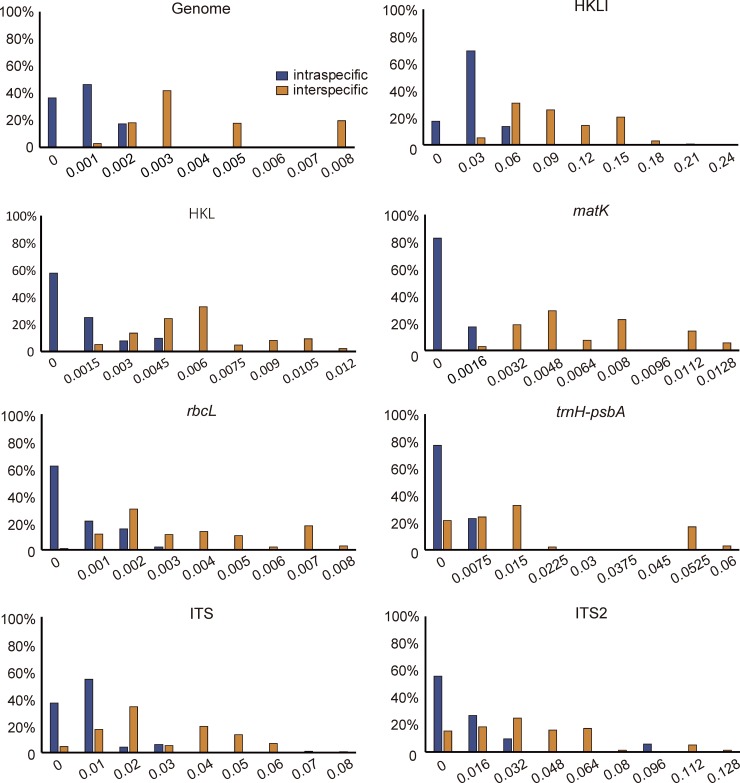
The inter- and intraspecific distances were calculated for each of the 14 datasets (Table 3). In these datasets, the ITS2 region exhibited the highest inter- and intraspecific distances (0.0326 and 0.0094, respectively), followed by the ITS (0.0235 and 0.0044), trnH-psbA (0.0147 and 0.0010) and ndhF-trnL-UAG (0.0112 and 0.0008), whereas these distances were the lowest for the rbcL (0.0033 and 0.0005). Meanwhile, the inter- and intraspecific distances of the complete plastid genome showed relatively low values (0.0037 and 0.0005, respectively) compared with those calculated for other datasets. Furthermore, the barcoding gap between inter- and intraspecific distances based on the p-distance model revealed that matK had the highest interspecific gap (divergence), but overlap between inter- and intraspecific distances existed for almost all single regions and their combinations, except for matK (Fig 3, S2 Fig).
Fig 3. Histograms of the frequencies (y-axes) of pairwise inter- and intraspecific divergences calculated based on the p-distances (x-axes) of each single regions and their combinations.
(H: trnH-psbA; K: matK; L: rbcL; I: ITS).
Discriminatory powers of all regions and their combinations
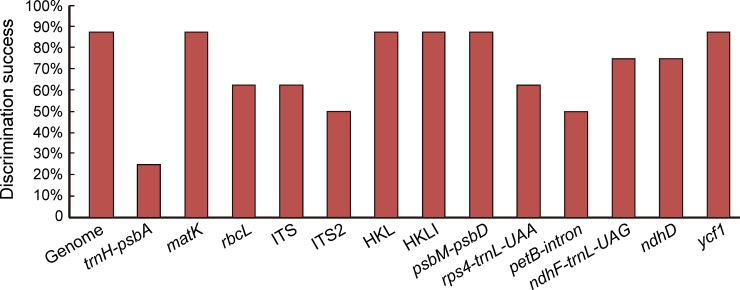
We calculated the species discrimination ability of each region and their combinations based on 14 datasets using tree-building methods (Fig 4). The super-barcode, comprised of complete plastid genomes, showed the highest power for species identification (87.5%), with strongest bootstrap values (Fig 5), except for F. cirrhosa, which is probably due to its polyphyletic nature. Among the universal DNA barcodes tested here, matK had the highest discriminatory power (87.5%), same as that of the super-barcode (Fig 4), followed by rbcL and ITS (both 62.5%). In contrast, trnH-psbA and ITS2, with relatively short DNA sequences (253 bp and 239 bp, respectively) and heavy overlaps between inter- and intraspecific distances, had relatively low success rates in distinguishing Fritillaria species (25% and 37.5%, respectively). Moreover, the combinations of the four barcodes (HKLI) or of the three plastid DNA regions (HKL) also showed the highest power for species discrimination (both 87.5%). Among the highly variable loci, psbM-psbD and ycf1 had the highest species discrimination rates (both 87.5%), followed by rps4-trnL-UAA (75%), ndhF-trnL-UAG ndhD, and petB-intron (both 50%) (Fig 4).
Fig 4. Species discrimination rate of all single fragments and their combinations based on the tree-building method.
(H: trnH-psbA; K: matK; L: rbcL; I: ITS).
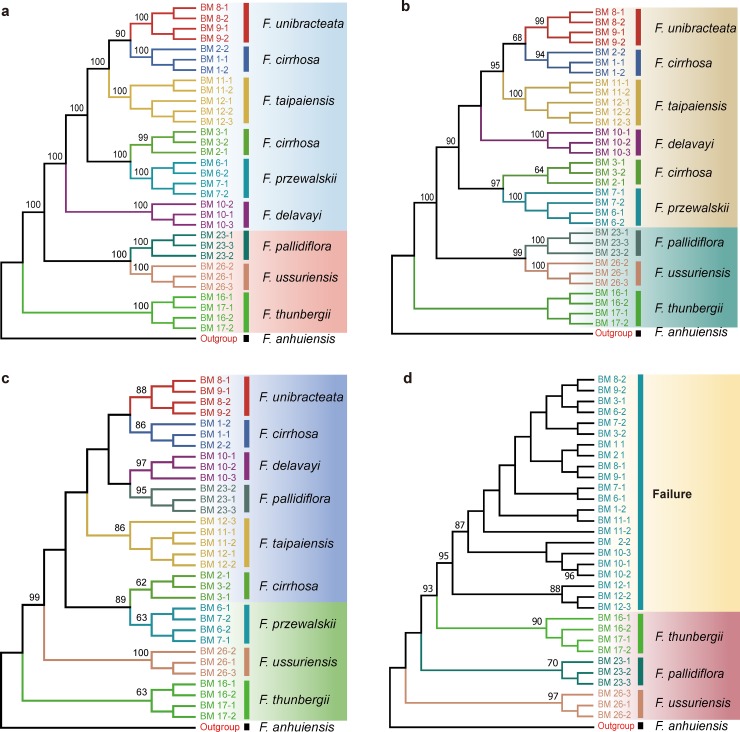
Fig 5.
Neighbor-joining (NJ) tree of 33 specimens of Fritillaria, comprising 32 individuals of Fritillariae cirrhosae bulbus and its adulterants based on p-distances calculated for each barcode region: a. whole plastid genome; b. ycf1; c. matK; d. ITS2.
According to the NJ trees, each of the DNA regions psbM-psbD, ycf1, and matK alone could be used to efficiently identify the original plants of Fritillariae cirrhosae bulbus and its adulterants. In contrast, the ITS, ndhD, ndhF-trnL-UAG, and rps4-trnL-UAA regions identified only the adulterant species from the genuine ones in the medicine. Moreover, the ITS2 could discriminate the adulterant from the genuine medicine, but could not effectively identify any of the original species of Fritillariae cirrhosae bulbus. Finally, the trnH-psbA, rbcL, and petB-intron regions could neither discriminate the adulterant medicine from the genuine nor their original species.
Definition of Fritillaria cirrhosa
In the NJ trees inferred from the whole plastid genomes, other DNA regions, or combinations, the six individuals of F. cirrhosa collected from three locations were clustered into two distinct clades: one was placed close to F. unibracteata and the other was close to F. przewalskii with strong support values (Fig 5).
Discussion
DNA barcoding has been demonstrated to be an efficient tool for identifying traditional medicines as well as their original species [23,58–60]. In the present study, this tool was used to discriminate among the five original species of Fritillariae cirrhosae bulbus and three of its adulterants.
Performance of universal DNA barcodes
In plants, the term universal DNA barcodes, which are identified based on Sanger sequencing, generally refers to rbcL and matK, to be supplemented with trnH-psbA and ITS/ITS2 [21,22]. Among the four universal DNA barcodes, matK had the highest species discriminatory power and successfully distinguished all the original species of Fritillariae cirrhosae bulbus, except F. cirrhosa. In contrast, ITS2 and trnH-psbA showed the lowest species resolution and failed to correctly identify the original species of Fritillariae cirrhosae bulbus, although these two barcodes were previously suggested as core barcode for species identification in traditional Chinese medicines [22,61]. ITS/ITS2 correctly discriminated the genuine medicine from the adulterants (Fig 5, S3 Fig). This result was nearly consistent with that of our previous study [24]. In fact, ITS/ITS2 were rich in variable sites and nucleotide diversity (Pi) compared with the studied plastid DNA regions (Table 3), but the obvious overlap between inter- and intraspecific distances in such regions among and within Fritillaria species might impact their discriminating ability (Fig 3).
It should be noted that the matK sequences used for analysis were obtained from the complete plastid genome, not by Sanger sequencing. If the obstacles to Sanger sequencing of matK are resolved, matK will become the ideal candidate barcode for identifying the original species of Fritillariae cirrhosae bulbus, despite its lower nucleotide diversity (Pi) when compared with that of other regions. The best performance of this barcode might be attributed to the existence of a clear barcoding gap (Table 3, Fig 3). The matK gene has been confirmed to perform poorly in discriminating species of Fritillaria and other genera such as Primula [62] and Garcinia [63] because of its low success rates in PCR amplification and Sanger sequencing [24]. However, next-generation sequencing (NGS) technology could resolve the aforementioned deficiency of the Sanger method, as it was revealed in the present study that this marker has satisfactory discriminatory power. Therefore, we propose that NGS technology should be adopted for DNA sequencing of matK to overcome the disadvantages of the Sanger sequencing method for such fragments.
Furthermore, combined barcodes generally perform better in species discrimination than single barcodes [23]. In the present study, combinations of the four barcodes (HKLI) or three plastid regions (HKL) also showed strong species discrimination abilities (87.5%) (Fig 5, S3 Fig). In short, multi-locus combination could raise discrimination ability for Fritillaria species and improve the reliability.
Super-barcode–a crucial candidate DNA barcode in Fritillaria
Because of the low power for species discrimination of universal DNA barcodes, new methods are necessary to discriminate closely related species [23,37]. Complete plastid genomes are extremely rich in genetic variations and have been shown to be powerful tools for resolving the phylogenetic relationships of complex groups [30,42,43,64,65]. Their use can greatly improve the resolution at lower taxonomic levels in plant phylogeny, phylogeography, and population genetics; therefore, they were also proposed as a type of super DNA barcode that is likely to resolve the defects of the universal DNA barcodes [23,66]. In this study, plastid genomes of Fritillaria species, with lengths from 151,518 to 152,073 bp, provided abundant informative sites for species identification. As a result, this super-barcode identified almost all of the original species of Fritillariae cirrhosae bulbus and its adulterants with high bootstrap values, except for F. cirrhosa because of its possible polyphyletic nature (Fig 5). Herein, the complete plastid sequences in the present study were obtained from dried leaf materials using NGS methods. Nevertheless, the use of this super-barcode might face extreme challenges if one needed to extract DNA from specific materials, such as kiln-dried specimens or medicines. However, recent procedures have been developed that can use total DNA as a template for genome skimming to assemble plastid genome, which not only solves the problem of extracting plastid DNA from dried or even degraded materials but also simplifies the whole process [23,67,68]. As sequencing technology and bioinformatics continue to improve rapidly, super DNA barcode will become more popular and may eventually replace Sanger-based DNA barcoding. Thus, super DNA barcodes could be adopted as useful complements to universal DNA barcodes, especially in identifying closely related species.
Specific DNA barcode–a trade-off between universal and super-barcode
The present study revealed that, when adopting the NGS method, the universal DNA barcodes were limited in their ability to discriminate species, except matK region, which showed high success rate for identifying species in Fritillaria (87.5%). Although the super-barcode exhibited high discriminatory power and sufficient reliability in this study, its use might be limited due to complications in data analyses and expensive sequencing costs. Therefore, it is of great importance to search for specific barcodes from highly variable regions that can be used as a trade-off between universal and super DNA barcodes.
Highly variable regions in the plastome could help to effectively resolve phylogenetic relationships and identify species within complicated groups [30,69]. In the present study, six highly variable regions were extracted from the complete plastid genomes. Among these regions, ycf1 had the same high species discriminating ability (87.5%) as the combination of four markers (HKLI); similarly, psdM-psbD showed the strikingly high discriminatory power (87.5%) (Fig 4, Fig 5). In fact, ycf1 had been successfully used to reconstruct the phylogenies of the family Orchidaceae [70] and the genus Astragalus [71], and it showed an extremely strong power to resolve interspecific relationships.
Therefore, we conclude that ycf1 and psdM-psbD can be used as potential specific barcodes for Fritillaria. However, the overly long DNA sequences of these regions result in some difficulties during PCR amplification and DNA Sanger sequencing [72]. Therefore, designing suitable primers to amplify shorter sections of those regions (about 1000 bp) with more variation might be another approach to be used in Sanger sequencing.
Delimitation of F. cirrhosa and its effect on species discrimination abilities of DNA barcodes
In the current study, the NJ trees generated using plastid genomes, universal barcodes, specific barcodes, and their combinations failed to resolve the samples of F. cirrhosa from different locations into one clade (Fig 5, S3 Fig). In the tree based on genome sequences, six individuals were divided into two different clades, one (BM1-1, BM1-2, and BM 2–2) was sister to F. unibracteata, but the other (BM2-1, BM 3–1, and BM 3–2) was placed close to F. przewalskii. These results indicated that F. cirrhosa is not a “good” species but contains multiple different lineages, which was also supported by the previous analysis of population genetics data based on amplified fragment length polymorphism (AFLP) markers (unpublished data). It is well known that F. cirrhosa has extremely complex variations in its morphology, especially its floral characteristics. According to the field survey, individuals from Lijiang (ZDQ15019) possess yellow-green tepals, but they are dark purple in individuals from Shangri-La (ZDQ13053). The complex morphology of this species unavoidably causes confusion in its taxonomy. Delimitation, as well as phylogeny, of F. cirrhosa and its closely related species is still controversial [8] and requires further study based on more samples and better markers. Thus, if the polyphyletic condition of this species is not considered, we could conclude that all of the examined fragments, including the whole plastid genomes, matK, ycf1, and psbM-psbD, as well as the combination HKLI, were able to discriminate all the original species of Fritillariae cirrhosae bulbus and its adulterants with a success rate of 100%.
Conclusion
In the present study, 32 individuals from eight species, representing five species of the original plants of Fritillariae cirrhosae bulbus and three of its adulterants, were employed to compare the species discriminatory powers of universal, super, and specific DNA barcodes. The results revealed that the whole plastid genome used as a super-barcode exhibited a powerful ability to identify Fritillaria species, with high reliability. Among the universal barcodes, only matK could discriminate almost all the original species when NGS methods are employed. It should be noted that ITS2 separated genuine Fritillariae cirrhosae bulbus from its adulterants, but it could not correctly identify the original species. Among the highly variable regions examined, ycf1 and psbM-psbD are considered the primary potential specific barcodes for Fritillaria species, but their successful sequencing using the Sanger method will depend on developing primers that will amplify the barcodes in sections. Moreover, NJ analysis based on complete plastid genomes, as well as other regions, revealed that F. cirrhosa was polyphyletic and the variations in its morphology requires further research at the population level. As the costs of NGS continue to decrease and data analysis methods are simplified, the use of super-barcodes might become the primary method for species discrimination in plants. Overall, the results in this study help to recognize species discrimination ability of super, universal, and specific barcodes in complex groups, and provide new knowledge to accurately identify the original plants of Fritillariae cirrhosae bulbus and its adulterants.
Supporting information
Genes shown on the outside of the circle are transcribed clockwise, and genes shown on the inside of the circle are transcribed counter-clockwise. Genes belonging to different functional groups are color-coded. The darker gray color in the inner circle corresponds to the GC content, and the lighter gray color corresponds to the AT content. (PDF)
(PDF)
(H: trnH-psbA; K: matK; L: rbcL; I: ITS) (PDF)
(PDF)
(H: trnH-psbA; K: matK; L: rbcL; I: ITS) (PDF)
(PDF)
(DOCX)
Acknowledgments
We thank Junbo Yang, Zhirong Zhang, Rong Zhang, Tingshuang Yi, and Shudong Zhang in Kunming Institute of Botany, Chinese Academy Science (CAS) for their help in the molecular experiment and data analysis.
Data Availability
All sequences files are available from the NCBI database (accession number(s) listed in Table 1).
Funding Statement
Funded by National Natural Science Foundation of China (31660081); Yunnan Provincial Science and Technology Department (Grant No. 2016FB144).
References
- 1.Tekşen M, Aytaç Z, Pınar NM. Pollen morphology of the genus Fritillaria L. (Liliaceae) in Turkey. Turk J Bot. 2010; 34(5):397–416. 10.3906/bot-0907-93 [DOI] [Google Scholar]
- 2.Day PD, Berger M, Hil L, Fay MF, Leitch AR, Leitch IJ, et al. Evolutionary relationships in the medicinally important genus Fritillaria L. (Liliaceae). Mol Phylogenet Evol. 2014; 80:11–9. 10.1016/j.ympev.2014.07.024 [DOI] [PubMed] [Google Scholar]
- 3.Rix EM. Fritillaria: a revised classification together with an updated list of species. Publication of the Fritillaria Group of the Alpine Garden Society, UK; 2001. [Google Scholar]
- 4.Chen SC, Helen VM. Fritillaria L In: Wu Z.Y & Raven PH (Eds.) Flora of China (24th). Beijing: Science Press; 2000. [Google Scholar]
- 5.Wang SJ, Gao WY, Yu L, Xiao PG. Progress of taxonomic study on Fritillaria (Liliaceae) medicinal plant. China J Chin Mater Med. 2007; 32(16):1609–14. [PubMed] [Google Scholar]
- 6.Xiao PG, Jiang Y, Li P, Luo YB, Liu Y. The botanical origin and pharmacophylogenetic treatment of Chinese materia medica Beimu. Acta Phytotaxon Sin. 2007; 45(4):473–87. 10.1360/aps06113 [DOI] [Google Scholar]
- 7.National Pharmacopoeia Committee. Pharmacopoeia of the People' s Republic of China, 2015ed, vol. 1. Beijing, China: Chemical Industry Press; 2015. [Google Scholar]
- 8.Luo YB, Chen SC. A revision of Fritillaria L. (Liliaceae) in the Hengduan mountains and adjacent regions, China (1)—A study of Fritillaria cirrhosa D. Don and its related spcies. Acta Phytotaxon Sin. 1996. a; 34(3):304–12. [Google Scholar]
- 9.Luo YB, Chen SC. A revision of Fritillaria L. (Liliaceae) in the Hengduan mountains and adjacent regions, China (2). Acta Phytotaxon Sin. 1996. b; 34(3):547–53. [Google Scholar]
- 10.Li J. Identification of character and chemical composition of Sichuan Fritillaria between true and counterfei. J Chengdu Univ TCM. 2009; 32(2):83–84. [Google Scholar]
- 11.Zhang YC, Wang YH. Fritillaria the authenticity of the identification. J Liaoning univ TCM. 2011; 13(11):220–1. [Google Scholar]
- 12.Hebert PD, Ratnasingham S, Dewaard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci. 2003; 270 (Suppl-1):S96–S9. 10.1098/rsbl.2003.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chase MW, Salamin N, Wilkinson M, Dunwell JM, Rao PK, Haidar N, et al. Land plants and DNA barcodes: short-term and long-term goals. Philos Trans R Soc Lond B Biol Sci. 2005; 360(1462):1889–95. 10.1098/rstb.2005.1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, et al. DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci U S A. 2008; 105(8):2923–8. 10.1073/pnas.0709936105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newmaster SG, Fazekas AJ, Steeves RA, Janovec J. Testing candidate plant barcode regions in the Myristicaceae. Mol Ecol Res. 2010; 8(3):480–90. 10.1111/j.1471-8286.2007.02002.x [DOI] [PubMed] [Google Scholar]
- 16.Hebert PD, Stoeckle MY, Zemlak TS, Francis CM. Identification of birds through DNA barcodes. Plos Biol. 2004;2(10):e312 10.1371/journal.pbio.0020312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hebert PD, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci U S A. 2004; 101(41):14812–7. 10.1073/pnas.0406166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollingsworth PM. Refining the DNA barcode for land plants. Proc Natl Acad Sci U S A. 2011; 108(49):19451–2. 10.1073/pnas.1116812108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purty RS, Chatterjee S. DNA barcoding: an effective technique in molecular taxonomy. Austin J Biotechnol Bioeng.2016; 3(1):1059. [Google Scholar]
- 20.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci U S A. 2005; 102(23):8369–74. 10.1073/pnas.0503123102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CBOL Plant Working Group. A DNA barcode for land plants. Proc Natl Acad Sci U S A.2009; 106(31):12794–12797. 10.1073/pnas.0905845106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.China Plant BOL Group, Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ,Chen ZD,Zhou SL,Chen SL. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci U S A.2011; 108(49):19641–6. 10.1073/pnas.1104551108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li XW, Yang Y, Henry RJ, Rossetto M, Wang YT, Chen SL. Plant DNA barcoding: from gene to genome. Biol Rev. 2015; 90(1):157–66. 10.1111/brv.12104 [DOI] [PubMed] [Google Scholar]
- 24.Zhang DQ, Mo XC, Xiang JY, Zhou N. Molecular identification of original plants of Fritillariae cirrhosae bulbus, a tradtional chinese medicine (TCM) using plant dna barcoding. Afr J Tradit Complem Altern Med. 2016; 13(6):74–82. 10.21010/ajtcam.v13i6.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu FH, Chan MT, Liao DC, Hsu CT, Lee YW, Daniell H, et al. Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol. 2010; 10(1):68 10.1186/1471-2229-10-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li PB, Li ZH, Liu HM, Hua JP. Cytoplasmic diversity of the cotton genus as revealed by chloroplast microsatellite markers. Genet Resour Crop Ev. 2014; 61(1):107–19. 10.1007/s10722-013-0018-9 [DOI] [Google Scholar]
- 27.Carbonell-caballero J, Alonso R, Ibañez V, Terol J, Talon M, Dopazo J. A phylogenetic analysis of 34 chloroplast genomes elucidates the relationships between wild and domestic species within the genus Citrus. Mol Phylogenet Evol. 2015; 32(8):2015–35. 10.1093/molbev/msv082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayly MJ, Rigault P, Spokevicius A, Ladiges PY, Ades PK, Anderson C, et al. Chloroplast genome analysis of Australian eucalypts–Eucalyptus, Corymbia, Angophora, Allosyncarpia and Stockwellia (Myrtaceae). Mol Phylogenet Evol. 2013; 69(3):704–16. 10.1016/j.ympev.2013.07.006 [DOI] [PubMed] [Google Scholar]
- 29.Henriquez CL, Arias T, Pires JC, Croat TB, Schaal BA. Phylogenomics of the plant family Araceae. Mol Phylogenet Evol. 2014; 75(1):91–102. 10.1016/j.ympev.2014.02.017 [DOI] [PubMed] [Google Scholar]
- 30.Du YP, Bi Y, Yang FP, Zhang MF, Chen XQ, Xue J, et al. Complete chloroplast genome sequences of Lilium: insights into evolutionary dynamics and phylogenetic analyses. Sci Rep. 2017; 7(1):5751 10.1038/s41598-017-06210-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugiura M. The chloroplast genome. Plant Mol Biol. 1992; 19:149–57. 10.1007/bf00015612 [DOI] [PubMed] [Google Scholar]
- 32.Yurina NP, Odintsova MS. Compsrative structural organization of plant chloroplast and mitochondrial genomes. Genetika. 1998; 34:5–22. [Google Scholar]
- 33.Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci U S A. 2007; 104(49):19363–8. 10.1073/pnas.0708072104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang MX, Cui LC, Feng KW, Deng PC, Du XH, Wan FH, et al. Comparative analysis of Asteraceae chloroplast genomes: structural organization, RNA editing and evolution. Plant Mol Biol Rep.2015; 33(5):1526–38. 10.1007/s11105-015-0853-2 [DOI] [Google Scholar]
- 35.Maliga P. Engineering the plastid genome of higher plants. Curr Opin plant Biol. 2002; 5(2):164–72. 10.1016/s1369-5266(02)00248-0 [DOI] [PubMed] [Google Scholar]
- 36.Tangphatsornruang S, Sangsrakru D, Chanprasert J, Uthaipaisanwong P, Yoocha T, Jomchai N, et al. The chloroplast genome sequence of Mungbean (Vigna radiata) determined by High-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res. 2009; 17(1):11–22. 10.1093/dnares/dsp025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci U S A. 2010; 107(10):4623–8. 10.1073/pnas.0907801107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krawczyk K, Nobis M, Myszczyński K, Klichowska E, Sawicki J. Plastid super-barcodes as a tool for species discrimination in feather grasses (Poaceae: Stipa). Sci Rep. 2018; 8(1):1924 10.1038/s41598-018-20399-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen XL, Zhou JG, Cui YX, Wang Y, Duan BZ, Yao H. Identification of Ligularia herbs using the complete chloroplast genome as a super-barcode. Front Pharmacol. 2018;9:695 10.3389/fphar.2018.00695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma SJ, Zhou JG, Li Y, Chen XL, Wu ML, Sun W, et al. Complete chloroplast genomes of Dioscorea opposite and D. collettii and screening specific DNA barcodes (in Chinese). Sci Sin Vitae. 2018; 48:571–82. 10.1360/N052017-00160 [DOI] [Google Scholar]
- 41.Park I, Kim WJ, Yeo SM, Choi G, Kang YM, Piao RZ, et al. The complete chloroplast genome sequences of Fritillaria ussuriensis Maxim. and Fritillaria cirrhosa D. Don, and comparative analysis with other Fritillaria species. Molecules. 2017; 22(6):982 10.3390/molecules22060982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bi Y, Zhang MF, Xue J, Dong R, Du YP, Zhang XH. Chloroplast genomic resources for phylogeny and DNA barcoding: a case study on Fritillaria. Sci Rep. 2018; 8(1):1184 10.1038/s41598-018-19591-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Zhang ZR, Yang JB, Lv GH. Complete chloroplast genome of seven Fritillaria species, variable DNA markers identification and phylogenetic relationships within the genus. Plos One. 2018; 13(3):e0194613 10.1371/journal.pone.0194613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doyle J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987; 19(1):11–5. [Google Scholar]
- 45.Yang JB, Li DZ, Li HT. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Res. 2014; 14(5):1024–31. 10.1111/1755-0998.12251 [DOI] [PubMed] [Google Scholar]
- 46.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30(15):2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 2018:1–11. 10.1101/256479 [DOI] [Google Scholar]
- 48.Li QS, Li Y, Song JY, Xu HB, Xu J, Zhu YJ, et al. High-accuracy de novo assembly and SNP detection of chloroplast genomes using a SMRT circular consensus sequencing strategy. New Phytol. 2014; 204(4):1041–9. 10.1111/nph.12966 [DOI] [PubMed] [Google Scholar]
- 49.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012; 19(5):455–77. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015; 31(20):3350–2. 10.1093/bioinformatics/btv383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh K, Standley D. MAFFT multiple sequence alignment software version improvements in performance and usability. Mol Biol Evol. 2013; 30(4):772–80. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kearse M, Moir R, Wilson A, Stoneshavas S, Cheung M, Sturrock S, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012; 28(12):1647–9. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lohse M, Drechsel O, Kahlau S, Bock R. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013; 41: W575–W81. 10.1093/nar/gkt289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 1999; 41(41):95–8 [Google Scholar]
- 55.Rozas J, Ferrermata A, Sánchezdelbarrio JC, Guiraorico S, Librado P, Ramosonsins SE, et al. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol Biol Evol. 2017; 34(12):3299–302. 10.1093/molbev/msx248 [DOI] [PubMed] [Google Scholar]
- 56.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; 33(7):1870–4. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hollingsworth ML, Andra CA, Forrest LL, Richardson J, Pennington RT, Long DG, et al. Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol Ecol Resour. 2010; 9(2):439–57. 10.1111/j.1755-0998.2008.02439.x [DOI] [PubMed] [Google Scholar]
- 58.Zhang DQ, Yang YS, Jiang B, Duan LZ, Zhou N. How to correctly identify herbal materials in market: A case study based on DNA barcodes. Afr J Tradit Complem.2014; 11(6):66–76. 10.4314/ajtcam.v11i6.7 [DOI] [Google Scholar]
- 59.Mei QX, Chen XL, Xiang L, Liu Y, Su Yy, Gao YQ, et al. DNA barcode for identifying Folium Artemisiae Argyi from counterfeits. Biol Pharm Bull. 2016; 39(9):1531 10.1248/bpb.b16-00336 [DOI] [PubMed] [Google Scholar]
- 60.Xiong C, Sun W, Li JJ, Yao H, Shi YH, Wang P, et al. Identifying the species of seeds in Traditional Chinese Medicine using DNA barcoding. Front Pharmacol. 2018; 9:701 10.3389/fphar.2018.00701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen SL, Yao H, Han JP, Liu C, Song JY, Shi LC, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One. 2010; 5(1):e8613 10.1371/journal.pone.0008613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan HF, Hao G, Hu CM, Ge XJ. DNA barcoding in closely related species: A case study of Primula L. sect. Proliferae Pax (Primulaceae) in China. J Diabetes Invest. 2011; 49(3):225–36. 10.1111/j.2040-1124.2010.00046.x PMID: 24843434 [DOI] [Google Scholar]
- 63.Costion C, Ford A, Cross H, Crayn D, Harrington M, Lowe A. Plant DNA barcodes can accurately estimate species richness in poorly known floras. PLoS One. 2011; 6(11):e26841 10.1371/journal.pone.0026841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu RS, Li P, Qiu YX. The complete chloroplast genomes of three Cardiocrinum (Liliaceae) species: comparative genomic and phylogenetic analyses. Front Plant Sci. 2016; 7:2054 10.3389/fpls.2016.02054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim JH, Lee SI, Kim BR, Choi IY, Ryser P, Kim NS. Chloroplast genomes of Lilium lancifolium, L. amabile, L. callosum, and L. philadelphicum: Molecular characterization and their use in phylogenetic analysis in the genus Lilium and other allied genera in the order Liliales. PLoS One. 2017; 12(10):e0186788 10.1371/journal.pone.0186788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fu CN, Wu CS, Ye LJ, Mo ZQ, Liu J, Chang YW, et al. Prevalence of isomeric plastomes and effectiveness of plastome super-barcodes in yews (Taxus) worldwide. Sci Rep. 2019; 9:2773 10.1038/s41598-019-39161-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nock CJ, Waters DL, Edwards MA, Bowen SG, Rice N, Cordeiro GM, et al. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol J. 2011;9(3):328 10.1111/j.1467-7652.2010.00558.x [DOI] [PubMed] [Google Scholar]
- 68.Zeng CX, Hollingsworth PM, Yang J, He ZS, Zhang ZR, Li DZ, et al. Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods. 2018; 14:43 10.1186/s13007-018-0300-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dong WP, Liu J, Yu J, Wang L, Zhou SL. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One. 2012; 7(4):e35071 10.1371/journal.pone.0035071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neubig KM, Whitten WM, Carlsward BS, Blanco MA, Endara L, Williams NH, et al. Phylogenetic utility of ycf1 in orchids: a plastid gene more variable than matK. Plant Syst Evol. 2009; 277(1–2):75–84. 10.1007/s00606-008-0105-0 [DOI] [Google Scholar]
- 71.Dastpak A, Osaloo SK, Maassoumi AA, Safar KN. Molecular phylogeny of Astragalus sect. Ammodendron (Fabaceae) inferred from chloroplast ycf1 gene. Ann Bot Fenn. 2018; 55(1–3):75–82. 10.5735/085.055.0108 [DOI] [Google Scholar]
- 72.Dong WP, Xu C, Li CH, Sun JH, Zuo YJ, Shi S, et al. ycf1, the most promising plastid DNA barcode of land plants. Sci Rep. 2015; 5:8348 10.1038/srep08348 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes shown on the outside of the circle are transcribed clockwise, and genes shown on the inside of the circle are transcribed counter-clockwise. Genes belonging to different functional groups are color-coded. The darker gray color in the inner circle corresponds to the GC content, and the lighter gray color corresponds to the AT content. (PDF)
(PDF)
(H: trnH-psbA; K: matK; L: rbcL; I: ITS) (PDF)
(PDF)
(H: trnH-psbA; K: matK; L: rbcL; I: ITS) (PDF)
(PDF)
(DOCX)
Data Availability Statement
All sequences files are available from the NCBI database (accession number(s) listed in Table 1).