Abstract
Background/aim
Gastric cancer (GC) is one of the major causes of cancer mortality worldwide. As a novel type of endogenous noncoding RNAs, circular RNAs (circRNAs) are formed by a covalent link between 5’ and 3’ ends. They are very stable and abundant in eukaryotes. As there were no reported studies on the expression profiles of circular RNA ITCH (cir-ITCH) and circHIPK3in GC, in the current study, we aimed to delineate the expression profiles and clinicopathological relevance of these two circRNAs in GC tissues compared to their paired adjacent noncancerous tissues.
Materials and methods
Quantitative real-time polymerase chain reaction was performed to evaluate cir_ITCH and circHIPK3 expression in 30 paired gastric cancer tissues. The clinicopathological relevance of these two circular RNAs’ expression levels with gastric cancer was further examined.
Results
Our results showed that the expression of cir_ITCH and circHIPK3 were significantly downregulated in GC tumoral tissues compared with their paired adjacent nonneoplastic counterparts. Further analyses showed that cir_ITCH and circHIPK3 expression levels were related with numerous clinicopathological featuresof tumoral tissues.
Conclusion
Cir_ITCH and circHIPK3 may have imperative roles in GC and serve in the future as potential prognostic biomarkers in GC.
Keywords: Gastric cancer, circular RNAs, cir_ITCH, circHIPK3, gene expression
1. Introduction
Gastric cancer (GC) is one of the unresolved causes of cancer mortality worldwide with a high rate of incidence and death (1,2). It is the second and fourth most common cancer leading to a high death rate in men and women, respectively (3,4). Lack of reliable diagnostic methods in early stages of GC denies the majority of patients an effective treatment (3,5). Accordingly, to overcome these problems, it is critical to find novel biomarkers to improve the chance to diagnose GC in its early stages.
As a novel type of endogenous noncoding RNAs, circular RNAs (circRNAs) are formed by a covalent link between 5’ and 3’ ends (6–8). They are very stable and abundant in eukaryotes (9–11). Recent reports show that circRNAs can function as microRNA sponges, regulate gene expression, linear RNA transcription and protein production (7). Association between circRNAs and several diseases like atherosclerotic vascular disease (12), Alzheimer (13), and various cancer types (14) has been documented and could represent these molecules as attractive novel biomarkers.
cir-ITCH is a circular RNA which is derived from Itchy E3 ubiquitin protein ligase (ITCH) gene (10,15). Deregulation of cir-ITCH has been recently reported in esophageal squamous cell carcinoma (ESCC) (16), colorectal cancer (17), and lung cancer. Further functional studies showed that cir-ITCH can act as microRNA sponge thus increasing the level of parental gene, ITCH (15).
In 2016, by characterizing circRNA, transcripts using RNA-sequencing (RNA-seq) from six normal tissues (brain, colon, heart, liver, lung, and stomach) and seven cancerous tissues including gastric cancer, Zheng et al. introduced an abundant circRNA derived from Exon2 of the HIPK3 gene, termed circHIPK3. Their functional assays revealed that circHIPK3 may function to modulate the growth of human cells (18).
As there were no reported studies on the expression profiles of these two circular RNAs in gastric cancer, in the current study, we aimed to delineate the expression profiles of cir-ITCH and circHIPK3 in gastric cancer tissues compared to their paired adjacent noncancerous tissues. Then, the clinicopathological relevance of these two circular RNAs with gastric cancer was further examined.
2. Materials and methods
2.1. Clinical specimens
A total of 30 pairs of gastric cancer and matched adjacent nontumoral tissues were obtained from patients with gastric cancer. The specimens were collected by the Iran National Tumor Bank, which is funded by the Cancer Institute of Tehran University, for Cancer Research (19–21). There, the tissues are immediately snap-frozen in the liquid nitrogen. Informed written consent was taken from the patients by Iran National Tumor Bank. The study protocol was approved by the Ethics Committee of Isfahan University of Medical Sciences and was in accordance with the Helsinki Declaration. The clinical staging of the tumor samples was based on the seventh edition of the American Joint Committee on Cancer classification (AJCC) cancer staging manual for stomach (22).
2.2. Total RNA extraction and complementary DNA (cDNA) synthesis
TRIzol® reagent (Invitrogen, California, USA) was used to extract total RNA from powdered gastric cancer tissues, following the manufacturer’s instructions. One percent agarose gel electrophoresis was used to assess the quality of the RNA. Purity and quantity of the total RNA were determined with Nanodrop instrument (Nanolytik, Düsseldorf, Germany). DNase treatment was performed by using DNase set (Fermentas, Vilnius, Lithuania) for eliminating genomic DNA. cDNA was synthesized by using PrimeScriptTM RT reagent Kit (TaKaRa, Kusatsu, Shiga, Japan) according to manufacturer’s protocol.
2.3. Quantitative real-time PCR and DNA sequencing
Applying the relative quantitative real-time RT-PCR, the expression levels of cir_ITCH and circHIPK3 were assessed compared to GUSB (β-Glucuronidase) as an internal control (23). Divergent primers, rather than convergent primers, were designed with GeneRunner software, version 4.0 to amplify the circular RNAs. Basic local alignment search tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for confirmation of unity attachment of divergent primers to genome. A set of convergent primers in an opposite direction were designed to amplify only the linear forms. The sequences of primers are listed in Table 1. RealQ Plus 2x Master Mix, green (high ROX) (AMPLIQON, Odense M, Denmark) was applied in an Applied Biosystems StepOnePlus™ instrument for PCR amplification. The amplification conditions consisted of an initial denaturation at 95 °C for 2 min, then 30 cycles of denaturation at 95 °C for 30 s, annealing at 55.9 °C for cir_ITCH and circHIPK3 and at 60 °C for GUSB genes for 60 s, and the extension step for 30 s at 72 °C. Furthermore, the PCR products of some samples were sequenced with an Applied Biosystems 3730XL sequencer (Macrogen, Seoul, South Korea) to verify the specific amplification of circular RNAs.
Table 1.
The sequences of primers used in the current study.
| Characteristics | Numbers(#30)(%) | circHIPK3(mean ± SEM*) | P-value |
| Gender Male Female | 17(56.67)13(44.33) | 0.29 ± 0.660.37 ± 0.46 | 0.25 |
| Age (years) ≥70 <70 | 15(50.00)15(50.00) | 1.93 ± 0.44−1.28 ± 0.55 | 0.002** |
| Depth of invasion T2 T3-T4 | 1(3.33.00)29(96.67) | −3.21 ± 0.45 ± 0.57 | 0.14 |
| N classification NX-N0 N1 N2-N3 | 6(20.00)11(36.67)13(43.33) | −1.01 ± 0.811.52 ± 0.44−0.07 ± 0.53 | 0.16 |
| M classification MX M0 M1 | 5(16.67)19(63.33)6(20.00) | −0.47 ± 0.231.31 ± 0.54−1.89 ± 0.66 | 0.045**(M0 vs. M1) |
| TNM stage I-II III IV | 16(53.33)8(26.67)6(20.00) | 0.86 ± 0.560.92 ± 0.44−1.89 ± 0.66 | 0.16 |
| Perineural invasion Negative Positive | 12(40.00)18(60.00) | 0.08 ± 0.660.57 ± 0.51 | 0.28 |
| Lymphatic invasion Negative Positive | 6(20.00)24(80.00) | 0.91 ± 0.530.24 ± 0.46 | 0.20 |
| Tumor size (cm) ≥5 <5 | 25(83.33)5(16.67) | 0.23 ± 0.570.79 ± 0.64 | 0.40 |
| Tumor grades I II III | 9(30.00)8(26.67)13(43.33) | −0.84 ± 0.670.98 ± 0.610.73 ± 0.48 | 0.25 |
| Tumor types Diffuse Intestinal | 14(46.67)16(53.33) | 0.78 ± 0.49−0.07 ± 0.64 | 0.38 |
2.4. Statistical gene expression analysis
The ΔCt method was applied to relatively quantify the levels of gene expressions. All experiments were performed at least three times and expressed as means ± standard error of mean (SEM). To check the normal distribution of samples, Kolmogorov–Smirnov test was applied. Student’s t-test, analysis of variance (ANOVA), and chi-square tests were performed to examine statistical significances. Data were analyzed by SPSS software, version 16.0 (SPSS, Chicago, IL, USA) and P-values less than 0.05 were considered statistically significant.
3. Results
3.1. PCR optimization of the cir_ITCH and circHIPK3
We designed two sets of primer pairs: a divergent primer set for only amplification of the circular form and a convergent one to amplify the linear form. We utilized the cDNA and DNA of a human lung adenocarcinoma epithelial cell line (A540) for PCR optimization. As expected, no amplification was seen with divergent primers on DNA template, whereas we could detect the expected bands corresponding to circular forms of RNAs (Figure 1). Products giving a band were sequenced by conventional Sanger capillary methods and compared to the reference sequence. Sanger sequencing of the RT-PCR products of cir_ITCH and circHIPK3 showed that convergent primers could specifically amplify the circular forms of RNAs (data not shown).
Figure 1.
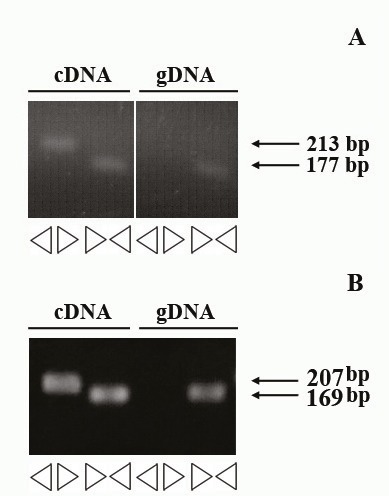
Identification of circHIPK3 and cir_ITCH in a human lung adenocarcinoma epithelial cell line (A540). A) RT-PCR products with divergent and convergent primers of circHIPK3 verified that specificity of designed primers. B) Specific divergent primers of cir_ITCH could amplify a 207 bp PCR product using a cDNA as a PCR template but not a genomic DNA (gDNA).
3.2. cir_ITCH and circHIPK3 expression levels were significantly downregulated in GC tumoral tissues and were significantly correlated with various clinicopathological parameters
The results of real-time qRT-PCR experiments showed that expression of both cir_ITCH and circHIPK3 were significantly downregulated (P = 0.015 and P= 0.046, respectively) in GC tumoral tissues compared with their paired adjacent nontumoral tissues (Figure 2). Analyzing melting curves showed single peaks with neither primer dimers nor nonspecific amplification, further confirming the specificity of cir_ITCH and circHIPK3 amplifications.
Figure 2.
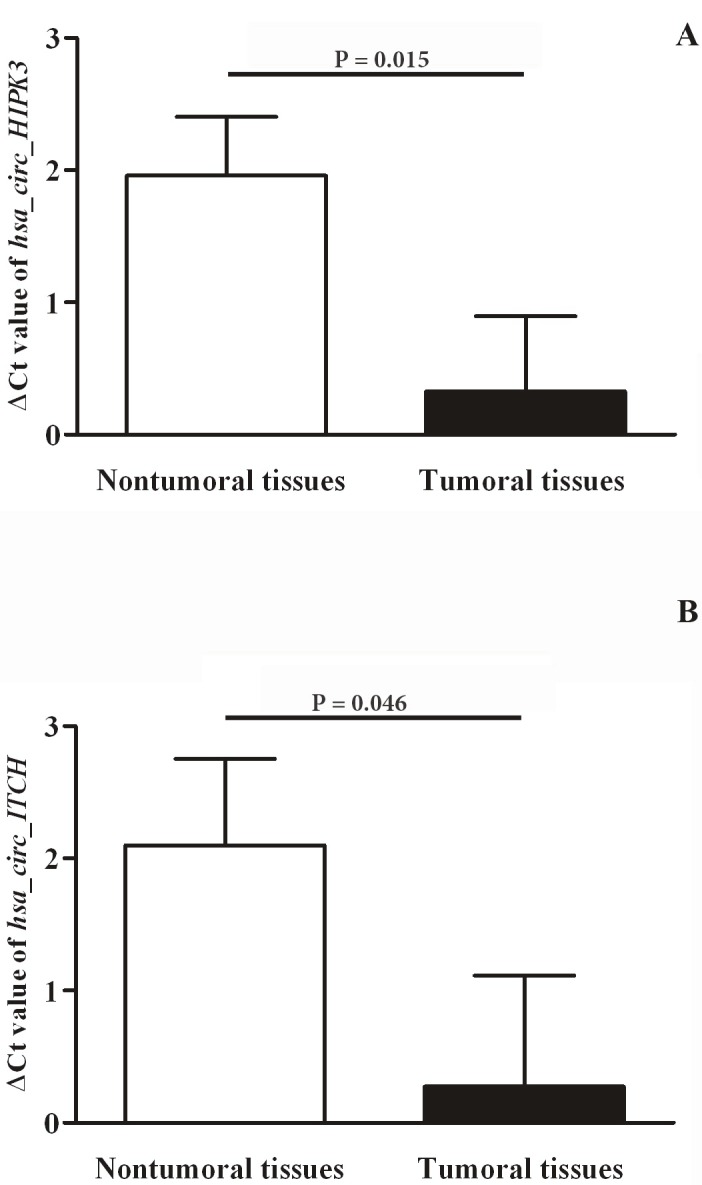
Relative expression of circHIPK3 and cir_ITCH in tumoral and nontumoral gastric tissue samples. A higher ΔCt value shows higher relative expression levels. Error bars stand for standard error of mean (SEM). A) circHIPK3, B) cir_ITCH.
Further analyses showed that circHIPK3 expression was related with numerous clinicopathological featuresof tumoral tissues. As shown in Table 2, circHIPK3 expression level was significantly associated with age (P = 0.002) and M classification (P = 0.045). GC patients were further classified into two groups based on the median value of circHIPK3 expression in tumoral tissues. circHIPK3 expression levels stratified based on median value showed a significant association with age (P = 0.01) (Table 3). Same analyses on cir_ITCH showed a significant correlation between circRNA expression levels and age (P = 0.0005) and tumor grades (P = 0.02) (Tables 4 and 5). We furthermore tested if there is any correlation between gene expression levels and the anatomical location of tumors within the stomach. Although the stomach is anatomically divided into four regions, i.e. the cardia, fundus, body, and pylorus, the two main regions in our patient cohort were the cardia and the main body. Therefore, we compared the gene expressions between these two groups (body vs. cardia) and found no statistically significant difference in gene expressions of the two examined circular RNAs between tumors in the main body of the stomach and the ones in cardia (1.06 ± 1.03 vs. −1.54 ± 1.67 for cir_ITCH (P= 0.20) and 1.14 ± 0.62 vs. −0.57 ± 1.18 for circHIPK3 (P= 0.22)).
Table 2.
The relationship between circHIPK3 expression level (based on mean ± SEM gene expression level) in GC tissues with clinicopathological parameters.
| Characteristic | Number (#30) | circHIPK3 expression | P-value | |
| Low (#15) | High (#15) | |||
| Gender Male Female | 1713 | 96 | 87 | 0.50 |
| Age (years) ≥70 <70 | 1515 | 411 | 114 | 0.01** |
| Depth of invasion T2 T3-T4 | 129 | 114 | -15 | 0.50 |
| N classification NX-N0 N1 N2-N3 | 61113 | 348 | 375 | 0.23 |
| M classification MX M0 M1 | 5196 | 474 | 1122 | 0.09(M0 vs. M1) |
| TNM stage I-II III IV | 1686 | 744 | 942 | 0.31 |
| Perineural invasion Negative Positive | 1218 | 69 | 69 | 0.50 |
| Lymphatic invasion Negative Positive | 624 | 213 | 411 | 0.32 |
| Tumor size (cm) ≥5 <5 | 255 | 123 | 132 | 0.50 |
| Tumor grades I II III | 9813 | 645 | 348 | 0.21 |
| Tumor types Diffuse Intestinal | 1416 | 69 | 87 | 0.35 |
* A higher ΔCt value indicates higher expression. ** Statistically significant.
Table 3.
The relationship between circHIPK3 expression level (as divided into two groups based on the median of ΔCt) in GC tissues with clinicopathological parameters
| Characteristic | Number (#30)(%) | cir_ITCH expression | P-value | |
| Low (#15) | High (#15) | |||
| Gender Male Female | 1614 | 78 | 96 | 0.36 |
| Age (years) ≥70 <70 | 1416 | 213 | 123 | 0.0005** |
| Depth of invasion T2 T3-T4 | 129 | 114 | 015 | 0.50 |
| N classification NX-N0 N1 N2-N3 | 61113 | 555 | 168 | 0.08 |
| M classification MX M0 M1 | 5196 | 573 | 0123 | 0.28(M0 vs. M1) |
| TNM stage I-II III IV | 1686 | 744 | 942 | 0.31 |
| Perineural invasion Negative Positive | 1218 | 510 | 78 | 0.35 |
| Lymphatic invasion Negative Positive | 624 | 213 | 411 | 0.34 |
| Tumor size (cm) ≥5 <5 | 255 | 123 | 132 | 0.50 |
| Tumor grades I II III | 9813 | 744 | 249 | 0.04** |
| Tumor types Diffuse Intestinal | 1416 | 510 | 96 | 0.13 |
* A higher ΔCt value indicates higher expression. ** Statistically significant.
Table 4.
The relationship between cir_ITCH expression level (based on mean ± SEM gene expression level) in GC tissues with clinicopathological parameters
| Characteristics | Numbers (#30)(%) | cir_ITCH (mean±SEM*) | P-value |
| Gender Male Female | 16(53.33)14(46.66) | 0.31 ± 0.930.23 ± 0.75 | 0.42 |
| Age (years) ≥70 <70 | 14(46.66)16(53.33) | 2.66 ± 0.76−2.11 ± 0.69 | 0.0005** |
| Depth of invasion T2 T3-T4 | 1(3.33)29(96.67) | 0.45 ± 0.27 ± 0.85 | 0.50 |
| N classification NX-N0 N1 N2-N3 | 6(20.00)11(36.67)13(43.33) | −1.18 ± 1.250.89 ± 0.870.42 ± 0.61 | 0.45 |
| M classification MX M0 M1 | 5(16.67)19(63.33)6(20.00) | −2.24 ± 0.331.76 ± 0.63−1.62 ± 1.34 | 0.16(M0 vs. M1) |
| TNM stage I-II III IV | 16(53.33)8(26.67)6(20.00) | 1.19 ± 0.680.41 ± 0.60−2.35 ± 1.35 | 0.23 |
| Perineural invasion Negative Positive | 12(40.00)18(60.00) | −0.34 ± 0.960.68 ± 0.77 | 0.44 |
| Lymphatic invasion Negative Positive | 6(20.00)24(80.00) | 3.28 ± 1.60−0.04 ± 0.89 | 0.08 |
| Tumor size (cm) ≥5 <5 | 25(83.33)5(16.67) | −0.02 ± 0.891.72 ± 0.49 | 0.19 |
| Tumor grades I II III | 9(30.00)8(26.67)13(43.33) | −2.36 ± 0.84−0.77 ± 0.612.74 ± 0.77 | 0.02** |
| Tumor types Diffuse Intestinal | 14(46.66)16(46.66) | 1.68 ± 0.82−0.96 ± 0.82 | 0.06 |
* A higher ΔCt value indicates higher expression. ** Statistically significant.
Table 5.
The relationship between cir_ITCH expression level (as divided into two groups based on the median of ΔCt) in GC tissues with clinicopathological parameters.
| Name | Sequence (5’--> 3’) | Ta for PCR (°C) | Amplicon size | |||||||||
| Divergentprimers | hcircITCH-F1 | GTCCGGAACTATGAACAATG | 55.9 | 207 bp | hcircITCH-R1 | CTCTGTTGGCTCTTTGTCAC | hcircHIPK3-F1 | TATGTTGGTGGATCCTGTTC | 55.9 | 213 bp | hcirc HIPK3-R1 | AACTGCTTGGCTCTACTTTG |
| Convergent primers | hlinITCH-F1 | GGTTCACCATCTGCCACTTC | 60.7 | 169 bp | hlinITCH-R1 | AGGGAGCTTGAGTTACAGGATT | hlin HIPK3-F1 | GAAAGAAACTATCCACGGAC | 54.4 | 177 bp | hlin HIPK3-R1 | TATGACCTTTGTAGCACCTG |
* A higher ΔCt value indicates higher expression. ** Statistically significant.
4. Discussion
In the current study, we explored the expression levels of circHIPK3 and cir_ITCH in gastric cancer tissues compared to their paired adjacent noncancerous tissues as well as their clinicopathological significance.
Our results showed a significant underexpression of circHIPK3 in GC tumoral tissues. Using RNA sequencing (RNA-seq), Zheng et al. (18) characterized an abundant circular RNA derived from the HIPK3 gene, termed circHIPK3. They reported the significant overexpression of circHIPK3 in liver cancer compared with their matched normal tissues. In 2017, Li et al. (24) measured the relative expression of circHIPK3 in 44 pairs of bladder cancer and normal bladder tissues. Consistent with their RNA-seq results, they observed a significant decrease of circHIPK3 levels in 79.5% of bladder tumoral tissues compared to their normal counterparts. Overexpression of circHIPK3 in bladder cancer cell lines could suppress migration, invasion, and angiogenesis in vitro and inhibit bladder cancer growth and metastasis in vivo. Their findings support the tumor-suppressive activity of circHIPK3 in bladder cancer. In agreement with the results of expression of this circular RNA in bladder cancer (24), we also observed the relative downregulation of circHIPK3 in gastric cancer tumoral tissues. The discrepancy between Li et al. (24) and our results vs. findings of Zheng et al. (18) on liver cancer may be ascribed to the potential tissue-specific expression and function of circHIPK3.
We furthermore analyzed the relative expression of cir_ITCH in GC and found that it was significantly underexpressed in tumoral tissues compared to the nontumoral adjacent tissues. The association between cir_ITCH and cancer was firstly described in a study on esophageal squamous cell carcinoma conducted by Li et al. (16). cir_ITCH was highly and significantly expressed in around 70.61% of tumoral tissues compared to their paired nontumoral tissues (16). Evaluation of cir_ITCH expression in colorectal cancer (17) showed its underexpression in 75.6% of cancerous tissues compared with the adjacent noncancerous tissues. In 2016, Wan et al. (15) evaluated the expression levels of cir_ITCH in a cohort of lung cancer compared to their paired adjacent normal tissues. Compared to non-tumoral lung tissues, cir_ITCH showed a significantly lower expression in 73.08% of tumoral tissues (15). Taken together, our result on gastric cancer is consistent with those of previous reports on ESCC (16), colorectal cancer (17), and lung cancer (15).
In 2017, Li et al. (24) reported a negative correlation of circHIPK3 expression levels with bladder cancer grade, invasion, stage and the lymph nodes metastasis. In the same vein, we also observed that those patients with metastasis had a significant lower expression levels for this circular RNA. Gastric cancer patients with higher TNM stage or positive lymphatic invasion had lower levels of circHIPK3, although these correlations were not significant.
Furthermore, we found that the cir_ITCH expression level was significantly associated with age, metastasis, and cancer grades. Consistently, Wan et al. (15) reported a significant association between age and expression levels of this circular RNA in a cohort of lung cancer tissues.
5. Conclusions
In summary, our results showed that the expression of cir_ITCH and circHIPK3 were significantly down regulated in GC tumoral tissues compared with their paired adjacent non-neoplastic counterparts. Further investigation of these preliminary results is needed to elucidate the precise functional roles of these two circular RNAs in gastric cancer pathogenesis. cir_ITCH and circHIPK3 may serve in future as potential prognostic biomarkers in cancer.
Acknowledgments
This original article was derived from the master’s thesis of Sara GHASEMI and was supported in part by a research grant number 395259 from Isfahan University of Medical Sciences, Isfahan, Iran. The funding body had no role in the design of the study and collection, analysis, and interpretation of the data and in writing the manuscript.
References
- Keeney S Bauer TL Epidemiology of adenocarcinoma of the esophagogastric junction. Surg Oncol Clin N Am. 2006;15:687. doi: 10.1016/j.soc.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Malekzadeh R Derakhshan MH Gastric cancer in Iran: epidemiology and risk factors. Arch Iran Med. 2009;12:576. [PubMed] [Google Scholar]
- Mousavi SM Gouya MM Ramazani R Davanlou M Hajsadeghi N Cancer incidence and mortality in Iran. Ann Oncol. 2009;20:556. doi: 10.1093/annonc/mdn642. [DOI] [PubMed] [Google Scholar]
- Kolahdoozan S Sadjadi A Radmard AR Khademi H Five common cancers in Iran. Arch Iran Med. 2010;13:143. [PubMed] [Google Scholar]
- Riquelme I Saavedra K Espinoza JA Weber H García P Nervi B Garrido M Corvalán AH Roa JC Bizama C Molecular classification of gastric cancer: towards a pathway-driven targeted therapy. Oncotarget. 2015;6:24750. doi: 10.18632/oncotarget.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL Yang L Regulation of circRNA biogenesis. RNA Biol. 2015;12:381. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S Yang X Li X Wang J Gao Y Shang R Sun W Dou K Li H. Circular RNA a new star of noncoding RNAs. Cancer Lett. 2015;365:141. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Sanger HL Klotz G Riesner D Gross HJ Kleinschmidt AK Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. P Natl Acad Sci USA. 1976;73:3852. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y Zheng Q Bao C Li S Guo W Zhao J Chen D Gu J He X Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–981. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S Jens M Elefsinioti A Torti F Krueger J Rybak A Maier L Mackowiak SD Gregersen LH Munschauer M Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Salzman J Chen RE Olsen MN Wang PL Brown PO Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777–e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CE Jeck WR Liu Y Sanoff HK Wang Z Sharpless NE Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233–e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ Circular RNA (circRNA) in Alzheimer’s disease (AD) Front Genet. 2013;4:307–307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J Xie Q Xu H Li J Li Y Circular RNAs and cancer. Cancer Lett. 2017;396:138. doi: 10.1016/j.canlet.2017.03.027. [DOI] [PubMed] [Google Scholar]
- Circular RNA-ITCH Suppresses lung cancer proliferation via inhibiting the Wnt/beta-catenin pathway. Biomed Res Int. 2016. pp. 1579490–1579490. [DOI] [PMC free article] [PubMed]
- Li F Zhang L Li W Deng J Zheng J An M Lu J Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G Zhu H Shi Y Wu W Cai H Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PloS one. 2015;10:e0131225–e0131225. doi: 10.1371/journal.pone.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q Bao C Guo W Li S Chen J Chen B Luo Y Lyu D Li Y Shi G Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215–11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahari F Emadi-Baygi M -92 host gene, uderexpressed in gastric cancer and its expression was negatively correlated with the metastasis. Indian J Cancer. 2015;52:22. doi: 10.4103/0019-509X.175605. [DOI] [PubMed] [Google Scholar]
- Emadi-Baygi M Nikpour P Emadi-Andani E. SIX1 overexpression in diffuse-type and grade III gastric tumors: features that are associated with poor prognosis. Adv Biomed Res. 2015;4:139–139. doi: 10.4103/2277-9175.161540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baratieh Z Khalaj Z Honardoost MA Emadi-Baygi M Khanahmad H Salehi M Nikpour P Aberrant expression of PlncRNA-1 and TUG1: potential biomarkers for gastric cancer diagnosis and clinically monitoring cancer progression. Biomark Med. 2017;11:1077. doi: 10.2217/bmm-2017-0090. [DOI] [PubMed] [Google Scholar]
- 7th Edition of the AJCC cancer staging manual: stomach. Ann Surg Oncol. 2010;17:3077. doi: 10.1245/s10434-010-1362-z. [DOI] [PubMed] [Google Scholar]
- Baygi ME Soheili ZS Schmitz I Sameie S Schulz WA Snail regulates cell survival and inhibits cellular senescence in human metastatic prostate cancer cell lines. Cell Biol Toxicol. 2010;26:553. doi: 10.1007/s10565-010-9163-5. [DOI] [PubMed] [Google Scholar]


