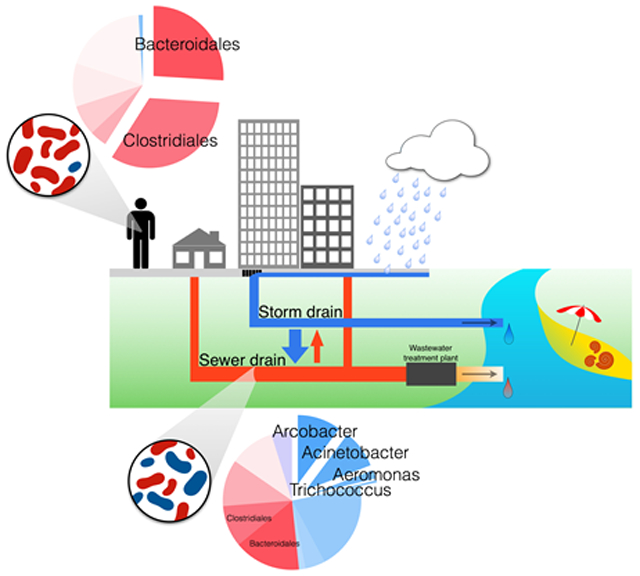
Abstract
Modern urban sewer pipe infrastructure is a unique niche where microbes can thrive. Arcobacter, Acinetobacter, Aeromonas, and Trichococcus are among the organisms that dominate the microbial community of sewage influent, but are not major members of human fecal microbiome, drinking water, or groundwater. Pipe resident communities in untreated sewage are distinct from sewer biofilm communities. Because of their high biomass, these organisms likely have a role in biotransformation of waste during conveyance and could represent an important inoculum for treatment plants. Studies demonstrate stormwater systems act as direct conduits for sewage to surface waters, releasing organisms propagated in sewer pipes. Frequent occurrence of these pipe residents, in particular Arcobacter, demonstrates the extent that urban infrastructure impacts rivers, lakes, and urban coasts worldwide.
Graphical abstract
Introduction
Sewer infrastructure is a unique manmade environment that is a relatively new habitat for microbes in comparison to soil or oceans that have been around for millions of years. Only recently have the indigenous communities within these systems been defined [1-4]. Sewer pipe networks have been recognized as propagating rather than only conveying microorganisms [4,5]. Human fecal-associated bacteria account for only 15–20% of the sewage influent community which reflects an integrated profile of the human population microbiome within a city [6]. In contrast, approximately 80% of the total community appears to be residents within the sewer pipe and is dominated by a few genera that include Arcobacter, Acinetobacter, Aeromonas, and Trichococcus [4,5]. The sheer size of cities, and the thousands of kilometers of pipes within urban areas, suggest that the organisms that proliferate within pipes can have a large influence on the overall microbiome of the urban environment. This review will examine the environmental conditions and selective pressures within sewer pipes and the bacterial community composition. This review will also highlight some of the dominant organisms that thrive within sewer conveyance systems. The extensive release of pipe-derived organisms to the environment is illustrated by their imprint on receiving waters and urban coastal systems worldwide.
The environment within pipes, organic matter transformation, and selective pressures
The sewer conveyance system is an aqueous, dark, high nutrient environment with fluctuating oxygen conditions. Gravity sewers can be aerobic, but become anaerobic or microaerophilic depending on flow, turbulence, and organic matter [7,8]. Because these systems are generally buried two meters below the surface, temperatures are less variable then the daily fluctuating air temperatures. Temperature is a major factor that can select for organisms with different growth optimums. Colder temperatures also reduce microbial activity and favor aerobic conditions [7].
Sanitary sewer pipes contain both biofilm and sediment [9] with distinct microbial assemblages in each compartment involved in the metabolism of organic compounds [2,8]. Organic matter is deposited into sediments during transport where it can be transformed into more readily degradable products and released back into sewage as it moves down the system [8,10]. The biotransformation of nutrients and other contaminants involves complex bacterial processes in both biofilm and sediments, which are shaped by dynamic environmental conditions [11,12]. Heterotrophic bacterial activity determines dissolved oxygen levels [7] and facilitates anaerobic conditions, which favor methanogenesis and sulfate reduction, two undesirable processes leading to methane production and sewer corrosion. In a pilot scale system, Shi et al. [13] demonstrated that fermentative bacteria were major members in the top layer sediments, while methanogens and sulfate reducing bacteria were more prominent in deeper layers. In this modelled system, abundant bacterial members linked to carbon transformation included Trichococcus, Cloacibacterium, Aeromonas, Paludibacter, Arcobacter, and Moraxellaceae (presumably Acinetobacter) [13]. Bacteria that can thrive in a broad range of conditions may have an advantage in an environment with changing oxygen availability and metabolism byproducts from other community members. By understanding the complex, interrelated conversions that take place in conveyance pipes, systems could be designed to favor beneficial microbes and limit undesirable reactions [10,13].
The ability to effectively compete with the large densities of viable bacteria, 10–100 times higher than surface fresh or saltwater [14], may also determine what organisms are dominant in the sewage. In addition, there is a nearly constant flow in sewer systems, and bacteria that are efficient at filling open niches may dominate. These organisms have been termed “microbial weeds” because of their rapid growth, their tolerance to diverse stresses, and inhibitory effect on other bacteria, e.g. production of surfactants or antimicrobial toxins, acidification of the environment, etc. [15].
Microbial communities in sewer systems can also be shaped by selective pressures including antimicrobial agents, disinfectants and heavy metals that create harsh conditions favoring well-adapted or specialized organisms [16,17]. Antibiotics directly favor the accumulation of antibiotic resistance genes, however, co-selection exerted by heavy metals or nutrients might also facilitate the acquisition and maintenance of antibiotic resistance genes in bacterial populations (see [18] and reference therein). The high biomass of organisms may be an important reservoir for resistance genes, making sewers a hot spot for antibiotic resistance accumulation and spread [19]. A large range of antibiotic gene resistance traits have been associated with Acinetobacter [16,20] and Arcobacter [20,21]. In one study, a metagenomic assembly of A. cryaerophilus from sewage recovered 25 categories of antibiotic resistance genes [21]. Arcobacter has also been suggested as a keystone species in shuttling antibiotic resistance genes among a broad group of phylogenetically distant organisms [16]. Metagenomics studies of untreated sewage detected 381 different bacterial resistance genes in China [22], and 303 different resistance subtypes in Singapore [23], which highlights the complex reservoirs of resistance genes within these systems.
Community structure in sewer pipe conveyance systems
The microbial community in wastewater treatment plant influent contains a unique assemblage of bacteria that are distinct from the human fecal microbiome (Figure 1), treated potable water [24], or urban groundwater [25] that may infiltrate the system. As sewage is transported from upstream portions of the conveyance system to the treatment plant, the non-fecal component increases and is dominated by Arcobacter, Acinetobacter and Aeromonas [5]. In addition, some facultative anaerobes associated with the human microbiome such as Lactoccocus, Enterococcus, or Enterobacteriaceae (the family that holds Klebsiella, Enterobacter, and Escherichia coli) have been observed 10–100 time more abundant in untreated sewage relative to human gut-microbiota [26]. The enrichment in sewage of these taxa, commonly associated with fecal pollution, suggests either these fecal-originated strains can thrive and grow in the pipe environment, or there are non-fecal, free living strains that colonized sewer pipes. The bacterial community in both biofilm and sediment share members but with distinct abundance patterns compared to wastewater [19,27]. The sewage influent community profiles suggest resident bacteria with the sediment can be mobilized, whereas the biofilm may be harder to detach. Many of the community members recovered from sewage appear in activated sludge [28], where they may be inoculum for wastewater treatment plants [29].
Figure 1.
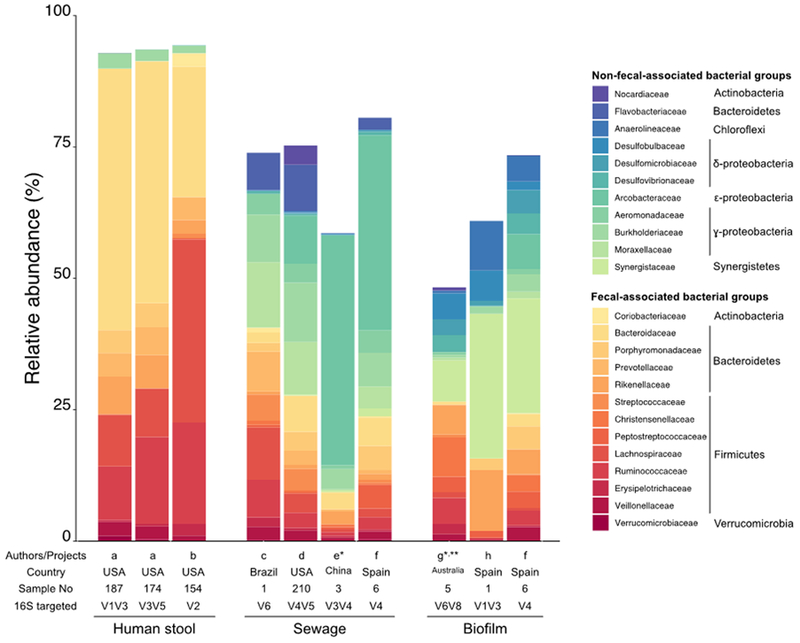
Dominant bacterial families in fecal human microbiome, untreated sewage and sewer biofilm based on 16S rRNA gene amplicon data. Only the top 10 dominant families among at least one of the three environments are displayed. Fecal-derived taxa are represented using red/orange hue colors, while non-fecal-derived taxa are colored in blue/green hue. List of the publications: a [26], b [66], c [67], d [6], e [3], f [19], g [68], h [69]. *: data reprocessed from the SRA files using the 454 Mothur SOP and Silva132 database. **: sewer pilot.
Notable traits of dominant sewer pipe resident organisms
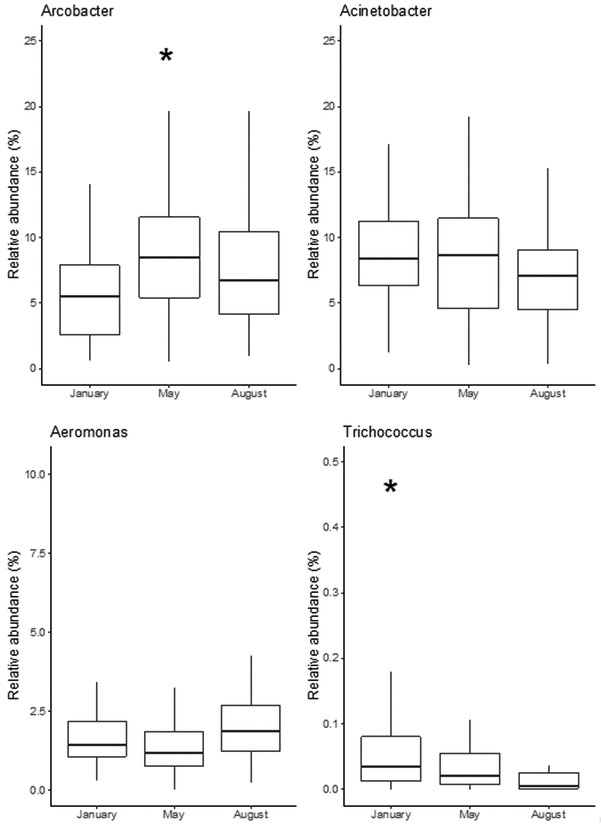
A study involving 71 U.S. cities sampled three times across a year [6] highlighted the consistency of the bacterial communities in sewers, which are dominated by a few members. Across 213 samples, Arcobacter averaged 9.2%, and Acinetobacter 8.1% of the community. Aeromonas was the third most abundant taxa comprising 1.8% of the community. Trichococcus represented on average 0.05% of the total community, but was found to comprise up to 1% for some cities. In this study, few spatio-temporal differences in the relative abundance of the most dominant genera were seen except for Trichococcus that appears more abundant in colder climates and in cooler months (Figure 2). This result is consistent with the optimal low-temperature growth reported for this genus [30,31]. Worldwide, these four genera have been reported to be prevalent in sewage [1,3,19,20,32] suggesting that urban sewer infrastructure provides a consistent environment in which they can thrive despite different physicochemical environments of specific systems or climates.
Figure 2.
Box plot depicting the relative abundance of the bacterial groups Arcobacter, Acinetobacter, Aeromonas and Trichococcus within sewage influent from 71 U.S. cities [6]. A total of 31 cold cities (blue dots, average air temperature over the campaigns ≤ 10°C), and 40 warm cities (red dots, average air temperature over the campaigns > 10°C) were sampled in January, May and August between 2012 and 2013. *Six outliers were removed from the plot for Arcobacter (88.9, 85.5, 84.4, 73.1, 58.9, and 44.6%) and one for Trichococcus (1.5%).
In sewer pipes, Arcobacter have been recovered from both sediment and biofilm communities [2,19], but comparatively, are major members in wastewater influent, suggesting Arcobacter are primarily planktonic. The water-sediment interface, so-called near-bed solids, may be a potential reservoir for members of this genus [33], but to date this has not been investigated. Arcobacter may be particularly suited to tolerate conditions near sediments. Arcobacter can use nitrate as a terminal electron acceptor [34,35]. They have been observed to compete efficiently with other sulfur-oxidizing bacteria by being able to tolerate high concentrations of hydrogen sulfide, and to grow in low or the absence of oxygen [35,36].
Arcobacter belong to the family Arcobacteraceae within the newly proposed phylum Epsilonbacteraeota [37] (formerly Campylobacteraceae within the Epsilon subdivision of the Proteobacteria). This genus holds about thirty species, among which, A. butzleri, A. cryaerophilus and A. skirrowii, are considered as potential human pathogens (see review [38] and references therein). Genome sequencing of A. butzleri demonstrated a large part of the genome is devoted to genes involved in growth and survival under diverse conditions [39]. Although Arcobacter are ubiquitous motile bacteria in terrestrial and aquatic habitats [38], they are not dominant members in the bacterial community in these environments. In contrast, in untreated sewage, Arcobacter are found with a density estimated around one million of cells per milliliter of wastewater [40]. The predominant species isolated from sewage include A. butzleri and A. cryaerophilus following by A. thereius, A. defluvii, A. skirrowii, A. ellisii, A cloacae and A. nitrofigilis [40,41]. In contrast, using 16S rDNA gene amplicon sequencing, the Arcobacter assemblage were found to be primarily assigned to A. cryaerophilus, while A. butzleri was only the eleventh most abundant member after A. suis; A. cloacae/A. defluvii ranked twenty-third [42]. Genome sequencing of 52 strains of A. cryaerophilus revealed four genomvars clusters, with human isolates only found in cluster 1 [43]. A. butzleri has been reported to be more easily cultured that A. cryaerophilus from the environment [44], and both have been found to be viable after wastewater treatment [40,45]. Temperature has been observed to shape Arcobacter densities and assemblages with warm and cold ecotypes observed across multiple cities [42,46]. Warm water seems to favor A. butzleri survival in comparison to A. cryaerophilus [47].
Similar to Arcobacter, Acinetobacter are motile and appear to be primarily planktonic as they are in higher proportions in influent samples relative to sediment or biofilm community members. Acinetobacter, within the family Moraxellaceae, holds about fifty species that are widely distributed in the environment from soil and water to humans [48], likely because of its ability to use a wide range of carbon sources. Sewage isolates were found to be more limited in the range of compounds they can utilize as sole carbon sources compared to soil isolates, suggesting they have been selected for with the sewer environment [49]. Acinetobacter is denoted as a “bacterial weed” and has developed strategies to inhibit other species though creation of an acid environment and production of biosurfactants [15]. Strong seasonal shifts in the relative abundance and density of Acinetobacter ecotypes have been observed across a 4-year survey of U.S. sewage [5]. Full length cloned 16S rRNA gene sequences suggested that changes were due to reoccurring cycles of dominant ecotypes within the genus rather than seasonal replacement by species [5].
Arcobacter as a tracer of urban sewer impacts on receiving water.
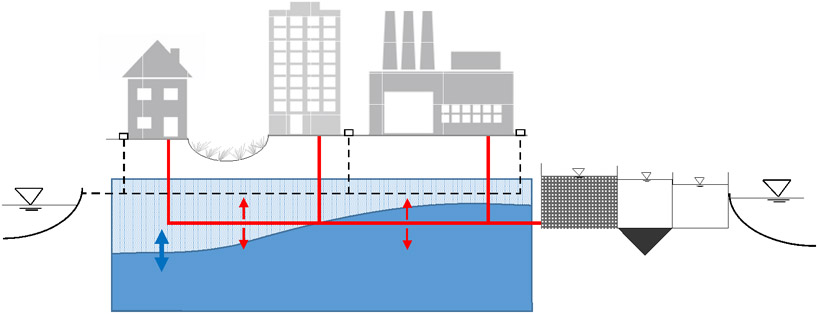
Urban sewer infrastructure is designed to carry harmful waste to the treatment plant. Over time, these systems deteriorate causing sewage to be released directly into the environment and groundwater, or indirectly through stormwater systems that act as a conduit to surface waters (Box 1). In addition, each year, millions of liters of untreated sewage are released during combined sewer overflows. Sewage contamination is evident after rainfall in the absence of known sewage overflows [50], and microbial communities from pipes can serve as an early warning that pipe are failing [2].
Box 1. Sewage exfiltration from urban infrastructure.
Box 1 Figure.
Illustration of the dynamic between urban sanitary sewers and stormwater systems with changing groundwater levels.
The two most dominant organisms in untreated sewage, i.e. Arcobacter and Acinetobacter, are prevalent in lakes, rivers, and coastal waters in or near urban areas. Several studies reported the presence of Arcobacter spp. in environmental waters was associated with high levels of fecal pollution [46]. In coastal waters where dilution reduces signals, Arcobacter was a sensitive measure of microbial footprint of the urban area [42,51]. Arcobacter may be the more specific indicator of sewage contamination given that Acinetobacter has also been observed to be prevalent in stormwater when there was no other evidence of sewage [52]. Worldwide, Arcobacter has been found in contaminated urban waters [53-57].
Conclusions
Modern sewer infrastructure has created a new habitat for microbial communities to develop and reach high biomass at a city-wide scale. The genomic potential and ecology of the dominant organisms within these communities may provide clues to the nutrient conversions and antibiotic resistance reservoirs within pipes, as well as illustrate more basic themes of fitness strategies in complex and highly competitive environments. These communities are not contained to the pipes of a city, and the impact due to the release of sewer-pipe organisms has not yet been explored. Further, these organisms are potentially ideal tracers of urban sewage contamination that release a plethora of chemicals, nutrients, and harmful microorganisms carried by sewage. Without question, increasing urbanization and hydrological pressures on urban infrastructure from climate change will make this a more relevant issue in the future.
Sewage exfiltration from pipes had previously been considered contained to surrounding soils and relatively harmless [58]. However, studies demonstrate sewage can be mobilized during rain events and reach groundwater and stormwater systems [59-62]. Sewage is released through joints or deteriorating pipes and exfiltration can be as high as 10% of dry weather flows [58,61,63]. If deteriorating sewage pipes are above groundwater elevations, the positive pressure in the pipe causes sewage to be released [64,65] (See Box 1 Figure). Researchers have developed a probability based model to predict exfiltration that relies on pipe attributes and groundwater elevation without prior knowledge of the locations of defects [61]. In other studies, Ly et al. [63] modelled local-scale migration of sewage leakage from a sewage pipe to nearby stormwater drains. Given the thousands of miles in cities though the U.S., modeling is useful to identify area for more labor intensive testing with closed circuit televising, dye flooding, or by molecular methods [59,60].
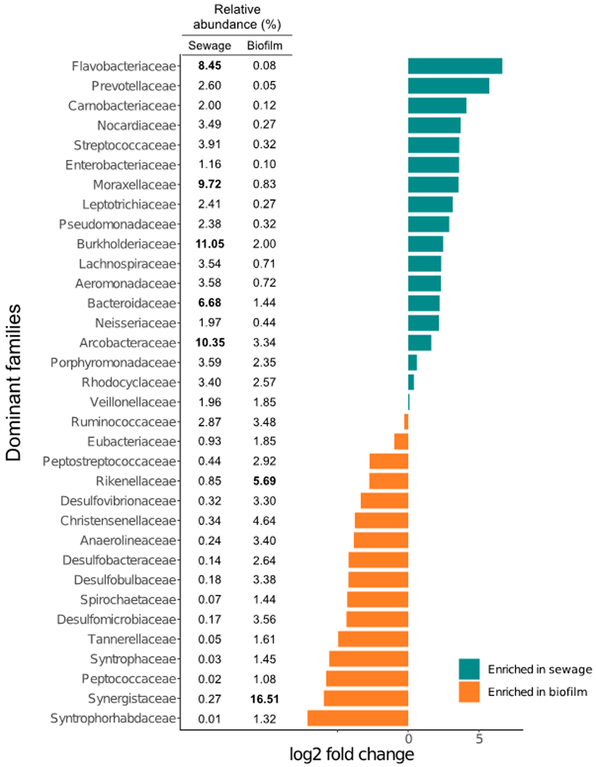
Figure 3.
Log2 fold change comparison between the relative abundance of bacterial families in untreated sewage and biofilm. Families more abundant in sewage are represented using green bars, while families more abundant in biofilm are in orange. Relative abundance of the families per compartment are listed on the left of the plot. The relative abundance per family was computed by averaging the abundances pondered with the number of sample from the publications described in the Figure 1. Relative abundance higher than 10% are in bold. Only the families with a relative abundance higher than 1% in at least one of the two compartments were considered.
Highlights.
Arcobacter, Acinetobacter, and Aeromonas dominate communities in sewage.
Pipe organisms in sewage influent have abundance patterns distinct from biofilm.
The high stress environment selects for organisms with unique fitness strategies.
Sewer pipe organisms carry a wide range of antibiotic resistance genes.
Urban sewer pipes leak and during rain, sewage is mobilized to stormwater systems.
Arcobacter is an indicator of urban impact and readily detected in waters worldwide.
Acknowledgments
We would like to thank Maria José Figueras and Patricia Holden for insightful discussion on this topic. We would also like to thank Patricia Holden for providing the conceptual illustration for Box 1. This effort was supported by the National Institutes of Health grant number R01AI091829.
References
- 1.Buelow E, Bayjanov JR, Majoor E, Willems RJL, Bonten MJM, Schmitt H, van Schaik W: Limited influence of hospital wastewater on the microbiome and resistome of wastewater in a community sewerage system. FEMS Microbiol Ecol 2018, doi: 10.1093/femsec/fiy087. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, Dong Q, Shi H: Distribution and population structure characteristics of microorganisms in urban sewage system. Appl Microbiol Biotechnol 2015, 99:7723–7734. [DOI] [PubMed] [Google Scholar]
- 3.Tang J, Bu Y, Zhang XX, Huang K, He X, Ye L, Shan Z, Ren H: Metagenomic analysis of bacterial community composition and antibiotic resistance genes in a wastewater treatment plant and its receiving surface water. Ecotoxicol Environ Saf 2016, 132:260–269. [DOI] [PubMed] [Google Scholar]
- 4.McLellan SL, Huse SM, Mueller-Spitz SR, Andreishcheva EN, Sogin ML: Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ Microbiol 2010, 12:378–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vandewalle JL, Goetz GW, Huse SM, Morrison HG, Sogin ML, Hoffmann RG, Yan K, McLellan SL: Acinetobacter, Aeromonas and Trichococcus populations dominate the microbial community within urban sewer infrastructure. Environ Microbiol 2012, 14:2538–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newton RJ, McLellan SL, Dila DK, Vineis JH, Morrison HG, Murat Eren A, Sogin ML: Sewage reflects the microbiomes of human populations. MBio 2015, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudjonsson G, Vollertsen J, Hvitved-Jacobsen T: Dissolved oxygen in gravity sewers - measurement and simulation. Water Sci Technol 2002, 45:35–44. [PubMed] [Google Scholar]
- 8.Shi X, Sang L, Wang XC, Jin P: Pollutant exchange between sewage and sediment in urban sewer systems. Chem Eng J 2018, 351:240–247.●● In a long-term monitoring of a pilot sewer system, the authors investigated the pollutant exchange between sewage and sediment, and how it affects the water influent quality.
- 9.Rocher V, Azimi S, Moilleron R, Chebbo G: Biofilm in combined sewers: wet weather pollution source and/or dry weather pollution indicator? Water Sci Technol 2003, 47:35–43. [PubMed] [Google Scholar]
- 10.Jin P, Shi X, Sun G, Yang L, Cai Y, Wang XC: Co-variation between distribution of microbial communities and biological metabolization of organics in urban sewer systems. Environ Sci Technol 2018, 52:1270–1279.● The authors investigated the environmental factors and substrates that shape the microbial community composition distribution in a pilot sewer system.
- 11.Nielsen PH, Raunkjær K, Norsker NH, Jensen NA, Hvitved-Jacobsen T: Transformation of wastewater in sewer systems - a review. Water Sci Technol 1992, 25:17–31. [Google Scholar]
- 12.Tanaka N, Hvitved-Jacobsen T: Transformations of wastewater organic matter in sewers under changing aerobic/anaerobic conditions. Water Sci Technol 1998, 37:105–113. [Google Scholar]
- 13.Shi X, Ngo HH, Sang L, Jin P, Wang XC, Wang G: Functional evaluation of pollutant transformation in sediment from combined sewer system. Environ Pollut 2018, 238:85–93. [DOI] [PubMed] [Google Scholar]
- 14.Ma L, Mao G, Liu J, Yu H, Gao G, Wang Y: Rapid quantification of bacteria and viruses in influent, settled water, activated sludge and effluent from a wastewater treatment plant using flow cytometry. Water Sci Technol 2013, 68:1763–1769. [DOI] [PubMed] [Google Scholar]
- 15.Cray JA, Bell ANW, Bhaganna P, Mswaka AY, Timson DJ, Hallsworth JE: The biology of habitat dominance; can microbes behave as weeds? Microb Biotechnol 2013, 6:453–492.● The authors discuss in a review the concept of microbial weeds that can easily fill open ecological niches.
- 16.Jacquiod S, Brejnrod A, Morberg SM, Abu Al-Soud W, Sørensen SJ, Riber L: Deciphering conjugative plasmid permissiveness in wastewater microbiomes. Mol Ecol 2017, 26:3556–3571. [DOI] [PubMed] [Google Scholar]
- 17.Gillings MR, Stokes HW: Are humans increasing bacterial evolvability? Trends Ecol Evol 2012, 27:346–352. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q-Q, Tian G-M, Jin R-C: The occurrence, maintenance, and proliferation of antibiotic resistance genes (ARGs) in the environment: influencing factors, mechanisms, and elimination strategies. Appl Microbiol Biotechnol 2018, doi: 10.1007/s00253-018-9235-7. [DOI] [PubMed] [Google Scholar]
- 19.Auguet O, Pijuan M, Borrego CM, Rodriguez-Mozaz S, Triadó-Margarit X, Giustina SV Della, Gutierrez O: Sewers as potential reservoirs of antibiotic resistance. Sci Total Environ 2017, 605-606:1047–1054. [DOI] [PubMed] [Google Scholar]
- 20.Hultman J, Tamminen M, Pärnänen K, Cairns J, Karkman A, Virta M: Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent. FEMS Microbiol Ecol 2018, 94:1–10.●● A metagenonic analysis highlighted that Arcobacter and Acinetobacter were carried the four antibiotic resistance genes investigated in the untreated sewage tested.
- 21.Millar JA, Raghavan R: Accumulation and expression of multiple antibiotic resistance genes in Arcobacter cryaerophilus that thrives in sewage. PeerJ 2017, 5:e3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su JQ, An XL, Li B, Chen QL, Gillings MR, Chen H, Zhang T, Zhu YG: Metagenomics of urban sewage identifies an extensively shared antibiotic resistome in China. Microbiome 2017, 5:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng C, Tay M, Tan B, Le TH, Haller L, Chen H, Koh TH, Barkham TMS, Thompson JR, Gin KYH: Characterization of metagenomes in urban aquatic compartments reveals high prevalence of clinically relevant antibiotic resistance genes in wastewaters. Front Microbiol 2018, 9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang C, Ling F, Andersen GL, LeChevallier MW, Liu WT: Microbial community dynamics of an urban drinking water distribution system subjected to phases of chloramination and chlorination treatments. Appl Environ Microbiol 2012, 78:7856–7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajasekar A, Sekar R, Medina-Roldán E, Bridge J, Moy CKS, Wilkinson S: Next-generation sequencing showing potential leachate influence on bacterial communities around a landfill in China. Can J Microbiol 2018, 549:1–13. [DOI] [PubMed] [Google Scholar]
- 26.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI: The human microbiome project. Nature 2007, 449:804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen HS, Sekar R, Shepherd WJ, Osborn AM, Tait S, Biggs CA: Spatial and temporal variability of bacterial communities within a combined sewer system. Microbiologyopen 2016, 5:616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SH, Kang HJ, Park HD: Influence of influent wastewater communities on temporal variation of activated sludge communities. Water Res 2015, 73:132–144. [DOI] [PubMed] [Google Scholar]
- 29.Frigon D, Wells GF: Microbial immigration in wastewater treatment systems: analytical considerations and process implications. Curr Opin Biotechnol [this issue] [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez-Martinez A, Sihvonen M, Muñoz-Palazon B, Rodriguez-Sanchez A, Mikola A, Vahala R: Microbial ecology of full-scale wastewater treatment systems in the Polar Arctic Circle: Archaea, Bacteria and Fungi. Sci Rep 2018, 8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pikuta E V, Hoover RB: The genus Trichococcus In Lactic Acid Bacteria: Biodiversity and Taxonomy. Edited by Holzapfel WH, Wood BJB. John Wiley & Sons; 2014:135–145. [Google Scholar]
- 32.Lu X, Zhang XX, Wang Z, Huang K, Wang Y, Liang W, Tan Y, Liu B, Tang J: Bacterial pathogens and community composition in advanced sewage treatment systems revealed by metagenomics analysis based on high-throughput sequencing. PLoS One 2015, 10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Zhou X, Shi H: Sulfur cycle by in situ analysis in the sediment biofilm of a sewer system. J Environ Eng 2016, 142:C4015011. [Google Scholar]
- 34.Gevertz D, Telang AJ, Voordouw G, Jenneman GE: Isolation and characterization of strains CVO and FWKO B, two novel nitrate-reducing, sulfide-oxidizing bacteria isolated from oil field brine. Appl Environ Microbiol 2000, 66:2491–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Gusseme B, De Schryver P, De Cooman M, Verbeken K, Boeckx P, Verstraete W, Boon N: Nitrate-reducing, sulfide-oxidizing bacteria as microbial oxidants for rapid biological sulfide removal. FEMS Microbiol Ecol 2009, 67:151–161. [DOI] [PubMed] [Google Scholar]
- 36.Sievert SM, Wieringa EBA, Wirsen CO, Taylor CD: Growth and mechanism of filamentous-sulfur formation by Candidatus Arcobacter sulfidicus in opposing oxygen-sulfide gradients. Environ Microbiol 2007, 9:271–276. [DOI] [PubMed] [Google Scholar]
- 37.Waite DW, Vanwonterghem I, Rinke C, Parks DH, Zhang Y, Takai K, Sievert SM, Simon J, Campbell BJ, Hanson TE, et al. : Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front Microbiol 2017, 8:682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collado L, Figueras MJ: Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin Microbiol Rev 2011, 24:174–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller WG, Parker CT, Rubenfield M, Mendz GL, Wösten MMSM, Ussery DW, Stolz JF, Binnewies TT, Hallin PF, Wang G, et al. : The complete genome sequence and analysis of the epsilonproteobacterium Arcobacter butzleri. PLoS One 2007, 2:e1358.●● First metagenomc analysis focusing on Arcobacter butzleri isolated from human to investigated the metabolism properties of this strain compared with other bacterial species.
- 40.Levican A, Collado L, Figueras MJ: The use of two culturing methods in parallel reveals a high prevalence and diversity of Arcobacter spp. in a wastewater treatment plant. Biomed Res Int 2016, 2016:8132058.● This study established the prevalence of Arcobacter spp. in a wastewater treatment plan using in parallel two culturing methods and a direct detection by m-PCR.
- 41.Collado L, Inza I, Guarro J, Figueras MJ: Presence of Arcobacter spp. in environmental waters correlates with high levels of fecal pollution. Environ Microbiol 2008, 10:1635–1640. [DOI] [PubMed] [Google Scholar]
- 42.Fisher JC, Levican A, Figueras MJ, McLellan SL: Population dynamics and ecology of Arcobacter in sewage. Front Microbiol 2014, 5:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pérez-Cataluña A, Collado L, Salgado O, Lefiñanco V, Figueras MJ: A polyphasic and taxogenomic evaluation uncovers Arcobacter cryaerophilus as a species complex that embraces four genomovars. Front Microbiol 2018, 9.●● The authors evaluated for the first time the polyphasic and taxogenomic divergeance uncovered by Arcobacter cryaerophilus. They observed that A. cryaerophilus embraces four genomovars.
- 44.Fera MT, Maugeri TL, Gugliandolo C, Beninati C, Camera E La, Carbone M, Giannone M: Detection of Arcobacter spp . in the coastal environment of the Mediterranean Sea. Appl Environ Microbiol 2004, 70:1271–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webb AL, Taboada EN, Selinger LB, Boras VF, Inglis GD: Efficacy of wastewater treatment on Arcobacter butzleri density and strain diversity. Water Res 2016, 105:291–296. [DOI] [PubMed] [Google Scholar]
- 46.Lee C, Agidi S, Marion JW, Lee J: Arcobacter in Lake Erie beach waters: An emerging gastrointestinal pathogen linked with human-associated fecal contamination. Appl Environ Microbiol 2012, 78:5511–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levican A, Collado L, Yustes C, Aguilar C, Figueras MJ: Higher water temperature and incubation under aerobic and microaerobic conditions increase the recovery and diversity of Arcobacter spp. from shellfish. Appl Environ Microbiol 2014, 80:385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atrouni A Al, Joly-Guillou ML, Hamze M, Kempf M: Reservoirs of non-baumannii Acinetobacter species. Front Microbiol 2016, 7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warskow AL, Juni E: Nutritional requirements of Acinetobacter strains isolated from soil, water, and sewage. J Bacteriol 1972, 112:1014–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olds HT, Corsi SR, Dila DK, Halmo KM, Bootsma MJ, McLellan SL: High levels of sewage contamination released from urban areas after storm events: A quantitative survey with sewage specific bacterial indicators. PLoS Med 2018, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newton RJ, Bootsma MJ, Morrison HG, Sogin ML, McLellan SL: A microbial signature approach to identify fecal pollution in the waters off an urbanized coast of Lake Michigan. Microb Ecol 2013, 65:1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLellan SL, Fisher JC, Newton RJ: The microbiome of urban waters. Int Microbiol 2015, 18:141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghaju Shrestha R, Tanaka Y, Malla B, Tandukar S, Bhandari D, Inoue D, Sei K, Sherchand JB, Haramoto E: Development of a quantitative PCR assay for Arcobacter spp. and its application to environmental water samples. Microbes Environ 2018, 00:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghaju Shrestha R, Tanaka Y, Malla B, Bhandari D, Tandukar S, Inoue D, Sei K, Sherchand JB, Haramoto E: Next-generation sequencing identification of pathogenic bacterial genes and their relationship with fecal indicator bacteria in different water sources in the Kathmandu Valley, Nepal. Sci Total Environ 2017, 601-602:278–284. [DOI] [PubMed] [Google Scholar]
- 55.Brown PC, Borowska E, Schwartz T, Horn H: Impact of the particulate matter from wastewater discharge on the abundance of antibiotic resistance genes and facultative pathogenic bacteria in downstream river sediments. Sci Total Environ 2019, 649:1171–1178. [DOI] [PubMed] [Google Scholar]
- 56.Sun H, He X, Ye L, Zhang X-X, Wu B, Ren H: Diversity, abundance, and possible sources of fecal bacteria in the Yangtze River. Appl Microbiol Biotechnol 2017, 101:2143–2152. [DOI] [PubMed] [Google Scholar]
- 57.Salas-Massó N, Figueras MJ, Andree KB, Furones MD: Do the Escherichia coli European Union shellfish safety standards predict the presence of Arcobacter spp., a potential zoonotic pathogen? Sci Total Environ 2018, 624:1171–1179. [DOI] [PubMed] [Google Scholar]
- 58.Reynolds JH, Barrett MH: A review of the effects of sewer leakage on groundwater quality. Water Environ J 2003, 17:34–39. [Google Scholar]
- 59.Sercu B, Van De Werfhorst LC, Murray JLS, Holden PA: Sewage exfiltration as a source of storm drain contamination during dry weather in urban watersheds. Environ Sci Technol 2011, 45:7151–7157. [DOI] [PubMed] [Google Scholar]
- 60.Sauer EP, Vandewalle JL, Bootsma MJ, McLellan SL: Detection of the human specific Bacteroides genetic marker provides evidence of widespread sewage contamination of stormwater in the urban environment. Water Res 2011, 45:4081–4091. [DOI] [PubMed] [Google Scholar]
- 61.Roehrdanz PR, Feraud M, Lee DG, Means JC, Snyder SA, Holden PA: Spatial models of sewer pipe leakage predict the occurrence of wastewater indicators in shallow urban groundwater. Environ Sci Technol 2017, 51:1213–1223.●● The authors developed and tested a spatially explicit model of exfiltration probability based on pipe attributes and groundwater elevation without prior knowledge of exfiltrating defect locations.
- 62.Gotkowitz MB, Bradbury KR, Borchardt MA, Zhu J, Spencer SK: Effects of climate and sewer condition on virus transport to groundwater. Environ Sci Technol 2016, 50:8497–8504. [DOI] [PubMed] [Google Scholar]
- 63.Ly DK, Chui TFM: Modeling sewage leakage to surrounding groundwater and stormwater drains. Water Sci Technol 2012, 66:2659–2665.● The authors created a model to describe sewage infiltration into storm drains through weep holes in the dense urban are of Singapore. In these simulations, the amount of sewage exfiltration and degraded water quality were higher in dry years because groundwater levels were lower allowing more exfiltration from sewer pipes and less dilution of contamination in storm drains.
- 64.Selvakumar A, Field R, Burgess E, Amick R: Exfiltration in sanitary sewer systems in the US. Urban Water J 2004, 1:227–234. [Google Scholar]
- 65.Amick RS, Burgess EH, Selvakumar A: Exfiltration in sewer systems. 2000. [Google Scholar]
- 66.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. : A core gut microbiome in obese and lean twins. Nature 2009, 457:480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koskey AM, Fisher JC, Eren AM, Ponce-Terashima R, Reis MG, Blanton RE, Mclellan SL: Blautia and Prevotella sequences distinguish human and animal fecal pollution in Brazil surface waters. Environ Microbiol Rep 2014, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun J, Hu S, Sharma KR, Ni BJ, Yuan Z: Stratified microbial structure and activity in sulfide- and methane- producing anaerobic sewer biofilms. Appl Environ Microbiol 2014, 80:7042–7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Auguet O, Pijuan M, Batista J, Borrego CM, Gutierrez O: Changes in microbial biofilm communities during colonization of sewer systems. Appl Environ Microbiol 2015, 81:7271–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]