Abstract
Tissue residency is considered a defining feature of the innate lymphoid cell (ILC) populations located within mucosal and adipose tissues. ILCs are also present within all lymphoid tissues, but whether ILCs migrate between lymphoid and non-lymphoid sites and in what context is poorly understood. To determine whether migratory ILCs exist within peripheral lymph nodes (LNs), all cells within the brachial LN (bLN) of transgenic mice expressing a photoconvertible fluorescent protein were labelled by direct exposure to light. Tracking of cellular changes in the labelled LN revealed the gradual migration of new ILCs into the tissue, balanced by egress of ILCs dependent upon sphingosine-1-phosphate receptors. Most of the migratory ILCs were ILC1s, entering LNs directly from the circulation in a CD62L and CCR7-dependent manner and thus behaving like conventional (c)NK cells. Upon egress, both ILC1s and cNK cells were found to recirculate through peripheral LNs. A distinct population of migratory ILC2s were detected in the LN, but the majority of ILC3s were tissue resident. Functionally, both migratory and resident ILC1s within LNs were able to rapidly produce IFN-γ to support the generation of robust Th1 T cell responses after immunization. Thus, migratory and resident ILC populations exist within peripheral LNs, with ILC1s, akin to cNK cells, able to traffic into these tissues where they can contribute to the initiation of adaptive immunity.
Introduction
ILCs have been identified in most tissues in the body where they act as sentinels, orchestrating homeostasis, but able to dynamically respond to a wide range of signals indicative of infection or danger (1–3). As ILC populations were discovered, it became evident that these cells were most numerous at barrier tissues such as the gastrointestinal tract and lung and the majority of studies have focused on their roles at these sites (4–8). To address whether ILCs were tissue resident, several studies have used parabiosis models which indicate that the vast majority of ILCs at these barrier sites, as well as in adipose tissue, permanently reside within these tissues and do not traffic between host mice when the circulatory system is shared (9, 10), (11, 12). Current models of ILC development propose the seeding of tissues with progenitors that develop from both fetal liver and bone marrow and then expand to provide tissue resident populations (13–17).
Although less numerous than at mucosal sites, ILCs are also present within all lymphoid tissues, suggesting roles influencing the adaptive immune responses initiated in these structures (18, 19). Crucially, how ILC populations within lymphoid tissue relate to those within barrier tissues is poorly understood. Analysis of the mesenteric LN (mLN) and spleen of parabiotic mice also suggested that most ILCs within these secondary lymphoid tissues were tissue resident (9). Furthermore, given the role of the lymphoid tissue inducer subset of ILC3s in LN and Peyer’s patch organogenesis (20), it is possible that such LN resident populations may be established very early in life. To date, the lack of mouse models that specifically and completely delete ILC populations has hindered our ability to clearly define their functions within lymphoid tissues (21). Beyond their ability to rapidly produce effector cytokines, investigations of how ILCs might regulate adaptive immune responses have identified that both ILC2s (22) and ILC3s (19, 23) are able to process and present peptides via MHCII and modulate CD4 T cell responses. Where and how ILCs might acquire such antigen is unresolved, but relevant to understanding the role such antigen presentation plays within the immune system. It is conceivable that as a consequence of antigen uptake, ILCs migrate from tissue to draining LNs via the lymphatics. Such trafficking would not be evident within the parabiosis studies used to identify tissue residency. In support of ILC migration from tissues to their draining LNs, direct trafficking of ILCs from the intestine to the mLN has been described (18) and analysis of the mLNs of mixed bone marrow chimeras using WT and CCR7-/- donors, indicated a cell-intrinsic requirement for CCR7 in establishing normal ILC populations within LNs (18). Since many studies of human ILCs rely on isolating these cells from blood, this suggests that circulatory ILCs exist in human blood at least, however, recent data indicate that the human ILCs isolated from blood include progenitor populations (16, 24, 25). In addition, with better understanding of ILC subsets, a highly migratory ILC2 population was recently described (26) suggesting that the tissue resident description of ILCs may not be true of all subsets in all circumstances. Finally, cNK cells which share many similarities with ILC1s, are thought to be highly migratory, entering LNs directly from the circulation via high endothelial venules (HEV) (27). Thus, the dogma of ILC tissue residency needs to be further tested for lymphoid ILC populations. As we seek to better understand the roles ILCs play in secondary lymphoid tissue, how and when ILCs establish residency within LNs will help to clarify their functions the generation and regulation of adaptive immunity.
To test the hypothesis that ILCs in peripheral LNs are migratory, we have used Kaede photoconvertible mice (28), having established methods to label all cells within a peripheral LN by illumination following surgical exposure (29). This approach offers direct in vivo assessment of the migration of new cells into the tissue, identification of resident populations and tracking of cells leaving the LN to new locations. Our data reveal that LNs contain both migratory and resident ILC populations, with the ILC1 subset highly migratory and able to recirculate through peripheral lymphoid tissue, where it can contribute to supporting the priming of CD4+ T cells after immunization.
Results
ILCs migrate into peripheral LNs
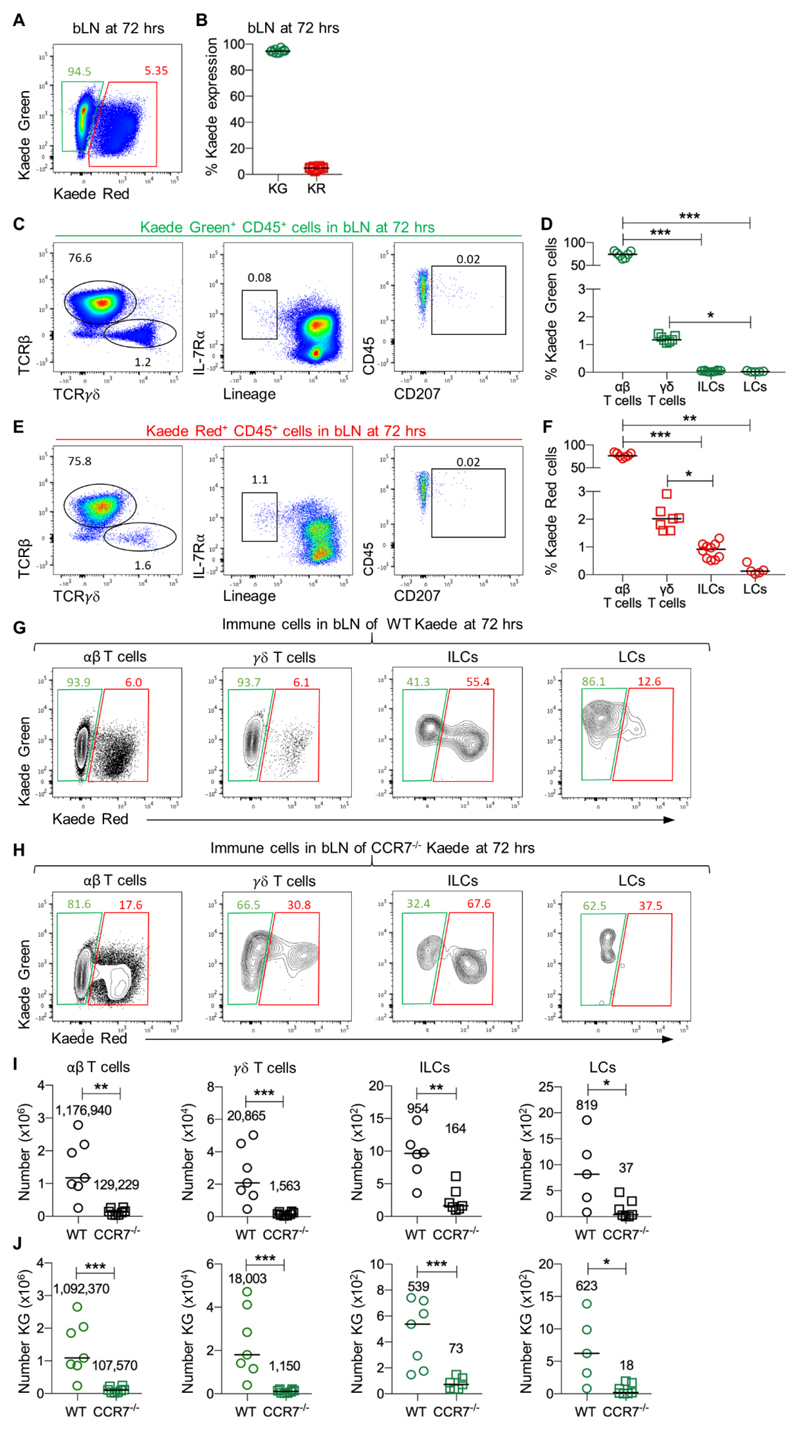
To test the hypothesis that migratory ILCs exist in peripheral LNs, the bLN of Kaede mice was exposed through a small incision in the skin and photoconverted as described previously, changing the cellular fluorescence from Kaede Green to Kaede Red (29). While cells expressing the Kaede Red form of the fluorochrome frequently also express the Kaede Green form (due to a combination of incomplete conversion of total Kaede protein within the cell and the generation of new protein post-photoconversion), to simplify the description of the data, here these cells are termed ‘Kaede Red+’. Analysis of Kaede expression confirmed that the entire hematopoietic compartment of the LN was converted to Kaede Red+ cells (Fig. S1). Thus, analysis of the labelled bLN at subsequent time points after photoconversion allows identification of cells newly entering the tissue from unexposed sites (Kaede Green+) and those that have remained within the LN (Kaede Red+). A fraction of cells identified as Kaede Red+ resident cells may be migratory cells that have recirculated back to the initially photoconverted LN. This experimental approach enables the simple but direct assessment of residency within the LN for any cell type. After 72 hrs, approximately 95% of the CD45+ cells in the labelled LN were Kaede Green+ cells (Fig. 1A, B), consistent with the constant circulation of naïve lymphocytes through secondary lymphoid tissue. The majority of the Kaede Green+ cells within LNs were αβ T cells although some γδ T cells were also detected (Fig. 1C, D). Notably, a small but distinct population of IL-7Rα+Lin (B220, CD11b, CD11c, CD3, CD5, CD19, Ter119, Gr1, CD49b, F4/80 and FcεRI)- ILCs was evident and comparable in frequency to Langerhans cells (LCs, Fig. 1C, D). These data indicate that some ILCs had entered the tissue subsequent to photoconversion of the LN 72 hrs previously. Amongst the Kaede Red+ cells that had remained within the LN since labelling, ILCs were also detected and appeared enriched compared with the proportion of ILCs amongst the Kaede Green+ population (Fig. 1E, F). Thus, our initial analysis suggested the presence of both migratory and resident ILC populations within peripheral LNs.
Figure 1. CCR7-dependent entry of ILCs into peripheral LNs.
To assess the migratory kinetics of different cellular populations within peripheral LNs, the bLN of Kaede mice was photoconverted and analysed 72 hrs later. (A) Expression of Kaede Green versus Kaede Red protein by CD45+ cells isolated from the bLN at 72 hrs post photoconversion. (B) Percentage of CD45+ cells expressing Kaede Green or Kaede Red protein at 72 hrs post photoconversion (n=10). (C) Characterisation of Kaede Green+ CD45+ cells isolated from the bLN, showing αβ T cells (CD45+ CD3+ TCRβ+ TCRγδ-), γδ T cells (CD45+ CD3+ TCRβ- TCRγδ+), ILCs (CD45+ IL-7Rα+ Lin{B220, CD11b, CD11c, CD3, CD5, CD19, Ter119, Gr1, CD49b, F4/80, FcεRI}-) and LCs (CD45+ MHCII+ CD11c+ CD207+). (D) Percentage of αβ T cells (n=7), γδ T cells (n=7), ILCs (n=10) or LCs (n=5) amongst CD45+ Kaede Green+ cells. (E) Characterisation of Kaede Red+ cells isolated from the bLN, showing αβ T cells, γδ T cells, ILCs and LCs. (F) Percentage of αβ T cells (n=7), γδ T cells (n=7), ILCs (n=10) and LCs (n=5) amongst CD45+ Kaede Red+ cells. Expression of Kaede Green versus Kaede Red protein by αβ T cells, γδ T cells, ILCs and LCs isolated from the bLN of (G) Kaede and (H) CCR7-/- Kaede mice. (I) Number of αβ T cells (WT n=7, CCR7-/- n=8), γδ T cells (WT n=7, CCR7-/- n=8), ILCs (WT n=6, CCR7-/- n=8) and LCs (WT=5, CCR7-/- n=7) isolated from the bLN of WT Kaede versus CCR7-/- Kaede mice. (J) Number of Kaede Green αβ T cells (WT n=7, CCR7-/- n=8), γδ T cells (WT n=7, CCR7-/- n=8), ILCs (WT n=7, CCR7-/- n=8) and LCs (WT n=5, CCR7-/- n=7) isolated from the bLN. Data are pooled from a minimum of 2 independent experiments. Values on flow cytometry plots represent percentages, bars on scatter plots represents the median, which is also shown numerically. Statistical significance was tested using an unpaired, non-parametric, Mann-Whitney two-tailed U test: *p≤0.05, **p≤0.01, ***p≤0.001.
Whilst the majority of CD45+ population in LNs are naïve lymphocytes and thus expected to be highly migratory, the CD45- stromal compartment would be expected to be obligatorily tissue-resident. Therefore we assessed Kaede expression in LN stromal cells to further confirm that our experimental model properly distinguished between resident and migratory populations. Blood endothelial cells, fibroblastic reticular cells and lymphatic endothelial cells were all found to remain >96% Kaede Red+ at 72 hrs post photoconversion (Fig. S1) consistent with being resident populations.
To specifically assess the relative migratory abilities of individual cell types, different immune cell populations were first identified, and then Kaede Green versus Kaede Red expression was analysed. These data showed that for αβ and γδ T cells, the vast majority (>93%) had trafficked into the LN within the preceding 72 hrs (Fig. 1G). Surprisingly, analysis of the ILC compartment revealed that approximately half the population had newly arrived into the tissue within the 72 hr time frame, arguing that a substantial proportion of ILCs in the LN were not tissue-resident (Fig. 1G). To better understand the kinetics of ILC trafficking into LNs, T cell and ILC populations were assessed at different times post photoconversion (Fig. S2). Whilst over 90% of the LN T cell compartment were Kaede Green+ 24 hrs after photoconversion, only 20% of the ILCs were Kaede Green+, suggesting that the rate of replacement of LN ILCs within peripheral LNs with newly migrating cells, was substantially slower than for naïve T cells. Given the importance of CCR7 in the recruitment of cells into LNs, either via the lymphatics or from the blood (30), the bLN of CCR7-/- Kaede mice was assessed at 72 hrs post-photoconversion (Fig. 1H). For both T cells and ILCs, the proportion of Kaede Red+ cells increased compared to WT Kaede controls, however this reflected the significantly reduced cellularity of the CCR7-/- tissue due to the inability of cells to enter and exit many tissues in the body (Fig. 1I). Nonetheless, consistent with a requirement for CCR7 for T cell and ILC entry into LNs, the number of Kaede Green+ T cells or ILCs was substantially reduced (Fig. 1J).
Combined, these data reveal that both migratory and resident ILC populations exist within peripheral LNs and the entry of migratory ILCs into these tissues is CCR7-dependent.
ILCs egress from LNs in an S1P-dependent manner
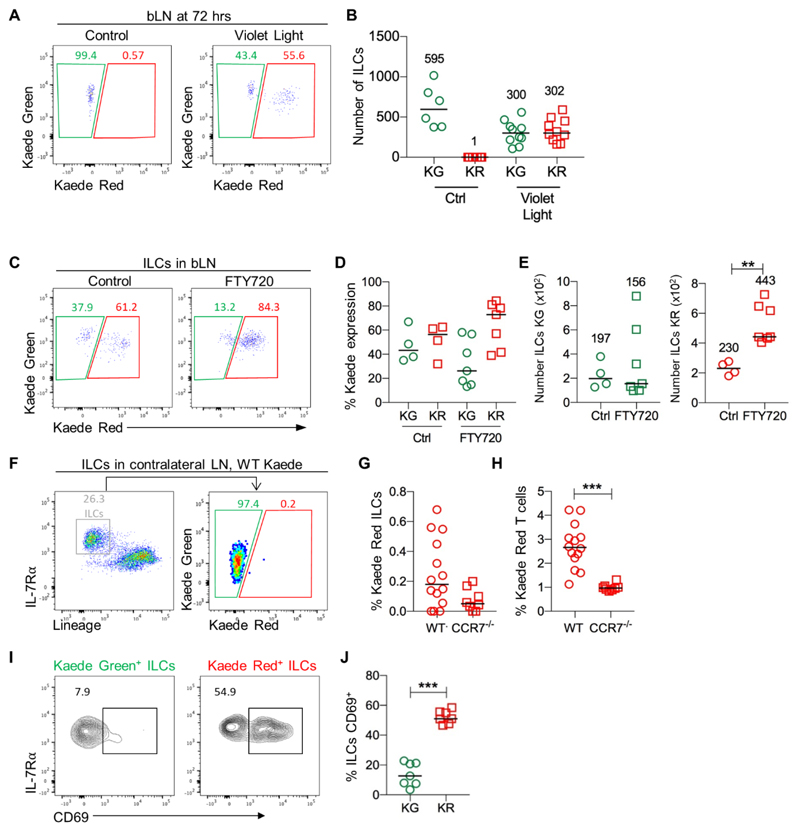
Since labelling of the LN was performed following a small excision in the skin, it was possible that the Kaede Green+ ILCs observed in the LN reflected additional migration of ILCs in response to local tissue damage, rather than normal homeostatic trafficking of these cells into the tissue. Enumeration of the number of Kaede Green+ versus Kaede Red+ ILCs in control mice (no photoconversion) compared with 72 hrs post-photoconversion indicated that the total number of ILCs in the LN was not increased by the process of labelling (Fig. 2A, B), suggesting new cells had not migrated into the tissue. Furthermore, quantitation of ILC numbers in the bLN of WT mice that had never undergone surgery versus those that had experienced a skin incision and photoconversion 72 hrs previously, revealed no differences in ILC numbers (Fig. S3).
Figure 2. ILC frequency in LNs is maintained by balanced ingress and egress.
To further investigate the migratory ILCs identified within the labelled bLN, the effect of photoconversion on the number of ILCs was assessed. (A) Expression of Kaede Green versus Kaede Red protein by ILCs (CD45+ IL-7Rα+ Lin{B220, CD11b, CD11c, CD3, CD5, CD19, Ter119, Gr1, CD49b, F4/80, FcεRI}-) isolated from the bLN of control Kaede mice (no photoconversion) and Kaede mice at 72 hrs post photoconversion. (B) The number of Kaede Green+ and Kaede Red+ ILCs recovered from the bLN of control (n=6) versus photoconverted bLNs at 72 hrs post labelling (n=10). To investigate egress of ILCs from the bLN, Kaede mice were maintained on vehicle control (H2O) or FTY720, the bLN was photoconverted and then analyzed 72 hrs later. (C) Representative flow cytometry plots showing expression of Kaede Green versus Kaede Red protein by ILCs in the bLN 72 hrs after photoconversion. (D) Percentage of Kaede Green+ or Kaede Red+ ILCs in the bLN of Ctrl (n=4) and FTY720 (n=7) mice. (E) Total numbers of Kaede Green+ and Kaede Red+ ILCs in the bLN. To assess whether ILCs recirculate through peripheral LNs, the contralateral LNs of Kaede mice were analyzed at 72 hrs post photoconversion of the bLN, with combined B- and T-cell depletion used to enrich for ILCs. (F) Expression of Kaede Green versus Kaede Red protein by ILCs (CD45+ IL-7Rα+ Lin{B220, CD11b, CD11c, CD3, CD5, CD19, Ter119, Gr1, CD49b, F4/80, FcεRI}-) isolated from contralateral LNs. Percentage of Kaede Red+ ILCs (G) and T cells (H) in WT Kaede (n=14) and CCR7-/- Kaede (n=8) contralateral LNs. (I) Representative flow cytometry plots of Kaede Green+ (left) and Kaede Red+ (right) ILC expression of CD69. (J) Percentage of Kaede Green+ and Kaede Red+ ILCs expressing CD69 (n=7). Data are pooled from 2 independent experiments. Bars on scatter plots represent the median, which is also shown numerically. Statistical significance was tested using an unpaired, non-parametric, Mann-Whitney two-tailed U test: **p≤0.01, ***p≤0.001.
These data indicate that ILC migration into the tissue was matched by the loss of ILCs either through cell death or egress from the LN. The egress of B and T cells from LNs is dependent upon the sensing of sphingosine-1-phosphate (S1P) by S1P receptor 1 (S1P1) (31), whilst the egress of cNK cells from LNs is dependent upon S1P receptor 5 (S1P5) (32, 33). Expression of S1P1 was detected on a proportion of ILC1s, ILC2s and ILC3s isolated from the bLN (Fig. S4), suggesting that ILCs might use S1P:S1P1 interactions to mediate egress from peripheral LNs, as described for T cells. To test whether ILC egress from LNs, photoconversion of the bLN was performed on mice maintained on vehicle control (H2O) or the S1P receptor agonist FTY720, a small molecule inhibitor that blocks lymphocyte recirculation through inhibiting three of the five S1P receptors: S1P1, S1P3 and S1P4 (31, 34). The total number of T cells within the LN was not significantly altered by FTY720 treatment (Fig. S5), however, analysis of Kaede Red expression confirmed this was due to the expected retention of T cells within the LNs (Fig. S5), which in turn meant that far fewer Kaede Green+ T cells were migrating into the tissue (Fig. S5). Analysis of ILCs revealed an increase in the proportion of Kaede Red+ ILCs (Fig. 2C, D) and a significant increase in the total number of Kaede Red+ ILCs in the LN whilst the number of Kaede Green+ ILCs remained unchanged through FTY720 treatment (Fig. 2E). Thus, inhibition of S1P receptor-mediated exit from the tissue resulted in the accumulation of ILCs within the LN, arguing that under normal conditions, these cells egress from the tissue. To further investigate expression of S1P receptors by ILCs, gene expression analysis was performed on sorted ILC1 (NK1.1+NKp46+) and non-ILC1 (NK1.1-NKp46-) ILC populations isolated from peripheral LNs by FACS. S1P1 and S1P4, both of which are inhibited by FTY720, were observed to be most highly expressed by LN ILC populations, while relatively little expression of S1P5 was detected (Fig. S6). Interestingly, the sorted ILC1 population, which expressed significantly higher levels of IFNγ and T-bet than the other LN ILC, also expressed significantly higher levels of CD62L, required for entry into LNs from HEV (Fig. S6).
Since ILCs that have left the photoconverted LN can be identified as Kaede Red+, the ability of ILCs to recirculate through peripheral LNs was tested by assessing Kaede Red+ ILCs in a pool of contralateral LNs. ILCs were enriched (through depletion of B and T cells) to aid identification of rare Kaede Red+ ILCs and this approach could detect a small Kaede Red+ ILC population that constituted only a very small proportion of the ILCs isolated from the contralateral LNs (0-0.7% of the population, Fig. 2F, G) although this was more than detected in the contralateral LNs of CCR7-/- Kaede mice. This low frequency of recirculating ILCs may represent either the limitations of tracking rare populations, alongside some ILCs migrating to tissues other than LNs. T cells recirculating from the photoconverted LN comprised approximately 2.5% of the T cell population in contralateral LNs (Fig. 2H and Fig. S5). To further assess expression of molecules associated with retention of cells within tissues, expression of CD69 was assessed on Kaede Green+ and Kaede Red+ ILCs. Notably, Kaede Green+ ILCs expressed very little CD69, whilst the majority of Kaede Red+ ILCs were CD69+, consistent with a tissue resident phenotype (Fig. 2I, J).
In summary, these data show that the migration of ILCs into peripheral LNs is accompanied by the egress of these cells in an S1P receptor-dependent manner and at least some of these ILCs then recirculate through other peripheral LNs.
ILC1s are the migratory ILC subset
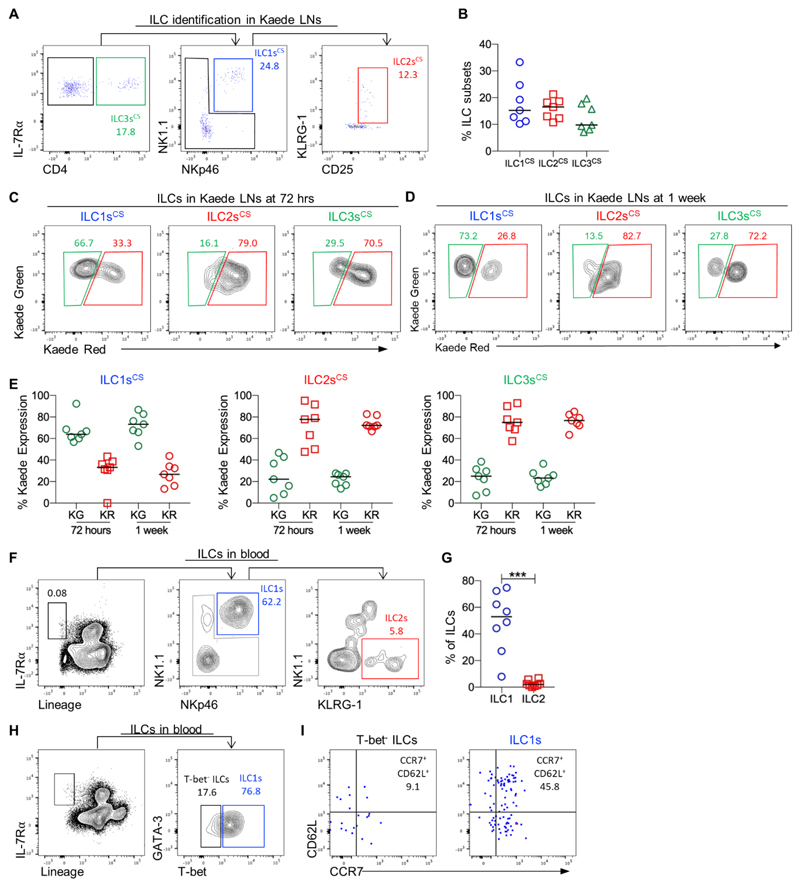
To determine whether all ILC populations were equally migratory we sought to define the Kaede Green+ and Kaede Red+ ILCs within the bLN. ILC subsets in the bLN can be readily distinguished by staining for transcription factors (TFs) (Fig. S7) (18). Importantly, T-bet+ ILC1s in the bLN lacked expression of Eomes (i.e. T-bet+Eomes-) demonstrating that the ILC1s we had identified, were not misidentified cNK cells (Fig. S7). In photoconverted LNs, methods to preserve Kaede protein expression resulted in suboptimal TF detection. Therefore, a panel of surface markers that identified the ILC subsets in the bLN of WT mice was devised (Fig. S7) and used to identify different ILC subsets only by surface markers (Fig. 3A, B). Analysis of Kaede Green versus Kaede Red protein expression in bLN ILCs at 72 hrs post-photoconversion revealed that there were relatively few migratory ILC2s and ILC3s (~20% Kaede Green+), whilst ILC1s were much more migratory since over 60% were Kaede Green+ (Fig. 3C). Similar proportions of Kaede Green+ ILCs were also observed at 1 wk post photoconversion (Fig. 3D), indicating that that LNs contained both migratory and resident ILC populations, with different subsets showing distinct migratory properties (Fig. 3E). ILC tissue residency within the LN was then further assessed using a second photoconvertible mouse strain, the H2B-Dendra2 model (35). The fusion of the Dendra protein to histones enabled intracellular staining of TFs to be used to more definitively identify ILC subsets. In H2B-Dendra2 mice, ILC subsets could be split into resident and migratory populations in a comparable manner to that observed in the Kaede mice (Fig. S8). Notably, identification of ILC2s based upon GATA-3 expression, rather than surface markers used in Kaede mice, indicated a larger migratory ILC2 population within the LN (Fig. S9). Since expression of Kaede Red protein is lost over several days in rapidly proliferating cells due to label dilution, the emergence of Kaede Green+ ILCs could theoretically be explained by highly proliferative populations in the tissue (28). Analysis of Ki-67 expression within the bLN revealed that the majority of each ILC subset was Ki-67- and the proportions of Ki-67+ ILC1s and ILC2s were comparable, despite very different patterns of Kaede Red and Kaede Green expression at 72 hrs post photoconversion (Fig. S10). Thus the high proportion of Kaede Green+ ILC1s cannot be explained by Kaede red protein dilution through proliferation.
Figure 3. ILC1s traffic through peripheral LNs from the circulation.
To test whether all ILC subsets showed an equal ability to traffic through the peripheral LNs, an established panel of surface markers was used to identify which ILC subsets were migrating into the bLN. (A) Representative flow cytometry plots showing identification of ILC1 (CD45+ Lin{B220, CD11b, CD11c, CD3, CD5, CD19, Ter119, Gr1, CD49b, F4/80, FcεRI}- IL-7Rα+ CD4- (NK1.1, NKp46)+), ILC2 (CD45+ Lin{B220, CD11b, CD11c, CD3, CD5, CD19, Ter119, Gr1, CD49b, F4/80, FcεRI}-IL-7Rα+ CD4- (NK1.1, NKp46)-(KLRG-1, CD25)+), and ILC3 (CD45+ Lin{B220, CD11b, CD11c, CD3, CD5, CD19, Ter119, Gr1, CD49b, F4/80, FcεRI}-IL-7Rα+ CD4+) subsets in the bLN. (B) The percentage of each ILC subset identified within the bLN of Kaede mice using the surface marker staining. (C-D) Kaede Green versus Kaede Red protein expression by ILC1s, ILC2s and ILC3s at 72 hrs and 1 week after photoconversion of the bLN. (E) The percentage of Kaede Green+ and Kaede Red+ ILC1s, ILC2s and ILC3s in the bLN at 72 hrs and 1 week post photoconversion. Data are pooled from 2 independent experiments, n = 7. To investigate the source of ILC1s migrating into the bLN, ILC populations in murine blood were analysed. (F) Representative flow cytometry plots showing identification of putative ILC1s (CD45+ IL-7Rα+ (NKp46NK1.1)+Lin{B220, CD11b, CD11c, CD3, CD5, CD19, Ter119, Gr1, CD49b, F4/80, FcεRI}-) and ILC2s (CD45+ IL-7Rα+ NK1.1- NKp46- KLRG-1+ Lin-). (G) The proportion of ILC1s and ILC2s within the ILC population isolated from the blood, n = 8. (H) Representative flow cytometry plots showing expression of T-bet versus GATA-3 by blood ILCs. To isolate sufficient numbers of ILCs, blood samples from 3 mice were enriched for the CD45+ fraction separately and then pooled for staining and analysis. (I) Representative flow cytometry plots showing expression of CD62L and CCR7 by T-bet+ ILC1s versus T-bet- ILCs isolated from blood. All data are pooled from 2 independent experiments. Values on flow cytometry plots represent percentages, bars on scatter plots represents the median. Statistical significance was tested using an unpaired, non-parametric, Mann-Whitney two-tailed U test: *p≤0.05, **p≤0.01, ***p≤0.001.
The IL-7Rα+T-bet+Eomes- ILC1s assessed within the LN are developmentally distinct from cNK cells, despite both being grouped in the ILC1 subset and cNK cells having been previously shown to enter LNs direct from the circulation (27). To directly compare ILC1 and cNK cell migration, these two populations were identified within the photoconverted bLN at 72 hrs post labelling and in a pool of contralateral LNs (Fig. S11). Notably, while ILC1s were 60% Kaede Green+, cNK cells were 90% Kaede Green+, indicating that while the LN compartment of ILC1s contains both migratory and resident populations, the vast majority of cNK cells are highly migratory. Analysis of contralateral LNs identified a small Kaede Red+ ILC1 population and a larger Kaede Red+ cNK cell population, demonstrating that both populations recirculate through peripheral LNs.
Given that cNK cells are reported to enter LNs via HEV (27), ILC populations in mouse blood were assessed to investigate whether ILC1s in the murine circulation might be the source of the ILCs trafficking into the bLN. A very small (<0.05 – 0.011 % total CD45+ cells) IL-7Rα+Lin- population was detected in the circulation, the majority of which expressed both NK1.1 and NKp46 at steady state alongside a few putative ILC2s (NK1.1-KLRG-1+) (Fig. 3F, G). These ILCs also expressed T-bet (Fig. 3H) providing further evidence of an ILC1 phenotype and consistent with previous data suggesting ILC1s were present in the circulation (9, 11). To investigate whether ILC1s in the blood were equipped to enter LNs directly from the circulation via HEV, expression of CD62L and CCR7 were assessed. Notably, approximately half of blood ILC1s expressed both CD62L and CCR7, whereas the few other ILCs (IL-7Rα+Lin- cells) detectable in blood appeared to lack expression of these molecules (Fig. 3I). Combined, our analysis of ILC subsets revealed that ILC1s show behavioral similarities to cNK cells and a circulating ILC1 population in the blood is able to traffic into and out of peripheral LNs.
ILCs do not migrate from the skin to peripheral LNs under steady state conditions
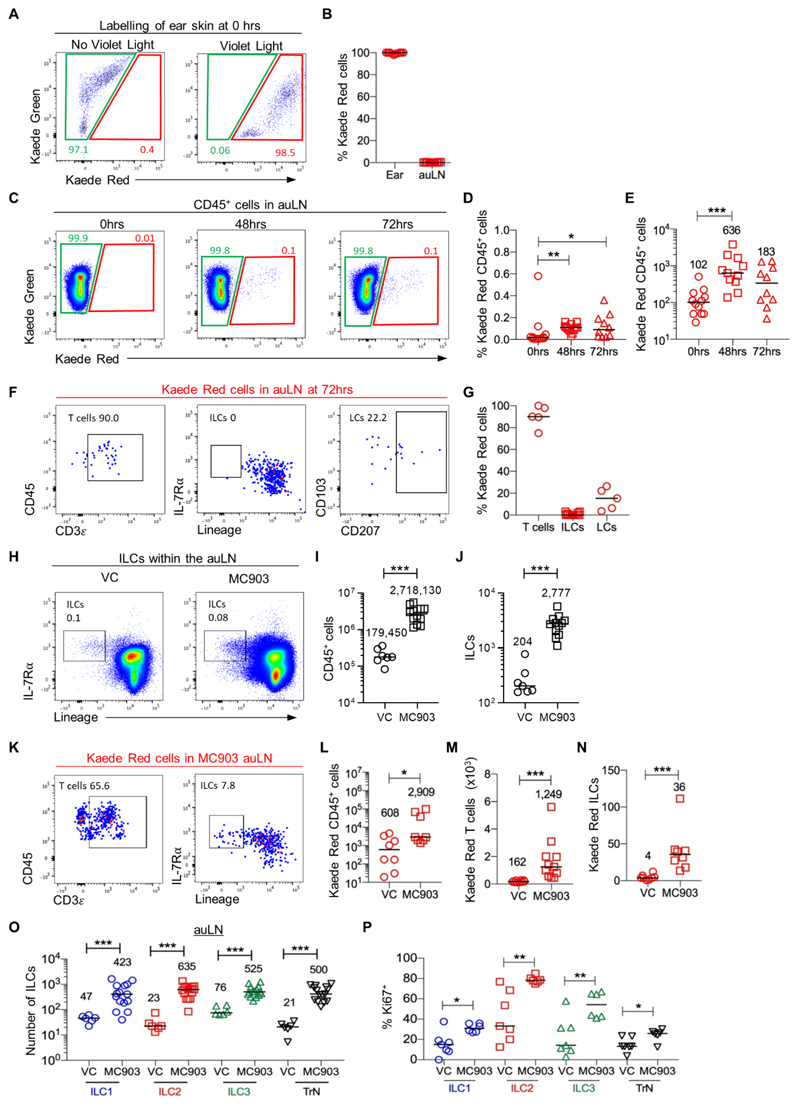
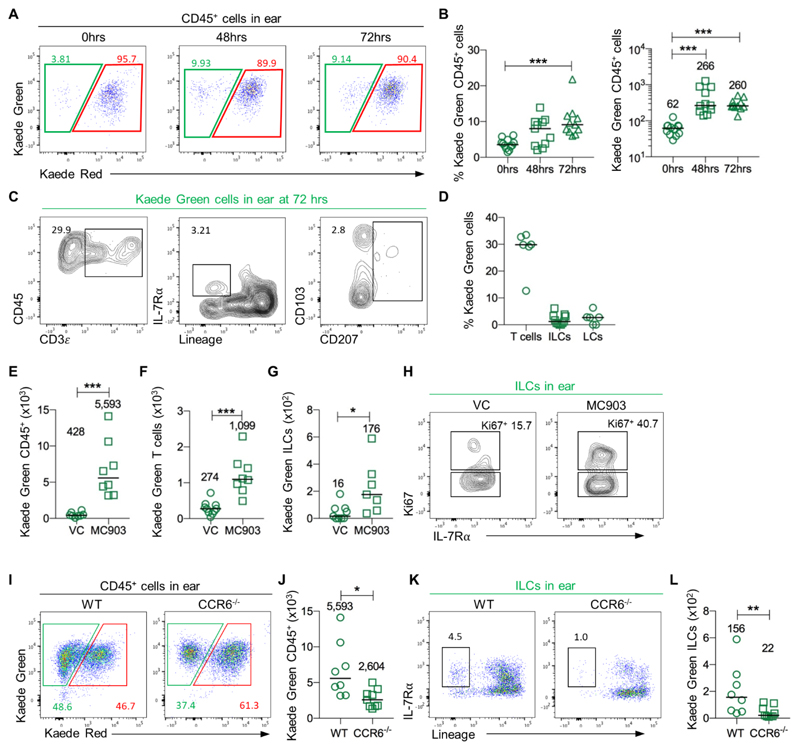
Whilst our data was consistent with ILC migration into LNs arising from the circulation, it was possible that ILCs also trafficked from skin (36) to peripheral LNs via the lymphatics, as observed for dermal DCs and LCs (37). To directly test this, photoconversion of the entire CD45+ compartment of ear skin was established, enabling the analysis of cellular migration into the draining auricular LN (auLN) as well as identification of new cells moving into the ear skin (Fig. 4A, B). To directly test the migration of cells from the ear skin to the auLN, the ear was photoconverted and the auLN analysed 48 and 72 hrs later. A modest, but distinct Kaede Red+ population was detected within the auLN (Fig. 4C-E), with T cells, but not ILCs detected amongst this small migratory population (Fig. 4F, G). These data argue that under steady state conditions ILCs do not migrate from skin to peripheral LNs. To determine whether ILCs in skin only migrate to peripheral LNs during inflammation, the MC903-induced atopic dermatitis model (38) was established in Kaede mice, combining 5 days of MC903 ear painting with photoconversion of the skin. The short five day treatment was chosen to enable early changes in ILC populations to be assessed and caused a substantial increase in the size of the auLN, significant increases in ear thickness, and increased numbers of basophils and other hematopoietic cells in the ear skin including ILC2s (Fig. S12) (39). The draining auLN had increased substantially in cellularity, including the number of ILCs which had increased approximately 15-fold (Fig. 4H-J). Analysis of Kaede Red+ cells in the auLN revealed increased numbers of cells trafficking to the draining LN (Fig. 4K, L), which were mostly T cells but also ILCs (Fig. 4M, N). Detection of such few migratory ILCs compared with the substantial increases in all ILC subsets observed in the auLN (Fig. 4O), argued that migration through the lymphatics was unlikely to be the main mechanism increasing ILC numbers in the LN. Notably, ILCs had significantly increased their expression of Ki-67 (Fig. 4P) suggesting at least that the increased numbers of ILCs in the auLN reflect local expansion in situ.
Figure 4. Minimal ILC migration from skin to the draining LN even under conditions of inflammation.
To directly test whether ILCs migrate from skin to peripheral LNs, the ears of Kaede mice were photoconverted and the presence of labelled cells assessed in the auLN. (A) Expression of Kaede Green versus Kaede Red protein by CD45+ cells isolated from ear skin without (left panel) or immediately after (right panel) photoconversion. (B) Percentage of CD45+ cells expressing Kaede Red protein in the ear and auLN immediately after photoconversion (n=12). (C) Expression of Kaede Green versus Kaede Red protein by CD45+ cells in the auLN at 0, 48 and 72 hrs after photoconversion of the ear skin. The percentage (D) and total number (E) of Kaede Red+ CD45+ cells in the auLN over the time course of 0 hrs (n=12), 48 hrs (n=11) and 72 hrs (n=10). (F) Representative flow cytometry plots assessing the phenotype of Kaede Red+ CD45+ cells in the auLN, with identification of T cells (CD3+), ILCs (IL-7Rα+ Lin{B220, CD11b, CD11c, CD3, CD5, CD19, Ter119, Gr1, CD49b, F4/80, FcεRI}-) and LCs (CD45+ MHCII+ CD11c+ CD207+). (G) Percentage of Kaede Red+ CD45+ cells in the auLN that were T cells (n=5), ILCs (n=12) and LCs (n=5). To determine whether inflammation in the skin resulted in the migration of ILCs to the draining LN, the ears of mice were treated with vehicle control (VC) or MC903, with photoconversion of the ear after 48 hrs of treatment. (H) Representative flow cytometry plots showing ILCs within VC (left) and MC903 (right) treated auLN. Enumeration of CD45+ cells (I) and ILC (J) recovered from the auLN of mice treated with VC (n=7) or MC903 (n=12). (K) Representative flow cytometry plots assessing the phenotype of Kaede Red+ CD45+ cells in the auLN of MC903 treated mice, with the identification of T cells and ILCs. Enumeration of Kaede Red+ (L) CD45+ cells in VC (n=8) or MC903 (n=8), (M) T cells in VC (n=10) or MC903 (n=11) and (N) ILC in VC (n=8) or MC903 (n=8) within the auLN. (O) Enumeration of ILC subsets in the auLN of mice treated with vehicle control (VC, n=6) and MC903 (n=15). (P) Expression of Ki-67 by ILCs in the auLN of mice treated with VC (n=6) or MC903 (n=7). Data are pooled from a minimum of 2 independent experiments. Values on flow cytometry plots represent percentages; bars on scatter plots represent the median, which is also shown numerically. Pairs of samples were compared using an unpaired Mann-Whitney U test. When comparing more than two sets of data, statistical significance was tested using Kruskal-Wallis one-way ANOVA with post hoc Dunn’s test: *p≤0.05, **p≤0.01, ***p≤0.001.
Combined, these data reveal that under steady state ILCs traffic from the circulation into peripheral LNs rather than through lymphatic drainage from the skin. Whilst inflammation in the skin can cause some ILCs to migrate to the draining LN, the substantial increase in ILC numbers reflects other mechanisms such as local proliferation and recruitment from the blood.
Recruitment of ILCs into inflamed skin is dependent upon CCR6
Simultaneous photolabelling of all the hematopoietic cells in the ear enabled migration of cells into the ear skin to be assessed. At 48 and 72 hrs post photoconversion, the proportion of Kaede Green+ CD45+ cells in the ear had significantly increased compared with 0 hr controls, indicating the trafficking of new cells into this site at steady state (Fig. 5A, B), of which ILCs were a small (~3%) proportion (Fig. 5C, D). This may represent redistribution of ILCs within the skin, or trafficking of these cells from the following education in the draining LN (40). To test whether the recruitment of ILCs into skin was enhanced during inflammation, Kaede Green+ cells migrating into the ear after administration of MC903 was assessed. A significant increase in the number of Kaede Green+ CD45+ cells (Fig. 5E), T cells (Fig. 5F) and ILCs (Fig. 5G) were observed in MC903-treated mice versus vehicle control, suggesting that ILCs were recruited into the inflamed ear tissue. Over half of the population of Kaede Green+ ILCs lacked Ki-67 expression (Fig. 5H), arguing that the increase in Kaede Green+ ILCs was not solely attributable to proliferation. To investigate the potential mechanism regulating ILC recruitment to the inflamed tissue, we assessed CCR6 expression on ILCs given the proposed role of its ligand, CCL20 in regulating homing to the skin (41). Strikingly, while expression of CCR6 is restricted to ILC3s in the mLN, all ILC subsets isolated from ear skin or the auLN expressed CCR6, indicating that this might be a key mechanism by which these cells are recruited to the skin (Fig. S13). To directly test this CCR6-/- Kaede mice and WT Kaede mice were treated with MC903 as described above. Analysis of Kaede Green+ cells in the ear of CCR6-/- Kaede mice revealed a significant drop in the total number of CD45+ cells which included a significant decrease in the number of ILCs (Fig. 5I-L), indicating that CCR6 is important for the recruitment of ILCs into inflamed skin.
Figure 5. CCR6-dependent recruitment of ILCs into the inflamed ear.
To investigate mechanisms controlling the recruitment of ILCs into the ear, the entire ear CD45+ population was labelled through photoconversion, with Kaede Green+ cells identified at subsequent time points as cells that had newly entered the tissue. (A) Expression of Kaede Green versus Kaede Red protein by CD45+ cells in the ear at 0, 48 and 72 hrs after photoconversion of the ear skin. (B) The percentage (left) and total number (right) of Kaede Green+ CD45+ cells in the auLN over the time course of 0 hrs (n=12), 48 hrs (n=11) and 72 hrs (n=11). (C) Representative flow cytometry plots assessing the phenotype of Kaede Green+ CD45+ cells in the ear with identification of T cells (CD3+), ILCs (IL-7Rα+ Lin{B220, CD11b, CD11c, CD3, CD5, CD19, Ter119, Gr1, CD49b, F4/80, FcεRI}-) and LCs (CD45+ MHCII+ CD11c+ CD207+). (D) Percentage of Kaede Green+ CD45+ cells in the ear that were T cells (n=6), ILCs (n=11) and LCs (n=6). To assess migration under inflammatory conditions MC903 was applied to the ears for 5 consecutive days to induce atopic dermatitis, with photoconversion of the ear 48 hrs after treatment was started. Number of Kaede Green+ (E) CD45+ cells in VC (n=9) or MC903 (n=7), (F) T cells in VC (n=10) or MC903 (n=8) and (G) ILCs in VC (n=9) or MC903 (n=7) within the ear. (H) Representative flow cytometry plots of Kaede Green+ ILCs Ki-67 expression within the ear of VC and MC903 treated mice. To test the requirement for CCR6 in ILC recruitment into the ear, MC903 was applied to the ears of Kaede and CCR6-/- Kaede mice for 5 consecutive days to induce atopic dermatitis, with photoconversion of the ear 48 hrs after treatment was started. (I) Expression of Kaede Green and Kaede Red protein by CD45+ cells within MC903-treated ears. (J) Number of Kaede Green+ CD45+ cells (Kaede n=8, CCR6-/- Kaede n=9). (K) Representative flow cytometry plots showing Kaede Green+ ILCs. (L) Number of Kaede Green+ ILC (Kaede n=8, CCR6-/- Kaede n=9). Each data point represents cells isolated from 1 ear and 1 auLN. Data are pooled from a minimum of 2 independent experiments. Values on flow cytometry plots represent percentages, bars on scatter plots represents the median, which is also shown numerically. Pairs of samples were compared using an unpaired Mann-Whitney U test. When comparing more than two sets of data, statistical significance was tested using Kruskal-Wallis one-way ANOVA with post hoc Dunn’s test: *p≤0.05, **p≤0.01, ***p≤0.001.
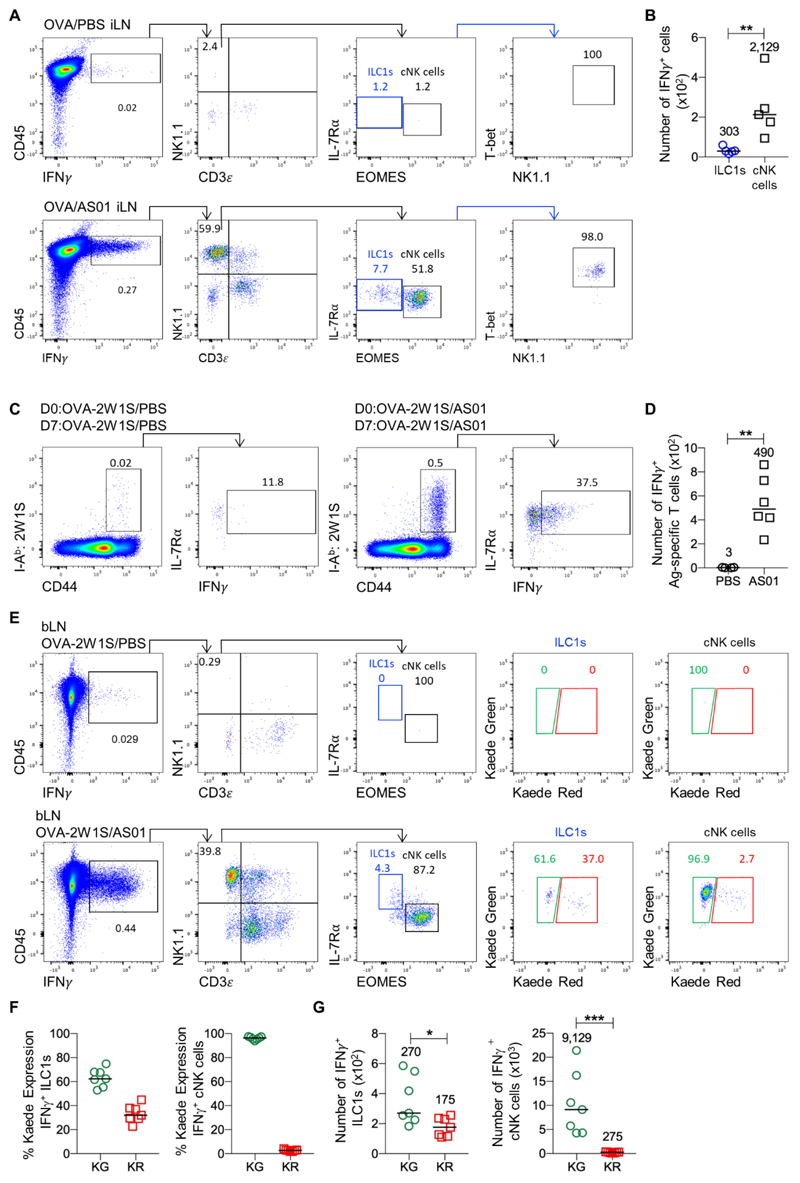
ILC1s are a rapid source of IFN-γ in the draining LN after immunisation
Combined, our data revealed that a migratory ILC1 population was present within peripheral LNs, alongside a population that remained resident in the tissue for at least 1 wk. The ability of cNK cells to rapidly produce effector cytokines is thought to modulate the microenvironment of the LN and shape the T cell responses generated in these tissues (42). Thus, we asked whether ILC1s in peripheral LNs also contributed to the rapid early release of cytokines in response to immunization. NK cell production of IFN-γ within the draining peripheral LNs was recently identified as a key mechanism through which the adjuvant AS01 drives activation of DCs and the development of Th1 immunity (43). To assess whether the ILC1 population in peripheral LNs can also contribute to this early IFN-γ response, WT mice were immunized intramuscularly with the antigen OVA-2W1S, in the presence of PBS or with AS01. Consistent with the previous study, AS01 induced the rapid production of IFN-γ by NK1.1+CD3- cells in the draining LN within hrs of immunization (Fig. 6A). While the majority of the NK1.1+CD3- cells expressed Eomes, a distinct population of NK1.1+CD3-IL-7Rα+Eomes- cells expressing T-bet were evident, thus revealing that ILC1s in the LN also release IFN-γ in response to AS01 (Fig. 6A, B). To confirm that this response had resulted in a Th1-skewed T cell response, analysis of 2W1S-specific CD4 T cells in the draining LN at 7 days post immunization using MHCII tetramers revealed that the AS01 adjuvant had generated a robust Th1 effector T cell response (Fig. 6C, D). Finally, to determine whether the migratory and resident ILC1 populations showed functional differences in their production of IFN-γ, Kaede mice were immunized with OVA-2W1S in the presence of PBS or AS01, 72 hrs post photoconversion of the draining bLN. Both Kaede Green+ and Kaede Red+ ILC1 in the draining LN expressed IFN-γ (Fig. 6E), although numerically, there were significantly more Kaede Green+ ILC1s amongst the IFN-γ producing cells than Kaede Red+ ILC1 (Fig. 6F). The vast majority of IFN-γ produced by cNK cells came from Kaede Green+ cells (Fig. 6G). Therefore, both migratory and resident ILC1s appear able to functionally contribute to the shaping of adaptive immune responses within LNs through rapid effector cytokine release.
Figure 6. ILC1s in the draining LN are a rapid source of IFN-γ after immunization.
To investigate the function of ILC1 within peripheral LNs, mice were injected intramuscularly with 20 μg OVA-2W1S/PBS or 20 μg OVA-2W1S/AS01 and culled either 6 hrs post immunization to assess the early innate IFN-γ response or after 7 days following a second immunization performed 6 hrs prior to analysis. (A) Representative flow cytometry plots showing inguinal LN cells from mice given OVA-2W1S/PBS (top) or OVA-2W1S/AS01 (bottom), with identification of different IFN-γ producing populations including cNK cells (CD45+ IFN-γ+ CD3ε- NK1.1+EOMES+) and ILC1s (CD45+ IFNγ+ CD3ε- NK1.1+ EOMES- T-bet+) shown. (B) Total number of IFN-γ+ ILC1s and cNK cells in OVA/ASO1 treated mice (n=5). (C) Representative flow cytometry plots showing IFN-γ production by 2W1S-specific CD4+ T cells at day 7 post initial immunization with (AS01) or without (PBS) the adjuvant AS01. (D) Total number of IFN-γ producing 2W1S-specific CD4 T cells in the draining inguinal LN of PBS (n=4) and AS01 mice (n=6). To test whether both migratory and resident ILC1 subsets were able to produce IFN-γ, the bLN of Kaede mice was photoconverted and 3 days later mice were then immunized in the paw pad with either 20 μg OVA-2W1S/PBS or 20 μg OVA-2W1S/AS01 and analyzed 6 hrs later. (E) Representative flow cytometry plots showing bLN cells from mice given OVA-2W1S/PBS (top) or OVA-2W1S/AS01 (bottom), with identification of ILC1 and cNK populations producing IFN-γ and their expression of Kaede Green versus Kaede Red protein. (F) Percentage Kaede Green (migratory) and Kaede Red (resident) ILC1s and cNK cells amongst IFN-γ producing cells. (G) Total numbers of IFN-γ+ ILC1s and cNK cells expressing either Kaede Green (migratory) or Kaede Red (resident). All sets of data are pooled from 2 independent experiments. Values on flow cytometry plots represent percentages, bars on scatter plots represents the median, which is also shown numerically. Statistical significance was tested using an unpaired, non-parametric, Mann-Whitney two-tailed U test: *p≤0.05, **p≤0.01, ***p≤0.001.
Discussion
Here we provide compelling evidence that peripheral LNs contain migratory ILC populations. Our studies revealed the balanced ingress and egress of ILCs from LNs with the majority of migratory ILCs belonging to the ILC1 subset over the time frame assessed. ILC1s are the most prevalent subset in murine circulation and like cNK cells, recirculate through peripheral LNs utilising CCR7 and CD62L to enter the tissue, whilst egressing in a S1P1 dependent manner. Functionally, ILC1s in peripheral LNs were able to rapidly produce effector cytokines in response to immunization, as observed for cNK cells, supporting the initial priming of CD4 T cells. Combined, these data provide critical further insight into the behavior of ILCs within lymphoid tissues.
Following their relatively recent identification (3), understanding the functions of ILCs within the immune system has become a key area of immunological research. Despite ILCs being present within all lymphoid tissues, direct evidence of their function here is very limited. Having established techniques to track the movement of cells into and out of peripheral LNs, we sought to understand more of the basic behavior of ILCs within these tissues in an effort to better understand their roles within such tissue. Photoconvertible mice provide a means of site specifically labelling cells and the direct assessment of cellular migration in vivo. Tracking ILCs within the photoconverted bLN over time revealed that within 72 hrs, approximately 50% of the ILCs had migrated into the tissue since photoconversion. Whilst the fluorescence level of Kaede Red protein will dilute as cells proliferate, the majority of ILCs within the LN were Ki-67- and expression of Kaede Red did not correlate with proliferation amongst ILC subsets. Although a clear caveat of the photoconvertible mouse models is the potential loss of the light-induced label, studying ILC migration under steady state conditions over a time frame of up to 1 wk helped to mitigate this problem. Treatment with the S1P1 agonist FTY720 resulted in the accumulation of ILCs within the LN, consistent with blocked egress. These data strongly indicated the balanced migration of ILCs into and out of peripheral LNs. Combined with our data detecting minimal ILC trafficking from skin to the draining peripheral LN, the detail of which ILC subsets migrate really reflects the composition of the ILC compartment within murine blood. Here, the majority of ILCs were of an ILC1 phenotype and expression of the key mechanisms required to enter LNs direct from the circulation (CD62L and CCR7) was detected. Only a minority of the ILC2 and ILC3 populations within the LN were found to be migratory over 1 wk, indicating that bona fide LN resident ILC populations exist. These observations would have been strengthened by approaches facilitating the tracking of ILC populations for longer periods of time, but concerns regarding the potential loss of the photoconverted red form of the fluorochromes inhibited such studies. In the future, novel mice enabling ate mapping of cre recombinase activity induced through photoactivation might provide a superior approach to the long term tracking of tissue residency.
Whilst the IL-7Rα+T-bet+Eomes- ILC1 population detected within LNs is developmentally distinct from cNK cells (44), it is striking that the data provided here reveals a similar migratory behavior. Direct comparison of the ‘helper’ ILC1 (17) and cNK cell migration indicated that cNK cells were more migratory, since a higher proportion of the cNK cells had newly entered the tissue within 72 hrs of labelling. Our data also provides direct evidence that both cNK cells and ILC1s recirculate through peripheral LNs, a behavior demonstrable in photoconvertible mice. While only a few labelled ILCs were detected within contralateral LNs this likely reflects the technical limitations of trying to track a population of a few hundred cells within the entire mouse and the larger cNK cell population was more readily detected, as were T cells. It remains possible however, that some ILCs exit peripheral LNs and traffic to tissues that were not assessed. The data presented here do not contradict published studies of parabiotic mouse models since here ILCs in the mLN and spleen, but not peripheral LNs, were assessed and lymphatic migration would not be captured (9). Consistent with these previous studies, our data also indicate that the ILC2 and ILC3 compartments of peripheral LNs contain resident populations. Interestingly, while both the Kaede and H2B-Dendra2 mouse models indicated that the majority of ILC2s and ILC3s were tissue-resident, identification of ILC2s by GATA-3 expression in the H2B-Dendra2 mice did identify a sizeable migratory ILC2 population in the LNs (approximately 40% of total LN ILC2). Few ILC2 are present in LNs compared to tissues such as the lung and the role of these cells is unclear. Our data indicates that there is some movement of these cells between the circulation and peripheral LNs. The ILC3 subset displayed the least evidence of migratory behavior within the timeframe assessed and it remains possible that the cells present here are the remnants of the lymphoid tissue inducer population that initially seeded the LN anlagen in the embryo.
The function of the helper ILC populations in LNs is poorly understood, whilst cNK cells have been considered to be key early producers of effector cytokines within the LN to support the initiation of T cell responses. NK cells were initially described as being specifically recruited into draining LNs (42) in response to immunization. Subsequent studies identified that cNK cells enter peripheral LNs via the HEVs in a CD62L- and CCR7-dependent manner (27) and egress dependent upon S1P5 (32, 33). Thus increases in cNK cell numbers in the draining LN may reflect the restricted exit of cells to drive their accumulation, rather than enhanced recruitment. Here we demonstrate that both resident and migratory ILC1s rapidly produced IFN-γ in response to immunization, as described for cNK cells. Questions remain as to how migratory and resident ILC1s differ in their functions and whether LN ILC1s contribute to peripheral LNs beyond the roles described for cNK cells. Further studies and the degree to which ILC1s can contribute to Th1 priming in the absence of cNK cells needs to be tested. Given the location of cNK cells within LNs (45), it is likely that ILC1s may reside away from the interfollicular zone and border between the B and T cell areas, where ILC2s and ILC3s have been described (18).
In summary, the data presented here sheds new light on the relationship between ILC populations within the circulation and peripheral lymphoid tissues, revealing that a murine ILC1 subset continuously traffics between the blood and peripheral LNs. Despite rapid advances in our understanding of how ILCs develop, much of their basic biology remains poorly understood and the dynamic changes in LN ILCs described here provides further insight into how these cells might contribute to the regulation of adaptive immunity in lymphoid tissues.
Materials and Methods
Study Design
The main aims of this study were to understand the extent of ILC migration into and out of peripheral LNs and determine whether ILCs trafficked from skin into LNs. We sought to address these aims through specifically labelling ILCs in peripheral LNs and skin, testing the extent to which ILCs entered and left these tissues, and identifying the sites to which these cells migrated. These was done using transgenic mice expressing green photoconvertible proteins which, when exposed to violet light, switched to a red fluorescence that could then be detected for a number of days post photoconversion. A key feature of the photoconversion performed was that this was optimised such that all cells in the target tissue (e.g. LN, or ear skin) simultaneously converted to the red fluorescent form. This enabled both the tracking of labelled red cells migrating to different sites as well as detection of new cells (expressing the native green form of the photoconvertible) entering the tissue after labelling. The migration of ILCs was assessed under steady state conditions as well as in response to inflammation.
Mice
Animal husbandry and experiments were ethically reviewed and carried out in accordance with UK Home Office guidelines. C57BL/6, Kaede (28), C57BL/6 H2B-Dendra2 (35), Kaede CCR6-/-, and Kaede CCR7-/- mice were maintained and bred at the University of Birmingham Biomedical Services Unit. Mice were culled between the ages of 6-15 weeks.
Experimental procedures
To establish inflammation in the ear (MC903 AD model) 4 nmol of Calcipotriol (Tocris), in absolute ethanol (VWR Chemicals), was applied to the dorsal and ventral area of the ear for 5 consecutive days. As a vehicle control absolute ethanol was used.
Mice were injected intraperitoneally (i.p) with 1 mg/mL of FTY720 (in 200 μL H20) (Sigma). As a vehicle control mice were injected i.p. with H20. When FTY720 was administered in conjunction with bLN surgery or the MC903 model, mice were injected one day prior to the start of the procedure and every day during the procedure.
For photoconversion of the bLN, mice were subcutaneously injected with 100 μL of analgesic buprenorphine 1 hr prior to surgery (Vetergesic, Reckitt Benckiser Healthcare). Under anesthesia via gaseous isoflurane, a small incision was made exposing the bLN to violet light (365 nm UV LED) for 3 minutes and 30 seconds. The skin was sutured together, and mice were subcutaneously injected with 500 μL of saline solution. 0 hr time points were culled immediately after surgery.
For violet light conversion of the ear, under anaesthesia ears were exposed to a violet light (405nm UV laser) for 40 seconds with periodic breaks. Black cardboard was used to shield the rest of the mouse and 0 hr time points were culled immediately after photoconversion.
Immunizations were performed under anesthesia, via gaseous isoflurane, using 2W1S peptide conjugated to OVA protein. The 2W1S peptide is a variant of peptide 52-68 from the I-Eα chain that is highly immunogenic in C57BL/6 mice (46). The antigen and adjuvant were administered through intramuscular injections according to GSK standards, or through paw pad injections. Upon intramuscular injections the site of the injection, the gastrocnemius muscle, was shaved and approximately 3mm of the needle was inserted into the muscle. The injection volume of 10μl was administered into gastrocnemius muscle of both hind legs. The injections contained 20 μg OVA-2W1S (Proimmune) and 1/50 Human Dose (HD) AS01B (i.e. 1 μg of each MPL and QS-21) or 20 μg OVA-2W1S and PBS. AS01 is a proprietary GSK adjuvant system containing MPL, QS-21 and liposome. QS-21 (Quillaja saponaria Molina, fraction 21) is licensed by GSK from Antigenics LLC, a wholly owned subsidiary of Agenus Inc., a Delaware, USA corporation. For analysis at day 7, a second injection of OVA-2W1S with AS01 or PBS was administered 6 hrs prior to analysis of the response.
Cell Culture
In AS01 experiments, assessing the production of IFN-γ, cell suspensions were incubated for 3h in culture media in the presence of brefeldin A (BFA) (10 μg/ml, BD Biosciences) and monensin (10 μg/ml, BD Biosciences) at 37°C 5% C02.
Flow Cytometry
Tissues were prepared using the same methods in Dutton et al. (47). LNs were cleaned and teased in Roswell Park Memorial Institute (RPMI) 1640 Medium (ThermoFisher Scientific) and crushed through a 70μm filter. Samples were centrifuged (5 minutes, 1,500 rpm, 4°C), supernatant removed and resuspended in appropriate amount of DPBS supplemented with 2% FBS and 0.5% EDTA. Ears were cut into small sections and agitated whilst incubated at 37°C in a thermomixer (Eppendorf) for 30 minutes in 1 ml of Liberase TM/DNase digestion solution (3 mls per ear; 3 ml Dulbecco’s Modified Eagle Media (DMEM, ThermoFisher Scientific) 75 μL Liberase TM (0.28 Wunsch units/mL) and 75μL DNase (200 mg/mL)). Tissue was then filtered through a 70 μm filter and washed with DMEM. The ear was then removed from the filter and this process repeated twice more. Following the third incubation the ear was crushed through the filter and washed with DMEM. Samples were centrifuged and suspended in an appropriate amount of DPBS supplemented with 2% FBS and 0.5% EDTA. 1-2mLs of blood was harvested, via cardiac puncture. Samples were centrifuged (5minutes, 2,000 rpm), supernatant removed and incubated in 500 μL of Gey’s solution (47) on ice for 5minutes. This was repeated twice more, before being resuspended in appropriate amount of DPBS supplemented with 2% FBS and 0.5% EDTA. Samples enriched for T and B cells were stained for B220 (APC), CD3 (APC) and CD5 (APC) for 30 minutes on ice followed by an enrichment with anti-APC MicroBeads (MACS). Samples enriched for CD45+ hematopoietic cells were enriched using CD45 MicroBeads (MACS). All samples were stained with a LIVE/DEAD cell viability assay in APC-Cy7 in PBS (1:1000) for 15 minutes at 4°C. Extracellular staining was conducted with antibodies diluted in DPBS supplemented with 2% FBS and 0.5% EDTA for 30 minutes on ice. The Foxp3 fixation and permeabilization kit (eBioscience) was used to fix and intracellularly stain samples. Kaede samples did not undergo fixation and intracellular staining. The antibodies raised against the following mouse agents were used: B220 (clone RA3-6B2, 1:300 or 1:200, eBioscience), CCR6 (clone 29-2L17, 1:100, BioLegend), CCR7 (clone 4B12, 1:100, eBioscience), CD3ε (clone 145-2C11 or eBio500A2 or 17A2, 1:100 or 1:200, eBioscience or BD Horizon or BioLegend), CD4 (clone RM4-5, 1:100, BioLegend), CD5 (clone 53-7.3, 1:100 or 1:200, eBioscience), CD11b (clone M1/70, 1:300 or 1:200, eBioscience), CD11c (clone N418, 1:300 or 1:200, eBioscience or BioLegend), CD19 (clone eBio1D3, 1:200, eBioscience), CD25 (clone PC61, 1:50, eBioscience), CD31 (clone, 1:400, eBioscience), CD45.2 (clone 104, 1:100, BioLegend), CD49b (clone DX5, 1:200, eBioscience), CD62L (clone MEL/14, 1:1500 or 1:500 or 1:100, eBioscience or BioLegend), CD69 (clone H1.2F3, 1:100, eBioscience), CD103 (clone, 1:200, BioLegend) CD123 (clone 5B11, 1:500, eBioscience), CD207 (clone 4C7, 1:100, BioLegend), c-kit (clone H57-597, 1:100, eBioscience), EOMES (clone Dan11mag, 1:50, eBioscience), F4/80 (clone BM8, 1:200, eBioscience), FcεRI (clone MAR-1, 1:200, eBioscience), GATA-3 (clone TWAJ, 1:75, eBioscience), gp38 (clone, 1:1000, BioLegend), Gr-1 (clone RB6-8C5, 1:2000, eBioscience), IL-7Rα (clone A7R34, 1:50, BioLegend), Ki-67 (clone SolA15, 1:400, eBioscience), KLRG1 (clone 2F1, 1:200, BioLegend or eBioscience), MHCII (clone M5/114.15.2, 1:100, eBioscience), NK1.1 (clone, 1:100 or 1:200, eBioscience or BD Horizon) NKp46 (clone 29A1.4, 1:50 or 1:100, BioLegend or eBioscience), RORγ (clone AFKJS-9, 1:100, eBioscience), T-bet (clone eBio4B10, 1:50, eBioscience), TCRβ (clone H57-597, 1:100, BioLegend), TCRγδ (clone GL3, 1:200, BD Horizon) and TER119 (clone TER-119, 1:200 or 1:100, eBioscience). To identify ILCs the lineage channel included (B220, CD11b, CD11c, CD3, CD5, CD19, TER119, Gr-1, CD123, F4/80, FcεRI, CD49b). CD123 was not included in the lineage channel when identifying ILCs in Kaede mice. Samples were run on the BD LSRFortessa™ X-20 (BD Biosciences) and data collected using BD FACSDiva Software (BD biosciences).
Gene expression analysis
FACS-sorted NK1.1+/- ILCs were lysed in RLT buffer (Qiagen) and total RNA was subsequently isolated using Quick-RNA Microprep kit according to the manufacturer’s instructions (Zymo Research). cDNA was generated from total RNA in a reverse transcription reaction using random primers. TaqMan gene expression assays for selected genes (Applied Biosystems; S1pr1 (Mm00514644_m1), S1pr2 (custom plus TaqMan RNA assay) S1pr3 (Mm04229896_m1), S1pr4 (Mm00468695_s1) and S1pr5 (Mm02620565_s1) were prepared using the Taqman PreAmp Master Mix Kit. Samples were then analysed on QuantStudio 12K Flex RT-PCR system. qPCR results were analysed using the R environment for statistical computing. Delta-delta (ΔΔCT) values were calculated using the mean of three reference genes (b-actin, glyceraldehyde 3-phosphate dehydrogenase, and 18S ribosomal DNA). Fold changes were determined by calculating power 2-ΔΔCT.
Statistical analysis
Data collected were analyzed using Flow Jo 10.4.2 software (Treestar) and GraphPad Prism 7. Normal distribution of the data was not assumed. Pairs of samples were compared using an unpaired two-tailed Mann-Whitney U test. When comparing more than two sets of data, statistical significance was tested using Kruskal-Wallis one-way ANOVA with post hoc Dunn’s test: *p≤0.05, **p≤0.01, ***p≤0.001. The bar represents the median and where appropriate the median has been shown numerically. Where a p value is not indicated, no statistical difference was observed.
Supplementary Material
One sentence summary.
Innate lymphoid cells recirculate through peripheral lymph nodes and contribute to early IFN-γ production following immunization.
Acknowledgements
The 2W1S:I-Ab tetramer was obtained from the NIH Tetramer Facility. We thank Dr Y. Miwa (Tsukuba University) and Dr O. Kanagawa (RCAI, RIKEN) for the Kaede mice. We thank Professor O. Pabst (RWTH Aachen University) for kindly providing the H2B-Dendra2 mice. We thank the Biomedical Services Unit at the University of Birmingham for all their help with in vivo experimental work. We are very grateful to Dr M. Hepworth and Professor S. Milling for critical reading of the manuscript.
Funding
This work was supported by a Senior Research Fellowship from the Wellcome Trust to D.R.W (Grant Number 110199/Z/15/Z). Emma Dutton is a BBSRC CASE Student sponsored by Medimmune LLC, who partially funded the studentship (BB/L016893/1).
Footnotes
Author Contributions: EED designed and performed experiments, analyzed data, performed statistical analysis and wrote the manuscript; DWG, CW, RF, ELB designed and performed experiments, FP, CAM performed experiments and analyzed data, AC, MAS, and GC designed experiments, analyzed data and critiqued data, MC and AMD provided key reagents, designed experiments and critiqued data; MT provided key reagents and expert advice in experimental design and methodology; DRW designed and performed experiments, analyzed data, performed statistical analysis and wrote the manuscript.
Competing Interests: A. Camelo, G. Carlesso F. Pilataxi and C.A. Morehouse are full time employees of MedImmune LLC. M.A. Sleeman is a former employee of MedImmune and a full time employee of Regeneron. All past or current MedImmune coauthors own or owned Astra Zeneca stock and/or stock options. M. Coccia and A.M. Didierlaurent are full time employees of the GSK group of companies. A.M. Didierlaurent reports ownership of shares and/or restricted shares in GSK. The other authors declare that they have no competing interests.
Data and Materials Availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References
- 1.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 2.Walker JA, Barlow JL, McKenzie AN. Innate lymphoid cells--how did we miss them? Nat Rev Immunol. 2013;13:75–87. doi: 10.1038/nri3349. [DOI] [PubMed] [Google Scholar]
- 3.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, Powrie F, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 4.Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TK, Bucks C, Kane CM, Fallon PG, Pannell R, Jolin HE, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, Diefenbach A. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O, Eberl G, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, Powrie F. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, Angelosanto JM, Laidlaw BJ, Yang CY, Sathaliyawala T, Kubota M, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gasteiger G, Fan X, Dikiy S, Lee SY, Rudensky AY. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science. 2015;350:981–985. doi: 10.1126/science.aac9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Sullivan TE, Rapp M, Fan X, Weizman OE, Bhardwaj P, Adams NM, Walzer T, Dannenberg AJ, Sun JC. Adipose-Resident Group 1 Innate Lymphoid Cells Promote Obesity-Associated Insulin Resistance. Immunity. 2016;45:428–441. doi: 10.1016/j.immuni.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boulenouar S, Michelet X, Duquette D, Alvarez D, Hogan AE, Dold C, O'Connor D, Stutte S, Tavakkoli A, Winters D, Exley MA, et al. Adipose Type One Innate Lymphoid Cells Regulate Macrophage Homeostasis through Targeted Cytotoxicity. Immunity. 2017;46:273–286. doi: 10.1016/j.immuni.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Moro K, Kabata H, Tanabe M, Koga S, Takeno N, Mochizuki M, Fukunaga K, Asano K, Betsuyaku T, Koyasu S. Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat Immunol. 2016;17:76–86. doi: 10.1038/ni.3309. [DOI] [PubMed] [Google Scholar]
- 13.Baerenwaldt A, von Burg N, Kreuzaler M, Sitte S, Horvath E, Peter A, Voehringer D, Rolink AG, Finke D. Flt3 Ligand Regulates the Development of Innate Lymphoid Cells in Fetal and Adult Mice. J Immunol. 2016;196:2561–2571. doi: 10.4049/jimmunol.1501380. [DOI] [PubMed] [Google Scholar]
- 14.Bando JK, Liang HE, Locksley RM. Identification and distribution of developing innate lymphoid cells in the fetal mouse intestine. Nat Immunol. 2015;16:153–160. doi: 10.1038/ni.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim AI, Li Y, Lopez-Lastra S, Stadhouders R, Paul F, Casrouge A, Serafini N, Puel A, Bustamante J, Surace L, Masse-Ranson G, et al. Systemic Human ILC Precursors Provide a Substrate for Tissue ILC Differentiation. Cell. 2017;168:1086–1100 e1010. doi: 10.1016/j.cell.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 17.Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, Fabiunke C, Pfeifer D, Sexl V, Fonseca-Pereira D, Domingues RG, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 18.Mackley EC, Houston S, Marriott CL, Halford EE, Lucas B, Cerovic V, Filbey KJ, Maizels RM, Hepworth MR, Sonnenberg GF, Milling S, et al. CCR7-dependent trafficking of RORgamma(+) ILCs creates a unique microenvironment within mucosal draining lymph nodes. Nat Commun. 2015;6 doi: 10.1038/ncomms6862. 5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, Mantegazza AR, Ma HL, Crawford A, Angelosanto JM, Wherry EJ, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mebius RE. Organogenesis of lymphoid tissues. Nat Rev Immunol. 2003;3:292–303. doi: 10.1038/nri1054. [DOI] [PubMed] [Google Scholar]
- 21.Withers DR. Innate lymphoid cell regulation of adaptive immunity. Immunology. 2016;149:123–130. doi: 10.1111/imm.12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliphant CJ, Hwang YY, Walker JA, Salimi M, Wong SH, Brewer JM, Englezakis A, Barlow JL, Hams E, Scanlon ST, Ogg GS, et al. MHCII-Mediated Dialog between Group 2 Innate Lymphoid Cells and CD4(+) T Cells Potentiates Type 2 Immunity and Promotes Parasitic Helminth Expulsion. Immunity. 2014;41:283–295. doi: 10.1016/j.immuni.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Burg N, Chappaz S, Baerenwaldt A, Horvath E, Bose Dasgupta S, Ashok D, Pieters J, Tacchini-Cottier F, Rolink A, Acha-Orbea H, Finke D. Activated group 3 innate lymphoid cells promote T-cell-mediated immune responses. Proc Natl Acad Sci U S A. 2014;111:12835–12840. doi: 10.1073/pnas.1406908111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villanova F, Flutter B, Tosi I, Grys K, Sreeneebus H, Perera GK, Chapman A, Smith CH, Di Meglio P, Nestle FO. Characterization of innate lymphoid cells in human skin and blood demonstrates increase of NKp44+ ILC3 in psoriasis. J Invest Dermatol. 2014;134:984–991. doi: 10.1038/jid.2013.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim AI, Menegatti S, Bustamante J, Le Bourhis L, Allez M, Rogge L, Casanova JL, Yssel H, Di Santo JP. IL-12 drives functional plasticity of human group 2 innate lymphoid cells. J Exp Med. 2016;213:569–583. doi: 10.1084/jem.20151750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Mao K, Chen X, Sun MA, Kawabe T, Li W, Usher N, Zhu J, Urban JF, Jr, Paul WE, Germain RN. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science. 2018;359:114–119. doi: 10.1126/science.aam5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bajenoff M, Breart B, Huang AY, Qi H, Cazareth J, Braud VM, Germain RN, Glaichenhaus N. Natural killer cell behavior in lymph nodes revealed by static and real-time imaging. J Exp Med. 2006;203:619–631. doi: 10.1084/jem.20051474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomura M, Yoshida N, Tanaka J, Karasawa S, Miwa Y, Miyawaki A, Kanagawa O. Monitoring cellular movement in vivo with photoconvertible fluorescence protein "Kaede" transgenic mice. Proc Natl Acad Sci U S A. 2008;105:10871–10876. doi: 10.1073/pnas.0802278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marriott CL, Dutton EE, Tomura M, Withers DR. Retention of Ag-specific memory CD4+ T cells in the draining lymph node indicates lymphoid tissue resident memory populations. Eur J Immunol. 2017;47:860–871. doi: 10.1002/eji.201646681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Girard JP, Moussion C, Forster R. HEVs, lymphatics and homeostatic immune cell trafficking in lymph nodes. Nat Rev Immunol. 2012;12:762–773. doi: 10.1038/nri3298. [DOI] [PubMed] [Google Scholar]
- 31.Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 32.Walzer T, Chiossone L, Chaix J, Calver A, Carozzo C, Garrigue-Antar L, Jacques Y, Baratin M, Tomasello E, Vivier E. Natural killer cell trafficking in vivo requires a dedicated sphingosine 1-phosphate receptor. Nat Immunol. 2007;8:1337–1344. doi: 10.1038/ni1523. [DOI] [PubMed] [Google Scholar]
- 33.Jenne CN, Enders A, Rivera R, Watson SR, Bankovich AJ, Pereira JP, Xu Y, Roots CM, Beilke JN, Banerjee A, Reiner SL, et al. T-bet-dependent S1P5 expression in NK cells promotes egress from lymph nodes and bone marrow. J Exp Med. 2009;206:2469–2481. doi: 10.1084/jem.20090525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chun J, Hartung HP. Mechanism of action of oral fingolimod (FTY720) in multiple sclerosis. Clin Neuropharmacol. 2010;33:91–101. doi: 10.1097/WNF.0b013e3181cbf825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ugur M, Kaminski A, Pabst O. Lymph node gammadelta and alphabeta CD8(+) T cells share migratory properties. Sci Rep. 2018;8 doi: 10.1038/s41598-018-27339-8. 8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salimi M, Barlow JL, Saunders SP, Xue L, Gutowska-Owsiak D, Wang X, Huang LC, Johnson D, Scanlon ST, McKenzie AN, Fallon PG, et al. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J Exp Med. 2013;210:2939–2950. doi: 10.1084/jem.20130351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomura M, Hata A, Matsuoka S, Shand FH, Nakanishi Y, Ikebuchi R, Ueha S, Tsutsui H, Inaba K, Matsushima K, Miyawaki A, et al. Tracking and quantification of dendritic cell migration and antigen trafficking between the skin and lymph nodes. Sci Rep. 2014;4 doi: 10.1038/srep06030. 6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li M, Hener P, Zhang Z, Kato S, Metzger D, Chambon P. Topical vitamin D3 and low-calcemic analogs induce thymic stromal lymphopoietin in mouse keratinocytes and trigger an atopic dermatitis. Proc Natl Acad Sci U S A. 2006;103:11736–11741. doi: 10.1073/pnas.0604575103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim BS, Siracusa MC, Saenz SA, Noti M, Monticelli LA, Sonnenberg GF, Hepworth MR, Van Voorhees AS, Comeau MR, Artis D. TSLP elicits IL-33-independent innate lymphoid cell responses to promote skin inflammation. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3005374. 170ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Hu S, Zhao L, Kaplan DH, Perdew GH, Xiong N. Selective programming of CCR10(+) innate lymphoid cells in skin-draining lymph nodes for cutaneous homeostatic regulation. Nat Immunol. 2016;17:48–56. doi: 10.1038/ni.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schutyser E, Struyf S, Van Damme J. The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev. 2003;14:409–426. doi: 10.1016/s1359-6101(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 42.Martin-Fontecha A, Thomsen LL, Brett S, Gerard C, Lipp M, Lanzavecchia A, Sallusto F. Induced recruitment of NK cells to lymph nodes provides IFN-gamma for T(H)1 priming. Nat Immunol. 2004;5:1260–1265. doi: 10.1038/ni1138. [DOI] [PubMed] [Google Scholar]
- 43.Coccia M, Collignon C, Herve C, Chalon A, Welsby I, Detienne S, van Helden MJ, Dutta S, Genito CJ, Waters NC, Deun KV, et al. Cellular and molecular synergy in AS01-adjuvanted vaccines results in an early IFNgamma response promoting vaccine immunogenicity. NPJ Vaccines. 2017;2:25. doi: 10.1038/s41541-017-0027-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serafini N, Vosshenrich CA, Di Santo JP. Transcriptional regulation of innate lymphoid cell fate. Nat Rev Immunol. 2015;15:415–428. doi: 10.1038/nri3855. [DOI] [PubMed] [Google Scholar]
- 45.Fang V, Chaluvadi VS, Ramos-Perez WD, Mendoza A, Baeyens A, Rivera R, Chun J, Cammer M, Schwab SR. Gradients of the signaling lipid S1P in lymph nodes position natural killer cells and regulate their interferon-gamma response. Nat Immunol. 2017;18:15–25. doi: 10.1038/ni.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rees W, Bender J, Teague TK, Kedl RM, Crawford F, Marrack P, Kappler J. An inverse relationship between T cell receptor affinity and antigen dose during CD4(+) T cell responses in vivo and in vitro. Proc Natl Acad Sci U S A. 1999;96:9781–9786. doi: 10.1073/pnas.96.17.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dutton EE, Camelo A, Sleeman M, Herbst R, Carlesso G, Belz GT, Withers DR. Characterisation of innate lymphoid cell populations at different sites in mice with defective T cell immunity. Wellcome Open Res. 2017;2:117. doi: 10.12688/wellcomeopenres.13199.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.