Abstract
Rationale:
Studies identified kininogen as a potential biomarker of preeclampsia, a major cause of adverse maternal outcomes. High-molecular-weight kininogen (HK) and its activated form (HKa) participate in numerous pathways associated with establishing and maintaining pregnancy. However, dynamic changes in HK and naturally occurring HK-derived peptides during natural course of pregnancy are largely unknown.
Methods:
Longitudinal serum samples during course of normal pregnancy (Trimester [T]1, 2, 3) from 60 pregnant women were analyzed by western blot with an anti-HK antibody. Circulating peptides in longitudinal serum specimens derived from 50 participants were enriched using nanoporous silica thin films. Peptides were identified by LC-MS/MS and database searching. Relative quantification was performed by MaxQuant and in-house scripts. Normality was evaluated by either ANOVA or Friedman tests with p-value < 0.05 for statistical significance.
Results:
Western blotting revealed that HK significantly decreased during normal pregnancy (T1 vs T2, p<0.05; T1 vs T3, p<0001). A 100 KD intermediate increased during pregnancy (T1 vs T2, p<0.005; T1 vs T3, p<0.01). Moreover, the heavy chain (T1 vs T2, p<.0001; T1 vs T3, p<.0001; T2 vs T3, p<0.01) and light chain (T1 vs T2, p<0.0001; T1 vs T3, p<.0001; T2 vs T3, p<0.05) significantly increased during pregnancy. LC-MS/MS analysis identified 180 kininogen-1 peptides, of which 167 mapped to domain 5 (D5). 73 peptides with ten or more complete data sets were included for further analysis. 70 peptides mapped to D5, and 3, 24, and 43 peptides showed significant decrease, no trend, and significant increase, respectively, during pregnancy.
Conclusions:
This study demonstrates dynamic changes in HK and naturally occurring HK-derived peptides during pregnancy. Our study shed lights on the gestational changes of HK and its peptides for further validation of them as potential biomarkers for pregnancy related complications.
Keywords: Kininogen, pregnancy, mass spectrometry, peptide, proteomics
1. Introduction
In the United States, hypertensive disorders affect 6–8% of pregnancies and are the second leading cause of maternal mortality. Among the various etiologies leading to pregnancy-induced hypertension (PIH), pre-eclampsia (PE) is a major maternal morbidity with immediate complications for the pregnant mother and her baby and increases risk of later developing cardiovascular disease, chronic kidney disease and diabetes. Early detection of PE can aid in reducing maternal mortalities 1,2. Several studies have discovered and validated PE biomarkers, including kininogen-1 and high molecular weight kininogen (HK) and their proteolytic fragments/peptides3–9.
The kininogen-1 (KNG-1) gene is located on chromosome three. KNG-1 mRNA is alternatively spliced to produce two structurally and functionally distinct proteins, HK and low molecular weight kininogen (LK). HK is a 120 kDa cysteine protease inhibitor with six functional domains, labeled D1 to D6 10. HK and LK share identical D1–4; however, LK contains an alternate, 4 kDa D5 and lacks D6 entirely 11. The function of LK remains largely unknown. HK and three serine proteases, FXII, FXI, and plasma prekallikrein, comprise the plasma kallikrein-kinin system (KKS), which regulates coagulation, inflammation, and blood pressure 12. Kallikrein cleaves HK to release bradykinin (BK, D4) and generate activated HK (HKa), composed of the 65 kDa heavy chain (HC, D1-D3) joined by disulfide bond to the 56 kDa light chain (LC, D5-D6) 11,13.
Ovulation, endometrial proliferation, and implantation are inflammatory processes that locally activate the KKS 14–16. Kallikrein protein is detectable in rat uterus, placental vessels, and amniotic fluid and in porcine endometrium and in human placental villous capillaries 15–18. HK is detectable in porcine endometrium and human placenta 17,19. HK and its D5 peptides are implicated in the regulation of diverse functions relevant to normal pregnancy and PE pathogenesis including blood pressure regulation, smooth muscle contraction, coagulation, angiogenesis, immune response, immunity, and endothelial cell migration, proliferation, and apoptosis 11,20,21. HK is highly concentrated in the female reproductive tract and may be an important regulator of placental vascularization and nutrient transport to the fetus 22.
Previous biomarkers studies compared HK protein and/or HK-derived peptide levels in normal and hypertensive pregnancies, but interpretation of the results is confounded by differing methodologies and a lack of knowledge regarding kininogen dynamics in the course of normal pregnancy. To the best of our knowledge, no study has systematically examined HK protein and peptide dynamics during pregnancy. Here we performed a longitudinal analysis on serum samples using western blot and unbiased proteomic approaches to understand the natural processing of HK protein during the course of normal pregnancy.
2. Experimental
2.1. Samples
In this retrospective study, coded serum specimens were obtained from the Washington University Women and Infant’s Health Specimen Consortium (WIHSC). 354 specimens were collected from 118 women receiving prenatal care at Washington University/Barnes Jewish Hospital, St Louis MO. The patients were recruited from October 19, 2010 to August 26, 2013. Specimens from subjects ≥18 years with singleton/viable pregnancies were utilized. Subjects had sequential samples of venous blood drawn in the first trimester (T1, <13 weeks), second trimester (T2, 13–27 weeks), and third trimester (T3, ≥ 27 weeks). Samples were collected and stored in liquid nitrogen for up to 4 years and then transferred to −80°C freezer for up to 2 years before analysis.
2.2. Western Blot
Western blotting was performed on 180 serum samples from 60 participants. Serum was diluted four-fold and equal volumes were size fractionated on Novex WedgeWell 10% Tris-Glycine polyacrylamide reducing gels (Invitrogen) and then transferred to PVDF membranes. Membranes were incubated for 30 minutes at room temperature in Odyssey Blocking Buffer TBS (Li-Cor) then incubated at room temperature for 2.5 hours with 1:500 dilution of rabbit HMW Kininogen Antibody (GeneTex, GTX100833). Membranes were washed three times in TBST for ten minutes. Then membranes were incubated at room temperature in 1:10,000 dilution of IRDye 800CW Donkey anti-Rabbit IgG (Li-Cor) secondary antibody for one hour. Membranes were washed twice in TBST for ten minutes and once in TBS for five minutes before imaging on an Odyssey Classic Imaging System with densitometry quantification by Image Studio v.3.1. Relative intensity was calculated by normalizing to trimester one intensity values.
2.3. Liquid chromatography– high resolution tandem mass spectrometry (LC-HRMS/MS)
150 longitudinal serum samples from 50 participants were enriched for naturally occurring, circulating peptides using nanoporous silica thin films (NanoTraps) 23. A Thermo Fisher Scientific EASY-nLC 1000 coupled on-line to a Fusion Lumos high resolution mass spectrometer (Thermo Fisher Scientific) was used for comprehensive peptide analysis. Buffer A (0.1% FA in water) and buffer B (0.1% FA in ACN) were used as mobile phases for gradient separation. A 100 µm x 15 cm chromatography column (ReproSil-Pur C18-AQ, 3 µm, Dr. Maisch GmbH, German) was packed in-house for peptide separation. Peptides were separated with a gradient of 3–30% buffer B over 25 min, 30%−90% B over 10 min at a flow rate of 600 nL/min. The Fusion Lumos mass spectrometer was operated in data dependent mode. Full MS scans were acquired in the Orbitrap mass analyzer over a range of 300–1500 m/z with resolution 120,000 at m/z 200. Time between master scans was set at 1s, and the top most abundant precursors with charge states between 2 and 10 were selected with an isolation window of 1.6 Thomsons and fragmented by higher-energy collisional dissociation with normalized collision energy of 35. MS/MS scans were acquired in the Orbitrap mass analyzer with resolution 30,000 at m/z 200. The automatic gain control target value was 1e6 for full scans and 5e4 for MS/MS scans respectively, and the maximum ion injection time is 60 ms for both. The raw files were processed using the MaxQuant computational proteomics platform version 1.5.5.1 (Max Planck Institute, Munich, Germany) for protein identification. The fragmentation spectra were used to search the UniProt human protein database (downloaded August 22, 2016) containing 70,630 protein sequences. Oxidation of methionine and protein N-terminal acetylation were used as variable modifications for database searching. The precursor and fragment mass tolerances were set to 7 and 20 ppm, respectively. Both peptide and protein identifications were filtered at 5% false discovery rate based on decoy search using a database with the protein sequences reversed. Peptide quantitation was based on the peptide intensity values reported by MaxQuant.
2.4. Statistics
Analysis of peptide intensities was performed in GraphPad Prism 6. Normality was assessed with the D’Agostino-Pearson omnibus normality test. Normally distributed data was analyzed using the repeated-measures one-way ANOVA and Tukey’s multiple comparison test. Non-normally distributed data was analyzed using the Friedman test and Dunn’s multiple comparisons. p < .05 was considered significant.
2.5. Protease Prediction
The amino acid sequence of HK was retrieved from UniProt (entry P01042) and protease predictions were obtained from PROSPER 24,25. All amino acid numbers match the UniProt sequence.
3. Results
3.1. Western blot reveals increased HKa
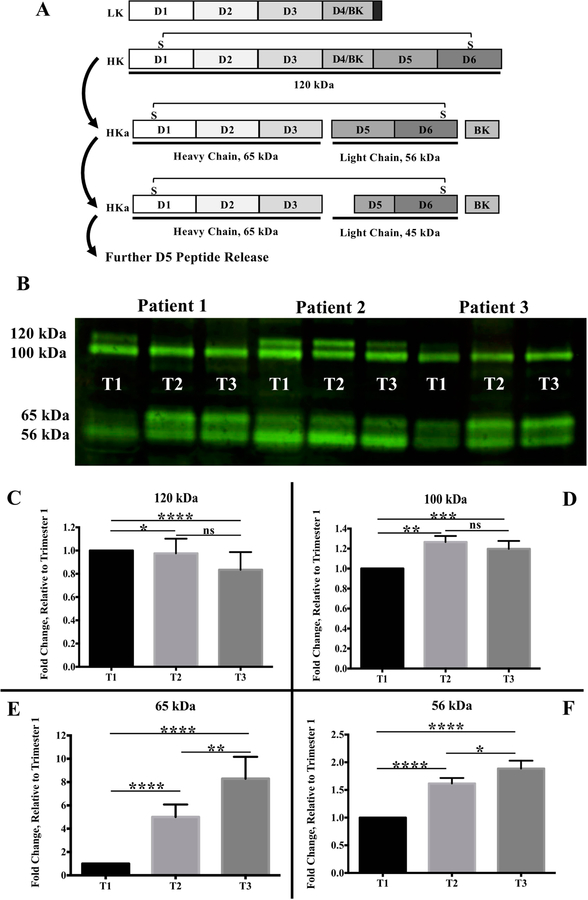
T1, T2, and T3 serum samples from 60 women were diluted and size-fractionated on Tris-Glycine SDS gels before blotting with a polyclonal antibody that specifically detects HK, but not LK. Figure 1a is diagram depicting LK, HK, and some known steps of HK processing. Our western blot revealed four bands, 120 kDa, 100 kDa, 65 kDa, and 56 kDa, as shown in the representative blot (Fig 1b). Figure 1c–f displays bar graphs, showing the means and SEMs of densitometry values relative to T1, where T1 values were set equal to one. Densitometry values displayed a non-normal distribution. Since the experiment is a repeated-measures design the Friedman test with Dunn’s multiple comparison was used to calculate p-values. The 120 kDa band represents inactive HK, which significantly decreased in abundance from T1 to T2 and T3 (T1 vs T2, p<0.05; T1 vs T3, p<0001). The decrease between T2 and T3 was not statistically significant. (Fig 1c) The 100 kDa band increased in abundance during the course of pregnancy with significant increases observed from T1 to T2 and T3 (T1 vs T2, p<0.005; T1 vs T3, p<0.01) (Fig 1d). The 65 kDa HC (T1 vs T2, p<.0001; T1 vs T3, p<.0001; T2 vs T3, p<0.01) and 56 kDa LC (T1 vs T2, p<0.0001; T1 vs T3, p<.0001; T2 vs T3, p<0.05) increased progressively during pregnancy with statistical significance found with every trimester to trimester comparison (Fig 1e–f). Further processing of HKa by kallikrein at Arg419-Lys420 creates a 45 kDa LC 26. However, neither the 45 kDa band nor any smaller HK fragments were detected by our antibody.
Figure 1. Western Blot Reveals Increased HKa.
A. Schematic comparing the structure of LK to HK. LK and HK share identical D1 to D4. LK contains an alternate D5 and lacks D6. 120 kDa HK contains six functional domains. D4/BK release generates HKa, composed of the 65 kDa HC connected by disulfide bond to the 56 kDa LC. Further proteolysis produces a 45 kDa LC and D5 peptide release. B. Representative blot showing four bands (120, 100, 65, 56 kDa) derived from HK. C–F. Bar graphs showing mean densitometry values with SEM. T2 and T3 values are relative to T1, which is set equal to one. Friedman’s test for repeated measurements of non-parametric data was used to obtain p-values. C. 120 kDa band representing intact HK significantly decreases during T1 to T2 and T1 to T3. D. 100 kDa band increases T1 to T2 and T1 to T3. E. 65 kDa band representing the HC increases significantly during pregnancy. F. 56 kDa band representing the LC increases significantly during pregnancy. * p < 0.05; ** p < 0.01; **** p < .0001; ns not significant.
3.2. LC-MS/MS analysis reveals differential processing of Domain 5
Longitudinal serum samples from 50 participants were enriched for small, naturally occurring peptides using nanoporous silica thin films 23. Enriched samples were analyzed by LC-MS/MS and database sequencing, which revealed 180 peptides that mapped to KNG-1. Six peptides mapped uniquely to LK. Three peptides mapped to D4, and four peptides mapped to D1 or D2 and could be derived from either LK or HK. The remaining 167 peptides mapped uniquely to HK D5. Peptides with less than ten complete data sets, defined as a value recorded in all three longitudinal T1, T2, and T3 samples for ten participants, were excluded from statistical analysis, leaving 73 analyzable HK peptides. 70 of these 73 peptides mapped to D5. Table 1 summarizes the 73 HK peptides and their statistics. Three of the D5 peptides significantly decreased during the course of normal pregnancy while 24 peptides displayed no significant trend, and 43 peptides significantly increased.
Table 1.
Table of all HK derived peptides presenting ten or more complete data sets.
| Multiple Comparison | Median (log2) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mass | n | Start | End | Trend | Normal | P-value | T1 vs T2 | T1 vs T3 | T2 vs T3 | T1 | T2 | T3 | Prev Rep Seq | Domain | Sequence |
| 1233.626 | 48 | 381 | 391 | + | N | 0.0005 | ns | *** | ns | 17.77 | 18.17 | 18.41 | D4 | RPPGFSPFRSS | |
| 1347.655 | 25 | 428 | 437 | - | Y | .0004 | * | **** | ns | 20.38 | 19.32 | 18.82 | D5 | RHDWGHEKQR | |
| 1357.758 | 47 | 185 | 196 | 0 | Y | ns | ns | ns | ns | 18.1 | 18.14 | 17.74 | D2 | AQRQVVAGLNFR | |
| 1389.727 | 49 | 381 | 392 | - | N | 0.0004 | * | *** | ns | 20.84 | 20.49 | 20.07 | D4 | RPPGFSPFRSSR | |
| 1510.718 | 21 | 465 | 477 | + | N | 0.0119 | * | ns | ns | 17.66 | 19.16 | 18.93 | D5 | HEQQHGLGHGHKF | |
| 1599.74 | 25 | 459 | 473 | 0 | Y | ns | ns | ns | ns | 20.35 | 20.79 | 21.03 | D5 | HGLGHGHEQQHGLGH | |
| 1704.798 | 41 | 463 | 477 | + | Y | 0.001 | ** | ** | ns | 22.02 | 22.91 | 23.23 | D5 | HGHEQQHGLGHGHKF | |
| 1761.82 | 25 | 462 | 477 | 0 | Y | ns | ns | ns | ns | 20 | 20.32 | 20.8 | D5 | GHGHEQQHGLGHGHKF | |
| 1793.821 | 36 | 459 | 475 | 0 | Y | ns | ns | ns | ns | 22.28 | 22.86 | 22.56 | D5 | HGLGHGHEQQHGLGHGH | |
| 1850.842 | 17 | 458 | 475 | 0 | Y | ns | ns | ns | ns | 21.06 | 21.82 | 21.55 | X | D5 | GHGLGHGHEQQHGLGHGH |
| 1874.904 | 39 | 461 | 477 | + | Y | 0.003 | ** | ns | ns | 18.63 | 19.75 | 19.72 | D5 | LGHGHEQQHGLGHGHKF | |
| 1931.925 | 45 | 460 | 477 | + | N | 0.0003 | * | *** | ns | 21.13 | 22.45 | 22.72 | D5 | GLGHGHEQQHGLGHGHKF | |
| 2006.943 | 21 | 457 | 475 | + | N | 0.0006 | * | *** | ns | 19.58 | 21.55 | 21.56 | X | D5 | RGHGLGHGHEQQHGLGHGH |
| 2068.984 | 50 | 459 | 477 | + | N | <.0001 | *** | **** | ns | 24.24 | 25.67 | 25.79 | D5 | HGLGHGHEQQHGLGHGHKF | |
| 2126.006 | 49 | 458 | 477 | + | N | <.0001 | *** | **** | ns | 25.16 | 26.24 | 26.26 | X, A | D5 | GHGLGHGHEQQHGLGHGHKF |
| 2254.101 | 50 | 458 | 478 | + | N | 0.0002 | *** | *** | ns | 24.15 | 25.14 | 25.48 | X, A | D5 | GHGLGHGHEQQHGLGHGHKFK |
| 2282.107 | 43 | 457 | 477 | + | N | <.0001 | *** | **** | ns | 19.97 | 21.64 | 22.51 | X, A | D5 | RGHGLGHGHEQQHGLGHGHKF |
| 2286.089 | 30 | 478 | 497 | + | N | 0.0177 | ns | * | ns | 17.68 | 18.46 | 18.4 | D5 | KLDDDLEHQGGHVLDHGHKH | |
| 2367.185 | 46 | 458 | 479 | + | Y | <.0001 | **** | **** | ns | 20.78 | 22.32 | 22.68 | X, A | D5 | GHGLGHGHEQQHGLGHGHKFKL |
| 2403.158 | 26 | 460 | 481 | 0 | Y | ns | ns | ns | ns | 18.86 | 19.03 | 19.09 | D5 | GLGHGHEQQHGLGHGHKFKLDD | |
| 2410.165 | 19 | 456 | 477 | + | Y | 0.016 | ns | * | ns | 18.98 | 20.22 | 20.87 | X, A | D5 | QRGHGLGHGHEQQHGLGHGHKF |
| 2482.212 | 34 | 458 | 480 | + | Y | <.0001 | ** | **** | ** | 17.76 | 19.19 | 19.28 | X, A | D5 | GHGLGHGHEQQHGLGHGHKFKLD |
| 2547.224 | 21 | 455 | 477 | + | Y | 0.004 | * | * | ns | 17.53 | 18.73 | 19.06 | X, A | D5 | HQRGHGLGHGHEQQHGLGHGHKF |
| 2597.238 | 48 | 458 | 481 | + | Y | 0.0005 | * | ** | ns | 21.31 | 22.11 | 22.09 | X, A | D5 | GHGLGHGHEQQHGLGHGHKFKLDD |
| 2698.311 | 19 | 475 | 497 | + | N | 0.0294 | * | ns | ns | 18.5 | 19.67 | 19.23 | D5 | HKFKLDDDLEHQGGHVLDHGHKH | |
| 2785.363 | 19 | 438 | 462 | + | Y | 0.0403 | ns | ns | ns | 18.18 | 18.88 | 19.67 | D5 | KHNLGHGHKHERDQGHGHQRGHGLG | |
| 2892.392 | 25 | 473 | 497 | + | N | 0.0027 | * | ** | ns | 18.32 | 19.76 | 19.99 | D5 | HGHKFKLDDDLEHQGGHVLDHGHKH | |
| 2996.425 | 28 | 478 | 504 | 0 | Y | ns | ns | ns | ns | 19.27 | 19.6 | 19.57 | D5 | KLDDDLEHQGGHVLDHGHKHKHGHGHG | |
| 2996.425 | 25 | 479 | 505 | + | Y | 0.006 | * | * | ns | 18.46 | 19.87 | 19.34 | D5 | LDDDLEHQGGHVLDHGHKHKHGHGHGK | |
| 3020.4 | 22 | 480 | 506 | 0 | N | ns | ns | ns | ns | 20.48 | 22.03 | 21.31 | D5 | DDDLEHQGGHVLDHGHKHKHGHGHGKH | |
| 3041.412 | 45 | 450 | 477 | + | N | <.0001 | ** | **** | ns | 20.59 | 21.74 | 22.06 | X, A | D5 | DQGHGHQRGHGLGHGHEQQHGLGHGHKF |
| 3133.484 | 46 | 479 | 506 | + | N | <.0001 | ** | **** | ns | 19.9 | 20.9 | 21.75 | D5 | LDDDLEHQGGHVLDHGHKHKHGHGHGKH | |
| 3197.513 | 34 | 449 | 477 | + | Y | 0.0004 | ** | ** | ns | 20.87 | 22.54 | 22.39 | X, A | D5 | RDQGHGHQRGHGLGHGHEQQHGLGHGHKF |
| 3217.6 | 13 | 482 | 510 | 0 | Y | ns | ns | ns | ns | 21.25 | 21.14 | 20.52 | D5 | DLEHQGGHVLDHGHKHKHGHGHGKHKNKG | |
| 3219.51 | 17 | 458 | 486 | 0 | Y | ns | ns | ns | ns | 17.4 | 17.38 | 17.6 | X, A | D5 | GHGLGHGHEQQHGLGHGHKFKLDDDLEHQ |
| 3261.579 | 34 | 478 | 506 | + | Y | 0.002 | ** | ** | ns | 19.13 | 20.63 | 20.66 | D5 | KLDDDLEHQGGHVLDHGHKHKHGHGHGKH | |
| 3262.538 | 11 | 480 | 508 | + | Y | 0.01 | ns | * | ns | 21.49 | 23.26 | 22.98 | D5 | DDDLEHQGGHVLDHGHKHKHGHGHGKHKN | |
| 3276.531 | 20 | 458 | 487 | 0 | Y | ns | ns | ns | ns | 17.54 | 17.8 | 18.01 | X, A | D5 | GHGLGHGHEQQHGLGHGHKFKLDDDLEHQG |
| 3326.555 | 25 | 448 | 477 | 0 | Y | ns | ns | ns | ns | 18.22 | 18.84 | 19.38 | X, A | D5 | ERDQGHGHQRGHGLGHGHEQQHGLGHGHKF |
| 3375.622 | 39 | 479 | 508 | + | Y | .002 | ** | * | ns | 21.11 | 21.8 | 22 | D5 | LDDDLEHQGGHVLDHGHKHKHGHGHGKHKN | |
| 3381.597 | 43 | 429 | 456 | 0 | N | ns | ns | ns | ns | 25.62 | 25.59 | 25.38 | D5 | HDWGHEKQRKHNLGHGHKHERDQGHGHQ | |
| 3408.647 | 14 | 477 | 506 | 0 | N | ns | ns | ns | ns | 17.44 | 18.63 | 18.64 | D5 | FKLDDDLEHQGGHVLDHGHKHKHGHGHGKH | |
| 3447.654 | 28 | 480 | 510 | + | Y | 0.023 | ** | ns | ns | 21.87 | 24.09 | 24.24 | D5 | DDDLEHQGGHVLDHGHKHKHGHGHGKHKNKG | |
| 3463.614 | 37 | 447 | 477 | + | Y | 0.009 | * | * | ns | 20.92 | 22.14 | 21.97 | X, A | D5 | HERDQGHGHQRGHGLGHGHEQQHGLGHGHKF |
| 3503.717 | 42 | 478 | 508 | 0 | Y | ns | ns | ns | ns | 20.44 | 21.11 | 20.93 | D5 | KLDDDLEHQGGHVLDHGHKHKHGHGHGKHKN | |
| 3537.699 | 49 | 428 | 456 | - | N | 0.0026 | ns | ** | ns | 28.66 | 28.3 | 27.63 | D5 | RHDWGHEKQRKHNLGHGHKHERDQGHGHQ | |
| 3560.738 | 48 | 479 | 510 | + | Y | <.0001 | **** | **** | ns | 22.36 | 24.11 | 24.39 | D5 | LDDDLEHQGGHVLDHGHKHKHGHGHGKHKNKG | |
| 3638.721 | 22 | 438 | 469 | 0 | Y | ns | ns | ns | ns | 20.82 | 18.69 | 20.84 | D5 | KHNLGHGHKHERDQGHGHQRGHGLGHGHEQQH | |
| 3650.785 | 15 | 477 | 508 | 0 | N | ns | ns | ns | ns | 18.43 | 19.09 | 19.46 | D5 | FKLDDDLEHQGGHVLDHGHKHKHGHGHGKHKN | |
| 3688.833 | 45 | 478 | 510 | + | Y | <.0001 | **** | *** | ns | 20.75 | 22.03 | 22.37 | D5 | KLDDDLEHQGGHVLDHGHKHKHGHGHGKHKNKG | |
| 3688.833 | 37 | 479 | 511 | + | Y | <.0001 | **** | *** | ns | 19.55 | 21.56 | 21.54 | D5 | LDDDLEHQGGHVLDHGHKHKHGHGHGKHKNKGK | |
| 3693.8 | 12 | 428 | 457 | - | Y | 0.027 | * | ns | ns | 24.49 | 23.07 | 22.26 | D5 | RHDWGHEKQRKHNLGHGHKHERDQGHGHQR | |
| 3816.928 | 23 | 479 | 512 | + | N | 0.0005 | ** | *** | ns | 18.64 | 20.06 | 20.02 | D5 | LDDDLEHQGGHVLDHGHKHKHGHGHGKHKNKGKK | |
| 3835.902 | 33 | 477 | 510 | + | Y | <.0001 | ** | **** | ns | 18.32 | 20.06 | 20.55 | D5 | FKLDDDLEHQGGHVLDHGHKHKHGHGHGKHKNKG | |
| 3865.848 | 27 | 438 | 472 | 0 | Y | ns | ns | ns | ns | 21.29 | 22.15 | 21.85 | D5 | KHNLGHGHKHERDQGHGHQRGHGLGHGHEQQHGLG | |
| 3987.993 | 19 | 479 | 514 | + | Y | <.0001 | *** | ** | ns | 19.06 | 20.84 | 20.64 | ^ | D5 | LDDDLEHQGGHVLDHGHKHKHGHGHGKHKNKGKKNG |
| 4002.907 | 35 | 438 | 473 | 0 | N | ns | ns | ns | ns | 22.5 | 23.27 | 23.18 | X | D5 | KHNLGHGHKHERDQGHGHQRGHGLGHGHEQQHGLGH |
| 4196.987 | 45 | 438 | 475 | + | N | <.0001 | ** | **** | ns | 24.98 | 26.61 | 26.37 | X | D5 | KHNLGHGHKHERDQGHGHQRGHGLGHGHEQQHGLGHGH |
| 4344.056 | 23 | 439 | 477 | + | N | 0.0006 | ns | *** | ns | 22.55 | 23.91 | 24.49 | X, A | D5 | HNLGHGHKHERDQGHGHQRGHGLGHGHEQQHGLGHGHKF |
| 4394.084 | 48 | 458 | 497 | + | Y | <.0001 | **** | **** | * | 21.22 | 22.61 | 23.04 | X, $, A | D5 | GHGLGHGHEQQHGLGHGHKFKLDDDLEHQGGHVLDHGHKH |
| 4472.151 | 41 | 438 | 477 | + | N | 0.0001 | ns | **** | ns | 26.55 | 28.04 | 28.46 | X, A | D5 | HNLGHGHKHERDQGHGHQRGHGLGHGHEQQHGLGHGHKFK |
| 4472.151 | 48 | 439 | 478 | + | N | 0.0011 | ns | *** | ns | 26.2 | 27.66 | 27.94 | X, A | D5 | KHNLGHGHKHERDQGHGHQRGHGLGHGHEQQHGLGHGHKF |
| 4522.179 | 38 | 458 | 498 | + | N | <.0001 | ** | **** | ns | 19.02 | 20.46 | 20.88 | X, $, A | D5 | GHGLGHGHEQQHGLGHGHKFKLDDDLEHQGGHVLDHGHKHK |
| 4600.246 | 36 | 438 | 478 | 0 | N | ns | ns | ns | ns | 24.01 | 24.07 | 23.5 | X, A | D5 | KHNLGHGHKHERDQGHGHQRGHGLGHGHEQQHGLGHGHKFK |
| 4713.33 | 30 | 438 | 479 | 0 | Y | ns | ns | ns | ns | 21.4 | 22.42 | 22.6 | X, A | D5 | KHNLGHGHKHERDQGHGHQRGHGLGHGHEQQHGLGHGHKFKL |
| 4943.384 | 21 | 438 | 481 | 0 | N | ns | ns | ns | ns | 20.32 | 20.46 | 20.64 | X, A | D5 | KHNLGHGHKHERDQGHGHQRGHGLGHGHEQQHGLGHGHKFKLDD |
| 5104.42 | 35 | 458 | 504 | + | Y | 0.028 | ns | *** | * | 19.87 | 20.29 | 20.84 | X, $, A | D5 | GHGLGHGHEQQHGLGHGHKFKLDDDLEHQGGHVLDHGHKHKHGHGHG |
| 5232.515 | 23 | 458 | 505 | 0 | Y | ns | ns | ns | ns | 19.9 | 20.21 | 20.65 | X, $, A | D5 | GHGLGHGHEQQHGLGHGHKFKLDDDLEHQGGHVLDHGHKHKHGHGHGK |
| 5369.574 | 10 | 458 | 506 | 0 | Y | ns | ns | ns | ns | 19.42 | 19.78 | 20.16 | X, $, A | D5 | GHGLGHGHEQQHGLGHGHKFKLDDDLEHQGGHVLDHGHKHKHGHGHGKH |
| 5611.712 | 48 | 458 | 508 | 0 | N | ns | ns | ns | ns | 20.94 | 21.52 | 21.64 | X, $, A | D5 | GHGLGHGHEQQHGLGHGHKFKLDDDLEHQGGHVLDHGHKHKHGHGHGKHKN |
| 5796.828 | 36 | 458 | 510 | + | Y | <.0001 | *** | **** | ns | 21.61 | 23.17 | 23.68 | X, $, A | D5 | GHGLGHGHEQQHGLGHGHKFKLDDDLEHQGGHVLDHGHKHKHGHGHGKHKNKG |
| 5924.923 | 26 | 458 | 511 | + | N | 0.0002 | ns | **** | ns | 19.78 | 20.67 | 21.4 | X, $, A | D5 | GHGLGHGHEQQHGLGHGHKFKLDDDLEHQGGHVLDHGHKHKHGHGHGKHKNKGK |
| 6740.229 | 31 | 438 | 497 | + | Y | <.0001 | *** | **** | ** | 20.58 | 22.04 | 22.83 | X, $, A | D5 | KHNLGHGHKHERDQGHGHQRGHGLGHGHEQQHGLGHGHKFKLDDDLEHQGGHVLDHGHKH |
n = number of complete data sets. “Start” and “End” are the positions of the starting and ending amino acids, numbering corresponds to UniProt sequence. “Trend” indicates a peptide’s change in abundance during pregnancy, where “+” indicates an increase, “-” indicates a decrease, and “0” indicates no significant trend. Normality was tested using the D’Agostino-Pearson omnibus normality test; the result is indicated in the “Normal” column, where “Y” = normal and “N” = not normal. For normally distributed data, p-values were determined by repeated-measurements one-way ANOVA and Tukey’s multiple comparison. For non-normally distributed data, p-values were determined by Friedman test and Dunn’s multiple comparison. p-values derived from multiple comparison tests are represented by stars according to Prism 6’s output, where * is p < 0.05; ** is p < 0.01; *** is p < .001; **** is p < .0001; and ns is not significant. In the “Prev Rep Seq” column, “X” indicates a peptide that contains the previously reported Gly58-His47333 sequence, “$” indicates His459-Asp49233, A indicates Gly458-Phe47727, and “^” indicates Lys498-Asn51330.
3.3. Analysis of proteolysis patterns and potential protease cleavage sites
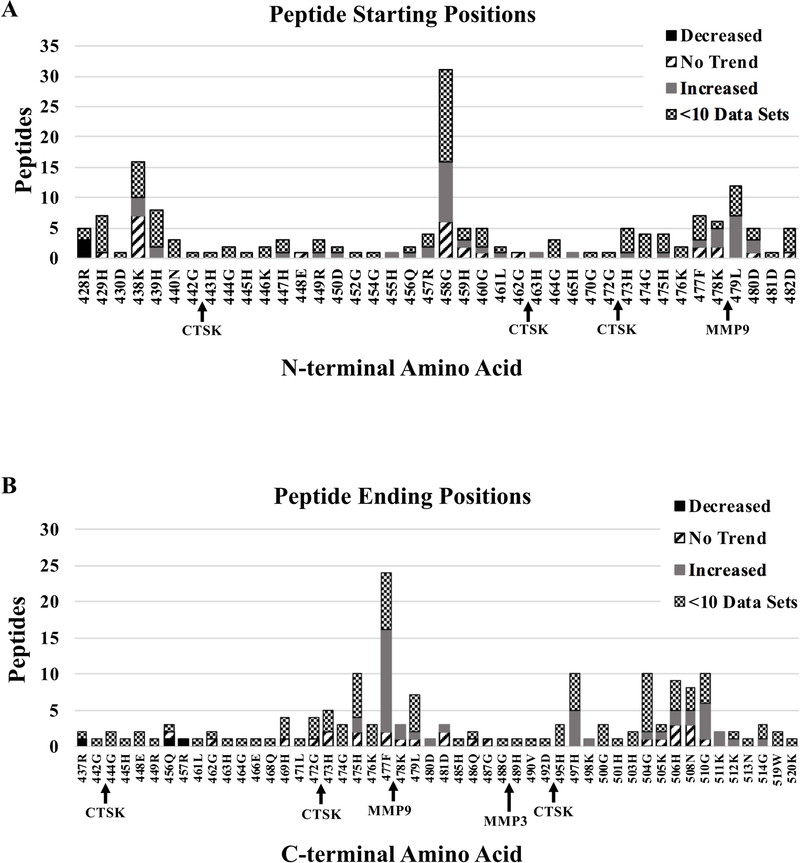
The N- and C- terminal positions of the 167 peptide sequences that mapped to D5 were counted. Figure 2a shows the number of D5 peptides that share the same N-terminal amino acid. The top three most-common N-terminal amino acids are 458G, 438K, and 479L, which revealed that the most common N-terminal proteolytic sites were between 457R-458G, 437R-438K, and 479L-480D. Figure 2b shows the number of D5 peptides that share the same C-terminal amino acid. In our study, the most common C-terminal amino acid is 477F, and the most common C-terminal cleavage site is 477F-478K. Four additional positions, 475H, 497H, 504G, and 510G, are tied for the second-most common C-terminal amino acid.
Figure 2.
Analysis of D5 Proteolysis. A. Bar graph showing the number of peptides sharing the same N-terminal amino acids. B. Bar graph showing the number of peptides sharing the same C-terminal amino acid. A–B. Bars are subdivided and color coded according to legend. “Decreased” indicates peptides that significantly decreased during the course of normal pregnancy. “Increased” indicates peptides that significantly increased during normal pregnancy while “no trend” indicates peptides that displayed no significant trend. “<10 Data sets” indicates peptides that presented less than 10 complete datasets and were therefore not statistically analyzed. Arrows underneath the bar graphs indicate protease cleavage sites predicted by PROSPER. Arrows in A indicate the protease is predicted to generate the start site immediately to the right of the arrow. Arrows in B indicates the protease is predicted to generate the end site immediately to the left of the arrow. CTSK = Cathepsin K, MMP9 = Matrix Metalloprotease 9, MMP3 = Matrix Metalloprotease 3.
Although many proteases are known to cleave HK, their proteolytic sites and the resulting HK fragments are not well defined. Therefore, we utilized PROSPER to predict which proteases could cleave HK. PROSPER proteolytic site predictions are indicated under the bar graphs with an arrow (Fig 2a–b). Cathepsin K (CTSK), matrix metalloproteinase 9 (MMP9), and matrix metalloproteinase 3 (MMP3) were predicted to cleave within D5; however, these predictions almost exclusively explain minor cleavage events in our cohort. The exception is a predicted MMP9 site that explains the third-most common N-terminal site, 478K-479L, in our cohort.
4. Discussion
This study is the first to systematically examine both HK protein and its natural occurring peptide dynamics in longitudinal serum samples during the course of normal pregnancy. Our results revealed decreased 120 kDa HK, increased 100 kDa intermediate, and increased HC and LC that comprise HKa. The 100 kDa band was previously reported to be an intermediate cleavage product created by the action of plasma kallikrein 13. Using LC-MS/MS analysis, we also revealed dynamic, gestational changes of naturally occurring HK-derived peptides. The majority of these peptides mapped to D5 and significantly increased in intensity during pregnancy. Overall, our results suggest increased proteolysis of HK to HKa as pregnancy progresses. HK D5 has been implicated in specific, pregnancy-related processes. D5 peptides have been shown to down-regulate angiogenesis, inhibit endothelial cell proliferation, and exert anti-microbial effects 11,27. Additionally, D5 binds heparin, induces vascular permeability, and binds to endothelial cells (EC) 28–30. Using LC-MS/MS analysis, we identified 174 naturally occurring HK-derived peptides. 167 of those peptides mapped to D5. This suggests that the LC is specifically targeted for additional proteolysis to release potentially biologically active peptides or to degrade and regulate HKa/D5 activity. Although prior studies designed overlapping synthetic peptides to completely cover domain 5 and used these in functional studies, to the best of our knowledge, we show for the first time that these sequences are naturally produced, with increased release of domain 5, in maternal circulation.
Many of the D5 peptides detected in our study contain previously reported functional synthetic peptides. Activation of HK occurs in circulation and on the EC surface. HKa maintains anti-proliferative and anti-adhesive signaling in HUVECs 31. Recombinant D5 was shown to induce apoptosis of cultured EC 32. Colman et al. synthesized D5 peptides and showed that synthetic peptides, His459-Asp492 and Gly458-His473, could inhibit in vivo angiogenesis and EC proliferation and migration 33. The His459-Asp492 sequence was detected in our serum samples with an m/z = 3797.7908. An additional 17 larger peptides in our cohort contained the His459-Asp492 sequence. Three peptides with ten or more complete data sets showed no trend, and six peptides significantly increased during normal pregnancy. The second sequence Gly458-His473 occurred in our samples with an m/z = 1656.762. The Gly458-His473 sequence was contained within 68 larger peptides. 34 of these peptides presented 10 or more complete data sets, with 11 peptides showing no trend and 23 peptides significantly increasing during the natural course of pregnancy. Zhang et al. showed that Lys498-Asn513, a synthetic peptide containing an EC binding site, inhibits EC proliferation 30. In our cohort, seven peptides contained this synthetic sequence, and one of significantly increased during pregnancy. Our results demonstrate a temporal increase of numerous D5 peptides with the potential to inhibit EC function and angiogenesis.
D5 shares both structural and functional features with antimicrobial peptides. The synthetic, Gly458-Phe477, showed potent antibacterial properties by radial diffusion assay27. We detected this sequence within 51 naturally occurring peptides. Of peptides with 10+ complete data sets, nine showed no trend, and twenty significantly increased during pregnancy. Our results confirm the natural production and temporal increase of numerous peptides containing a functional antimicrobial sequence in the serum of pregnant women. The diverse actions of D5 combined with the large number of peptides significantly changing during pregnancy strongly suggest that D5 is an important regulatory domain that requires further investigation. In contrast, three D5 peptides were identified that significantly decreased during pregnancy; however, these three peptides contain none of the previously annotated functional sequences.
Acute phase proteins, the complement pathway, and coagulation are important to the pathology of PE, making HK a potential target for understanding PE 5. Using a BK-release assay, Mohamed et al. compared five sites within placentas obtained from healthy and hypertensive pregnancies and found decreased LK and HK in PIH 6. Using western blot, Blumenstein et al. detected decreased HK in the plasma from women with PE complicated by small for gestational age at 20 weeks of gestation compared to healthy controls 7. However, LC-MS/MS analysis of serum samples revealed that peptides mapping to KNG-1 were increased in severe PE 5. Likewise, two-dimensional gel electrophoresis and MALDI-TOF, detected increased abundance of KNG-1 peptides in the plasma of HELLP patients 3. Wen et al. compared serum samples from healthy and late-term PE pregnancies and developed a diagnostic panel of 19 differentially expressed peptides, which included two HK peptides 4. Finally using BLOTCHIP and MALDI-TOF, Araki et al. reported a D5 peptide with an m/z = 2126.006 was significantly decreased in serum from PIH compared to healthy pregnancy 9. In our cohort, this peptide significantly increased during normal pregnancy.
Plasma kallikrein accounts for the two most frequent N-terminal proteolytic sites in our cohort. While numerous proteases are known to cleave HK, most of the proteolytic sites in our cohort remain unexplained because prior studies focused on BK release and HK activation. Proteolysis of HK produces highly vasoactive and pro-inflammatory kinins at sites of tissue injury and inflammation 27. Proteolysis by kallikrein dramatically changes the conformation of HK and exposes D5 34. FXI releases a large D5 fragment, Arg428-Lys520 35. Recently studied HK-interacting proteases include: MASP-1 and MASP-2 (lectin pathway of complement activation), neutrophil elastase (increased in PE), FXII (increased in response to estrogen and implicated in recurrent pregnancy loss), tissue kallikrein, plasmin, and calpains 36–40. Human mast cell tryptase was shown to cleave multiple sites within the LC of HK 41. Additionally, HK bound to ECs is internalized and fused to lysosomes, containing proteases such as cathepsins B and L 40.
PROSPER analysis predicted proteolysis by Cathepsin K, MMP-9, and MMP-3, which have been implicated in normal placentation, trophoblast invasion, and PE 42–45. However, this analysis is limited due to PROSPER’s exclusion of many known HK-interacting proteases from its database. For example, although kallikrein and MASP-1 are excluded from PROSPER, kallikrein has been shown to cleave the 457R-458G site while both kallikrein and MASP-1 can cleave the 437R-438K site, which were the two most common N-terminal cleavage sites in our cohort 37,46. Future studies should determine which proteases produce D5 peptides to clarify the role HK and its interacting proteases play in normal and pathogenic pregnancy.
Our data demonstrate the importance of studying dynamic, gestational changes of HK and HK-derived peptides. Further investigation is required to determine the mechanism of D5 peptide release, biological activity, and worth as a disease biomarker. Prior studies have not elaborated on the diversity of D5 peptides in serum despite interest in using HK peptides as disease biomarkers. Current challenges in developing a viable PE biomarker may be the result of comparing PE and control samples at a specific time point rather than considering differences in the trends of peptide abundance. Future studies comparing PE to healthy pregnancy might be clarified by considering peptide dynamic changes during the natural course of pregnancy.
Acknowledgements
The authors would like to thank Dr. Ann Gronowski from Washington University in St Louis for helping obtain the specimens, Drs. Zhe Cheng and Guoan Zhang from Proteomics and Metabolomics Core Facility at Weill Cornell Medicine for LC-MS/MS analysis and Dr. Christopher J. Lyon from Arizona State University for carefully editing the manuscript.
Funding
KRC, SD, SKD, WW and ZZ were supported by the Intermural Research Program at the National Institutes of Health Clinical Center.
List of Abbreviations
- BK
Bradykinin
- D
Domain
- EC
Endothelial cell
- HC
Heavy chain
- HK
High molecular weight kininogen
- HKa
Activated high molecular weight kininogen
- KKS
Kallikrein-kinin system
- LC
Light Chain
- LK
Low molecular weight kininogen
- PIH
Pregnancy-induced hypertension
- T
Trimester
Footnotes
Declarations
Ethics Approval and Consent to Participate
Not applicable. This study used de-identified specimens/data and investigators cannot readily ascertain the identities of the individuals to whom the data or samples belong. This study is not human subjects research.
Consent for Publication
Not applicable
Availability of Data and Material
All data generated or analyzed during this study are included in the manuscript. Any further information is available from the corresponding author on request.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Report of the National High Blood Pressure Education Program Working Group on High Blood Pressure in Pregnancy. Am J Obstet Gynecol 183, S1–S22, doi: 10.1067/mob.2000.107928 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Mannisto T et al. Elevated blood pressure in pregnancy and subsequent chronic disease risk. Circulation 127, 681–690, doi: 10.1161/CIRCULATIONAHA.112.128751 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heitner JC et al. Differentiation of HELLP patients from healthy pregnant women by proteome analysis--on the way towards a clinical marker set. J Chromatogr B Analyt Technol Biomed Life Sci 840, 10–19, doi: 10.1016/j.jchromb.2006.06.002 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Wen Q et al. Peptidomic Identification of Serum Peptides Diagnosing Preeclampsia. PLoS One 8, e65571, doi: 10.1371/journal.pone.0065571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu C et al. Proteomic analysis of human serum for finding pathogenic factors and potential biomarkers in preeclampsia. Placenta 32, 168–174, doi: 10.1016/j.placenta.2010.11.007 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohamed M, Larmie ET, Singh HJ & Othman MS Tissue kallikrein and kininogen levels in fetoplacental tissues from normotensive pregnant women and women with pregnancy-induced hypertension. Eur J Obstet Gynecol Reprod Biol 134, 15–19, doi: 10.1016/j.ejogrb.2006.09.004 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Blumenstein M, Prakash R, Cooper GJ, North RA & Consortium S Aberrant processing of plasma vitronectin and high-molecular-weight kininogen precedes the onset of preeclampsia. Reprod Sci 16, 1144–1152, doi: 10.1177/1933719109342756 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Hamamura K et al. Simple quantitation for potential serum disease biomarker peptides, primarily identified by a peptidomics approach in the serum with hypertensive disorders of pregnancy. Ann Clin Biochem 53, 85–96, doi: 10.1177/0004563215583697 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Araki Y et al. Quantitative peptidomic analysis by a newly developed one-step direct transfer technology without depletion of major blood proteins: Its potential utility for monitoring of pathophysiological status in pregnancy-induced hypertension. Proteomics 11, 2727–2737, doi:doi: 10.1002/pmic.201000753 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Kaplan AP & Silverberg M The coagulation-kinin pathway of human plasma. Blood 70, 1–15 (1987). [PubMed] [Google Scholar]
- 11.Lalmanach G, Naudin C, Lecaille F & Fritz H Kininogens: More than cysteine protease inhibitors and kinin precursors. Biochimie 92, 1568–1579, doi: 10.1016/j.biochi.2010.03.011 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Colman RW & Schmaier AH Contact system: a vascular biology modulator with anticoagulant, profibrinolytic, antiadhesive, and proinflammatory attributes. Blood 90, 3819–3843 (1997). [PubMed] [Google Scholar]
- 13.Schiffman S, Mannhalter C & Tyner KD Human high molecular weight kininogen. Effects of cleavage by kallikrein on protein structure and procoagulant activity. J Biol Chem 255, 6433–6438 (1980). [PubMed] [Google Scholar]
- 14.Gao X, Greenbaum LM, Mahesh VB & Brann DW Characterization of the kinin system in the ovary during ovulation in the rat. Biology of reproduction 47, 945–951 (1992). [DOI] [PubMed] [Google Scholar]
- 15.Valdes G, Figueroa CD & Corthorn J Temporospatial changes of kallikrein-like enzymes during the estrous cycle and pregnancy in the rat uterus. Biology of reproduction 55, 236–245 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Miatello R, Lama M, Gonzalez S, Damiani T & Nolly H Biochemical evidence of a kallikrein-like activity in rat reproductive tissues. Hypertension (Dallas, Tex. : 1979) 23, I193–197 (1994). [DOI] [PubMed] [Google Scholar]
- 17.Hermann A, Buchinger P, Somlev B & Rhebock J High and low molecular weight kininogen and plasma prekallikrein/plasma kallikrein in villous capillaries of human term placenta. Placenta 17, 223–230, doi: 10.1016/S0143-4004(96)90042-9 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Vonnahme KA et al. Detection of kallikrein gene expression and enzymatic activity in porcine endometrium during the estrous cycle and early pregnancy. Biology of reproduction 61, 1235–1241 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Vonnahme KA et al. Porcine endometrial expression of kininogen, factor XII, and plasma kallikrein in cyclic and pregnant gilts. Biology of reproduction 70, 132–138, doi: 10.1095/biolreprod.103.020412 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Weidmann H et al. The plasma contact system, a protease cascade at the nexus of inflammation, coagulation and immunity. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1864, 2118–2127, doi: 10.1016/j.bbamcr.2017.07.009 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Colman RW Biologic activities of the contact factors in vivo--potentiation of hypotension, inflammation, and fibrinolysis, and inhibition of cell adhesion, angiogenesis and thrombosis. Thromb Haemost 82, 1568–1577 (1999). [PubMed] [Google Scholar]
- 22.Sugi T & Makino T Factor XII, kininogen and plasma prekallikrein in abnormal pregnancies. Curr Drug Targets 6, 551–557 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Fan J et al. Low molecular weight protein enrichment on mesoporous silica thin films for biomarker discovery. Journal of visualized experiments : JoVE, doi: 10.3791/3876 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.UniProt Consortium, T. UniProt: the universal protein knowledgebase. Nucleic Acids Res 46, 2699, doi: 10.1093/nar/gky092 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song J et al. PROSPER: an integrated feature-based tool for predicting protease substrate cleavage sites. PLoS One 7, e50300, doi: 10.1371/journal.pone.0050300 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mori K & Nagasawa S Studies on human high molecular weight (HMW) kininogen. II. Structural change of HMW kininogen by the action of human plasma kallikrein. J Biochem 89, 1465–1473 (1981). [DOI] [PubMed] [Google Scholar]
- 27.Nordahl EA, Rydengard V, Morgelin M & Schmidtchen A Domain 5 of high molecular weight kininogen is antibacterial. J Biol Chem 280, 34832–34839, doi: 10.1074/jbc.M507249200 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Pixley RA, Lin Y, Isordia-Salas I & Colman RW Fine mapping of the sequences in domain 5 of high molecular weight kininogen (HK) interacting with heparin and zinc. J Thromb Haemost 1, 1791–1798 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Hasan AA, Cines DB, Herwald H, Schmaier AH & Muller-Esterl W Mapping the cell binding site on high molecular weight kininogen domain 5. J Biol Chem 270, 19256–19261 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Zhang JC et al. Two-chain high molecular weight kininogen induces endothelial cell apoptosis and inhibits angiogenesis: partial activity within domain 5. FASEB J 14, 2589–2600, doi: 10.1096/fj.99-1025com (2000). [DOI] [PubMed] [Google Scholar]
- 31.Tesfay L, Huhn AJ, Hatcher H, Torti FM & Torti SV Ferritin blocks inhibitory effects of two-chain high molecular weight kininogen (HKa) on adhesion and survival signaling in endothelial cells. PLoS One 7, e40030, doi: 10.1371/journal.pone.0040030 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo YL, Wang S & Colman RW Kininostatin, an angiogenic inhibitor, inhibits proliferation and induces apoptosis of human endothelial cells. Arterioscler Thromb Vasc Biol 21, 1427–1433 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Colman RW, Jameson BA, Lin Y, Johnson D & Mousa SA Domain 5 of high molecular weight kininogen (kininostatin) down-regulates endothelial cell proliferation and migration and inhibits angiogenesis. Blood 95, 543–550 (2000). [PubMed] [Google Scholar]
- 34.Weisel JW et al. The shape of high molecular weight kininogen. Organization into structural domains, changes with activation, and interactions with prekallikrein, as determined by electron microscopy. J Biol Chem 269, 10100–10106 (1994). [PubMed] [Google Scholar]
- 35.Mauron T, Lammle B & Wuillemin WA High molecular weight kininogen is cleaved by FXIa at three sites: Arg409-Arg410, Lys502-Thr503 and Lys325-Lys326. Thromb Haemost 83, 709–714 (2000). [PubMed] [Google Scholar]
- 36.Wiggins RC Kinin release from high molecular weight kininogen by the action of Hageman factor in the absence of kallikrein. J Biol Chem 258, 8963–8970 (1983). [PubMed] [Google Scholar]
- 37.Dobo J et al. Cleavage of kininogen and subsequent bradykinin release by the complement component: mannose-binding lectin-associated serine protease (MASP)-1. PLoS One 6, e20036, doi: 10.1371/journal.pone.0020036 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta AK, Gebhardt S, Hillermann R, Holzgreve W & Hahn S Analysis of plasma elastase levels in early and late onset preeclampsia. Archives of gynecology and obstetrics 273, 239–242, doi: 10.1007/s00404-005-0093-z (2006). [DOI] [PubMed] [Google Scholar]
- 39.Citarella F et al. Estrogen induction and contact phase activation of human factor XII. Steroids 61, 270–276 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Motta G & Tersariol ILS Modulation of the Plasma Kallikrein-Kinin System Proteins Performed by Heparan Sulfate Proteoglycans. Front Physiol 8, 481, doi: 10.3389/fphys.2017.00481 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coffman LG et al. Cleavage of high-molecular-weight kininogen by elastase and tryptase is inhibited by ferritin. Am J Physiol Lung Cell Mol Physiol 294, L505–L515, doi: 10.1152/ajplung.00347.2007 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Christensen J & Shastri VP Matrix-metalloproteinase-9 is cleaved and activated by cathepsin K. BMC Res Notes 8, 322, doi: 10.1186/s13104-015-1284-8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matjila M, Millar R, van der Spuy Z & Katz A The differential expression of Kiss1, MMP9 and angiogenic regulators across the feto-maternal interface of healthy human pregnancies: implications for trophoblast invasion and vessel development. PLoS One 8, e63574, doi: 10.1371/journal.pone.0063574 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Husslein H et al. Expression, regulation and functional characterization of matrix metalloproteinase-3 of human trophoblast. Placenta 30, 284–291, doi: 10.1016/j.placenta.2008.12.002 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plaks V et al. Matrix metalloproteinase-9 deficiency phenocopies features of preeclampsia and intrauterine growth restriction. Proc Natl Acad Sci U S A 110, 11109–11114, doi: 10.1073/pnas.1309561110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parthasarathy N, Torti SV & Torti FM Ferritin binds to light chain of human H-kininogen and inhibits kallikrein-mediated bradykinin release. Biochem J 365, 279–286, doi: 10.1042/BJ20011637 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]