Abstract
Iron is an essential element to all living organisms and plays a vital role in many cellular processes, such as DNA synthesis and energy production. The Mdm2 oncogene is an E3 ligase and known to promote tumor growth. However, the role of Mdm2 in iron homeostasis is not certain. Here, we showed that Mdm2 expression was increased by iron depletion but decreased by iron repletion. We also showed that Iron Regulatory Protein 2 (IRP2) mediated iron-regulated Mdm2 expression. Specifically, Mdm2 expression was increased by ectopic IRP2 but decreased by knockdown or knockout of IRP2 in human cancer cells as well as in mouse embryonic fibroblasts. Additionally, we showed that IRP2-regulated Mdm2 expression was independent of tumor suppressor p53. Mechanistically, we found that IRP2 stabilized Mdm2 transcript via binding to an iron response element (IRE) in the 3’UTR of Mdm2 mRNA. Finally, we showed that Mdm2 is required for IRP2-mediated cell proliferation and Mdm2 expression is highly associated with IRP2 in both normal and cancerous liver tissues. Together, we uncover a novel regulation of Mdm2 by IRP2 via mRNA stability and the IRP2-Mdm2 axis may play a critical role in cell growth.
Introduction
Iron is an essential nutrient for all living organisms. The most important feature of iron is that it acts as an elector donor or acceptor through interchanging between ferrous (Fe(II), Fe2+) and ferric (Fe(III), Fe3+) states. As a result, iron plays a vital role in many fundamental biochemical processes including oxygen transport, energy metabolism, and DNA synthesis [1]. However, iron can be toxic to the cells since it participates in Fenton reaction and generates hydroxyl radical, a reactive oxygen species (ROS) [2]. Indeed, numerous studies suggest that dysregulated iron metabolism occurs frequently in tumor cells and contributes to tumorigenesis [3]. Thus, the uptake, storage, and usage of iron need to be exquisitely tightly controlled at both systemic and cellular levels.
Iron regulatory protein 1 (IRP1) and IRP2, also known as ACO1 and IREB2, respectively, are RNA-binding proteins and play a fundamental role in regulating cellular iron homeostasis. IRP1/2 posttrancriptionally regulate a set of mRNAs encoding proteins involved in iron metabolism via binding to iron-responsive elements (IRES). Generally, the binding of IRPs to an IRE in the 5′ UTR of an mRNA represses its translation, while binding to an IRE in the 3′ UTR of an mRNA enhances its stability. Interestingly, the activities of IRP1/2 are also controlled by iron levels. When intracellular levels of iron are low, both IRP1/2 stabilize mRNAs for proteins involved in iron uptake, but represses the translation of mRNAs for proteins involved in iron storage and export. When intracellular levels of iron are high, both IRP1 and IRP2 are inactive: IRP1 functions as an aconitase and loses its RNA-binding ability whereas IRP2 undergoes iron-dependent protein degradation mediated by FBXL5 [4]. The biological significance of IRPs in cellular iron homeostasis has been underscored by mouse models. Genetic ablation of both IRPs in the mouse leads to embryonic lethality [5, 6]. However, mice deficient in either IRP1 or IRP2 are viable and fertile. Mice deficient in IRP2 develop abnormal body iron distribution, mild microcytic anemia, and neurodegeneration [7, 8]. IRP1 deficiency was initially found to be asymptomatic in mice [9], but later found to result in age-dependent erythropoietic abnormalities and dysregulation of body iron metabolism [10, 11]. Nevertheless, despite the role of IRP1/2 in iron homeostasis has been very well established, very litter is known about their roles in tumorigenesis.
The murine double minute-2 (MDM2) gene was originally identified as a gene that is located on double minute chromosomes (acentromeric extrachromosomal nuclear bodies) in a spontaneously transformed mouse cell line, 3T3-DM [12] and subsequently found to induce tumorigenicity when overexpressed [13]. Later, Mdm2 was found to be a negative regulator of tumor suppressor p53 by either inhibiting p53 transcriptional activities or targeting p53 for proteasomal degradation [14]. The importance of Mdm2 as a negative regulator of p53 is exemplified by elegant mouse models. Homozygous deletion of the MDM2 gene leads to embryonic lethality due to inappropriate apoptosis, which can be rescued by concomitant deletion of the p53 gene [15]. Additionally, MDM2 was found to exert p53-independent activities by interacting with an array of proteins, such as RB, Numb, and p21, and modulates their expression and/or activities [16, 17]. However, very little is known whether Mdm2 plays a role in iron homeostasis.
In this study, we examined whether Mdm2 is involved in iron metabolism. We found that Mdm2 expression is regulated by iron levels, which is mediated by IRP2 via mRNA stability. We also found that an IRE in the 3’UTR of Mdm2 mRNA is required for IRP2 to regulate Mdm2 expression. Furthermore, we showed that Mdm2 is required for IRP2-regulated cell proliferation. Thus, our data suggest a role of Mdm2 in iron homeostasis and the IRP2-Mdm2 axis may play a role in promoting tumor cell growth.
Materials and Methods
Mice and MEF isolation
IRP2+/− mice (ID: MMRRC:030490-MU) were obtained from Mutant Mouse Resource and Research Center (MMRRC) at the University of Missouri. p53+/− mice were obtained from Jackson Laboratory as described previously [18]. All animals and use protocols were approved by the University of California at Davis Institutional Animal Care and Use Committee. MEFs are isolated from 12.5 to 13.5 postcoitum (p.c.) mouse embryos as described previously [19]. IRP2+/− mice were intercrossed to generate WT and IRP2−/− MEFs. p53+/−; Irp2+/− mice were intercrossed to generate WT, Irp2−/−, p53+/−, p53−/−, p53+/−;Irp2−/−, and p53−/−;Irp2−/− MEFs. MEFs were cultured in DMEM supplemented with 10% FBS (HyClone), 55 μM β-mercaptoethanol, and 1× non-essential amino acids (NEAA) solution (Cellgro).
Cell culture, cell line generation and reagents
HCT116, MCF7, HepG2, H1299, and Mia-PaCa2 cells were obtained from ATCC. p53−/−HCT116 cells were obtained from Vogelstein’s group [20]. All the cells in this study were used at below passage 20. All the cells and their derivatives were cultured in DMEM (Dulbecco’s modified Eagle’s medium, Invitrogen) supplemented with 10% fetal bovine serum (Gibco). H1299 cells that can inducibly express IRP1 or IRP2 under a tetracycline inducible system was previously generated [21, 22]. Isogenic control and IRP2-KO p53−/−HCT116 and Mia-PaCa2 cells were previously generated [23]. Diisopropyl fluorophosphate (DFP) and Deferoxamine mesylate (DFO) were purchased from Sigma.
Plasmids
pcDNA3-HA-EGFP vector was previously generated [23]. pcDNA3 vector expressing IRP2 were previously generated [22]. pcDNA3 vector expressing Mdm2 were previously generated [24]. To generate pcDNA3-HA-EGFP vector containing A, B, C, and D fragment of Mdm2 3’UTR, a DNA fragment was digested from pcDNA4 vectors containing A, B, C, or D fragment of Mdm2 3’UTR [25] and the resulted DNA fragments were then inserted into pcDNA3-HA-EGFP vector via NotI. To generate pcDNA3-HA-EGFP vector containing A1, A2, or A3 fragment of Mdm2 3’UTR, a DNA fragment was amplified using A fragment of Mdm2 3’UTR as a template and the resulted DNA was then cloned into pcDNA3-HA-EGFP vector via NotI and XhoI sites. The primers for A1 fragment were forward primer, 5’-AAT GCG GCC GCT TGA CCT GTC TAT AAG AGA ATT ATA TAT TTC-3’, and reverse primer, 5’-ATC TCG AGC ACG GTG AAA CCC TGT CTC TAC-3’. The primers for A2 fragment were forward primer, 5’-AAT GCG GCC GCT TAG CCA GGA TGG TCT CGA TC-3’, and reverse primer, 5’-ATC TCG AGC ACC ATG TTG GCC AGG CTG G-3’. The primers for A3 fragment were forward primer, 5’-AAT GCG GCC GCT CAC GAA CTC CTG ACC TCA AG-3’and reverse primer, 5’-ATC TCG AGT TTG GGA GGC TGA GGT GAG TAG-3’. To generate pcDNA3-HA-EGFP vector with point mutation for IRE A2, two steps PCR were used. The first step PCR was to amplify two DNA fragments (#1 and #2) using A2 fragment as a template. To amplify DNA fragment#1, the forward primer was 5’-AAT GCG GCC GCT TAG CCA GGA TGG TCT CGA TC-3’ and the reverse primer was 5’TAG ATC ATG ACA CTT CAC TCC AG 3’. To amplify DNA fragment #2, the forward primer was 5’CTG GAG TGA AGT GTC ATG ATC TA 3’ and the reverse primer was 5’-ATC TCG AGC ACC ATG TTG GCC AGG CTG G-3’. The second step PCR was performing by using DNA fragment #1 and #2 as templates along with the forward primer for DNA fragment #1 and the reverse primer for DNA fragment #2. The resulting PCR product was then cloned into pcDNA3-HA-EGFP via NotI and XhoI sites.
RNAi
Scrambled siRNA(5′-GCA GTG TCT CCA CGT ACT A-3’), IRP2 siRNA (5’-GCA AAC ATG TGT CCG GAA T −3’) and IRP1 siRNA (5’-GAA CGA UAC ACU AUC AUU A-3’) were purchased from Dharmacon. SiRNA transfection were performed using RNAiMAX according to the User’s manual (Life Technology).
Western blot analysis
Western blot analysis was performed as previously described [26]. Briefly, whole cell lysates were harvested by 2×SDS/sample buffer. Proteins were separated in 7-13% SDS-polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with indicated antibodies, followed by detection with enhanced chemiluminescence. The antibodies against Mdm2, actin, vinculin, TFR and IRP2 were purchased from Santa Cruz biotechnology. The antibody against HA was purchased from Covance. The mouse IRP2 antibody [27] was a gift from Dr. Leibold at the University of Utah.
RNA isolation, RT-PCR, and quantitative PCR
Total RNA was isolated with Trizol reagent as described according to the user’s manual. cDNA was synthesized with Reverse Transcriptase (Promega) and used for RT-PCR. The PCR program used for amplification was (i) 94°C for 5 minutes, (ii) 94°C for 45 seconds, (iii) 58°C for 45 seconds, (iv) 72°C for 30 seconds, and (v) 72°C for 10 minutes. From steps 2 to 4, the cycle was repeated 22-30 times depend on the transcripts amplified. For Quantitative PCR (qPCR) was performed in 20-μl reactions using 2X qPCR SYBR Green Mix (ABgene, Epsom, UK) with 5 μM primers. Reactions were run on a StepOnePlus Real-Time PCR System (Life Technology) using a two-step cycling program: 95°C for 15 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, 68°C for 30 s. A melt curve (57-95°C) was generated at the end of each run to verify the specificity. The primers for human Mdm2 were forward primer, 5’-GAA CTT GGT AGT AGT CAA TCA GC −3’, and reverse primer, 5’-GCC TGA TAC ACA GTA ACT TGA TA −3’. The primers for mouse Mdm2 were forward primer, 5’-ATG AGG TCT ATC GGG TCA CAG T-3’, and reverse primer, 5’-CAC ATC CAA GCC TTC TTC TGC-3’. The primers for human IRP1 were forward primer, 5’ ATA AAC CCT GTC TGC CCT GC 3’, and reverse primer, 5’ AGC CAA TCA CCT GAG GAA GC 3’. The primers for Human TFR were forward primer, 5’-ACG CCA GAC TTT GCT GAG TT −3’, and reverse primer, 5’-GAG GAG CCA GGA GAG GAC TT −3’. The primers for human GAPDH were forward primer, 5’-AGC CTC AAG ATC ATC AGC AAT G-3’, and reverse primer, 5’-ATG GACTGT GTC ATG AGT CCT T-3’. The primers for both human and mouse actin were forward primer, 5’-TCC ATC ATG AAG TGT GAC GT-3’, and reverse primer, 5’-TGA TCC ACA TCT GCT GGA AG −3’. The primers for mouse IRP2 were forward primer, 5’-TTT GCA ACC AGT GCC TGA AC-3’, and reverse primer, 5’-CCA TTC CAG TTC CAG GAG GG −3’. The primers for mouse GAPDH were forward primer, 5’-AAC TTT GGC ATT GTG GAA GG-3’, and reverse primer, 5’-GGA TGC AGG GAT GAT GTT CT-3’.
RNA-ChIP
RNA-chromatin immunoprecipitation (RNA-ChIP) was performed as described [28]. Briefly, H1299 cells were uninduced or induced to express HA-tagged IRP2 for 24 hours and cells were lysed in a buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPEs, 1 mM 1,4-Dithiothreitol (DTT), and 0.5% NP-40). The cell lysates were incubated with 1 μg of anti-HA or isotype control IgG at 4 °C overnight. The RNA–protein immunocomplexes were brought down by magnetic protein A/G beads (MCE), followed by RT-PCR analysis.
mRNA half-life assay
The stability of Mdm2 mRNA was examined as previously describe [25]. Briefly, cells were treated with 5, 6-dichloro-1--D-ribofuranosylbenzimidazole (DRB), an inhibitor of transcription, for various time. The total RNAs were isolated and the levels of Mdm2 mRNA were measured by qRT-PCR. The relative levels of Mdm2 were normalized by the levels of actin mRNA, which were then plotted versus time to calculate the half-life of Mdm2 mRNA.
Colony formation assay
1x103 cells were seeded in triplicate in a well of the six-well plate. Colonies were then fixed with methanol/glacial acetic acid (7:1) and stained with 0.1% of crystal violet. To quantify the colony results, Image J software was used with the ColonyArea plugin as previously described [29].
Results
Mdm2 expression is regulated by iron.
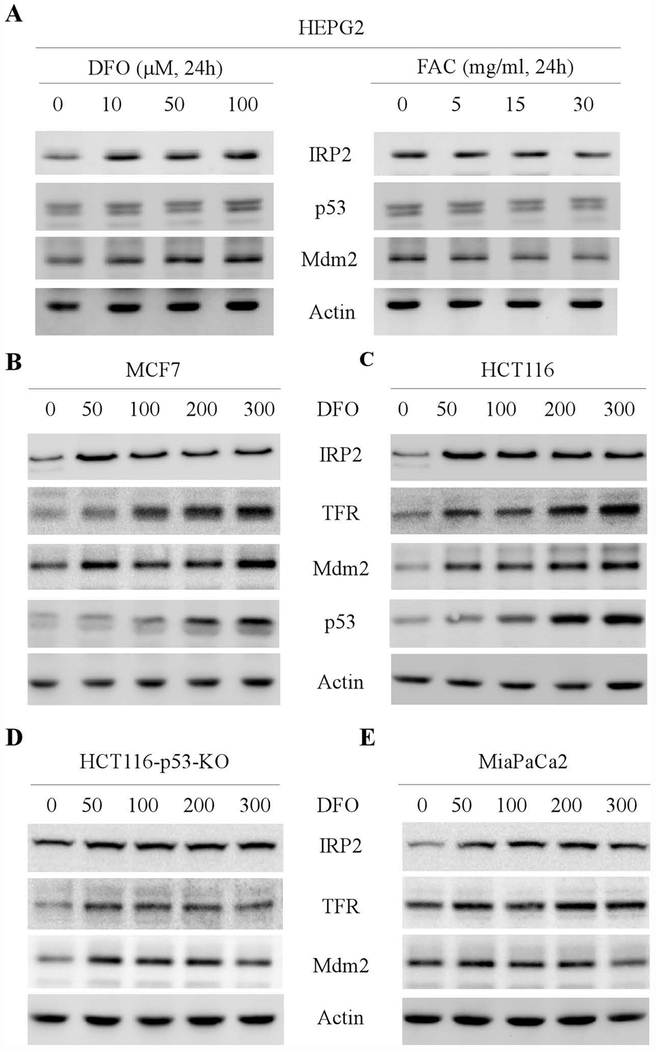
To determine whether Mdm2 plays a role in iron homeostasis, HEPG2 cells were treated with various amounts of deferoxamine (DFO), an iron chelator, or ferric ammonium acetate (FAC), an iron source. We found that IRP2 was increased by DFO but decreased by FAC (Fig. 1A, IRP2 panel), consistent with previous report [22]. Interestingly, the levels of Mdm2 proteins were also increased by DFO treatment but reduced by FAC (Fig. 1A, Mdm2 panels), suggesting that Mdm2 expression is regulated by iron levels. To verify this, MCF7 and HCT116 cells were mock-treated or treated with various amount of DFO and the levels of IRP2, TFR, Mdm2 and p53 were examined by western blot analysis. TFR is a known target of IRP2 and thus was used as a positive control [30]. Indeed, we found that DFO treatment resulted in increased expression of Mdm2 in both MCF7 and HCT116 cells, concomitantly with an increase in IRP2, TFR, and p53 (Fig. 1B–C). Since Mdm2 is a target of p53, which was also increased by DFO, it is thus necessary to examine whether wild-type p53 is required for iron-regulated Mdm2 expression. In this regard, p53-KO HCT116 cells and mutant p53(R248W)-containing Mia-PaCa2 cells were used. We found that in response to DFO treatment, Mdm2 expression was increased in both p53-KO HCT116 and MiaPaC2 cells, which was accompanied with an increased expression of IRP2 and TFR (Fig. 1D–E). Together, these data suggest that Mdm2 expression is regulated by iron in a p53-independent manner.
Figure 1.
Mdm2 expression is regulated by iron. (A) HEPG2 cells were mock-treated or treated with various amounts of DFO or FAC, followed by western blot analysis to examine the levels of IRP2, Mdm2 and actin proteins. (B-C) MCF7 (B) and HCT116 (C) cells were mock-treated or treated with DFO (0-300 μM, 24h) for 24 hours and the levels of IRP2, TFR, Mdm2, p53, and actin proteins were examined by western blot analysis. (D-E) HCT116-p53-KO (D) and Mia-PaCa2 (E) cells were mock-treated or treated with DFO (0-300 μM, 24h) for 24 hours and the levels of IRP2, TFR, Mdm2, and actin proteins were examined by western blot analysis.
IRP2, but not IRP1, is required for iron-mediated Mdm2 expression
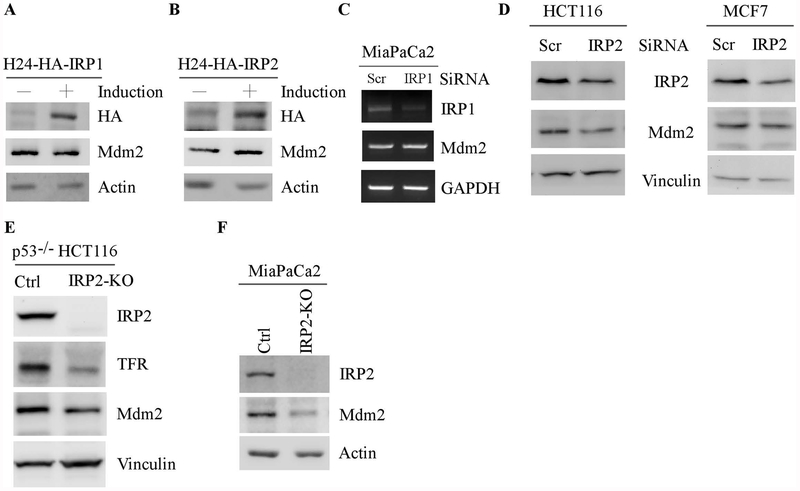
To identify a regulator(s) that mediates iron-regulated Mdm2 expression, we examined whether Mdm2 is regulated by IRP1 or IRP2. H1299 cells that can inducibly express HA-tagged IRP1 and IRP2 were used. As shown in Fig. 2A–B, IRP1 and IRP2 were expressed upon induction (Fig. 2A–B). Interestingly, we found that ectopic IRP2, but not IRP1, was capable of increasing Mdm2 expression (Fig. 2A–B, Mdm2 panels). Next, to verify this, siRNAs against IRP1 or IRP2 were used. We found that knockdown of IRP1 had very little effect on the levels of Mdm2 transcripts in Mia-PaCa2 cells (Fig. 2C). By contrast, knockdown of IRP2 led to reduced levels of Mdm2 proteins in both HCT116 and MCF7 cells (Fig. 2D, Mdm2 panels). To further verify this, IRP2-KO p53−/− HCT116 and Mia-PaCa2 cells in that the IRP2 gene was deleted by CRISPR/Cas9 were used to examine Mdm2 expression along with isogenic control cells. As expected, IRP2 protein was undetectable in IRP2-KO p53−/− HCT116 and Mia-PaCa2 cells (Fig 2E–F, IRP2 panels). Importantly, the level of Mdm2 protein was reduced by IRP2-KO in both p53−/− HCT116 and Mia-PaCa2 cells (Fig, 2D–E, Mdm2 panels). Together, these data suggest that IRP2 is required for Mdm2 expression.
Figure 2.
IRP2, but not IRP1, is required for iron-mediated Mdm2 expression. (A-B) H1299 cells were uninduced or induced to express HA-tagged IRP1 (A) or IRP2 (B) for 24 hours, the levels of IRP1 (A), IRP2 (B), Mdm2, and actin proteins were examined by western blot analysis. (C) The level of IRP1, Mdm2, and GAPDH transcripts was measured in Mia-PaCa2 cells transfected with a scrambled or IRP1 siRNA for 3 days. (D) HCT116 and MCF7 cells were transfected with a scrambled or IRP2 siRNA for 3 days, and the levels of IRP2, Mdm2 and vinculin were examined by western blot analysis. (E-F) Isogenic control and IRP2-KO p53−/−HCT116 (D) and Mia-PaCa2 (E) cells were used to examine the levels of IRP2, Mdm2, vinculin (D) and actin (E) proteins by western blot analysis.
Loss of IRP2 leads to reduced Mdm2 expression in primary mouse embryonic fibroblasts independent of p53
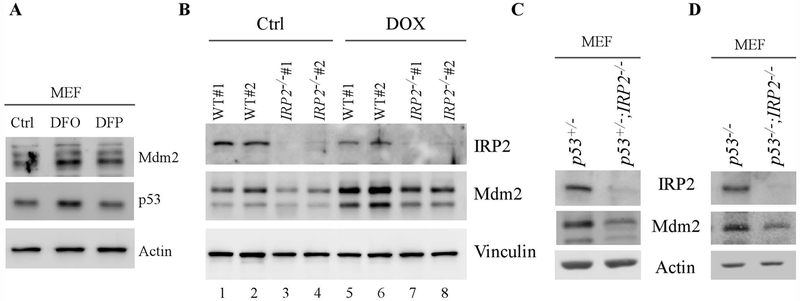
To gain more insight into the regulation of Mdm2 by iron, primary mouse embryonic fibroblasts (MEFs) were mock-treated or treated with iron chelator, DFO or DFP. We showed that the levels of both Mdm2 and p53 proteins were increased by DFO and, to a less extent, by DFP (Fig. 3A, Mdm2 and p53 panels), consistent with the data obtained from human cancer cells (Fig. 1). Next, we examined whether loss of IRP2 has an effect on Mdm2 expression in MEFs. As shown in Fig. 3B, two sets of WT and IRP2−/− MEFs were generated and used to examine Mdm2 expression in the presence or absence of doxorubincin, a DNA damage agents known to increase Mdm2 expression. Indeed, we found that the levels of Mdm2 proteins were reduced by knockout of IRP2 regardless of doxorubicin treatment (Fig. 3B, Mdm2 panel, compare lanes 1-2 and 5-6 with 3-4 and 7-8, respectively). Additionally, we found that IRP2 protein was decreased by doxorubicin in MEFs (Fig. 3B, IRP2 panel, compare lanes 1-2 with 3-4), suggesting a role of IRP2 in DNA damage response. To verify that the regulation of Mdm2 by IRP2 is independent of p53, a cohort of p53+/−, p53+/−;IRP2−/−, p53−/−, and p53−/−;IRP2−/− MEFs were generated. We showed that the level of Mdm2 was decreased in p53+/−;IRP2−/− and p53−/−;IRP2−/− MEFs when compared to that in p53+/− and p53−/− MEFs, respectively (Fig. 3C–D). Together, these data indicate that IRP2 is required for Mdm2 expression in MEFs in a p53-independent manner.
Figure 3.
Loss of IRP2 leads to reduced Mdm2 expression in primary mouse embryonic fibroblasts independent of p53. (A) The levels of Mdm2, p53 and actin proteins were examined in wild-type MEFs treated with or without DFO or DFP. (B) Wild-type and IRP2−/− MEFs were mock-treated or treated with doxorubicin for 18 hours and the levels of IRP2, Mdm2, and vinculin proteins were determined by western blot analysis. (C) The levels of IRP2, Mdm2, and vinculin proteins were examined by western blot analysis in p53+/− and p53+/−;IRP2−/− MEFs. (D) The levels of IRP2, Mdm2, and actin proteins were examined by western blot analysis in p53−/− and p53−/−;IRP2−/− MEFs.
The level of Mdm2 transcripts is reduced by loss of IRP2
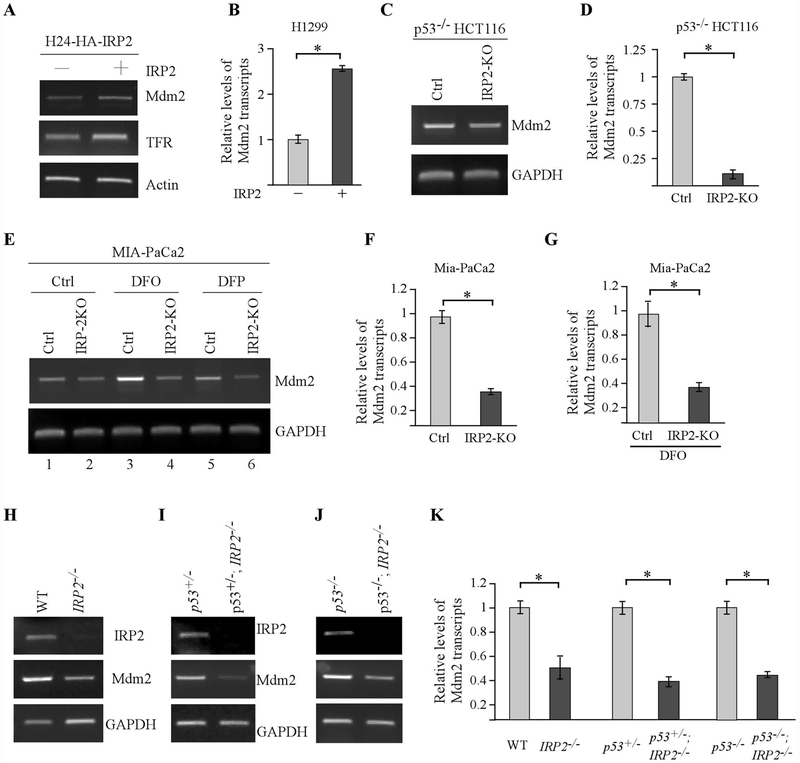
To understand how IRP2 regulates Mdm2 expression, H1299 cells that were uninduced or induced to express IRP2 were used to examine the levels of Mdm2 transcripts by semi-quantitative and quantitative RT-PCR. We found that upon induction of IRP2, Mdm2 transcripts were up-regulated (Fig. 4A–B). To verify that Mdm2 transcripts is regulated by endogenous IRP2, isogenic control and IRP2-KO p53−/− HCT116 cells were used to measure the levels of Mdm2 transcripts by semi-quantitative and quantitative RT-PCR. We found that loss of IRP2 decreased the level of Mdm2 transcripts in p53−/− HCT116 (Fig. 4C–D). In line with this, knockout of IRP2 decreased the level of Mdm2 transcripts in Mia-PaCa2 cells mock-treated or treated with DFO or DFP (Fig.4E, compare lanes 1, 3, and 5 with 2, 4 and 6, respectively). We also noticed that the level of Mdm2 transcripts was elevated by DFO or DFP in Mia-PaCa2 (Fig. 4E, compare lane1 with lanes 3 and 5), consistent with the observation that iron chelator augments the level of Mdm2 proteins (Fig. 1). Additionally, quantitative RT-PCR analysis confirmed that loss of IRP2 led to reduced levels of Mdm2 transcripts in Mia-PaCa2 cells mock-treated or treated by DFO (Fig. 4F–G). Furthermore, we examined the level of Mdm2 transcript in three sets of MEFs: WT vs IRP2−/−, p53+/− vs IRP2−/−;p53+/−, and p53−/− vs IRP2−/−;p53−/−. We found that the level of Mdm2 transcript was much lower in IRP2−/−, IRP2−/−;p53+/− and IRP2−/−;p53+/− MEFs than that in WT, p53+/− and p53−/− MEFs, respectively (Fig. 4H–K). Together, these data indicate that IRP2 regulates Mdm2 expression at the level of mRNA.
Figure 4.
The level of Mdm2 transcripts is reduced by loss of IRP2. (A-B) H1299 cells were uninduced or induced to express HA-tagged IRP2 for 24 hours and the levels of Mdm2, TFR, and actin transcripts were examined by semi-quantitate (A) or quantitative (B) RT-PCR analysis. * indicates p < 0.05 by paired Student’s t test. (C-D) The levels of Mdm2 and GAPDH transcripts were examined in isogenic control and IRP2-KO p53−/−HCT116 cells by semi-quantitate (C) or quantitative (D) RT-PCR analysis. * indicates p < 0.05 by paired Student’s t test. (E-G) The levels of Mdm2 and GAPDH transcripts were examined in isogenic control and IRP2-KO Mia-PaCa2 cells treated with or without DFO or DFP by semi-quantitate (E) or quantitative (F-G) RT-PCR analysis. * indicates p < 0.05 by paired Student’s t test. (H-J) The levels of IRP2, Mdm2 and GAPDH transcripts were examined in wild-type and IRP2−/− MEFs (H), p53+/− and p53+/−;IRP2−/− (I), and p53−/− and p53−/−;IRP2−/− MEFs (J). (K) Quantitative RT-PCR analysis was performed with the samples in H-J. * indicates p < 0.05 by paired Student’s t test.
IRP2 is required for Mdm2 mRNA stability via binding to the IRE in its 3’UTR
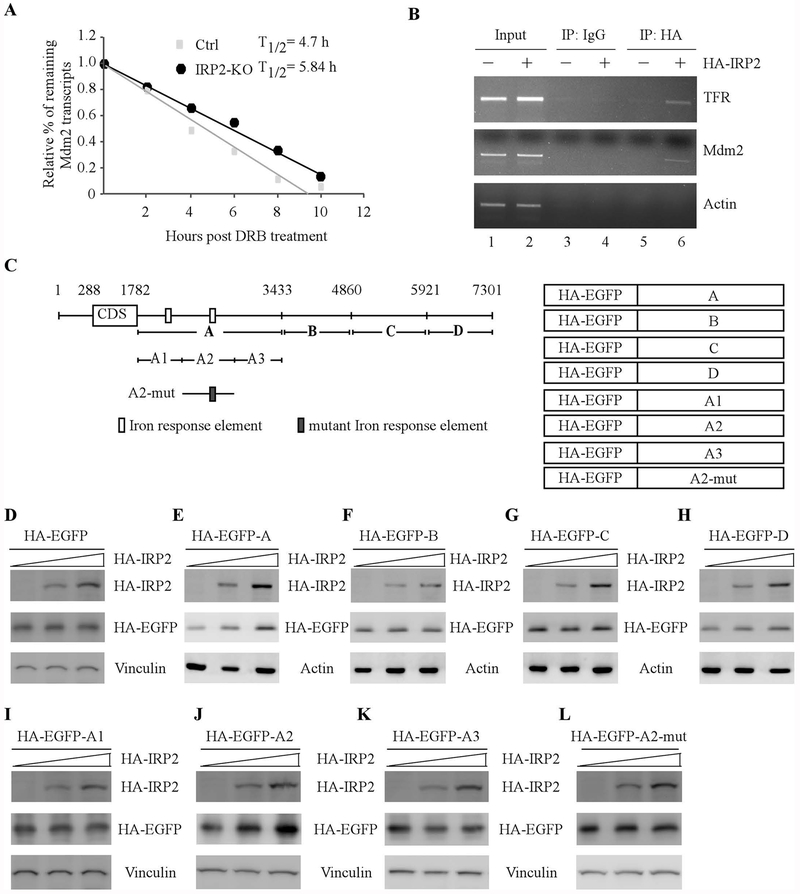
Since Mdm2 transcripts are altered by IRP2 (Fig. 4), we asked whether IRP2 regulates Mdm2 mRNA stability. To test this, the half-life of Mdm2 mRNA was measured in isogenic control and IRP-KO p53−/− HCT116 cells treated with or without DRB for various time. DRB inhibits transcription elongation mediated by RNA polymerase II, and thus inhibiting mRNA synthesis in eukaryotic cells [31]. Indeed, we found that the half-life of Mdm2 transcripts was decreased from 5.84 h in isogenic control cells to 4.72 h in IRP-KO cells (Fig. 5A). Next, RNA-Chip assay was performed to examine whether IRP2 binds to Mdm2 transcripts by using H1299 cells with or without IRP2 expression. We found that Mdm2 mRNA was detectable in IRP2, but not in control IgG, immunoprecipitates (Fig. 5B, Mdm2 panel, compare lane 5 and 6 with lane 3 and 4, respectively). Additionally, we showed that IRP2 bound to TFR, a known target of IRP2 [30], but not actin mRNA (Fig. 5B, TFR and actin panels, compare lane 5 with 6).
Figure 5.
IRP2 is required for Mdm2 mRNA stability via binding to the IRE in its 3’UTR. (A) The half-life of Mdm2 transcripts was measured in isogenic control and IRP2-KO p53−/−HCT116 cells treated with DRB for various times. The level of MDM2 transcript was normalized to that of GAPDH control and the relative half-life of MDM2 was calculated. (B) IRP2 associates with MDM2 transcript in vivo. H1299 cells were uninduced or induced to express HA-tagged IRP2, followed by immunoprecipitation with anti-HA or an isotype control IgG. RT-PCR analysis was performed to measure the level of TFR, MDM2 and GAPDH transcripts in the control and IRP2-RNA immunocomplexes. (C) Schematic representation of the Mdm2 transcript, the locations of IREs as well as the reporters containing various lengths of Mdm2 3’UTRs. (D-H) p53−/− HCT116 cells were transiently transfected with various amounts of IRP2 expression vector and a fixed amount of EGFP expression vector alone (D) or a EGFP vector fused with Mdm2-3’UTR-A (E), Mdm2-3’UTR-B (F), Mdm2-3’UTR-C (G), or Mdm2-3’UTR-D (H). Twenty-four hours posttransfection, the levels of IRP2 (D-H), EGFP (D-H), vinculin (D), and actin (E-H) proteins were examined by western blot analysis. (I-L) The experiments were performed as in (D-H) except that an EGFP vector fused with Mdm2-3’UTR-A1 (I), Mdm2-3’UTR-A2 (J), Mdm2-3’UTR-A3 (K), or Mdm2-3’UTR-A2-M (L) were used.
To explore the mechanism by which IRP2 stabilizes Mdm2 transcripts, we sought to delineate the binding site of IRP2 in Mdm2 transcript. To address this, we generated several EGFP expression vectors fused with various length of Mdm2 3’UTR fragments that span the entire Mdm2 3’UTR (Fig. 5C). Next, these vectors were transfected into p53−/− HCT116 cells along with various amounts of IRP2 expression vector. We showed that IRP2 had very little effect on EGFP expression when co-transfected with a vector only containing EGFP coding region (Fig. 5D). Interestingly, the levels of EGFP proteins were increased by IRP2 when co-transfected with a vector containing the A fragment of Mdm2 3’UTR (Fig. 5E). By contrast, IRP2 did not alter EGFP expression when co-transfected with a vector containing the B, C, or D fragment of Mdm2 3’UTR (Fig. 5F–H). To further map the binding site of IRP2 in the A fragment of Mdm2 3’UTR, three more reporters were generated and named as A1, A2 and A3 (Fig. 5C). We found that IRP2 was able to enhance EGFP expression when co-transfected with a vector containing A2 fragment, but not A1 or A3 fragment (Fig. 5I–K). Interestingly, we found that there is one potential IRE located in the A2 fragment (Fig. 5C). To determine whether the IRE in the A2 fragment is required for IRP2 to bind to Mdm2 transcripts, a reporter in that the “C” nucleotide in the IRE loop sequence was substituted with “A” was generated and named as A2-M. Consequently, we found that A2-M was not responsible to IRP2 (Fig. 5L). Together, our data suggest that IRP2 stabilizes Mdm2 transcript via binding to the IRE in the 3’UTR of Mdm2 transcript.
The IRP2-Mdm2 axis plays a role in cell proliferation
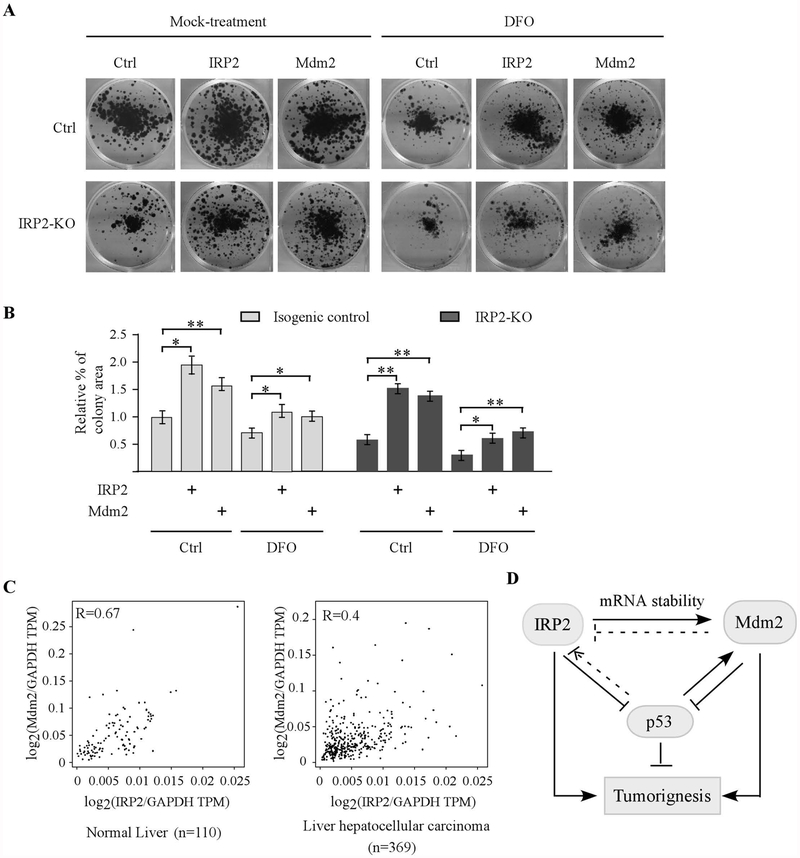
Both Mdm2 and IRP2 are found to promote cell proliferation [32, 33]. Thus, to determine the biological significance of IRP2-mediated Mdm2 expression, colony formation assay was performed with isogenic control and IRP2-KO p53−/− HCT116 cells transiently transfected with a control vector or a vector expressing IRP2 or Mdm2, followed with or without DFO treatment. We found that under mock-treatment condition, loss of IRP2 led to reduced number of colonies, which can be rescued by ectopic IRP2 or Mdm2 (Fig. 6A–B and Supplemental Figure 1). Upon DFO treatment, loss of IRP2 led to a further reduction in the number of colonies, which was then attenuated by ectopic IRP2 or Mdm2 (Fig. 6A–B and Supplemental Figure 1). Furthermore, we searched the GEPIA database (www.gepia.cancer-pku-cn) for the expression profile of IRP2 and Mdm2 in both normal and cancer tissues. Interestingly, we found that IPR2 expression was associated with Mdm2 expression in both normal (Pearson’s r=0.67) and cancerous liver tissues (Pearson’s r=0.4) (Fig. 6C). We also showed that unlike IRP2 and Mdm2, the correlation between IRP1 and Mdm2 is much less significant in the same samples (Pearson’s r=0.3 in normal liver tissues and Pearson’s r=0.31 in cancerous liver tissues) (Supplemental Fig. 2). Together, these data suggest that IRP2-regualted Mdm2 expression is critical for cell growth.
Figure 6.
The IRP2-Mdm2 axis plays a role in cell proliferation. (A) Isogenic control and IRP2-KO p53−/− HCT116 cells were transiently transfected with a control vector or a vector expressing IRP2 or Mdm2 for 24 hours, followed by colony formation with or without treatment of DFO (100 μM, 24 hours). (B) Quantification of colony formation assays in (A). * indicates p < 0.05 and ** indicates p< 0.01 by paired Student’s t test. (C) Mdm2 expression is associated with IRP2 expression in normal liver tissues (left panel) and liver hepatocellular carcinomas (right panel). The analysis was performed using the GEPIA2 database (http://gepia2.cancer-pku.cn/#correlation). Statistical analysis suggests a strong correlation between IRP2 and Mdm2 expression in normal liver tissues (Pearson’s r=0.67) and cancerous liver tissues (Pearson’s r=0.4). (D) A model proposed to elucidate the interplay among IRP2, Mdm2, and p53.
Discussion
Dysregulation in iron homeostasis leads to a variety of disorders, such as anemia and cancers. Recent studies indicate that cancer cells rely on a large amount of iron for proliferation and as a result, iron has been found to play a critical role in tumor initiation, progression, and metastasis [1, 3]. However, the underlying mechanism how irons controls cancer cell growth remains to be elucidated. In this study, we showed that the expression of Mdm2 oncogene is altered by iron depletion and repletion. To understand the mechanism, we showed that IRP2 is the major regulator involved in iron-regulated Mdm2 expression. We showed that over-expression of IRP2 stabilizes, whereas knockdown or knockout of IRP2 destabilizes, Mdm2 transcripts via the IRE in its 3’UTR. Furthermore, we showed that Mdm2 is required for IRP2-regulated cell growth and that the level of Mdm2 expression is associated with the level of IRP2 expression in both normal and cancerous liver tissues. Together, these data indicate a novel regulation of Mdm2 by IRP2 and the IRP2-Mdm2 axis may play a critical role in cell growth by modulating iron homeostasis. A model is proposed in Fig. 6C.
Several studies have suggested that IRP2 has an oncogenic activity. For example, IRP2 was found to promote breast and prostate cancer cell growth [33–35]. In addition, IRP2 was found to be over-expressed in several types of cancer including liver and lung [36, 37]. However, the signaling pathway mediated by IRP2 to promote tumorigenesis was not fully understood. In our study, we found that IRP2 is required for Mdm2 expression via mRNA stability. Thus, the enhanced Mdm2 expression mediated by IRP2 may contribute to the oncogenic activities of IRP2. In support of this notion, we found that high levels of Mdm2 transcripts are associated with the levels of IRP2 transcripts in normal and cancerous liver tissues (Fig. 6C). Additionally, both Mdm2 and IRP2 were found to be over-expressed in HCC patients and high levels of Mdm2 and IRP2 were associated with poor prognosis of HCC patients [36, 38]. These data suggest that the IRP2-Mdm2 axis contributes to development of HCC. Furthermore, we showed previously that IRP2 represses p53 expression via mRNA translation [23]. Since Mdm2 is a negative regulator of p53, it is possible that IRP2 suppresses p53 activity via enhancing Mdm2 expression, thus adding another layer of regulation of p53. Therefore, future studies are needed to examine the interplay among IRP2, Mdm2, and p53 in tumorigenesis, which may be explored as a strategy for cancer therapy by altering iron metabolism.
In this study, we found that Mdm2 expression is altered by iron in a p53-independent manner (Fig. 1). Consistent with our observation, a previous report showed that Mdm2 expression is increased by DFO but decreased by FAC in hepatocytes [39]. However, the role of Mdm2 in iron homeostasis remains to be elucidated. Does Mdm2 play a role in regulating labile iron pool and whether it modulates intracellular iron level? Is there a target regulated by Mdm2 involved in iron homeostasis? Would iron-mediated Mdm2 expression contribute to its oncogenic activity? Interestingly, we also found that IRP2 expression is repressed by doxorubicin (Fig. 3B), suggesting that a role of IRP2 in DNA damage response. Thus, it will be interesting to determine whether and how IRP2 expression is modulated by DNA damage. Since Mdm2 functions as E3 ligase, it will be interesting to determine whether Mdm2 can target IRP2 for degradation during DNA damage response. Addressing these questions would help us further understand the biological functions of Mdm2 and IRP2 as well as the role of the IRP2-Mdm2 axis in iron metabolism and DNA damage response.
Supplementary Material
Acknowledgements
This work is supported in part by National Institutes of Health R01 grants (CA224433 and CA081237).
Abbreviations:
- IRE
iron response element
- IRP2
Iron Responsive Protein 2
- DFO
Deferoxamine
- DFP
Deferiprone
- FAC
ferric ammonium acetate
- TFR
transferrin receptor
- FBXL5
F-box/LRR-repeat protein 5
- DRB
5,6-Dichlorobenzimidazole 1-β- D –ribofuranoside
- HCC
Hepatocellular Carcinoma
Footnotes
Conflict of Interest The authors declared no conflict of interest.
References
- 1.Torti SV, et al. , Iron and Cancer. Annu Rev Nutr, 2018. 38: p. 97–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inoue S and Kawanishi S, Hydroxyl radical production and human DNA damage induced by ferric nitrilotriacetate and hydrogen peroxide. Cancer Res, 1987. 47(24 Pt 1): p. 6522–7. [PubMed] [Google Scholar]
- 3.Torti SV and Torti FM, Iron and cancer: more ore to be mined. Nat Rev Cancer, 2013. 13(5): p. 342–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moroishi T, et al. , The FBXL5-IRP2 axis is integral to control of iron metabolism in vivo. Cell Metab, 2011. 14(3): p. 339–51. [DOI] [PubMed] [Google Scholar]
- 5.Smith SR, et al. , Complete loss of iron regulatory proteins 1 and 2 prevents viability of murine zygotes beyond the blastocyst stage of embryonic development. Blood Cells Mol Dis, 2006. 36(2): p. 283–7. [DOI] [PubMed] [Google Scholar]
- 6.Galy B, et al. , Iron regulatory proteins are essential for intestinal function and control key iron absorption molecules in the duodenum. Cell Metab, 2008. 7(1): p. 79–85. [DOI] [PubMed] [Google Scholar]
- 7.Cooperman SS, et al. , Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood, 2005. 106(3): p. 1084–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galy B, et al. , Altered body iron distribution and microcytosis in mice deficient in iron regulatory protein 2 (IRP2). Blood, 2005. 106(7): p. 2580–9. [DOI] [PubMed] [Google Scholar]
- 9.Meyron-Holtz EG, et al. , Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J, 2004. 23(2): p. 386–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh MC, et al. , Deletion of iron regulatory protein 1 causes polycythemia and pulmonary hypertension in mice through translational derepression of HIF2alpha. Cell Metab, 2013. 17(2): p. 271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson SA, et al. , The IRP1-HIF-2alpha axis coordinates iron and oxygen sensing with erythropoiesis and iron absorption. Cell Metab, 2013. 17(2): p. 282–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cahilly-Snyder L, et al. , Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somat Cell Mol Genet, 1987. 13(3): p. 235–44. [DOI] [PubMed] [Google Scholar]
- 13.Fakharzadeh SS, Trusko SP, and George DL, Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J, 1991. 10(6): p. 1565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haupt Y, et al. , Mdm2 promotes the rapid degradation of p53. Nature, 1997. 387(6630): p. 296–9. [DOI] [PubMed] [Google Scholar]
- 15.Jones SN, et al. , Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature, 1995. 378(6553): p. 206–8. [DOI] [PubMed] [Google Scholar]
- 16.Bouska A and Eischen CM, Murine double minute 2: p53-independent roads lead to genome instability or death. Trends Biochem Sci, 2009. 34(6): p. 279–86. [DOI] [PubMed] [Google Scholar]
- 17.Ganguli G and Wasylyk B, p53-independent functions of MDM2. Mol Cancer Res, 2003. 1(14): p. 1027–35. [PubMed] [Google Scholar]
- 18.Zhang J, et al. , Genetic Ablation of Rbm38 Promotes Lymphomagenesis in the Context of Mutant p53 by Downregulating PTEN. Cancer Res, 2018. 78(6): p. 1511–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, et al. , Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes Dev, 2011. 25(14): p. 1528–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunz F, et al. , Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science, 1998. 282(5393): p. 1497–501. [DOI] [PubMed] [Google Scholar]
- 21.Wang J and Pantopoulos K, Conditional derepression of ferritin synthesis in cells expressing a constitutive IRP1 mutant. Mol Cell Biol, 2002. 22(13): p. 4638–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, et al. , Iron-mediated degradation of IRP2, an unexpected pathway involving a 2-oxoglutarate-dependent oxygenase activity. Mol Cell Biol, 2004. 24(3): p. 954–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, et al. , Ferredoxin reductase is critical for p53-dependent tumor suppression via iron regulatory protein 2. Genes Dev, 2017. 31(12): p. 1243–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung YS, Qian Y, and Chen X, DNA polymerase eta is targeted by Mdm2 for polyubiquitination and proteasomal degradation in response to ultraviolet irradiation. DNA Repair (Amst), 2012. 11(2): p. 177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu E, Zhang J, and Chen X, MDM2 expression is repressed by the RNA-binding protein RNPC1 via mRNA stability. Oncogene, 2013. 32(17): p. 2169–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dohn M, Zhang S, and Chen X, p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene, 2001. 20(25): p. 3193–205. [DOI] [PubMed] [Google Scholar]
- 27.Guo B, et al. , Characterization and expression of iron regulatory protein 2 (IRP2). Presence of multiple IRP2 transcripts regulated by intracellular iron levels. J Biol Chem, 1995. 270(28): p. 16529–35. [DOI] [PubMed] [Google Scholar]
- 28.Peritz T, et al. , Immunoprecipitation of mRNA-protein complexes. Nat Protoc, 2006. 1(2): p. 577–80. [DOI] [PubMed] [Google Scholar]
- 29.Guzman C, et al. , ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One, 2014. 9(3): p. e92444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erlitzki R, Long JC, and Theil EC, Multiple, conserved iron-responsive elements in the 3’-untranslated region of transferrin receptor mRNA enhance binding of iron regulatory protein 2. J Biol Chem, 2002. 277(45): p. 42579–87. [DOI] [PubMed] [Google Scholar]
- 31.Zandomeni R, et al. , Casein kinase type II is involved in the inhibition by 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole of specific RNA polymerase II transcription. J Biol Chem, 1986. 261(7): p. 3414–9. [PubMed] [Google Scholar]
- 32.Deb SP, Singh S, and Deb S, MDM2 overexpression, activation of signaling networks, and cell proliferation. Subcell Biochem, 2014. 85: p. 215–34. [DOI] [PubMed] [Google Scholar]
- 33.Wang W, et al. , IRP2 regulates breast tumor growth. Cancer Res, 2014. 74(2): p. 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng Z, et al. , Iron-responsive element-binding protein 2 plays an essential role in regulating prostate cancer cell growth. Oncotarget, 2017. 8(47): p. 82231–82243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maffettone C, et al. , Tumorigenic properties of iron regulatory protein 2 (IRP2) mediated by its specific 73-amino acids insert. PLoS One, 2010. 5(4): p. e10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muto Y, et al. , Disruption of FBXL5-mediated cellular iron homeostasis promotes liver carcinogenesis. J Exp Med, 2019. 216(4): p. 950–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khiroya H, et al. , IRP2 as a potential modulator of cell proliferation, apoptosis and prognosis in nonsmall cell lung cancer. Eur Respir J, 2017. 49(4). [DOI] [PubMed] [Google Scholar]
- 38.Azer SA, MDM2-p53 Interactions in Human Hepatocellular Carcinoma: What Is the Role of Nutlins and New Therapeutic Options? J Clin Med, 2018. 7(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dongiovanni P, et al. , Iron-dependent regulation of MDM2 influences p53 activity and hepatic carcinogenesis. Am J Pathol, 2010. 176(2): p. 1006–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.