Abstract
Using a systems biology approach to prioritize potential points of intervention in ovarian cancer, we identified the lysine rich coiled-coil 1(KRCC1), as a potential target. High-grade serous ovarian cancer patient tumors and cells express significantly higher levels of KRCC1 which correlates with poor overall survival and chemo-resistance. We demonstrate that KRCC1 is predominantly present in the chromatin-bound nuclear fraction, interacts with HDAC1, HDAC2 and with the serine-threonine phosphatase PP1CC. Silencing KRCC1 inhibits cellular plasticity, invasive properties and potentiates apoptosis resulting in reduced tumor growth. These phenotypes are associated with increased acetylation of histones and with increased phosphorylation of H2AX and CHK1, suggesting modulation of transcription and DNA damage that may be mediated by the action of HDAC and PP1CC respectively. Hence, we address an urgent need to develop new targets in cancer.
Keywords: Ovarian Cancer, Systems Biology, KRCC1
Introduction
Identifying and establishing novel targets for therapy in cancer is a priority, particularly, in ovarian cancer (OvCa) because despite high response rates to initial chemotherapy, up to 70% of patients relapse with a median progression-free survival of only 12–18 months (1). Here, we used a systems biology approach to identify and subsequently establish a new target for OvCa.
In previous studies, we reported that the microRNAs, miR-15a and miR-16 were under-expressed in high-grade serous ovarian cancer patient tumors (2), and co-delivery of miR-15a and miR-16 reduced the growth of chemo-resistant orthotopic ovarian tumors to a greater degree than cisplatin alone (3). We inferred that the higher efficacy of the microRNA combination resulted from their cumulative disruption of a network of proteins encoded by the targeted mRNAs. MicroRNA therapy, however, presents challenges of off-target toxicity, delivery and promiscuity. To overcome these challenges, we employed a systems biology approach to identify gene/s which might be most influential in this phenotype.
Systems biology construct a network of influences based upon public datasets such as protein-protein interactions, gene expression correlations, and microRNA-RNA inhibition studies. Systems biology additionally integrates information within the dataset and responses (metabolic and signaling pathway) to generate computational models of structure and function for the molecule(s) of interest (4). We reasoned that a biological network based on protein-protein interactions, co-expression, co-localization and shared pathways represented by targets of miR-15a and miR-16 would identify highly connected key hub nodes (proteins); disruption of which is predicted to affect the maximum possible number of target proteins in the network and thus may serve as viable therapeutic targets (5, 6).
Herein we used the 72 common validated targets of miR-15a and miR-16 to generate a seed network of protein-protein interactions with the aim of identifying key interaction nodes. Expansion of the seed network by ~25%, using proteins that were highly connected to the seed network predicted new targets for OvCa (7–10). Based on systematic network analyses that computed various topological parameters, we identified KRCC1, a protein with unknown biological function, as one of the most connected proteins in the expanded network.
We report that KRCC1 is a nuclear protein that is clinically significant because it is frequently over-expressed in high-grade serous OvCa patient tumor tissues which correlate with poor overall survival and with chemo-resistance. Interestingly, KRCC1 via its interactions with the Serine/threonine-protein phosphatase PP1-gamma catalytic subunit (PP1CC) and histone deacetylases (HDACs) regulates cellular plasticity and apoptosis that impact ovarian tumor growth. Our results endorse a systems biology approach for the identification of novel targets in clinically challenging chemo-resistant sub-types of cancer.
Materials and Methods
Construction of target networks of miR-15a/16:
We identified the set of miR-15a/16 gene targets from miRTarBase (11) retaining only those gene targets validated using proteomics or gene-expression experiments (microarray, qRT-PCR, proteomics or luciferase reporter assays). We used the resulting miR-15a/16 target set as input to the GeneMANIA database (version 3.3.2; accessed April 15, 2014) (12) to seed construction of target networks. Networks were constructed by using genes targeted by miR-15a/16 as “nodes”, and “edges” were defined based on experimental or computational evidence and weighted function as explained in Mostafavi et.al (12). For example, the Jun proto-oncogene (cJUN) was an interacting partner of the modulator of VRAC current 1 (MCL1) in the network based on evidence from co-expression studies (13), and the cJUN and cyclin D1 (CCND1) interaction are based on pathway databases and evidence from human functional protein interaction networks (7) (Supplementary Table 2). If an edge is derived based on the interaction between two proteins, the Pearson correlation coefficient is calculated for the interaction as the correlation between two genes using available evidence (for example shared protein domains get a lower score compared to co-expression of two genes with the same direction of expression in a microarray experiment). Interactions attributed to the edges were weighted using the Pearson correlation coefficient. For experimentally validated yeast-two-hybrid data, an interaction will have a high correlation score of 1. Two separate networks were constructed as follows: the “seed-network” was constructed by deriving interactions among the query genes that are targeted by miR-15a/16, and the “expanded network” was constructed by adding 25% of genes that have high interaction with genes in seed-network. The weights were defined using linear regression, and an “expanded-network” was derived (Supplementary Table 5). Genes were added to the seed-network systematically to make genes in the seed-network interact as much as possible with each other, and as little as possible with additional genes, not on the expanded-network; 20 genes that met the inclusion criteria were added using the weighting approach.
Prioritizing hub-proteins using network topology properties
Quantitative investigation of network topologies can aid in obtaining biological insights from functional networks (8, 9). Biological systems often exhibit a scale-free network topology characterized by highly connected or central or regulatory hubs. We used different topological parameters, degree and radiality, to prioritize genes for function test experiments from the target network of miR-15a/16. Genes with high consensus degree, radiality and eccentricity were defined as hubs and prioritized as candidates for functional studies (8–10). Previously, we have shown that perturbing hub nodes in a biological network is an effective approach to identifying new targets for both ovarian and pancreatic cancer (14, 15).
To derive topological parameters, we defined the functional network as a graph (G), let G = (V, E), an undirected graph based on seed-network or expanded-network with n = |V | vertexes (or nodes). Topological index degree of the graph was computed using the equation G = deg (v). Here, all edges that are directly connected to the edge v were counted. Parameters related to the degree of the graph were defined as: in-degree = deg(v); out-degree = deg + (v); shortest path between two genes v and w = dist(v, w) is the shortest path.
Radiality, a node centrality index was computed as node-based index using the following equation.
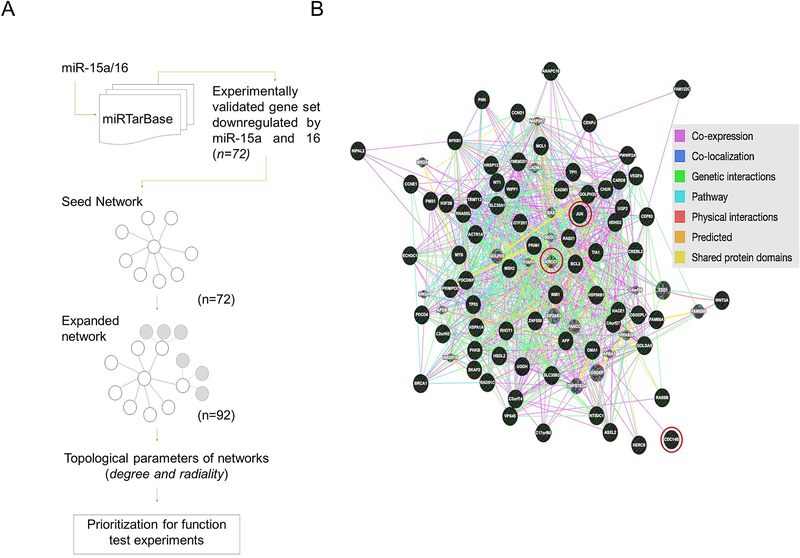
Network parameters were computed using R package igraph (16) and visualized using the Cytoscape based NetworkAnalyzer plugin (17). A consensus function based on two topological parameters was used to prioritize genes for function test experiments. Fig 1 summarizes the methodology used for gene prioritization.
Figure 1: Functional protein interaction network and target discovery approach.
(A) Systems biology flow-diagram of the approach used for construction of networks (seed and expanded) and target prioritization using GeneMania and STRING databases (B) Expanded network with nodes from seed network (black) and expanded network (grey); three nodes used for experimental perturbation highlighted.
Cell Culture
The SV40 transformed primary normal ovarian epithelial cell line (OSE tsT/hTERT, henceforth OSE) (18) was a kind gift from Dr. V. Shridhar, Mayo Clinic, Rochester, MN, USA. The SV40 transformed primary normal fallopian tube epithelial cells (henceforth FTE188) (19) were kindly provided by Dr. Jinsong Liu, MD Anderson Cancer Center, Houston, TX, USA. The CP20 cell line was a kind gift from Dr. Anil K. Sood, MD Anderson Cancer Center, Houston, TX, and was authenticated by the STR profiling facility at MD Anderson Cancer Center. The OV90, and OVCAR4 cell lines were purchased from ATCC and NCI respectively and OVSAHO, TYKNU, KURAMOCHI, OVKATE were purchased from JCRB Cell Bank (Japanese Collection of Research Bioresources Cell Bank). OSE cells were routinely cultured in 1:1 MCDB 105 and Medium 199 (Corning, Corning, NY, USA) + 10% FBS (Gibco, Grand Island, NY); FTE188 cells were cultured in 1:1 MCDB 105 and Medium 199 + 10% FBS + 0.01ug/ml EGF; CP20, OV90 and OVCAR4 were routinely cultured in RPMI + 10% FBS. All the cells were cultured with 1× penicillin-streptomycin (Gibco, Grand Island, NY) in a 5% CO2 humidified atmosphere and tested for mycoplasma contamination prior to any experiment.
siRNA Transfection
Gene silencing was performed in 60 mm culture dish containing 5×105 cells in suspension using Hiperfect (Qiagen) and 10 picomoles siRNA (scrambled control D-001206-13-20, Dharmacon) Si-KRCC-1 (SASI-HS01–00181201 and SASI-HS01–00181202 sigma) si-cJUN (SASI_Hs02_00333461, sigma) and cell division cycle 14B (CDC14B) (SASI_Hs02_00312084, Sigma) in OPTIMEM (Invitrogen). Effective silencing was achieved after 48h of transfection determined by qRT-PCR (real-time reverse transcription-PCR).
Micro-RNA transfection
miR transfection was done in 60 mm culture dish containing 5×105 cells in suspension using Hiperfect (Qiagen) and miRIDIAN Mimic Negative Control (Dharmacon, CN-001000–01) or Pre-miR precursor hsa-miR-15a or hsa-miR-16 (Ambion®).
Soft agar assay
For the soft agar colony assay, 24h post miR transfection 1.2×103 OvCa cells in RPMI medium (Corning) containing 10% FBS with 0.3% agar (Sea Plaque GTG agarose) were seeded on top of 0.6% agar in the same medium in 24 well plates. After two weeks of incubation, the colonies were counted using a GelCount™ colony counter.
RNA isolation, Reverse Transcription and Analysis of Gene expression using quantitative real time PCR (RTqPCR)
Total RNA was extracted using quick RNA miniprep kit (Zymo) following the manufacturer’s protocol. 1μg of isolated RNA was reverse transcribed (iScript cDNA Synthesis kit, Bio-Rad) using random hexamer primers. Quantitative real-time RT-PCR analysis was carried out in a 10μl reaction volume including 15ng cDNA, iTaq™ universal SYBR® Green supermix, and IDT primers. The comparative Ct method was used to calculate the relative abundance of the mRNA and compared with that of 18s rRNA.
Cell Lysis and Western Blotting
Formalin fixed paraffin-embedded (FFPE) patient samples were prepared as previously described (20). briefly, samples were deparaffinized with 1 ml of xylene; vortexed and stored for 10 min, followed by centrifugation at 14,000g for 5 minutes, and then the supernatant was removed. The deparaffinization process was repeated thrice following which samples were hydrated using an ethanol gradient, starting at 100% ethanol followed with 80% and 50% ethanol. After each step, the samples were centrifuged at 14,000g for 5 minutes and the supernatant was removed. Samples were stored overnight at 4 °C in 1 ml of DEPC-treated water and then centrifuged at 14,000 g for 15 min and the pellet was resuspended in 2% SDS buffer with a brief pulse of sonication (10 s × 3 times), centrifuged (14,000x g for 15 min) and the supernatant was collected for downstream assay.
Total cell lysate was prepared in RIPA (Boston Bioproducts) containing protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). The cell lysate was quantified using BCA assay (Thermo Fisher Scientific) and proteins separated on an SDS-PAGE gel and transferred to a PVDF membrane (BioRad). Membranes were blocked in 5% nonfat milk in TBS with 0.1% Tween-20 (TBST) for 1 h at room temperature followed by incubation with primary antibodies in TBST with 4% BSA overnight. The following primary antibodies were used - Bmi-1 (Millipore, 1:1000), PARP, Caspase 8, cJUN, pCHK1, pCHK2, γH2AX(Cell Signaling Technology, 1:1000), KRCC-1 (proteintech, 1:1000), PP1CA, PP1CB, PP1CC (Bethyl) and α tubulin and β actin (Sigma, 1:15,000). Secondary antibodies were used at a concentration of 1:10,000. Equal loading was verified by immunoblotting with α Tubulin/ β Actin.
Preclinical model of ovarian cancer
Female athymic nude mice (NCr-nu/nu; 5–6 weeks old) were purchased from ENVIGO. All mice were housed and maintained under specific pathogen-free conditions in facilities approved by the American Association for Accreditation of Laboratory Animal Care and in accordance with current regulations and standards of the US Department of Agriculture, US Department of Health and Human Services and National Institutes of Health. All studies were approved and supervised by the OUHSC Institutional Animal Care and Use Committee (IACUC). Tumors were established by intra-bursal injection of either 1.0 × 106 OV90 shCTL (20 animals) or 1.0 × 106 OV90 shKRCC1 (20 animals) cells. Once established, this tumor model reflects the growth pattern of advanced OvCa. One week later mice were randomized into four groups of 10 mice each, and cisplatin or vehicle administered intraperitoneally at 5mg/kg/weekly for 3 weeks. Mice were euthanized four weeks after tumor cell inoculation, or earlier if IACUC approved humane endpoint criteria were satisfied.
Immunohistochemistry
Immunohistochemistry was performed according to the manufacturer’s protocol using the Leica Bond-III™ Polymer Refine Detection system (DS 9800). In brief, FFPE tissues were sectioned at 4μm and were mounted on positively charged slides. The slides were dried overnight at room temperature and incubated at 60°C for 45 minutes followed by deparaffinization and rehydration in an automated Multistainer (Leica ST5020). Subsequently, these slides were transferred to the Leica Bond-III™, and treated for target retrieval at 100°C for 20 minutes in a retrieval solution, at pH 6.0. The sections were incubated with 5% goat serum (01–6201, ThermoFisher Scientific) for 30 minutes. Endogenous peroxidase was blocked using peroxidase-blocking reagent, followed by the selected primary antibody (Ki67 1:1500, Abcam) incubation for 60 minutes. For the secondary antibody, post-primary IgG-linker and/or Poly-HRP IgG reagents were used. Detection was done using 3, 3′-diaminobenzidine tetrahydrochloride (DAB), as chromogen and counter stained with hematoxylin. Completed slides were dehydrated (Leica ST5020), and mounted (Leica MM24). Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining was performed after deparaffinization of 4 μM sections using In-situ Cell Death Detection Kit, AP (Roche Diagnostics GmbH, Manheim, Germany) following the manufacturer’s protocol. Images were taken using a Nikon Eclipse Ni microscope.
Data analysis and Statistics
All the experiments were performed in triplicate and repeated independently three times. Data are expressed as means ± standard deviation (SD). For animal experiments, 10 mice were assigned per treatment group. This sample size gave 80% power to detect a 50% reduction in tumor weight with 95% confidence. ANOVA was performed to compare the mean among three or more groups and Student t test was performed to compare the mean between two groups. Statistical significance was set at P <0.05, using GraphPad Prism 6 software.
Results
Functional protein interaction network and target discovery
Here, we used a combination of network biology based predictive modeling and experimental validation to uncover a novel target and its mechanistic associations. Micro-RNA targets that were common to miR-15a and miR-16 were downloaded from the miRTarBase (11) retaining only those that were validated by microarray, qRT-PCR, western blot or luciferase reporter assays (Supplementary Table 1). Using this process a total of 72 proteins were identified and used as input within GeneMANIA, a database of functional and network level annotations of proteins (ver:3.3.2) to generate a “seed network” (Fig 1A and Supplementary Table 2 &3). In the seed network, the validated common microRNA targets served as nodes, and edges were defined based on experimental evidence and weighted function (12). In a discovery approach, 20 proteins which may or may not be microRNA targets but were most highly connected (based on their weightage), to the proteins in the seed network were added to construct an “expanded network” of 92 proteins (7–10) (Fig 1A & B and Supplementary Table 4, 5 & 6). Eight different Kyoto Encyclopedia of Genes and Genome (KEGG) pathways were significantly associated with the represented proteins in the network. Five key pathways involved in cancer were commonly enriched among protein sets from the seed and expanded networks (Supplementary Fig S1 and Supplementary Table 7). For example, the NOD-like receptor signaling pathway (21) involved in inflammation was significantly enriched among both the seed and the expanded networks. The expanded network was also enriched for the apoptosis and WNT signaling pathways, both of which play a key role in cancer including the pathophysiological basis for OvCa (22, 23).
Our group and others have shown that perturbation of a hub node with high connectivity within a functional interaction network may lead to the identification of novel targets and mechanisms (14, 15, 24–26). Herein, for functional studies candidate hub proteins were prioritized based on topological parameters, consensus high degree, radiality and eccentricity (8–10) (Fig 1B & Table 1); from these analyses, KRCC1 which was not a target of miR-15a or miR-16 (Supplementary Fig S2), emerged as the top-prioritized protein in the extended network (degree=39, radiality=0.846).
Table 1.
Based on the topological parameters the genes from the extended network were prioritized for network validation. Top six and bottom three genes based on degree and radiality are shown.
| S. no | Gene Name | Degree | Radiality |
|---|---|---|---|
| 1 | KRCC1 | 39 | 0.85 |
| 2 | SLC35A1 | 36 | 0.84 |
| 3 | PHKB | 35 | 0.83 |
| 4 | C14orf109 | 33 | 0.83 |
| 5 | MAP3K7 | 33 | 0.81 |
| 6 | JUN | 32 | 0.82 |
| == | == | == | == |
| 90 | TPI1 | 3 | 0.57 |
| 91 | CDC14B | 2 | 0.59 |
| 92 | FAM122C | 2 | 0.58 |
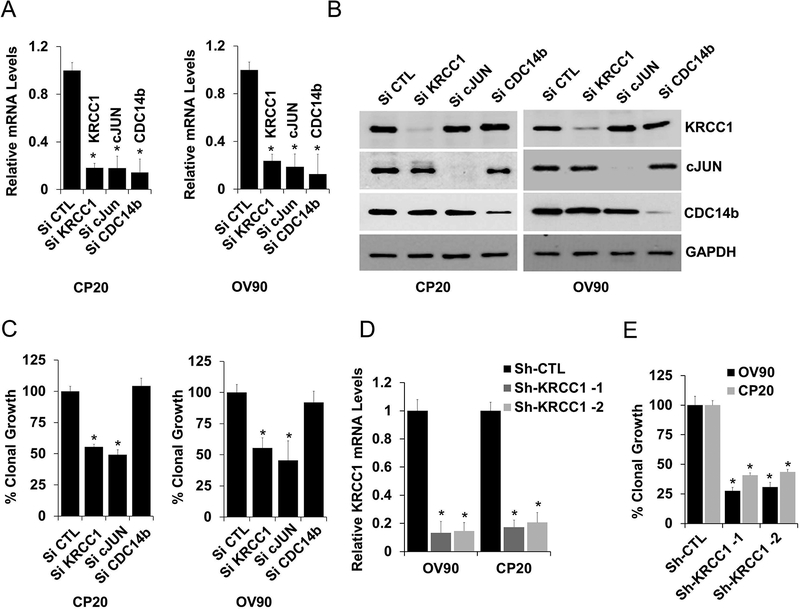
To determine the biological relevance of the hub nodes, two of the most connected genes, KRCC1, and the known cancer oncogene c-JUN, and one least connected node, CDC14b was silenced using siRNA. Effective silencing was verified at both the mRNA and protein levels in CP20 and OV90 OvCa cells (Fig 2A and 2B). Silencing KRCC1 and c-JUN, significantly inhibited anchorage-independent clonal growth (~50%) a characteristic property of cancer cells, while silencing CDC14b had no effect (Fig 2C). The effect of KRCC1 inhibition on clonal growth was further verified in an extended panel of OvCa cell lines using siRNA (Supplementary Fig S3A). While silencing KRCC1 by two different shRNAs (Fig 2D and Supplementary Fig S3B) confirmed inhibition (Fig 2E), overexpression of KRCC1 (Supplementary Fig S3C) increased the clonal growth potential (Supplementary Fig S3D). These results validated the network analysis in that perturbation of hub proteins, importantly, including the previously uncharacterized KRCC1, significantly impacted clonal growth.
Figure 2: Biological relevance of the hub proteins.
Based on Nodal connectivity KRCC1, cJUN and CDC14b genes were selected. To evaluate their role in OvCa, these genes were silenced using siRNA, Effective silencing was verified at both the (A) mRNA (mean ± SD) and (B) protein levels and their (C) percent clonal growth compared to control is plotted (mean ± SE). (D&E) Stable cell lines for CP20 and OV90 expressing sh-RNA targeting KRCC1 (sh-KRCC1–1 & sh-KRCC1–2) or non-target shRNA (shCTL) were generated, efficient silencing of KRCC1 by two different shRNA was confirmed at mRNA (RT-qPCR, mean ± SD). Anchorage independent clonal potential of these were evaluated and percent clonal growth along with SE is plotted. P<0.05 was considered significant; Experiments were repeated independently at least 3 times.
Expression and significance of KRCC1 in ovarian cancer
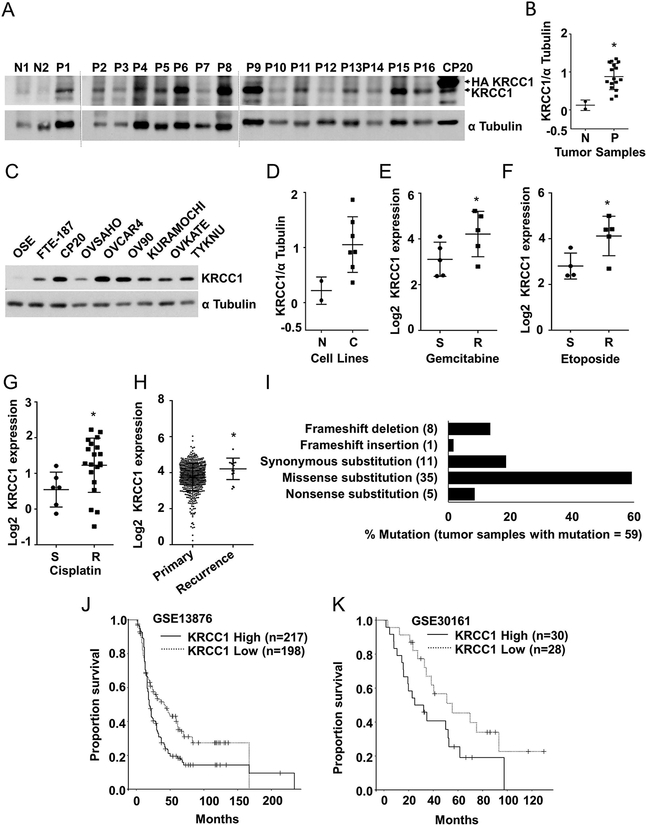
To analyze disease relevance of KRCC1, we determined its expression in high-grade serous ovarian cancer (HGSOC) tumor tissues and ovarian cancer cell lines. Interestingly, while the expression of KRCC1 was minimal in two normal epithelial fallopian tube tissue samples, its expression in tumor tissues was significantly higher (Fig 3A and B). Similarly compared to the non-malignant ovarian surface epithelial cell line (OSE) or the fallopian tube epithelium derived cell line FTE187, the expression of KRCC1 was notably higher in the majority of the OvCa cell lines (Fig 3C and D). Analysis of Oncomine datasets revealed that transcript level expression of KRCC1 was significantly higher in OvCa cell lines that were resistant to gemcitabine (27) (Fig 3E), etoposide (27) (Fig 3F) or cisplatin resistant cell lines from OvCa as well as other cancer types (28) (Fig 3G). In the Cancer Genome Atlas (TCGA), compared to the primary tumor tissues, KRCC1 transcript levels were significantly higher in the recurrent ovarian serous cystadenocarcinoma patient tumor samples (Fig 3H). According to the Catalogue of Somatic Mutations in Cancer (COSMIC), KRCC1 alleles are mutated in only 0.12% of tumor tissues, i.e., 59 unique mutations out of 47480 samples with a preponderance of missense mutations suggest an important cellular function of the protein (Fig 3I). Analysis of Gene Expression Omnibus (GEO) datasets indicated a significant correlation between high expression of KRCC1 and reduced overall survival of OvCa patients (Fig 3J and K). These results suggest an important role for KRCC1 in OvCa. To investigate the role of KRCC1 in OvCa, we selected CP20, OV90, OVCAR4, and OVSAHO cell lines for further studies. The chemo resistant, CP20 cell line was developed by sequential exposure of cisplatin on A2780 and the OV90 cell line was derived from ascites of a grade 3, stage IIIC HGSOC patient (29). OVCAR4 and OVSAHO cell lines were characterized by Domcke et al. as the most clinically relevant models of HGSOC (30).
Figure 3: Expression and significance of KRCC1 in ovarian cancer.
(A). Expression of KRCC1 in tumor tissues: Protein was extracted from the 16 primary tumors (P1–P16) and 2 normal fallopian tube epithelial (N 1 and N2) samples as described in the material and methods section, CP20 cells overexpressed with HA-KRCC1 were used as control. (B) Expression of KRCC1 in individual samples was quantified by densitometry analysis using Image J and normalized to their respective α-tubulin levels, Data represent mean ± SD. (C, D) Immunoblot showing expression of KRCC1 in OvCa cell lines (C) along with normal (N) OSE and FTE cells, Expression of KRCC1 were individually normalized to their respective α-tubulin levels and plotted, Data represent mean ± SD. (E, F) Levels of KRCC1 mRNA in gemcitabine (E) and etoposide, (F) sensitive (S) and resistant (R) OvCa cell lines. Data represent mean ± SD. (G) Levels of KRCC1 mRNA in cisplatin sensitive (S) and resistant (R) cancer cell lines including OvCa. Data represent mean ± SD. (H) KRCC1 mRNA expression levels in primary vs recurrent ovarian serous cystadenocarcinoma tumor samples from oncomine data base (reporter 218303_x_at). Data represent mean ± SD. *P<0.05; (I) % mutation in KRCC1 gene from COSMIC database; out of 47480 tumor samples, 59 samples had mutations. Type of mutations (%) in 59 samples were plotted. Number of tumor samples with mutations are shown on X axis. (J, K) Kaplan-Meier overall survival curves in KRCC1 low and high expression OvCa cases from GEO databases on GSE13876 (-log10 (p value) = 3.43661) and GSE30161 (-log10 (p value) = 1.32055). The proportion survival is plotted versus time (since diagnosis in month). Kaplan-Meier curves with a log-rank test where P value<0.05 considered significant.
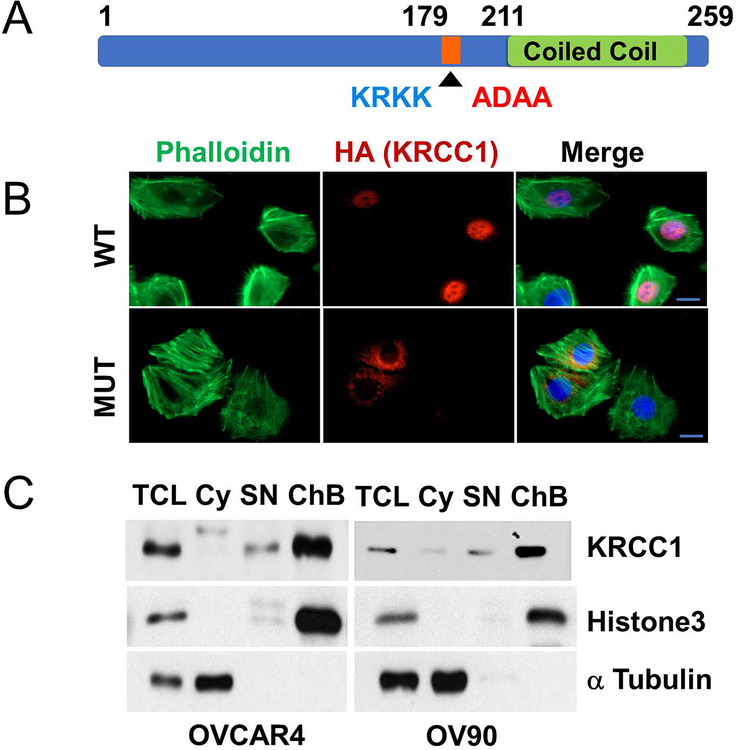
KRCC1 is a nuclear protein
The protein KRCC1 comprises of 259 amino acids and besides a lysine rich-coiled coil region at the C-terminus, it does not appear to possess any strictly defined sequence motifs except two computationally predicted nuclear localization signals (NLS) at residues 159–162 (KRKR) and 179–182 (KRKK) respectively (Fig 4A). The human protein is 99% similar to that in Chimpanzee and ~75% similar to that in mouse, rat, hamster, dog, pig and horse (Supplementary Fig S4A & B). To determine sub-cellular localization of KRCC1, we expressed either HA-tagged wild-type KRCC1 (WT-KRCC1), or a mutant where the NLS sequence KRKK was replaced with ADAA (Mut-KRCC1) in OVCAR4 cells. Prominent nuclear immunofluorescence (IF) was observed in WT-KRCC1 expressing cells while the signal in the Mut-KRCC1 expressing cells was extra-nuclear suggesting that KRCC1 is a nuclear protein and that KRKK represents the NLS (Fig 4B). In the same field, the absence of staining in un-transfected cells confirmed specificity of the HA antibody, while phalloidin and DAPI served as markers for cellular actin and the nuclei respectively. To confirm localization of the endogenous protein we performed sub-cellular fractionation (31), and determined that KRCC1 was predominantly present in the nuclear chromatin-bound fraction with minimum expression in the soluble nuclear fraction (Fig 4C). Histone3 (H3) and α tubulin served as nuclear and cytoplasmic fraction purity controls. These results indicate that KRCC1 is a chromatin-bound nuclear protein.
Figure 4: KRCC1 is a nuclear protein.
(A) Schematic representation of KRCC1 with nuclear localization signal sequence (NLS, native sequence is shown in blue and the mutated sequence is shown in red). (B) Immunofluorescence of HA-tagged wild type (WT) or NLS mutant KRCC1 with phalloidin (PH) and DAPI, (Scale Bar=10 μm). (C) Sub-cellular fractionation of OVCAR4 and OV90 was done to detect the localization of endogenous KRCC1, presence of KRCC1 in different fractions were evaluated using immunoblotting along with markers of fraction purity. Total cell lysate (TCL), cytoplasm (Cy), soluble nuclear (SN), chromatin bound fraction (ChB).
KRCC1 regulates plasticity and apoptosis in ovarian cancer cells
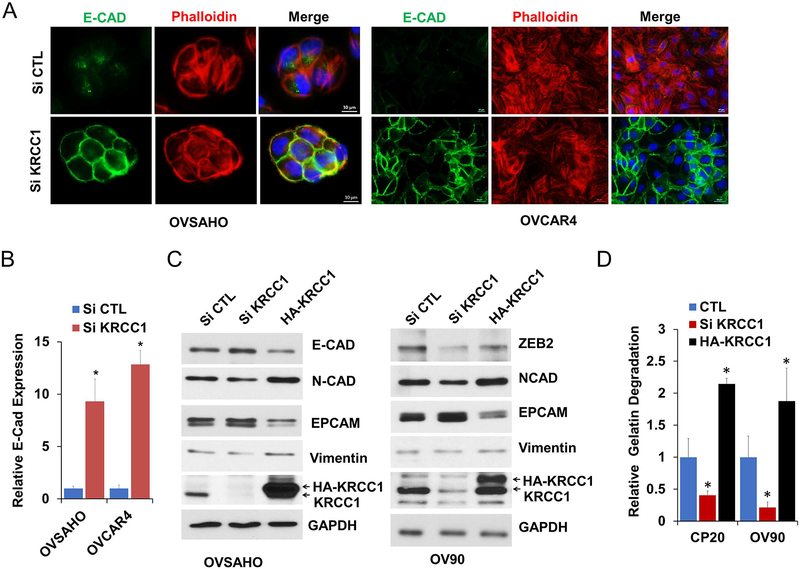
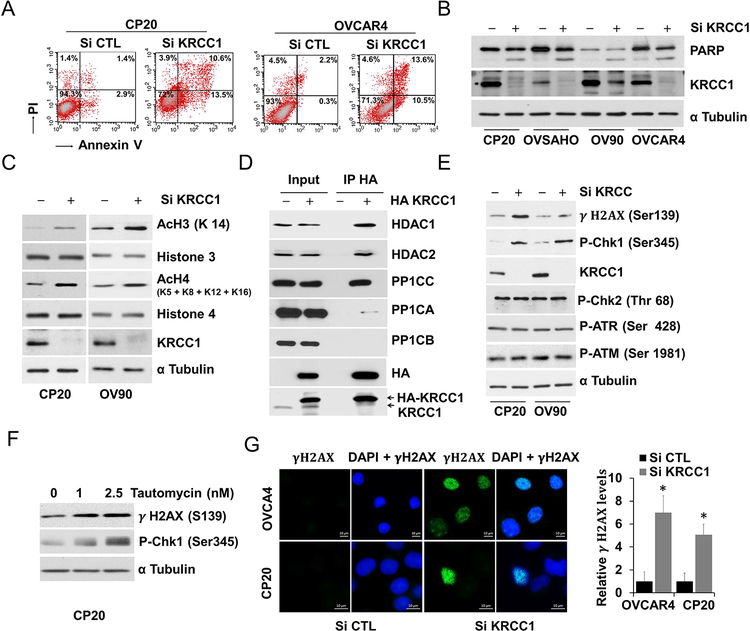
An exceptionally plastic, more mesenchymal state facilitates peritoneal dissemination and invasion of OvCa (32). Therefore, we determined the expression levels of epithelial and mesenchymal markers in control versus KRCC1 silenced cells. Compared to the control, silencing of KRCC1 increased the expression of epithelial E-cadherin (E-Cad) which was recruited to the cell junctions (Fig 5A and 5B). The mesenchymal markers N-cadherin (N-Cad) and to some extent vimentin were downregulated while epithelial markers E-Cad and the epithelial cell adhesion molecule (EPCAM) were upregulated in KRCC1 silenced cells and overexpression of KRCC1 reversed this effect (Fig 5C). Importantly zinc finger E-box binding homeobox 2 (ZEB-2), but not other transcriptional repressors of E-Cad, were downregulated in KRCC1 silenced cells (Fig 5C and Supplementary Fig S5). These results suggest that KRCC1 promotes a more mesenchymal state in OvCa cells. In the process of migration and invasion, tumor cells degrade the extra-cellular matrix via invasive protrusions that are sites for targeted secretion of metalloproteases. We next addressed the consequences of KRCC1 silencing and over-expression using a FITC-Gelatin matrix degradation assay that allows quantitation of the invasive capability of cells. While silencing KRCC1 significantly inhibited the matrix degrading ability of OV90 and CP20 cells, over-expression enhanced the capacity of degradation (Fig 5D and Supplementary Fig S6), suggesting an important role for KRCC1 in cellular invasion. The striking decrease in clonal growth prompted us to evaluate cell death upon KRCC1 silencing. Using AnnexinV/PI based flow cytometry, we observed that compared to the control, apoptosis was significantly enhanced in KRCC1 silenced CP20 and OVCAR4 cells by 5.6 and 9.6-fold respectively. Apoptotic cell death by KRCC1 inhibition was further confirmed by enhanced PARP cleavage (Fig 6A–6B) in OvCa cells. Together these results suggest that loss of KRCC1 promotes a more epithelial, less-invasive state alongside increased apoptosis in OvCa cells.
Figure 5: KRCC1 regulates cellular plasticity.
(A) OvCa cells were transfected with either non-target control siRNA (si CTL) or siRNA targeting KRCC1 (si KRCC1). 72 h post transfection, cells were fixed and stained with DAPI (blue), anti-E-cadherin (E-CAD, green), and phalloidin (Tubulin, red). The localization of E-cadherin and F-actin were visualized by immunofluorescence microscopy (Scale Bar=10μm). (B) Relative E-cadherin expression were quantified using image J and normalized with cell counts. Data represent mean ± SD, *= P value<0.05. (C) OVSAHO and OV90 cells were transfected as shown in image and immunoblotted for respective EMT markers in KRCC1 silenced or overexpressing OvCa Cells, (D) Relative FITC-gelatin matrix degradation i.e., ability to migrate and invade, in non-target control siRNA (CTL), siRNA targeting KRCC1 (si KRCC1) or HA-KRCC1 transfected CP20 and OV90 cells; >100 cells were evaluated, Data represent mean ± SD; * = P value<0.05.
Figure 6: KRCC1 inhibition results in DNA damage and Apoptosis.
(A) Non-target control siRNA (si CTL) or si KRCC1 transfected CP20 and OVCAR4 cells were stained by Annexin V/PI and analyzed by flow cytometry, representative quadrant analysis of Annexin V/PI stained cells after 72h KRCC1 silencing are shown. (B) OvCa cells after 72h KRCC1 silencing were immunoblotted for PARP, efficient KRCC1 inhibition was verified by immunoblotting for KRCC1, α Tubulin was used as loading control. (C) Acetylation changes of Histone 3 (H3) and Histone 4 (H4) by KRCC1 inhibition was evaluated using immunoblotting for H3K-14, H4K 5/8/12/16, total H3 and H4. α Tubulin was used as loading control and efficient KRCC1 inhibition was confirmed by immunoblotting. (D) KRCC1 interaction with HDACs and PP1 was evaluated using co-immunoprecipitation in empty vector (EV) or pCDAN3-KRCC1-HA (HA-KRCC1) overexpressed OV90 cells. (E, F) Immunoblotting for markers of DNA damage after 72h KRCC1 silencing or 48h tautomycin treatment. (G) OVCAR4 and CP20 cells were transfected with si CTL or si KRCC1. 48h post transfection cells were fixed and stained with gamma H2AX. Representative images of gamma H2AX foci at 40× magnification are shown along with quantitation using Image J (Fold ± SE). *, P value<0.05 considered significant.
Molecular determinants of the KRCC1 phenotype
As shown above, KRCC1 was enriched in the chromatin-bound fraction; since histones are a major component of the chromatin, we assessed the effect of KRCC1 on the histone acetylation status. In OvCa cells, silencing KRCC1 increased acetylation of H3 at K14 and H4 at K5/8/12/16 (Fig 6C). Interestingly, a previous yeast-two-hybrid study demonstrated the interaction of the catalytic subunit of the protein phosphatase 1 with KRCC1 (33) and Class I HDACs reportedly associate with PP1 complexes (34, 35). We therefore probed for an association between KRCC1 and the nuclear Class I histone deacetylases (HDACs). KRCC1 consistently co-precipitated with HDAC1, HDAC2 and with the gamma catalytic subunit of PP1 (PP1CC) but not with the alpha or beta isoforms (Fig 6D). Specificity of the PP1CC antibody was confirmed by immunoblotting (Supplementary Fig S7). Several reports have implicated PP1 in the DNA damage checkpoint activation and in DNA repair (36–39). Interestingly, silencing KRCC1 increased phosphorylation of H2AX at Ser139 (γH2AX), and CHK1 at Ser 345 while CHK2, ATM and ATR were unchanged (Fig 6E,). Similar results were obtained with tautomycin (Fig 6F), a protein phosphatase inhibitor, with specificity towards PP1 at less than 10 nM (40) concentration. Additionally, a significant increase in γH2AX positive foci in KRCC1 silenced cells indicated enhanced DNA damage in these cells (Fig 6G). These results suggest that the KRCC1/PP1/HDAC axis is important for regulation of cellular plasticity and apoptosis.
Silencing KRCC1 inhibits tumor growth in vivo
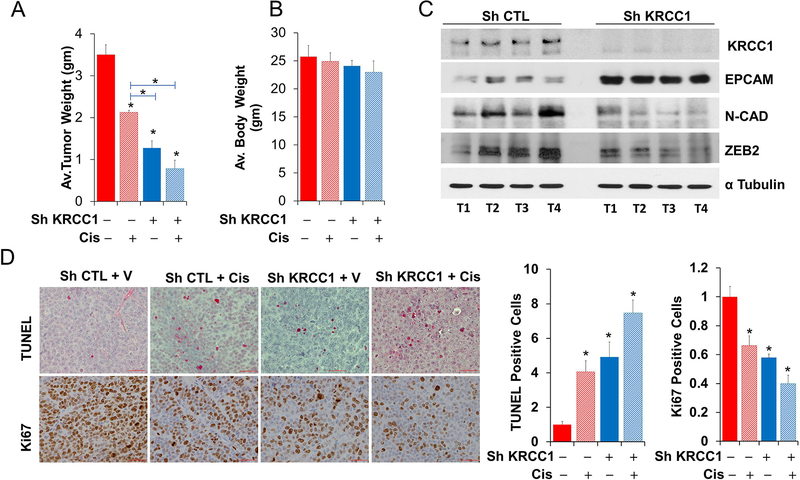
The role of KRCC1 in tumor growth was evaluated in an orthotopic xenograft model using stable OV90-shRNA expressing cells. Compared to the control, we observed significant, ~64% reduction in tumor weight in the shKRCC1 group while cisplatin treatment reduced tumor growth by ~39%. In the shKRCC1 group, cisplatin treatment decreased tumor growth by an additional ~14% compared to shKRCC1 alone (Fig 7A). As assessed by body-weight no major toxicity was noted due to treatment (Fig 7B). Immunoblotting of individual tumors demonstrated decreased ZEB2 and N-Cad with increased EPCAM in the shKRCC1 group indicating a reduced mesenchymal phenotype and corroborating our in vitro results (Fig 7C). Immunohistochemical quantitation further confirmed enhanced TUNEL positivity, by ~5 fold in shKRCC1 group and by ~7 fold in the cisplatin treated shKRCC1 group compared to the control (Fig 7D). Concurrently, immunostaining for Ki67 was significantly reduced by ~0.42 fold in the shKRCC1 group and by ~0.6 fold in the cisplatin treated shKRCC1 group compared to the control (Fig 7D). The decreased proliferative index and increased apoptosis in the KRCC1 silenced groups corroborate our in vitro observations and suggest that combination with cisplatin may have additional benefit.
Figure 7: Silencing KRCC1 inhibits tumor growth in vivo.
Silencing KRCC1 reduces orthotopic ovarian tumor burden and enhances cisplatin sensitivity. 1×106, KRCC1-ShRNA or Control-ShRNA OV90 cells were injected into the ovarian bursa of 6 week-old female athymic nude mice. One-week later mice were randomized into 4 groups of 10 mice each and cisplatin or vehicle administered intraperitoneally at 5mg/kg/weekly for 3 weeks; mice were euthanized 4 weeks after inoculation. (A) Average tumor weight and (B) body weight across the groups. (C) Immunoblotting for KRCC1 and respective EMT markers in 4 tumors each from sh control (shCTL) or Sh-KRCC1 group. (D) Representative IHC for TUNEL and Ki67 along with IHC quantitation for all animals per groups are shown. Data represent average ± SD; *, P value<0.05 considered significant.
Discussion
Our study leverages translational bioinformatics in identifying and establishing KRCC1 as a potential target in OvCa. We demonstrate that KRCC1 with a confirmed nuclear localization signal is a nuclear protein that associates with PP1CC, HDAC1, and HDAC2. Our results, support that KRCC1 regulated essential cancer cell phenotypes may be mediated by PP1CC and Class 1 HDACs. Furthermore, analysis of several publicly available datasets link expression of KRCC1 with poor prognosis in OvCa and we speculate that an expansive analysis across pan-cancer data may additionally implicate KRCC1 in other malignancies. Thus, perturbing second-degree interactions or expanded networks could facilitate the discovery of novel proteins involved in the pathophysiology of cancer.
PP1 is a major serine/threonine phosphatase that regulates a wide range of cellular processes through the interaction of its core catalytic subunit with several different targeting subunits (41–43). Interestingly like PP1 interacting proteins (PIPs), KRCC1 is intrinsically disordered (Supplementary Fig S8) and specifically associates with the catalytic subunit PP1CC and with HDAC1/2 but not with PP1CA or PP1CB, the other closely related isoforms that are expressed in mammalian cells but differ primarily in their N- and C-termini (41). While the interaction of PP1CC with HDAC1/2 is well-documented the precise biological implications are emerging (34, 35) and suggest regulation of gene transcription (44) and replication stress (37).
Increased acetylation of histone 3/4 and phosphorylation of CHK1 in KRCC1 silenced cells suggest reduced deacetylase and phosphatase activities at these respective sites, which may indicate the inability of PP1CC and HDAC to localize to specific chromatin in the absence of KRCC1. Concurrently increased γH2AX in KRCC1 silenced cells is indicative of endogenous DNA damage that is consistent with a role for PP1 in the DNA damage response, DNA repair and in replication stress (45). Inhibition of yeast PP1, enhances phosphorylation of H2AX and CHK1 (46, 47); an observation we substantiated in mammalian cells with tautomycin, a PP1 inhibitor. In Xenopus oocytes, inhibition of Repo-Man increased phosphorylation of H2AX and CHK1 in the absence of exogenous DNA damage, which is purportedly mediated by activation of ATM (48). Similarly, although H2AX and CHK1 were phosphorylated in KRCC1 silenced cells, there were no changes in phospho-ATM or ATR levels compared to the control.
The respective histone modifications and associated phenotypes are further substantiated by studies of nuclear targeting subunits of PP1, including NIPP1, PNUTS, and Repo-Man (36, 37, 48). Expression of NIPP1, a nuclear targeting subunit that inhibits PP1 induced mitotic catastrophe, apoptosis, and reduced xenograft tumor growth (38). Inhibition of Repo-Man, another chromatin targeting subunit of PP1CC decreased clonal growth and increased apoptosis, a phenotype recapitulated by KRCC1 silenced cells (39).
The significant reduction in cellular plasticity, migration and invasive potential in KRCC1 silenced cells could be attributed to epigenetic regulation by PP1CC and HDAC that either directly or indirectly affect E-Cadherin and ZEB transcription. Similar to KRCC1, inhibition of HDAC restores epithelial differentiation and abolishes anchorage independent growth in cancer cells (49). Genome-wide binding studies have established a link between PP1 phosphatase and HDAC complexes that bind chromatin regions in an overlapping fashion (44). Moreover, genome-wide promoter binding studies in HeLa have identified ~500 promoters that bind PP1 (50). However, the nuclear PIPs, PNUTS, NIPP1 and Repo-Man taken together account for only ~24% overlap with this promoter dataset (50) raising the possibility that a percentage of these and other promoters may be co-occupied and transcriptionally regulated by the KRCC1-PP1CC-HDAC axis ultimately affecting important processes including cellular plasticity.
Targeting nuclear proteins for cancer therapy remains a viable and interesting avenue (51). Poly ADP ribose polymerase (PARP) inhibition exemplifies the promise of nuclear protein targeting (52). Additionally, in recent years, chromatin modifying agents have gained attention since epigenetic modifications are at the forefront of several malignancies (53). While, HDAC inhibitors are being rigorously pursued for clinical translation (54), inhibitors of the PP1 phosphatase regulatory complexes are being developed (55, 56). In summary, we speculate that KRCC1 localizes PP1CC and HDAC1/2 to specific chromatin regions, which is instrumental in transcriptional regulation and in ameliorating DNA damage. In the absence of KRCC1, persistent DNA damage further sensitizes tumors to DNA damaging agents such as cisplatin as demonstrated in the animal model. We envision that leveraging results from our study and target-based drug repositioning to inhibit KRCC1 can open up new avenues for therapy in OvCa.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health Grant CA157481 to R.B., We thank the Peggy and Charles Stephenson Cancer Center at the University of Oklahoma Health Sciences Center for a seed grant and also acknowledge an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (P20 GM103639) for the use of Histology and Immunohistochemistry Core, which provided immunohistochemistry and image analysis service. KS, and JTD acknowledge support from the National Institutes of Health: National Center for Advancing Translational Sciences (NCATS, grant number UL1TR000067); and Clinical and Translational Science Awards (CTSA) grant. JW acknowledge support from 5U54GM104938.
Abbreviations
- ATM
ATM serine/threonine kinase
- ATR
ATR serine/threonine kinase
- CCND1
Cyclin D1
- CHK
Checkpoint kinase
- cJUN
Jun proto-oncogene
- COSMIC
Catalogue of Somatic Mutations in Cancer
- DAB
3, 3′-diaminobenzidine tetrahydrochloride
- DAPI
4′,6-diamidino-2-phenylindole
- FFPE
Formalin-fixed, Paraffin-embedded
- GEO
Gene Expression Omnibus
- HDAC1
Histone deacetylase1
- HDAC2
Histone deacetylase2
- KRCC1
Lysine rich coiled-coil 1
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MCL1
MCL1 apoptosis regulator
- miR
Micro RNA
- NIPP1
protein phosphatase 1 regulatory subunit 8
- NLS
Nuclear Localization Signal
- OvCa
ovarian cancer
- PIPs
PP1 interacting proteins
- PNUTS
Protein phosphatase 1 regulatory subunit 10
- PP1CA
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit
- PP1CB
Serine/threonine-protein phosphatase PP1-beta catalytic subunit
- PP1CC
Serine/threonine-protein phosphatase PP1-gamma catalytic subunit
- qRT-PCR
Real-Time Quantitative Reverse Transcription Polymerase chain reaction
- Repo-Man
cell division cycle associated 2
- SD
standard deviation
- TCGA
The Cancer Genome Atlas
Footnotes
Conflicts of Interest
JTD has received consulting fees or honoraria from Janssen Pharmaceuticals, GlaxoSmithKline, AstraZeneca and Hoffman-La Roche. JTD is a scientific advisor to LAM Therapeutics and holds equity in NuMedii, Ayasdi and Ontomics. KS has received consulting fees or honoraria from Kencor Health, OccamzRazor, Philips Healthcare, Alphabet, McKinsey and Company, LEK Consulting and Parthenon-EY.
References-
- 1.Cortez AJ, Tudrej P, Kujawa KA, and Lisowska KM (2018) Advances in ovarian cancer therapy. Cancer Chemother Pharmacol 81, 17–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharya R, Nicoloso M, Arvizo R, Wang E, Cortez A, Rossi S, Calin GA, and Mukherjee P (2009) MiR-15a and MiR-16 control Bmi-1 expression in ovarian cancer. Cancer Res 69, 9090–9095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dwivedi SK, Mustafi SB, Mangala LS, Jiang D, Pradeep S, Rodriguez-Aguayo C, Ling H, Ivan C, Mukherjee P, Calin GA, Lopez-Berestein G, Sood AK, and Bhattacharya R (2016) Therapeutic evaluation of microRNA-15a and microRNA-16 in ovarian cancer. Oncotarget 7, 15093–15104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yadav BS, and Tripathi V (2018) Recent Advances in the System Biology-based Target Identification and Drug Discovery. Curr Top Med Chem 18, 1737–1744 [DOI] [PubMed] [Google Scholar]
- 5.Zhu X, Gerstein M, and Snyder M (2007) Getting connected: analysis and principles of biological networks. Genes & development 21, 1010–1024 [DOI] [PubMed] [Google Scholar]
- 6.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, and Barabasi AL (2007) The human disease network. Proceedings of the National Academy of Sciences of the United States of America 104, 8685–8690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu G, Feng X, and Stein L (2010) A human functional protein interaction network and its application to cancer data analysis. Genome Biol 11, R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barabasi AL, and Albert R (1999) Emergence of scaling in random networks. Science 286, 509–512 [DOI] [PubMed] [Google Scholar]
- 9.Barabasi AL, and Oltvai ZN (2004) Network biology: understanding the cell’s functional organization. Nat Rev Genet 5, 101–113 [DOI] [PubMed] [Google Scholar]
- 10.Maslov S, and Sneppen K (2002) Specificity and stability in topology of protein networks. Science 296, 910–913 [DOI] [PubMed] [Google Scholar]
- 11.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, Chiew MY, Tai CS, Wei TY, Tsai TR, Huang HT, Wang CY, Wu HY, Ho SY, Chen PR, Chuang CH, Hsieh PJ, Wu YS, Chen WL, Li MJ, Wu YC, Huang XY, Ng FL, Buddhakosai W, Huang PC, Lan KC, Huang CY, Weng SL, Cheng YN, Liang C, Hsu WL, and Huang HD (2018) miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res 46, D296–D302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mostafavi S, Ray D, Warde-Farley D, Grouios C, and Morris Q (2008) GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol 9 Suppl 1, S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, Olson JA Jr., Marks JR, Dressman HK, West M, and Nevins JR (2006) Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439, 353–357 [DOI] [PubMed] [Google Scholar]
- 14.Giri K, Shameer K, Zimmermann MT, Saha S, Chakraborty PK, Sharma A, Arvizo RR, Madden BJ, McCormick DJ, Kocher JP, Bhattacharya R, and Mukherjee P (2014) Understanding protein-nanoparticle interaction: a new gateway to disease therapeutics. Bioconjug Chem 25, 1078–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saha S, Xiong X, Chakraborty PK, Shameer K, Arvizo RR, Kudgus RA, Dwivedi SK, Hossen MN, Gillies EM, Robertson JD, Dudley JT, Urrutia RA, Postier RG, Bhattacharya R, and Mukherjee P (2016) Gold Nanoparticle Reprograms Pancreatic Tumor Microenvironment and Inhibits Tumor Growth. ACS Nano 10, 10636–10651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Csardi G, and Nepusz T (2006) The igraph software package for complex network research. InterJournal Complex Systems 1695, 1695 [Google Scholar]
- 17.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chien J, Narita K, Rattan R, Giri S, Shridhar R, Staub J, Beleford D, Lai J, Roberts LR, Molina J, Kaufmann SH, Prendergast GC, and Shridhar V (2008) A role for candidate tumor-suppressor gene TCEAL7 in the regulation of c-Myc activity, cyclin D1 levels and cellular transformation. Oncogene 27, 7223–7234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shan W, Mercado-Uribe I, Zhang J, Rosen D, Zhang S, Wei J, and Liu J (2012) Mucinous adenocarcinoma developed from human fallopian tube epithelial cells through defined genetic modifications. Cell Cycle 11, 2107–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee Mustafi S, Chakraborty PK, Dwivedi SK, Ding K, Moxley KM, Mukherjee P, and Bhattacharya R (2017) BMI1, a new target of CK2alpha. Mol Cancer 16, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxena M, and Yeretssian G (2014) NOD-Like Receptors: Master Regulators of Inflammation and Cancer. Frontiers in immunology 5, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyer A, Goff AK, and Boerboom D (2010) WNT signaling in ovarian follicle biology and tumorigenesis. Trends in endocrinology and metabolism: TEM 21, 25–32 [DOI] [PubMed] [Google Scholar]
- 23.Gatcliffe TA, Monk BJ, Planutis K, and Holcombe RF (2008) Wnt signaling in ovarian tumorigenesis. International journal of gynecological cancer : official journal of the International Gynecological Cancer Society 18, 954–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong H, Mason SP, Barabasi AL, and Oltvai ZN (2001) Lethality and centrality in protein networks. Nature 411, 41–42 [DOI] [PubMed] [Google Scholar]
- 25.Mistry D, Wise RP, and Dickerson JA (2017) DiffSLC: A graph centrality method to detect essential proteins of a protein-protein interaction network. PLoS One 12, e0187091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters LA, Perrigoue J, Mortha A, Iuga A, Song WM, Neiman EM, Llewellyn SR, Di Narzo A, Kidd BA, Telesco SE, Zhao Y, Stojmirovic A, Sendecki J, Shameer K, Miotto R, Losic B, Shah H, Lee E, Wang M, Faith JJ, Kasarskis A, Brodmerkel C, Curran M, Das A, Friedman JR, Fukui Y, Humphrey MB, Iritani BM, Sibinga N, Tarrant TK, Argmann C, Hao K, Roussos P, Zhu J, Zhang B, Dobrin R, Mayer LF, and Schadt EE (2017) A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat Genet 49, 1437–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, Liu Q, Iorio F, Surdez D, Chen L, Milano RJ, Bignell GR, Tam AT, Davies H, Stevenson JA, Barthorpe S, Lutz SR, Kogera F, Lawrence K, McLaren-Douglas A, Mitropoulos X, Mironenko T, Thi H, Richardson L, Zhou W, Jewitt F, Zhang T, O’Brien P, Boisvert JL, Price S, Hur W, Yang W, Deng X, Butler A, Choi HG, Chang JW, Baselga J, Stamenkovic I, Engelman JA, Sharma SV, Delattre O, Saez-Rodriguez J, Gray NS, Settleman J, Futreal PA, Haber DA, Stratton MR, Ramaswamy S, McDermott U, and Benes CH (2012) Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gyorffy B, Surowiak P, Kiesslich O, Denkert C, Schafer R, Dietel M, and Lage H (2006) Gene expression profiling of 30 cancer cell lines predicts resistance towards 11 anticancer drugs at clinically achieved concentrations. Int J Cancer 118, 1699–1712 [DOI] [PubMed] [Google Scholar]
- 29.Provencher DM, Lounis H, Champoux L, Tetrault M, Manderson EN, Wang JC, Eydoux P, Savoie R, Tonin PN, and Mes-Masson AM (2000) Characterization of four novel epithelial ovarian cancer cell lines. In Vitro Cell Dev Biol Anim 36, 357–361 [DOI] [PubMed] [Google Scholar]
- 30.Domcke S, Sinha R, Levine DA, Sander C, and Schultz N (2013) Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun 4, 2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendez J, and Stillman B (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol 20, 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klymenko Y, Kim O, and Stack MS (2017) Complex Determinants of Epithelial: Mesenchymal Phenotypic Plasticity in Ovarian Cancer. Cancers (Basel) 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fardilha M, Esteves SL, Korrodi-Gregorio L, Vintem AP, Domingues SC, Rebelo S, Morrice N, Cohen PT, da Cruz e Silva OA, and da Cruz e Silva EF (2011) Identification of the human testis protein phosphatase 1 interactome. Biochem Pharmacol 82, 1403–1415 [DOI] [PubMed] [Google Scholar]
- 34.Brush MH, Guardiola A, Connor JH, Yao TP, and Shenolikar S (2004) Deactylase inhibitors disrupt cellular complexes containing protein phosphatases and deacetylases. J Biol Chem 279, 7685–7691 [DOI] [PubMed] [Google Scholar]
- 35.Canettieri G, Morantte I, Guzman E, Asahara H, Herzig S, Anderson SD, Yates JR 3rd, and Montminy M (2003) Attenuation of a phosphorylation-dependent activator by an HDAC-PP1 complex. Nat Struct Biol 10, 175–181 [DOI] [PubMed] [Google Scholar]
- 36.Wang F, Zhu S, Fisher LA, Wang L, Eurek NJ, Wahl JK 3rd, Lan L, and Peng A (2019) Phosphatase 1 Nuclear Targeting Subunit Mediates Recruitment and Function of Poly (ADP-Ribose) Polymerase 1 in DNA Repair. Cancer Res 79, 2526–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winkler C, Rouget R, Wu D, Beullens M, Van Eynde A, and Bollen M (2018) Overexpression of PP1-NIPP1 limits the capacity of cells to repair DNA double-strand breaks. J Cell Sci 131 [DOI] [PubMed] [Google Scholar]
- 38.Winkler C, De Munter S, Van Dessel N, Lesage B, Heroes E, Boens S, Beullens M, Van Eynde A, and Bollen M (2015) The selective inhibition of protein phosphatase-1 results in mitotic catastrophe and impaired tumor growth. J Cell Sci 128, 4526–4537 [DOI] [PubMed] [Google Scholar]
- 39.Trinkle-Mulcahy L, Andersen J, Lam YW, Moorhead G, Mann M, and Lamond AI (2006) Repo-Man recruits PP1 gamma to chromatin and is essential for cell viability. J Cell Biol 172, 679–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ju J, Li W, Yuan Q, Peters NR, Hoffmann FM, Rajski SR, Osada H, and Shen B (2009) Functional characterization of ttmM unveils new tautomycin analogs and insight into tautomycin biosynthesis and activity. Org Lett 11, 1639–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen PT (2002) Protein phosphatase 1--targeted in many directions. J Cell Sci 115, 241–256 [DOI] [PubMed] [Google Scholar]
- 42.Trinkle-Mulcahy L, Sleeman JE, and Lamond AI (2001) Dynamic targeting of protein phosphatase 1 within the nuclei of living mammalian cells. J Cell Sci 114, 4219–4228 [DOI] [PubMed] [Google Scholar]
- 43.Andreassen PR, Lacroix FB, Villa-Moruzzi E, and Margolis RL (1998) Differential subcellular localization of protein phosphatase-1 alpha, gamma1, and delta isoforms during both interphase and mitosis in mammalian cells. J Cell Biol 141, 1207–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Castro IJ, Amin HA, Vinciotti V, and Vagnarelli P (2017) Network of phosphatases and HDAC complexes at repressed chromatin. Cell Cycle 16, 2011–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freeman AK, and Monteiro AN (2010) Phosphatases in the cellular response to DNA damage. Cell Commun Signal 8, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bazzi M, Mantiero D, Trovesi C, Lucchini G, and Longhese MP (2010) Dephosphorylation of gamma H2A by Glc7/protein phosphatase 1 promotes recovery from inhibition of DNA replication. Mol Cell Biol 30, 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.den Elzen NR, and O’Connell MJ (2004) Recovery from DNA damage checkpoint arrest by PP1-mediated inhibition of Chk1. EMBO J 23, 908–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng A, Lewellyn AL, Schiemann WP, and Maller JL (2010) Repo-man controls a protein phosphatase 1-dependent threshold for DNA damage checkpoint activation. Curr Biol 20, 387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang HM, Kuay KT, Koh PF, Asad M, Tan TZ, Chung VY, Lee SC, Thiery JP, and Huang RJ (2016) An epithelial marker promoter induction screen identifies histone deacetylase inhibitors to restore epithelial differentiation and abolishes anchorage independence growth in cancers. Cell Death Discov 2, 16041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verheyen T, Gornemann J, Verbinnen I, Boens S, Beullens M, Van Eynde A, and Bollen M (2015) Genome-wide promoter binding profiling of protein phosphatase-1 and its major nuclear targeting subunits. Nucleic Acids Res 43, 5771–5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao YL, and Yang WM (2005) Nuclear proteins: promising targets for cancer drugs. Curr Cancer Drug Targets 5, 595–610 [DOI] [PubMed] [Google Scholar]
- 52.Jiang X, Li W, Li X, Bai H, and Zhang Z (2019) Current status and future prospects of PARP inhibitor clinical trials in ovarian cancer. Cancer Manag Res 11, 4371–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seidel C, Florean C, Schnekenburger M, Dicato M, and Diederich M (2012) Chromatin-modifying agents in anti-cancer therapy. Biochimie 94, 2264–2279 [DOI] [PubMed] [Google Scholar]
- 54.Eckschlager T, Plch J, Stiborova M, and Hrabeta J (2017) Histone Deacetylase Inhibitors as Anticancer Drugs. Int J Mol Sci 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vagnarelli P, and Alessi DR (2018) PP1 Phosphatase Complexes: Undruggable No Longer. Cell 174, 1049–1051 [DOI] [PubMed] [Google Scholar]
- 56.Krzyzosiak A, Sigurdardottir A, Luh L, Carrara M, Das I, Schneider K, and Bertolotti A (2018) Target-Based Discovery of an Inhibitor of the Regulatory Phosphatase PPP1R15B. Cell 174, 1216–1228 e1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.