Abstract
Background
Genome‐wide profiling to examine brain transcriptional features associated with excessive ethanol (EtOH) consumption has been applied to a variety of species including rodents, nonhuman primates (NHPs), and humans. However, these data were obtained from cross‐sectional samples which are particularly vulnerable to individual variation when obtained from small outbred populations typical of human and NHP studies. In the current study, a novel within‐subject design was used to examine the effects of voluntary EtOH consumption on prefrontal cortex (PFC) gene expression in a NHP model.
Methods
Two cohorts of cynomolgus macaques (n = 23) underwent a schedule‐induced polydipsia procedure to establish EtOH self‐administration followed by 6 months of daily open access to EtOH (4% w/v) and water. Individual daily EtOH intakes ranged from an average of 0.7 to 3.7 g/kg/d. Dorsal lateral PFC area 46 (A46) brain biopsies were collected in EtOH‐naïve and control monkeys; contralateral A46 biopsies were collected from the same monkeys following the 6 months of fluid consumption. Gene expression changes were assessed using RNA‐Seq paired analysis, which allowed for correction of individual baseline differences in gene expression.
Results
A total of 675 genes were significantly down‐regulated following EtOH consumption; these were functionally enriched for immune response, cell adhesion, plasma membrane, and extracellular matrix. A total of 567 genes that were up‐regulated following EtOH consumption were enriched in microRNA target sites and included target sites associated with Toll‐like receptor pathways. The differentially expressed genes were also significantly enriched in transcription factor binding sites.
Conclusions
The data presented here are the first to use a longitudinal biopsy strategy to examine how chronic EtOH consumption affects gene expression in the primate PFC. Prominent effects were seen in both cell adhesion and neuroimmune pathways; the latter contained both pro‐ and antiinflammatory genes. The data also indicate that changes in miRNAs and transcription factors may be important epigenetic regulators of EtOH consumption.
Keywords: EtOH, Brain Gene Expression, Cynomolgus Macaque, Cortex
Walter et al. used a unique within subject design to detect changes in the brain prefrontal cortex transcriptome associated with chronic ethanol exposure in non‐human primates. The data obtained showed that broad categories of genes were affected, especially those associated with neuro‐immune function, cell adhesion and the extracellular matrix. The data suggest that chronic ethanol exposure has marked functional and structural effects on the prefrontal cortex.

An estimated 16 million people in the United States suffer from an alcohol use disorder (AUD; Substance Abuse and Mental Health Administration (SAaMHS) 2015). Identifying the genetic effects of AUD is important to advance prevention and treatment options in the human population. Beginning with Lewohl and colleagues (2000), there are more than 100 studies using genome‐wide profiling to examine the brain transcriptional features associated with excessive ethanol (EtOH) consumption (see, e.g., Contet, 2012; Farris et al., 2018; Mulligan et al., 2006). These studies have focused on factors associated with risk, individual variation, dependence, and/or chronic consumption. The transcriptional features associated with each of these factors can generally be well isolated in rodent studies; for example, selective breeding can be used to detect risk features in the absence of EtOH exposure (see, e.g., Colville et al., 2017). There are, however, substantial caveats associated with extending the rodent data to the human condition; while the homology between rodent and human for coding genes is greater than 90%, the homology for noncoding RNAs and especially long noncoding RNAs is substantially less (Roux et al., 2017). Further, there are substantial differences between the rodent and human brain including marked differences in proportional cortical volume and the lack of granular cortex in the rodent brain (Saleem et al., 2007 and references therein). A macaque model of excessive alcohol self‐administration (Baker et al., 2014; Grant et al., 2008) can provide increased homology to humans in transcriptional, neuroanatomical, and behavioral features. In contrast to postmortem human studies (Farris and Mayfield, 2014; Lewohl et al., 2000; Warden and Mayfield, 2017), the nonhuman primate (NHP) model (e.g., Iancu et al., 2018a) allows 1 to examine features associated with the initiation and relatively early phase of excessive EtOH consumption, a time window when therapeutic intervention is likely to have the greatest benefit.
Our NHP model of EtOH consumption (Baker et al., 2014; Grant et al., 2008) produces long‐term voluntary consumption over months and individual differences in drinking that reflect a wide range of intoxication and daily drinking patterns similar to humans. Compared to humans, macaques have similar alcohol absorption and metabolism rates (Green et al., 1999; Jimenez et al., 2017). To our knowledge, only 1 NHP brain genome‐wide transcriptome analysis has been completed (Iancu et al., 2018a). This analysis looked at the transcriptional effects of chronic EtOH self‐administration in animals that underwent daily drinking over 12 months of the 22 h/d self‐administration protocol. Iancu and colleagues (2018a) observed postmortem gene coexpression and cosplicing patterns correlated to EtOH intake in the central nucleus of the amygdala and in prefrontal cortical area 32. Membrane and synaptic annotations were significantly overrepresented in the network modules associated with EtOH consumption. What was not known was the expression patterns in these brain areas prior to alcohol ingestion. In the current study, we identified longitudinal transcriptional changes in prefrontal cortex (PFC) area 46 (A46), important in decision‐making in macaques (Gerbella et al., 2013 and references therein). This is the first within‐subject study of gene expression changes in cortical neural circuitry and implicates cellular adaptations in neuroimmune, stress, and extracellular matrix processes in the development of excessive alcohol drinking.
Materials and Methods
Animals
Two consecutive, replicate cohorts of 11 (cohort 9) and 12 (cohort 13) young adult, male cynomolgus macaques (Macaca fascicularis) were housed in quadrant cages (0.8 × 0.8 × 0.9 m) with constant temperature (20 to 22°C), humidity (65%), and an 11‐hour light cycle. Animals had visual, auditory, and olfactory contact with other animals in the protocol. All animals were maintained on positive caloric and fluid balance throughout the experiment, and body weights were recorded weekly. All procedures were conducted in accordance with the Guide for the Care and Use of Laboratory Animals and the NIH guidelines for the care and use of laboratory animal resources and approved by the Oregon National Primate Research Center IACUC. Cynomolgus macaques were ordered from SNBL USA (Everett, WA) and were of Cambodian origin. Monkeys were 5 to 7 years of age (equivalent to 20 to 28 human years) when they first had access to EtOH (See Fig. 1 for overview of procedural timeline). Cohort 9 had 3 controls and 8 EtOH drinkers, and cohort 13 had 3 controls and 9 EtOH drinkers. Detailed drinking data and additional tissue resources are available for these cohorts via the Monkey Alcohol and Tissue Resource Center (Daunais et al., 2014).
Figure 1.
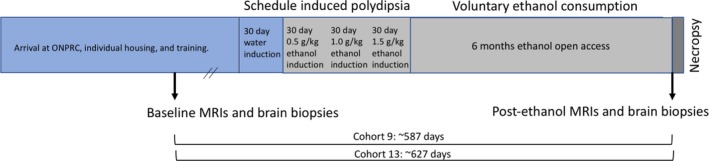
Voluntary EtOH consumption and sample collection timeline.
EtOH Subjects
Monkeys were moved into individual housing and then were trained to use operant drinking panels. Once trained, they underwent 3 months of schedule‐induced polydipsia (SIP) to induce EtOH self‐administration in daily 16‐hour sessions, as previously described (Grant et al., 2008). SIP is an effective protocol to establish voluntary EtOH consumption through the use of interval schedules of food delivery. Briefly, banana‐flavored food pellets (1 g) were delivered every 300 seconds (for a fixed time of 300 seconds) until the intended volume of fluid was consumed. After water only SIP, monkeys were induced to increasing volumes of EtOH in 30‐day increments: 0.5, 1.0, and 1.5 g/kg/d (Fig. 1). Following induction, open access (or EtOH self‐administration) began. For each 22‐hour session, water and 4% (w/v) EtOH were concurrently available for 6 months. The subjects received 3 meals (approximately 2 hours apart) of banana pellets each session (day). Each daily session of 22 hours began in the late morning and ended early morning the following day. EtOH was removed for 2 hours each day for cage cleaning, blood draws, and other daily routine procedures. The lighting schedule in the room was 11‐hour light, 13‐hour dark; the lights went off 7 hours after the start of the open‐access session. The details of the open‐access protocol are discussed in Grant and colleagues (2008). The 2 cohorts were run consecutively. EtOH intake in the macaque model is recorded daily with a 0.5‐second resolution.
Control Subjects
Control subjects were housed in the same room as the EtOH drinking subjects and participated in all experimental manipulations. For the controls, SIP and self‐administration conditions were identical, with the exception that both spouts dispensed water. A maltose dextrin solution (10% in water) was given to the controls to calorically match the drinkers and controls. The dextrin solution was given at the beginning of each daily session by attaching a bottle to the front of the housing cage.
Biopsy Samples
MRI was used to obtain the stereotaxic coordinates for prefrontal cortical A46 biopsies from the contralateral dominant hemisphere (determined by handedness) from 6 controls and 10 drinkers. Seven drinkers were not biopsied but had sham surgeries. Biopsy samples were 35 to 45 mg wet weight. Samples from cohort 9 were homogenized immediately in Trizol and then frozen at −80°C for future processing; samples from cohort 13 were fresh, frozen on dry ice, and stored at −80°C for future processing. Contralateral A46 biopsy samples after 6 months of drinking were taken from anesthetized animals immediately before necropsy. Before and after samples for cohort 9 were processed simultaneously using a standard Trizol protocol. Before and after samples for cohort 13 were processed simultaneously using a standard Qiagen AllPrep DNA/RNA/miRNA Universal Kit protocol.
RNA‐Seq
Libraries without strand orientation were prepared using the TruSeq RNA Sample Preparation Kit version 2 (Illumina, San Diego, CA). Libraries were sequenced according to specification on a HiSeq 2500 (Illumina) at the Oregon Health and Science University Massively Parallel Sequencing Shared Resource. PolyA‐selected libraries were multiplexed 4 per lane, yielding an average of 44 million total single‐end reads per sample. FastQC (Babraham Bioinformatics, Babraham Institute, Cambridge, UK) was used for quality checks on the raw sequence data. Reads were aligned to NCBI assembly MF5 (accession GCA_000364345.1) using STAR version 2.5.2b (Dobin et al., 2013) with default parameters except for the following: outFilterMismatchNmax = 2 and outFilterMultimapNmax = 1. Using HTSeq version 0.6.1p1 (Anders et al., 2015) and the MF5 GTF annotation file (33,107 genes), read counts were summarized at the gene level. On average, 85% of reads uniquely mapped to the MF5 genome. Generalized linear models, using DESeq2 (Love et al., 2014), were used to detect differential expression utilizing a blocking design to account for baseline differences within individual animals and individual cohorts (see “Results” for threshold details). RNA‐Seq data are available through NCBI GEO database (accession # pending).
Gene Ontology Enrichment
GO enrichment was conducted on the EtOH‐affected differentially regulated genes using GOrilla (Eden et al., 2009) with human annotations and TPM ≥ 1 as the background. Significance level was set at FDR q < 0.05.
miRNA Target Enrichment
Enrichment of human miRNA target sites in the up‐ and down‐regulated genes was conducted using miRTarBase (Chou et al., 2018) which is a database of validated microRNA–target interactions. Enrichr (Chen et al., 2013; Kuleshov et al., 2016) was used for the specific enrichment analysis. Subsequently, miRNA Enrichment Analysis and Annotation tool (miEAA; Backes et al., 2016) was used to assess overrepresentation of gene ontology categories in the most connected miRNAs (i.e., those with target sites in the most up‐regulated genes). The 2263 human miRNAs in miRTarBase were used as the reference dataset (i.e., the full human dataset queried for target site enrichment), as well as the full human gene list in miRTarBase.
Transcription Factor Enrichment
Enrichr (Chen et al., 2013; Kuleshov et al., 2016) and position weighted matrices (PWM) from TRANSFAC (Matys et al., 2006) and JASPAR (Sandelin et al., 2004) were used to assess enrichment of human transcription factor (TF) binding sites in the promoter regions of the identified DEGs. The reference datasets used for enrichment were promoter regions of all human genes in the TRANSFAC and JASPAR databases and the reference TF binding sites therein.
Results
Voluntary EtOH Consumption
During 6 months of open access to EtOH, the subjects across both cohorts consumed an average of 0.7 to 3.7 g/kg/d of EtOH (Fig. 2) or an overall average of 2.3 g/kg/d of EtOH. This resulted in average blood EtOH concentrations (BECs) ranging from 6 to 172 mg% (Fig. 2). BECs were measured approximately every 5 days during the 6‐month open‐access period at 7 hours into the daily 22 hours of open access. Due to the timing of the blood sampling, the BECs do not necessarily reflect the total daily intake, but rather the intake pattern over the first 7 hours of daily access. One of 4 drinking categories was assigned to each subject following 6 months of collected data: low, binge, heavy, or very heavy (Fig. 2). These drinking categories are based on data‐driven definitions extracted from previous cohorts that underwent this same consumption protocol (Baker et al., 2014). None of the subjects in these 2 cohorts fit the category for a binge drinker. By chance, most of the low drinkers (LD) were in cohort 9 and most of the high drinkers were in cohort 13. We did account for cohort effect in the analyses. This type of grouping is unpredictable, and there is not a straightforward explanation for this difference. Therefore, the cohorts were analyzed together. The drinking results of the subjects that received a sham surgery rather than a biopsy are included here because it is important to report the drinking results of the full cohort of animals. The gene expression results of these animals are not analyzed here because they did not have a paired before‐EtOH sample; however, the results are included in the GEO submission for further analyses of these cohorts in the future.
Figure 2.
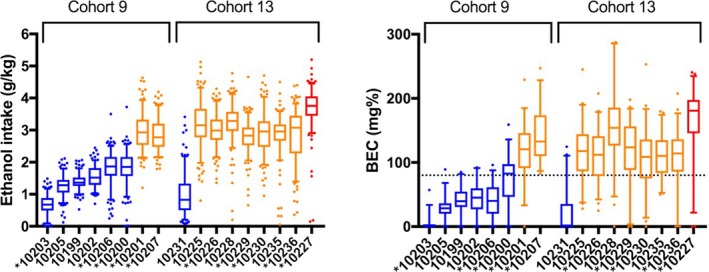
Daily EtOH intake (g/kg) of all the EtOH drinkers in cohorts 9 and 13 over 6 months of EtOH self‐administration. Individual days that fall outside of the 5 to 95% confidence interval are shown. Individuals are colored by drinking category: low (blue), heavy (orange), and very heavy (red). BECs (mg%) were measured every 5 to 7 days at 7 hours into the open‐access session. Dotted line at 80 mg%, which is intoxication level. For both plots, box and whiskers indicate 25 to 75 percentiles and 5 to 95 percentiles, respectively, with line at the median. Subject IDs with an asterisk (*) had brain biopsy samples collected prior to open access.
Differential Gene Expression Following 6 Months of Voluntary EtOH Consumption
A total of 10 drinkers and 6 controls across 2 cohorts were biopsied in A46 prior to EtOH induction (preEtOH biopsy) and again after 6 months (postEtOH biopsy) of voluntary EtOH consumption (or maltose dextrin for the controls; Fig. 1), resulting in a total of 32 RNA‐Seq samples. Utilizing a block design to account for individual and cohort baseline differences, differentially expressed genes identified after alcohol exposure were identified by comparing preEtOH biopsy and postEtOH biopsy samples. In an effort to eliminate confounding effects due to aging, environment, surgeries, etc., differentially expressed genes identified from the comparison of pre‐ and postEtOH biopsies from nonalcohol exposed animals (controls) were subtracted from those identified above. A total of 29,961 coding and noncoding genes are annotated in the cyno MF5 genome. The genes queried were limited to the 15,344 that have average transcripts per million (TPM) ≥1 across all samples. Using FDR q < 0.05 and a gene expression fold change (FC) of at least 1.5, we identified 567 up‐regulated and 675 down‐regulated DEGs in the EtOH drinkers before versus after EtOH open access (Fig. 3). These genes were not differentially expressed in the controls. Thirty‐nine up‐regulated and 38 down‐regulated DEGs specific to the controls were identified. A total of 57 up‐regulated and 42 down‐regulated DEGs were differentially expressed in both the controls and the drinkers (Fig. 3). A full list of gene counts (transcripts per million) and differential expression calculations for the controls and the EtOH drinkers are included in Table S1. Only the DEGs specific to EtOH drinkers were further analyzed.
Figure 3.
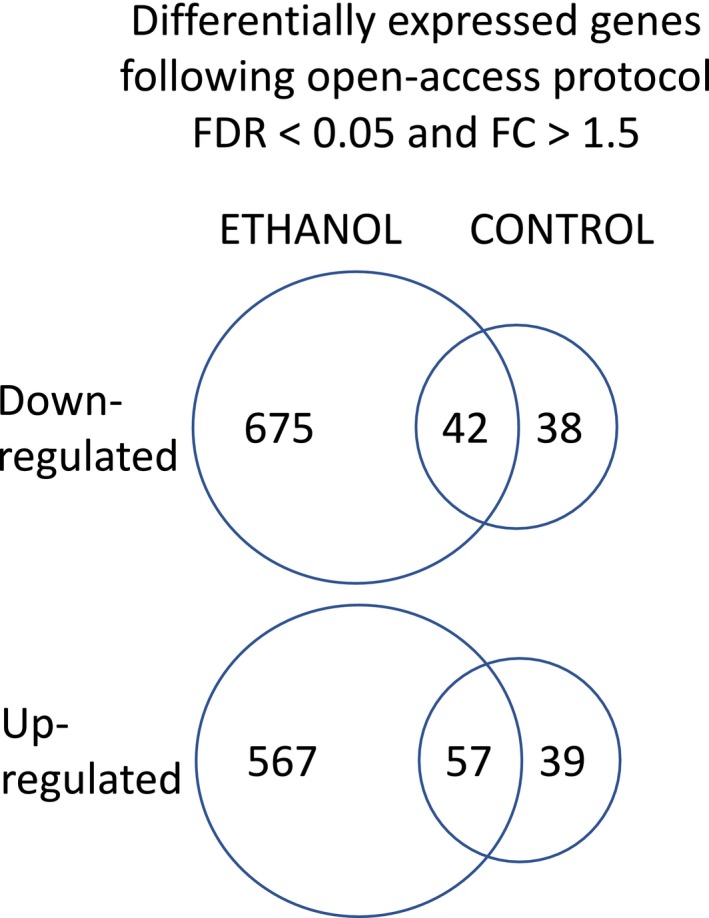
Venn diagram indicating differentially expressed genes (DEGs) following open‐access protocol that reached criteria (q < 0.05 and fold change (FC) >1.5) and whether they were identified in EtOH subjects, controls, or both.
GO Enrichment of DEGs
The 1,242 DEGs following EtOH consumption were enriched in GO Process [GO:0006955 immune response (q = 1.7 × 10−13), GO:0006954 inflammatory response (q = 1.5 × 10−11), GO:0007155 cell adhesion (q = 1.0 × 10−5)], GO Function [GO:0038023 signaling receptor activity (q = 1.9 × 10−10), GO:0019955 cytokine binding (q = 1.8 × 10−4)], and GO Component [GO:0005886 plasma membrane (q = 3.7 × 10−9), GO:0031012 extracellular matrix (q = 2.1 × 10−2)]. The gene lists for each ontology category are primarily down‐regulated genes following EtOH consumption. The full list of GO enrichment categories is found in Table S2, which includes the genes in each annotation.
Transcription Enrichment of DEGs
We examined the transcriptional changes related to the up‐ or down‐regulated genes. Because miRNA target sites for rhesus and human are ~90% conserved (Xu et al., 2013), we used experimentally validated target sites for human microRNAs (miRNAs). The 567 up‐regulated genes were enriched in target sites for 197 human miRNAs (q < 0.05). The top 10 miRNAs (enriched based on q value) are listed in Table 1a as examples of miRNAs potentially affected by chronic EtOH exposure. The full list of significantly enriched miRNA target sites, including gene lists, is found in Table S3. There was no significant enrichment for miRNA target sites in the down‐regulated genes.
Table 1.
(a) Top 10 (Based on Adjusted p Value) Human miRNAs That Have Significant Number of Target Sites in Our Up‐Regulated Genes. Overlap = [Number of Genes Up‐regulated in Our Dataset With a Binding Site for This miRNA)]/[Total Number of Genes in miRTarBase With Target Sites for This miRNA in the 3′ Untranslated Region]. (b) Top 11 Most Connected miRNAs With the Number of Genes Having 3′UTR Binding Sites for the miRNAs
| (a) Most enriched miRNAs | (b) Most connected miRNAs | |||
|---|---|---|---|---|
| miRNA | Overlap | Q value | miRNA | # of genes |
| hsa‐miR‐192‐5p | 65/993 | 2.53E−07 | hsa‐miR‐192‐5p | 65 |
| hsa‐miR‐30b‐5p | 38/416 | 2.53E−07 | hsa‐miR‐93‐5pa | 67 |
| hsa‐miR‐30a‐5p | 53/733 | 2.97E−07 | hsa‐miR‐30a‐5p | 53 |
| hsa‐miR‐30e‐5p | 32/347 | 2.85E−06 | hsa‐miR‐16‐5p | 63 |
| hsa‐miR‐215‐5p | 50/755 | 4.76E−06 | hsa‐miR‐215‐5p | 50 |
| hsa‐miR‐190a‐3p | 45/648 | 4.76E−06 | hsa‐miR‐106b‐5pa | 60 |
| hsa‐miR‐5011‐5p | 45/653 | 4.76E−06 | hsa‐miR‐17‐5pa | 60 |
| hsa‐miR‐1277‐5p | 44/624 | 4.76E−06 | hsa‐miR‐20a‐5pa | 56 |
| hsa‐miR‐101‐3p | 33/393 | 4.76E−06 | hsa‐miR‐155‐5p | 53 |
| hsa‐miR‐548i | 23/202 | 4.76E−06 | hsa‐miR‐519d‐3p | 51 |
| hsa‐miR‐20b‐5pa | 51 | |||
Part of the mir‐17 family. The 3 miRNAs in bold are included in both lists.
The most connected miRNAs have validated target sites in 50 or more genes up‐regulated in the EtOH drinkers (Table 1b). A total of 222 (40%) of the 567 up‐regulated genes have a 3′UTR target site for at least one of the 11 most connected miRNAs, and 52 of the genes are associated with 5 or more of the miRNAs (see Table S3). The top 10 overrepresented genes with binding sites for the top connected miRNAs are DNAJB4(10), KLF10(9), PHF6(9), ZBTB35(9), FAM126B(8), HSPA4L(8), ITGA2(8), SPRED1(8), VPS13C(8), and XIAP(8), with the number of top miRNA binding sites in parentheses. Overall, for the 197 miRNAs that have significantly represented target sites (q < 0.05) in our 567 EtOH up‐regulated genes, each miRNA has validated target sites for a minimum of 5 and a maximum of 67 genes. A total of 385 (68%) of the up‐regulated genes have at least 1 overrepresented miRNA target site.
Overrepresentation analysis of gene ontology categories using miEAA for the 11 most connected miRNAs are listed in Table S3; the annotations associated with 9 or more of the miRNAs are listed (N = 465). Annotations of interest are highlighted in Table S3 and include GO0002224 Toll‐like receptor signaling pathway (q = 6.0 × 10−4), GO0007155 cell adhesion (q = 6.5 × 10−4), GO0001666 response to hypoxia (q = 1.8 × 10−3), GO0006965 immune response (q = 1 × 10−3), GO0005518 collagen binding (q = 6.5 × 10−4), and GO0005525 GTP binding (q = 6 × 10−3).
Significant enrichment for transcription factor binding sites in DEGs was also detected with binding motifs for 14 and 6 human transcription factors (TF) significantly (q < 0.05) enriched in the down‐ and up‐regulated genes, respectively (Table 2). The list of significantly enriched (q < 0.05) TF binding sites, including gene lists, is found in Table S4. Of the 6 TF sites enriched in up‐regulated genes, none were differentially expressed. The most enriched transcription factor (KLF11) in the down‐regulated genes was down‐regulated only in the controls (DE q = 4.9 × 10−4). The TF associated with the down‐regulated genes clustered into 2 groups (Table 2), those significant at 10−8 or better (N = 4) and those significant at ≥10−4 (N = 10). Associated with the first group were 149 unique genes, and 50 of these genes were associated with all 4 TF. Annotations for the genes associated with the 149 unique genes are found in Table S4. Annotations for the 149‐gene grouping included cell adhesion (q < 1 × 10−5), immune response (q < 6 × 10−5), cell motility (q < 2 × 10−2), plasma membrane (q < 1 × 10−3), and extracellular matrix (q < 3 × 10−2; Table S4).
Table 2.
Transcription Factors (TFs) That Have Significant Number of Target Sites in Our Up‐ or Down‐Regulated Genes
| Term | Overlap | Adjusted p‐value |
|---|---|---|
| TF binding sites overrepresented in up‐regulated genes | ||
| JUND (human) | 80/1380 | 1.93E−07 |
| MYB (human) | 76/1409 | 5.98E−06 |
| POU1F1 (human) | 70/1408 | 2.83E−04 |
| TBP (human) | 104/2486 | 1.82E−03 |
| HNF1B (human) | 10/90 | 1.30E−02 |
| CBEPB (human) | 102/2598 | 1.67E−02 |
| TF binding sites overrepresented in down‐regulated genes | ||
| Group 1 | ||
| KLF11 (human) | 103/1388 | 6.53E−12 |
| KLF4 (human) | 104/1485 | 9.47E−11 |
| ZNF148 (human) | 104/1591 | 4.01E−09 |
| CACYBP (human) | 91/1350 | 1.25E−08 |
| Group 2 | ||
| MZF1 (human) | 79/1389 | 1.72E−04 |
| SNAI1 (human) | 78/1428 | 5.68E−04 |
| SNAI2 (human) | 78/1428 | 5.68E−04 |
| TCF3 (human) | 78/1428 | 5.68E−04 |
| NFE2 (human) | 127/2681 | 1.16E−03 |
| IKZF1 (human) | 73/1373 | 2.10E−03 |
| ELF3 (human) | 76/1488 | 4.84E−03 |
| SREBF2 (human) | 70/1361 | 6.65E−03 |
| KLF13 (human) | 72/1453 | 1.45E−02 |
| SP1 (human) | 69/1406 | 2.27E−02 |
Overlap = [number of genes up or down‐regulated in our dataset (FDR < 0.005 + Fold Change > 1.5)]/[total number of genes in TRANSFAC and JASPAR with motifs for this TF in the promoter region].
The second grouping of 10 TF was associated with 329 unique genes. A total of 135 of these genes are also in the group of 149 noted above. Annotations for the 194 genes found only in the second grouping of TF included immune system process (q < 4 × 10−12), regulation of immune response (q < 9 × 10−12), cytokine‐mediated signaling pathway (q < 2 × 10−6), signaling receptor activity (q < 4 × 10−6), Toll‐like receptor binding (q < 7 × 10−3), and extracellular region (q < 6 × 10−5; Table S4).
Discussion
Two cohorts of young adult, male cynomolgus macaque monkeys achieved a range of stable, daily voluntary EtOH consumption with 6 months of open access to EtOH. Based on body weight and volume consumed, the drinkers ranged from a low average of 3 (human equivalent) drinks per day to a high average of 15 drinks per day. The overall average across the 2 cohorts was about 9 drinks per day. These animals established drinking patterns similar to previous monkey cohorts, and these patterns mimic those found in the humans with AUD (Baker et al., 2014). Detailed daily data on these cohorts and previous cohorts are available on the Monkey Alcohol and Tissue Resource Center Web site (MATRR.com). Based on previously defined categories (Baker et al., 2014), these 2 cohorts had a distribution of 7 LD, 9 heavy drinkers (HD), and 1 very heavy drinker.
Because we collected longitudinal samples from the same animals before and after voluntary EtOH consumption, we were able to adjust for individual baseline differences in gene expression. This paired longitudinal analysis in controls (n = 6) and drinkers (n = 10) allowed us to identify baseline differences in gene expression and to identify those gene expression changes that are specifically the result of voluntary EtOH consumption. This approach allowed the identification of 1242 DEGs specifically associated with EtOH consumption, highlighting the neuroplasticity of the brain following 6 months of voluntary EtOH consumption. The up‐regulated genes were a diverse set showing a significant enrichment in 2 membrane‐associated annotations (Table S2). Genes in these categories included cadherins and protocadherins (CDH10, CDH19, PCDH18, and PCDH20), receptors and receptor subunits (CHRM2, DRD5, GABRA1, GABRA4, GABRG2, GLRB, HTR2A, HTR2C, and NPY5R), and a cluster of G‐protein receptors (GPR19, 22, 52, and 63). Cadherins and protocadherins have been implicated as having key roles in EtOH preference (Colville et al., 2017). Variants in CHRM2 have been implicated as correlates to the age of onset of AUDs in adolescents and young adults (Chorlian et al., 2013). The evidence that DRD5 has a role in AUDs is limited (Hack et al., 2011). Enoch and colleagues (2012) found that the expression of GABRG2 was down‐regulated in both the brains of alcoholics and cocaine addicts. Some but not all studies suggest an association between polymorphisms in HTR2A and AUDs (see Cao et al., 2014). Mottagui‐Tabar and colleagues (2004) found some evidence of HTR2C promotor variants as alcoholism risk factors; however, to our knowledge this has not been confirmed or refuted in more contemporary studies. The cluster of orphan G‐protein receptors has no known association with EtOH phenotypes. GPR19 is a potential receptor for adropin which is involved in the regulation of water intake. GPR22 has linked to the regulation of ciliary function which has been shown to be associated with binge EtOH consumption (Iancu et al., 2018a; Verleyen et al., 2014). GPR52 has been linked to the regulation of striatal function (Song et al., 2018). A deletion on human chromosome 6 that affects GPR63, NOUFA4, and KLHL32 has been associated with Tourette’s syndrome and obsessive–compulsive disorder (Hooper et al., 2012).
The up‐regulated genes were significantly enriched in a number of miRNA binding sites, an association that was absent in the down‐regulated genes. Eleven of the miRNAs were the most connected and had validated target sites in 50 or more up‐regulated genes. Overrepresentation analysis of gene ontology categories using miRNA enrichment analysis revealed a broad representation of GO annotation categories including the extracellular matrix which has been previously identified as a risk category for binge EtOH consumption (Iancu et al., 2018a); however, none of the drinkers in this study met the criteria for binge consumption. Of particular interest was the observation that the overrepresentation analysis revealed an association with immune process‐related categories including Toll‐like receptor signaling pathway and immune response. Four previous studies identified EtOH specific miRNA changes in human alcoholics compared to control subjects (Lewohl et al., 2011; Mamdani et al., 2015; Manzardo et al., 2013; Wang et al., 2013). Specifically, Manzardo and colleagues (2013) looked at miRNA expression from frontal cortex of human alcoholics and identified 13 miRNAs up‐regulated in subjects with AUD and 2 miRNAs down‐regulated. In our 567 genes that were up‐regulated following 6 months of EtOH consumption, we identified enrichment in target sites for 2 miRNAs also identified by Manzardo and colleagues (2013): hsa‐miR‐375 and hsa‐miR‐379. These were both found to be up‐regulated in alcoholic humans. Lewohl and colleagues (2011) identified 35 up‐regulated miRNAs in human alcoholic PFC samples. We identified 8 of those miRNAs (hsa‐miR‐1, hsa‐miR‐144, hsa‐miR‐153, hsa‐miR‐101, hsa‐miR‐374b, hsa‐miR‐140, hsa‐miR‐586, and hsa‐miR‐580) as having target sites in our list of up‐regulated genes. Mamdani and colleagues (2015) performed coexpression analyses on mRNAs and miRNAs in nucleus accumbens of postmortem alcoholic subjects. Of the 25 miRNA hubs they identified, we found enrichment for target sites in our up‐regulated genes for 4 of them (hsa‐miR‐375, hsa‐miR‐132, hsa‐miR‐361, and hsa‐miR‐4760). Several of the most connected miRNAs also have previous evidence of being affected by EtOH in rodent studies (e.g., mmu‐miR‐16‐5p, mmu‐miR‐16‐5p, mmu‐miR‐93‐5p, mmu‐miR‐17‐5p, mmu‐miR‐155‐5p, mmu‐miR‐30a‐5p; Darcq et al., 2015; Gorini et al., 2013; Lippai et al., 2013; Nunez et al., 2013; Osterndorff‐Kahanek et al., 2018).
It is interesting to note that for the 3 human studies with overlap, the miRNAs were up‐regulated, which usually leads to down‐regulation of mRNA expression; however, the genes we identified were up‐regulated. Temporal shifts in expression in miRNAs affected by EtOH have been observed in mouse at 0 hour versus 8 hour versus 120 hour postEtOH exposure (Osterndorff‐Kahanek et al., 2018). The temporal variation in the miRNAs could explain the difference in direction of the gene expression changes we observe with regard to previously identified EtOH‐affected human miRNAs. In our protocol, the postEtOH biopsies were collected on what would have been a regular voluntary drinking day.
The genes differentially expressed specifically in the drinking macaques have a rich annotation structure and include immune response, inflammatory response, cell adhesion, signaling receptor activity, cytokine binding, plasma membrane, and extracellular matrix. There is now robust evidence of relationships between neuroimmune signaling and AUDs (e.g., Coleman and Crews, 2018; Cui et al., 2014; Harris and Koob, 2017). Neuro‐inflammatory mechanisms are associated with both the risk for and individual variation in excessive EtOH consumption (e.g., Hitzemann et al., 2017; Iancu et al., 2018a). Although these immune categories were primarily associated with the down‐regulated genes, this does not necessarily imply that chronic EtOH consumption has down‐regulated the neuroimmune system and in fact the available data would suggest otherwise (Erickson et al., 2019 and references therein). Examining the genes associated with the annotation of cytokine signaling reveals both proinflammatory (IL1R1, IL2RG, IL17RA, and CD74) and antiinflammatory (IL6R and IL10RA) receptor genes suggesting that chronic EtOH consumption may have established a new balance between the opposing forces. The actual inflammatory status in A46 at the time of sample acquisition is unknown. Nor it is known what cell type(s) are associated with a particular cytokine signal. However, there is ample evidence that chronic EtOH consumption has a marked effect on the expression of cytokine ligands and receptors (Erickson et al., 2019; Montesinos et al., 2016; Warden et al., 2019).
As noted above, the differentially expressed genes were enriched in genes (N = 47) associated with the extracellular matrix (ECM). Genes with this annotation included 4 collagens (COL1A1, COL6A2, COL18A1, and COL23A1) and 2 matrix metaloproteases (MMP9 and MMP25). Collagens and the ECM were previously identified as associated with the selection for the High‐Drinking‐In‐the Dark mouse lines (Iancu et al., 2018b). Alcohol and other drugs of abuse can have marked effects on ECM constituents (reviewed in Lubbers et al., 2014). EtOH is known to affect the brain expression of tPA (or PLAT), MMP9, BCAN & NCAN, and TSP2 & TSP4 (Bahi and Dreyer, 2012; Coleman et al., 2011; Pawlak et al., 2005; Risher et al., 2015; Wright et al., 2003). Previously, we emphasized (Iancu et al., 2018a) the potential interactions between the ECM and neuroimmune processes, and we repeat that emphasis here. For example, Seo and colleagues (2008) have shown that collagens can induce an inflammatory response in microglia.
Although the down‐regulated genes were not enriched in miRNA binding sites, there was a significant enrichment in TF binding sites. The TF associated with the down‐regulated genes clustered into 2 groups: those significant at 10−8 or better (N = 4) and those significant at ≥10−4 (N = 10). None of the TFs associated with these binding sites met our criteria for significant down‐regulation; however, for many they were trending in the right direction (See Table S1). Not surprisingly, the annotations for the down‐regulated genes associated with these TFs (75% of the total) showed significant enrichment in genes associated with immune processes and the ECM.
Some of the TF identified in the current study have been previously found to be associated with alcohol‐related phenotypes. For example, protein levels of Kruppel‐like factor 11 (KLF11) have been shown to increase in the postmortem PFC of subjects with alcohol dependence (Udemgba et al., 2014). Further binge drinking increased KLF11 expression in the rat frontal cortex (Duncan et al., 2016). EtOH has been shown to decrease myeloid zinc finger 1 (MZF1) transcription, and MZF1 is a transcription factor involved in EtOH induced decreases in bone remodeling and fracture healing (Driver et al., 2015). MZF1 was significantly down‐regulated following 6 months of EtOH consumption, and the set of down‐regulated genes were significantly enriched in MZF1 binding sites with 83 down‐regulated genes containing a MZF1 binding site(s) in their promoter regions.
In conclusion, the data presented here are, to our knowledge, the first example of using a within‐subject design to assess the effects of chronic EtOH consumption on brain gene expression. The statistical advantages of this approach are obvious and especially welcome when working with NHPs. The small sample sizes precluded dividing the drinkers according to their average daily consumption; however, we retain the expectation that the transcriptional signature for the low and HD will differ and is likely to differ across some of the dimensions described here such as neuroimmune response. A key observation noted here, as well as previously (Iancu et al., 2018a), is that both pro and antiinflammatory immune‐related genes are affected. In addition, we have again noted a strong effect on ECM‐related elements. Finally, we consider that an interaction between immune and ECM gene networks is likely critical to understanding the transcriptional effects of chronic EtOH exposure.
Funding
This work was supported by National Institute of Health grants P60AA010760 (RH, KG, SM, CZ, RS, NW), R24AA019431 (KG), and AA013484 (RH).
Conflict of interest
The authors declare that they have no competing financial interests.
Author contributions
NW, KG, and RH conceived and designed the study. RS contributed to RNA‐Seq via MPSSR Core at OHSU. SM, CZ, and NW contributed to gene expression analyses. NW, RH, and CZ prepared the manuscript.
Supporting information
Table S1. Summary of Gene Expression Data.
Table S2. Gene Ontology (GO) of Alcohol Affected Genes.
Table S3. miRNAs Associated with Alcohol Affected Genes.
Table S4. Transcription Factors Associated with Alcohol affected Genes.
Acknowledgments
The authors wish to thank Denesa Lockwood for her assistance in preparing this manuscript.
References
- Anders S, Pyl PT, Huber W (2015) HTSeq–a Python framework to work with high‐throughput sequencing data. Bioinformatics 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes C, Khaleeq QT, Meese E, Keller A (2016) miEAA: microRNA enrichment analysis and annotation. Nucleic Acids Res 44:W110–W116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahi A, Dreyer J‐L (2012) Involvement of tissue plasminogen activator “tPA” in ethanol‐induced locomotor sensitization and conditioned‐place preference. Behav Brain Res 226:250–258. [DOI] [PubMed] [Google Scholar]
- Baker EJ, Farro J, Gonzales S, Helms C, Grant KA (2014) Chronic alcohol self‐administration in monkeys shows long‐term quantity/frequency categorical stability. Alcohol Clin Exp Res 38:2835–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Liu X, Han S, Zhang CK, Liu Z, Li D (2014) Association of the HTR2A gene with alcohol and heroin abuse. Hum Genet 133:357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles G, Clark NR, Ma’ayan A (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform 14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorlian DB, Rangaswamy M, Manz N, Wang J‐C, Dick D, Almasy L, Bauer L, Bucholz K, Foroud T, Hesselbrock V, Kang SJ, Kramer J, Kuperman S, Nurnberger J, Rice J, Schuckit M, Tischfield J, Edenberg HJ, Goate A, Bierut L, Porjesz B (2013) Genetic and neurophysiological correlates of the age of onset of alcohol use disorders in adolescents and young adults. Behav Genet 43:386–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C‐H, Shrestha S, Yang C‐D, Chang N‐W, Lin Y‐L, Liao K‐W, Huang W‐C, Sun T‐H, Tu S‐J, Lee W‐H, Chiew M‐Y, Tai C‐S, Wei T‐Y, Tsai T‐R, Huang H‐T, Wang C‐Y, Wu H‐Y, Ho S‐Y, Chen P‐R, Chuang C‐H, Hsieh P‐J, Wu Y‐S, Chen W‐L, Li M‐J, Wu Y‐C, Huang X‐Y, Ng FL, Buddhakosai W, Huang P‐C, Lan K‐C, Huang C‐Y, Weng S‐L, Cheng Y‐N, Liang C, Hsu W‐L, Huang H‐D (2018) miRTarBase update 2018: a resource for experimentally validated microRNA‐target interactions. Nucleic Acids Res 46:D296–D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman LG, Crews FT (2018) Innate immune signaling and alcohol use disorders. Handb Exp Pharmacol 248:369–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman LG, He J, Lee J, Styner M, Crews FT (2011) Adolescent binge drinking alters adult brain neurotransmitter gene expression, behavior, brain regional volumes, and neurochemistry in mice. Alcohol Clin Exp Res 35:671–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colville AM, Iancu OD, Oberbeck DL, Darakjian P, Zheng CL, Walter NAR, Harrington CA, Searles RP, McWeeney S, Hitzemann RJ (2017) Effects of selection for ethanol preference on gene expression in the nucleus accumbens of HS‐CC mice. Genes Brain Behav 16:462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contet C (2012) Gene expression under the influence: transcriptional profiling of ethanol in the brain. Curr Psychopharmacol 1:301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Shurtleff D, Harris RA (2014) Neuroimmune mechanisms of alcohol and drug addiction. Int Rev Neurobiol 118:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darcq E, Warnault V, Phamluong K, Besserer GM, Liu F, Ron D (2015) MicroRNA‐30a‐5p in the prefrontal cortex controls the transition from moderate to excessive alcohol consumption. Mol Psychiatry 20:1219-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daunais JB, Davenport AT, Helms CM, Gonzales SW, Hemby SE, Friedman DP, Farro JP, Baker EJ, Grant KA (2014) Monkey alcohol tissue research resource: banking tissues for alcohol research. Alcohol Clin Exp Res 38:1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver J, Weber CE, Callaci JJ, Kothari AN, Zapf MA, Roper PM, Borys D, Franzen CA, Gupta GN, Wai PY, Zhang J, Denning MF, Kuo PC, Mi Z (2015) Alcohol inhibits osteopontin‐dependent transforming growth factor‐β1 expression in human mesenchymal stem cells. J Biol Chem 290:9959–9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JW, Zhang X, Wang N, Johnson S, Harris S, Udemgba C, Ou X‐M, Youdim MB, Stockmeier CA, Wang JM (2016) Binge ethanol exposure increases the Krüppel‐like factor 11‐monoamine oxidase (MAO) pathway in rats: examining the use of MAO inhibitors to prevent ethanol‐induced brain injury. Neuropharmacology 105:329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z (2009) GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch M‐A, Zhou Z, Kimura M, Mash DC, Yuan Q, Goldman D (2012) GABAergic gene expression in postmortem hippocampus from alcoholics and cocaine addicts; corresponding findings in alcohol‐naïve P and NP rats. PLoS ONE 7:e29369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson EK, Grantham EK, Warden AS, Harris RA (2019) Neuroimmune signaling in alcohol use disorder. Pharmacol Biochem Behav 177:34–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris SP, Mayfield RD (2014) RNA‐Seq reveals novel transcriptional reorganization in human alcoholic brain. Int Rev Neurobiol 116:275–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris SP, Riley BP, Williams RW, Mulligan MK, Miles MF, Lopez MF, Hitzemann R, Iancu OD, Colville A, Walter NAR, Darakjian P, Oberbeck DL, Daunais JB, Zheng CL, Searles RP, McWeeney SK, Grant KA, Mayfield RD (2018) Cross‐species molecular dissection across alcohol behavioral domains. Alcohol 72:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbella M, Borra E, Tonelli S, Rozzi S, Luppino G (2013) Connectional heterogeneity of the ventral part of the macaque area 46. Cereb Cortex 23:967–987. [DOI] [PubMed] [Google Scholar]
- Gorini G, Nunez YO, Mayfield RD (2013) Integration of miRNA and protein profiling reveals coordinated neuroadaptations in the alcohol‐dependent mouse brain. PLoS ONE 8:e82565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant KA, Leng X, Green HL, Szeliga KT, Rogers LSM, Gonzales SW (2008) Drinking typography established by scheduled induction predicts chronic heavy drinking in a monkey model of ethanol self‐administration. Alcohol Clin Exp Res 32:1824–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KL, Szeliga KT, Bowen CA, Kautz MA, Azarov AV, Grant KA (1999) Comparison of ethanol metabolism in male and female cynomolgus macaques (Macaca fascicularis). Alcohol Clin Exp Res 23:611–616. [PubMed] [Google Scholar]
- Hack LM, Kalsi G, Aliev F, Kuo P‐H, Prescott CA, Patterson DG, Walsh D, Dick DM, Riley BP, Kendler KS (2011) Limited associations of dopamine system genes with alcohol dependence and related traits in the Irish Affected Sib Pair Study of Alcohol Dependence (IASPSAD). Alcohol Clin Exp Res 35:376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RA, Koob GF (2017) The future is now: a 2020 view of alcoholism research. Neuropharmacology 122:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzemann R, Oberbeck D, Iancu O, Darakjian P, McWeeney S, Spence S, Schlumbohm J, Metten P, Crabbe J (2017) Alignment of the transcriptome with individual variation in animals selectively bred for High Drinking‐In‐the‐Dark (HDID). Alcohol 60:115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper SD, Johansson ACV, Tellgren‐Roth C, Stattin E‐L, Dahl N, Cavelier L, Feuk L (2012) Genome‐wide sequencing for the identification of rearrangements associated with Tourette syndrome and obsessive‐compulsive disorder. BMC Med Genet 13:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu OD, Colville A, Walter NAR, Darakjian P, Oberbeck DL, Daunais JB, Zheng CL, Searles RP, McWeeney SK, Grant KA, Hitzemann R (2018a) On the relationships in rhesus macaques between chronic ethanol consumption and the brain transcriptome. Addict Biol 23:196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu OD, Colville AM, Wilmot B, Searles R, Darakjian P, Zheng C, McWeeney S, Kawane S, Crabbe JC, Metten P, Oberbeck D, Hitzemann R (2018b) Gender‐specific effects of selection for drinking in the dark on the network roles of coding and noncoding RNAs. Alcohol Clin Exp Res 42:1454–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez VA, Allen DC, McClintick MN, Grant KA (2017) Social setting, social rank and HPA axis response in cynomolgus monkeys. Psychopharmacology 234:1881–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44:W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, Nunez YO, Dodd PR, Tiwari GR, Harris RA, Mayfield RD (2011) Up‐regulation of microRNAs in brain of human alcoholics. Alcohol Clin Exp Res 35:1928–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA (2000) Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res 24:1873–1882. [PubMed] [Google Scholar]
- Lippai D, Bala S, Csak T, Kurt‐Jones EA, Szabo G (2013) Chronic alcohol‐induced microRNA‐155 contributes to neuroinflammation in a TLR4‐dependent manner in mice. PLoS ONE 8:e70945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubbers BR, Smit AB, Spijker S, van den Oever MC (2014) Neural ECM in addiction, schizophrenia, and mood disorder. Prog Brain Res 214:263–284. [DOI] [PubMed] [Google Scholar]
- Mamdani M, Williamson V, McMichael GO, Blevins T, Aliev F, Adkins A, Hack L, Bigdeli T, van der Vaart AD, Web BT, Bacanu S‐A, Kalsi G, COGA Consortium , Kendler KS, Miles MF, Dick D, Riley BP, Dumur C, Vladimirov VI (2015) Integrating mRNA and miRNA weighted gene co‐expression networks with eQTLs in the nucleus accumbens of subjects with alcohol dependence. PLoS ONE 10:e0137671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzardo AM, Gunewardena S, Butler MG (2013) Over‐expression of the miRNA cluster at chromosome 14q32 in the alcoholic brain correlates with suppression of predicted target mRNA required for oligodendrocyte proliferation. Gene 526:356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel‐Margoulis OV, Fricke E, Liebich I, Land S, Barre‐Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki‐Potapov B, Saxel H, Kel AE, Wingender E (2006) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34:D108–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos J, Alfonso‐Loeches S, Guerri C (2016) Impact of the innate immune response in the actions of ethanol on the central nervous system. Alcohol Clin Exp Res 40:2260–2270. [DOI] [PubMed] [Google Scholar]
- Mottagui‐Tabar S, McCarthy S, Reinemund J, Andersson B, Wahlestedt C, Heilig M (2004) Analysis of 5‐hydroxytryptamine 2c receptor gene promoter variants as alcohol‐dependence risk factors. Alcohol Alcohol 39:380–385. [DOI] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE (2006) Toward understanding the genetics of alcohol drinking through transcriptome meta‐analysis. Proc Natl Acad Sci USA 103:6368–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez YO, Truitt JM, Gorini G, Ponomareva ON, Blednov YA, Harris RA, Mayfield RD (2013) Positively correlated miRNA‐mRNA regulatory networks in mouse frontal cortex during early stages of alcohol dependence. BMC Genom 14:725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterndorff‐Kahanek EA, Tiwari GR, Lopez MF, Becker HC, Harris RA, Mayfield RD (2018) Long‐term ethanol exposure: Temporal pattern of microRNA expression and associated mRNA gene networks in mouse brain. PLoS ONE 13:e0190841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak R, Melchor JP, Matys T, Skrzypiec AE, Strickland S (2005) Ethanol‐withdrawal seizures are controlled by tissue plasminogen activator via modulation of NR2B‐containing NMDA receptors. Proc Natl Acad Sci USA 102:443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risher M‐L, Sexton HG, Risher WC, Wilson WA, Fleming RL, Madison RD, Moore SD, Eroglu C, Swartzwelder HS (2015) Adolescent intermittent alcohol exposure: dysregulation of thrombospondins and synapse formation are associated with decreased neuronal density in the adult hippocampus. Alcohol Clin Exp Res 39:2403–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux BT, Heward JA, Donnelly LE, Jones SW, Lindsay MA (2017) Catalog of differentially expressed long non‐coding RNA following activation of human and mouse innate immune response. Front Immunol 8:1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem KS, Price JL, Hashikawa T (2007) Cytoarchitectonic and chemoarchitectonic subdivisions of the perirhinal and parahippocampal cortices in macaque monkeys. J Comp Neurol 500:973–1006. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B (2004) JASPAR: an open‐access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 32:D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M‐C, Kim S, Kim S‐H, Zheng LT, Park EK, Lee W‐H, Suk K (2008) Discoidin domain receptor 1 mediates collagen‐induced inflammatory activation of microglia in culture. J Neurosci Res 86:1087–1095. [DOI] [PubMed] [Google Scholar]
- Song H, Li H, Guo S, Pan Y, Fu Y, Zhou Z, Li Z, Wen X, Sun X, He B, Gu H, Zhao Q, Wang C, An P, Luo S, Hu Y, Xie X, Lu B (2018) Targeting Gpr52 lowers mutant HTT levels and rescues Huntington’s disease‐associated phenotypes. Brain J Neurol 141:1782–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Substance Abuse and Mental Health Administration (SAaMHS) (2015) National Survey on Drug Use and Health Available at: https://www.samhsa.gov\x9BNSDUH-DetTabs-2015\x9BNSDUH-DetTabs-2015 [PubMed] [Google Scholar]
- Udemgba C, Johnson S, Stockmeier CA, Luo J, Albert PR, Wang J, May WL, Rajkowska G, Harris S, Sittman DB, Ou X‐M (2014) The expression of KLF11 (TIEG2), a monoamine oxidase B transcriptional activator in the prefrontal cortex of human alcohol dependence. Alcohol Clin Exp Res 38:144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verleyen D, Luyten FP, Tylzanowski P (2014) Orphan G‐protein coupled receptor 22 (Gpr22) regulates cilia length and structure in the zebrafish Kupffer’s vesicle. PLoS ONE 9:e110484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Gelernter J, Zhang H (2013) Differential expression of miR‐130a in postmortem prefrontal cortex of subjects with alcohol use disorders. J Addict Res Ther 4:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden AS, Azzam M, DaCosta A, Mason S, Blednov YA, Messing RO, Mayfield RD, Harris RA (2019) Toll‐like receptor 3 activation increases voluntary alcohol intake in C57BL/6J male mice. Brain Behav Immun 77:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden AS, Mayfield RD (2017) Gene expression profiling in the human alcoholic brain. Neuropharmacology 161-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JW, Masino AJ, Reichert JR, Turner GD, Meighan SE, Meighan PC, Harding JW (2003) Ethanol‐induced impairment of spatial memory and brain matrix metalloproteinases. Brain Res 963:252–261. [DOI] [PubMed] [Google Scholar]
- Xu J, Zhang R, Shen Y, Liu G, Lu X, Wu C‐I (2013) The evolution of evolvability in microRNA target sites in vertebrates. Genome Res 23:1810–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of Gene Expression Data.
Table S2. Gene Ontology (GO) of Alcohol Affected Genes.
Table S3. miRNAs Associated with Alcohol Affected Genes.
Table S4. Transcription Factors Associated with Alcohol affected Genes.


