Abstract
Many of the 20-member mucin family are evolutionarily conserved proteins that are often aberrantly expressed and glycosylated in various benign and malignant pathologies including oncogenic signaling leading to tumor invasion, metastasis, and immune evasion. Large size and extensive glycosylation present challenges to study mucin structure using traditional methods, including crystallography. We offer the hypothesis that the functional versatility of mucins may be attributed to the presence of intrinsically disordered regions (IDRs), which provide dynamism and flexibility; further, that these sites offer potential therapeutic targets. Herein, we examined the links between mucin structure and function based on IDRs, post-translational modifications (PTMs), and potential impact on their interactome. Using sequence-based bioinformatics tools, we observed that mucins are predicted to be moderately (20–40%) to highly (>40%) disordered and many conserved mucin domains could be disordered. Phosphorylation sites overlap with IDRs throughout the mucin sequences. Additionally, the majority of predicted O- and N- glycosylation sites in the tandem repeat regions occur within IDRs, and these IDRs contain a large number of functional motifs, i.e. molecular recognition features (MoRFs), which directly influence PPIs. This investigation provides a novel perspective and offers an insight into the complexity and dynamic nature of mucins.
Keywords: intrinsically disordered protein, glycoprotein, protein-protein interaction, protein structure
INTRODUCTION
MUCIN PROTEIN FAMILY
Mucins (MUCs) are heavyweight (over 106 Dalton) glycoproteins that are expressed by epithelial cells in many organs throughout the body (1). In humans, the mucin protein family contains over twenty members and is subdivided, based on structural differences, into transmembrane and secretory mucins. The primary distinction between these two groups is the presence or absence of a transmembrane domain (TM), which anchors them to the cell membrane. Mucins contain a characteristic large polymorphic variable number of tandem repeat domain (VNTR) that is rich in proline, threonine and serine residues (PTS). The VNTR is susceptible to enzymatic modification by O-linked and N-linked oligosaccharides (2). All mucins harbor one or more domains with high sequence similarity to a known functional domain present in other proteins. These include the EGF-like domain (EGF), sea-urchin sperm protein, enterokinase and agrin (SEA) domain, von Willebrand factor D domain (vWD), nidogen-like domain (NIDO), the adhesion-associated domain in MUC4 and other proteins (AMOP), and the D-domain (3). These domains have been implicated in several biological processes such as cell-to-cell interaction (4), cell-to-ECM interaction (5), apoptosis inhibition (6), and cell signaling complexes (7).
Aberrant expression, splicing, and glycosylation in various members of the mucin family is a characteristic feature of several malignancies including pancreatic ductal adenocarcinoma (1, 8, 9), colorectal (10, 11), lung (12, 13) and ovarian cancer (14). Further, tumor cells exploit mucin differential localization, alternative splicing, cellular adhesive/anti-adhesive properties, and alterations in glycosylation profile, to metastasize to distant locales and survive in hostile environments (1, 15).
INTRINSICALLY DISORDERED PROTEINS
Until recently, it was general held that the three-dimensional structure of a protein defined its function (16). However, it is now well established that intrinsically disordered proteins (IDPs) and regions (IDRs), are complete proteins (or segments of proteins) that lack a traditional globular secondary or tertiary structure, yet are fully functional (17–22). Disordered regions generally are sequences of low complexity with a low proportion of hydrophobic residues and a high number of repeating residues with a preponderance of polar and charged residues (23, 24). This lack of bulky hydrophobic amino acids prevents the formation of an ordered core that comprises a traditional structured domain (25). Disorder is ubiquitous throughout the human proteome. A study estimated that 30% of all proteins harbor some degree of disorder with a majority of these proteins containing disorder ranging between 20–40% of their total sequence (25–27)
IDPs/IDRs have wide-ranging implications in various physiological and biological processes such as transcription, splicing, translation, signaling (20, 28–31), scaffolding (32, 33), cell cycle regulation (34–36), protein-protein interactions (PPIs) (19, 37–41), chaperoning (17), and phenotypic plasticity (42, 43) (that is the ability to switch phenotypes). Further, IDP/IDR-mediated modulations are implicated in the pathogenesis of various diseases such as cancer, diabetes, cardiovascular defects, amyloidosis, and neurodegeneration (44). More specifically, many cancer-associated proteins have been shown to have a higher percentage of IDRs relative to the rest of the human proteome (44–47).
This functional versatility of IDPs/IDRs encourages us to investigate their presence in known cancer-associated proteins like mucins. Molecular events such as mutations that increase protein hydrophilicity, or alter protein splicing, can lead to changes in IDR length and affect protein-protein interactions, leading to pathological properties. This often affects protein solubility and aggregation, leading to nonproductive or over-productive complexes that disturb regulatory networks (25, 48).
Considering the significant role of mucins in normal physiology as well as pathological conditions, hub protein characteristics, and their simple abundance and aberrant expression tendency in a variety of cancers, IDR/IDP presence within mucins could have important clinical implications. Mucins are also prime targets for IDP/IDR analyses because conventional methods of structural delineation are limited by large size, the high number of PTMs, and the presence of multiple splice variants.
For many proteins implicated in cancer, structural biology information has proven invaluable for understanding their functional implications as well as discovering novel therapeutic modalities (49–52). Unfortunately, it is difficult to study mucins structurally by traditional methods. Crystallographic methods falter because of the sheer size of mucins (up to 14,000 kDa), extensive variation in the number of tandem repeats within the VNTR (up to 120), sequence variation, and inability to clone, express, and purify fully folded and glycosylated forms. While specific domains have been cloned, purified, and studied (i.e. SEA (53)), domain homology between mucins and other proteins varies. What structural analyses have been accomplished (via x-ray crystallography) were conducted domain-by-domain and not as a part of the complete protein (54). In addition, improper refolding of solitary domains is a constraint on these experiments. This dearth of advanced structural knowledge constrains the investigation of mucins as possible therapeutic targets.
Based on the earlier studies, we hypothesized that the functional versatility of mucins may be attributed to the presence of intrinsically disordered regions (IDRs); further, that these sites offer potential therapeutic targets.
To support this hypothesis, we analyzed the protein sequences of mucins using the Database of Disordered Protein Predictions (D2P2) to predict disorder based on a ≥75% consensus between the nine disorder prediction models incorporated within the tool itself (55). The presence of IDRs was determined within conserved mucin domains, inter-domain sequences, C-terminal, and transmembrane domains. Next, we assessed the relationships of IDRs with curated phosphorylation sites and predicted N- and O-glycosylation sites, to discern whether posttranslational modifications occurred preferentially in IDRs within the mucin sequences. Finally, we also assessed the effect of conformational disorder on the mucin family interactome.
ASSESSMENT METHODS
Mucin disorder prediction with D2P2
Mucin sequences were searched for predicted disorder using the text search option provided on the web-based D2P2 (version v0.3–689) (55) portal (http://d2p2.pro/). D2P2 comprises nine different disorder prediction tools involving a variety of prediction methods (Espritz-D, Espritz-X, Espritz-N, IUPred-L, IUPred-S, PV2, PrDOS, VSL2b, VLXT). Due to variable length and high sequence ambiguity for tandem repeat domains of mucins, the longest available human mucin transcripts that were present in D2P2 were used for our analysis. Disorder was predicted based upon 75% consensus of all nine predictors, and high confidence disorder regions were then obtained. The percentage disorder was computed by dividing the total length of disordered regions by the protein sequence length. For our study, mucins with < 20% disorder will be considered to have low levels of disorder, those with >20% and <40% will be defined as moderately disordered, and those with >40% will be considered as highly disordered.
Mucin disorder prediction with FoldIndex
An algorithm originally developed by Uversky and colleagues (56) was implemented using an in-house python script that predicts if a region in a protein sequence would assume a folded or intrinsically unfolded state. This algorithm works on two properties of an amino acid: net charge and hydrophobicity of amino acids. The net charge represents the difference between the positive and negative amino groups at a physiological pH = 7.0, and the mean hydrophobicity is the sum of the individual hydrophobicity of each residue divided by the total number of residues. The Kyte-Doolittle scale was used to determine the hydrophobic propensities of amino acids in the protein sequence, and the following equation was used to calculate the disorder score (I):
In the above equation, <H> represents the mean hydrophobicity, i.e. the sum of hydrophobicity of all residues, and |<R>| is the absolute difference between positively and negatively charged residues. The protein sequences were inputted to the python script, which calculates the score for each residue in the sequence. It is noteworthy to mention that this algorithm assumes that different regions of a protein vary in their folding properties, so a sliding window scores specific regions of proteins rather than the whole protein. Note, the length of the sliding window was 51 aa, as used in the original study.
Domain-wise disorder prediction of mucins
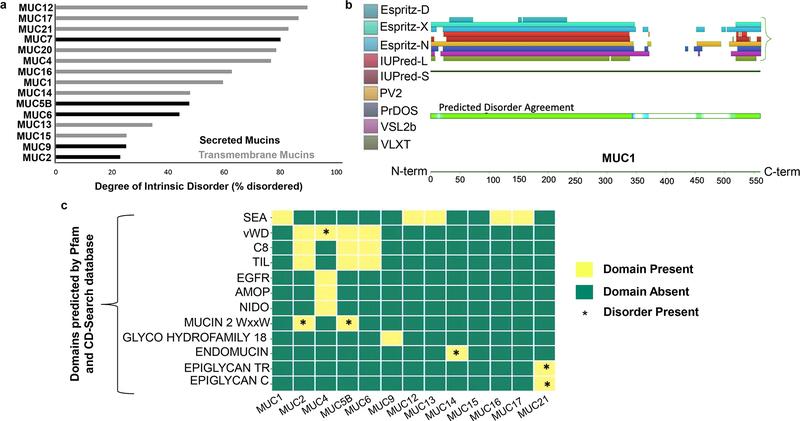
Protein domains were predicted on mucin sequences using the open-access Pfam (version 32.0 produced at the European Bioinformatics Institute, September 2018) (https://pfam.xfam.org/) and CD-Search (57) (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) interfaces. Only those with significant E-values (a parameter denoting significance of the actual number of sequences aligned compared to the number expected by chance) were utilized. Domain coordinates for each mucin were compared with predicted disordered regions (D2P2). Figure 1c represents the presence or absence of these domains across mucins (x-axis) and their corresponding disorder (represented by *) across mucins (y-axis).
Fig. 1: Intrinsic disorder across mucins determined using the Database of Disordered Protein Predictions (D2P2).
D2P2 is a database of pre-computed disorder predictions on a large library of proteins from completely sequenced genomes. a. Bar graph displaying the percentage of intrinsic disorder across transmembrane (grey) and secreted (black) mucins. All mucins analyzed were either highly disordered (defined as >40% disorder) or moderately disordered (>20% and <40% disorder). The disorder was calculated by dividing the total number of 75% consensus disordered residues, by the total mucin length to obtain a percentage disorder for each mucin. b. Pictorial representation of intrinsic disorder observed in MUC1 (length=550 amino acids) with a truncated tandem repeat region, as available within D2P2. The portion of the protein sequence with a high degree of consensus between tools (at least 6 of 9) is demarcated by green coloring. The regions with a lower degree of consensus (3–5 of 9 tools) but still predicted as disordered, are demarcated by shades of blue (darker blue denotes higher consensus). Both the N-terminus and C-terminus contain disordered sequences, as does much of the extracellular domain. c. Intrinsic disorder observed within different mucin domains as predicted by Pfam and CD-search databases. The presence of intrinsic disorder is denoted by (*). D2P2 data analyses suggested that disorder is present within the vWD domain of MUC4, WxxW domain of MUC2 and MUC5B, endomucin domain of MUC14, and the Epiglycan TR and C domains of MUC21
Disorder prediction in the cytoplasmic tail and transmembrane domains of mucins
Literature searches provided the starting point for the cytoplasmic tail (CT) sequences of mucins (3). Furthermore, the transmembrane (TM) sequences were obtained from Mucin database 2.0, 2015 (58) (http://www.medkem.gu.se/mucinbiology/databases/). Disorder predicted by D2P2 was compared within the transmembrane and cytoplasmic region to determine the specific residues disordered in these regions.
MoRFs prediction
The D2P2 database also identifies molecular recognition features (MoRFs) across proteins by using ANCHOR (59). This web server predicts disordered binding regions using protein sequences. The total number of MoRFs in each mucin was divided by its length. For instance, MUC12 has 145 predicted MoRFs and a length of 5478 aa, yielding a representative value of ~0.026. This assessment helped us identify the relative MoRFs per base pair in each of the mucins, enabling a relative assessment across mucins.
PhosphoSitePlus® curated phosphorylation site
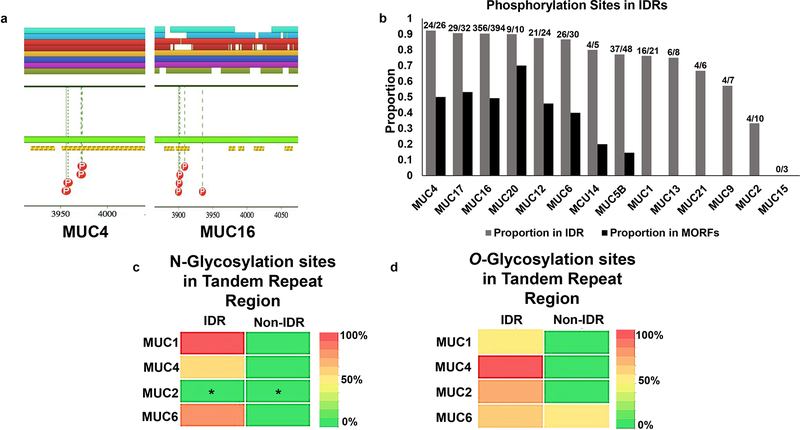
PhosphoSitePlus® is a database of mammalian post-translational modification sites curated from the scientific literature. Over 95% of the presented PTM sites have been elucidated by tandem mass spectrometry experimentation requiring a P <.05 for each site assignment (http://www.phosphosite.org/) (60). Phosphorylation sites curated by PhosphoSitePlus® were presented as a part of D2P2 analyses. These were subsequently aligned with the predicted IDRs and MoRFs for each mucin individually, and the proportion found within IDRs is presented in Figure 4a.
Fig. 4: Association of Post Translational Modifications, namely phosphorylation, and glycosylation, with IDRs and MoRFs in mucins.
Disordered regions are shown to be amenable to various forms of post-translational modification such as phosphorylation and glycosylation. We assessed entire mucin sequences to determine the proportion of phosphorylation sites within IDRs (and MoRFs). VNTR regions were further assessed for the overlap of predicted glycosylation sites with IDRs. a. Pictorial representation of phosphorylation sites found in D2P2 along with the IDR and MoRF predictions. MoRFs observed in transmembrane mucin MUC4 and MUC16 determined using ANCHOR as a part of D2P2. The yellow and black bar represents IDR-associated MoRFs predicted by ANCHOR (a tool that determines sequence motifs within an IDR that have a decrease in free energy upon binding with another protein). Curated phosphorylation sites (PhosphoSitePlus®) are displayed by red dots with “P” inside. Of those pictured, all reside within IDRs and some are found within MoRF sequences. b. Bar graph showing the proportion of (PhosphoSitePlus®) curated phosphorylation sites found in D2P2 that are inside regions of predicted disorder (grey bars) as well as in regions predicted as MoRFs (black bars). The numbers on top of each grey bar are the actual number of sites found in IDRs out of the total number of phosphorylation sites assessed. c. Heatmap representing percentage occurrence of predicted N-glycosylation sites within IDR and Non-IDR across the VNTR domain of representative transmembrane mucins, MUC1 and MUC4, and representative secreted mucins, MUC2 and MUC6. The analysis was conducted by NetNGlyc 1.0 server. N-glycosylation occurs almost exclusively in IDRs as compared to non-IDR regions within the tandem repeat domain for MUC1, MUC4, and MUC6. No prediction was made for MUC2 (represented by *) due to lack of Asparagines (Asn) in the input tandem repeat domain of MUC2, required for NetNGlyc prediction. d. Heatmap representing percentage occurrence of O-glycosylation sites within IDR and Non-IDR across tandem repeat domain. Representative transmembrane mucins, MUC1 and MUC4, representative secreted mucin, MUC2 and MUC6 were analyzed using NetOGlyc 4.0 server. O-glycosylation occurs almost exclusively in IDRs compared to Non-IDR regions within the tandem repeat domains of mucins. Note: A cutoff of > 0.5, (with a value of 0.0 being least likely and 1.0 most likely amenable to glycosylation) for a site prediction was considered as a potential glycosylation site by both servers. The amino acid sequences and the residue-by-residue disorder prediction that were utilized for N- and O- glycosylation analyses can be found in Supplementary Table 1.
Predicted O- and N-linked Glycosylation sites
N- and O-linked glycosylation sites were predicted for all mucin sequences using NetNGlyc 1.0 (61) server and NetOGlyc 4.0 (62) server, respectively. The NetNGlyc 1.0 server is an artificial neural network-based program that examines the Asn-Xaa-Ser/Thr sequons, AA sequence at which an N glycosylation can occur and predicts if a residue can act as a potential N glycosylation site with an accuracy of 77%. The NetOGlyc 4.0 server was derived by a ‘bottom-up’ ETD-based mass spectrometric analysis of 12 human cancer cell lines to develop a training set of the human O-glycoproteome. Mucin sequences from D2P2 were queried into these tools and potential N- and O-glycosylation sites were predicted. These predicted sites were then compared with the pre-computed disordered regions across mucins. We performed these predictions for two transmembrane mucins: MUC1 and MUC4 and two secretory mucins: MUC2 and MUC6. A threshold of 0.5 was used to predict a potential N- and O-glycosylation site within the tandem repeat domains of these mucins and individually analyzed the ability of these domains to serve as potential sites for N- and O-glycosylation post-translational modifications.
PONDR-VSL2
To further analyze the disorder regions in mucins and assess their inter-species pattern, the PONDR (63) based tool VSL2 was used. Mucin sequences from D2P2 were loaded into the online tool and the graphical representation was analyzed. With this method, an amino acid residue with a score close to 0 is considered to be ordered whereas the score approaching 1 is considered as disordered with a defined cutoff of ordered vs. disordered at a value of 0.5.
Mucin interactome and functional annotation of mucin interaction partners
All mucins that were assessed for intrinsic disorder (MUC1, MUC2, MUC3, MUC4, MUC5B, MUC6, MUC7, MUC9, MUC12, MUC13, MUC14, MUC15, MUC16, MUC17, MUC18, MUC19, MUC20, MUC21, and MUC22) were included for the Reactome functional analysis. We obtained 25 major mucin pathways and retained the pathway names along with the adjusted p-value. The protein-protein interaction information was extracted using the Biological General Repository for Interaction Datasets (BioGRID) Homo sapiens database (64). Duplicate edges emerging due to different validation methods were removed from the network to prevent redundancy. A total of 144 unique interactions between mucins and other proteins were obtained. BioGRID interactions are either peer-reviewed or experimentally validated by empirical protein-protein interaction methods. Further, we performed GO (65) analysis for mucins to illustrate the functional versatility and the implications in cancer-associated pathways. To further elucidate the pathways to which mucins contribute, we next used the FunSet webserver (66) to cluster and visualize the enriched GO pathway terms. This technique detects semantic similarity between terms and spectrally clusters them into neighborhoods in a bubble chart format.
RESULTS
INTRINSIC DISORDER ANALYSIS ACROSS MUCINS
A substantial portion of mucin sequences lacks meaningful structural annotation. In order to analyze the degree of disorder and their location across mucins, the percentage of disorder was calculated with 75% prediction consensus by nine disorder predictors present in D2P2 database (i.e. 7 out of 9 tools in agreement). Therefore, these meta-prediction tools provide more reliable predictions than a single disorder prediction tool (55).
The longest available protein sequence of mucins present within D2P2 was used in this analysis. The mucins analyzed included MUC1, MUC2, MUC4, MUC5B, MUC6, MUC7, MUC9, MUC12, MUC13, MUC14, MUC15, MUC16, MUC17, MUC20, and MUC21. Due to the unavailability of the sequences within D2P2, some mucins such as MUC3, MUC18, MUC19, and MUC22, were not included in the analysis. Also, MUC5AC (with an extremely short transcript available within the database), MUC8 (which is not a mucin protein although called MUC8 (58)), MUC10 (which is found only in mice) and MUC11 (which is part of MUC12 (58)) were all excluded from the analysis.
The disorder prediction analysis of mucin sequences showed that all mucins were moderately (20% - 40%) to highly (40%−90%) disordered. MUC12 (90%), MUC17 (87%), and MUC21 (83%) were the most disordered transmembrane mucins (Fig. 1a). Of the other transmembrane members, MUC20 (79%) and MUC4 (77%) were more disordered compared to MUC1 (60%) and MUC16 (63%). All transmembrane mucins were considered highly disordered with the exception of MUC13 (34%) and MUC15 (25%) which were moderately disordered. For secreted mucins, MUC7 (80%), MUC5B (48%), and MUC6 (44%) were highly disordered, whereas MUC9 (25%) and MUC2 (23%) were moderately disordered (Fig. 1b). We observed that disorder occurs more in transmembrane mucins compared to secreted mucins with the exception of MUC7 (Fig. 1a).
Next, we determined the domains for all mucins using Pfam (67) and the Conserved Domain Database (CDD)(68). These sequences were subsequently analyzed for the presence of IDRs. Domains determined using two databases allowed higher confidence predictions with a significant E-value (E). We observed that the SEA domain was correctly predicted across transmembrane mucins including MUC1, MUC12, MUC13, MUC16 and MUC17 as confirmed by the Mucin Biology Database (Fig. 1c). Similarly, vWD was predicted to be present in MUC2, MUC4, MUC5B, and MUC6 (Fig. 1c)
Combined analyses of the mucin domain sequence and disorder prediction identified the vWD domain of MUC4 to be disordered (Fig. 1c). In addition to the vWD domain, MUC4 also contains AMOP and NIDO domains (Fig. 1c), but the disorder predicted by D2P2 did not reach the 75% consensus pre-set cut-off. Three of the nine tools predicted sections of these domains to be disordered. The Mucin2_WxxW (a.k.a. CysD) domain known to contain a conserved repeat sequence motif (WxxW) of at least six conserved cysteine residues, also displayed a high level (>40%) of disorder; CysD was also predicted to be present in MUC2 and MUC5B. The Endomucin domain in MUC14, a highly O-glycosylated region that affects cell adhesion, was predicted to be disordered as well (Fig. 1c). MUC21 contained repeating motifs, represented by epiglycan TR and epiglycan C, which were also found to contain disorder (Fig. 1c).
INTRINSIC DISORDER IN TRANS-MEMBRANE AND INTRACELLULAR C-TERMINAL DOMAINS OF MUCINS
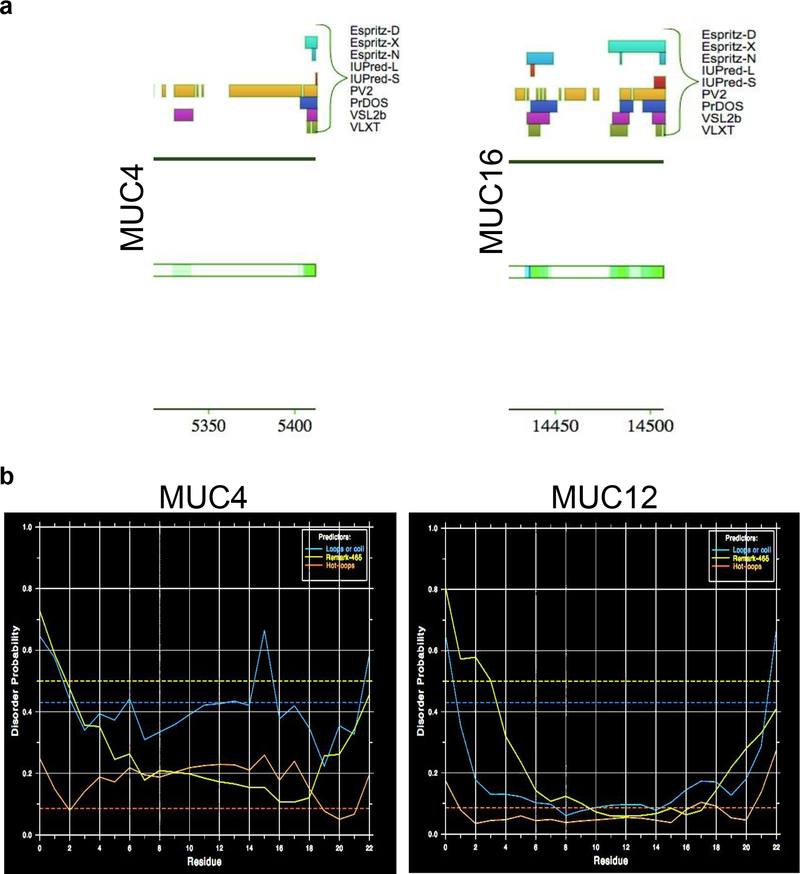
Next, we used D2P2 to predict disorder in the transmembrane and C-terminal domains of mucins. The cytoplasmic tail protein sequences of MUC1, MUC4, MUC12, MUC13, MUC15, MUC16, MUC17 and MUC20 (Table 1) were obtained from earlier published findings (3). Though the majority of the cytoplasmic tail sequences of MUC4 and MUC16 did not reach the pre-set 75% disorder consensus cutoff, a high level of agreement between tools, 6 out of 9, found them to be disordered (66%) (Fig. 2a).
Table 1.
Residues within the cytoplasmic domains of membrane-tethered mucin predicted to be disordered
| MUCIN | TRANSMEMBRANE DOMAIN WITH MARKED DISODER |
|---|---|
| MUC1 | RRKNYGQLDIFPARDTYHPMSEYPTYHTHGRYVPPSSTDRSPYEKVSAGNGGSSLSYTNPAVAATSANL |
| MUC4 | GCSGARFSYFLNSAEALP |
| MUC12 | SQRKRHREQYDVPQEWRKEGTPGIFQKTAIWEDQNLESRFGLENAYNNFRPTLETVDSGTEKHIQRPEMVASTV |
| MUC13 | TARSNNKTKHIEEENLIDEDFQNLKLRSTGFTNLGAEGSVFPKVRITASRDSQMQNPYSRHSSMPRPDYDIPPLRTSV |
| MUC15 | CGKAKTDSFSHRRLYDDRNEPVLRLDNAPEPYDVSFGNSSYYNPTLNDSAMPESEENARDGIPMDDIPPLRTSV |
| MUC16 | VTTRRRKKEGEYNVQQQCPGYYQSHLDLEDLQ |
| MUC17 | RSKREVKRQKYRLSQLYKWQEEDSGPAPGTFQNIGFDICQDDSIHLESIYSNFQPSLRHIDPETKIRIQRPQVMTTSF |
| MUC22 | RNSLSLRNTFNTAVYHPHGLNHGLGPGGNHGAPHRPRWSPNWFWRRPVSSIAMEMSGRNSGP |
Intrinsically disordered residues (bold/underlined) observed within cytoplasmic tails of MUC1, MUC12, MUC13, MUC15, and MUC20 with a 75% consensus of D2P2. The entirety of the MUC16 and MUC4 CT domains are predicted to be disordered by 6 of 9 tools reaching 66% consensus.
Fig. 2: Assessment of intrinsic disorder in cytoplasmic and transmembrane domains of membrane-tethered mucins.
Considering the differential sequence attributes for transmembrane (hydrophobic and lacking protein-protein interacting sites) and cytoplasmic domains (sites for purported signaling functions of mucins), we assessed intrinsic disorder regions across these domains. a.. Pictorial representation from the D2P2 disorder predicted in the cytoplasmic tails of MUC4 and MUC16. b. Representative figure of MUC4 and MUC12 transmembrane domain displaying extremely low disorder probability as determined by DisEMBL. Disorder probability increases as values approach 1 and decreases as it approaches 0. The greenish-yellow curve is the disorder prediction for missing residues (those lacking crystal structure), the red curve indicates residues predicted as hot loops, and the blue curve indicates those predicted as coils. Of note, loops and coils are considered necessary but not sufficient for disorder and the lack of these features as predicted by DisEMBL is indicative of a low level of disorder
Similarly, we obtained transmembrane domain sequences from the Mucin Biology Database (Human) and assessed if those transmembrane sequences were disordered in the global disorder prediction of mucins by D2P2. No disorder was observed within the transmembrane domains of mucins (Table 2). It is established that disorder prediction consensus approaches are generally more accurate (69), however, to verify our observation, we analyzed the transmembrane sequences with DisEMBL for further confirmation of our predictions. Similarly to D2P2, no disorder was observed for the transmembrane domain of mucins with DisEMBL. Representative figures for MUC4 and MUC12 show low disorder probability within the transmembrane domains (Fig. 2b). This is in line with the fact that transmembrane regions are largely hydrophobic static regions and are not involved in dynamic protein-protein interactions, thus decreasing the probability of disorder.
Table 2.
Residues within the transmembrane domains of membrane-tethered mucins predicted to be disordered.
| MUCIN | TRANSMEMBRANE DOMAINS OF MUCINS |
|---|---|
| MUC1 | WGIALLVLVCVLVALAIVYLIAL |
| MUC4 | IFFGALGGLLLLGVGTFVVLRFW |
| MUC12 | GIVGAVMAVLLLALIILIILTMFSL |
| MUC13 | LILTIVGTIAGIVILSMIIALIV |
| MUC14 | LPVVIALIVITLSVFVLVTMGLY |
| MUC15 | IVFGAILGAILGVSLLTLVGYLL |
| MUC16 | VILIGLAGLLGLITCLICGVLTMVT |
| MUC17 | YGLVGAGVVLMLIILVALLMLVF |
| MUC22 | WAIILISLAAVVAAVGLSVGTML |
No residues within the transmembrane region of any mucin were predicted to be disordered.
ASSESSMENT OF MOLECULAR RECOGNITION FEATURES (MORFS) IN MUCIN IDRS
Disorder-to-order transition of IDRs is facilitated by stretches of amino acids known as MoRFs, which facilitate molecular recognition and signal transduction (70). MoRFs undergo a conformational change to a lower energy state when interacting with an appropriate binding partner and are thus one of the keys to the executioner function of IDRs (70).
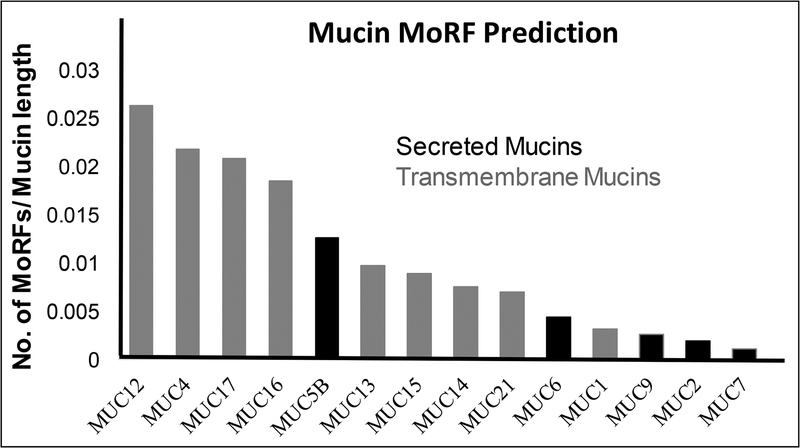
The presence of MoRFs was determined via ANCHOR, as a component of D2P2 that determines sequence motifs within an IDR that have a decrease in free energy upon binding with another protein (71). Interestingly, mucins contain a large number of predicted MoRFs within their IDRs (Table 3). Transmembrane mucins, particularly MUC12 and MUC16, were predicted to contain greater numbers of MoRFs within their IDRs compared to other mucins (145 and 413, respectively, Table 3). Further analysis showed that mucins implicated in multiple human cancers particularly MUC4, MUC17, and MUC16 contain a large number of MoRFs >30 residues (38, 46, and 46, respectively, Table 3). When the number of MoRFs are normalized to mucin protein length by dividing with the total number of residues in each mucin, MUC12 and MUC4 have the greatest number of MoRFs, at a ratio of 0.026 and 0.022, respectively (Fig. 3). Within secreted mucin members, MUC5B has the highest total number of MoRFs with 72, as well as the highest normalized quantity at 0.013 (Fig. 3).
Table 3.
Number and size of MoRFs observed in transmembrane and secreted mucins.
| MUCIN | TOTAL NO. OF MoRFs | TOTAL NO. OF MoRFs > 30 RESIDUES | LENGTH OF LONGEST MoRFS | |
|---|---|---|---|---|
| MEMBRANE BOUND MUCIN(S) | ||||
| MUC1 | 4 | 1 | 214 | |
| MUC4 | 116 | 38 | 7 | |
| MUC12 | 145 | 28 | 133 | |
| MUC13 | 5 | 0 | 28 | |
| MUC14 | 2 | 0 | 16 | |
| MUC15 | 3 | 0 | 10 | |
| MUC16 | 413 | 46 | 64 | |
| MUC17 | 94 | 46 | 28 | |
| MUC21 | 4 | 0 | 11 | |
| SECRETED MUCINS | ||||
| MUC2 | 10 | 0 | 27 | |
| MUC5B | 72 | 9 | 54 | |
| MUC6 | 25 | 5 | 82 | |
| MUC7 | 6 | 3 | 70 | |
| MUC9 | 7 | 3 | 62 | |
Membrane-bound mucins harbor more total MoRFs and many of greater length than secreted mucins, conferring greater probability of influencing interaction properties and binding partners.
Fig. 3. Prediction of Molecular Recognition Features (MoRFs) within Intrinsically Disordered Regions (IDRs) in mucins.
MoRFs are disorder-to-order (order upon binding) recognition motifs that influence and participate in protein-protein interactions (PPIs). Considering this, determining the prevalence and location of such motifs would improve our understanding of mucin function and interaction. We found mucins contain many large-sized MoRFs that have a greater propensity to affect PPIs. The bar graph shows the number of MoRFs normalized to mucin length by dividing MoRFs with the number of residues for each mucin (available lengths in D2P2). Interestingly, MUC4 and MUC16, two transmembrane mucins that are differentially expressed in multiple malignancies, have a high MoRF/length ratio.
DELINEATING THE ASSOCIATION OF MUCIN IDRS & PTMS
Many post-translational modifications (PTMs) that modulate protein actions and interactions are predicted within disordered regions of mucins and, more specifically, some reside within IDRs that harbor MoRFs (Fig. 4a, yellow and black bars are predicted as MoRFs). The predicted presence of smaller MoRFs within IDRs provides structural insight into the function of each mucin. Further investigation in this regard would help to associate the presence of disorder and MoRFs with domain and inter-domain functionality.
IDRs within structured protein domains are preferred and accessible sites for a variety of PTMs, including glycosylation and phosphorylation (72). With this in mind, we compared the phosphorylation sites to the regions of disorder for each mucin. For this, we utilized curated phosphorylation sites provided by PhosphoSitePlus® as a part of D2P2 as well as direct inquiry via PhosphoSitePlus® and subsequent sequence alignment. We found that the majority of phosphorylation sites were found within the predicted disordered regions when assessing entire mucin sequences (Fig. 4b). The proportion of phosphorylation sites found within IDRs for each mucin are as follows: MUC15 – 1.0, MUC4 – 1.0, MUC14 – 1.0, MUC20 – 1.0, MUC12 – 0.97, MUC17 – 0.91, MUC6 – 0.89, MUC16 – 0.86, MUC5B - 0.76, MUC13 – 0.75, MUC21 – 0.71, MUC9 – 0.5, MUC9 – 0.5, MUC2 – 0.44, and MUC1 – 0.36 (Fig. 4b). MUC7 did not have any curated phosphorylation sites presented within D2P2 nor with direct inquiry via PhosphoSitePlus®. The proportions for phosphorylation sites found to reside in MoRFs within IDRs are as follows: MUC15 – 0.0, MUC4 – 1.0, MUC14 – 0.2, MUC20 – 0.57, MUC12 – 0.42, MUC17 – 0.59, MUC6 – 0.38, MUC16 – 0.49, MUC5B - 0.14, MUC13 – 0.0, MUC21 – 0.0, MUC9 – 0.0, MUC2 – 0.0, and MUC1 – 0.0 (Fig. 4b).
We then specifically analyzed the tandem repeat regions of mucins to correlate N- and O- glycosylation, and high level of disorder predicted by D2P2. The predicted N- and O- glycosylation sites reside almost exclusively in IDRs compared to the ordered regions within tandem repeat regions (representative transmembrane mucins, MUC1 & MUC4, representative secreted mucin, MUC6, Fig. 4c and d). No prediction was made for MUC2 tandem repeat sequence by the NetNGlyc tool due to the absence of asparagine residues in the input sequence, as indicated by (*) in Fig. 4c. The amino acid sequences and the residue-by-residue disorder prediction that were utilized for N- and O- glycosylation analyses can be found in Supplementary Table 1.
ASSESSMENT OF IDR CONSERVATION ACROSS MUCINS
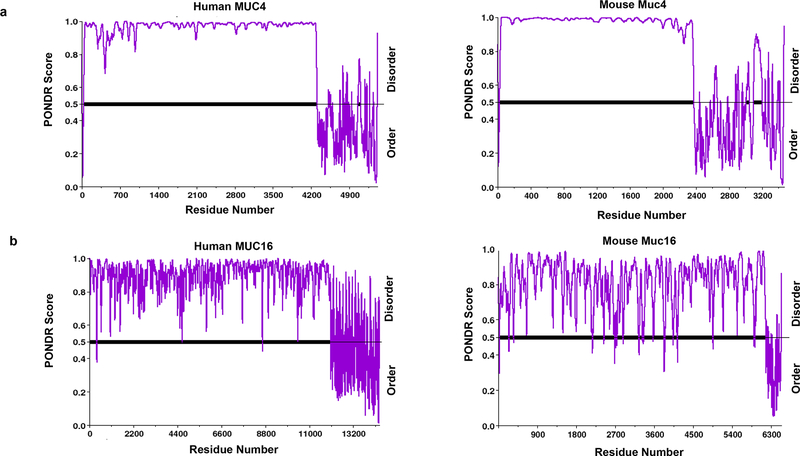
To evaluate the potential functional significance of IDRs in mucins, we examined the evolutionarily conserved regions between mouse and human. Interestingly, MUC4 and MUC16, each with important implications in oncogenic development, were found to have similar patterns of disorder across human and mouse (Fig. 5a and b). N-terminal residues in the protein sequences of MUC4 and MUC16, in both human and mouse, have a consistently high degree of disorder, while the C-terminal residues fluctuated between order and disorder.
Fig. 5.
Intrinsic disorder patterns in human and mouse MUC4 and MUC16. Evolutionary conservation of protein structure/sequence highlights the preservation of necessary biological functions. Though the actual interspecies sequence homology for both of these proteins is minimal, we speculated that the pattern of disorder may be conserved. We analyzed the IDR homology between mouse and human MUC4 and MUC16. Predictors of Natural Disordered Regions (PONDR) is an online compilation of five artificial intelligence tools that utilize previously defined structures for predicting intrinsic disorder. Due to the lack of mice full-length sequences in D2P2, PONDR was chosen for an inter-species comparison of disorder. Mice and human sequences (longest transcripts) were assessed with VSL2, a tool within PONDR, which makes a length-dependent prediction of protein intrinsic disorder to facilitate inter-species comparisons. a. Disorder prediction in human MUC4 and mouse MUC4 by PONDR VSL2 (a tool that predicts disorder and addresses protein length bias). b. PONDR VSL2 disorder prediction across human MUC16 and mouse MUC16. Residue values above 0.5 are predicted to be disordered. Both mucins displayed a significantly high degree of interspecies IDR pattern conservation
MUCIN INTERACTOMES
Disorder allows for rapid on/off binding kinetics with other proteins because of high specificity yet low affinity for their partners, frequently observed in hub and signaling proteins (20, 28–31). Considering this attribute of IDRs, we asked if mucins with high predicted disorder can interact with multiple partners or occupy hub positions. Using BioGRID, interacting partners of MUC1, MUC2, MUC3A, MUC5B, MUC7, MUC9 (OVGP1), MUC12, MUC13, MUC14, MUC15, MUC16, and MUC20 were retrieved. No interaction partners were found for MUC3B, MUC4, MUC6, MUC8, MUC10, MUC17, MUC19, MUC21 and MUC22. Interaction partners for MUC4 and MUC17 were identified from a literature search.
We next associated the number of interacting partners of mucins with the percentage of disorder present within the mucins. We observed that most transmembrane mucins including MUC1, MUC16, MUC13, and MUC20, which tend to have more IDRs and the longest MoRFs, had a higher number of interacting partners (Fig. 6a and Supplementary Fig. 1a). MUC20, which is highly disordered at 79%, has interactions with 24 other proteins involved in various pathways. Similarly, MUC16 (63% disorder) has 10 interacting partners (Fig. 6a and Supplementary Fig. 1a) and 46 MoRFs (Table 3). However, transmembrane mucin MUC12, the most disordered protein in our analysis at 89%, has only two interacting partners. This is likely due to few studies on MUC12, and a dearth of knowledge regarding its interactome. (Fig. 6a and Supplementary Fig. 1a).
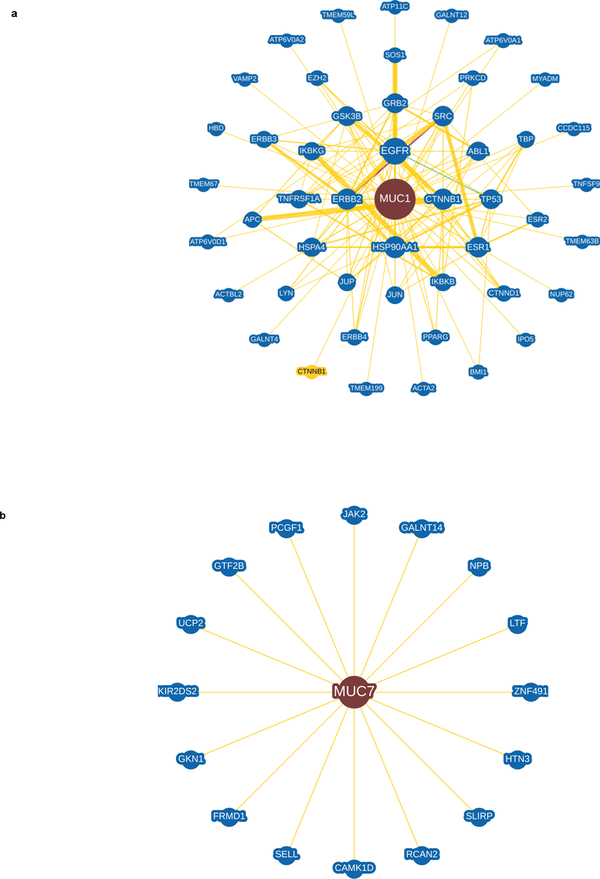
Fig. 6. Transmembrane mucin MUC1 and secretory mucin MUC7 interacting partners determined using the BioGRID database.
To assess if the quantity and prevalence of intrinsic disorder affect mucin interactomes, we utilized BioGrid, an online repository of protein chemical and genetic interactions. Physical interactions of all mucins were assessed using BioGrid to identify the interactions and the functional implications thereof. a. The network of the physical interactions of representative transmembrane mucin MUC1. b. The network of the physical interactions of representative secreted mucin, MUC7. Each Mucin is in the center of each interactome. The edge thickness connecting the mucin with its partners is linked to the number of times the interaction has been experimentally verified. Interactions with proteins not observed in humans, but in other organisms, are indicated by a yellow node. All other mucins with interactomes can be found in Supplementary Fig. 1
For secreted mucins, the most disordered family members, MUC7 (80%) and MUC5B (48%), had a greater number of interactions when compared to the other secreted mucins with lower levels of predicted disorder, MUC9 (25%) and MUC2 (23%) (Fig. 6a and Supplementary Fig. 1b). Unfortunately, in the case of other membrane-bound mucins, due to lack of information on their interactome, it was difficult to discern an accurate overall representation.
FUNCTIONAL DIVERSITY OF MUCINS AND THEIR INTERACTOME
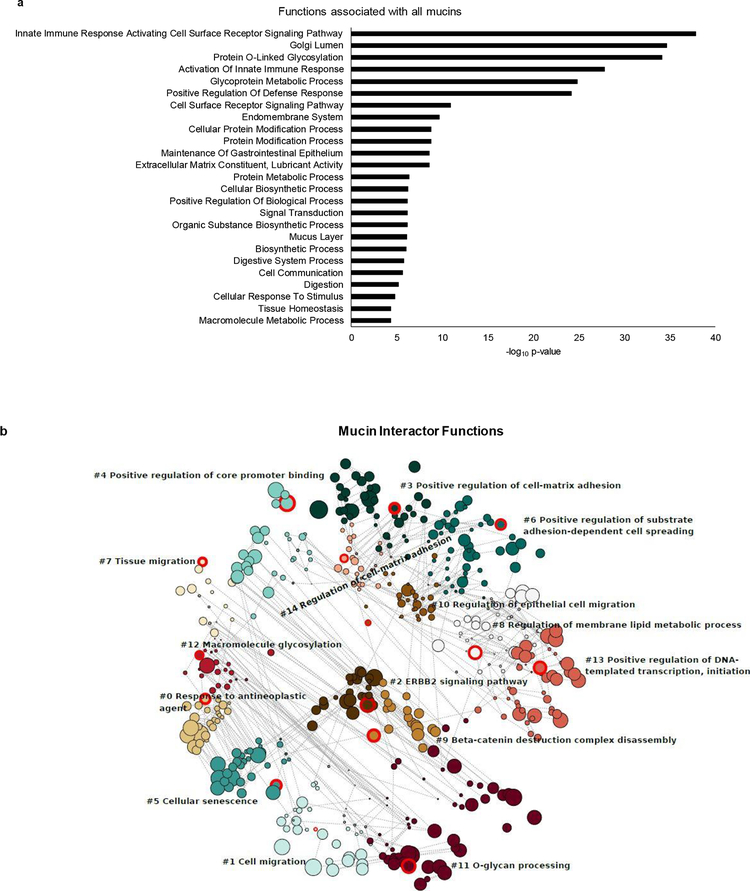
We next explored the functional significance of the mucin interactome. Reactome pathway analysis of the entire mucin family revealed significant involvement in a variety of important mechanisms including immune function, protein metabolism and signal transduction (Fig. 7a). Additionally, the mucin interacting partners generated from the BioGRID database were subsequently analyzed for their contribution to functional pathways. These interactome members are involved in a variety of functions associated with cancer including response to antineoplastic agents, cell migration, ERBB2 signaling, cell adhesion, and protein glycosylation (Fig. 7b). These pathways are directly involved in many aspects of oncogenesis, invasion, metastasis, and response to treatment. As mentioned, mucins have been shown to impact these cancer-associated pathways, further corroborating our findings.
Fig. 7. Functional annotation of mucins and their interacting partners.
To further assess the functional implications of the previously identified interacting partners a gene ontology (GO) based functional enrichment analysis was carried out. GO enrichment analyses are used to assess the functional implications of a gene or set of genes. The mucins and the interacting partners were compared to the GO database to assess the enriched terms and the enriched pathways assessed more closely. a. Graphical representation of GO Reactome pathway analysis of the mucin family (false discovery rate (FDR) < 0.05). The bar length is indicative of -log10 p-value. b. A bubble plot depicting the functions of the mucin interacting partners grouped into 15 neighborhoods. Each circle represents a GO pathway term to which mucin interacting partners contribute. The size of each circle is representative of the enrichment score of each pathway. Each numbered circle cluster (0–14) is demarcated into neighborhoods by color. Within each neighborhood is a circle highlighted in red, labeled with a pathway term. This shows mucin partners are involved in a variety of functions associated with cancer including response to antineoplastic agents, cell migration, ERBB2 signaling, cell adhesion, and protein glycosylation
DISCUSSION
The hypothesis was supported by the predicted prevalence of intrinsic disorder across the entire mucin family. The presence of IDRs within the functional domains, between domains, extracellular, and cytoplasmic tail regions were analyzed. We observed that all mucins were predicted to have high (>40%) to moderate levels (>20% and <40%) of disorder regions. Indeed, 11 out of 15 of the mucins assessed were >40% disordered. Transmembrane mucins were more disordered compared to secreted mucins with an exception of MUC7. The average predicted disorder across all assessed mucins (58%) far exceeds the average of what is present throughout the human (30%) and eukaryotic (32%) proteomes (25–27, 73). In fact, this would place the mucin family in the top 10–15% most disordered proteins found in the human Ensemble database analyzed by D2P2 (25) with the majority of individual mucins harboring a far greater amount of disorder.
Apart from the nine predictors included in D2P2, we assessed mucin disorder with FoldIndex and PONDR CH plots, which are tools based on the dual assumption that IDPs/IDRs are generally enriched in polar and charged residues and depleted in the hydrophobic regions of proteins (17, 56). The mucin sequences used for the disorder analysis in D2P2 were used for FoldIndex disorder. This method predicted mucins to be far more ordered as compared to the D2P2 consensus results (Supplemental Fig. 2). Confirmation with PONDR CH plots corroborates the FoldIndex findings and predicts native folding considering sequence charge and net hydrophobicity (Supplemental Fig. 2a & b).
Though these findings seem incongruous with the other prediction methods, we postulate that charge hydropathy is not the best method to predict disorder in mucins for two reasons. First, FoldIndex has been shown to be the least accurate predictor of transmembrane protein disorder (74), and secondly, high number, variability, and degeneration of the tandem repeat sequences present in mucins are not accurately characterized leading to falsely low disorder prediction.
Correlations between the organization and evolution of chromosomes and chromosomal gene congregation with the extent of disorder have previously been evaluated. A study by Rajagopalan et al. found that cancer/testis antigens, a family of proteins often aberrantly expressed in cancer, are highly disordered (46). Further, they found that CTAs located on the X-chromosome (CT-Xs) displayed the largest extent of disorder compared to the family members located on other chromosomes (46). To help bolster the disorder prediction for mucins, we assessed if any correlation existed between their degree of disorder and chromosomal location. Interestingly, MUC4 and MUC20 with a similar percentage of disorder are located at the same 3q29 locus. Also, MUC12 and MUC17, which are predicted as the most disordered transmembrane mucins, cluster at 7q22 locus. Among secreted mucins, MUC5B and MUC6 predicted as almost equally disordered, are located at 11p15.5 locus.
Conserved regions of disorder contribute to myriad biological activities (75). These activities have been categorized into six functional classes by Tompa et al (76) and highlighted in comprehensive review articles on disordered proteins by the Uversky and Babu groups (17, 25). Entropic chain classifiers (does not acquire ordered confirmation for their functioning) is the first class where IDRs can act as flexible inter-domain linkers and spacers necessary for appropriate functional-domain activity Effectors, where IDR act to modulate (inhibit or activate) interaction partner activity. A third class is assembler functioning when IDRs facilitate and provide scaffolding for large multi-protein complexes including signaling complexes. They can also have scavenger functions, where IDPs/IDRs interact with small ligands and capture, neutralize, or store them for later release. In chaperone class disordered proteins facilitate the folding of various molecules into their functional conformation. Finally, IDRs also harbor display site functions, where they provide conformational flexibility allowing PTM enzymes access to the protein backbone, thus facilitating their action including phosphorylation and glycosylation. Based on our data, we speculate that many of these functional classifications of IDRs will hold true for IDR present within mucins.
Many of these attributed IDR functions overlap with mucin activities. For example, MUC15 is moderately disordered (25%). MUC15/EGFR interaction is shown to diminish the aggressiveness of hepatocellular carcinoma by preventing EGFR dimerization (77). EGFR dimerization promotes the loss of intrinsic disorder in its kinase domain (disorder-to-order transition) leading to an increase in kinase signaling activity and the presence of an L834R mutation facilitates this dimerization by suppression of disorder within the kinase domain (48). MUC15/EGFR interaction and subsequent inhibition of EGFR dimerization may be facilitated by an effector function of these disordered sequences present in both proteins or even MUC15 prevention of EGFR kinase domain loss of disorder. Another mucin with effector type functioning is MUC4. It has been shown that MUC4 interacts and stabilizes the receptor tyrosine kinase HER2 in the setting of pancreatic cancer, thus promoting cell proliferation (78). Interestingly, our analysis showed that MUC4 is over 76% disordered. Given the impact EGFR and HER2 signaling have in many cancers, the precise mechanism of these interactions, their effect on disorder, and the respective inhibition or activation of kinase capability warrants further study.
Other functional contributions of mucin IDRs could exist. For example, salivary proline-rich glycoproteins (much like mucins) contain high levels of IDRs and have been shown to have scavenger functions and bind small ligands such as ions and organic compounds either for disposal, sequestration, or later release (79) Congruently, our results show salivary MUC7 contains the highest percentage of disorder among all secreted mucins, at 80% predicted disorder. With this, many IDPs, or ordered proteins rich in IDRs, can form proteinaceous membrane‐less organelles (PMLOs) via a liquid-liquid phase separation (80). Given the prevalence of intrinsic disorder in mucins, it is conceivable that mucins could contribute to PMLO formation and scavenger functioning within the local tumor milieu trapping nutrients, growth factors, and various other cytokines, thus forming a synergistic oncogenic environment. Another possible function of mucin IDRs arises in light of the observation that viruses with a greater number of IDRs in their coat proteins are able to evade memory T-cells (81–83). Mucin polymerization and bulky glycosylation could also form an immunomodulating “glycoblanket” that shields cancer cells from detection and killing by leukocytes, thereby facilitating the unchecked growth of the disease. Along with this, our results show that IDRs are present in the cytoplasmic tail of transmembrane mucins as well as the extracellular region. Many oncogenic molecules involved in signaling are enriched in IDRs in their cytoplasmic tail (84, 85), thus, their existence in mucin CTs could impact mucin activity.
Due to the inherent ability to engage in promiscuous interactions, and the ability of rapid on/off binding, IDP/IDR ensembles are associated with dosage sensitivity. The higher the protein concentration, the larger the interaction pool. This, in turn, can lead to a dose-dependent non-specific response (86, 87). In conjunction, proteins with the most disorder are associated with hub positions in cancer-associated protein-protein interaction networks (19, 46). The combination of these two IDR aspects may explain why mucins can bind with and activate a great number of surface receptors (88), signaling molecules (3), and transcription factors (89, 90).
It is known that protein-protein interactions involving IDPs are influenced by molecular recognition features (MoRFs) (67). We found mucins contain higher numbers and many large-sized MoRFs, suggesting that they may participate in a variety of mucin interactions including the aforementioned effector activities of MUC15 and MUC4. The presence of MoRFs and their ability to undergo rapid binding events further insinuates mucins in a myriad of cellular functions including cell signaling as well as rheostat (on/off) functioning. For example, MUC16 has the highest number of MoRFs that are >30 residues in length (413 and 46, respectively). Consistently, MUC16 has multiple interaction partners (91–94). MUC1 has the highest number of interaction partners as it is the best characterized of all mucins and the most studied. While MUC1 does not contain a large number of MoRFs, it contains the largest found in all mucins, at 214 residues in length.
MUC12 was predicted to have a large number of MoRFs with 145 but has only two interacting partners (Supplementary Fig. 1a). This could be due to the fact that few studies have characterized MUC12 interactions. However, both of the MUC12 interaction partners (MAPK14 and CDC42) are involved in MAPK signaling and cell division, indicating that MUC12 harbors certain motifs/IDRs that contribute to oncogenic signaling and may impact cancer progression.
Though IDRs have been implicated in PTMs for years, a concept of an IDR-PTM-AS (alternative splicing) toolkit has recently been proposed (95). This toolkit allows for a single protein-coding gene to produce multiple disparate functional units, predicated in tissue or cell-specific manner. This also correlates with observed site-specific and context-dependent signaling of mucins under normal physiological as well as pathological conditions (e.g. MUC5AC deleterious effects in pancreatic cancer (96) and its protective role in the lung epithelium (97)). Specific tissues or cell types are able to rewire/remodel protein pathways and gene expression patterns (via transcription factors) through changes in PTMs and alternative splicing, which are impacted by the presence, prevalence, and size of IDRs (95). Mucins exhibit a high level of alternative splicing thus warranting further investigation into the effect of disorder on splicing events and impact on PTMs.
In addition to splice events, numerous structural modifications of mucins drive protein-protein interactions which serve as a key to their oncogenic role in cancer progression (98). The flexibility of IDRs facilitates access to enzymes involved in PTM (19) and the ability of an IDR to interact with target proteins is dramatically altered by the presence of these PTMs (95). Our findings show a high degree of overlap between the PhosphoSitePlus® curated phosphorylation sites found in D2P2 and IDRs, throughout the mucin protein family. The proportions of phosphorylation sites residing in IDRs are extremely high (ranging from 1.0 to .86) for each mucin with the lone exception of MUC9 (.5). This finding corroborates the amenability of mucin IDRs to PTM and further underscores the importance of these regions in signaling and interaction dynamics due to phosphorylation events.
Another PTM assessed in conjunction with disorder was glycosylation, specifically within the VNTR region. We found that predicted mucin glycosylation sites within the VNTR, overlap markedly with IDRs. A specific signaling motif (e.g. Proline-Threonine-Serine PTS sequence) could be working in combination with disorder, to facilitate glycosylation of this region. Disordered regions lying outside of the tandem repeats may not harbor this signaling motif allowing a variety of other PPIs, thus conferring the aforementioned hub protein characteristics of mucins. Alteration in the amount of disorder present within the VNTR including expression, mutations, and repeat expansion, could augment susceptibility to enzymatic modification and dramatically affect the glycome, thus altering mucin interactomes. As aberrant glycosylation has long been a hallmark of cancer (9), understanding the amount and variety of disorder present within the system is incredibly important.
There is also overwhelming evidence that glycosylation is associated with protein stability (99). Contrarily, the presence and length of IDRs are negatively correlated with protein half-life, due to facile interaction with ubiquitin ligases (100). Thus, the balance between the presence of IDRs and their glycosylation status may act as a homeostatic mechanism to modulate mucin turnover and associated signaling pathways. Aberrant glycosylation of mucins, like what is observed in cancer, could alter their half-life and thus facilitate dose-dependent promiscuous binding and subsequent increases in pro-growth cellular signaling. Altogether, these observations indicate that IDRs may influence mucin protein-protein interactions as well as half-life. Future studies characterizing the complete mucin interactome and the mucin functional life-spans could enhance the associations reported here and further elucidate the relationship between IDRs, MoRFs, and PTMs.
Our assessments also show that IDR patterns are conserved between human and mouse, even though the number and order of tandem repeats within the PTS domain of mucins vary between species. We speculate that this IDR pattern conservation between human and mouse mucins is evidence that these regions preserve an important biological function despite underlying genetic variations and limited sequence homology. Additionally, IDRs may also explain the expansion of the repeat sequence in human mucins compared to mice. Their lack of structural rigidity and the conferred decrease in evolutionary constraint can allow for replication slippage. If mucin function is predicated not only on their structure but also their ability to undergo plastic and dynamic structural changes, understanding of mucin IDRs becomes incredibly valuable for the assessment of their biological functioning and oncogenic implications.
IDRs are excellent cancer therapeutic targets for a variety of unique reasons. Disordered proteins can be sensitive to modulation through a variety of methods and mechanisms of action. IDRs can be targeted directly via small molecules which can affect the affinity of the parent protein for binding partners, thus altering specific protein-protein interactions. Along with this, small molecule binding to IDRs can act through a variety of mechanisms including steric and/or allosteric hindrance, induced order upon binding, dimerization prevention, and conformation “locking” which decreases the dynamism of the protein. A specific unique advantage associated with IDRs is that since dynamism is key to their interactions, a small molecule that is able to diminish this dynamism, could have dramatic effects on the function throughout the entire region, regardless of where the binding occurs (i.e. not necessarily at the site of parent-partner interaction). Converse to this but equally as effective, small molecules or peptides can be used to target the IDR interactor proteins in (referred to as a “clamp”) and prevent the undesired PPI. Along with this, binding regions within IDRs can be predicted by utilizing MoRFs and computer-aided drug design to identify binding partners and subsequently substituted for small molecules allowing for a facile and high-throughput method of discovery (101).
IDR therapeutic relevance is corroborated by the fact that many oncogenic molecules involved in signaling are enriched in IDRs. Importantly, IDRs have been confirmed in many cancer-associated proteins including p53, BRACA1, PAGE4, and PTEN (82, 102–105) and successful attempts have been made to target these regions. For example, a small molecule inhibitor was used to lock the normally dynamic IDR in the MYC protein in a static conformation that was unable to bind MAX, thereby preventing its oncogenic signaling (106). In another study, an alpha-helix-stapled peptide was engineered to interact with an IDR in P53, preventing its activation and subsequent anti-apoptotic effects (107).
Mucins, like the aforementioned proteins, are cancer-associated molecules that have eluded traditional therapeutic modalities. The development of compounds to target mucins is still in its infancy partly because detailed structures of this family are unavailable. We hypothesize that IDRs could serve as novel drug target sites for mucins, but this requires a detailed elucidation of their location and functional contributions. For instance, MUC16 cleavage and shedding of the EC domain is a major barrier to efficient MUC16 targeting in cancers (108). Where antibody therapy has failed, the IDRs present within the remaining membrane-bound MUC16 could be utilized as a means of targeting cancer cells. Alternatively, MUC16 has a cleavage site within the cytoplasmic tail region (109), and the sequence distal to the cleavage is predicted as completely disordered (6 out of 9 tools (66% consensus) in D2P2). When the intracellular cleaved portion of MUC16 is released, it increases cell proliferation, prevents apoptosis and influences the transcription of oncogenic genes (109). A cleaved MUC16 IDP would present a valuable therapeutic target for disruption of these functions and further investigation is warranted.
Another mucin that holds therapeutic relevance is MUC1. Monoclonal antibody (Mab) intervention attempts have been of limited success for MUC1 (84). In one study, Raina et al. were unsuccessful in attempts to crystalized the MUC1 CT and subsequent structural analysis with ROBETTA (110) and IGB-SSPro (111) revealed no identifiable secondary structure. Given these results, they determined that the MUC1 CT has features characteristic of an IDP. Notably, despite a lack of structure, IDPs are emerging as attractive drug targets (112–117). Further investigation into these MUC1 disordered regions is warranted which could provide insight into their relevance to its oncogenic signaling. In turn, this opens up a new possibility for therapeutic intervention by providing new targets for small molecule inhibitors or stapled peptides that can bind to MUC1 IDRs and inhibit its oncogenic function.
As mentioned prior, many studies are warranted to validate these in-silico findings as well as accurate attribution of IDR functions in mucins. Experimental characterization methods including (but not limited to) nuclear magnetic resonance (NMR) spectroscopy (including in-cell NMR), small-angle X-ray scattering (SAXS), 20S proteasomal degradation, and single-molecule fluorescence resonance energy transfer (smFRET) are necessary to determine the accuracy of the D2P2 disorder prediction. These techniques will help to elucidate the overall degree of disorder, mucin structure and dynamics, and conformation changes upon partner interaction. In addition, experimental strategies to investigate the role of IDRs on mucin function are required as well. For example, selective mutations to residues that alter overall order in CT regions of MUC1 and MUC16 could be utilized to assess the effects of disorder on dimerization, proliferation, and/or oncogene transcription. Phage displays could also be utilized for the CT region of these mucins to determine what peptides bind and could be used as a therapeutic strategy. Another strategy to determine disorder effect on PPIs would be to use various isoforms of MUC4 (i.e. MUC4X, MiniMUC4, MUC4β, and WT MUC4) with different lengths of the tandem repeat regions (found to be highly disordered in our analysis) in pulldown assays or SPR based studies. Furthermore, disordered links between mucin domains could be deleted to determine if these are required for adequate domain functioning.
The prevalence of IDRs within mucins could have vast clinical potential. Though we have utilized multiple prediction tools to determine the level of disorder, however, these computational findings must be validated experimentally. These studies would provide validation of the predictions and hypotheses presented herein, and justify a new and alternative perspective when assessing mucin structure.
Supplementary Material
ACKNOWLEDGMENTS
The authors in this article were supported, in parts, by the following grants from the National Institutes of Health (R01 CA183459, RO1 CA195586, RO1 CA210637, UO1 CA200466, UO1 CA210240 and P01 CA217798, R44CA224619 and UO1 CA210240) and the Nebraska Department of Health and Human Services LB595. JC and PA are supported by the UNMC Graduate fellowships.
NON-STANDARD ABBREVIATIONS LIST
- IDR
intrinsically disordered region
- IDP
intrinsically disordered protein
- PTM
post-translational modification
- MoRF
molecular recognition feature
- PPI
protein-protein interaction
- TM
transmembrane
- VNTR
variable number of tandem repeat domain
- PTS
sequence, proline, threonine and serine sequence
- EGF
epidermal growth factor-like domain
- SEA
sea-urchin sperm protein enterokinase and agrin module
- vWD
von Willebrand factor D domain
- NIDO
nidogen-like domain
- AMOP
adhesion-associated domain in MUC4 and other proteins domain
- ECM
extracellular matrix
- D2P2
Database of Disordered Protein Predictions
- CH
charge hydropathy
Footnotes
CONFLICT OF INTEREST STATEMENT
SKB is one of the co-founders of Sanguine Diagnostics and Therapeutics, Inc. The other authors disclosed no potential conflicts of interest.
REFERENCES
- 1.Kaur S, Kumar S, Momi N, Sasson AR, Batra SK. Mucins in pancreatic cancer and its microenvironment. Nat Rev Gastroenterol Hepatol. 2013;10(10):607–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silverman HS, Parry S, Sutton-Smith M, Burdick MD, McDermott K, Reid CJ, et al. In vivo glycosylation of mucin tandem repeats. Glycobiology. 2001;11(6):459–71. [DOI] [PubMed] [Google Scholar]
- 3.van Putten JPM, Strijbis K. Transmembrane Mucins: Signaling Receptors at the Intersection of Inflammation and Cancer. J Innate Immun. 2017;9(3):281–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quin RJ, McGuckin MA. Phosphorylation of the cytoplasmic domain of the MUC1 mucin correlates with changes in cell-cell adhesion. Int J Cancer. 2000;87(4):499–506. [DOI] [PubMed] [Google Scholar]
- 5.Wesseling J, van der Valk SW, Vos HL, Sonnenberg A, Hilkens J. Episialin (MUC1) overexpression inhibits integrin-mediated cell adhesion to extracellular matrix components. J Cell Biol. 1995;129(1):255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bafna S, Kaur S, Batra SK. Membrane-bound mucins: the mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene. 2010;29(20):2893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pai P, Rachagani S, Dhawan P, Batra SK. Mucins and Wnt/beta-catenin signaling in gastrointestinal cancers: an unholy nexus. Carcinogenesis. 2016;37(3):223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaur S, Smith LM, Patel A, Menning M, Watley DC, Malik SS, et al. A Combination of MUC5AC and CA19–9 Improves the Diagnosis of Pancreatic Cancer: A Multicenter Study. Am J Gastroenterol. 2017;112(1):172–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan S, Chen R, Tamura Y, Crispin DA, Lai LA, May DH, et al. Quantitative glycoproteomics analysis reveals changes in N-glycosylation level associated with pancreatic ductal adenocarcinoma. J Proteome Res. 2014;13(3):1293–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishn SR, Kaur S, Smith LM, Johansson SL, Jain M, Patel A, et al. Mucins and associated glycan signatures in colon adenoma-carcinoma sequence: Prospective pathological implication(s) for early diagnosis of colon cancer. Cancer Lett. 2016;374(2):304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niv Y, Rokkas T. Mucin Expression in Colorectal Cancer (CRC): Systematic Review and Meta-Analysis. J Clin Gastroenterol. 2019;53(6):434–440. [DOI] [PubMed] [Google Scholar]
- 12.Lakshmanan I, Salfity S, Seshacharyulu P, Rachagani S, Thomas A, Das S, et al. MUC16 Regulates TSPYL5 for Lung Cancer Cell Growth and Chemoresistance by Suppressing p53. Clin Cancer Res. 2017;23(14):3906–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauer AK, Umer M, Richardson VL, Cumpian AM, Harder AQ, Khosravi N, et al. Requirement for MUC5AC in KRAS-dependent lung carcinogenesis. JCI Insight. 2018;3(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baert T, Van Camp J, Vanbrabant L, Busschaert P, Laenen A, Han S, et al. Influence of CA125, platelet count and neutrophil to lymphocyte ratio on the immune system of ovarian cancer patients. Gynecol Oncol. 2018;150(1):31–7. [DOI] [PubMed] [Google Scholar]
- 15.Singh AP, Senapati S, Ponnusamy MP, Jain M, Lele SM, Davis JS, et al. Clinical potential of mucins in diagnosis, prognosis, and therapy of ovarian cancer. Lancet Oncol. 2008;9(11):1076–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181(4096):223–30. [DOI] [PubMed] [Google Scholar]
- 17.Habchi J, Tompa P, Longhi S, Uversky VN. Introducing protein intrinsic disorder. Chem Rev. 2014;114(13):6561–88. [DOI] [PubMed] [Google Scholar]
- 18.Kjaergaard M, Kragelund BB. Functions of intrinsic disorder in transmembrane proteins. Cell Mol Life Sci. 2017;74(17):3205–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu G, Wu Z, Uversky VN, Kurgan L. Functional Analysis of Human Hub Proteins and Their Interactors Involved in the Intrinsic Disorder-Enriched Interactions. Int J Mol Sci. 2017;18(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgi J, Xue B, Uversky VN, van der Goot FG. Intrinsic Disorder in Transmembrane Proteins: Roles in Signaling and Topology Prediction. PLoS One. 2016;11(7):e0158594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boomsma W, Nielsen SV, Lindorff-Larsen K, Hartmann-Petersen R, Ellgaard L. Bioinformatics analysis identifies several intrinsically disordered human E3 ubiquitin-protein ligases. PeerJ. 2016;4:e1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larion M, Miller B, Bruschweiler R. Conformational heterogeneity and intrinsic disorder in enzyme regulation: Glucokinase as a case study. Intrinsically Disord Proteins. 2015;3(1):e1011008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uversky VN. What does it mean to be natively unfolded? Eur J Biochem. 2002;269(1):2–12. [DOI] [PubMed] [Google Scholar]
- 24.Lise S, Jones DT. Sequence patterns associated with disordered regions in proteins. Proteins. 2005;58(1):144–50. [DOI] [PubMed] [Google Scholar]
- 25.van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114(13):6589–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pentony MM, Jones DT. Modularity of intrinsic disorder in the human proteome. Proteins. 2010;78(1):212–21. [DOI] [PubMed] [Google Scholar]
- 27.Edwards YJ, Lobley AE, Pentony MM, Jones DT. Insights into the regulation of intrinsically disordered proteins in the human proteome by analyzing sequence and gene expression data. Genome Biol. 2009;10(5):R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41(21):6573–82. [DOI] [PubMed] [Google Scholar]
- 29.Shammas SL. Mechanistic roles of protein disorder within transcription. Curr Opin Struct Biol. 2017;42:155–61. [DOI] [PubMed] [Google Scholar]
- 30.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18(5):343–84. [DOI] [PubMed] [Google Scholar]
- 31.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16(1):18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Astro V, de Curtis I. Plasma membrane-associated platforms: dynamic scaffolds that organize membrane-associated events. Sci Signal. 2015;8(367):re1. [DOI] [PubMed] [Google Scholar]
- 33.Guharoy M, Szabo B, Contreras Martos S, Kosol S, Tompa P. Intrinsic structural disorder in cytoskeletal proteins. Cytoskeleton (Hoboken). 2013;70(10):550–71. [DOI] [PubMed] [Google Scholar]
- 34.Galea CA, Wang Y, Sivakolundu SG, Kriwacki RW. Regulation of cell division by intrinsically unstructured proteins: intrinsic flexibility, modularity, and signaling conduits. Biochemistry. 2008;47(29):7598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitrea DM, Yoon MK, Ou L, Kriwacki RW. Disorder-function relationships for the cell cycle regulatory proteins p21 and p27. Biol Chem. 2012;393(4):259–74. [DOI] [PubMed] [Google Scholar]
- 36.Yoon MK, Mitrea DM, Ou L, Kriwacki RW. Cell cycle regulation by the intrinsically disordered proteins p21 and p27. Biochem Soc Trans. 2012;40(5):981–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272(20):5129–48. [DOI] [PubMed] [Google Scholar]
- 38.Gsponer J, Babu MM. The rules of disorder or why disorder rules. Prog Biophys Mol Biol. 2009;99(2–3):94–103. [DOI] [PubMed] [Google Scholar]
- 39.Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, et al. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol. 2006;2(8):e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14–3-3 with their partners. BMC Genomics. 2008;9 Suppl 1:S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patil A, Kinoshita K, Nakamura H. Hub promiscuity in protein-protein interaction networks. Int J Mol Sci. 2010;11(4):1930–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue B, Oldfield CJ, Van YY, Dunker AK, Uversky VN. Protein intrinsic disorder and induced pluripotent stem cells. Mol Biosyst. 2012;8(1):134–50. [DOI] [PubMed] [Google Scholar]
- 43.Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30(2):137–49. [DOI] [PubMed] [Google Scholar]
- 44.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–46. [DOI] [PubMed] [Google Scholar]
- 45.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323(3):573–84. [DOI] [PubMed] [Google Scholar]
- 46.Rajagopalan K, Mooney SM, Parekh N, Getzenberg RH, Kulkarni P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J Cell Biochem. 2011;112(11):3256–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uversky VN, Dave V, Iakoucheva LM, Malaney P, Metallo SJ, Pathak RR, et al. Pathological unfoldomics of uncontrolled chaos: intrinsically disordered proteins and human diseases. Chem Rev. 2014;114(13):6844–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shan Y, Eastwood MP, Zhang X, Kim ET, Arkhipov A, Dror RO, et al. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell. 2012;149(4):860–70. [DOI] [PubMed] [Google Scholar]
- 49.Dong G, Chen W, Wang X, Yang X, Xu T, Wang P, et al. Small Molecule Inhibitors Simultaneously Targeting Cancer Metabolism and Epigenetics: Discovery of Novel Nicotinamide Phosphoribosyltransferase (NAMPT) and Histone Deacetylase (HDAC) Dual Inhibitors. J Med Chem. 2017;60(19):7965–83. [DOI] [PubMed] [Google Scholar]
- 50.Merino F, Raunser S. Electron Cryo-microscopy as a Tool for Structure-Based Drug Development. Angew Chem Int Ed Engl. 2017;56(11):2846–60. [DOI] [PubMed] [Google Scholar]
- 51.Goyal S, Jamal S, Shanker A, Grover A. Structural investigations of T854A mutation in EGFR and identification of novel inhibitors using structure activity relationships. BMC Genomics. 2015;16 Suppl 5:S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verkhivker GM. Computational Modeling of the Hsp90 Interactions with Cochaperones and Small-Molecule Inhibitors. Methods Mol Biol. 2018;1709:253–73. [DOI] [PubMed] [Google Scholar]
- 53.Pelaseyed T, Zach M, Petersson AC, Svensson F, Johansson DG, Hansson GC. Unfolding dynamics of the mucin SEA domain probed by force spectroscopy suggest that it acts as a cell-protective device. FEBS J. 2013;280(6):1491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maeda T, Inoue M, Koshiba S, Yabuki T, Aoki M, Nunokawa E, et al. Solution structure of the SEA domain from the murine homologue of ovarian cancer antigen CA125 (MUC16). J Biol Chem. 2004;279(13):13174–82. [DOI] [PubMed] [Google Scholar]
- 55.Oates ME, Romero P, Ishida T, Ghalwash M, Mizianty MJ, Xue B, et al. D(2)P(2): database of disordered protein predictions. Nucleic Acids Res. 2013;41(Database issue):D508–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, et al. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21(16):3435–8. [DOI] [PubMed] [Google Scholar]
- 57.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32(Web Server issue):W327–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lang T, Klasson S, Larsson E, Johansson ME, Hansson GC, Samuelsson T. Searching the Evolutionary Origin of Epithelial Mucus Protein Components-Mucins and FCGBP. Mol Biol Evol. 2016;33(8):1921–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma R, Kumar S, Tsunoda T, Patil A, Sharma A. Predicting MoRFs in protein sequences using HMM profiles. BMC Bioinformatics. 2016;17(Suppl 19):504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43(Database issue):D512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chuang GY, Boyington JC, Joyce MG, Zhu J, Nabel GJ, Kwong PD, et al. Computational prediction of N-linked glycosylation incorporating structural properties and patterns. Bioinformatics. 2012;28(17):2249–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32(10):1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta. 2010;1804(4):996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45(D1):D369–D79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hale ML, Thapa I, Ghersi D. FunSet: an open-source software and web server for performing and displaying Gene Ontology enrichment analysis. BMC Bioinformatics. 2019;20(1):359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32(Database issue):D138–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45(D1):D200–D3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walsh I, Giollo M, Di Domenico T, Ferrari C, Zimmermann O, Tosatto SC. Comprehensive large-scale assessment of intrinsic protein disorder. Bioinformatics. 2015;31(2):201–8. [DOI] [PubMed] [Google Scholar]
- 70.Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, et al. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res. 2007;6(6):2351–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Malhis N, Wong ET, Nassar R, Gsponer J. Computational Identification of MoRFs in Protein Sequences Using Hierarchical Application of Bayes Rule. PLoS One. 2015;10(10):e0141603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kurotani A, Sakurai T. In Silico Analysis of Correlations between Protein Disorder and Post-Translational Modifications in Algae. Int J Mol Sci. 2015;16(8):19812–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Basile W, Salvatore M, Bassot C, Elofsson A. Why do eukaryotic proteins contain more intrinsically disordered regions? PLoS Comput Biol. 2019;15(7):e1007186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pryor EE Jr., Wiener MC. A critical evaluation of in silico methods for detection of membrane protein intrinsic disorder. Biophys J. 2014;106(8):1638–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen JW, Romero P, Uversky VN, Dunker AK. Conservation of intrinsic disorder in protein domains and families: II. functions of conserved disorder. J Proteome Res. 2006;5(4):888–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27(10):527–33. [DOI] [PubMed] [Google Scholar]
- 77.Wang RY, Chen L, Chen HY, Hu L, Li L, Sun HY, et al. MUC15 inhibits dimerization of EGFR and PI3K-AKT signaling and is associated with aggressive hepatocellular carcinomas in patients. Gastroenterology. 2013;145(6):1436–48 e1–12. [DOI] [PubMed] [Google Scholar]
- 78.Chaturvedi P, Singh AP, Chakraborty S, Chauhan SC, Bafna S, Meza JL, et al. MUC4 mucin interacts with and stabilizes the HER2 oncoprotein in human pancreatic cancer cells. Cancer Res. 2008;68(7):2065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boze H, Marlin T, Durand D, Perez J, Vernhet A, Canon F, et al. Proline-rich salivary proteins have extended conformations. Biophys J. 2010;99(2):656–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin Y, Currie SL, Rosen MK. Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J Biol Chem. 2017;292(46):19110–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tarakhovsky A, Prinjha RK. Drawing on disorder: How viruses use histone mimicry to their advantage. J Exp Med. 2018;215(7):1777–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tamarozzi ER, Giuliatti S. Understanding the Role of Intrinsic Disorder of Viral Proteins in the Oncogenicity of Different Types of HPV. Int J Mol Sci. 2018;19(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Charon J, Barra A, Walter J, Millot P, Hebrard E, Moury B, et al. First Experimental Assessment of Protein Intrinsic Disorder Involvement in an RNA Virus Natural Adaptive Process. Mol Biol Evol. 2018;35(1):38–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raina D, Agarwal P, Lee J, Bharti A, McKnight CJ, Sharma P, et al. Characterization of the MUC1-C Cytoplasmic Domain as a Cancer Target. PLoS One. 2015;10(8):e0135156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Machkalyan G, Trieu P, Petrin D, Hebert TE, Miller GJ. PPIP5K1 interacts with the exocyst complex through a C-terminal intrinsically disordered domain and regulates cell motility. Cell Signal. 2016;28(5):401–11. [DOI] [PubMed] [Google Scholar]
- 86.Marcotte EM, Tsechansky M. Disorder, promiscuity, and toxic partnerships. Cell. 2009;138(1):16–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138(1):198–208. [DOI] [PubMed] [Google Scholar]
- 88.Lakshmanan I, Seshacharyulu P, Haridas D, Rachagani S, Gupta S, Joshi S, et al. Novel HER3/MUC4 oncogenic signaling aggravates the tumorigenic phenotypes of pancreatic cancer cells. Oncotarget. 2015;6(25):21085–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yamashita MSA, Melo EO. Mucin 2 (MUC2) promoter characterization: an overview. Cell Tissue Res. 2018;374(3):455–63. [DOI] [PubMed] [Google Scholar]
- 90.Singh PK, Hollingsworth MA. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 2006;16(9):467–76. [DOI] [PubMed] [Google Scholar]
- 91.Muniyan S, Haridas D, Chugh S, Rachagani S, Lakshmanan I, Gupta S, et al. MUC16 contributes to the metastasis of pancreatic ductal adenocarcinoma through focal adhesion mediated signaling mechanism. Genes Cancer. 2016;7(3–4):110–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Das S, Rachagani S, Torres-Gonzalez MP, Lakshmanan I, Majhi PD, Smith LM, et al. Carboxyl-terminal domain of MUC16 imparts tumorigenic and metastatic functions through nuclear translocation of JAK2 to pancreatic cancer cells. Oncotarget. 2015;6(8):5772–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rump A, Morikawa Y, Tanaka M, Minami S, Umesaki N, Takeuchi M, et al. Binding of ovarian cancer antigen CA125/MUC16 to mesothelin mediates cell adhesion. J Biol Chem. 2004;279(10):9190–8. [DOI] [PubMed] [Google Scholar]
- 94.Chen SH, Dallas MR, Balzer EM, Konstantopoulos K. Mucin 16 is a functional selectin ligand on pancreatic cancer cells. FASEB J. 2012;26(3):1349–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou J, Zhao S, Dunker AK. Intrinsically Disordered Proteins Link Alternative Splicing and Post-translational Modifications to Complex Cell Signaling and Regulation. J Mol Biol. 2018;430(16):2342–59. [DOI] [PubMed] [Google Scholar]
- 96.Hoshi H, Sawada T, Uchida M, Iijima H, Kimura K, Hirakawa K, et al. MUC5AC protects pancreatic cancer cells from TRAIL-induced death pathways. Int J Oncol. 2013;42(3):887–93. [DOI] [PubMed] [Google Scholar]
- 97.Ridley C, Thornton DJ. Mucins: the frontline defence of the lung. Biochem Soc Trans. 2018;46(5):1099–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Senapati S, Das S, Batra SK. Mucin-interacting proteins: from function to therapeutics. Trends Biochem Sci. 2010;35(4):236–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lo PW, Shie JJ, Chen CH, Wu CY, Hsu TL, Wong CH. O-GlcNAcylation regulates the stability and enzymatic activity of the histone methyltransferase EZH2. Proc Natl Acad Sci U S A. 2018;115(28):7302–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van der Lee R, Lang B, Kruse K, Gsponer J, Sanchez de Groot N, Huynen MA, et al. Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell Rep. 2014;8(6):1832–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheng Y, Oldfield CJ, Meng J, Romero P, Uversky VN, Dunker AK. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry. 2007;46(47):13468–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Uversky VN. p53 Proteoforms and Intrinsic Disorder: An Illustration of the Protein Structure-Function Continuum Concept. Int J Mol Sci. 2016;17(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yadav LR, Rai S, Hosur MV, Varma AK. Functional assessment of intrinsic disorder central domains of BRCA1. J Biomol Struct Dyn. 2015;33(11):2469–78. [DOI] [PubMed] [Google Scholar]
- 104.Malaney P, Uversky VN, Dave V. Identification of intrinsically disordered regions in PTEN and delineation of its function via a network approach. Methods. 2015;77–78:69–74. [DOI] [PubMed] [Google Scholar]
- 105.Salgia R, Jolly MK, Dorff T, Lau C, Weninger K, Orban J, et al. Prostate-Associated Gene 4 (PAGE4): Leveraging the Conformational Dynamics of a Dancing Protein Cloud as a Therapeutic Target. J Clin Med. 2018;7(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Metallo SJ. Intrinsically disordered proteins are potential drug targets. Curr Opin Chem Biol. 2010;14(4):481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chang YS, Graves B, Guerlavais V, Tovar C, Packman K, To KH, et al. Stapled alpha-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc Natl Acad Sci U S A. 2013;110(36):E3445–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Balachandran VP, Luksza M, Zhao JN, Makarov V, Moral JA, Remark R, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551(7681):512–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Aithal A, Rauth S, Kshirsagar P, Shah A, Lakshmanan I, Junker WM, et al. MUC16 as a novel target for cancer therapy. Expert Opin Ther Targets. 2018;22(8):675–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chivian D, Kim DE, Malmstrom L, Bradley P, Robertson T, Murphy P, et al. Automated prediction of CASP-5 structures using the Robetta server. Proteins. 2003;53 Suppl 6:524–33. [DOI] [PubMed] [Google Scholar]
- 111.Pollastri G, Przybylski D, Rost B, Baldi P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins. 2002;47(2):228–35. [DOI] [PubMed] [Google Scholar]
- 112.Wojcik S, Birol M, Rhoades E, Miranker AD, Levine ZA. Targeting the Intrinsically Disordered Proteome Using Small-Molecule Ligands. Methods Enzymol. 2018;611:703–34. [DOI] [PubMed] [Google Scholar]
- 113.Neira JL, Bintz J, Arruebo M, Rizzuti B, Bonacci T, Vega S, et al. Identification of a Drug Targeting an Intrinsically Disordered Protein Involved in Pancreatic Adenocarcinoma. Sci Rep. 2017;7:39732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yu C, Niu X, Jin F, Liu Z, Jin C, Lai L. Structure-based Inhibitor Design for the Intrinsically Disordered Protein c-Myc. Sci Rep. 2016;6:22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jung KY, Wang H, Teriete P, Yap JL, Chen L, Lanning ME, et al. Perturbation of the c-Myc-Max protein-protein interaction via synthetic alpha-helix mimetics. J Med Chem. 2015;58(7):3002–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ambadipudi S, Zweckstetter M. Targeting intrinsically disordered proteins in rational drug discovery. Expert Opin Drug Discov. 2016;11(1):65–77. [DOI] [PubMed] [Google Scholar]
- 117.Tsafou K, Tiwari PB, Forman-Kay JD, Metallo SJ, Toretsky JA. Targeting Intrinsically Disordered Transcription Factors: Changing the Paradigm. J Mol Biol. 2018;430(16):2321–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.