Abstract
Introduction
Distinguishing between hypertrophic cardiomyopathy and other causes of left ventricular hypertrophy can be difficult in pediatric patients. We hypothesized that cardiac MRI T1 mapping could improve diagnosis of pediatric hypertrophic cardiomyopathy and that measures of myocardial function would correlate with T1 times and extracellular volume fraction.
Methods
Thirty patients with hypertrophic cardiomyopathy completed MRI with tissue tagging, T1-mapping, and late gadolinium enhancement. Left ventricular circumferential strain was calculated from tagged images. T1, partition coefficient, and synthetic extracellular volume were measured at base, mid, apex and thickest area of myocardial hypertrophy. MRI measures compared to cohort of 19 healthy children and young adults. Mann-Whitney U, Spearman’s rho, and multivariable logistic regression used for statistical analysis.
Results
Hypertrophic cardiomyopathy patients had increased left ventricular ejection fraction and indexed mass. Hypertrophic cardiomyopathy patients had decreased global strain and increased native T1 (−14.3% IQR[−16.0,−12.1] vs. −17.3%[−19.0,15.7], p<0.001 and 1015ms[991,1026] vs 990ms[972,1001], p=0.019). Partition coefficient and synthetic extracellular volume were not increased in hypertrophic cardiomyopathy. Global native T1 correlated inversely with ejection fraction (rho=−0.63, p=0.002) and directly with global strain (rho=0.51, p=0.019). A logistic regression model using ejection fraction and native T1 distinguished between hypertrophic cardiomyopathy and control with an area under the receiver operating characteristic curve of 0.91.
Conclusion
In this cohort of pediatric hypertrophic cardiomyopathy, strain was decreased and native T1 was increased compared with controls. Native T1 correlated with both ejection fraction and strain and a model using native T1 and ejection fraction differentiated patients with and without hypertrophic cardiomyopathy.
Keywords: Pediatric Cardiology, Hypertrophic Cardiomyopathy, Cardiac MRI, T1 mapping, Myocardial strain
Introduction
Hypertrophic cardiomyopathy is the leading cause of sudden cardiac death in healthy young athletes.[1] Hallmarks of the disease include ventricular hypertrophy, myocyte disarray, and both replacement and interstitial fibrosis. Distinguishing between hypertrophic cardiomyopathy and other causes of left ventricular hypertrophy can be difficult. Cardiac MRI has significant advantages over echocardiography for assessment of adults with hypertrophic cardiomyopathy, and newer methods of tissue characterization may aid in the diagnosis of pediatric patients with hypertrophic cardiomyopathy.[2]
The longitudinal relaxation time constant, or T1, can be measured non-invasively in myocardium using cardiac MRI. Native T1 maps, or maps obtained prior to contrast administration, can be analyzed separately or combined with post-contrast T1 maps to derive either a partition coefficient or an extracellular volume fraction. Native T1 and extracellular volume are surrogates of extracellular matrix expansion[3] and have been validated histologically.[4–7]
Studies demonstrate lower myocardial strain in areas of replacement fibrosis in pediatric hypertrophic cardiomyopathy.[8] Interstitial fibrosis also seems to have an effect on myocardial mechanics in multiple cardiovascular disease states, but this has never been studied in pediatric hypertrophic cardiomyopathy.[9–11] A better understanding of the underlying relationship between myocardial mechanics and markers of fibrosis and myocyte hypertrophy may help clarify disease progression and inform future treatment strategies. The objectives of this study were: 1) to evaluate whether T1 mapping, partition coefficient, and extracellular volume mapping improve accuracy of diagnosis in patients with hypertrophic cardiomyopathy; 2) to define the relationship between T1 and extracellular volume mapping and measures of myocardial function, particularly myocardial strain and left ventricular ejection fraction.
METHODS
Enrollment
The Vanderbilt Institutional Review Board approved this retrospective study and the study was performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. Patients who underwent cardiac MRI between 2015 and 2018 were identified from the pediatric cardiac MRI database. Inclusion criteria were: 1) pediatric patients with a diagnosis of hypertrophic cardiomyopathy and 2) prior cardiac MRI that included a modified Look-Locker inversion recovery. All hypertrophic cardiomyopathy patients had a definitive diagnosis, either with: 1) family history and either a positive genotype or a phenotype consistent with hypertrophic cardiomyopathy (n=11); 2) clinical diagnosis with pathologic hypertrophy and known hypertrophic cardiomyopathy causing mutation (n=5); or 3) Definitive clinical diagnosis of hypertrophic cardiomyopathy with negative genetic testing (n=14). Subjects were excluded if they had: 1) an underlying neurologic, metabolic, or other secondary cause of hypertrophic cardiomyopathy, 2) inadequate modified Look-Locker inversion recovery image quality for analysis. Genotype positive phenotype negative was defined as a patient with a known positive genotype but with clinical testing that would not meet criteria for definitive diagnosis of hypertrophic cardiomyopathy.
Control images for our study were acquired from 19 patients 12–30 years of age from previous cohorts of either clinically indicated cardiac MRIs or research cardiac MRIs. All controls had normal cardiac MRIs. Exclusion criteria for control cohort were: cardiovascular disease, risk factors for cardiovascular disease, muscular dystrophy or unexplained skeletal muscle weakness, any diagnosis that could affect cardiac function or lead to myocardial fibrosis, and contraindication to cardiac MRI with gadolinium. All patients undergoing research cardiac MRI were aged 18–30 and previously healthy. For patients undergoing clinically indicated cardiac MRIs, indications were: 1) concern for possible arrhythmogenic right ventricular cardiomyopathy due to either an abnormal electrocardiogram or history of premature ventricular contractions with normal work up, including normal cardiac MRI, and arrhythmogenic right ventricular cardiomyopathy ruled out; 2) concern for abnormal left ventricular function on echocardiogram with normal function on cardiac MRI; 3) evaluation for possible apical hypertrophic cardiomyopathy with normal cardiac MRI; 4) evaluation of atypical chest pain with normal work up.
The electronic medical record was reviewed for: genetic test results, Holter monitor results, treatment interventions (pharmacologic management, placement of implantable cardioverter defibrillator or pacemaker, or septal myectomy), and risk stratification criteria for hypertrophic cardiomyopathy (septal thickness greater than 30mm, history of ventricular tachycardia or unexplained syncope, family history of hypertrophic cardiomyopathy with sudden cardiac death or aborted sudden cardiac death, and inadequate blood pressure response during treadmill test).
Cardiac MRI Acquisition
Cardiac MRI was performed on a 1.5 Tesla Siemens Avanto. Functional imaging was performed as previously described using balanced steady state free-precession images in a short axis stack.[12] Intravenous gadolinium contrast (gadopentate dimeglumine, Magnevist®, Bayer Healthcare Pharmaceuticals, Wayne, NJ, USA or gadobutrol, Gadovist®, Bayer Healthcare Pharmaceuticals, Wayne, NJ, USA) was administered through a peripheral intravenous line at a dose of 0.2mmol/kg. Late gadolinium enhancement imaging was performed using: 1) single shot (balanced steady state free-precession) and segmented (turboflash) inversion recovery with optimized inversion recovery to null the signal from the myocardium, as well as phase sensitive inversion recovery balanced steady state free-precession with an inversion time of 300ms.
Myocardial tagging was performed in the short axis at the base, level of the papillary muscles, and apex using a segmented k-space fast gradient echo sequence with electrocardiogram triggering. Grid tagging was performed with a spacing of 8 mm and 9–13 phases (Siemens). Typical imaging parameters included: slice thickness 6–8 mm, field of view 340 mm × 340 mm, matrix size 256 × 192, and minimum echo time and repetition time. The sequences were breath-holds and parallel imaging with generalized autocalibrating partially parallel acquisition with an acceleration factor of two was used. One patient had individual horizontal and vertical tagging performed and in one patient, tagged images were not analyzable.
Breath-held modified Look-Locker inversion recovery sequences were performed prior to and 15 minutes after contrast administration at the left ventricular base, mid left ventricle, and apex in the short axis plane.[13] Modified Look-Locker inversion recovery sequences were motion-corrected, electrocardiogram-triggered images obtained in diastole with typical imaging parameters: non-selective inversion with a 35 degree flip angle, single shot stead state free-precession imaging, initial inversion time of 120ms with 80ms increments, field of view 340 × 272 mm2, matrix size 256 × 144, slice thickness 8mm, voxel size 1.3 × 1.9 × 8.0 mm3, repetition time of 2.6ms, echo time of 1.1ms, parallel imaging factor of 2. The matrix size was decreased to 192 × 128 for heart rates >90 (approximate voxel size 1.8 × 2.1 × 8 mm3). The pre-contrast modified Look-Locker inversion recovery acquired 5 images after the first inversion with the equivalent of a 3 second pause followed by 3 images after the second inversion, or 5(3s)3 (as a true 3 second pause is not possible with the current software package, the number of heartbeats used for recovery was varied depending on the average heart rate measured just prior to T1 mapping, with 3 beats used for a heart rate of 60, 4 beats used for a heart rate of 80, 5 beats for a heart rate of 100, and 6 beats for a heart rate of 120; no patient had a heart rate over 120 in this study). The post-contrast protocol was acquired at a 4(1)3(1)2, or 4 images acquired after the first inversion with a 1 beat pause followed by a second inversion after which 3 images were acquired, an additional 1 beat pause, then a final inversion after which 2 images were acquired.[14] Motion correction as described by Xue, et al. was performed and a T1 map was generated on the scanner.[15] A goodness of fit map was also performed at the time of the scan to evaluate data quality. In 6 hypertrophic cardiomyopathy patients, the modified Look-Locker inversion recovery was only performed at the mid-LV slice; the basilar slice was deemed inadequate for analysis in 1 patient and the apical slice in 2 patients with hypertrophic cardiomyopathy.
Cardiac MRI Post-processing
Left ventricular volume, mass, and function were calculated as previously described.[16] The presence or absence of late gadolinium enhancement was qualitatively assessed. Percent of scar was calculated using the 5 standard deviation method on Medis (Medis Medical Imaging Systems, Leiden, The Netherlands) on the phase sensitive inversion recovery images as per our labs standard protocol. Analysis of myocardial tagged images was performed as previously described using harmonic phase methodology (Diagnosoft Inc., Morrisville, NC).[17] In brief, a contour or mesh was drawn over the tagged image at peak systole by outlining the epicardium and endocardium. The superior right ventricular insertion was identified manually. The contours were performed by the same reader (SS) with verification of each contour by a second reader (JHS) with more than 7 years of experience using the software. The software then calculated the global peak circumferential strain and the circumferential strain values for each segment (16 segment model) and slice (base, mid, and apex).
Using software programmed in MATLAB 2014a (The MathWorks, Natick, MA, USA), regions of interest were manually drawn on native and post-contrast T1 maps within the left ventricular mesocardium in 16 segments using the standard American Heart Association model of segmentation.[18] These regions of interest were contoured by one reader with experience analyzing T1 maps and confirmed by a second reader with 6 years of experience in analyzing T1 mapping. To evaluate reproducibility, a second reader with experience analyzing T1 maps repeated the analysis at the base, mid, and apex in a random sample of 10 hypertrophic cardiomyopathy subjects on both the native and post-contrast maps. In addition, hypertrophic cardiomyopathy patients with significant left ventricular hypertrophy (left ventricular thickness >15mm) had a region of interest drawn in the region of thickest hypertrophy. Only 10 hypertrophic cardiomyopathy patients had concurrent hematocrit levels, so extracellular volume maps were not analyzed; instead, synthetic extracellular volume was calculated as described below. Regions of interest were carefully traced to avoid partial volume averaging with blood-pool or epicardial fat. Based on the T1 mapping consensus statement, areas of late gadolinium enhancement were not excluded as these areas were felt to be the most focal areas in a continuum of diffuse extracellular matrix expansion.[19]
Partition coefficient was calculated from the native and post-contrast T1 using the following equation:
Synthetic hematocrit was calculated from the following equation optimized to this magnet:[20]
The basal, mid, and apical synthetic extracellular volume fraction was calculated from the native and post-contrast T1 and the synthetic hematocrit using the following equation:
In patients with adequate maps at all 3 slices, global myocardial T1, global partition coefficient, and global synthetic extracellular volume were calculated. Imaging artifact was not contoured. Segments were not included in the analysis if the bounds of the myocardium could not be distinguished from surrounding tissue and blood pool or if image registration was inadequate in those segments.
Statistical analysis
Categorical variables were compared using a Chi-square or Fisher’s exact test and continuous variables were compared using a Wilcoxon rank-sum test. Correlations between continuous variables were obtained using a Spearman’s rho. Reproducibility of native and post-contrast T1 mapping was assessed using intraclass correlation coefficient for absolute agreement. We fit univariate logistic regression models to estimate the probability of hypertrophic cardiomyopathy evaluating the following pre-specified predictors: left ventricular ejection fraction, indexed left ventricular mass, percent late gadolinium enhancement, circumferential strain at mid left ventricle, native T1 at mid left ventricle, partition coefficient at mid left ventricle, and synthetic extracellular volume at mid left ventricle. The mid left ventricle slice was used for circumferential strain and T1 mapping because some subjects did not have adequate image quality to calculate these measures at either the apex or the base (and thus inadequate images to calculate global values). Multivariable analysis was then performed using those predictors that were significant to determine the best predictors of hypertrophic cardiomyopathy (a total of 6 models evaluated). Analyses were performed with IBM SPSS statistics, version 25.0 (Armonk, NY: IBM Corp). Study data were collected and managed using REDCap (Research Electronic Data Capture) electronic data capture tools hosted at Vanderbilt.[21]
RESULTS
Demographics
Thirty hypertrophic cardiomyopathy subjects met inclusion and exclusion criteria. The mean age of hypertrophic cardiomyopathy diagnosis was 14.0 ± 2.9 years and the mean age at cardiac MRI was 15.8 ± 2.2. Nineteen hypertrophic cardiomyopathy patients (63%) were male while 17 controls (89%) were male. Further demographics can be found in Table 1.
Table 1.
Demographics
| Control N=19 | Hypertrophic cardiomyopathy N=30 | |
|---|---|---|
| Age at cardiac MRI (years) | 21.2 ± 5.3 (range 12–30) | 15.8 ± 2.2 (range 10–19) |
| Male Gender | 17 (89%) | 19 (63%) |
| Height (cm) | 176 ± 13 | 170 ± 11 |
| Weight (kg) | 71 ± 12 | 85 ± 28 |
| Body surface area (m2) | 1.9 ± 0.2 | 2.0 ± 0.4 |
| Race | ||
| Caucasian | 11 (58%) | 21 (70%) |
| African American | 3 (16%) | 3 (10%) |
| Asian | 1 (5%) | 1 (3%) |
| Other | 0 | 2 (7%) |
| Mixed | 1 (5%) | 0 |
| Unknown/unwilling to report | 3 (16%) | 3 (10%) |
| Hispanic/Latino | 1 (5%) | 4 (13%) |
Data presented as mean ± standard deviation or N (percent).
Genetic testing was completed in 28 patients, with 16 testing positive for known hypertrophic cardiomyopathy mutations (Table 2), 10 with negative testing, and 2 with variants of unknown significance. Six patients had an implantable cardioverter defibrillator placed and 3 had a history of implantable cardioverter defibrillator shock. One patient who did not meet criteria for implantable cardioverter defibrillator placement died during sleep. Thirteen patients had received pharmacotherapy at some point during their medical course, with betablockade the most common therapy. Risk factors for sudden cardiac death are listed in Table 2.
Table 2:
Clinical characteristics of hypertrophic cardiomyopathy patients
| N=30 | |
|---|---|
| Age at diagnosis (years) | 14.0 ± 2.9 |
| Alive | 29 (97%) |
| Genetic Testing | 28 (93%) |
| Positive for known genetic mutation | 16 (57%) |
| Implantable Cardioverter Defibrillator (ICD) placed | 6 (20%) |
| Patients with ICD shocks delivered | 3 (50%) |
| History of septal myectomy | 3 (10%) |
| Use of medications | 16 (53%) |
| Angiotensin converting enzyme inhibitor | 1 (3%) |
| Beta-blocker | 15 (50%) |
| Calcium channel blocker | 1 (3%) |
| Aspirin | 1 (3%) |
| Other | 2 (10%) |
| Risk Factors for Sudden Cardiac Death | |
| Maximal septal thickness > 30mm | 3 (10%) |
| Syncope | 6 (20%) |
| Family history of sudden cardiac death | 11 (37%) |
| History of ventricular tachycardia | 3 (10%)* |
| Inadequate blood pressure response to exercise | 8 (27%) |
| History of aborted sudden death | 2 (7%) |
All non-sustained ventricular tachycardia
Standard cardiac measures
Hypertrophic cardiomyopathy subjects had increased indexed left ventricular mass (median 79g/m2 interquartile range[65,91] vs 57 g/m2 [54,62], p<0.001) and increased left ventricular ejection fraction (69% [63,71] vs. 60% [58,64], p=0.001) in comparison to controls. The right ventricular ejection fraction was also higher in hypertrophic cardiomyopathy (64% [60,68] vs. 57% [55,60], p<0.001). The median maximal wall thickness of the left ventricle in the hypertrophic cardiomyopathy group was 18 mm [16, 22.5]. A total of 18 hypertrophic cardiomyopathy subjects (65%) had late gadolinium enhancement, almost exclusively in areas of pathologic hypertrophy, though some patients had mild late gadolinium enhancement at the right ventricular insertion points. Patients with late gadolinium enhancement had a median percent scar of 3.3% [1.7,5.1].
Myocardial strain and T1 mapping
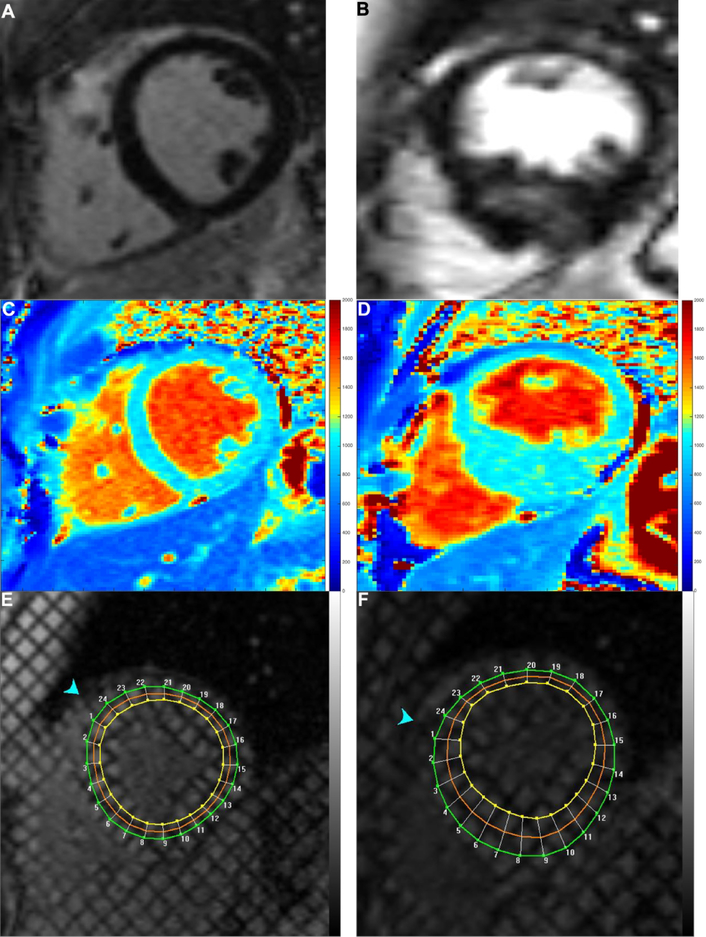
The native and post-contrast T1 maps had good reproducibility (native T1: base intraclass correlation coefficient 0.89 p=0.001, mid intraclass correlation coefficient 0.91 p<0.001, apex intraclass correlation coefficient 0.90 p=0.001; post contrast T1: base intraclass correlation coefficient 0.95 p<0.001, mid intraclass correlation coefficient 0.96 p<0.001, apex intraclass correlation coefficient 0.94 p<0.001). Hypertrophic cardiomyopathy subjects had decreased global circumferential strain compared with controls as well as decreased circumferential strain at the base, mid left ventricle, and apex (Table 3). Example T1 maps and tagged images are demonstrated in Figure 1. Hypertrophic cardiomyopathy patients had significantly increased global and mid native T1 compared with controls (Table 3 for all results, including basal and apical). The global and mid partition coefficient and synthetic extracellular volume were not significantly different between hypertrophic cardiomyopathy and control.
Table 3:
Comparison of cardiovascular magnetic resonance imaging in control and hypertrophic cardiomyopathy patients.
| Control | Hypertrophic cardiomyopathy | p-value | |||
|---|---|---|---|---|---|
| n | Median (IQR) | n | Median (IQR) | ||
| Left ventricular ejection fraction (%) | 19 | 60 (58, 64) | 30 | 69 (63, 71) | 0.001 |
| Left ventricular mass index (g/m2) | 19 | 57 (54, 62) | 30 | 79 (65, 91) | <0.001 |
| Right ventricular ejection fraction (%) | 12 | 57 (55, 60) | 30 | 64 (60, 68) | <0.001 |
| Base circumferential strain (%) | 19 | −16.9 (−18.4, −16.7) | 29 | −14.0 (−15.7, −12.8) | <0.001 |
| Mid circumferential strain (%) | 19 | −16.8 (−19.5, −15.2) | 29 | −14.3 (−16.0, −12.5) | 0.001 |
| Apex circumferential strain (%) | 19 | −18.4 (−20.5, −15.9) | 28 | −14.7 (−17.4, −10.7) | 0.001 |
| Global circumferential strain (%) | 18 | −17.3 (−19.9, −15.7) | 28 | −14.3 (−16.0, −12.1) | <0.001 |
| Base T1 (ms) | 19 | 996 (981, 1007) | 23 | 1026 (989, 1039) | 0.013 |
| Mid T1 (ms) | 19 | 990 (964, 1004) | 30 | 1013 (986, 1036) | 0.007 |
| Apex T1 (ms) | 18 | 983 (968, 1005) | 23 | 996 (969, 1032) | 0.237 |
| Global T1 (ms) | 18 | 990 (972, 1001) | 22 | 1015 (991, 1026) | 0.019 |
| Base partition coefficient | 19 | 0.39 (0.36, 0.39) | 23 | 0.39 (0.35, 0.45) | 0.850 |
| Mid partition coefficient | 19 | 0.39 (0.36, 0.43) | 28 | 0.39 (0.37, 0.43) | 0.948 |
| Apex partition coefficient | 17 | 0.42 (0.40, 0.45) | 21 | 0.41 (0.37, 0.45) | 0.437 |
| Global partition coefficient | 17 | 0.40 (0.39, 0.42) | 19 | 0.39 (0.37, 0.46) | 0.812 |
| Base synthetic extracellular volume | 19 | 0.23 (0.21, 0.23) | 23 | 0.22 (0.20, 0.26) | 0.950 |
| Mid synthetic extracellular volume | 19 | 0.23 (0.21, 0.25) | 28 | 0.23 (0.21, 0.25) | 0.931 |
| Apex synthetic extracellular volume | 17 | 0.25 (0.23, 0.26) | 21 | 0.24 (0.21, 0.27) | 0.271 |
| Global synthetic extracellular volume | 17 | 0.23 (0.22, 0.25) | 19 | 0.23 (0.21, 0.27) | 0.763 |
Figure 1:
Example in healthy control (left column) and patient with hypertrophic cardiomyopathy (right column). A) and B) late gadolinium enhancement images, C) and D) native T1 maps, and E) and F) tagged images.
Native T1 in the area of maximal myocardial hypertrophy was increased compared to the median mid left ventricle T1 of the control population (1027 ms [1004,1047] vs. 990 [964, 1004], p= 0.001). The partition coefficient and synthetic extracellular volume in the region of maximal hypertrophy were not statistically different between hypertrophic cardiomyopathy and control, suggesting the primary pathology in these regions may be due to myocyte hypertrophy and not extracellular matrix expansion.
Within the hypertrophic cardiomyopathy group, patients with late gadolinium enhancement had significantly increased mid native T1 compared to those without late gadolinium enhancement; global native T1 was higher but did not reach statistical significance (Table 4 for all results, including basal and apical). The global and mid synthetic extracellular volume were also increased in hypertrophic cardiomyopathy patients with late gadolinium enhancement as were the global and mid partition coefficients (Table 4).
Table 4:
Comparison of native T1 mapping, partition coefficient, and synthetic extracellular volume in patients with hypertrophic cardiomyopathy with and without late gadolinium enhancement.
| Hypertrophic cardiomyopathy with late gadolinium enhancement | Hypertrophic cardiomyopathy without late gadolinium enhancement | p-value | |||
|---|---|---|---|---|---|
| n | Median (IQR) | n | Median (IQR) | ||
| Base T1 (ms) | 14 | 1032 (1006,1042) | 9 | 1002 (971,1029) | 0.038 |
| Mid T1 (ms) | 18 | 1019 (1009,1045) | 12 | 986 (960,1034) | 0.034 |
| Apex T1 (ms) | 14 | 1006 (983,1038) | 9 | 969 (957,1011) | 0.044 |
| Global T1 (ms) | 13 | 1017 (1006,1027) | 9 | 990 (962,1029) | 0.10 |
| Base partition coefficient | 14 | 0.39 (0.37,0.46) | 9 | 0.36 (0.34,0.38) | 0.059 |
| Mid partition coefficient | 17 | 0.41 (0.38,0.45) | 11 | 0.37 (0.34,0.39) | 0.037 |
| Apex partition coefficient | 14 | 0.43 (0.40,0.47) | 7 | 0.37 (0.36,0.39) | 0.035 |
| Global partition coefficient | 12 | 0.42 (0.39,0.46) | 7 | 0.37 (0.34,0.38) | 0.023 |
| Base synthetic extracellular volume | 14 | 0.23 (0.21,0.27) | 9 | 0.21 (0.19,0.22) | 0.014 |
| Mid synthetic extracellular volume | 17 | 0.24 (0.22,0.26) | 11 | 0.21 (0.20,0.22) | 0.025 |
| Apex synthetic extracellular volume | 14 | 0.25 (0.23,0.27) | 7 | 0.21 (0.21,0.23) | 0.028 |
| Global synthetic extracellular volume | 12 | 0.25 (0.22,0.27) | 7 | 0.20 (0.20,0.22) | 0.038 |
Within the hypertrophic cardiomyopathy group, left ventricular ejection fraction had a weak inverse correlation with percent late gadolinium enhancement (rho=−0.39, p=0.035) and a strong inverse correlation with global native T1 (rho=−0.63, p=0.002). Maximal wall thickness correlated with percent late gadolinium enhancement (rho=0.63, p<0.001). Global T1 correlated with percent late gadolinium enhancement (rho=0.44, p=0.041). Global circumferential strain correlated with global T1 (rho=0.51, p=0.019) and indexed mass (rho=0.63, p=0.004) but not with percent late gadolinium enhancement; however, a sub analysis comparing all segments with and without strain demonstrated worse strain in segments with late gadolinium enhancement (−11.1% [−8.1,−14.2] vs −14.9 [−11.0,−17.8], p<0.001).
Global synthetic extracellular volume and partition coefficient did not correlate with left ventricular ejection fraction (rho=−0.3, p=0.211 and rho=−0.19, p=0.432) or global circumferential strain (rho=0.09, p=0.723 and rho=0.069, p=0.785). Both global synthetic extracellular volume and partition coefficient correlated with percent late gadolinium enhancement (rho=0.51, p=0.03 and rho=0.48, p=0.039).
Modeling
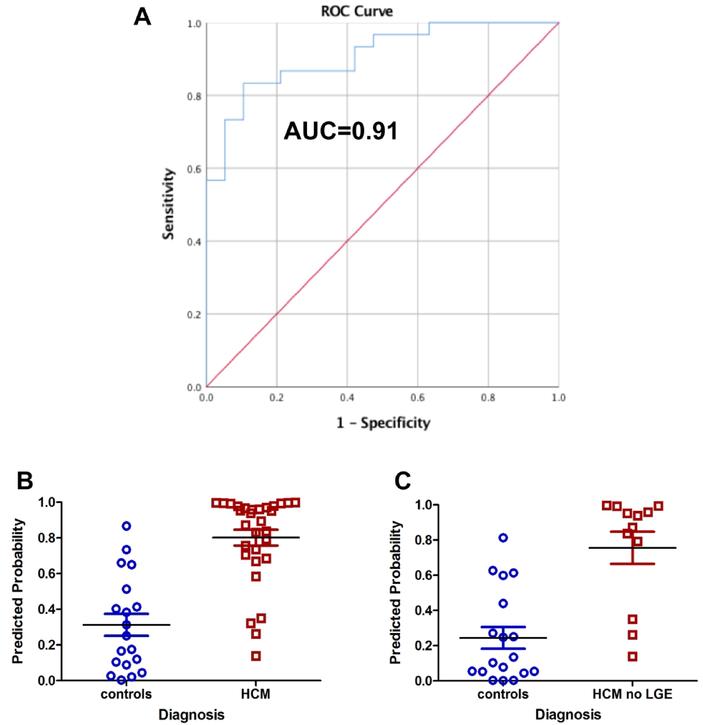
Univariate logistic regression analysis demonstrated that left ventricular ejection fraction, circumferential strain at the mid left ventricle, native T1 at the mid left ventricle, and indexed left ventricular mass were statistically significant in predicting presence of hypertrophic cardiomyopathy (p<0.05 for all) (Table 5). Percent late gadolinium enhancement, partition coefficient at the mid left ventricle, and synthetic extracellular volume at the mid left ventricle were not significant in predicting hypertrophic cardiomyopathy. Six models were evaluated based on univariate regression results and the model with the highest receiver operating characteristic for prediction of hypertrophic cardiomyopathy included left ventricular ejection fraction and native T1 at the mid left ventricle (p=0.001 for left ventricular ejection fraction and p=0.003 for native T1 at mid left ventricle); this model had an area under the receiver operating characteristic curve of 0.91 (Figure 2A). However, a scatterplot displaying the predicted probabilities for this model demonstrated some overlap between patients with and without hypertrophic cardiomyopathy (Figure 2B).
Table 5:
Univariate logistic regression modeling with diagnosis of hypertrophic cardiomyopathy as outcome
| Univariate analysis | |
|---|---|
| Left ventricular ejection fraction | 0.005 |
| Indexed left ventricular Mass | 0.004 |
| Percent late gadolinium enhancement | 0.990 |
| Circumferential strain at mid left ventricle | 0.003 |
| Native T1 at mid left ventricle | 0.012 |
| Partition coefficient at mid left ventricle | 0.952 |
| Synthetic extracellular volume fraction at mid left ventricle | 0.927 |
Figure 2:
A) Receiver operating characteristic curve for model using left ventricular ejection fraction and native T1 at the mid left ventricle to predict a diagnosis of hypertrophic cardiomyopathy. B) Scatterplot of model results demonstrates some overlap in patients with and without hypertrophic cardiomyopathy. C) Scatterplot of same model only in patients without late gadolinium enhancement (milder disease) again demonstrates some overlap of patients with an without hypertrophic cardiomyopathy.
DISCUSSION
The primary findings of this study are: 1) hypertrophic cardiomyopathy patients have increased native T1 compared with controls; 2) Native T1 correlates strongly with global circumferential strain and left ventricular ejection fraction; 3) modeling using multiple cardiac imaging markers may aid in diagnosis of hypertrophic cardiomyopathy.
While native T1 mapping correlated strongly with circumferential strain and left ventricular ejection fraction, the partition coefficient and synthetic extracellular volume did not. Even in the thickest areas of pathologic hypertrophy, partition coefficient and synthetic extracellular volume were not significantly increased compared with controls. These findings may reflect an overall mild phenotype in this cohort, which would also be supported by the relatively low percent late gadolinium enhancement. As expected, the native T1, partition coefficient, and synthetic extracellular volume were increased in hypertrophic cardiomyopathy patients with late gadolinium enhancement. However, native T1 mapping detects abnormalities of both the extracellular matrix and the myocytes, while partition coefficient and synthetic extracellular volume are more specific for extracellular matrix expansion.[22] We hypothesize that the increased native T1 times are primarily reflecting underlying structural abnormalities in the myocytes, not the extracellular matrix. Indeed, a study by Swaboda et al used multivariable modeling in adult hypertrophic cardiomyopathy patients to demonstrate a correlation between native T1 and myocardial mechanics but not extracellular volume fraction, suggesting that the structural abnormalities resulting in impaired function were cellular.[22]
Previous studies in adult hypertrophic cardiomyopathy have demonstrated a statistically significant difference in native T1 between hypertrophic cardiomyopathy and controls.[23] Indeed, some reports have suggested that native T1 can be used to distinguish between patients with and without hypertrophic cardiomyopathy.[24,25] The only pediatric study of which we are aware also demonstrated a significant difference between hypertrophic cardiomyopathy and controls.[26] While use of native T1 in isolation predicts a diagnosis hypertrophic cardiomyopathy, there was significant overlap between groups. A model combining native T1 with left ventricular ejection fraction provided the highest area under the curve for diagnosis of hypertrophic cardiomyopathy. It is possible that a more comprehensive model that also includes circumferential strain and indexed left ventricular mass would improve segregation of groups, though we were unable to adequately evaluate a more comprehensive model due to sample size limitations.
Partial volume averaging, particularly at the apex or base, can lead to inaccuracies in T1 maps.[27] While we repeated and/or eliminated poor quality maps, these data could skew the results at these slices. Of note, the model only included T1 mapping at the mid-left ventricle, so is much less likely to be affected by partial volume averaging.
Our patient cohort only included 4 patients who were genotype positive and phenotype negative and it is unclear whether this model can distinguish between patients with milder forms of hypertrophic cardiomyopathy and other causes of left ventricular hypertrophy. Evaluation of model performance in patients without late gadolinium enhancement demonstrated relatively good separation between hypertrophic cardiomyopathy and control (Figure 2C), but future studies should be performed comparing mild hypertrophic cardiomyopathy to patients with other causes of left ventricular hypertrophy. While this model is promising, the small sample size necessitates validation in a larger cohort of patients before using clinically. Our intent is to use this model as pilot data for future analyses of larger cohorts of patients with hypertrophic cardiomyopathy. We caution the use of this model in clinical practice at this time.
Left ventricular ejection fraction, indexed left ventricular mass, native T1, and circumferential strain are of particular interest for hypertrophic cardiomyopathy diagnosis because all measures can be obtained without the use of contrast, allowing for shorter scan times and decreasing exposure to gadolinium. Moreover, these measures provide a combination of structural and functional measures for diagnosis. Further studies in a larger cohort could help clarify the diagnostic utility in patients with mild disease and allow for expansion of the number of predictors, potentially improving the model results. These studies should include patients with other causes of hypertrophy, such as hypertension or athlete’s heart, to further evaluate whether native T1 can effectively distinguish between patients with and without hypertrophic cardiomyopathy.
Limitations
This study is limited by a small sample size. In addition, only a small number of patients had hematocrit values drawn at the same time as the cardiac MRIs, so calculation of the extracellular volume was not possible. We elected to use a locally derived model to calculate synthetic extracellular volume. While our previous data suggested that synthetic extracellular volume can lead to clinical errors in individual patients, our data demonstrated that synthetic extracellular volume correlates well with extracellular volume and can be used in research cohorts such as this one.[20] Given the difficulty in obtaining pediatric control values, particularly for extracellular volume, our age and gender distribution did not match perfectly between hypertrophic cardiomyopathy and controls.[28] Per protocol, the modified Look-Locker inversion recovery sequences were performed in one slice at the base, mid left ventricle, and apex. Because of this, it was possible that the thickest segment on the T1 maps did not perfectly correspond to the patient’s thickest segment on cine imaging. Placement of modified Look-Locker inversion recovery sequences through the segments with largest pathological hypertrophy could increase the difference we detected between native T1 of the thickest segments and controls.
Some patients with hypertrophic cardiomyopathy did not have either basal (n=7) or apical (n=8) T1 mapping analyzed as the maps were either not performed or were inadequate. These patients were included in the analysis for all available slices but the missing data could have skewed the results. The change in contrast agents, which have different relaxivity, may have affected late gadolinium enhancement or extracellular volume assessment but was unavoidable as the contrast change was made institution-wide. While some studies suggest that late gadolinium enhancement is more apparent with Gadovist than Magnevist, others suggest similar images for the two contrast agents.[29–31] Though there are minimal available data comparing extracellular volume values with Gadovist and Magnevist, the extracellular volume appears to be relatively contrast independent, with no significant difference or differences of questionable clinical significance between contrast agents.[32–34]
CONCLUSIONS
In this cohort of hypertrophic cardiomyopathy, native T1 was increased in hypertrophic cardiomyopathy compared with control. Native T1 also correlated with circumferential strain, suggesting a relationship between structural and functional abnormalities in pediatric hypertrophic cardiomyopathy. A model including left ventricular ejection fraction and native T1 may aid in the diagnosis of pediatric patients with hypertrophic cardiomyopathy.
Acknowledgements
The authors would like to acknowledge the developers of the pulse sequence and reconstruction software: Bruce Spottiswoode, Andreas Greiser, Hui Xue, Christopher Glielmi, Shivraman Giri, and Randall Kroeker.
Financial Support:
Research reported in this publication was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under Award Number K23HL123938 (Bethesda, MD) (Soslow). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The project described was supported by CTSA award No. UL1 TR002243 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
This work was supported by Haley’s Heart Foundation.
Footnotes
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Ethical Standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national guidelines on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008, and has been approved by the Vanderbilt Institutional Review Board.
REFERENCES
- 1.Maron BJ, Doerer JJ, Haas TS, Tierney DM, Mueller FO (2009) Sudden deaths in young competitive athletes: analysis of 1866 deaths in the United States, 1980–2006. Circulation 119 (8):1085–1092. doi: 10.1161/CIRCULATIONAHA.108.804617 [DOI] [PubMed] [Google Scholar]
- 2.Patel AR, Kramer CM (2017) Role of Cardiac Magnetic Resonance in the Diagnosis and Prognosis of Nonischemic Cardiomyopathy. JACC Cardiovasc Imaging 10 (10 Pt A):1180–1193. doi: 10.1016/j.jcmg.2017.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kellman P, Wilson JR, Xue H, Ugander M, Arai AE (2012) Extracellular volume fraction mapping in the myocardium, part 1: evaluation of an automated method. J Cardiovasc Magn Reson 14:63. doi: 10.1186/1532-429X-14-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller CA, Naish JH, Bishop P, Coutts G, Clark D, Zhao S, Ray SG, Yonan N, Williams SG, Flett AS, Moon JC, Greiser A, Parker GJ, Schmitt M (2013) Comprehensive validation of cardiovascular magnetic resonance techniques for the assessment of myocardial extracellular volume. Circ Cardiovasc Imaging 6 (3):373–383. doi: 10.1161/CIRCIMAGING.112.000192 [DOI] [PubMed] [Google Scholar]
- 5.Flett AS, Hayward MP, Ashworth MT, Hansen MS, Taylor AM, Elliott PM, McGregor C, Moon JC (2010) Equilibrium contrast cardiovascular magnetic resonance for the measurement of diffuse myocardial fibrosis: preliminary validation in humans. Circulation 122 (2):138–144. doi: 10.1161/CIRCULATIONAHA.109.930636 [DOI] [PubMed] [Google Scholar]
- 6.Iles LM, Ellims AH, Llewellyn H, Hare JL, Kaye DM, McLean CA, Taylor AJ (2015) Histological validation of cardiac magnetic resonance analysis of regional and diffuse interstitial myocardial fibrosis. Eur Heart J Cardiovasc Imaging 16 (1):14–22. doi: 10.1093/ehjci/jeu182 [DOI] [PubMed] [Google Scholar]
- 7.Child N, Suna G, Dabir D, Yap ML, Rogers T, Kathirgamanathan M, Arroyo-Ucar E, Hinojar R, Mahmoud I, Young C, Wendler O, Mayr M, Sandhu B, Morton G, Muhly-Reinholz M, Dimmeler S, Nagel E, Puntmann VO (2017) Comparison of MOLLI, shMOLLLI, and SASHA in discrimination between health and disease and relationship with histologically derived collagen volume fraction. Eur Heart J Cardiovasc Imaging. doi: 10.1093/ehjci/jex309 [DOI] [PubMed] [Google Scholar]
- 8.Bogarapu S, Puchalski MD, Everitt MD, Williams RV, Weng HY, Menon SC (2016) Novel Cardiac Magnetic Resonance Feature Tracking (CMR-FT) Analysis for Detection of Myocardial Fibrosis in Pediatric Hypertrophic Cardiomyopathy. Pediatr Cardiol 37 (4):663–673. doi: 10.1007/s00246-015-1329-8 [DOI] [PubMed] [Google Scholar]
- 9.Wu LM, An DL, Yao QY, Ou YZ, Lu Q, Jiang M, Xu JR (2017) Hypertrophic cardiomyopathy and left ventricular hypertrophy in hypertensive heart disease with mildly reduced or preserved ejection fraction: insight from altered mechanics and native T1 mapping. Clin Radiol 72 (10):835–843. doi: 10.1016/j.crad.2017.04.019 [DOI] [PubMed] [Google Scholar]
- 10.Haggerty CM, Suever JD, Pulenthiran A, Mejia-Spiegeler A, Wehner GJ, Jing L, Charnigo RJ, Fornwalt BK, Fogel MA (2017) Association between left ventricular mechanics and diffuse myocardial fibrosis in patients with repaired Tetralogy of Fallot: a cross-sectional study. J Cardiovasc Magn Reson 19 (1):100. doi: 10.1186/s12968-017-0410-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siegel B, Olivieri L, Gordish-Dressman H, Spurney CF (2017) Myocardial Strain Using Cardiac MR Feature Tracking and Speckle Tracking Echocardiography in Duchenne Muscular Dystrophy Patients. Pediatr Cardiol. doi: 10.1007/s00246-017-1777-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulz-Menger J, Bluemke DA, Bremerich J, Flamm SD, Fogel MA, Friedrich MG, Kim RJ, von Knobelsdorff-Brenkenhoff F, Kramer CM, Pennell DJ, Plein S, Nagel E (2013) Standardized image interpretation and post processing in cardiovascular magnetic resonance: Society for Cardiovascular Magnetic Resonance (SCMR) board of trustees task force on standardized post processing. J Cardiovasc Magn Reson 15:35. doi: 10.1186/1532-429X-15-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP (2004) Modified Look-Locker inversion recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn Reson Med 52 (1):141–146. doi: 10.1002/mrm.20110 [DOI] [PubMed] [Google Scholar]
- 14.Kellman P, Hansen MS (2014) T1-mapping in the heart: accuracy and precision. J Cardiovasc Magn Reson 16:2. doi: 10.1186/1532-429X-16-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue H, Shah S, Greiser A, Guetter C, Littmann A, Jolly MP, Arai AE, Zuehlsdorff S, Guehring J, Kellman P (2012) Motion correction for myocardial T1 mapping using image registration with synthetic image estimation. Magn Reson Med 67 (6):1644–1655. doi: 10.1002/mrm.23153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soslow JH, Damon BM, Saville BR, Lu Z, Burnette WB, Lawson MA, Parra DA, Sawyer DB, Markham LW (2015) Evaluation of post-contrast myocardial t1 in duchenne muscular dystrophy using cardiac magnetic resonance imaging. Pediatr Cardiol 36 (1):49–56. doi: 10.1007/s00246-014-0963-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simpson SA, Field SL, Xu M, Saville BR, Parra DA, Soslow JH (2017) Effect of Weight Extremes on Ventricular Volumes and Myocardial Strain in Repaired Tetralogy of Fallot as Measured by CMR. Pediatr Cardiol. doi: 10.1007/s00246-017-1793-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerqueira MD, Weissman NJ, Dilsizian V, Jacobs AK, Kaul S, Laskey WK, Pennell DJ, Rumberger JA, Ryan T, Verani MS, American Heart Association Writing Group on Myocardial S, Registration for Cardiac I (2002) Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. A statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation 105 (4):539–542 [DOI] [PubMed] [Google Scholar]
- 19.Moon JC, Messroghli DR, Kellman P, Piechnik SK, Robson MD, Ugander M, Gatehouse PD, Arai AE, Friedrich MG, Neubauer S, Schulz-Menger J, Schelbert EB, Society for Cardiovascular Magnetic Resonance I, Cardiovascular Magnetic Resonance Working Group of the European Society of C (2013) Myocardial T1 mapping and extracellular volume quantification: a Society for Cardiovascular Magnetic Resonance (SCMR) and CMR Working Group of the European Society of Cardiology consensus statement. J Cardiovasc Magn Reson 15:92. doi: 10.1186/1532-429X-15-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raucci FJ Jr., Parra DA, Christensen JT, Hernandez LE, Markham LW, Xu M, Slaughter JC, Soslow JH (2017) Synthetic hematocrit derived from the longitudinal relaxation of blood can lead to clinically significant errors in measurement of extracellular volume fraction in pediatric and young adult patients. J Cardiovasc Magn Reson 19 (1):58. doi: 10.1186/s12968-017-0377-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG (2009) Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 42 (2):377–381. doi: 10.1016/j.jbi.2008.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swoboda PP, McDiarmid AK, Erhayiem B, Law GR, Garg P, Broadbent DA, Ripley DP, Musa TA, Dobson LE, Foley JR, Fent GJ, Page SP, Greenwood JP, Plein S (2017) Effect of cellular and extracellular pathology assessed by T1 mapping on regional contractile function in hypertrophic cardiomyopathy. J Cardiovasc Magn Reson 19 (1):16. doi: 10.1186/s12968-017-0334-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Boomen M, Slart R, Hulleman EV, Dierckx R, Velthuis BK, van der Harst P, Sosnovik DE, Borra RJH, Prakken NHJ (2018) Native T1 reference values for nonischemic cardiomyopathies and populations with increased cardiovascular risk: A systematic review and meta-analysis. J Magn Reson Imaging 47 (4):891–912. doi: 10.1002/jmri.25885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinojar R, Varma N, Child N, Goodman B, Jabbour A, Yu CY, Gebker R, Doltra A, Kelle S, Khan S, Rogers T, Arroyo Ucar E, Cummins C, Carr-White G, Nagel E, Puntmann VO (2015) T1 Mapping in Discrimination of Hypertrophic Phenotypes: Hypertensive Heart Disease and Hypertrophic Cardiomyopathy: Findings From the International T1 Multicenter Cardiovascular Magnetic Resonance Study. Circ Cardiovasc Imaging 8 (12). doi: 10.1161/CIRCIMAGING.115.003285 [DOI] [PubMed] [Google Scholar]
- 25.Puntmann VO, Voigt T, Chen Z, Mayr M, Karim R, Rhode K, Pastor A, Carr-White G, Razavi R, Schaeffter T, Nagel E (2013) Native T1 mapping in differentiation of normal myocardium from diffuse disease in hypertrophic and dilated cardiomyopathy. JACC Cardiovasc Imaging 6 (4):475–484. doi: 10.1016/j.jcmg.2012.08.019 [DOI] [PubMed] [Google Scholar]
- 26.Parekh K, Markl M, Deng J, de Freitas RA, Rigsby CK (2017) T1 mapping in children and young adults with hypertrophic cardiomyopathy. Int J Cardiovasc Imaging 33 (1):109–117. doi: 10.1007/s10554-016-0979-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Messroghli DR, Moon JC, Ferreira VM, Grosse-Wortmann L, He T, Kellman P, Mascherbauer J, Nezafat R, Salerno M, Schelbert EB, Taylor AJ, Thompson R, Ugander M, van Heeswijk RB, Friedrich MG (2017) Clinical recommendations for cardiovascular magnetic resonance mapping of T1, T2, T2* and extracellular volume: A consensus statement by the Society for Cardiovascular Magnetic Resonance (SCMR) endorsed by the European Association for Cardiovascular Imaging (EACVI). J Cardiovasc Magn Reson 19 (1):75. doi: 10.1186/s12968-017-0389-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosmini S, Bulluck H, Captur G, Treibel TA, Abdel-Gadir A, Bhuva AN, Culotta V, Merghani A, Fontana M, Maestrini V, Herrey AS, Chow K, Thompson RB, Piechnik SK, Kellman P, Manisty C, Moon JC (2018) Myocardial native T1 and extracellular volume with healthy ageing and gender. Eur Heart J Cardiovasc Imaging 19 (6):615–621. doi: 10.1093/ehjci/jey034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Cobelli F, Esposito A, Perseghin G, Sallemi C, Belloni E, Ravelli S, Lanzani C, Del Maschio A (2012) Intraindividual comparison of gadobutrol and gadopentetate dimeglumine for detection of myocardial late enhancement in cardiac MRI. AJR Am J Roentgenol 198 (4):809–816. doi: 10.2214/AJR.11.7118 [DOI] [PubMed] [Google Scholar]
- 30.Rudolph A, Messroghli D, von Knobelsdorff-Brenkenhoff F, Traber J, Schuler J, Wassmuth R, Schulz-Menger J (2015) Prospective, randomized comparison of gadopentetate and gadobutrol to assess chronic myocardial infarction applying cardiovascular magnetic resonance. BMC Med Imaging 15:55. doi: 10.1186/s12880-015-0099-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu D, Ma X, Liu J, Zhao L, Chen H, Xu L, Sun Z, Fan Z (2017) Quantitative analysis of late gadolinium enhancement in hypertrophic cardiomyopathy: comparison of diagnostic performance in myocardial fibrosis between gadobutrol and gadopentetate dimeglumine. Int J Cardiovasc Imaging 33 (8):1191–1200. doi: 10.1007/s10554-017-1101-7 [DOI] [PubMed] [Google Scholar]
- 32.Kawel N, Nacif M, Zavodni A, Jones J, Liu S, Sibley CT, Bluemke DA (2012) T1 mapping of the myocardium: intra-individual assessment of post-contrast T1 time evolution and extracellular volume fraction at 3T for Gd-DTPA and Gd-BOPTA. J Cardiovasc Magn Reson 14:26. doi: 10.1186/1532-429X-14-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahsepar AA, Ghasemiesfe A, Suwa K, Dolan RS, Shehata ML, Korell MJ, Naresh NK, Markl M, Collins JD, Carr JC (2019) Comprehensive evaluation of macroscopic and microscopic myocardial fibrosis by cardiac MR: intra-individual comparison of gadobutrol versus gadoterate meglumine. Eur Radiol. doi: 10.1007/s00330-018-5956-3 [DOI] [PubMed] [Google Scholar]
- 34.Kawel N, Nacif M, Zavodni A, Jones J, Liu S, Sibley CT, Bluemke DA (2012) T1 mapping of the myocardium: intra-individual assessment of the effect of field strength, cardiac cycle and variation by myocardial region. J Cardiovasc Magn Reson 14:27. doi: 10.1186/1532-429X-14-27 [DOI] [PMC free article] [PubMed] [Google Scholar]