Mucociliary clearance, the physiological process by which mammalian conducting airways expel pathogens and unwanted surface materials from the respiratory tract, depends on the coordinated function of multiple specialized cell types including basal stem cells, mucus-secreting goblet cells, motile ciliated cells, CFTR-rich ionocytes, and immune cells1,2. Bronchiectasis, a syndrome of pathological airway dilation associated with impaired mucociliary clearance, may occur sporadically or with Mendelian inheritance, such as in cystic fibrosis (CF), primary ciliary dyskinesia (PCD), and select immunodeficiencies3. Prior studies have identified mutations affecting ciliary structure and nucleation in PCD4, but the regulation of mucociliary transport remains incompletely understood and therapeutic targets for modulating it are lacking. Herein we identify a bronchiectasis syndrome caused by inactivating mutations in NEK10, a protein kinase with previously unknown in vivo function in mammals. Genetically modified primary human airway cultures establish NEK10 as a ciliated-cell specific kinase whose activity regulates the motile ciliary proteome to promote ciliary length and mucociliary transport, but which is dispensable for normal ciliary number, radial structure, and beat frequency. Together, these data identify a novel and likely targetable signaling axis which controls motile ciliary function in humans and has potential implications for other respiratory disorders characterized by impaired mucociliary clearance.
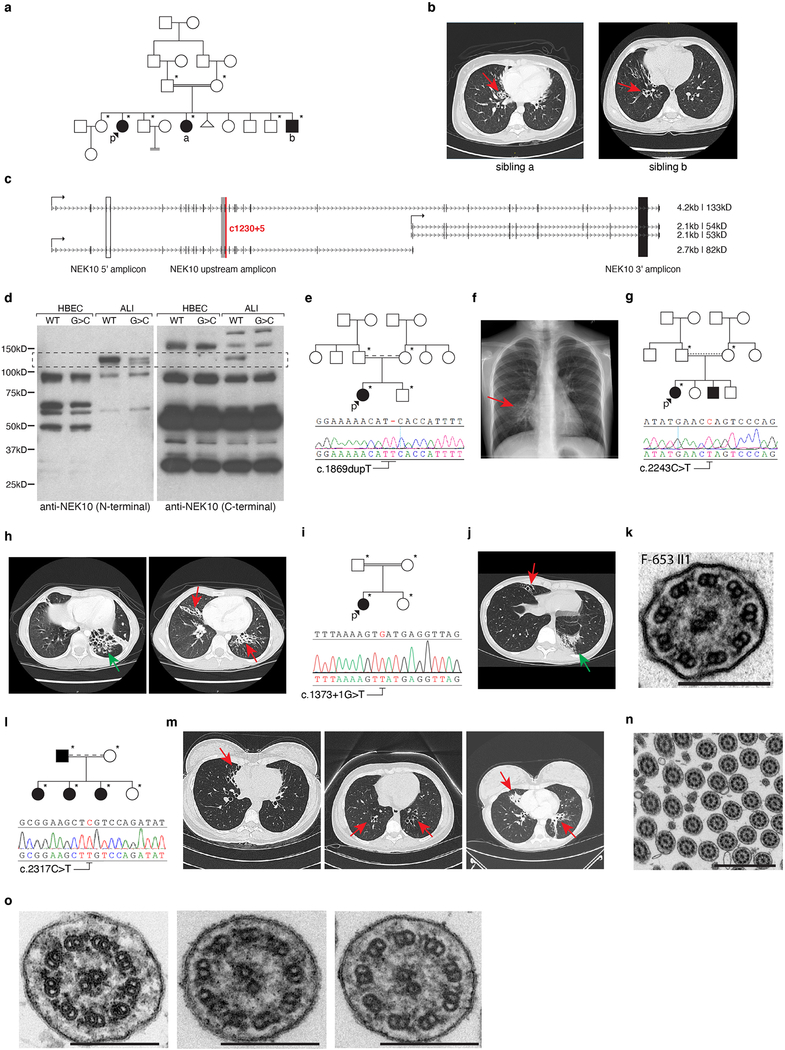
A 31-year-old consanguineous woman was evaluated for idiopathic respiratory failure characterized by neonatal respiratory distress and recurrent bacterial sinopulmonary infections (Extended Data Fig. 1a, Supplementary Table 1). Chest imaging demonstrated extensive pan-lobar bronchiectasis without heterotaxy and nasal biopsies revealed normal ciliary radial ultrastructure (Fig. 1a–1b). Cystic fibrosis and immunodeficiency were ruled out after extensive clinical and genetic testing. Similar but milder findings were present in 2 siblings (Extended Data Fig. 1a–1b, Supplementary Table 1), strongly suggestive of autosomal recessive inheritance. Whole exome sequencing of affected individuals unexpectedly revealed homozygous intronic splice site mutations (NM_152534:c.1230+5G>C, “NEK10G>C” hereafter) in NEK10, a serine/threonine kinase homologous to Aspergillus nidulans nimA (“never in mitosis a”) but not previously implicated in human disease5 (Fig. 1c). The functions of mammalian NEKs remain incompletely characterized; several, including NEK2 and NEK5/6/7/9 function like their fungal ortholog by regulating the cell cycle through phosphorylation of centrosome components and the mitotic spindle6. Mutations in NEK1 and NEK8 cause polycystic kidney phenotypes in mice7,8, in line with a role in regulation of primary cilia. Recent reports have proposed roles for NEK10 in cancer cell DNA damage response9 and in teleost fish nervous system and body axis specification10 but, to date, no published work suggests any roles for NEKs in the respiratory system.
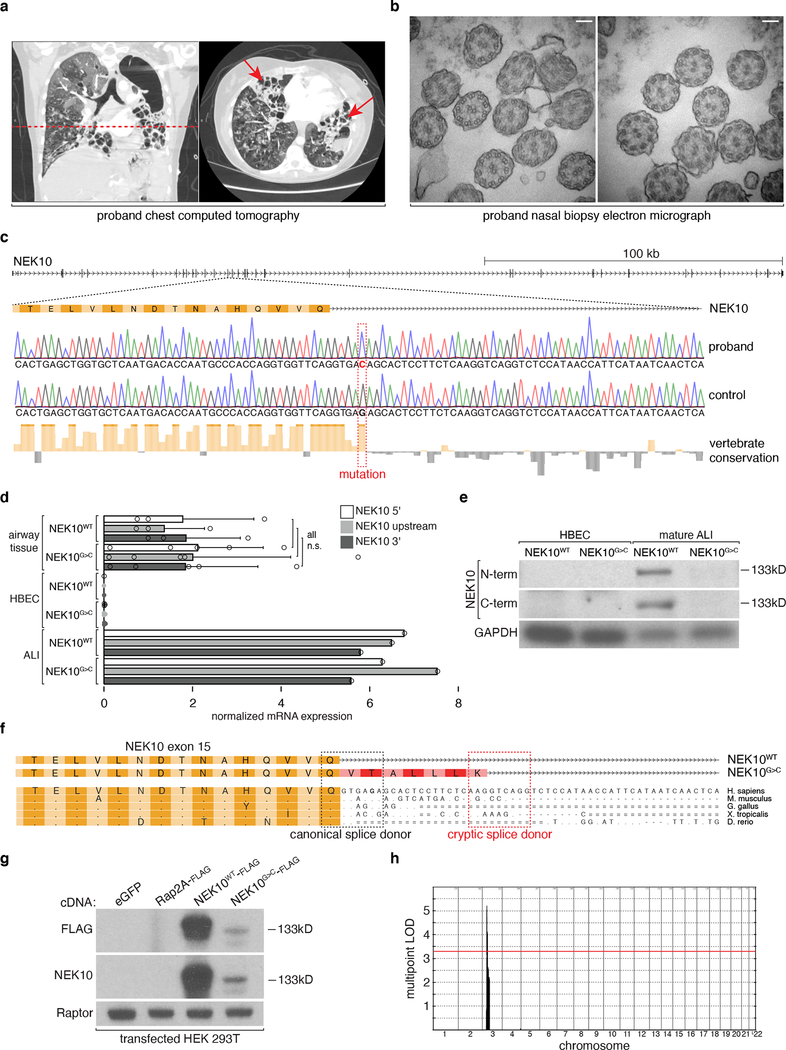
Fig. 1: Familial bronchiectasis associated with NEK10 loss-of-function.
a, Chest computed tomography (CT) imaging of proband 1 upon clinical presentation. Dashed line indicates level of cross-sectional imaging in right panel. Arrows highlight cystic bronchiectatic destruction of lung. b, transmission electron micrograph of proband 1 nasal biopsy specimen demonstrating normal radial ciliary ultrastructure, scale bars indicate 100nm. c, schematic depiction of 3’ terminus of NEK10 exon 15 and following intron, Sanger sequencing traces highlight G>C point mutation (red nucleotide) and high degree of conservation (red dashed box). d, 18S rRNA-normalized relative expression of indicated amplicons; n=3 independent lung tissue donors (controls), n=5 independently isolated lung regions (NEK10G>C), n=3 independently isolated HBEC lines for NEK10G>C, n=1 for remaining samples, mean ±S.D. e, immunoblotting against the indicated proteins from cultured HBECs and ALI, NEK10 immunogen indicated, representative of 3 experiments. f, schematic representation of NEK10 cDNA sequencing results from indicated genotypes, common (yellow) and NEK10G>C specific (red) residues indicated, canonical and cryptic splice donor motifs highlighted. g, immunoblotting after transient transfection of HEK293T cells with the indicated cDNAs, representative of 2 experiments. h, results of genome-wide linkage analysis incorporating individuals (n=15) highlighted with asterisks in (Extended Data Fig. 1a, 1e, 1g), peak bounded by marker SNPs rs13072262 and rs17798444, red line indicates LOD 3.3, equivalent to genome-wide p<0.05. Images in (c) and (f) generated from UCSC genome browser hg19 assembly (http://genome.ucsc.edu).
To begin to study the effects of NEK10G>C in the lung, we isolated and cultured control and proband bronchial epithelial cells (“HBECs”) obtained at the time of bilateral lung transplantation. Although NEK10 mRNA was robustly expressed in airway tissue it was essentially undetectable both in NEK10WT and NEK10G>C HBECs, suggesting its expression might be restricted to mature airway cells (Fig. 1d). We therefore differentiated control and patient-derived HBECs at an air-liquid interface (ALI), a well-validated method for generating airway epithelium in vitro11. Unexpectedly, both control and mutant ALI samples demonstrated robust NEK10 mRNA expression (Fig. 1d, Extended Data Fig. 1c) despite immunoblotting evidence that NEK10G>C encodes a loss of function allele (Fig. 1e, Extended Data Fig. 1d). To elucidate the mechanism by which NEK10G>C impairs protein expression we sequenced full-length cDNAs from mutant ALI, revealing the mutation-dependent in-frame insertion of 7 amino acids which we suspected rendered NEK10 unstable (Fig. 1f). To test this hypothesis we expressed epitope-tagged NEK10 constructs in HEK293T cells and found, as in ALI cultures, that the mutant protein was severely under-expressed - supporting a destabilizing effect of NEK10G>C causing loss-of-function (Fig. 1g).
These findings led us to wonder whether NEK10 mutations might underlie other cases of unexplained bronchiectasis. Indeed, further sequencing revealed 6 additional patients from 4 families harboring homozygous NEK10 mutation and exhibiting bronchiectasis (Extended Data Fig. 1e–1o). The first, an 11-year-old girl, had homozygous c.1869dupT mutation resulting in frameshift and premature stop (His624Serfs*4). The second, a 15-year-old girl, had homozygous c.2243C>T mutation causing substitution of leucine for a highly conserved proline (Pro748Leu) within the kinase domain. The third, a 23-year-old woman, had homozygous c.1373+1G>T mutation causing exon skipping, frameshift, and premature stop (Cys437Thrfs*9). The fourth, fifth, and sixth were siblings with homozygous c.2317C>T mutation resulting in mutation of a highly conserved arginine to cysteine (Arg773Cys). Notably, clinical assays revealed normal nasal ciliary ultrastructure by EM, normal nasal nitric oxide (nNO) levels, no evidence of heterotaxy, and only very subtle abnormalities by clinical high-speed video-microscopy (HSVM) in all tested individuals, suggesting such patients might escape detection during standard PCD evaluation12 (Supplementary Table 1). Linkage analysis incorporating kindreds 1–3 yielded a single, highly significant genome-wide linkage signal (pLOD 5.2), defining a 7.3Mb interval which includes the NEK10 locus (Fig. 1h). Conversely, healthy individuals with biallelic inactivating NEK10 mutation are completely absent from publicly available variant databases13. Together, these data provide strong genetic and clinical evidence that NEK10 represents a novel and bona fide autosomal recessive bronchiectasis locus.
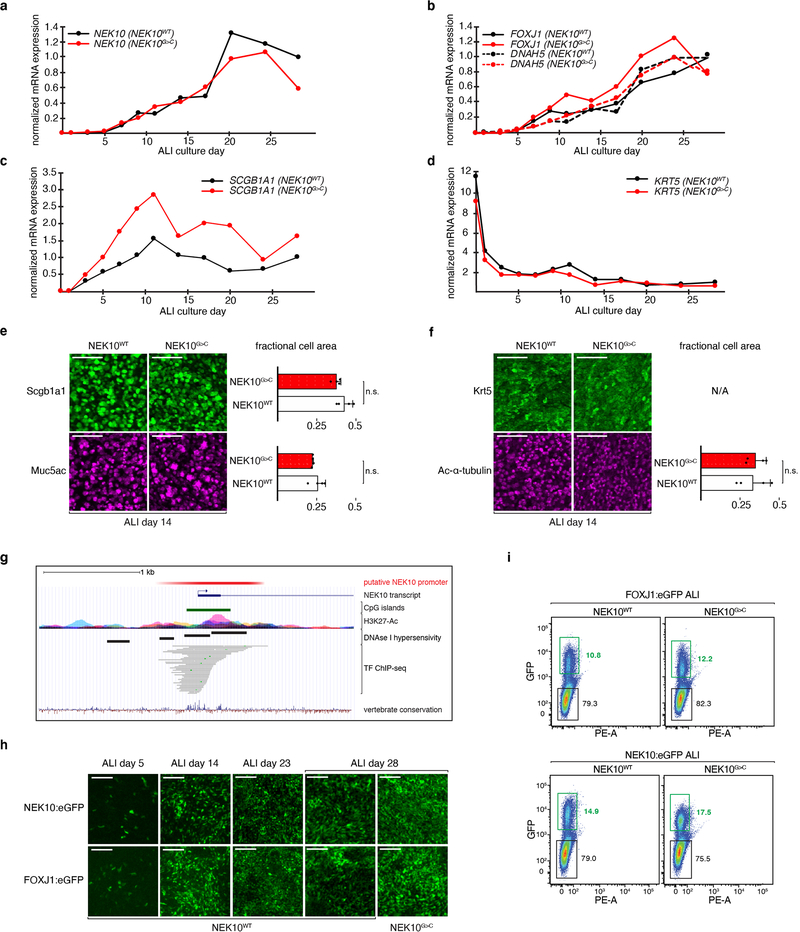
Given NEK roles in cell cycle regulation, we initially hypothesized that NEK10 is required for the specification of one or more cell types involved in mucociliary clearance. Quantitative reverse-transcriptase PCR (qRT-PCR) revealed that NEK10 is robustly induced during airway epithelial differentiation, coincident with the acquisition of markers for ciliated and secretory cells and the depletion of stem cell markers (Extended Data Fig. 2a–2d). However, NEK10G>C cultures induced and repressed these markers with identical kinetics as control and produced similar numbers of secretory, goblet, and multiciliated cells (MCCs, Extended Data Fig. 2e–2f). These data suggested NEK10 might instead function specifically within one of the specialized cells important for mucociliary clearance. To elucidate its expression domain, we generated ALI cultures in which eGFP is expressed under the control of the NEK10 promoter (NEK10:eGFP), which we found directs expression within a subset of ALI cells in a differentiation-dependent manner (Extended Data Fig. 2g–2i). FACS-purified GFP+ cells from mature NEK10:eGFP ALI revealed a 149-fold enrichment of the MCC marker FOXJ114 with reciprocal depletion of secretory and basal cell marker transcripts (Fig. 2a), while confocal imaging confirmed GFP positivity restricted to cells harboring apical cilia (Fig. 2b). Conversely, FACS-purified ciliated cells from FOXJ1:eGFP ALI enriched NEK10 152-fold (Extended Data Fig. 2h–2i, Fig. 2a). These findings establish NEK10 as a ciliated cell-specific gene in human airway induced during, but dispensable for, differentiation of this cell type.
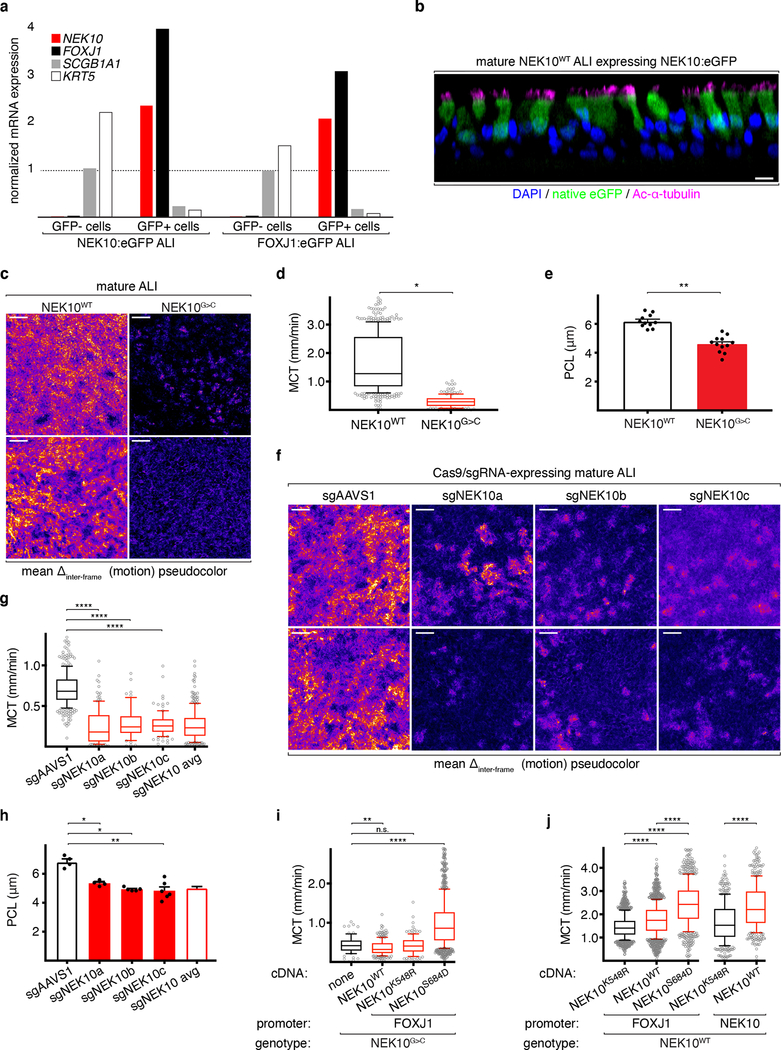
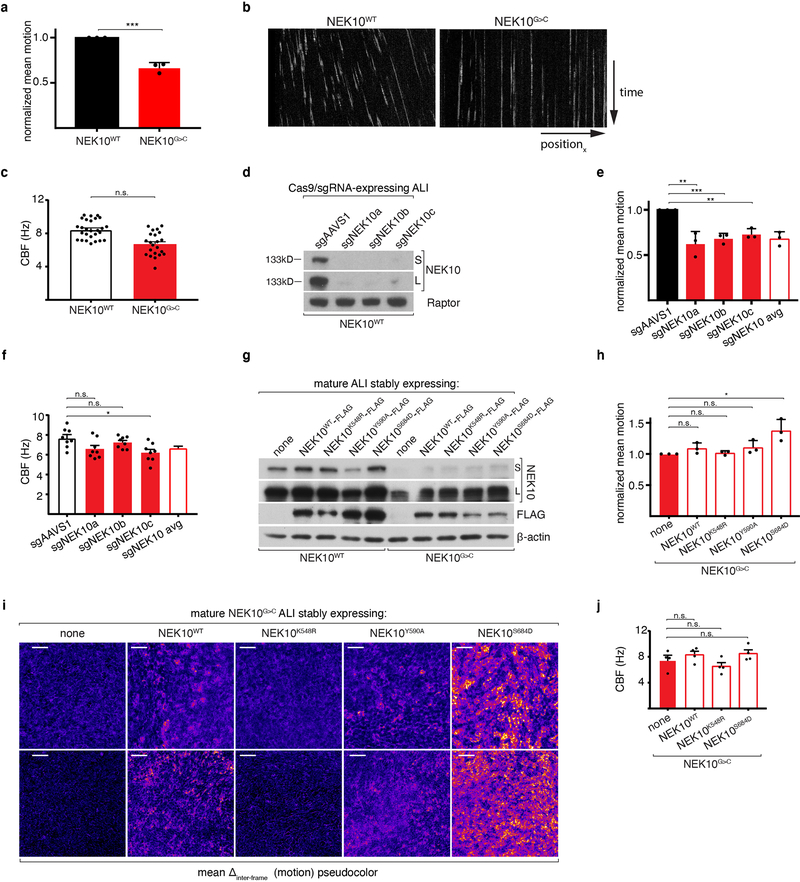
Fig. 2: NEK10 is a ciliated cell-specific gene required for effective mucociliary transport.
a, 18S rRNA-normalized relative expression of indicated transcripts from FACS-sorted ALI cells; dashed line indicates expression level from unsorted mature ALI. b, confocal immunofluorescence of GFP in ciliated cells in NEK10p:eGFP ALI, representative of 2 independent ALI differentiations, scale bar 10μm. c, pseudocolored video microscopy of ALI of the indicated genotypes, representative of 3 independent ALI differentiations, scale bars 50μm. d, MCT (μOCT) of mature ALI of the indicated genotypes, n=485 (NEK10WT), 180 (NEK10G>C) pooled from 3 independent ALI differentiations, plot indicates median (center line), 25th/75th percentiles (box), 10th/90th (whiskers) percentiles, and remaining points (open circles). e, PCL (μOCT) of ALI of the indicated genotypes, n=11 (NEK10WT), 12 (NEK10G>C) pooled from 3 independent ALI differentiations, mean ±S.E.M. f, pseudocolored video microscopy of CRISPR/Cas9-edited ALI, representative fields from 3 independent ALI differentiations, scale bars 50μm. g, MCT of CRISPR/Cas9-edited ALI, n=361 (sgAAVS1), 131 (sgNEK10a), 59 (sgNEK10b), 104 (sgNEK10c) pooled from 3 independent ALI differentiations, plotted as in (g). h, PCL of CRISPR/Cas9-edited ALI, n=4 (sgAAVS1), 4 (sgNEK10a), 5 (sgNEK10b), 6 (sgNEK10c) pooled from 3 independent ALI differentiations, mean ±S.E.M. i, MCT of NEK10G>C ALI expressing the indicated cDNAs, n=71 (no cDNA), 254 (NEK10WT), 129 (NEK10K548R), 1081 (NEK10S684D), pooled from 3 independent ALI differentiations, mean ±S.E.M. j, MCT of NEK10WT ALI expressing the indicated cDNAs, n=1385 (FOXJ1:NEK10K548R), 1624 (FOXJ1:NEK10WT), 728 (FOXJ1:NEK10S684D), 401 (NEK10:NEK10K548R), 426 (NEK10:NEK10WT) pooled from 3 independent ALI differentiations, plotted as in (g). *p≤0.05, **p≤0.01, ****p≤0.0001
To explore what functional roles NEK10 plays in MCCs, we imaged live ALI cultures with high-framerate phase contrast microscopy, exposing a striking reduction in overall ciliary motion in NEK10G>C cultures (Fig. 2c, Extended Data Fig. 3a, Supplementary Movie 1). Given this abnormality we performed advanced MCC functional phenotyping using micro-optical coherence tomography (μOCT), a state of the art high-resolution live imaging technique capable of quantitatively interrogating multiple ciliary parameters15. Unlike control cultures, which robustly transport polystyrene beads added to the apical ALI surface, we noted near-absence of mucociliary transport (MCT) in NEK10G>C ALI (Fig. 2d, Extended Data Fig. 3b, Supplementary Movie 2), as well as reduction in depth of the periciliary liquid layer (PCL, Fig. 2e). We were surprised to measure normal ciliary beat frequency in mutant ALI (Extended Data Fig. 3c), again suggesting a mode of dysfunction distinct from classical PCD. While these data suggested a causative role for NEK10 loss-of-function in MCC dysfunction, they could not rule out that secondary genetic or acquired changes in proband-derived cells were in fact responsible16. We therefore generated CRISPR/Cas9-mediated NEK10 loss-of-function ALI cultures (“NEK10KO” hereafter) by genetically disrupting the NEK10 locus in wild-type HBECs. Immunoblotting confirmed efficient depletion of NEK10 while live microscopy of NEK10KO ALI revealed, as in NEK10G>C, a dramatic reduction in ciliary motion (Fig. 2f, Extended Data Fig. 3d–3e). We also subjected NEK10KO ALI to μOCT imaging and found severe reduction in maximal particle transport velocity and thinning of the PCL, again without change in CBF (Fig. 2g–2h, Extended Data Fig. 3f).
Given a recent report of catalysis-independent roles for NEKs17, we sought to understand whether kinase activity per se is necessary for NEK10 function in MCCs. We therefore generated NEK10G>C ALI with FOXJ1 promoter-driven re-expression of NEK10WT or, alternatively, point mutants predicted to inactivate18 (NEK10K548R) or putatively hyperactivate (NEK10S684D, NEK10Y590A) activity based on prior studies of paralogous NEK kinases6,19,20. Despite sub-physiological expression of transduced NEK10 variants, live microscopy demonstrated a striking increase in motility upon expression of NEK10S684D, partial rescue with NEK10WT and NEK10Y590A, but no effect with catalytic-dead NEK10K548R (Extended Data Fig. 3g–3i). μOCT demonstrated that CBF was again unaffected by NEK10 status but that particle transport was significantly increased upon expression of NEK10S684D (Fig. 2i, Extended Data Fig. 3j). These data led us to ask whether ectopic activation of NEK10 signaling in wildtype ALI cultures could augment mucociliary transport to supraphysiological levels. Indeed, expression of NEK10WT or NEK10S684D under the control of the FOXJ1 promoter significantly increased transport velocity compared to catalytic-dead NEK10K548R, a phenotype also reproduced with the more transcriptionally active NEK10 promoter (Fig. 2j). Together, these data establish (1) a ciliated cell-specific NEK10 kinase activity requirement for effective airway mucociliary transport, (2) that NEK10 activity is constrained by the serine 684 activation loop residue, and (3) that potentiating NEK10 activity may represent a strategy for augmenting mucociliary transport.
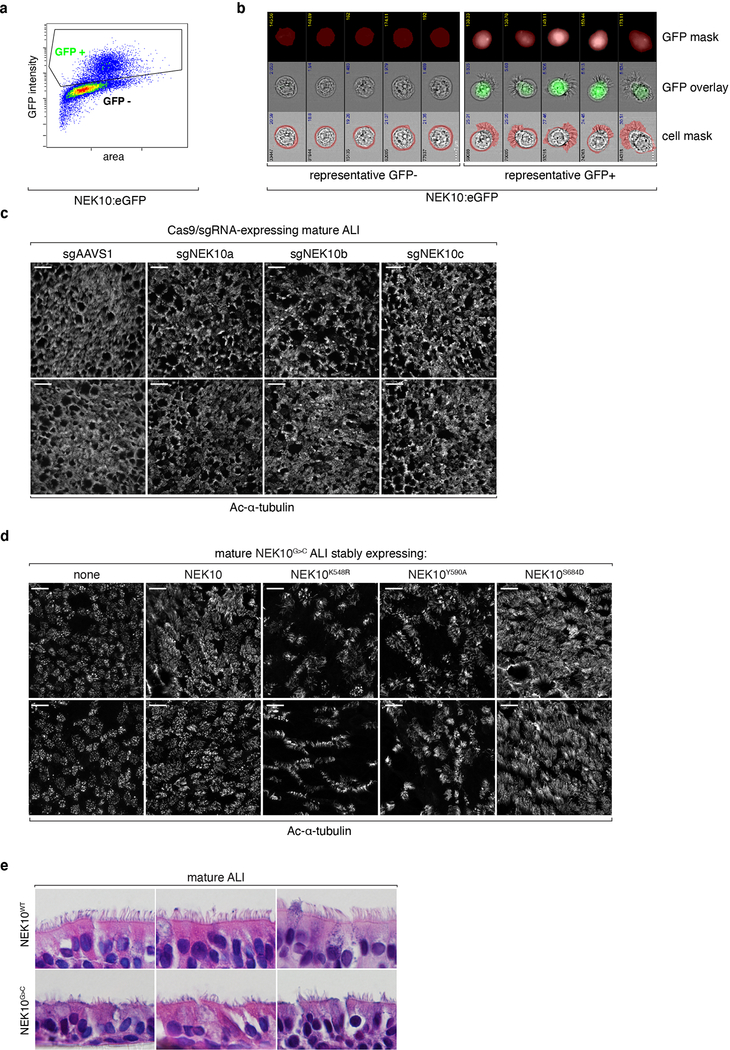
We next turned our attention to understanding the mechanisms by which NEK10 regulates MCC function. To evaluate whether mutant MCCs harbor some physical abnormality we employed imaging flow cytometry (IFC), which allows the capture of thousands of single cells for statistically robust morphology analysis21. After gating singlet MCCs from NEK10:eGFP ALI (Extended Data Fig. 4a–4b) we measured per-cell ciliary area as well as ciliary zone thickness, revealing a clear and statistically significant reduction in both parameters in mutant MCCs (Fig. 3a–3c). Representative single cells corroborated this finding, demonstrating a hypoplastic ciliary layer in NEK10G>C MCCs (Fig. 3d). To validate this finding using an orthogonal method, we subjected NEK10G>C ALI to confocal IF against acetylated α-tubulin. We again found mutant ALI harbored strikingly abnormal, hypoplastic-appearing cilia (Fig. 3e). NEK10KO ALI phenocopied this morphology while NEK10S684D-complemented NEK10G>C ALI reversed it (Extended Data Fig. 4c–4d), indicating this phenotype is specifically attributable to NEK10 kinase activity. As previously described human mutations22,23 are known to impair the nucleation and density of motile cilia, we evaluated MCC basal body density by IF, but found no reduction to account for the NEK10G>C phenotype (Fig. 3f). In light of links between planar cell polarity (PCP) and ciliogenesis24, we also confirmed that NEK10 activity is dispensable for MCC planar polarization (Fig. 3g). Finally, in order to validate ALI culture findings in human patients, we compared airway tissue from the explanted lungs of the proband with airway from non-bronchiectatic as well as end-stage CF patients. As in ALI cultures (Extended Data Fig. 4e), histological analysis revealed ciliary hypoplasia only in NEK10 mutant airway (Fig. 3h) – indicating this phenotype does not reflect a non-specific consequence of severe bronchiectasis and suggesting that NEK10 deficiency produces short motile cilia in vivo, a previously undescribed human genetic phenotype.
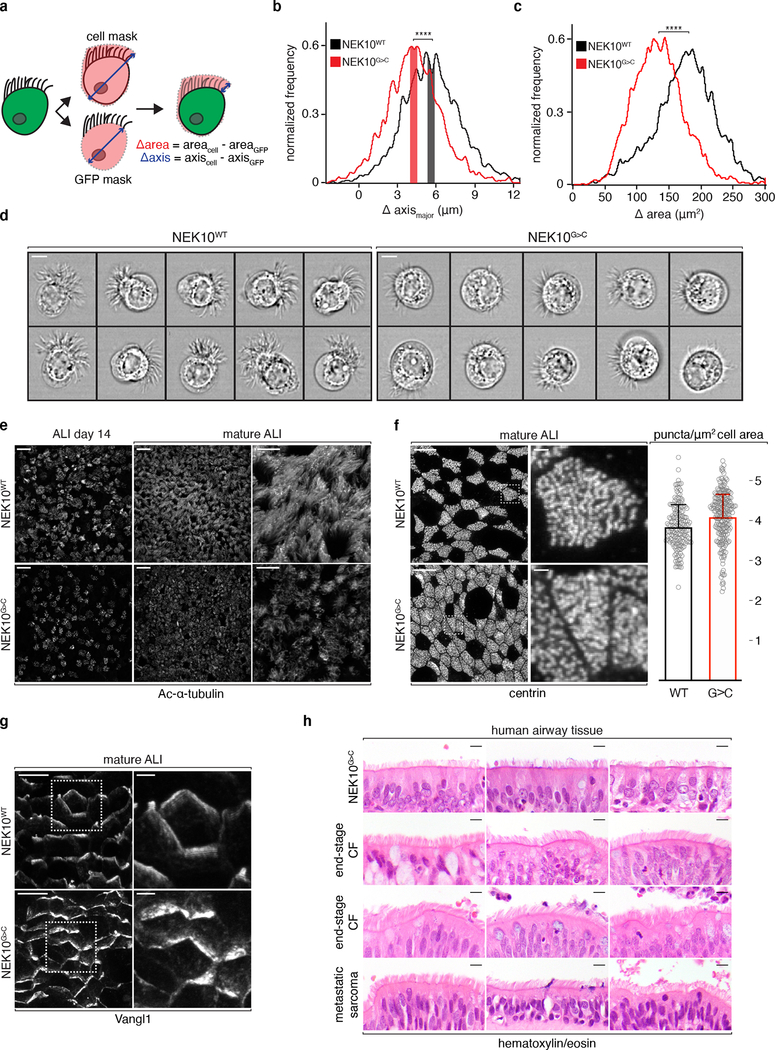
Fig. 3: Morphologically abnormal ciliated cells in NEK10-deficient airway.
a, schematic masking workflow for IFC morphological analysis. b, histogram of ciliary zone thickness of mature ALI MCCs of the indicated genotypes, n=4108 (NEK10WT), 3513 (NEK10G>C), shaded bars indicate medians ±0.25μm. c, histogram of ciliary area of mature ALI MCCs of the indicated genotypes, n=4108 (NEK10WT), 3513 (NEK10G>C). d, single cell images taken from the shaded regions in (b), scale bars 7μm. e, confocal maximum intensity projections (MIPs) of ALI of the indicated genotype and maturity following IF against Ac-α-tubulin, representative of 3 independent ALI differentiations, scale bars 25μm (left 4 panels) and 10μm (right 2 panels). f, confocal MIPs of mature ALI after IF against basal body marker centrin, dashed boxes mark full resolution regions in middle panels, scale bars 10μm (left 2 panels) and 1μm (middle 2 panels); column graph: centrin puncta per μm2 (mean ±S.D.) of ciliated cell surface area, n=71 cells and 10,855 puncta (NEK10WT), 38 cells and 5,369 puncta (NEK10G>C) pooled from 4 independent ALI differentiations. g, confocal MIPs of mature ALI after IF against PCP marker Vangl1, dashed boxes mark full resolution regions in right panel, scale bars 10μm (left panels) and 2.5μm (right panels), representative of 3 independent ALI differentiations. h, hematoxylin/eosin stained human large airway tissue; upper 3 samples taken from lung explants during transplantation for end-stage bronchiectasis due to the indicated etiologies, 4th sample from patient undergoing resection for an unrelated diagnosis, scale bars 5μm. ****p≤0.0001.
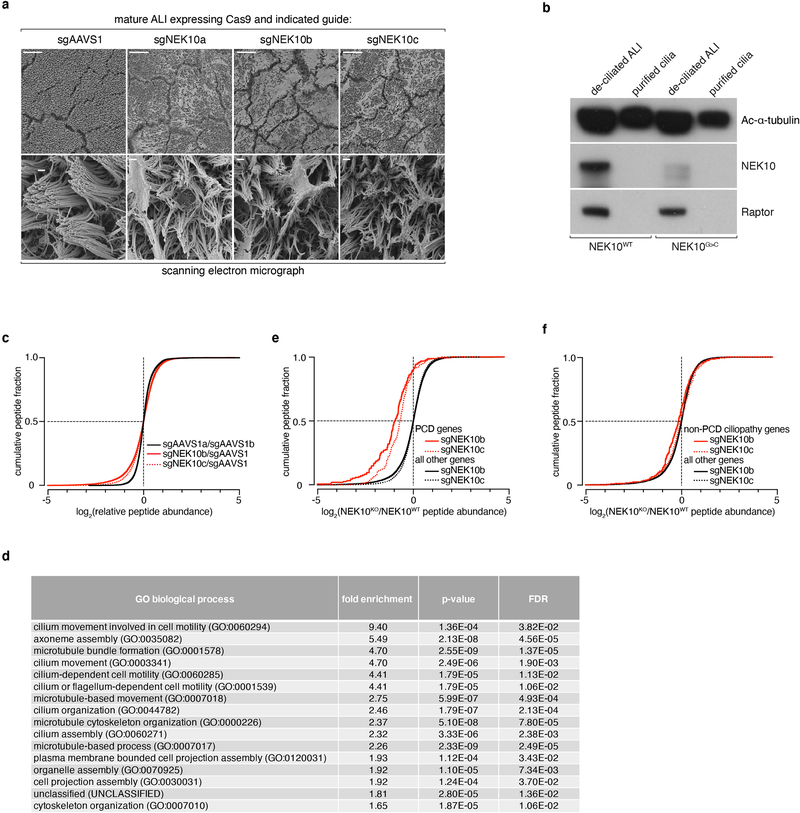
As the size of motile cilia limits their structural analysis by light microscopy, we next subjected ALI preparations to 3 complementary modes of electron microscopic (EM) analysis. Scanning electron microscopy (SEM) of intact ALI, like IF, revealed NEK10G>C and NEK10KO ALI to harbor morphologically abnormal cilia of heterogeneous length with excess mucus accumulation (Fig. 4a, Extended Data Fig. 5a). EM imaging of sectioned ALI cultures revealed an evident decrease in ciliary length in NEK10G>C ALI (Fig. 4b), but not undocked centrioles as in ciliary aplasia syndromes or ciliary tip “ballooning” as reported with certain intraflagellar transport (IFT) defects22,23. In order to precisely quantitate this length defect, we biochemically isolated axonemes from ALI cultures25 (Extended Data Fig. 5b) and subjected these preparations to negative stain EM followed by measurement of individual cilia. Isolated NEK10G>C cilia were indeed short (Fig. 4c–4d, 7.86±1.06μm vs. 6.24±1.26μm), providing at least a partial basis for ciliary transport failure based on biophysical models for mucociliary transport which include ciliary length as a critical parameter in force generation26–28.
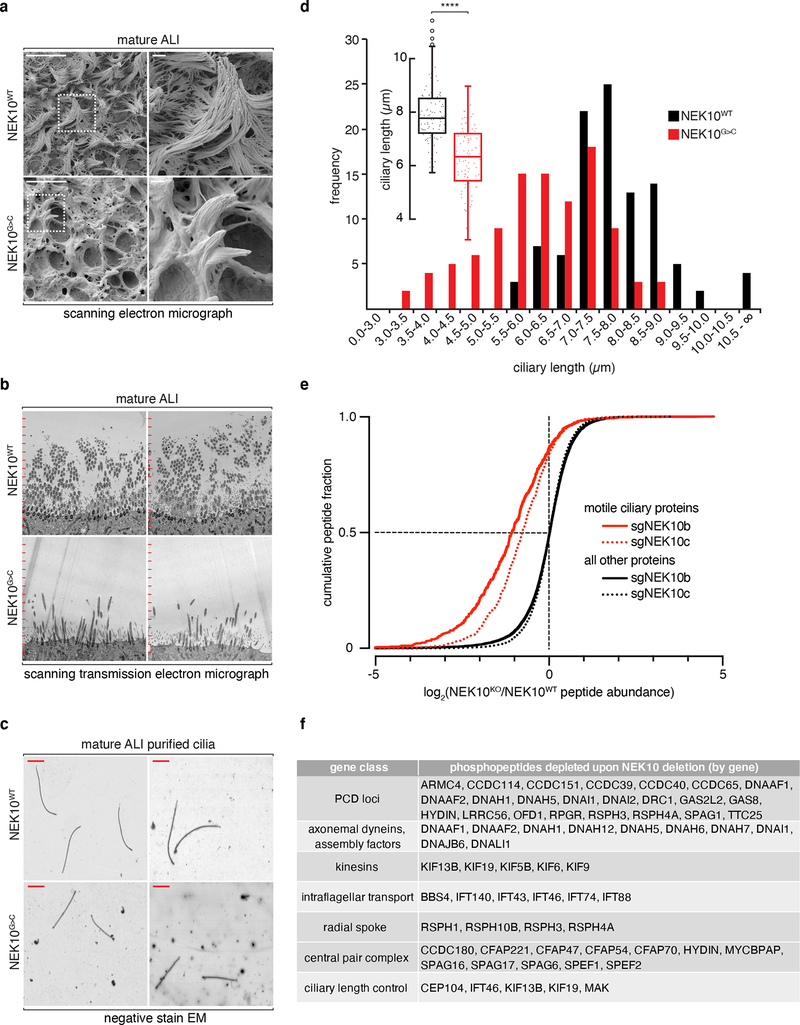
Fig. 4: NEK10 regulates ciliary length through widespread effects on the ciliary proteome.
a, SEM of mature ALI of the indicated genotype, dashed boxes mark full resolution regions in right panel, scale bars 10μm (left panels) or 1μm (right panels), representative of 3 independent ALI differentiations. b, STEM of mature ALI of the indicated genotype after embedding and sectioning orthogonal to the epithelial surface, tick marks spaced at 1μm, representative of 3 independent ALI differentiations. c, representative negative stain EM grids prepared from purified cilia of the indicated genotypes, red scale bar indicates 1μm, representative of 2 independent ALI differentiations. d, histogram of ciliary length from purified cilia of the indicated genotypes, n=101 (NEK10WT), 102 (NEK10G>C) pooled from 2 independent ALI differentiations; inset: box-and-whisker plot of these data, center-line indicates median, box bounds 25th and 75th percentile, whiskers indicate 1.5*IQR, circles indicate outliers. e, cumulative distribution of phosphopeptides by log2 fold change, previously identified motile ciliary proteins in red, all other detected proteins in black, sgNEK10b and sgNEK10c are independently targeting guide RNAs validated in (Extended Data Fig. 3d). f, Table of ciliary genes by functional class with phosphopeptides depleted ≥2-fold upon NEK10 deletion. ****p≤0.0001
In order to explore the biochemical functions of NEK10 in airway epithelium, we next performed iron-enrichment proteomics29 (LC-IMAC MS2), identifying a set of depleted phosphopeptides upon NEK10 inactivation in ALI (Extended Data Fig. 5c). Gene ontology analysis30,31 revealed these depleted peptides to be highly enriched for ciliary motility and axonemal assembly genes, suggesting a large complement of ciliary proteins are dysregulated upon NEK10 loss (Extended Data Fig. 5d). To test this hypothesis directly, we analyzed the effects of NEK10 deletion on the set of proteins previously proteomically identified in airway cilia32, exposing a striking and highly significant depletion of such ciliary phosphopeptides (median log2 fold change −1.06 (sgNEK10b), −0.771 (sgNEK10c), Fig. 4e). Notably, while we observed similar findings upon analysis of peptides mapping to annotated PCD genes, we saw little to no depletion of peptides from non-PCD ciliopathy genes, consistent with NEK10 specifically regulating motile ciliogenesis (Extended Data Fig. 5e–5f). Peptides from virtually all classes of motile ciliary genes were depleted in NEK10KO ALI (Fig. 4f) including axonemal motors, IFT components, central pair constituents, and ciliary length control proteins - indicating that a diverse array of the ciliary proteome is directly or indirectly dysregulated upon NEK10 loss and providing a rich data set for future efforts to dissect its target network.
In summary, here we describe a novel human disease caused by NEK10 deficiency and characterized by pathologically short motile cilia as evidenced by multiple orthogonal assays in physiologically-relevant human airway preparations. In light of NEK10’s highly cell-type restricted expression in our own data and that of recently generated cell atlases2,33,34 as well as the absence of extra-respiratory phenotypes in our patients, this gene does not appear essential for primary ciliogenesis in humans as has been previously proposed10, instead likely specifically controlling MCC function. Indeed, as protozoan beating cilia35 and flagella36–38 length are regulated by distant NEK homologs, analogous pathways likely operate in metazoan MCCs to tune ciliary function to physiological needs. While the central role of NEK10 in potentiating mucociliary clearance is clear from our study, the mechanistic basis for this activity remains to be fully elucidated as our current proteomic data cannot unambiguously distinguish direct phosphorylation effects from secondary protein abundance changes. We are optimistic that future work will clarify the full repertoire of direct and indirect NEK10 targets in ciliated cells.
In parallel with these scientific insights, our work has potential implications for the diagnosis and treatment of bronchiectasis. In particular, the identification of multiple NEK10-mutant kindreds in whom ciliary EM, nNO, and HSVM are all essentially normal suggests standard algorithms for exonerating PCD may fail to capture such patients and others with mechanistically similar motile ciliopathies. Indeed, as prior studies have suggested ciliary shortening (for example due to smoking26,39) may play a role in impaired mucociliary clearance in vivo, our results suggest this relatively subtle histological abnormality should be more regularly evaluated for in patients with otherwise unexplained mucus clearance deficiencies, as it may be more common than currently appreciated. Finally, as this report is the first directly implicating a kinase mutation in ciliary dyskinesia, it opens the door both to understanding new regulatory networks in ciliated cells and, based on this understanding, to potentially targeting this signaling axis in more common diseases of mucociliary clearance where promotion of mucociliary transport may be therapeutically beneficial26,40.
Methods
Whole exome sequencing and clinical phenotyping
Clinical WES was performed on kindreds 1–3 by the Molecular Diagnostics Laboratory of the King Faisal Specialist Hospital and Research Center (KFSHRC). Exome enrichment was performed using the Agilent SureSelect Target Enrichment workflow prior to high-throughput sequencing on the Illumina HiSeq 2500 system. Greater than 30X coverage of 95% of the target regions was obtained for all samples. Exome sequences were mapped to the UCSC hg19 reference sequence with a custom pipeline and interrogated for variants incorporating databases customized to Arab populations. Sequencing of proband siblings and additional kindreds was performed under a protocol approved by the KFSHRC IRB (REC#2121053). Sequencing and analysis of kindred 4 was performed under a protocol approved by University Children’s Hospital Muenster IRB AZ 2015–104-f-S. Sequencing and analysis of kindred 5 was performed under a protocol approved by UNC Chapel Hill IRBs 05–2979 and 13–2348.
Human bronchial epithelial cell (HBEC) and air-liquid interface (ALI) tissue culture
Control human samples were obtained from discarded lung allografts under a protocol approved by the Partners Human Research Committee (IRB #2012P001079). Proband samples were obtained at the time of bilateral lung transplantation under a protocol approved by the Partners Human Research Committee (IRB #2013P002332) with informed consent obtained prior to organ explantation. Airway cells were obtained as previously described41. Briefly, bronchial tube sections were rinsed in MEM supplemented with DTT and DNAse I prior to overnight incubation in MEM supplemented with pronase, DNAse I, antibiotics, and antifungals. Epithelial sheets were further dissociated with Accutase and plated into PneumaCult Ex-Plus expansion media (StemCell Technologies #05040). All plates were pre-coated with sterile-filtered, laminin-rich conditioned media (DMEM + 10% FBS) of the 804G rat bladder cell line to promote HBEC adhesion. HBECs were used between passage 2 and 5 for experiments and were dissociated for sub-culturing with TrypLE Select (Gibco 12563011). ALI cultures were established using 24-well (Corning 3470) or 6-well (Corning 3412) plates coated with 804G-conditioned media. At confluence, apical media was removed and basolateral media was changed to PneumaCult ALI (StemCell Technologies #05001) for 4 to 6 weeks of differentiation (“mature ALI”), except where stated otherwise in the text. Media was exchanged every 48 hours and cultures were washed with PBS weekly beginning on ALI day 14 on a plate shaker at 600rpm x2.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted with Trizol reagent (Invitrogen) according to the manufacturer’s protocol before first strand cDNA synthesis with the SuperScript VILO system (Invitrogen). Quantitative PCR was performed using PowerUp SYBR Green reagent (ABI) on an ABI QuantStudio6 instrument. All analyses were performed with standard curve-based quantitation and normalization to 18S rRNA abundance. 2-tailed Student’s t-test was performed for analysis of statistical significance. qRT-PCT primer sequences available in Supplementary Table 2 and were designed using Primer3Plus software to span exon-exon junctions in all cases.
Immunoblotting
Protein lysates were prepared in lysis buffer containing 1% Triton X-100, 10 mM β-glycerol phosphate, 10 mM pyrophosphate, 40 mM HEPES pH 7.4, 2.5 mM MgCl2, and one mini tablet of EDTA-free protease inhibitor (Roche) per 10mL. Lysates were subjected to SDS-PAGE electrophoresis and transferred to PVDF membranes before immunoblotting with the indicated antibodies. Primary antibodies and working dilutions were as follows: rabbit anti-NEK10 (Sigma HPA038941, lot R35857, 1:1000), mouse anti-NEK10 (Sigma WH0152110M1, lot 09058–1C9, 1:1000), rabbit anti-GAPDH (Abcam ab9485, 1:2500), mouse anti-FLAG M2 (Sigma F1804, lot SLBS3530V, 1:1000), rabbit anti-Raptor (Millipore 09–217, lot 3236353, 1:1000), mouse anti-β-actin (Santa Cruz sc-47778, lot K1718, 1:1000), mouse anti-acetylated α-tubulin (Sigma T7451, 1:1000). Secondary antibodies and dilutions were as follows: HRP-conjugated anti-rabbit IgG (Cell Signaling Technologies #7074, 1:3000), HRP-conjugated anti-mouse IgG (Cell Signaling Technologies #7076, 1:3000).
NEK10 cDNA cloning
Total RNA was extracted with Trizol reagent according to the manufacturer’s protocol before reverse transcription with the SuperScript IV system (Invitrogen) using oligo-dT priming. Full-length NEK10 cDNAs were amplified with Platinum SuperFi polymerase (Invitrogen), cloned into pCR-Blunt II-TOPO (Invitrogen) according to the manufacturer’s protocol, and individual clones sequenced completely. Consensus control and NEK10G>C sequences cloned from each genotype were utilized for further experiments as indicated in the text. Cloning primers available in Supplementary Table 2.
Linkage analysis
Genomic DNA was extracted from whole blood using a standard protocol. Genome-wide genotypes were obtained using an Affymetrix Axiom SNP Chip platform (Affymetrix, Santa Clara, CA) following the manufacturer’s instructions. Blocks of homozygosity were identified using AutoSNPa (dna.leeds.ac.uk/autosnpa/). Linkage analysis was performed on the SNP genotypes using the Allegro component of EasyLinkage software42. Statistical significance was assessed using a genome-wide LOD threshold of 3.3, corresponding to a type I error of 5%.
Immunofluorescence microscopy (IF)
ALI samples were washed in PBS, fixed for 15 minutes in 4% paraformaldehyde (PFA), washed thrice in PBS, and cut free from their plastic supports. For centrin staining only, samples were fixed instead in ice-cold methanol for 15 minutes at −20ºC but otherwise processed identically. ALI membranes were then blocked (5% donkey serum + 0.3% Triton X-100) for 1 hour at room temperature before incubation with the indicated primary antibodies overnight at 37ºC in dilution buffer (1% BSA + 0.3% Triton X-100). Membranes were washed 4 times in wash buffer (PBS + 0.1% Triton X-100) before probing with fluorophore conjugated secondary antibodies for 1 hour at 37ºC in dilution buffer. DAPI was added to the secondary antibody solution for nuclear counterstaining. Membranes were washed 4 additional times in wash buffer and once in PBS before mounting in Fluoromount-G (Southern Biotech). Confocal images were obtained with an Olympus FV10i confocal laser-scanning microscope with a 60× oil objective.
The following primary antibodies were used: mouse anti-acetyl-α-tubulin (gift from J. Rajagopal, Sigma T6793, 1:1000), mouse anti-MUC5AC (gift from J. Rajagopal, ThermoFisher MS-145, lot 145p1709c, 1:500), goat anti-CCSP (gift from B. Stripp, no lot data (non-commercial), 1:5000), chicken anti-KRT5 (gift from J. Rajagopal, Biolegend 905901, 1:500), rabbit anti-CETN1 (gift from I. Cheeseman, no lot data (non-commercial), 1:5000), rabbit anti-VANGL1 (gift from J. Rajagopal, Sigma HPA025235, lot c101664, 1:500). All secondary antibodies were Alexa Fluor conjugates used at 1:500 dilution (Life Technologies): gt anti-chicken 488 (A-11039, lot 1599396), dk anti-mouse 488 (A-21202, lot TF271737), dk anti-rabbit 488 (A-21206, lot TI271741), dk anti-mouse 594 (R37115), dk anti-rabbit 594 (R37119, lot TI271728).
Assessment of SCGB1A1-, MUC5AC-, and acetyl-α-tubulin-positive area was performed using automated scripts for object segmentation in ImageJ using the FIJI implementation43 and null hypothesis testing was performed using the 2-tailed Student’s t-test. KRT5-positive area could not be quantitated as basal cells form an essentially contiguous layer. Centrin puncta were quantitated using automated scripts in ImageJ/FIJI which counted local maxima within ciliated cells. Basal body density was then calculated by dividing total puncta by cell number. Source code for scripts available on request. Null hypothesis testing was performed using the 2-tailed Student’s t-test.
Lentivirus cloning, production, and HBEC infection
NEK10:eGFP and FOXJ1:eGFP vectors were generated by amplifying the respective promoter regions from human genomic DNA and, using NEBuilder Gibson assembly (New England Biolabs), replacing the existing CMV promoter of the pLJM1 construct (Addgene #19319). The putative NEK10 promoter was identified as a conserved ~1.6kb region bounding the NEK10 transcription start site and harboring epigenetic marks consistent with promoter function. The FOXJ1 promoter sequence was cloned using a previously described14 region as a guide. Gibson assembly was also used to generate FOXJ1 promoter-driven lentiviral expression vectors by replacing the pLJC2 vector (Addgene #87974) CMV promoter. NEK10 mutants were generated using a site-directed mutagenesis strategy followed by Gibson assembly into the aforementioned FOXJ1-driven expression vectors. Cas9/sgRNA-expressing lentiviral vectors were generated using the pLentiCRISPRv2 vector (Addgene #52961). Briefly, guide RNAs targeting NEK10 or the AAVS1 control locus were selected from our previously published44 guide library (Addgene #1000000100), synthesized from annealed DNA oligonucleotides, cloned into BsmbI-digested pLentiCRISPRv2 vector, screened, and sequenced to confirm identity. Cloning primers available in Supplementary Table 2.
Lentiviruses were produced as follows: HEK293T cells were transfected with lentiviral constructs and viral packaging plasmids psPAX2 (Addgene #12260) and pCMV-VSVG (Addgene #8454) using the X-tremeGeneHP reagent (Sigma). Viral supernatants were harvested and concentrated using the Lenti-X concentrator system (Takara) prior to use for HBEC infection. Low-passage HBECs were infected with concentrated lentivirus and, 48h later, selected with 1μg/ml puromycin for an additional 48 hours before ALI culture seeding.
Fluorescence-activated cell sorting (FACS)
Mature ALI cultures were washed with PBS and incubated in TrypLE Select (Gibco) for 1 hour at 37ºC to liberate single cells. Cells were strained through a 70um cell strainer to remove clumps and debris, washed in 1X PBS, and resuspended in a buffer containing 1% FBS, 1mM EDTA, and 25mM HEPES. Sorting was performed using a BD FACS Aria (BD Biosciences) running FACS Diva software and analysis was performed using FlowJo (version 10) software. GFP-positive cells were sorted after gating (Extended Data Fig. 2i) on viable (by exclusion of vital dye) singlets (by FSC/SSC) and harvested for RNA isolation in Trizol.
ALI live phase contrast imaging
Mature ALI transwells were removed from their media to glass-bottomed tissue culture dishes and imaged under 40X objective magnification with phase contrast optics at 30 frames per second for a total of 300 frames using a Zeiss Z1 AxioObserver inverted microscope. The resulting 8-bit monochrome videos were processed in ImageJ/FIJI as follows: each 300 frame stack was duplicated and a new 299 frame stack (the “difference stack”) generated by subtracting the (n+1)th frame from the nth frame. The mean of every pixel in the difference stack was then calculated and the resultant data output to a single TIF file visually representing the average pixel intensity change over the course of video, a surrogate for motion. This TIF file was pseudocolored using the “fire” lookup table to yield the final processed images. Data collected in a single experimental imaging session is displayed with identical brightness/contrast thresholds to allow direct visual comparison between experimental conditions. Source code for scripts available on request. For quantitation, mean intensity of TIF files were extracted in ImageJ, normalized per imaging session, and reported as relative intensity levels. Null hypothesis testing was performed using the 2-tailed Student’s t-test.
Micro-optical coherence tomography (μOCT)
μOCT was performed and analyzed as previously described15,45. In brief, periciliary liquid layer (PCL) depth and ciliary beat frequency (CBF) were directly assessed via cross-sectional images of the airway epithelium with high resolution (~1μm) and high acquisition speed (20,480 Hz A-line rate resulting in 40 frames/s at 512 A-lines per frame across 1mm lateral range). Quantitative analysis of images was performed in FIJI/ImageJ. To establish CBF, previously published custom code in Matlab (Mathworks) was used to quantify Fourier-transformed reflectance modulations of beating cilia. PCL depth was characterized directly by geometric measurement of the respective layers. For measurement of mucociliary transport (MCT), native mucus was washed away completely with PBS and 0.5μm polystyrene beads (Invitrogen F8813) were added to the apical ALI surface prior to imaging. Particle tracking was performed using MosaicSuite in ImageJ46 and the resulting particle tracks analyzed in Matlab. Particles included for analysis were within 90μm of the epithelial surface, present for at least 20 frames (0.5s), and had a velocity vector within 10º of horizontal. Hypothesis testing was performed by Mann Whitney U-test or, when necessary to control for environmental (temperature, humidity, etc.) and instrument changes between imaging sessions, by linear mixed effects model.
Imaging flow cytometry
Single cells were generated from mature ALI as above, fixed in 2% PFA, and resuspended in PBS before analysis on an ImageStreamX MkII instrument (Amnis). The gating strategy was as follows: Starting with unclipped events using a centroid X gate, we obtained in-focus cells using a gradient RMS gate. We next employed a liberal area vs. aspect ratio gate (R1) to exclude clumps, followed by an area vs. GFP intensity plot to gate on GFP+ cells (R2), followed by a GFP morphology mask vs aspect ratio gate (R3) to exclude GFP doublets, and followed again by a final doublet removal step again gated on brightfield (R4). We finally gated on R6, cells with a minimal mean pixel value of 45 in the GFP channel. This strategy yielded 4,108 imaged single cells from NEK10WT and 3,513 imaged single cells from NEK10G>C ALI samples. From this parent population of unclipped, singlet, in-focus, GFP-positive cells we defined masks of whole cells and GFP-positive cell bodies. Major axis difference was calculated by subtracting the major axis of the GFP mask from the whole cell mask, and area difference was likewise calculated by subtracting the GFP mask area from the whole cell mask area. Null hypothesis testing was performed using the 2-tailed Student’s t-test. For gallery images, we selected representative images centered on the medians of the indicated populations.
Clinical histopathology
Age-matched patients who underwent pneumonectomy (cases 1–3) or lobectomy (case 4) at Massachusetts General Hospital between 2018–2019 were identified and their H&E stained clinical slides photographed in compliance with IRB 2016P001475. Well-preserved and well-oriented airway was photographed at 400x magnification and processed identically for all panels.
Scanning, scanning/transmission electron microscopy (SEM, STEM), negative stain EM
ALI cultures were washed in PBS, fixed in 2%PFA/2.5% glutaraldehyde/0.1M sodium cacodylate for 15 minutes at room temperature, fixed in fresh fixative for 1 hour at 4ºC, washed thrice in 0.1M sodium cacodylate, dehydrated, then sputter coated with gold. SEM images were acquired on a Zeiss Crossbeam 540 using secondary electron imaging. STEM samples were prepared identically before resin embedding, ultrathin section cutting, and imaging on a Zeiss Crossbeam 540 operating in STEM mode. For negative stain EM, samples were adsorbed onto 200 mesh carbon film copper grids for 1 min, rinsed for 30 sec with distilled water, then stained with 2% uranyl acetate for 1 min, again washed with distilled water, and imaged on a Zeiss Crossbeam 540. Resultant micrographs were analyzed in FIJI/ImageJ where individual cilia were measured; null hypothesis testing was performed using the 2-tailed Student’s t-test.
Ciliary isolation
Cilia were purified according to a previously published protocol with minor modifications25. Mature ALI cultures were washed thrice in PBS before the addition of pre-chilled deciliation buffer (10mM Tris pH 7.5, 50mM NaCl, 10mM CaCl2, 1mM EDTA, 7mM β-mercaptoethanol, 0.1% Triton X-100, 1X Roche completeMINI protease inhibitor cocktail) and 1 minute of vigorous rocking at 4ºC. Supernatants, containing cilia, were removed to pre-chilled 1.5mL tubes and a second aliquot of deciliation buffer applied for 1 minute. Supernatants were then pooled and centrifuged 1min at 1000xg to pellet debris. Supernatants were carefully removed and centrifuged 5min at 12,000xg to pellet axonemes. Ciliary preparations were resuspended in chilled resuspension buffer (300mM HEPES pH 7.3, 250mM NaCl, 50mM MgSO4, 10mM EGTA, 1mM EDTA, 10mM DTT, 1X Roche completeMINI protease inhibitor cocktail) before use for EM or immunoblotting. De-ciliated ALI cultures were harvested in lysis buffer (composition as above) and used for immunoblotting.
Phosphoproteomics and analysis
ALI cultures were grown to maturity, washed in PBS, and lysed in urea lysis buffer containing: 20 mM HEPES (pH 8.0), 9.0 M urea, 1 mM sodium orthovanadate (activated), 2.5 mM sodium pyrophosphate, and 1 mM ß-glycerol-phosphate. Lysates were protease-digested prior to phosphopeptide enrichment using immobilized metal ion affinity chromatography (Fe-IMAC). Eluted peptides were then subjected to LC-MS/MS Analysis using an Orbitrap-Fusion Lumos (ESI-HCD dissociation) and the resulting spectra analyzed with the Thermo Scientific Proteome Discoverer implementation of the SEQUEST algorithm47. Each biological sample was run in duplicate injections. Searches were performed against the most recent update of the Uniprot Homo sapiens database with mass accuracy of +/−50ppm for precursor ions and 0.02Da for product ions. Results were filtered with mass accuracy of +/−5ppm on precursor ions and presence of the intended motif. This dataset yielded 56,540 redundant modified peptide assignments to 14,805 modified sites. Data were normalized by median offset correction based on all the relative abundance values in the data set and expressed as log2 fold changes for analysis. Publicly available data sets were used to establish gene lists for the motile ciliary proteome32, PCD genes12 (with Online Mendelian Inheritance in Man, OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: 244400: 05/30/2019. https://omim.org/), non-PCD ciliopathy genes48, axonemal dyneins and assembly factors (HGNC sets “Dyneins, axonemal” and “Axonemal dynein assembly factors”), kinesins (HGNC set “Kinesins”), intraflagellar transport49, radial spoke apparatus (HGNC RSPH gene family), central pair complex50–52, and ciliary length control53–59. Gene ontology (GO) analysis was performed using published methods30,31 utilizing the PANTHER overrepresentation test against the GO Ontology database (2019–10-08 release) and the “GO Biological Process Complete” annotation data set via publicly-available online interface.
Statistics and Reproducibility
Fig. 1: d, Statistical test: 2-tailed homoscedastic Student’s t-test. Exact p-values for comparisons of 5’, 3’ and upstream amplicons 0.77, 0.65, 0.99, respectively. Extended Data Fig. 2: e, f, 2-tailed homoscedastic Student’s t-test. Exact p-values for comparisons of Scgb1a1, Muc5ac, and Ac-α-tubulin 0.14, 0.21, 0.80, respectively. Fig. 2: d, Statistical test: Linear mixed effects model (fixed effect: MCT, random effect: μOCT instrument session), exact p= 0.0252. e, Linear mixed effects model (fixed effect: PCL, random effect: μOCT instrument session), exact p= 0.0059. g, Statistical test: Mann-Whitney U test, exact p-values all <0.0001 (minimum calculable by software). h, Statistical test: Mann-Whitney U test, exact p-values= 0.0286, 0.0159, 0.0095 for sgNEK10a, sgNEK10b, and sgNEK10c, respectively. i, Statistical test: Mann-Whitney U test, exact p-values= 0.0086, 0.8967, <0.0001 for NEK10WT, NEK10K548R, NEK10S684D, respectively. Extended Data Fig. 3: a, Statistical test: 2-tailed homoscedastic Student’s t-test. Exact p-value 0.00048. c, Statistical test: Linear mixed effects model (fixed effect: CBF, random effect: μOCT instrument session), exact p-value= 0.1109. e, Statistical test: 2-tailed homoscedastic Student’s t-test, exact p-values 0.0085, 0.0006, 0.0014 for sgNEK10a, sgNEK10b, and sgNEK10c, respectively. f, Statistical test: Mann-Whitney U test, exact p-values= 0.0535, 0.3947, 0.0112 for sgNEK10a, sgNEK10b, and sgNEK10c, respectively. h, Statistical test: 2-tailed homoscedastic Student’s t-test, exact p-values = 0.1374, 0.4176, 0.1688, 0.0220 for NEK10WT, NEK10K548R, NEK10Y590A, NEK10S684D respectively. j, Statistical test: Mann-Whitney U test, exact p-values= 0.3429, 0.4857, 0.8857 for NEK10WT, NEK10K548R, NEK10S684D respectively. Fig. 3: b, Statistical test: 2-tailed homoscedastic Student’s t-test, exact p-value = 1.802×10−124. c, Statistical test: 2-tailed homoscedastic Student’s t-test, exact p-value = 0.0000 (minimum calculable by Microsoft Excel). Fig. 4: d, Statistical test: 2-tailed homoscedastic Student’s t-test, exact p-value 3.005×10−19. Extended Data Fig. 5: d, Statistical test: Fisher’s Exact with false discovery rates listed for multiple hypothesis correction, p-values in table.
Data availability statement
Sequence data supporting the findings of this study have been deposited in NCBI GenBank, accession numbers MK806425 and MK806426 (embargoes released at publication). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD016600 (embargo released at publication)60. Plasmids pLRC1-NEK10p:NEK10–3XFLAG and pLRC1-FOXJ1p:NEK10–3XFLAG available for review and distribution at publication through Addgene (http://www.addgene.org, plasmid numbers 137030, 137031). All other data and code are provided within the paper or supplementary information. Raw data are included for statistical inputs (.xlsx form) and uncropped immunoblots (.pdf form) corresponding to Figures 1–4 and Extended Data Figures 2, 3, and 5.
Extended Data
Extended Data Fig. 1: Recurrent NEK10 mutations in familial bronchiectasis.
a, pedigree indicating affected siblings (filled), proband (“p”), and subjects from whom genomic DNA was available for analysis (asterisks). b, chest computed tomography (CT) of siblings “a” and “b” from panel (a), arrows indicate regions of bronchiectatic lung. c, RefSeq-annotated NEK10 variants annotated with transcription start sites, transcript sizes, predicted protein molecular weights, and exon-exon junctions assayed by qRT-PCR in (1d). d, immunoblotting against indicated NEK10 epitopes; HBEC bands are non-specific, full-length 133kDa NEK10 protein indicated with dashed box, representative of 3 experiments. e, pedigree of kindred 2, asterisks denote family members from whom genomic DNA was available; dashed line indicates consanguinity by shared tribal ancestry, Sanger sequencing trace confirming c.1869dupT. f, chest radiograph of proband 2, arrow highlights bronchiectasis. g, pedigree of kindred 3; dashed line indicates consanguinity by shared geographical ancestry, Sanger sequencing trace confirming c.2243C>T. h, CT from proband 3 demonstrating cystic (green arrow) and cylindrical (red arrow) bronchiectasis. i, pedigree of kindred 4; Sanger sequencing trace confirming c.1371+1G>T. j, CT from proband 4 indicating right middle lobe (red arrow) and left lower lobe (green arrow) bronchiectasis. k, proband 4 nasal biopsy TEM demonstrating normal radial ciliary ultrastructure, scale bar 200nm. l, pedigree of kindred 5; dashed line indicates consanguinity by shared tribal ancestry, Sanger sequencing trace confirming c.2317C>T. m, CTs of affected siblings in (l) demonstrating bronchiectasis. n-o, nasal biopsy TEM of affected siblings in (l), scale bars 1μm (n), 200nm (o).
Extended Data Fig. 2: NEK10 loss does not detectably alter airway epithelial differentiation.
a, 18S rRNA-normalized relative NEK10 expression during ALI differentiation; n=1 ALI culture per timepoint. b-d, 18S rRNA-normalized relative expression of ciliated cell markers FOXJ1, DNAH5 (b), secretory cell marker SCGB1A1 (c), and basal cell marker KRT5 (d); n=1 ALI culture per timepoint. e-f, whole-mount immunofluorescence microscopy against SCGB1A1 (e, upper panel), goblet cell marker MUC5AC (e, lower panel), KRT5 (f, upper panel), ciliated cell marker acetylated-α-tubulin (f, lower panel); scale bars 100μm; bar graphs indicate fraction of surface epithelium occupied by marker-positive cells, n=4 per condition representative of 6 ALI differentiations, mean ±S.D. g, schematic depiction of bioinformatic NEK10 promoter (red) identification using indicated UCSC genome browser (hg19) tracks: CpG islands, H3K27-Ac, DNAse I hypersensitivity clusters, transcription factor (TF) chromatin immunoprecipitation sequencing (ChIP-seq). h, live GFP imaging of ALI cultures of the indicated genotypes and maturity, representative of 3 independent ALI differentiations; scale bars 200μm. i, gating strategy for FACS sorting of GFP-labeled ALI cultures, numbers indicate percentage gated cells per population.
Extended Data Fig. 3: Functional consequences of NEK10 activity manipulation.
a, quantitation of analysis in (Fig. 2c), mean ±S.D. b, kymographs of μOCT-based particle tracking from mature ALI, representative of 3 independent ALI differentiations. c, CBF (μOCT) of mature ALI of the indicated genotypes, n=27 (NEK10WT), 22 (NEK10G>C) pooled from 3 independent ALI differentiations, mean ±S.E.M. d, immunoblotting of mature ALI lysates after CRISPR/Cas9-mediated gene editing with the indicated sgRNAs, representative of 2 experiments; short (S) versus long (L) exposures indicated. e, quantitation of analysis in (Fig. 2f), mean ±S.D. f, CBF of mature ALI edited with the indicated sgRNAs, n=8 per condition pooled from 3 independent ALI differentiations, mean ±S.E.M. g, immunoblotting of mature ALI lysates transduced with the indicated cDNAs, representative of 2 experiments; short (S) versus long (L) exposures indicated. h, quantitation of analysis in (i), mean ±S.D. i, pseudocolored video microscopy of mature ALI transduced with the indicated cDNAs, representative fields from 3 independent ALI differentiations, scale bars 50μm. j, CBF of mature ALI transduced with the indicated cDNAs, n=4 per condition pooled from 3 independent ALI differentiations, mean ±S.E.M. *p≤0.05, **p≤0.01, ****p≤0.0001.
Extended Data Fig. 4: Experimental manipulation of NEK10 activity alters ciliated cell morphology.
a, gating strategy for imaging flow cytometry analysis of MCCs. b, representative images and masking data of cells in (a), demonstrating ability to generate single NEK10:eGFP+ ciliated cells for analysis. c, confocal maximum intensity projections of mature ALI edited with the indicated sgRNAs after IF against Ac-α-tubulin, scale bars 25μm, representative of 2 independent ALI differentiations. d, confocal maximum intensity projections of mature ALI transduced with the indicated cDNAs after IF against Ac-α-tubulin, scale bars 25μm, representative of 2 independent ALI differentiations. e, H&E stained mature ALI samples of the indicated genotypes after sectioning orthogonal to the epithelial surface, representative of 3 independent ALI differentiations.
Extended Data Fig. 5: Structural and proteomic abnormalities in NEK10-deficient airway epithelium.
a, SEM of mature ALI edited with the indicated sgRNAs, scale bars 100μm (upper panels) and 1μm (lower panels), representative of 2 independent ALI differentiations. b, immunoblotting against the indicated proteins from lysates generated from purified cilia (lanes 2, 4) or remaining de-ciliated mature ALI (lanes 1, 3), representative of 2 experiments. c, cumulative distribution of phosphopeptides by log2 fold change between indicated conditions, solid (sgNEK10b) and dashed (sgNEK10c) red lines illustrate population of depleted phosphopeptides upon NEK10 deletion. d, table of gene ontology classes enriched among genes (n=395) whose peptides are depleted >1.5 fold (log2) after targeting with sgNEK10b, enrichment level, p-values, and false discovery rates (FDR) indicated. e, cumulative distribution of phosphopeptides by log2 fold change, previously validated PCD in red and all other detected proteins in black, as in (4e). f, cumulative distribution of phosphopeptides by log2 fold change, previously validated non-PCD ciliopathy loci in red and all other detected proteins in black, as in (4e).
Supplementary Material
Acknowledgements
We are grateful to the patients and family members who participated in this research. We thank J. Rajagopal for advice and reagents, B. Stripp (Cedars-Sinai) and I. Cheeseman (Whitehead Institute for Biomedical Research) for antibodies as cited in methods, the Koch Institute Nanomaterials core for electron microscopy services, S. Mordecai for advice and assistance with IFC, H. Zheng for assistance with statistical methods, M. Mino-Kenudson for pulmonary pathology assistance, KFSHRC genotyping and sequencing core facilities for technical help, M. Manion and the US PCD foundation for support, L. Ostrowski for assistance in ciliary motility studies, K. Sullivan and N. Capps for patient coordination and specimen collection, V. Madden, J. Stonebraker, R. Pace and K. Burns for technical assistance, H. Dang and H. Namkoong for bioinformatics assistance, and A. Lee and members of the Sabatini laboratory for critical reading of the manuscript.
This work was supported by grants from the NIH: T32HL116275 (to R.R.C), T32CA009216 (to M.S.T.), U54HL096458 and UL1TR000083 (to M.R.K., M.A.Z., M.L.D., P.R.S.), R01HL071798 (to M.R.K., M.L.D., M.A.Z.), R01HL117836-01 (to M.R.K., M.L.D., M.A.Z.), R01CA129105 and R37AI047389 (to D.M.S.). Additional funding support provided by Massachusetts General Hospital Department of Medicine Pathways program (to R.R.C) and by KSCDR and SHGP (to F.S.A.). The Amnis ImageStreamX MkII was purchased using an NIH Shared Instrumentation Grant 1S10OD012027-01A1 to Massachusetts General Hospital. D.M.S. is an investigator of the Howard Hughes Medical Institute and an American Cancer Society Research Professor.
Footnotes
Competing Interests Statement
The authors declare the following competing interests:
Provisional patent application in process: Applicants: Massachusetts General Hospital, Whitehead Institute for Biomedical Research, Massachusetts Institute of Technology; Inventors: Raghu R. Chivukula, David M. Sabatini; Specific aspects covered: Therapeutic augmentation of NEK10 signaling in disorders of mucociliary clearance.
Main Text References
- 1.Tilley AE, Walters MS, Shaykhiev R & Crystal RG Cilia Dysfunction in Lung Disease. Annu. Rev. Physiol 77, 379–406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montoro DT et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gould CM, Freeman AF & Olivier KN Genetic Causes of Bronchiectasis. Clinics in Chest Medicine 33, 249–263 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Zariwala MA, Knowles MR & Omran H Genetic defects in ciliary structure and function. Annu. Rev. Physiol 69, 423–450 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Online Mendelian Inheritance in Man, OMIM. omim.org Available at: https://www.omim.org/. (Accessed: 11 November 2019)
- 6.Moniz L, Dutt P, Haider N & Stambolic V Nek family of kinases in cell cycle, checkpoint control and cancer. Cell Division 2011 6:1 6, 18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiel C et al. NEK1 Mutations Cause Short-Rib Polydactyly Syndrome Type Majewski. The American Journal of Human Genetics 88, 106–114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith LA et al. Development of polycystic kidney disease in juvenile cystic kidney mice: insights into pathogenesis, ciliary abnormalities, and common features with human disease. J. Am. Soc. Nephrol 17, 2821–2831 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Moniz LS & Stambolic V Nek10 mediates G2/M cell cycle arrest and MEK autoactivation in response to UV irradiation. Mol Cell Biol 31, 30–42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porpora M et al. Counterregulation of cAMP-directed kinase activities controls ciliogenesis. Nat Comms 9, 1224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR & Randell SH in Human Cell Culture Protocols 107, 183–206 (Humana Press, 2005). [DOI] [PubMed] [Google Scholar]
- 12.MD MRK, PhD MZ & MD ML Primary Ciliary Dyskinesia. Clinics in Chest Medicine 37, 449–461 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karczewski KJ et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 49, 531210 (2019). [Google Scholar]
- 14.Ostrowski LE, Hutchins JR, Zakel K & O’Neal WK Targeting expression of a transgene to the airway surface epithelium using a ciliated cell-specific promoter. Mol. Ther 8, 637–645 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Liu L et al. Method for Quantitative Study of Airway Functional Microanatomy Using Micro-Optical Coherence Tomography. PLoS ONE 8, e54473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knowles MR, Daniels LA, Davis SD, Zariwala MA & Leigh MW Primary Ciliary Dyskinesia. Recent Advances in Diagnostics, Genetics, and Characterization of Clinical Disease. Am. J. Respir. Crit. Care Med. 188, 913–922 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He Y, Zeng MY, Yang D, Motro B & Nuñez G NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 530, 354–357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrera AC, Alexandrov K & Roberts TM The conserved lysine of the catalytic domain of protein kinases is actively involved in the phosphotransfer reaction and not required for anchoring ATP. PNAS 90, 442–446 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moniz L Characterization of NimA-related Kinase 10 (NEK10): A Role in Checkpoint Control. (2010). [Google Scholar]
- 20.Richards MW et al. An Autoinhibitory Tyrosine Motif in the Cell-Cycle-Regulated Nek7 Kinase Is Released through Binding of Nek9. Mol Cell 36, 560–570 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doan M et al. Diagnostic Potential of Imaging Flow Cytometry. Trends in Biotechnology 36, 649–652 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Wallmeier J et al. Mutations in CCNO result in congenital mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat Genet 46, 646–651 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Boon M et al. MCIDAS mutations result in a mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat Comms 5, 4418 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Vladar EK, Nayak JV, Milla CE & Axelrod JD Airway epithelial homeostasis and planar cell polarity signaling depend on multiciliated cell differentiation. JCI Insight 1, 183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostrowski LE in Cell Biology (ed. Celis JE) 99–102 (Elsevier, 2005). [Google Scholar]
- 26.Leopold PL, O’Mahony MJ, Lian XJ, Tilley AE & Crystal RG Smoking Is Associated with Shortened Airway Cilia. PLoS ONE 4, e8157 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oltean A, Schaffer AJ, Bayly PV & Brody SL Quantifying Ciliary Dynamics during Assembly Reveals Stepwise Waveform Maturation in Airway Cells. Am. J. Respir. Cell Mol. Biol 59, 511–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bottier M, Thomas KA, Dutcher SK & Bayly PV How Does Cilium Length Affect Beating? Biophysical Journal 116, 1292–1304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Block H et al. Chapter 27 Immobilized-Metal Affinity Chromatography (IMAC): A Review. Methods in Enzymology 463, 439–473 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Ashburner M et al. Gene Ontology: tool for the unification of biology. Nat Genet 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47, D330–D338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ostrowski LE et al. A Proteomic Analysis of Human Cilia Identification of Novel Components. Mol Cell Proteomics 1, 451–465 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Vieira Braga FA et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 25, 1153–1163 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Tabula Muris Consortium et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wloga D et al. Members of the NIMA-related Kinase Family Promote Disassembly of Cilia by Multiple Mechanisms. Molecular Biology of the Cell 17, 2799–2810 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradley BA & Quarmby LM A NIMA-related kinase, Cnk2p, regulates both flagellar length and cell size in Chlamydomonas. J Cell Sci 118, 3317–3326 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Hilton LK, Gunawardane K, Kim JW, Schwarz MC & Quarmby LM The Kinases LF4 and CNK2 Control Ciliary Length by Feedback Regulation of Assembly and Disassembly Rates. Current Biology 23, 2208–2214 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Lin H et al. A NIMA-Related Kinase Suppresses the Flagellar Instability Associated with the Loss of Multiple Axonemal Structures. PLOS Genetics 11, e1005508 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hessel J et al. Intraflagellar Transport Gene Expression Associated with Short Cilia in Smoking and COPD. PLoS ONE 9, e85453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Z-G et al. Aberrant epithelial remodeling with impairment of cilia architecture in non-cystic fibrosis bronchiectasis. Journal of Thoracic Disease 10, 1753–1764–1764 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 41.Neuberger T, Burton B, Clark H & Van Goor F in Cystic Fibrosis 39–54 (Humana Press, 2011). doi: 10.1007/978-1-61779-117-8_4 [DOI] [PubMed] [Google Scholar]
- 42.Hoffmann K & Lindner TH easyLINKAGE-Plus—automated linkage analyses using large-scale SNP data. Bioinformatics 21, 3565–3567 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang T et al. Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu L et al. Imaging the subcellular structure of human coronary atherosclerosis using micro-optical coherence tomography. Nat. Med 17, 1010–1014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sbalzarini IF & Koumoutsakos P Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol 151, 182–195 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Eng JK, McCormack AL & Yates JR An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5, 976–989 (1994). [DOI] [PubMed] [Google Scholar]
- 48.Reiter JF & Leroux MR Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 18, 533–547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishikawa H & Marshall WF Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol 12, 222–234 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Teves ME, Gude DRN, Zhang Z & Strauss JF Mammalian axoneme central pair complex proteins: Broader roles revealed by gene knockout phenotypes. Cytoskeleton 73, 3–22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osinka A et al. Ciliary Proteins: Filling the Gaps. Recent Advances in Deciphering the Protein Composition of Motile Ciliary Complexes. Cells 8, 730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao L, Hou Y, Picariello T, Craige B & Witman GB Proteome of the central apparatus of a ciliary axoneme. J Cell Biol 218, 2051–2070 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tammana TVS, Tammana D, Diener DR & Rosenbaum J Centrosomal protein CEP104 (Chlamydomonas FAP256) moves to the ciliary tip during ciliary assembly. J Cell Sci 126, 5018–5029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niwa S et al. KIF19A Is a Microtubule-Depolymerizing Kinesin for Ciliary Length Control. Dev. Cell 23, 1167–1175 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Lai CK et al. Functional characterization of putative cilia genes by high-content analysis. Molecular Biology of the Cell 22, 1104–1119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vasudevan KK et al. Kinesin-13 regulates the quantity and quality of tubulin inside cilia. Molecular Biology of the Cell 26, 478–494 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piao T et al. A microtubule depolymerizing kinesin functions during both flagellar disassembly and flagellar assembly in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A 106, 4713–4718 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L et al. Flagellar regeneration requires cytoplasmic microtubule depolymerization and kinesin-13. Journal of Cell Science 126, 1531–1540 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Broekhuis JR, Verhey KJ & Jansen G Regulation of Cilium Length and Intraflagellar Transport by the RCK-Kinases ICK and MOK in Renal Epithelial Cells. PLoS ONE 9, e108470 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perez-Riverol Y et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data supporting the findings of this study have been deposited in NCBI GenBank, accession numbers MK806425 and MK806426 (embargoes released at publication). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD016600 (embargo released at publication)60. Plasmids pLRC1-NEK10p:NEK10–3XFLAG and pLRC1-FOXJ1p:NEK10–3XFLAG available for review and distribution at publication through Addgene (http://www.addgene.org, plasmid numbers 137030, 137031). All other data and code are provided within the paper or supplementary information. Raw data are included for statistical inputs (.xlsx form) and uncropped immunoblots (.pdf form) corresponding to Figures 1–4 and Extended Data Figures 2, 3, and 5.