Abstract
Underwater adhesion in mussels (Bivalvia) is an extreme adaptation to achieve robust and firm wet adhesion in the freshwater/brackish/ocean, which biochemically shaped through millions of years. The protein-based adhesion has huge prospective in various fields like industry, medical, etc. Currently, no comprehensive records related to the systematic documentation of structural and functional properties of Mussel foot proteins (Mfps). In this study, we identified the nine species of bivalves in which the complete sequence of at least one adhesive protein is known. The insilico characterization revealed the specific physio-chemical structural and functional characters of each Mfps. The evolutionary analyses of selected bivalves are mainly based on Mfps, Mitogenome, and TimeTree. The outcome of the works has great applications for designing biomimetic materials in future.
Subject terms: Protein sequence analyses, Protein structure predictions
Introduction
Some groups of mussels are capable to produce proteinaceous glue- like sticky material known as byssus thread made by an array of foot proteins (fps). This byssus contains mainly four parts i.e. Plaque, thread, stem, and root. Individual threads proximally merged together to form stem and base of the stem (root) deeply anchored at the base of animal foot. Each byssus threads terminating distally with a flattened plaque which mediates adhesion to the substratum1–4. Each part of the byssus thread complex formed by the auto-assembly of secretory products originating from four distinct glands enclosed in the mussel foot4,5. These mussel foot protein (Mfps), mastered the ability to binding the diverse substratum by using adhesive plaques. 3,4- Dihydroxy phenylalanine (DOPA), is the core constituents in the Mfps, is formed by the post-translational hydroxylation of tyrosine. During the post- translational modification, polyphenol oxidases catalysis the o-hydroxylation of monophenols (tyrosine) to o-diphenols (DOPA), and the adhesion ability of Mfps are strongly correlated with the amount of DOPA2,5–7.
The byssal threads are engineered to withstand elevated mechanical loads applied by waves and currents in subtidal and intertidal zones.4,8. In recent decades, there has been significant understanding of Bivalvia origin, diversities and Mfps. The magnificent moisture-resistant adhesive property of Mfps has inspired to the development of a wide variety of functional materials2,4,9. Designing of mussel-mimetic adhesive materials, initially we need to understand the specific physio-chemical and functional property of each Mfps. This works aims to divulge the physio-chemical structural and functional characterization of currently available all Mfps of various species. And also disclose the evolutionary diversification and molecular clock level speciation of byssus thread producing bivalves and Mfps. The structural modeling and functional analysis of Mfps helps to understanding the which Mfps is highly promising for specific industrial and therapeutical applications.
Results and Discussion
Distribution frequency of available Mfps
A total of 78 Mfps are available in NCBI protein bank. Among these, 34 Mfps in Mytilus californianus Conrad,1837 (Mytilida: Mytilidae), 26 in Mytilus unguiculatus Valenciennes,1858 (Mytilida: Mytilidae) (Synonym of M.coruscus), five in Perna viridis (Linnaeus,1758) (Mytilida: Mytilidae), four in Perna canaliculus (Gmelin,1791) (Mytilida: Mytilidae), three in Mytilus galloprovincialis Lamarck,1819 (Mytilida: Mytilidae), two in Mytilus edulis Linnaeus,1758 (Mytilida: Mytilidae) and Mizuhopecten yessoensis (Jay,1857) (Pectinida: Pectinidae) and one in Atrina pectinata (Linnaeus,1767) (Pteriida: Pinnindae) and Dreissena polymorpha (Pallas,1771) (Myida: Dreissenidae). Scientific names of selected Bivalvia species were validated in Catalogue of Life:2019 Annual Checklist (http://www.catalogueoflife.org/annual- checklist/2019) and in World Register of Marine Species (WORMS) (http://www.marinespecies. org/index.php).
Molecular modeling of Mfps
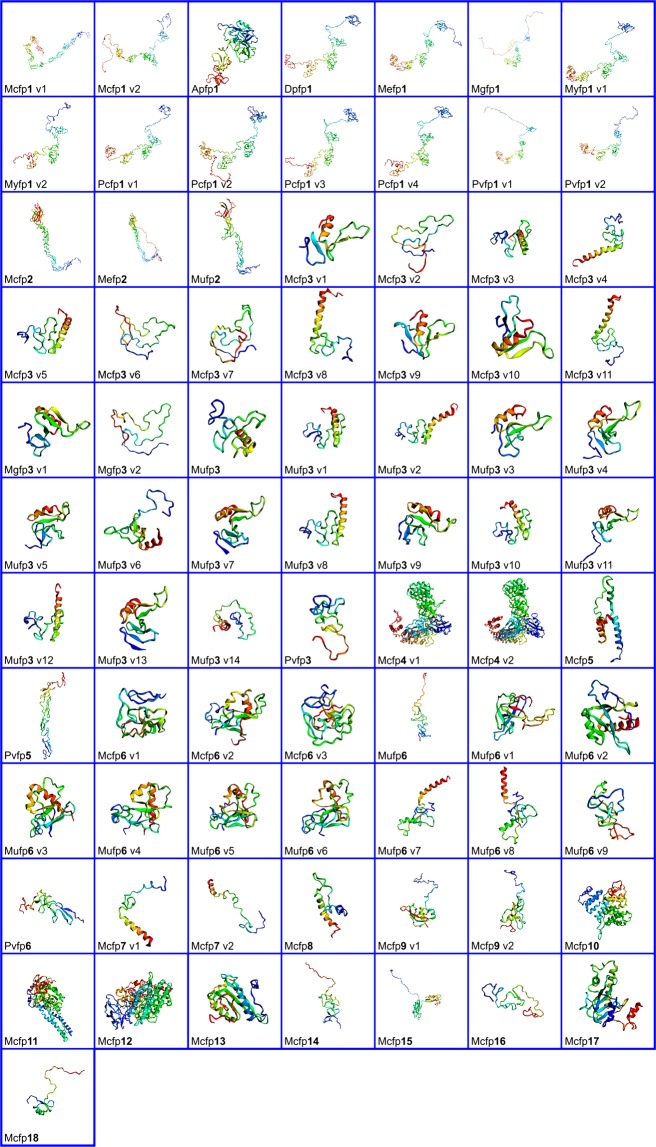
Structural information of Mfps is not available in PDB (Protein Data Bank). The complete structure of each Mfps is mandatory for analyzing their structural and functional aspects. Comparative homology modeling of Mfps was done by using the MUSTER server10. The best model template was selected to develop a full protein model (Table 1: Template used for each Mfps modelling). All protein models are visualized using PyMol tool and EzMol 2.1. (Fig.1 and Supplementary Data S2- Table 2).
Table 1.
List of Mfps used for In silico analysis with results of Ramachandran Plot analysis (Generated in PDBsum and PROCHECK).
| Sl. No |
Bivalve | GenBank Accession No. | Foot protein (fps) | Variant (v) | AA residues | Template | Ramachandran plot | ||
|---|---|---|---|---|---|---|---|---|---|
| Core region % | Additional allowed region % | Total % | |||||||
| 1 | Mytilus californianus | AAY29131.1 | Fp1 | Mcfp v1 | 763 | 3gavA | 67.4 | 23.7 | 91.1 |
| 2 | AAY29132.1 | Mcfp v2 | 672 | 2nbiA | 65.0 | 24.0 | 89.0 | ||
| 3 | AST36139.1 | FP2 | Mcfp2 | 431 | 4xbmB | 76.5 | 17.7 | 94.2 | |
| 4 | AAY29124.1 | FP3 | Mcfp3 v1 | 66 | 4ntqA | 83.7 | 12.2 | 95.9 | |
| 5 | AAY29125.1 | Mcfp3 v2 | 68 | 5w3nA | 82.4 | 15.7 | 98.1 | ||
| 6 | AAY29126.1 | Mcfp3 v3 | 78 | 5xn14 | 82.5 | 15.8 | 98.3 | ||
| 7 | AAY29127.1 | Mcfp3 v4 | 78 | 5xn14 | 81.0 | 15.5 | 96.5 | ||
| 8 | AAY29128.1 | Mcfp3 v5 | 78 | 5xn14 | 81.0 | 15.5 | 96.5 | ||
| 9 | AAY29129.1 | Mcfp3 v6 | 78 | 5w3nA | 77.6 | 17.2 | 94.8 | ||
| 10 | AAY29130.1 | Mcfp3 v7 | 75 | 5w3nA | 83.6 | 12.7 | 96.3 | ||
| 11 | AAZ94726.1 | Mcfp3 v8 | 69 | 6igz7 | 80.4 | 15.7 | 96.1 | ||
| 12 | AAZ94727.1 | Mcfp3 v9 | 69 | 4ntqA | 72.0 | 20.0 | 92.0 | ||
| 13 | AAZ94728.1 | Mcfp3 v10 | 69 | 4ntqA | 74.0 | 18.0 | 92.0 | ||
| 14 | AAZ94729 | Mcfp3 v11 | 69 | 6igz7 | 86.0 | 10.0 | 96.0 | ||
| 15 | ABC84184.1 | Fp4 | Mcfp4 v1 | 770 | 5a1uE | 83.7 | 13.3 | 97.0 | |
| 16 | ABC84185.1 | Mcfp4 v2 | 810 | 5a1uE | 82.0 | 13.5 | 95.5 | ||
| 17 | ABE01084.1 | Fp5 | Mcfp5 | 96 | 5yfpE1 | 74.7 | 18.7 | 93.4 | |
| 18 | ABC84186.1 | Fp6 | Mcfp6 v1 | 121 | 5ubmI | 73.0 | 23.0 | 96.0 | |
| 19 | ABC84187.1 | Mcfp6 v2 | 121 | 6ntwA | 82.2 | 15.8 | 98.0 | ||
| 20 | ABC84188.1 | Mcfp6 v3 | 121 | 5ubmI | 80.2 | 15.8 | 96.0 | ||
| 21 | AST36124.1 | Fp7 | Mcfp7 v1 | 59 | 6ijoH | 91.3 | 8.7 | 100.0 | |
| 22 | AST36126.1 | Mcfp7 v2 | 65 | 2rmsB | 89.4 | 8.5 | 97.9 | ||
| 23 | AST36125.1 | Fp8 | Mcfp8 | 68 | 3dl8E | 87.2 | 10.6 | 97.8 | |
| 24 | AST36127.1 | Fp9 | Mcfp9 v1 | 128 | 2kfwA2 | 78.4 | 15.5 | 93.9 | |
| 25 | AST36129.1 | Mcfp9 v2 | 125 | 2kfwA2 | 75.0 | 18.8 | 93.8 | ||
| 26 | AST36128.1 | Fp10 | Mcfp10 | 319 | 4eqfA | 83.6 | 12.9 | 96.5 | |
| 27 | AST36130.1 | Fp11 | Mcfp11 | 496 | 6fgzA | 87.8 | 8.3 | 96.1 | |
| 28 | AST36131.1 | Fp12 | Mcfp12 | 694 | 6hzeA | 79.8 | 15.6 | 95.4 | |
| 29 | AST36132.1 | Fp13 | Mcfp13 | 158 | 2vezA | 86.0 | 11.0 | 97.0 | |
| 30 | AST36133.1 | Fp14 | Mcfp14 | 120 | 2kraA | 83.0 | 14.0 | 97.0 | |
| 31 | AST36134.1 | Fp15 | Mcfp15 | 249 | 2zx0A | 86.6 | 10.2 | 96.8 | |
| 32 | AST36135.1 | Fp16 | Mcfp16 | 94 | 1n7dA | 62.5 | 28.8 | 91.3 | |
| 33 | AST36136.1 | Fp17 | Mcfp17 | 203 | 4ixjA | 81.4 | 14.1 | 95.5 | |
| 34 | AST36137.1 | Fp18 | Mcfp18 | 72 | 1c55A | 89.1 | 7.8 | 96.9 | |
| 35 | Atrina pectinata | AIW04139.1 | Fp1 | Apfp1 | 352 | 5xtsA | 78.1 | 18.6 | 96.7 |
| 36 | Dreissena polymorpha | AAF75279.1 | Fp1 | Dpfp1 | 430 | 2nbiA | 58.3 | 27.8 | 86.1 |
| 37 | Mytilus edulis | AAX23968.1 | Fp1 | Mefp1 | 565 | 2nbiA | 64.3 | 23.5 | 87.8 |
| 38 | AAX23970.1 | Fp2 | Mefp2 | 504 | 4xbmB | 82.9 | 14.5 | 97.4 | |
| 39 | Mytilus galloprovincialis | BAA09851.1 | Fp1 | Mgfp1 | 751 | 2nbiA | 70.3 | 18.6 | 88.9 |
| 40 | BAB16314.1 | Fp3 | Mgfp3 v1 | 70 | 4ntqA | 90.4 | 7.7 | 98.1 | |
| 41 | BAB16315.1 | Mgfp3 v2 | 77 | 5w3nA | 84.5 | 13.8 | 98.3 | ||
| 42 | Mytilus unguiculatus | ALA16015.1 | Fp2 | Mufp2 | 284 | 4xbmB | 79.1 | 17.7 | 96.8 |
| 43 | ACT66140.1 | Fp3 | Mufp3 | 77 | 5xnl4 | 75.0 | 19.6 | 94.6 | |
| 44 | ADB79738.1 | Mufp3 v1 | 78 | 5xnl4 | 82.5 | 14.0 | 96.5 | ||
| 45 | ADB79739.1 | Mufp3 v2 | 78 | 5xnl4 | 82.5 | 14.0 | 96.5 | ||
| 46 | ADB79740.1 | Mufp3 v3 | 66 | 4ntqA | 86.3 | 11.8 | 98.1 | ||
| 47 | ADB79741.1 | Mufp3 v4 | 78 | 4ntqA | 82.1 | 12.5 | 94.6 | ||
| 48 | ADB79742.1 | Mufp3 v5 | 78 | 4ntqA | 77.2 | 17.5 | 94.7 | ||
| 49 | ADB79743.1 | Mufp3 v6 | 77 | 3ck9B | 84.2 | 14.0 | 98.2 | ||
| 50 | ADB79744.1 | Mufp3 v7 | 78 | 4ntqA | 77.2 | 17.5 | 94.7 | ||
| 51 | ADB79745.1 | Mufp3 v8 | 78 | 5xnl4 | 81.1 | 13.2 | 94.3 | ||
| 52 | ADB79746.1 | Mufp3 v9 | 78 | 4ntqA | 77.2 | 14.0 | 91.2 | ||
| 53 | ADB79747.1 | Mufp3 v10 | 78 | 5xnl4 | 85.7 | 10.7 | 96.4 | ||
| 54 | ADB79748.1 | Mufp3 v11 | 80 | 4ntqA | 74.6 | 20.3 | 94.9 | ||
| 55 | ADB79749.1 | Mufp3 v12 | 78 | 5xnl4 | 82.5 | 12.3 | 94.8 | ||
| 56 | Mytilus unguiculatus | ADB79750.1 | Mufp3 v13 | 80 | 4ntqA | 75.0 | 17.9 | 92.9 | |
| 57 | ADB79751.1 | Mufp3 v14 | 72 | 5z62G | 80.8 | 15.4 | 96.2 | ||
| 58 | ALA16019.1 | Fp6 | Mufp6 | 122 | 2ahxB3 | 82.4 | 13.7 | 96.1 | |
| 59 | ADB79752.1 | Mufp6 v1 | 123 | 6jyxA | 74.5 | 19.6 | 94.1 | ||
| 60 | ADB79753.1 | Mufp6 v2 | 123 | 5uvdA1 | 87.1 | 12.9 | 100.0 | ||
| 61 | ADB79754.1 | Mufp6 v3 | 123 | 6ntwA2 | 89.2 | 9.8 | 99.0 | ||
| 62 | ADB79755.1 | Mufp6 v4 | 123 | 6ntwA2 | 87.4 | 11.7 | 99.1 | ||
| 63 | ADB79756.1 | Mufp6 v5 | 123 | 6ntwA2 | 81.4 | 11.8 | 93.2 | ||
| 64 | ADB79757.1 | Mufp6 v6 | 123 | 6ntwA2 | 81.4 | 15.7 | 97.1 | ||
| 65 | ADB79758.1 | Mufp6 v7 | 102 | 5x0mA | 77.4 | 20.2 | 97.6 | ||
| 66 | ADB79759.1 | Mufp6 v8 | 97 | 5x0mA2 | 74.7 | 20.3 | 95.0 | ||
| 67 | ADB79760.1 | Mufp6 v9 | 96 | 1yy9A3 | 68.4 | 25.3 | 93.7 | ||
| 68 | Mizuhopecten yessoensis | OWF35062.1 | Fp1 | Myfp1 v1 | 505 | 2nbiA | 60.2 | 27.0 | 87.2 |
| 69 | OWF42107.1 | Myfp1 v2 | 324 | 2nbiA | 65.5 | 25.0 | 90.5 | ||
| 70 | Perna canaliculus | AAY29133.1 | Fp1 | Pcfp1 v1 | 436 | 2nbiA | 57.6 | 29.7 | 87.3 |
| 71 | AAY29134.1 | Pcfp1 v2 | 432 | 2nbiA1 | 60.2 | 28.1 | 88.3 | ||
| 72 | AAY29135 | Pcfp1 v3 | 404 | 2nbiA | 59.0 | 25.2 | 84.2 | ||
| 73 | AAY29136.1 | Pcfp1 v4 | 388 | 2nbiA | 58.4 | 29.4 | 87.8 | ||
| 74 | Perna viridis | AAY46226.1 | Fp1 | Pvfp1 v1 | 561 | 2nbiA | 66.0 | 22.4 | 88.4 |
| 75 | AAY46227.1 | Pvfp1 v2 | 431 | 2nbiA1 | 61.7 | 25.3 | 87.0 | ||
| 76 | AGZ84285.1 | Fp3 | Pvfp3 | 70 | 3ds1a2 | 77.4 | 17.7 | 95.1 | |
| 77 | AGZ84279.1 | Fp5 | Pvfp5 | 176 | 5uk5A | 84.3 | 15.7 | 100.0 | |
| 78 | AGZ84283.1 | Fp6 | Pvfp6 | 122 | 2mhpA | 75.8 | 20.0 | 95.8 | |
Figure 1.
Ribbon diagram of the three-dimensional structure of mussel foot proteins (Mfps), visualized in EzMol 2.1.
Validation of Mfps model
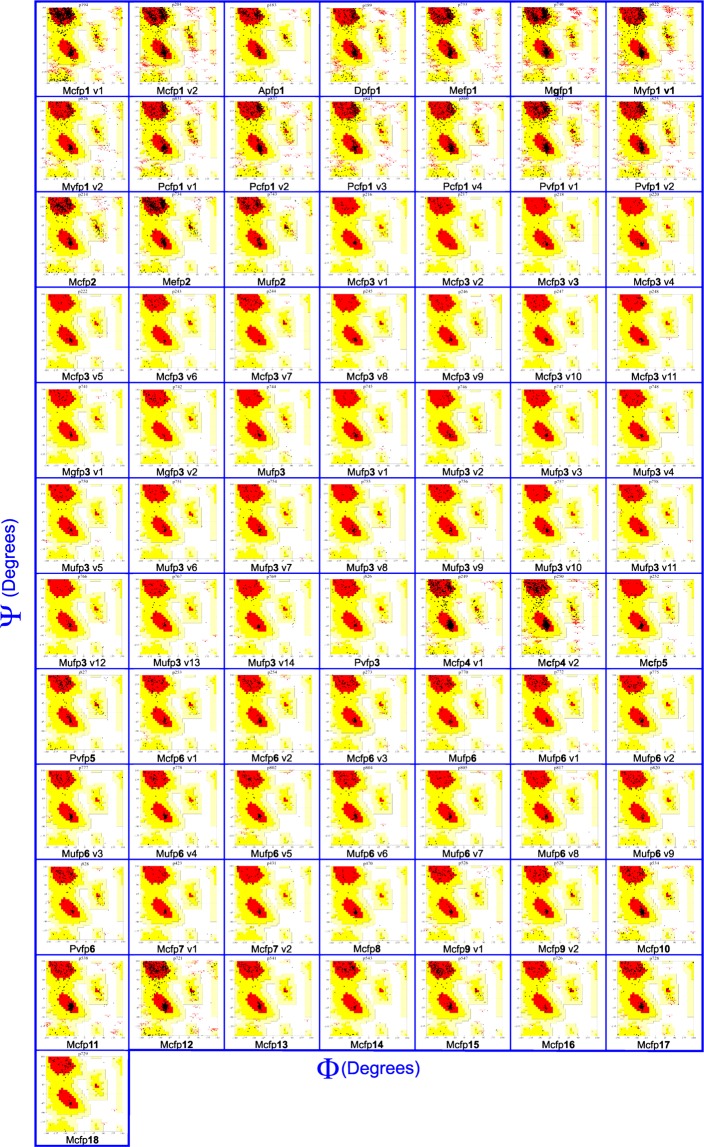
In a good protein model, is expected that there should be more than 90% of the residue in the core or favored region and additional allowed regions11. By analyzing the Ramachandran plot, among the 78 Mfps models, 67 protein models are highly stable because of the 90% residue occurred in core and additional allowed regions of Ramachandran plot. The other 11 protein models are moderately stable because the 85–90 % of residue occurred in core and additional allowed regions of Ramachandran plot (Table 1 and Fig. 2).
Figure 2.
Ramachandran plot of mussel foot proteins (Mfps), generated in PROCHECK, PDBSum.
Promotif documentation of Mfps
The simulated Mfps models were further analyzed using PDBsum for the promotif documentation. In PDBsum server12, analyze the secondary structure characterization of Mfps likes, sheets, beta-alpha beta units, beta hairpins, Psi loops, strands, helices, helix-helix interactions, beta turns, gamma turns and disulfides. The disulfide bond only present in Mcfp2, Mcfp6 v1, Mcfp6 v3, Mcfp14, Mcfp15, Mcfp16, Mcfp18, Apfp1, Mefp2, Mufp2, Mufp6 v9, Pvfp3, Pvfp5 and Pvfp6. The disulfide bond can be formed under oxidizing conditions and play an important role in the folding and stability of the extracellular proteins5,13–15. Normally the disulfide bonds are the crosslinking groups that contribute strength of the protein. All Mfps contained beta turns in varying numbers, but the psi loops present in Mcfp11 only. By analyzing the promotif of Mfps indicated as most of the Mfps exhibited the moderate structural complexity because of the limited number of secondary structural modifications (Table 2).
Table 2.
Promotif documentation of all Mfps, generated in PDBsum server.
| Sl. No | Bivalve | Variant | Sheets | Beta alpha beta unit | Beta hairpins | Psi loops | Beta bulge | Strand | helices | Helix-helix interactions | Beta turns | Gamma turns | Disulphides |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Mytilus californianus | Mcfp1 v1 | 7 | 0 | 6 | 0 | 1 | 15 | 2 | 0 | 97 | 11 | 0 |
| 2 | Mcfp1 v2 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 120 | 19 | 0 | |
| 3 | Mcfp2 | 12 | 0 | 11 | 3 | 27 | 2 | 0 | 71 | 5 | 17 | ||
| 4 | Mcfp3 v1 | 2 | 0 | 2 | 0 | 1 | 4 | 1 | 0 | 12 | 1 | 0 | |
| 5 | Mcfp3 v2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | |
| 6 | Mcfp3 v3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 20 | 1 | 0 | |
| 7 | Mcfp3 v4 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 21 | 0 | 0 | |
| 8 | Mcfp3 v5 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 17 | 0 | 0 | |
| 9 | Mcfp3 v6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | |
| 10 | Mcfp3 v7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 1 | 0 | |
| 11 | Mcfp3 v8 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 13 | 2 | 0 | |
| 12 | Mcfp3 v9 | 2 | 0 | 2 | 0 | 0 | 4 | 1 | 0 | 10 | 2 | 0 | |
| 13 | Mcfp3 v10 | 2 | 0 | 2 | 0 | 0 | 4 | 1 | 0 | 9 | 2 | 0 | |
| 14 | Mcfp3 v11 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 11 | 1 | 0 | |
| 15 | Mcfp4 v1 | 3 | 0 | 2 | 0 | 0 | 7 | 38 | 54 | 74 | 22 | 0 | |
| 16 | Mcfp4 v2 | 5 | 0 | 3 | 0 | 4 | 12 | 41 | 78 | 79 | 19 | 0 | |
| 17 | Mcfp5 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 4 | 19 | 1 | 0 | |
| 18 | Mcfp6 v1 | 2 | 0 | 2 | 0 | 1 | 4 | 1 | 0 | 26 | 1 | 2 | |
| 19 | Mcfp6 v2 | 2 | 1 | 2 | 0 | 1 | 6 | 4 | 2 | 20 | 3 | 0 | |
| 20 | Mcfp6 v3 | 2 | 0 | 2 | 0 | 1 | 4 | 3 | 0 | 27 | 2 | 2 | |
| 21 | Mcfp7 v1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 11 | 2 | 0 | |
| 22 | Mcfp7 v2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 12 | 1 | 0 | |
| 23 | Mcfp8 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 1 | 9 | 1 | 0 | |
| 24 | Mcfp9 v1 | 1 | 0 | 1 | 0 | 1 | 3 | 2 | 0 | 17 | 2 | 0 | |
| 25 | Mcfp9 v2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 21 | 1 | 0 | |
| 26 | Mcfp10 | 0 | 0 | 0 | 0 | 0 | 0 | 17 | 32 | 40 | 2 | 0 | |
| 27 | Mcfp11 | 2 | 2 | 1 | 2 | 1 | 8 | 22 | 33 | 33 | 5 | 0 | |
| 28 | Mcfp12 | 5 | 5 | 0 | 0 | 2 | 13 | 25 | 21 | 78 | 13 | 0 | |
| 29 | Mcfp13 | 1 | 1 | 2 | 0 | 2 | 4 | 6 | 2 | 25 | 4 | 0 | |
| 30 | Mcfp14 | 1 | 0 | 1 | 0 | 1 | 3 | 1 | 0 | 13 | 3 | 5 | |
| 31 | Mcfp15 | 3 | 0 | 1 | 0 | 3 | 7 | 6 | 1 | 25 | 5 | 2 | |
| 32 | Mcfp16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 4 | 2 | |
| 33 | Mcfp17 | 2 | 0 | 5 | 0 | 2 | 7 | 7 | 2 | 28 | 5 | 0 | |
| 34 | Mcfp18 | 1 | 0 | 1 | 0 | 0 | 2 | 1 | 0 | 3 | 5 | 3 | |
| 35 | Atrina pectinate | Apfp1 | 2 | 0 | 4 | 0 | 3 | 10 | 6 | 0 | 39 | 9 | 3 |
| 36 | Dreissena polymorpha | Dpfp1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 105 | 12 | 0 |
| 37 | Mytilus edulis | Mefp1 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 113 | 11 | 0 |
| 38 | Mefp2 | 10 | 0 | 10 | 0 | 4 | 23 | 2 | 0 | 74 | 7 | 17 | |
| 39 | Mytilus galloprovincialis | Mgfp1 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 127 | 9 | 0 |
| 40 | Mgfp3 v1 | 1 | 0 | 3 | 0 | 1 | 4 | 2 | 0 | 11 | 1 | 0 | |
| 41 | Mgfp3 v2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 1 | 0 | |
| 42 | Mytilus unguiculatus | Mufp2 | 8 | 0 | 8 | 0 | 2 | 16 | 1 | 0 | 46 | 1 | 12 |
| 43 | Mufp3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 22 | 1 | 0 | |
| 44 | Mufp3 v1 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 17 | 1 | 0 | |
| 45 | Mufp3 v2 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 2 | 19 | 1 | 0 | |
| 46 | Mufp3 v3 | 2 | 0 | 2 | 0 | 1 | 4 | 1 | 0 | 13 | 1 | 0 | |
| 47 | Mufp3 v4 | 1 | 0 | 1 | 0 | 0 | 2 | 2 | 0 | 12 | 2 | 0 | |
| 48 | Mufp3 v5 | 1 | 0 | 1 | 0 | 0 | 2 | 1 | 0 | 11 | 3 | 0 | |
| 49 | Mufp3 v6 | 1 | 0 | 1 | 0 | 0 | 2 | 3 | 0 | 11 | 1 | 0 | |
| 50 | Mufp3 v7 | 2 | 0 | 2 | 0 | 2 | 6 | 1 | 0 | 10 | 0 | 0 | |
| 51 | Mufp3 v8 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 24 | 0 | 0 | |
| 52 | Mufp3 v9 | 2 | 0 | 4 | 0 | 2 | 6 | 1 | 0 | 14 | 2 | 0 | |
| 53 | Mufp3 v10 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 19 | 1 | 0 | |
| 54 | Mufp3 v11 | 2 | 0 | 3 | 0 | 0 | 5 | 1 | 0 | 13 | 1 | 0 | |
| 55 | Mufp3 v12 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 22 | 0 | 0 | |
| 56 | Mytilus unguiculatus | Mufp3 v13 | 3 | 0 | 3 | 0 | 1 | 6 | 1 | 0 | 14 | 1 | 0 |
| 57 | Mufp3 v14 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 21 | 4 | 0 | |
| 58 | Mufp6 | 3 | 0 | 3 | 0 | 0 | 6 | 0 | 0 | 24 | 2 | 0 | |
| 59 | Mufp6 v1 | 4 | 0 | 3 | 0 | 1 | 9 | 0 | 0 | 14 | 4 | 0 | |
| 60 | Mufp6 v2 | 1 | 0 | 3 | 0 | 1 | 5 | 3 | 0 | 19 | 5 | 0 | |
| 61 | Mufp6 v3 | 2 | 1 | 2 | 0 | 0 | 5 | 3 | 1 | 18 | 3 | 0 | |
| 62 | Mufp6 v4 | 2 | 1 | 2 | 0 | 0 | 5 | 4 | 2 | 17 | 1 | 0 | |
| 63 | Mufp6 v5 | 2 | 1 | 2 | 0 | 0 | 5 | 4 | 1 | 16 | 2 | 0 | |
| 64 | Mufp6 v6 | 1 | 0 | 1 | 0 | 0 | 2 | 2 | 0 | 20 | 5 | 0 | |
| 65 | Mufp6 v7 | 1 | 0 | 1 | 0 | 0 | 2 | 3 | 0 | 12 | 2 | 0 | |
| 66 | Mufp6 v8 | 1 | 0 | 1 | 0 | 0 | 2 | 4 | 0 | 10 | 4 | 0 | |
| 67 | Mufp6 v9 | 2 | 0 | 2 | 0 | 2 | 4 | 1 | 0 | 23 | 4 | 1 | |
| 68 | Mizuhopecten yessoensis | Myfp1 v1 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 1 | 111 | 20 | 0 |
| 69 | Myfp1 v2 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 78 | 11 | 0 | |
| 70 | Perna canaliculus | Pcfp1 v1 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 103 | 9 | 0 |
| 71 | Pcfp1 v2 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 100 | 11 | 0 | |
| 72 | Pcfp1 v3 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 96 | 21 | 0 | |
| 73 | Pcfp1 v4 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 85 | 6 | 0 | |
| 74 | Perna viridis | Pvfp1 v1 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 120 | 16 | 0 |
| 75 | Pvfp1 v2 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 102 | 12 | 0 | |
| 76 | Pvfp3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 15 | 3 | 2 | |
| 77 | Pvfp5 | 5 | 0 | 5 | 0 | 2 | 10 | 2 | 0 | 27 | 0 | 11 | |
| 78 | Pvfp6 | 4 | 0 | 5 | 0 | 2 | 9 | 1 | 0 | 20 | 3 | 5 |
Signal peptide predictions of Mfps
To verify the signal peptide in Mfps, Phobius and SignaIP 5.0 server were used. Mizuhopecten yessoensis foot protein, Myfp1 V1 and V2 don’t contain any signal peptide region. Except Myfp, all other proteins have the signal peptide regions (~1–20 amino acid sequences), the efficiency of protein secretion in extracellular region is highly determined by the signal peptide and also the signal peptide are extremely heterogenous in nature16 (Table 3).
Table 3.
Physio-chemical characterization of Mfps (Generated in Expasy protparam) with signal peptide prediction (Generated in Phobius and SignaIP 5.0).
| Sl. No | Bivalve | Variant | Signal Peptide Prediction | pI | Mw (Dalton) | II | AI | EC | GRAVY | Half-life (in Hours) – in vivo | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SignaIP 5.0 | Phobius | Yeast | E. coli | |||||||||
| 1 | Mytilus californianus | Mcfp1 v1 | 1–24 | 1–20 | 10.04 | 85024.22 | 41.20 | 26.57 | 204255 | −1.357 | 20 | 10 |
| 2 | Mcfp1 v2 | 1–24 | 1–20 | 10.04 | 78048.08 | 40.93 | 28.07 | 186375 | −1.329 | 20 | 10 | |
| 3 | Mcfp2 | 1–27 | 1–17 | 9.04 | 46753.52 | 14.42 | 27.80 | 58545 | −0.890 | 20 | 10 | |
| 4 | Mcfp3 v1 | 1–24 | 1–22 | 10.09 | 7548.70 | 25.90 | 61.97 | 31400 | −0.658 | 20 | 10 | |
| 5 | Mcfp3 v2 | 1–26 | 1–19 | 10.05 | 7791.92 | 23.61 | 61.62 | 36900 | −0.629 | 20 | 10 | |
| 6 | Mcfp3 v3 | 1–24 | 1–22 | 8.80 | 8836.73 | 13.29 | 55.00 | 42860 | −0.505 | 20 | 10 | |
| 7 | Mcfp3 v4 | 1–24 | 1–22 | 9.07 | 8973.87 | 19.12 | 48.72 | 44350 | −0.649 | 20 | 10 | |
| 8 | Mcfp3 v5 | 1–24 | 1–22 | 9.07 | 9000.90 | 13.12 | 48.72 | 44350 | −0.683 | 20 | 10 | |
| 9 | Mcfp3 v6 | 1–24 | 1–22 | 9.70 | 9120.28 | 32.33 | 48.72 | 46870 | −0.791 | 20 | 10 | |
| 10 | Mcfp3 v7 | 1–24 | 1–22 | 7.88 | 8623.45 | 11.60 | 50.67 | 42860 | −0.631 | 20 | 10 | |
| 11 | Mcfp3 v8 | 1–24 | 1–22 | 8.86 | 7920.73 | 12.00 | 55.07 | 39880 | −0.626 | 20 | 10 | |
| 12 | Mcfp3 v9 | 1–24 | 1–22 | 7.94 | 7799.59 | 8.98 | 60.72 | 38390 | -0.501 | 20 | 10 | |
| 13 | Mcfp3 v10 | 1–24 | 1–22 | 7.94 | 7786.59 | 8.17 | 60.72 | 38390 | −0.461 | 20 | 10 | |
| 14 | Mcfp3 v11 | 1–24 | 1–22 | 7.94 | 7790.59 | 12.19 | 56.52 | 38390 | −0.439 | 20 | 10 | |
| 15 | Mcfp4 v1 | 1–26 | 1–19 | 10.20 | 90254.35 | 43.14 | 87.04 | 30955 | −0.835 | 20 | 10 | |
| 16 | Mcfp4 v2 | 1–26 | 1–19 | 10.48 | 95385.30 | 42.38 | 87.65 | 30955 | −0.853 | 20 | 10 | |
| 17 | Mcfp5 | 1–25 | 1–18 | 9.77 | 10928.51 | 20.17 | 47.71 | 29925 | −0.847 | 20 | 10 | |
| 18 | Mcfp6 v1 | 1–24 | 1–22 | 8.85 | 13928.70 | 45.50 | 41.07 | 33530 | −0.550 | 20 | 10 | |
| 19 | Mcfp6 v2 | 1–25 | 1–22 | 9.24 | 13911.86 | 57.41 | 42.73 | 30550 | −0.544 | 20 | 10 | |
| 20 | Mcfp6 v3 | 1–24 | 1–22 | 9.24 | 13824.73 | 55.42 | 42.73 | 30550 | −0.551 | 20 | 10 | |
| 21 | Mcfp7 v1 | 1–30 | 1–24 | 10.41 | 6333.36 | 23.99 | 71.02 | 8940 | −0.163 | 20 | 10 | |
| 22 | Mcfp7 v2 | 1–30 | 1–25 | 10.17 | 6688.79 | 7.99 | 62.92 | 8940 | −0.245 | 20 | 10 | |
| 23 | Mcfp8 | 1–26 | 1–26 | 10.00 | 7281.66 | −7.47 | 70.58 | 16390 | −0.122 | 20 | 10 | |
| 24 | Mcfp9 v1 | 1–21 | 1–19 | 9.74 | 13430.87 | 22.53 | 69.84 | 17420 | −0.330 | 20 | 10 | |
| 25 | Mcfp9 v2 | 1–22 | 1–19 | 9.74 | 13312.69 | 29.75 | 66.08 | 17420 | −0.466 | 20 | 10 | |
| 26 | Mcfp10 | 1–24 | 1–16 | 8.59 | 6316.67 | 29.41 | 68.68 | 82795 | −0.285 | 20 | 10 | |
| 27 | Mcfp11 | 1–30 | 1–20 | 9.89 | 58635.75 | 36.53 | 55.46 | 108585 | −0.914 | 20 | 10 | |
| 28 | Mcfp12 | 1–25 | 1–21 | 9.92 | 82004.53 | 41.55 | 55.55 | 144845 | −0.831 | 20 | 10 | |
| 29 | Mcfp13 | 1–30 | 1–22 | 10.35 | 15181.82 | 44.67 | 72.20 | 35425 | −0.482 | 20 | 10 | |
| 30 | Mcfp14 | 1–23 | 1–23 | 8.65 | 13006.00 | 41.16 | 75.50 | 10720 | −0.071 | 20 | 10 | |
| 31 | Mcfp15 | 1–21 | 1–18 | 9.52 | 2876.43 | 42.30 | 50.12 | 47635 | −0.794 | 20 | 10 | |
| 32 | Mcfp16 | 1–25 | 1–19 | 9.33 | 10630.48 | 33.36 | 50.74 | 3730 | −0.666 | 20 | 10 | |
| 33 | Mcfp17 | 1–27 | 1–27 | 9.21 | 23576.23 | 36.72 | 65.22 | 70205 | −0.446 | 20 | 10 | |
| 34 | Mcfp18 | 1–22 | 1–20 | 8.81 | 7925.25 | 47.24 | 63.75 | 9565 | −0.026 | 20 | 10 | |
| 35 | Atrina pectinate | Apfp1 | 1–25 | 1–18 | 9.47 | 40382.27 | 33.74 | 57.76 | 121185 | −0.662 | 20 | 10 |
| 36 | Dreissena polymorpha | Dpfp1 | 1–30 | 1–19 | 5.24 | 49361.70 | 53.65 | 31.40 | 122860 | −1.331 | 20 | 10 |
| 37 | Mytilus edulis | Mefp1 | 1–30 | 1–20 | 9.99 | 6467.22 | 36.51 | 26.02 | 152105 | −1.280 | 20 | 10 |
| 38 | Mefp2 | 1–23 | 1–17 | 9.14 | 54459.22 | 18.05 | 26.47 | 59630 | −0.896 | 20 | 10 | |
| 39 | Mytilus galloprovincialis | Mgfp1 | 1–34 | 1–34 | 9.99 | 85791.18 | 44.31 | 20.80 | 213195 | −1.353 | 20 | 10 |
| 40 | Mgfp3 v1 | 1–25 | 1–24 | 10.24 | 8003.02 | 48.41 | 55.71 | 31400 | −0.801 | 20 | 10 | |
| 41 | Mgfp3 v2 | 1–25 | 1–24 | 10.32 | 843.79 | 54.86 | 49.35 | 38390 | −0.908 | 20 | 10 | |
| 42 | Mytilus unguiculatus | Mufp2 | 1–26 | 1–17 | 9.23 | 31206.12 | 16.16 | 34.30 | 42145 | −0.842 | 20 | 10 |
| 43 | Mufp3 | 1–27 | 1–27 | 9.13 | 8769.96 | 6.05 | 63.72 | 42860 | −0.350 | 20 | 10 | |
| 44 | Mufp3 v1 | 1–26 | 1–19 | 9.13 | 8715.64 | 15.14 | 66.28 | 38850 | −0.426 | 20 | 10 | |
| 45 | Mufp3 v2 | 1–25 | 1–19 | 8.64 | 8701.63 | 13.24 | 66.28 | 44350 | −0.383 | 20 | 10 | |
| 46 | Mufp3 v3 | 1–25 | 1–19 | 10.33 | 7696.70 | 42.06 | 60.61 | 29910 | −0.889 | 20 | 10 | |
| 47 | Mufp3 v4 | 1–25 | 1–19 | 8.80 | 8729.62 | 18.18 | 61.28 | 44350 | −0.449 | 20 | 10 | |
| 48 | Mufp3 v5 | 1–25 | 1–19 | 8.66 | 8685.63 | 14.33 | 66.28 | 42860 | −0.331 | 20 | 10 | |
| 49 | Mufp3 v6 | 1–25 | 1–19 | 7.90 | 8689.55 | 22.48 | 67.14 | 38850 | −0.451 | 20 | 10 | |
| 50 | Mufp3 v7 | 1–25 | 1–19 | 7.93 | 8699.60 | 15.74 | 67.56 | 41370 | −0.428 | 20 | 10 | |
| 51 | Mufp3 v8 | 1–27 | 1–22 | 9.19 | 8612.64 | 15.36 | 60.00 | 32890 | −0.406 | 20 | 10 | |
| 52 | Mufp3 v9 | 1–27 | 1–24 | 7.90 | 8453.68 | 15.74 | 70.00 | 42860 | −0.373 | 20 | 10 | |
| 53 | Mufp3 v10 | 1–25 | 1–22 | 9.30 | 8609.75 | 17.49 | 66.28 | 36900 | −0.110 | 20 | 10 | |
| 54 | Mufp3 v11 | 1–26 | 1–22 | 8.83 | 9034.05 | 20.9 | 64.62 | 41370 | −0.310 | 20 | 10 | |
| 55 | Mufp3 v12 | 1–27 | 1–19 | 7.90 | 8725.63 | 15.74 | 67.56 | 42860 | −0.404 | 20 | 10 | |
| 56 | Mytilus unguiculatus | Mufp3 v13 | 1–27 | 1–19 | 9.63 | 8625.82 | 14.03 | 64.62 | 19940 | −0.184 | 20 | 10 |
| 57 | Mufp3 v14 | 1–26 | 1–19 | 10.05 | 8196.27 | 25.11 | 54.17 | 38390 | −0.772 | 20 | 10 | |
| 58 | Mufp6 | 1–22 | 1–22 | 9.05 | 14175.06 | 38.31 | 39.18 | 36510 | −0.678 | 20 | 10 | |
| 59 | Mufp6 v1 | 1–25 | 1–25 | 9.13 | 14410.36 | 39.18 | 34.07 | 36510 | −0.694 | 20 | 10 | |
| 60 | Mufp6 v2 | 1–25 | 1–22 | 9.13 | 14455.30 | 38.70 | 34.88 | 39490 | −0.750 | 20 | 10 | |
| 61 | Mufp6 v3 | 1–25 | 1–22 | 9.07 | 14392.29 | 40.29 | 37.24 | 36510 | −0.669 | 20 | 10 | |
| 62 | Mufp6 v4 | 1–25 | 1–25 | 9.18 | 14386.30 | 40.58 | 31.15 | 38000 | −0.788 | 20 | 10 | |
| 63 | Mufp6 v5 | 1–25 | 1–25 | 9.12 | 14454.42 | 36.96 | 34.07 | 38000 | −0.727 | 20 | 10 | |
| 64 | Mufp6 v6 | 1–25 | 1–25 | 9.07 | 14538.45 | 33.80 | 27.72 | 36510 | −0.677 | 20 | 10 | |
| 65 | Mufp6 v7 | 1–25 | 1–25 | 9.16 | 11972.66 | 36.17 | 37.25 | 30300 | −0.639 | 20 | 10 | |
| 66 | Mufp6 v8 | 1–21 | 1–17 | 9.08 | 11396.92 | 40.19 | 39.18 | 31790 | −0.668 | 20 | 10 | |
| 67 | Mufp6 v9 | 1–24 | 1–16 | 8.88 | 11138.42 | 33.51 | 30.52 | 30300 | −0.938 | 20 | 10 | |
| 68 | Mizuhopecten yessoensis | Myfp1 v1 | ND | ND | 5.12 | 56961.06 | 23.62 | 6.97 | 8480 | −2.038 | 20 | 10 |
| 69 | Myfp1 v2 | ND | ND | 8.66 | 33579.64 | 62.60 | 56.48 | 118370 | −0.615 | 20 | 10 | |
| 70 | Perna canaliculus | Pcfp1 v1 | 1–20 | 1–16 | 9.92 | 51565.91 | −10.03 | 65.96 | 131995 | −0.637 | 20 | 10 |
| 71 | Pcfp1 v2 | 1–20 | 1–16 | 9.92 | 51078.31 | −9.97 | 65.90 | 130505 | −0.637 | 20 | 10 | |
| 72 | Pcfp1 v3 | 1–20 | 1–16 | 9.97 | 48252.05 | −10.61 | 66.83 | 127150 | −0.673 | 20 | 10 | |
| 73 | Pcfp1 v4 | 1–20 | 1–16 | 9.97 | 46301.65 | −10.38 | 66.60 | 121190 | −0.674 | 20 | 10 | |
| 74 | Perna viridis | Pvfp1 v1 | 1–20 | 1–20 | 11.20 | 63247.45 | 11.20 | 32.57 | 510930 | −0.800 | 20 | 10 |
| 75 | Pvfp1 v2 | 1–28 | 1–20 | 11.15 | 48042.87 | 15.66 | 33.67 | 362430 | −0.780 | 20 | 10 | |
| 76 | Pvfp3 | 1–23 | 1–23 | 5.93 | 7919.17 | 44.26 | 64.00 | 10595 | 0.260 | 20 | 10 | |
| 77 | Pvfp5 | 1–18 | 1–18 | 9.29 | 19695.09 | 24.06 | 33.75 | 39000 | −0.514 | 20 | 10 | |
| 78 | Pvfp6 | 1–19 | 1–17 | 6.48 | 13262.41 | 39.62 | 57.46 | 12920 | 0.102 | 20 | 10 | |
pI (Isoelectric point), Mw (Molecular weight), II (Instability index), AI (Aliphatic index), EC (Extinction coefficient) and GRAVY (Grand average of hydropathicity). ND: Not detected.
Accessible surface area (ASA) of Mfps
ASA of each Mfps is extremely unique because the size of the fps varies from each species. In the wet adhesion, the hydrophobic nature is very important5, so that perspectives the percentage of side ASA hydrophobic analysis of all Mfps revealed. Among the all Mfps, Pvfp1 v1 is showed the highest percentage of side ASA hydrophobic and followed by Pvfp1 v2 (Table 4).
Table 4.
Accessible surface area of Mfps generated in VADAR server. (ND: Not detected).
| Sl. No | Bivalve | Variant | Total ASA (A2) | Exposed Nonpolar ASA (A2) | Exposed polar ASA (A2) | Exposed charged ASA (A2) | % side ASA Hydrophobic (A2) |
|---|---|---|---|---|---|---|---|
| 1 | Mytilus californianus | Mcfp1 v1 | 54354.3 | 38193.8 | 10977.8 | 5182.7 | 25.70 |
| 2 | Mcfp1 v2 | 61703.6 | 43389.1 | 13480.7 | 4833.9 | 28.23 | |
| 3 | Mcfp2 | 28106.8 | 17011.2 | 6720.1 | 4375.5 | 20.77 | |
| 4 | Mcfp3 v1 | 5209.0 | 3426.9 | 1196.6 | 585.5 | 26.86 | |
| 5 | Mcfp3 v2 | 7940.2 | 5438.8 | 1815.6 | 685.7 | 40.12 | |
| 6 | Mcfp3 v3 | 6817.9 | 4748.2 | 1700.9 | 368.8 | 40.25 | |
| 7 | Mcfp3 v4 | 6791.7 | 4804.3 | 1673.4 | 314.0 | 38.67 | |
| 8 | Mcfp3 v5 | 6589.2 | 4634.3 | 1576.7 | 378.2 | 37.06 | |
| 9 | Mcfp3 v6 | 8915.2 | 6049.7 | 2197.4 | 668.1 | 37.85 | |
| 10 | Mcfp3 v7 | 8316.2 | 5645.2 | 2294.8 | 376.2 | 39.56 | |
| 11 | Mcfp3 v8 | 6735.7 | 4668.2 | 1719.3 | 348.2 | 38.24 | |
| 12 | Mcfp3 v9 | 5298.4 | 3552.6 | 1417.1 | 328.7 | 37.24 | |
| 13 | Mcfp3 v10 | 5102.5 | 3400.6 | 1369.4 | 332.6 | 37.02 | |
| 14 | Mcfp3 v11 | 6827.5 | 4750.1 | 1644.7 | 432.8 | 39.74 | |
| 15 | Mcfp4 v1 | 36397.0 | 20650.4 | 5193.8 | 10552.8 | 26.84 | |
| 16 | Mcfp4 v2 | 37308.0 | 21249.2 | 5329.3 | 10729.5 | 27.11 | |
| 17 | Mcfp5 | 7366.2 | 4984.2 | 1362.8 | 1019.2 | 18.65 | |
| 18 | Mcfp6 v1 | 7754.1 | 4656.3 | 2288.2 | 809.6 | 24.16 | |
| 19 | Mcfp6 v2 | 8738.1 | 5182.1 | 2323.2 | 1232.8 | 21.21 | |
| 20 | Mcfp6 v3 | 7795.0 | 4889.5 | 1971.3 | 934.2 | 28.39 | |
| 21 | Mcfp7 v1 | 6455.1 | 4372.2 | 1094.8 | 988.2 | 32.13 | |
| 22 | Mcfp7 v2 | 6558.7 | 4620.7 | 1111.1 | 826.8 | 34.34 | |
| 23 | Mcfp8 | 6711.5 | 4944.5 | 1093.0 | 674.0 | 32.52 | |
| 24 | Mcfp9 v1 | 10823.5 | 6564.7 | 1960.0 | 2298.8 | 21.70 | |
| 25 | Mcfp9 v2 | 10833.7 | 6494.7 | 2013.4 | 2325.6 | 25.15 | |
| 26 | Mcfp10 | 16718.7 | 10454.6 | 3681.4 | 2582.7 | 34.18 | |
| 27 | Mcfp11 | 26224.0 | 15844.6 | 4453.3 | 5926.1 | 21.63 | |
| 28 | Mcfp12 | 32927.4 | 19211.7 | 6023.4 | 7692.3 | 19.09 | |
| 29 | Mcfp13 | 9718.7 | 5925.5 | 1991.9 | 1801.3 | 24.99 | |
| 30 | Mcfp14 | 9945.3 | 6185.1 | 2242.1 | 1518.1 | 40.15 | |
| 31 | Mcfp15 | 18644.2 | 11144.4 | 4544.9 | 2954.9 | 22.35 | |
| 32 | Mcfp16 | 8871.5 | 5481.7 | 1610.8 | 1779.0 | 24.91 | |
| 33 | Mcfp17 | 12578.3 | 7834.7 | 2931.0 | 1812.7 | 29.82 | |
| 34 | Mcfp18 | 7341.0 | 4762.4 | 1714.4 | 864.2 | 31.82 | |
| 35 | Atrina pectinate | Apfp1 | 21754.0 | 15043.6 | 3808.2 | 2902.2 | 38.67 |
| 36 | Dreissena polymorpha | Dpfp1 | 34184.7 | 22793.1 | 7307.0 | 4084.6 | 32.77 |
| 37 | Mytilus edulis | Mefp1 | 47802.4 | 33622.1 | 10934.8 | 3245.6 | 27.44 |
| 38 | Mefp2 | 37694.4 | 23355.4 | 9188.8 | 5150.1 | 24.66 | |
| 39 | Mytilus galloprovincialis | Mgfp1 | ND | ND | ND | ND | ND |
| 40 | Mgfp3 v1 | 5218.0 | 3288.9 | 1121.9 | 807.2 | 26.06 | |
| 41 | Mgfp3 v2 | 8730.4 | 5357.9 | 2293.6 | 1078.9 | 35.98 | |
| 42 | Mytilus unguiculatus | Mufp2 | 22841.5 | 14506.4 | 4889.4 | 3445.7 | 25.23 |
| 43 | Mufp3 | 6482.2 | 4683.3 | 1543.8 | 255.1 | 40.33 | |
| 44 | Mufp3 v1 | 6943.9 | 4860.6 | 1757.3 | 326.1 | 38.58 | |
| 45 | Mufp3 v2 | 6347.8 | 4474.4 | 1658.8 | 214.5 | 39.76 | |
| 46 | Mufp3 v3 | 5364.3 | 3081.3 | 1508.0 | 775.0 | 27.20 | |
| 47 | Mufp3 v4 | 5975.8 | 4043.0 | 1660.2 | 272.6 | 32.45 | |
| 48 | Mufp3 v5 | 5424.1 | 3661.8 | 1409.8 | 352.4 | 33.41 | |
| 49 | Mufp3 v6 | 6801.4 | 4421.9 | 1863.7 | 515.7 | 31.92 | |
| 50 | Mufp3 v7 | 5328.9 | 3451.3 | 1498.0 | 379.5 | 31.69 | |
| 51 | Mufp3 v8 | 6623.1 | 4863.7 | 1378.7 | 380.8 | 45.93 | |
| 52 | Mufp3 v9 | 5328.9 | 3451.3 | 1498.0 | 379.5 | 31.69 | |
| 53 | Mufp3 v10 | 6562.7 | 4872.7 | 1427.7 | 262.3 | 49.20 | |
| 54 | Mufp3 v11 | 6048.9 | 4037.5 | 1621.3 | 390.1 | 34.00 | |
| 55 | Mufp3 v12 | 6574.2 | 4638.2 | 1680.8 | 255.1 | 42.08 | |
| 56 | Mytilus unguiculatus | Mufp3 v13 | 5690.5 | 4228.7 | 1226.2 | 235.5 | 54.01 |
| 57 | Mufp3 v14 | 6830.6 | 4613.0 | 1679.1 | 538.4 | 37.47 | |
| 58 | Mufp6 | 11131.4 | 6985.8 | 3254.7 | 890.9 | 25.68 | |
| 59 | Mufp6 v1 | 9075.1 | 5278.1 | 2618.8 | 1178.2 | 16.89 | |
| 60 | Mufp6 v2 | 8518.2 | 5014.0 | 2389.0 | 1115.3 | 17.64 | |
| 61 | Mufp6 v3 | 8743.5 | 5300.6 | 2387.7 | 1055.3 | 20.17 | |
| 62 | Mufp6 v4 | 8680.7 | 5090.1 | 2460.18 | 1129.8 | 16.06 | |
| 63 | Mufp6 v5 | 8951.9 | 5493.0 | 2307.8 | 1151.1 | 18.11 | |
| 64 | Mufp6 v6 | 9017.3 | 5705.2 | 2288.0 | 1024.1 | 20.63 | |
| 65 | Mufp6 v7 | 8437.5 | 5233.0 | 2275.4 | 929.2 | 27.41 | |
| 66 | Mufp6 v8 | 7897.2 | 5104.9 | 1955.0 | 837.3 | 26.36 | |
| 67 | Mufp6 v9 | 7606.6 | 4752.6 | 2074.6 | 779.5 | 19.32 | |
| 68 | Mizuhopecten yessoensis | Myfp1 v1 | 37068.2 | 19088.8 | 13688.4 | 4291.0 | 11.35 |
| 69 | Myfp1 v2 | 27092.1 | 17700.9 | 6792.2 | 2599.0 | 37.77 | |
| 70 | Perna canaliculus | Pcfp1 v1 | 3554.1 | 25946.1 | 6726.4 | 2881.7 | 35.45 |
| 71 | Pcfp1 v2 | 35115.6 | 25368.2 | 7105.6 | 2641.8 | 34.81 | |
| 72 | Pcfp1 v3 | 32759.4 | 24128.3 | 6088.8 | 2542.2 | 35.47 | |
| 73 | Pcfp1 v4 | 31497.6 | 22851.4 | 5771.4 | 2874.8 | 36.18 | |
| 74 | Perna viridis | Pvfp1 v1 | 47417.8 | 36101.7 | 7917.3 | 3398.8 | 59.97 |
| 75 | Pvfp1 v2 | 35152.4 | 26011.5 | 6002.5 | 3138.4 | 57.11 | |
| 76 | Pvfp3 | 5458.0 | 3605.8 | 1299.7 | 552.5 | 47.02 | |
| 77 | Pvfp5 | 13685.6 | 9005.2 | 3104.4 | 1575.9 | 22.71 | |
| 78 | Pvfp6 | 9501.7 | 6390.9 | 2301.5 | 808.3 | 45.36 |
Functional characterization of Mfps
Functional characterization of Mfps, FFPred 3 server analyze the protein in three different categories like biological process prediction, cellular component prediction and molecular function prediction with GO (Gene Ontology) term17. This is the first attempt to revealing the molecular function, cellular process and biological activity of Mfps. By analyzing the biological process of Mfps, most of the Mfps show the cell surface receptor signaling pathway (GO:0007166) and cellular component prediction of Mfps shows that all Mfps is in the extracellular region (GO:0005576). The functional characterization of each Mfps showed unique functions. The wet adhesion is the core feature of all known Mfps5,9, other than this property the protein showed some different features also. G-protein coupled receptor activity (GO:0004930), is the common molecular function of all Mfps. (Supplementary data S3-Tables 3, 4 and 5).
Mytilus californianus foot protein (Mcfp), the fp1 showed the growth factor activity (GO:0008083). The mcfp2, exhibit the nine molecular function, and this protein also showed the zinc ion binding activity (GO:0008270) and endopeptidase activity (GO:0004175). In Mcfp3, contained 11 variants, all protein variants showed the receptor activity (GO:0004872), G-protein coupled receptor activity (GO:0004930) and Peptidase inhibitor activity (GO:0030414). Except for Mcfp3 v1, all other variants of Mcfp3 showed the enzyme inhibitory activity (GO:0004857). Purine nucleoside binding (GO:0001883) and catalytic activity (GO:0003824) are showed by Mcfp4 v1 and v2 respectively. Mcfp6 v2, Mcfp11, Mcfp15, Mcfp16 and Mcfp18 exhibit the zinc ion binding (GO:0008270) activity and Mcfp9 showed the co-factor binding activity (GO:0048037).
Atrina pectinata foot protein (Apfp1) showed the growth factor (GO:0008083) and cytokine activity (GO:0005125). Dreissena polymorpha foot protein (Dpfp1) showed the growth factor and G- protein-coupled receptor activity. In Mytilus edulis foot protein (Mefp), Mefp2, exhibit the zinc ion binding (GO:0008270) activity. Mefp2 showed the highest number of molecular functional activities comparing to the Mefp1. In Mytilus galloprovincialis foot protein (Mgfp), all proteins and their variant revealed the G-protein coupled receptor binding activity and Mgfp3 v1 and v2 showed the peptidase inhibitory activity.
The Mytilus unguiculatus foot protein (Mufp), contains three types of protein and their variant. Mufp2 showed the transmembrane signaling receptor activity, endopeptidase activity, signal transducer activity, serine hydrolase activity, cytokine, and zinc ion binding activity. In Mufp3 and their variants exhibit the peptidase inhibitory activity and G-protein coupled receptor activity. All proteins and their variables in Mufp6 showed the cytokine activity. Mufp6 and their variant like v3 and v9 exhibit the zinc ion binding activity. Except for Mufp6, all variants showed the cytokine receptor binding activity.
Mizuhopecten yessoensis foot protein (Myfp), showed the DNA binding (GO:0003677), cytoskeleton protein binding (GO:0008092) and nucleic acid binding (GO:0003676) activities. The Myfp1 v2 showed the sequence-specific DNA binding transcription factor activity. The Perna canaliculus foot protein (Pcfp), the fp1 have four variants and all variants showed the poly (A) RNA binding (GO:0044822) except Pcfp1 v1. Perna viridis foot protein (Pvfp), each protein has unique molecular functions. The Pvfp1 v1 and Pvfp6 showed the G-protein coupled receptor binding activity. Glycosaminoglycan binding (GO:0005539) activity observed in Pvfp1 v2. The variants like fp3 and fp5 exhibit the zinc ion binding activity.
Chemical structural evaluation of Mfps
Amino acid compositional analysis of Apfp1, the major amino acid composition is lysine (15.3%) and proline (15.1%). Most of the amino acid in neutral charge and with positive charge clusters from 121 to 146, (KKPPVYKPKKPVYKPKKRPAYKPKKK), mixed and negative charge clusters are absent in Apfp1. Core block tandem repeats like PPVD, KPPV and PDYKP repeated two times and YKPKK repeated three times. Dpfp1 showed the highest abundance of proline (22.3%) and tyrosine (14.9%), the charge cluster analysis revealed the absence of positive, negative and mixed charge clusters in Dpfp1. tandem repeated blocks, FTTK, PVYPT, PVYPY, PVYPP, PEYP repeated two times and a four-times repetition of PVYP are also observed.
Mcfp1 v1- 23.6 % of amino acid contributed by proline and followed by tyrosine (18.7%). And the absence of specific charge clusters like positive, negative and mixed. Interestingly the presence of 66 copy repetition of YK.K...YPP. the element from location 82–741. Comparing to the first variants of Mcfp1, the v2 was identified as 23.2% of proline and followed by lysine (20.1%) and tyrosine (18.6%). Same as the v1, v2 doesn′t contain any charge clusters. The KKSYPPAYK tandemly repeated four times and also 60 copies of YK.K...YPP. periodically present in-between location 82–681 of Mcfp1 v2. Mcfp2 contained the highest amount of cysteine (14.4%) and followed by glycine (13.5%). Absence of charge clusters and PCKN tandemly repeated five times. In Mcfp3 contained 11 variants, the highest abundance of glycine (18.2%) present in first variants (v1) and followed by tyrosine (15.2%). YPRG repeated two times and without any charge clusters. Glycine (17.6%), tyrosine (14.7%) is the abundant amino-acid present in v2, and this variant doesn′t contain any charge clusters. GWNK is the only tandem repeats present in v2, and it repeated by two times. The highest abundant amino acid in v3 is glycine (20.5%) and tyrosine (17.9%). This variant doesn′t contain any tandem repeated blocks and charge clusters. Tyrosine composition in each variant like v4, v5, v6, v7, v8, v9, v10 and v11 is 19.2, 19.2, 16.2, 18.7, 17.4, 15.9, 15.9, 15.9% respectively. And these variants don′t show any tandem repeated regions and charge clusters. Mcfp4 contained two variants, tyrosine content same in both variants (2.2%). Histidine is the most abundant amino acid in both variants, v1 (23.0%) and v2 (23.8%). Negative charge cluster region in v1, sequence from 486–536 and 565–628 (DLSNDLHPDNNIEQIANDHVNDIAQSTDGDINDFADTHYNDVA PIADVHVD) but in v2, the negative cluster range from 526–576 and 599–668 (DLSNDLHPDNN IEQIANDHVNDIAQSTDGDINDFADTHYNDVAPIADVHVD). HRHVH is the tandemly repeated six times in v1 and HVHRH tandemly repeated three times in v2 and also 39 copies of H.HVH.H.VL periodic element present in between 50–439. Tyrosine (20.8%) is abundantly present in Mcfp5 and also the presence of positive charge clusters from 53–89 (KGKYYGKGKKYYY KYKRTGKYKYLKKARKYHRKGYKK). In Mcfp6, 3 variants are currently reported. The abundancy of tyrosine in each variant is v1(18.2%), v2 (16.5%) and v3(15.5%), and the three variants don′t contain any charge clusters. Mcfp7 v1 and v2 are almost same amino acid compositions, and also showed the same chemical structural characterizations. The tyrosine amount is 10.2% in v1 and 9.2% in v2. Glycine (27.5%) is the most abundant amino acid present in Mcfp8 and without any charge clusters and specified tandem repeats. Glycine and histidine are the abundant amino acid in both variants of Mcfp9. GGHH repeated four times in v2 and two times in v1. By analyzing the amino acid composition in Mcfp10–18 variants, each variant exhibits the unique amino acid characterization. Mcfp11 contained the mixed charge clusters from 357–386 (ENQHKRHL REREYQNKRHLSNEEHLHNKHE), positive charge clusters in Mcfp12 (231–257, RFRRFKIRHGR FRYGGKYYKLSCNKRR) and other variants doesn′t exist any charge clusters.
Two fps reported in Mefp, comparing both variants tyrosine abundantly present in Mefp1 (18.2%) and Mefp2 (7.3%). The tandem repeat distribution in fp1 is PVYKP (two times), YKPKI (four times) and in fp2, GKTGYKC (two times), KPNPC (seven times), NACKPN (five times), VCSPNP (five times) and KPNPC (three times). And these variants don′t contain any charge clusters.
The three-foot proteins reported in Mgfp (Mgfp1, Mgfp3 v1, and v2). Tyrosine abundance same in both variants of fp3 (14.3%) and in fp1 is 19.0%. Absence of tandem repeats in the fp3 v1 and v2, tandem repeats in fp1 is TYKPKPSYPATYKSKSSY (three times) and TYKPKPSYPAT YKSKSSYPSSYKPKKTY (three times). The charge clusters are absent in Mgfps.
In Mufp, major foot protein is fp2, fp3 (15 variants) and fp6 (ten variants). Amino acid composition in fp2, lysine is the most abundant (14.1%) and followed by cysteine (13.4%). This protein contained negative charge clusters from 18–37 (TAPTTQYDDDEDDYKPDTAY) and tandem repeats are KPNPC (4 times). In fp3, glycine (20.8%) is the most abundant amino acid and followed by tyrosine (18.2%) and this protein doesn't contain any specified charge clusters and tandem repeated blocks. The variants of fp3 (v1 to v14), glycine is the most abundant amino acid present in each variants v1 (20.5%), v2 (20.5%), v3 (15.2%), v4 (20.5%), v5 (20.5%), v6 (19.5%), v7 (20.5%), v8 (21.8%), v9 (20.5%), v10 (20.5%), v11 (20.0%), v12 (20.5%), v13 (22.5%) and v14 (20.8%). The tyrosine composition in each variants is v1 (19.2%), v2 (19.2%), v3 (13.6%), v4 (19.2%), v5 (17.9%), v6 (19.5%), v7 (16.7%), v8 (14.1%), v9 (17.9%), v10 (12.8%), v11 (16.2%), v12 (17.9%), v13 (7.5%) and v14 (15.3%). These variants don't contain any specified charge clusters and tandem repeated blocks. Interestingly tyrosine (19.7%) abundantly observed in fp6 and followed by glycine (12.3%). Specified charge cluster absent in this protein and the tandem repeated sequences is NCNSYAGCCL (repeated 2 times) and YCTNKGC (2 times). Fp6 has 9 variants (v1 to v9), tyrosine is the most abundant amino acids in each variant in order to v1 (19.5%), v2 (21.1%), v3 (19.5%), v4 (20.3%), v5 (20.3%), v6 (19.5%), v7 (19.6%), v8 (21.6%) and v9 (20.8%). Perfectly matched tandem repeated blocks present in variants like v2 – RGYC (two times) and v5 – RGYC (two times). And charged cluster is absent in all variants of fp6.
Two variants present in Myfp1, threonine (37.4%) is most abundantly present in v1 and in the case of v2 is glycine (26.9%). The tyrosine concentration in each variant is v1 (0.4%) and v2 (4.0%). Only v2 contained the mixed charge clusters from 2–24 (DAGFEALKKIIVRMDETERY KRR). The specified tandem repeated blocks in v1 is TSQTDT (nine times), TDTTQN (ten times), TDTTQNT TSQ (five times), QNTTSQ (eight times), NTTSQT (nine times), TSQTDTR (three times), TSQTDTT (five times), RQNTTP (nine times), DITQN (two times) and TSQTDTK (two times). In the case of v2, YGLG (seven times), YGLGQSPG (six times), YGLGQSPGTGYWLGQ SPGTG (four times) and YGLGQSPGTGYWLGQSPGTGYGLGQSPGTVYGLGQSPGTGYWL G (three times).
Under the Pcfp1, currently four variants are reported (v1 to v4). The most abundant amino acid in v1 is lysine (23.4%), and in other v2 (lysine-23.4%), v3 (lysine-24.3%), v4 (24.2%). The tyrosine abundancy in each variant is v1 (20.2%), v2 (20.1%), v3 (21.0%) and v4 (20.9%). Tandem repeated elements in v1- KPYV (88 times), v2- KPYV (87 times), v3- KPYV (91 times) and v4 – KPYV (87 times). And this variant doesn't contain any charge clusters.
In Pvfp, currently available major foot protein is fp1, fp3, fp5, and fp6. Two variants present in fp1, proline (19.6%) is abundantly present in v1 and followed by alanine (18.7%) and in v2- proline (19.5%) and then alanine (18.6%). The tyrosine level in v1 is 1.4% and in v2 is 1.6%. The tandem repeated blocks in v1 are HPPSWTAWIA (4 times), WTAWKAHPPAWTAWK (5 times), PPPAWTAWK (8 times), GKPGKPG (3 times) and PPPAWTAWKATLKPWTAWKATPKPW TAWKATPKPWTAWKATPKPWTAWK (3 times). PPPAWTAWK (9 times), GKPG (4 times) and PPAWTAWKATPKPWTAWKAP (4 times) are the tandem repeated block present in v2. Both variants don't contain any charge cluster regions. In fp3, cysteine (15.7%) is the most abundantly present amino acid and followed by phenylalanine (8.6%). 4.3% of tyrosine present in fp3 and absence of specified charge cluster regions and tandem repeated blocks. Cysteine (16.5%) is the most abundant amino acid in the fp5 and tyrosine abundancy is 14.2%. The tandem repeated sequence is GYYGKNCQ (2 times), TCKC (2 times) and CLNGG (2 times) and without any charge cluster regions. Cysteine (13.9%) is the most abundant amino acid present in fp6 and followed by glycine (10.7%) and also the presence of 6.6% of tyrosine. Fp6 without any specified tandem repeated blocks and charge clusters.
The amino acid composition of each Mfps, glycine, and lysine are the major component of the Mfps other than tyrosine (Detailed data in Supplementary File S4). Recently discovered as multiple pairs of Dopa-lysine contribute to the critical underwater adhesion5,6,14,18. The polymorphism in Mfps, may indicated as the versatility of adhesion as variety forms of an adhesive protein can interact the various surfaces.
Physiochemical characterization of Mfps
Expasy protparam server revealed the physio-chemical properties of each protein (Table 3 and Fig. 3). This server helps to the grouping of Mfps, the all Mfps half-life more than 10 hours in E.coli and >20 hours in yeast. In Mcfp1 v1 and v2 have the same isoelectric point 10.04, molecular weight in v1 (85024.22 D) and in v2 (78048.08 D), and other characteristic features like Instability Index (II), AI (Aliphatic Index), Extinction Coefficient (EC) and Grand Average of Hydropathicities (GRAVY) of first variant is 41.20, 26.57, 204255 and −1.357. And the second variant characters are 40.93 (II), 28.07 (AI), 186375 (EC) and −1.329 (GRAVY). Mcfp2 physiochemical properties are 9.04 (pI), 46753.52 D (Mw), 14.42 (II), 27.80 (AI), EC (58545) and −0.890 (GRAVY). The 11 variants of fp3, the highest isoelectric point was observed in v1 (10.09) and followed by v2 (10.05) and the lowest pI in v7 (7.88). The other protein in Mcfp, all variants of fp4, fp7 and fp13 the pI value is higher than 10.00. Most of the fps, II values are below 40 except the variants of fp1, fp4, fp6, fp12, fp13, fp14, fp15 and fp18. By analyzing the hydropathicity values (GRAVY analysis), all fps in Mcfps showed the non-polar nature.
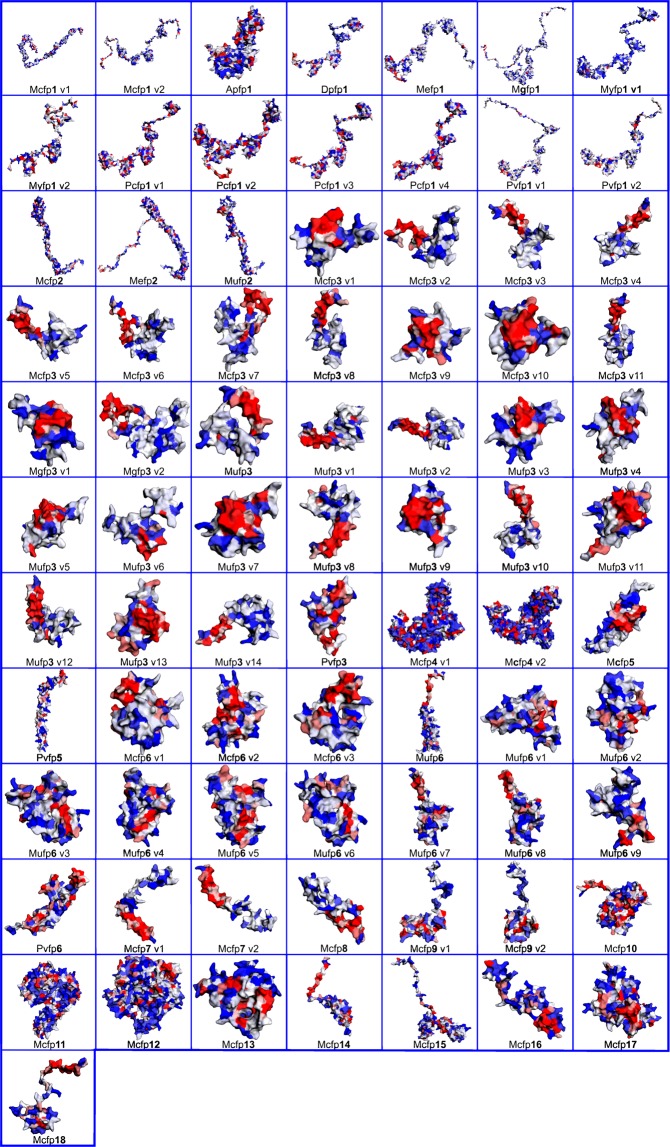
Figure 3.
Hydrophobic contour map of mussel foot proteins (Mfps), The color code indicated as Red: High/Positive, Light color: Neutral and Blue: low/negative hydrophobicity. Generated in EzMol 2.1.
Comparing the fp1 and fp2 of Mefp, only some slight difference is observed. The pI and AI of fp1 and fp2 almost similar. But the fp2 (54459.22 D) is higher molecular weight protein comparing to fp1 (6467.22 D). The hydropathicity nature of fp1 (−1.280) is higher than fp2 (−0.896).
The Mufp is the highly polymorphic group, the highest pI value observed in Mufp3 v3 (10.33) and followed by fp3 v14 (10.05). among the 26 Mfps, 15 fps molecular weight is below 10000 D and II values of fp3 v3 (42.06), fp6 v3 (40.29), fp6 v4 (40.58) and fp6 v8 (40.19) is moderately stable because the instability index is above 40. The highest hydropathicity observed in fp6 v9 (−0.938) and followed by fp3 v3 (−0.889) and fp2 (−0.842), and lowest GRAVY in fp3 v10 (−0.110).
Apfp1, Dpfp1 foot proteins are least polymorphic and less explored groups, the pI value in Apfp1 is 9.47 and pI of Dpfp1 is 5.24. In Apfp1 physiochemical features are Mw is 40382.27 D, II (33.47), AI (57.76), EC (121185) and GRAVY (−0.662). 49361.70 D molecular weighted Dpfp1, II is 53.65, AI (31.40), EC (122860) and GRAVY (−1.331). Myfp1 contained two variants (v1 and v2), but entirely different physiochemical characteristic features. The physiochemical features of v1, pI (5.12), Mw (56961.06 D), II (23.62), AI (6.97), EC (8480) and GRAVY (2.038). pI (8.66), Mw (33579.64 D),II (62.60), AI (56.48), EC (118370) and GRAVY (−0.615) is the physio-chemical feature of v2.
Comparing the all Mfps, instability index of four variants of Pcfp1 is negative (v1= −10.03, v2= −9.97, v3 = −10.61 and v4 = −10.38) and the AI is high (v1 = 65.96, v2 = 65.90, v3 = 66.83 and v4 = 66.60). Molecular weight is higher in v1 (51565.91) and lower in v4 (46301.65). Both v1 and v2 share the same hydropathicity nature and also in the case of v3 and v4. In Pvfp, the 2 variants of fp1 (v1 and v2) share the same physiochemical natures. Comparing the other fps, fp3 (0.260) and fp6 (0.102) is polar nature protein because the GRAVY value is positive in nature. Except for fp3 (44.26), other variants are highly stable because of the II is below 40.
The physiochemical structural and functional characterization of all Mfps is the first time. All protein is hydrophobic nature but except the Pvfp3 and Pvfp6 is polar but it is hydrophobic nature. In Mytilus sp. Mfp-3f is polar but hydrophobic nature, the protein may play vital role in metal and mineral surface adhesion7,18,19.
Ion ligand binding sites of Mfps
Understanding the general properties of the ligand-binding ability of the protein sites is the great importance to understand the functional diversity of the Mfps. One of the fundamental features of the Mfps receptor surface is the set of amino acids available for interactions with ligands. The stabilization and interlinking of Mfps mainly mediated by metal ions, by divulging the metal and acid radical ion binding ability helps to understanding the functional diversity of Mfps. (Detailed predicted binding residues (s) of each Mfps provided in Supplementary File S5).
Foot protein: Apfp1, mainly 3 ligand binding sites were identified in this protein i.e., Zn2+, Ca2+ and Na+. Among these ligand binding sites; 67 sites are available for zinc-binding and followed by 22 sites for sodium binding and three amino acid sites for calcium (I249 E255 E341). Except for calcium, the zinc and sodium bind to the tyrosine and they may associate with DOPA to help them interlinking. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−).
Foot protein: Dpfp1, the four metal ions binding sites were identified in this foot protein (Zn2+, Ca2+, Na+ and K+). The binding sites of each metal ions are, Zn2+ binding site contained 44 sites and followed by Ca2+ binding site (ten sites): P174 N243 D267 K269 D289 D293 G316 P317 P402 Y403, Na+ binding site: C8 and K+ binding site (three binding sites): D267 P272 I276. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, CO32−, NO2−, SO42−, PO43−).
Foot proteins: Mcfps, the ligand-binding sites of each Mfps and their variants showed extremely unique features. In Mcfp1 v1, 145 binding sites were predicted for Zn2+, among these sites most of the zinc ions prefer the histidine amino acid for binding. Only two binding sites were available in Ca2+ (D458 E461). Approximately 292 sites for Na+ metal ion binding are present in this protein, comparing the other metal-ligand ions 2/3rd portion of the amino acid in the protein capable to bind the Na+. The Na+ mostly prefer the histidine for the binding. The K+ metal ion binding sites are, M75 S79 I82 M83 H86 L99 H102 V103 V108. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, CO32−, NO2−, SO42−, PO43−). Mcfp1 v2, approximately 269 binding sites of Zn2+ is present, the histidine and tyrosine are the most common binding sites of Zn2+ metal ion. The other metal-binding sites are, Cu2+ binding site: H150 H152, Ca2+ binding site: D558 D561, Mn2+ binding site: H212 Q257 H260 H262, K+ binding site: H50 S51 and in Na+, approximately 186 metal ions sites were detected. No binding site detected for the following ions:(Fe2+, Fe3+, Mg2+, CO32−, NO2−, SO42−, PO43−). Comparing the two variants of Mcfp1, the v2 contained different ligand binding sites such as Zn2+, Cu2+, Ca2+, Mn2+, K+ and Na+. The metal ion Na+ is mostly preferring the v1 and the Zn2+ prefers the v2.
Mcfp2: in the foot protein only two metal ion binding sites are present and also one acid radical ion sites also detected. Comparing the Zn2+ and Na+ metal ion binding sites, Zn2+ (~221 sites) is widely distributed or binding the most of the regions and followed by Na+ (~103 sites) and Ca2+ (97 binding sites). The Zn2+, Na+ and Ca2+ randomly bind the different amino acids present in the protein. The acid radical ion, SO2− binding site: K206 R220 P221. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, PO43−).
The Mcfp3 contained 11 variants, each variant contained unique ligand binding sites were identified. In v1, mainly three metal ion binding sites and two acid radical ions were detected. The metal ion binding sites are, Zn2+ binding site (11 sites): Q21 D23 Y28 Y38 K39 N43 Y45 R47 Y50 W56 W61, Ca2+ binding site (Seven sites): L12 I15 G26 N27 G48 Y50 G51, Mg2+ binding site (two sites): R60 W61 and the acid radical binding sites are, CO32− binding site (five sites): L12 V13 I15 R63, SO42− binding site (11 sites): D23 R47 W52 W56 K57 K58 G59 R60 W61 K64 Y65. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mn2+, Na+, K+, NO2−, PO43−). Five different metal ions are ability to bind to the v2 (Zn2+, Ca2+, Mg2+, Na+, and K+). The metal ion binding sites of each metals are, Zn2+ binding site (14 sites): Q21 D23 K28 Y34 Y38 G39 Y42 Y48 R50 Y52 W54 K56 W58 W63, Ca2+ binding site (three site): G16 S22 G61, Mg2+ binding site (two sites): Y27 K41, Na+ binding site: Y48, K+ binding site (fives sites): K2 S3 S5 I6 L9, No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mn2+, CO32−, NO2−, SO42−, PO43−). In v3, metal ion ligand binding sites are, Zn2+ binding site (23 sites): K3 Q21 D23 Y26 Y28 Y38 N39 Y42 Y45 N46 G47 Y48 Y51 H52 Y55 G56 W57 K59 W61 N62 W66 Y70 Y71, Ca2+ binding site (six sites): G56 W57 N58 G60 W61 N62, Na+ binding site (three sites): Y42 Y48 Y51. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). The ligand-binding sites of v4: Zn2+ binding site (19 sites): K3 Q21 D23 D27 Y28 Y38 N39 Y45 Y48 Y51 H52 Y55 G56 K59 W61 N62 W66 Y70 Y71, Ca2+ binding site (six sites): D23 D27 W57 N58 W61 G64, Na+ binding site (two sites): Y48 Y51, K+ binding site (two sites): D23 D27, and the acid radical PO3−4 binding site (two sites): N58 G64. No binding site detected for the following ions: (Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, CO32−, NO2−, SO42−). v5, ligand binding sites are: Zn2+ binding site (20 sites): K3 Q21 D23 Y28 Y38 N39 Y43 Y45 Y48 Y51 H52 Y55 G56 W57 K59 W61 N62 W66 Y70 Y71, Ca2+ binding site (six sites): G56 W57 N58 G60 W61 N62 and Na+ binding site (two sites): Y48 Y51. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). v6, Zn2+ binding site (16 sites): K3 Q21 D23 Y28 Y44 G45 Y48 Y51 K52 Y54 R56 Y58 K62 W64 W68 W73, Ca2+ binding site (five sites): D29 G57 Y58 G59 N61, Mg2+ binding site (two sites): G46 K47, Na+ binding site: Y48 and acid radical ion, CO32− binding site (two sites): L30 Y32, SO42− binding site (six sites): Y28 N49 K52 G57 G71 R72, No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mn2+, K+, NO2−, PO43−). v7, Zn2+ binding site (20 sites): K3 Q21 D23 Y26 Y28 D29 Y34 Y44 N45 Y48 Y51 Y54 Y57 H58 Y61 K65 W67 N68 N69 G70, Ca2+ binding site (14 sites): F4 S5 T7 D23 N40 P41 W42 G53 N55 G56 Y57 W63 N64 W67, Mn2+ binding site (two sites): N69 G70, Na+ binding site (four sites): Q21 Y54 Y57 N64, and acid radical CO32− binding site: Y32, No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, K+, NO2−, SO42−, PO43−). v8, Zn2+ binding site (20 sites): K3 Q21 S22 D23 Y26 Y28 Y32 Y38 N39 Y43 Y45 Y48 Y51 H52 Y55 W57 K59 W61 N62 W66, Fe3+ binding site (three site): S22 D23 Y26, Mn2+ binding site (eight sites): L17 A19 V20 S22 D23 A24 Y26 Y28, Na+ binding site (three sites): Y42 Y48 Y51, K+ binding site (11 sites): I15 L17 F18 A19 V20 S22 D23 A24 Y26 Y28 Y32. No binding site detected for the following ions:(Cu2+, Fe2+, Ca2+, Mg2+, CO32−, NO2−, SO42−, PO43−). v9, Zn2+ binding site (20 sites): K3 Q21 D23 Y26 Y28 Y38 N39 Y42 Y43 Y45 N46 Y48 Y51 H52 Y55 W57 K59 W61 N62 W66, Mg2+ binding site (two sites): D23 H52, Mn2+ binding site (three sites): L9 L12 V13, Na2+ binding site (two sites): Y48 Y51, and acid radical SO42− binding site (four sites): L12 V13 G16 Y38, PO43− binding site (four sites): F18 A19 V20 Y42, No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, K+, CO32−, NO2−). v10, Zn2+ binding site (18 sites): K3 Q21 D23 Y26 Y28 Y38 N39 Y43 Y45 Y48 Y51 H52 Y55 W57 K59 W61 N62 W66, Ca2+ binding site (seven sites): V13 I15 N39 G44 N46 Y55 W57, Mg2+ binding site (two sites): D23 H52, Na+ binding site (three site): Y42 Y48 Y51, No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). v11, Zn2+ binding site (18 sites): K3 Q21 D23 Y28 Y38 N39 Y43 Y45 Y48 Y51 H52 Y55 G56 W57 K59 W61 N62 W66, Fe3+ binding site (two sites): D23 G27, Ca2+ binding site (two sites): D23 G27, Mg2+ binding site (two sites): D23 H52, Mn2+ binding site (two sites): D23 G27, Na+ binding site (five sites): Y42 Y43 Y45 Y48 Y51, No binding site detected for the following ions:(Cu2+, Fe2+, K+, CO32−, NO2−, SO42−, PO43−). Among the variants of Mcfp3, only v8 and v11 have the Fe3+ binding sites.
Ligand binding analysis of Mcfp5, a totally three metal ions and one acid radical ion binding site were predicted. The Zn2+ binding site (28 sites): K2 C5 C18 D20 S23 D26 Y28 D30 Y32 Y33 N39 Y40 P41 G43 H45 G46 Y47 H48 G49 H50 Y52 K53 Y57 K59 H83 Y87 Y90 Y91, Mg2+ binding site:K85 G86, Na+ binding site: N39 S44 K53, and acid radical CO32− binding site: H48 G51 K53 Y56, No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Mn2+, K+, NO2−, SO42−, PO43−).
Mcfp6 contained three fps variants, the first variant v1contained 37 binding sites for Zn2+ and followed by Fe3+ binding site (three sites): S32 Y79 N113, Na+ binding site (18 sites): K34 C36 R37 G39 Y40 A64 C67 R75 P87 D88 F107 N108 C109 S111 Y112 N113 C115 C116, and acid radical SO42− binding site (six sites): S22 N45 C49 Y51 G52 S53. No binding site detected for the following ions:(Cu2+, Fe2+, Ca2+, Mg2+, Mn2+, K+, CO32−, NO2−, PO43−). The v2, 35 binding sites for Zn2+ and followed by Na+ binding site (14 sites): C36 R37 G39 Y40 A64 C67 N75 P87 Y107 D108 C109 S111 Y112 N113 and acid radical ion SO42− binding site (six sites): F11 I13 T14 C17 G18 I19. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Mg2+, Mn2+, K+, CO32−, NO2−, PO43−). v3, 34 binding sites for Zn2+ and followed by Ca2+ binding site (two sites): Y79 Y99, Mg2+ binding site (three site): N71 S75 T81, Na2+ binding site (18 sites): K34 C36 R37 G39 Y40 A64 C67 S75 P87 F90 Y107 D108 C109 S111 Y112 N113 C115 C116. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). The first 18 amino acids of all residue showed similar ligand binding sites in the case of Zn2+.
Among the two variants of Mcfp7, showed similar ligand binding positions in the case of Zn2+. In v1, Zn2+ binding site (17 sites): Y28 R29 R30 Y32 K33 G34 S35 H36 S37 G39 G40 H41 H44 G45 H49 Y51 Y55, Ca2+ binding site (four sites): G34 S35 Y57 K58, Na+ binding site: S38 S42 Y51. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). v2, Zn2+ binding site (21 sites): K27 Y28 Y32 Y35 K36 G39 S40 H41 S42 G44 G45 H46 S47 G49 G50 H51 H54 G55 G56 K57 Y61, Mg2+ binding site (two sites): S43 G44, Na+ binding site: H51, and acid radical ion CO32− binding site: S42 G56. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Mn2+, K+, NO2−, SO42−, PO43−).
Mcfp8, Zn2+ binding site (18 sites): P21 Y25 Y28 K36 Y37 K39 Y41 Y44 Y48 R51 Y52 H53 G55 K56 Y57 K60 Y61 K64, Ca2+ binding site (three sites): G46 K47 G66, Mg2+ binding site (two sites): V22 Y25, Na+ binding site: Y41, No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−).
The Zn2+, Na+, and K+ capable to bind the various amino acid residue of the two variants, the Mg2+ only present in the v1 and Ca2+ is present in v2 only. Mcfp9 v1, 54 binding sites for Zn2+ and followed by Mg2+ binding site (four sites): G35 H36 H108 H111, Na+ binding site (14 sites): D27 G32 K34 Y53 H54 V57 H60 V64 G65 H67 W76 G78 P79 A91 and K+ binding site (nine sites): F10 G21 D29 G35 H36 V37 L38 I41 I70. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Mn2+, CO32−, NO2−, SO42−, PO43−). In v2, 57 binding sites of Zn2+ is predicted, Ca2+ binding site (two sites): V45 H47, Na+ binding site (12 sites): Y26 D27 G32 K34 G35 H36 L38 H54 G71 P72 S73 G93, K+ binding site (eight sites): Y23 G25 V37 L38 I41 V45 V62 I63 and acid radical ion PO43− binding site (three sites): H78 H80 V85, No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, CO32−, NO2−, SO42−). Zn2+ metal ion binding sites of two variants are almost the identical.
Mcfp10–17 doesn't contain any variants, each fps showed the unique spectacular metal ions and acid radical bindings. In v10, Zn2+ exhibit the 49 binding sites, Mg2+ binding site (eight sites): V45 S46 T169 D210 D220 D221 Y299 D300 and Na+ had 33 binding sites. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). The v11, 141 binding sites are available for Zn2+ metal ion, Cu2+ binding site (nine sites): H127 H129 H139 H143 H149 H159 H169 H173 H179. 41 amino acid is available for Na+ binding site. No binding site detected for the following ions:(Fe2+, Fe3+, Ca2+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). The v12 predicted ligand binding sites, approximately 139 amino acid can bind the Zn2+ metal ion. The other metal ion binding sites are, Cu2+ binding site (three sites): H59 H81 H93, Ca2+ binding site (eight sites): N196 R409 D464 H465 L473 H494 I498 K532, Mg2+ binding site (four sites): D28 D32 S305 I479, and in the case of Na+ metal ions 55 predicted binding sites are available. Only one acid radical ion can bind this variant protein, SO42− binding site (three sites): S34 V48 R91. No binding site detected for the following ions:(Fe2+, Fe3+, Mn2+, K+, CO32−, NO2−, PO43−). v13 had 23 amino acid residues showed the Zn2+ metal ion binding ability and followed by 18 sites available for Ca2+ binding. And other metal ions like Mn2+ binding site (six sites): L16 I21 N22 G24 R25 R85 and Na+ binding site (threes sites): D9 G71 G94. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, K+, CO32−, NO2−, SO42−, PO43−). v14 contained three metal ion binding sites (Zn2+, Ca2+ and Na+). In the case of Zn2+, it has 41 binding sites. And followed by 10 binding sites for each metal ions like Ca2+ and Na2+. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). v15, 70 amino acid residues for Zn2+ binding and Mg2+ binding site (four sites): S71 K166 K176 K180, K+ binding site (two sites): G15 S16 and Na+ showed the 13 amino acid binding sites. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Mn2+, CO32−, NO2−, SO42−, PO43−). v16, approximately 39 amino acid sites showed the Zn2+ metal ion binding ability and followed by Ca2+ binding site (five sites): V11 V13 E21 E31 G32, Mg2+ binding site (three sites): V49 R50 S64, Mn2+ binding site (two): K27 H29 and Na+ binding site (seven sites): D41 C42 C44 H45 N46 C48 D58. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, K+, CO32−, NO2−, SO42−, PO43−). v17 showed the 48 sites for Zn2+ binding. The other metal ions like Fe3+ binding site (nine sites): I61 D62 V63 G65 M66 E96 P97 Q98 W102, Ca2+ binding site (seven sites): R58 S60 D62 R69 K71 K72 S106, Mg2+ binding site: D62, Na+ binding site (five sites): G100 S114 P152 G194 C195 and K2+ binding site: C20. The two-acid radicals were identified as bind to this protein i.e., SO42− binding site (four sites): I61 M66 L67 P93 and PO43− binding site (four sites): T64 G65 M66 L67. No binding site detected for the following ions:(Cu2+, Fe2+, Mn2+, CO32−, NO2−).
Foot protein: Mgfp1, fps showed only two metal ions binding ability i.e. Zn2+ and Na+. Only one amino acid is available for Na2+ binding (S22) and in the case of Zn2+ had 21 binding sites. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Mg2+, Mn2+, K+, CO32−, NO2−, SO4 2−, P4O3−). The Mgfp3 contained two variants of fps. Only v2 showed the five-metal ion ligand binding site but in the case, v1 had only three metal ion binding been present. In v1, 12 sites for Zn2+ binding and followed by 20 sites for Ca2+ binding and in Na+ binding site (two sites): Y38 Y50. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO2−3, NO−2, SO42−, PO43−). In the case of v2, 18 sites were identified for Zn2+ binding and followed by Ca2+ binding site (seven sites): S25 D26 G54 Y55 G56 G57 Y58, Mg2+ binding site (four sites): N31 G33 G42 R46, Na+ binding site (two sites): W38 W61 and K+ binding site (two sites): D23 S25. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mn2+, CO32−, NO2−, SO42−, PO43−).
In Mefps-ligand binding analysis revealed the fp2 has shown the maximum ligand binding amino acid residue is present. Mefp1 had only three kinds of metal ions binding ability, Zn2+ binding site (15 sites): C9 C12 T15 D17 H30 Y34 Y94 Y163 Y193 P217 Y275 P279 Y311 P351 Y357, Fe3+ binding site (two sites): K324 S494 and Mg2+ binding site: G3. No binding site detected for the following ions:(Cu2+, Fe2+, Ca2+, Mn2+, Na+, K+, CO32−, NO2−, SO42−, PO43−). In Mefp2, ~262 amino acids exhibited the Zn2+ metal ion binding ability and followed by 135 binding sites for Na+ metal ion. The acid radical ion SO2−, binding site (five sites): S296 V302 C305 Y406 G408. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, PO43−).
In Mufp2, ~142 amino acids in this protein available for Zn2+ metal ion binding. Fe3+ binding site (two sites): P89 N94, 26 sites were identified for Ca2+ binding and followed by 46 binding sites for Na+ metal ion. The acid radical CO32− binding site (two sites): N130 R131. No binding site detected for the following ions:(Cu2+, Fe2+, Mg2+, Mn2+, K+, NO2−, SO42−, PO43−). The Mufp3, 20 amino acid residues showed the ability to bind the Zn2+ metal ion and Na+ binding site (two sites): Y47 Y50. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). The variants of fp3, the v1 and v2 had only metal ion binding ability but the v3 showed other than metal ion binding ability, also showed acid radical ion binding. The ligand-binding sites of v1 and v2 are almost similar. The metal ion binding sites of v1: 23 sites were available for Zn2+ binding and followed by Ca2+ binding site (10 sites): Y29 N39 Y42 Y43 Y45 N46 G47 G50 W57 W61 and two sites available for Na+ binding (Y48 Y51). No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). v2, contained 20 sites for Zn2+ binding and followed by 13 sites for Ca2+ binding and two binding sites for Na+ binding site (Y48 Y51). No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO42−). v3, Zn2+ binding site (12 sites): N3 Q21 D23 Y28 N32 Y38 K39 R47 Y50 W52 W56 W61, Mg2+ binding site (three sites): G62 R63 K64, Mn2+ binding site (three sites): A11 L14 I15 and acid radical CO32− binding site (two sites): I6 L10. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Na+, K+, NO2−, SO42−, PO43−). The ligand-binding sites of other variants of fp3 shared the similar ligand- binding sites. In v4, 25 binding sites are predicted in the case of Zn2+ metal ions and followed by the Ca2+ binding site (eight sites): G56 W57 N58 G60 W61 N62 Y70 L77, Mn2+ binding site (three sites): L9 L12 V13 and Na+ binding site (three sites): Y42 Y48 Y51. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, K+, CO32−, NO2−, SO42−, PO43−). The v5, contained 26 binding sites for Zn2+ binding and followed by Ca2+ binding site (seven sites): G56 W57 N58 G60 W61 N62 Y70. Na2+ binding site: Y42 Y48 Y51. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). In v6, 24 binding sites for Zn2+ metal ion, and followed by the Ca2+ binding site (six sites): G40 G43 Y44 S45 G46 G49, Na+ binding site: Y38. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). The v7, 23 binding sites are accessible for Zinc binding, and followed by Ca2+ binding site (three sites): W57 W61 Y70 L77, Na2+ binding site (three sites): Y42 Y48 Y51 and acid radical CO32− binding site (two sites): R54 G56 and SO42− binding site (three sites): N46 K59 G68. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, NO2−, PO43−). In v8, 19 predicted sites for Zn2+, and followed by Mg2+ binding site (two sites): Y51 K62, Na+ binding site (three sites): Y42 Y48 Y51. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). The v9, showed the five different metal ions has the ability to bind this protein (Zn2+, Ca2+, Mn2+, Na+, and K+). 20 amino acid residues capable to bind the Zn2+ metal ion and followed by Ca2+ binding site (three sites): W57 N58 W61, Mn2+ binding site (three sites): I6 L9 L10, Na+ binding site (two sites): Y48 Y51. K+ binding site (six sites): L12 L14 N39 N46 W57 K59. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, CO32−, NO2−, SO42−, PO43−). In v10, 17 sites for Zn2+ binding and two sites for Na+ binding (Y48 Y51). No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). The v11 contained 21 sites for Zn2+ binding and followed by five sites for Ca binding (G56 N58 G60 N62 W66), four sites for Na+ binding (Y48 Y51 G72 N73). No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). In v12, showed the 22 sites for Zn2+ binding, and followed by other elements like Ca2+ binding site (three sites): L17 V20 A24, Mg2+ binding site (two sites): Y48 Y51, Na+ binding site (two sites): Y48 Y51, K+ binding site: L17 V20 A24, and acid radical CO32− binding site (two sites): R54 G56. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mn2+, NO2−, SO42−, PO43−). In v13, 16 binding sites for Zn2+ binding, In the case of other metal ions, Na+ binding site (three sites): Y48 F51 G72 and K+ binding site (seven sites): L12 L14 G16 N39 N46 Y55 G57. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Mg2+, Mn2+, CO32−, NO2−, SO42−, PO43−). The ligand-binding sites of v14, 14 sites for Zn2+ binding and followed by Ca2+ binding site (six sites): G25 G26 G51 Y52 G53 N55, Na+ binding site: Y48 and K+ binding site (five sites): R50 K56 G57 W58 N63. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, CO32−, NO2−, SO42−, PO43−).
Mufp6 contained 34 sites for Zn2+ binding, two sites for the Ca2+ binding site (Y99 G111), and 16 sites for Na+ binding. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). The fp6 had nine variants almost all the ligand-binding sites are the same. v1, had 34 binding sites is present for Zn2+ metal ions and followed by 16 sites for Na+ binding. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). In v2, ligand binding sites of each element are, 42 sites for Zn2+ binding, three sites for Cu2+ binding,13 sites for Na+ binding. No binding site detected for the following ions:(Fe2+, Fe3+, Ca2+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). In v3, 35 binding sites for Zn2+, one binding site for Ca2+ (D28) and 15 binding sites for Na+. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). In v4, 35 binding sites for Zn2+ and 12 binding sites for Na+. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). The v5 contained 36 binding sites for Zn2+ and 13 sites for Na+ binding. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). In v6, 39 sites for Zn2+ binding and 13 sites for Na+ binding. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). v7 contained 31 binding sites for Zn2+, 10 binding sites for Na+ and three sites for K+ binding. No binding site detected for the following ions:(Cu2+, Fe2+, Fee+, Ca2+, Mg2+, Mn2+, CO32−, NO2−, SO42−, PO43−). In v8, 33 sites are available for Zn2+, four sites for Mg2+ binding and nine sites for Na+ binding. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Ca2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). The last variant v9, contained 32 binding sites for Zn2+, four sites for Ca2+ binding and nine sites for Na+ binding. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−).
Foot protein (Myfp1) contained two variants. In v1, 34 sites are mainly focused on the Zn2+ binding, 11 sites for Ca2+ binding and three sites for Na+ binding (N195 T197 S198). No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). The next variant v2, 31 binding sites available for Zn2+ binding, six sites for Ca2+ binding, two sites for Mn2+ binding (T319 Y321), Na+ binding site: P317 and K+ binding site: L233 Q235. No binding site detected for the following ions:(Cu2+ Fe2+, Fe3+, Mg2+, CO32−, NO2−, SO42−, PO43−). Comparing both variants of fp1, v2 showed the different types of metal ions binding capacities.
Foot protein, (Pcfp1) contained four variants of fp1. For each protein variant, some of the binding sites of metal ions are similar. In v1, 83 sites are available for Zn2+ binding, two sites for Fe3+ binding (K143 K323), six binding sites for Ca2+ (P236 Y237 P330 P350 Y351 H436) and three binding sites for Na+ (F10 K289 Y317). No binding site detected for the following ions:(Cu2+, Fe2+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). v2, 95 binding sites available for Zn2+, eight sites for Ca2+ binding (P338 P350 Y355 K367 P376 Y377 K379 H432), two sites for Na+ binding site (Y303 P306), and five sites for K+ binding (P174 K177 P178 V180 K181). No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, CO32−, NO2−, SO42−, PO43−). The variant v3 contained 67 binding sites for Zn2+, two binding sites for Fe3+ (K153 Y179, six binding sites for Ca2+ (P228 Y229 P322 P326 Y327 V328) and three sites for Mg2+ binding (K22 K23 P24). No binding site detected for the following ions:(Cu2+, Fe2+, Mn2+, Na+, K+, CO32−, NO2−, SO42−, PO43−). In v4, 87 binding sites for Zn2+, two sites for Fe3+ binding (Y217 K271) and four sites for Ca2+ binding (P224 Y225 P302 Y303). No binding site detected for the following ions:(Cu2+, Fe2+, Mg2+, Mn2+, Na+, K+, CO32−, NO2−, SO42−, PO43−). Except for variant v2, other variables have the Fe3+ binding sites.
Foot protein (Pvfp), a total of five different types of proteins present under the Pvfps, among the five two proteins are the variants of fp1. The ligand-binding characterization of these proteins showed distinct characteristic features. In fp1 v1, contained 38 sites for Zn2+, two sites for Fe3+ binding (H547 H549), two sites for Ca2+ binding (D517 E528), five sites for Mg2+ binding (G525 K526 G548 G550 A559) and four binding sites for Na+ (P443 G548 H549 W551). No binding site detected for the following ions:(Cu2+, Fe2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−). In fp1 v2, 27 binding sites for Zn2+, two sites for Fe3+ binding (H417 H419), two sites for Ca2+ binding (P123 K124) and five binding sites for Na+ (M5 P33 G418 H419 W421). No binding site detected for the following ions:(Cu2+, Fe2+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−).
Pvfp5, 80 sites are available for Zn2+ binding, 31 sites for Ca2+ binding, 22 sites for Na+ binding and acid radical CO32− binding site: C47 R48. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, NO2−, SO42−, PO43−). Pvfp6, 38 sites for Zn2+ binding (C6 E24 Q28 C29 I35 C38 C40 I41 E43 N44 S45 E46 C47 D50 N52 C53 A56 C59 C60 D61 F62 C64 C66 N67 C70 C80 G84 Y87 F93 D97 C99 C102 C104 N105 D107 C112 K115 C117) two sites for Ca2+ binding (C102 C112) and Na+ binding site: Q16 D50 S51 C53 C60 Y87 V96 D97 C99 N100. No binding site detected for the following ions:(Cu2+, Fe2+, Fe3+, Mg2+, Mn2+, K+, CO32−, NO2−, SO42−, PO43−).
The catechol containing polymers and peptides has the ability to bind various metal ions like Zn2+, Cu2+, Fe3+, Ca2+, Mg2+, Mn2+, Na+ and K+ and acid radical ions, CO32−, NO2−, SO42− and PO43−. The overall analysis most of the foot proteins showed the Zn2+ and Na+ metal ion binding ability and only few Mfps have the ability to bind acid radical ions.
Unbosoming the emergence of Bivalvia: Perspectives on mitogenome, TimeTree, and Mfps
The Bivalvia evolution is very complex in nature, the evolution of Bivalvia starts from Cambrian periods3. The bivalve origins, evolution of their phenotype and functional divergence of Mfps is largely remained unresolved. Evolutionary pattern of byssus thread producing Bivalvia in different perspectives revealed the functional divergence and speciation pattern.
Mitogenome – Phylogenetic construction analysis of bio-adhesive producing bivalves
The complete mitochondrial genome sequence analysis revealed the mitochondrial genome evolutionary pattern in the byssus thread producing bivalves. Based on the mitochondrial genome evolution, the speciation of all Mytilus species is from the same clad but interestingly founded that Perna perna is also originated from the speciation node of Mytilus. In the genus of Perna, currently, three living species only exist, the stem branch of P. canaliculus and P. viridis is entirely separated from P. perna. The monophyletic origin of mitogenome initially separated into two branches. The one branch contained three species of Bivalvia, with entirely different order taxa, the D. polymorpha under the order Myoida and another are Mytiloida (P.canaliculus and P.viridis). And another set of clad, the first separated taxa is Pectinoida and then followed by Ostreoida and Myoida. The Ostreoida is closely resembling the mitogenome of Mytiloida. Among the Mytilus species, the M.galloprovincialis is the first originated species and it is the ancestor of all other Mytilus sp. and P.perna also. The M.unguiculatus and M.californianus is existed in the same clad.
Phylogenetic speciation based on the mitochondrial genome, primitive to recently evolved byssus thread producing bivalve: Mytilus galloprovincialis → Mytilus californianus → Perna perna → Mytilus unguiculatus → Mytilus edulis → Atrina pectinata → Perna viridis → Perna canaliculus→ Dreissena polymorpha → Mizuhopecten yessoensis (Fig. 4F).
Figure 4.
TimeTree analysis of (A) Bivalvia (B) Mytilidae (C) Dreissena sp. (D) Perna sp. (E) Mytilus sp. generated in TimeTree.org tool. (F) Maximum likelihood phylogenetic construction based on mitogenome, MUSCLE alignment and Tamura-Nei model – generated in MEGA X tool.
Timetree of bivalvia
The evolutionary TimeTree of life (TTOL) it helps in understanding the origin and diversity of life forms. The clock like changes analysis, tracing out the speciation and diversification process and events. The diversification of Bivalvia order, TimeTree analysis interestingly revealed the ancestors of byssus thread producing bivalves. The evolution of Bivalvia starts from the Cambrian period (523 MYA), the first diversification/speciation (clad separation) occurred in the starting period of Ordovician (488 MYA). The majority of diversification observed from the Ordovician period. Nuculanoida order is the ancestral group, because without any diversification (clad separation) and followed by Limoida. The byssus thread producing bivalves mainly under the four orders ie, Myoida, Mytiloida, Ostreoida, and Pectinoida. The Myoida is the first diversified group (~465 MYA), evolutionarily it diverged from Mytiloida (~453 MYA). The other stem branch of Mytiloida, the diversification occurred in the Devonian periods, Ostreoida (~398 MYA) is the first diversified group comparing to the Pectinoida (~ 378 MYA). The Bivalvia evolutionary diversification analysis fascinatedly founded that only these two orders (Ostreoida and Pectinoida) are diversified during the Devonian period. After the major extinction (251 MYA), during the starting age of the Triassic period, diversification of Trigonioida and Unionoida has occurred, these are the latest evolved groups in Bivalvia (Fig. 4A).
In the case of Mytilidae time tree analysis, about 387 MYA, the initial diversification of Mytilidae started. During the Jurassic period, ~172 MYA, the genus of Mytilus and Perna is separated from the Perumytilus and Brachidontes. The diversification of clad separation of Mytilus and Perna in the Cretaceous period, upper Epochs. 88 MYA, speciation of Mytilus has occurred, comparing other species with M.californianus is present in a separate clade. The latest evolved species in the Mytilus sp. is M.galloprovincialis and M.trossulus. The M. edulis speciation occurred in 2 MYA. In about 15 MYA ago, the diversification and speciation of Perna are starts. The speciation of P.perna and P.canaliculus is observed in 6 MYA.
Based on the TimeTree analysis, it divulges the clock-like speciation and diversifications. The speciation event of byssus thread producing bivalve, ancestor to latest evolved order is Dreissena polymorpha → Mytilus californianus → Mytilus edulis → Mytilus galloprovincialis → Mytilus unguiculatus → Perna viridis → Perna perna → Perna canaliculus → Atrina pectinata → Mizuhopecten yessoensis (Fig. 4B–E).
The mitochondrial genome revealed the fascinating key of Mfps producing Bivalvia origin, M.galloprovincialis is the first evolved Bivalvia then followed by M.californianus. But, in the case of TimeTree analysis, the first evolved Bivalvia is D.polymorpha, it is an brackish/fresh water forms, then followed by M.califroninanus. Both mitogenome and TimeTree revealed the second evolved form is M.californianus. The natural selection behavior, molecular evolutionary clock speciation indicated as the fresh/brackish form bivalve is the first evolved form, is evidently supported by the Bivalvia taxa evolution and diversification. Contradiction outcome observed in the mitogenome analysis, under the natural selection pressure is marine bivalve to brackish/fresh aquatic forms of species diversification raised. The mitogenome is an important potential target of natural taxa selection spread across the gradients of the ecosystem20. The bivalve spread in the costal belt habitats with dynamic changes such as temperature fluctuation, salinity, dissolved oxygen, desiccation, UV- radiation and exposure to chemical pollutants etc., which can induce oxidative stress to them21, may influence the respiration of the mitochondria and cause irreversible damage to mtDNA22.
Intra-phyletic evolutionary relationship of Mfps
Insight the evolutionary pattern of foot proteins entirely different form the mitochondrial genome evolutionary pattern and TimeTree analysis. The first evolved Mfps is Mcfp3 V9 and followed by Mcfp3 v10, Mufp3, Mufp3 v7, Mufp3 v12, Mufp3 v9, Mufp3 v11, Mufp3 v8, Mcfp3 v7, Mcfp3 v3, Mcfp3 v11, Mcfp3 v8, Mcfp3 v5, Mcfp3 v4, Mufp3 v6, Dpfp1, Mufp3 v1, Mufp3 v4, Mufp3 v5, Mcfp6, Mufp3 v2, Mufp3 v10, Mufp3 v13, Mcfp3 v6, Mufp3 v14, Mcfp3 v2, Mcfp3 v1, Mufp3 v3, Myfp1 v2, Pvfp1 v1, Pvfp1 v2, Mgfp3 v2, Mgfp3 v1, Mufp6 v2, Mufp6 v4, Mufp6 v5, Mufp6 v6, Mufp6 v3, Mufp6 v8, Mufp6 v1, Mufp6 v7, Mufp6 v9, Mufp6, Mcfp6 v1, Mcfp5, Mcfp18, Mcfp6 v2, Mcfp6 v3, Mcfp10, Mcfp7 v1, Mcfp7 v2, Apfp1, Mcfp17, Mcfp9 v1, Mcfp9 v2, Mefp2, Mcfp14, Pvfp5, Mcfp12, Mcfp11, Mcfp13, Mcfp4 v2, Mufp2, Mcfp2, Mcfp8, Pcfp1 v3, Pcfp1 v4, Mcfp4 v1, Pvfp3, Pcfp1 v1, Pcfp1 v2, Mcfp1 v1, Mcfp1 v2, Pvfp6, Mefp1, Mgfp1, Mcfp16 and Myfp1 v1 is the latest evolved Mfps. The first foot protein is appeared in M.californianus and then followed by M.unguiculatus, D. polymorpha, M. yessoensis, P.viridis, M.galloprovincialis, A. pectinata, M.edulis, and P.californianus. Interestingly founded that natural selection divergence exists in Mfps. Because during the evolutionary patterns, the expression level of each Mfps has occurred at different time intervals based on their dynamic environmental conditions (Fig.5).
Figure 5.
Mussel foot proteins (Mfps) based phylogenetic analysis by using the maximum likelihood and the JTT matrix model. Sequence were aligned by using MUSCLE and tree generated in the MEGA X tool. Each color code indicated as the respective bivalves (Drawing: YSV).
The evolutionary and environmental forces equally blend together to tune the unique constitution, magnitude and function of each proteins in the adhesive secretion. The functional property of each protein in root, stem, thread and plaque of byssus thread are highly predisposed and extremely intricate2,4,23. The evolutionary lineage of Mfps revealed the sedentary mode of life style preference of an adult organisms. The Mfps property determined by the geographical habits of the organisms. These byssus threads producing bivalves is randomly distributed all over the world, each geographic zone has the specific dynamic characters are presents, in the case tidal power, salinity, temperature, wave actions etc. Based on this property the evolution and functional divergence of Mfps may evolved. The evolutionary divergence of Mfps producing bivalve, M.edulis is showed the highly complex geographical distribution pattern. The geographical distributional pattern of Mytilus sp. are widespread that exhibit an anti-tropical distribution with M.edulis, M.californianus, and M.unguiculatus occurring in the Northern Hemisphere and M.galloprovincialis distributed in Northern and Southern Hemispheres24. The geographical distribution of Perna sp. is mainly occurred in the tropical zone. P. canaliculus is randomly distributed in Southern temperate region (Fig. 6 and Supplementary data S1- Table 1).
Figure 6.
Approximate geographical distribution of selected Bivalvia species. Generated in – OBIS 2.0 server (2019) (https://mapper.obis.org/). [Available: Ocean biogeographical information system (OBIS). Intergovernmental oceanographic commission UNESCO. www.iobis.org. Accessed: 12th September 2019]. (Supplementary S1).
By analyzing all Mfps, the fp3 and then followed by fp5 and fp6 is the evolutionary lineage of foot proteins in selected bivalves species (all Mytilus sp. except M.edulis fp3 is not available) except P.viridis. Because the fp3,5 and 6 are predominantly found in plaque region of the byssus threads and it contribute to the wet adhesion2. It can be easily concluded that in all byssus thread producing bivalves, fp3 is the ancestor of all other existing Mfps and the wet adhesion property is the core phenomena of all Mfps. In Perna sp. first evolved Mfps is fp1, they actually provide the hydrophobic nature and act as protective varnish layer25. Comparing to the all Mfps, the fp1 and fp2 is the last evolved Mfps, because the fp1 mainly act as protective functions and fp2, provide the structural integrity to the adhesive plaques14. After the fp1, fp5 and followed by fp3 and fp6 is the evolutionary lineages of wet adhesive property of Mfps in Perna sp. except fp3, the fp5 has the specialized mechanism for ability to bind into the calcareous mineral substrate. Fp5 contained the phosphoserine, the post translational modification of phosphoserine gives the ability to bind calcareous materials. The fp5 is the first produced foot protein by P. viridis for surface water replacement and then followed by fp3 and fp6 it gives the stability to wet adhesion13. Mfps revealed functional evolutionary origin characterization and speciation of Perna sp26. The Darwin natural selection pressure is observed in the expression of Mfps diversification because the Mfps is played a vital role in wet adhesion and helps to development of the sedentary mode of lifestyle. The natural selection depends on the environment and requires existing heritable variation in a group. This is the first report of the phylogeny construction of all available Mfps and evolutionary analysis based on the functional divergence.
Conclusion
This is the first report by using the insilico methods to evaluate the physiochemical structural and functional characterization of all available Mfps revealed the unique characteristic features of each mussel foot proteins (Mfps). Required more than a thousand mussels for each Mfps extraction and characterization from different species of mussels in the aim of creating strong adhesives materials. In this works highlighted the several biochemicals, molecular, structural and functional features of the Mfps, these results help to the future development of bio-adhesives in different perspectives. We are not only revealed the bio-adhesive property of Mfps and also revealing the complex nature of evolutionary lineages and diversification of Mfps and selected Bivalvia species with geographical distributions.
Materials and Methods
Datasets
Bivalve Mfps (mussel foot proteins) sequences in FASTA format were retrieved from the NCBI protein database (August 2019) (http: www.ncbi.mlm.gov/protein). Selection criteria are mainly based on Mfps producing bivalves in which the complete sequence of at least one adhesive protein is identified.
Molecular modeling
The MUSTER algorithm used for protein modeling10,27. This server (https://zhanglab.ccmb.med.umich.edu/MUSTER/) analyzes the previous sequence profile-profile alignment (PPA) method and the best template used for the homology modeling of Mfps. The models were evaluated in PROCHECK27,28 and PDBsum server12,27, and the visualization of the protein model in PyMol27 and EzMol 2.129.
Signal peptide prediction
Phobius (http://phobius.sbc.su.se/)30 and SignaIP 5.0 (http://www.cbs. dtu.dk/services/SignalP/)31 servers were used to analyze the signal peptide topology prediction27 of Mfps.
Functional characterization of Mfps
FFPred 3 (http://bioinf.cs.ucl.ac.uk/psipred/)17 server used for functional characterization of Mfps. The predictions are made by scanning the input sequences against an array of Support Vector Machines (SVM). In this server, large SVM library that extends its coverage to the cellular component sub-ontology for the first time, prompted by the establishment of a dedicated evaluation category within the critical assessment of functional annotation. For further analysis of the functional characterization of Mfps, the probability range set to be above 0.800.
Chemical structural characterization of Mfps
SAPS32 (https://www.ebi.ac.uk/Tools/seqstats/saps/) server evaluates a wide variety of protein sequence properties using statistics. Properties considered include compositional biases, clusters and runs of charge and other amino acid types, different kinds and extents of repetitive structures, locally periodic motifs, and anomalous spacing between identical residue types.
Physio-chemical characterization of Mfps
Expasy protparam (https://web.expasy.org/protparam/) server33 analyze the physicochemical properties of Mfps likes, isoelectric point (pI), molecular weight (Mw), extinction coefficient (EC- quantitative study of protein-protein and protein-ligand interactions), instability index (II- stability of protein), aliphatic index (AI- relative volume of protein occupied by aliphatic side chains), and Grand Average of Hydropathicities (GRAVY – sum of all hydropathicity values of all amino acids divided by number of residues in a sequences).
Accessible surface area (ASA) analysis
VADAR (http://vadar.wishartlab.com/) server34 is a compilation of more than 15 different algorithms and programs for analyzing and assessing peptide and protein structures from their PDB coordinate data.
Ion ligand-binding site prediction
IonCom (https://zhanglab.ccmb.med.umich.edu/IonCom/)35 is a ligand-specific method for small ligand (including metal and acid radical ions) binding site prediction. Starting from given sequences or structures of the query proteins, IonCom performs a composite binding-site prediction that combines ab intio training and template-based transferals. The server focuses on binding site prediction of thirteen most important small ligand molecules, including nine metal ions (Zn2+, Cu2+, Fe2+, Fe3+, Ca2+, Mg2+, Mn2+, Na+, K+) and four acid radical ions (CO32−, NO2−,SO42−, PO43−).
Phylogeny construction of Mfps
Phylogenetic analysis of 78 Mfps were performed in MEGA X software36 and 78 Mfps were aligned by using MUSCLE software. The evolutionary history was inferred by using the Maximum Likelihood method and the JTT matrix-based model37. The tree with the highest log likelihood (−3861.65) and Initial tree(s) for the heuristic search was obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.
Ancestral analysis – mitogenome based
Ancestral states were inferred using the Maximum Likelihood method38 and the Tamura-Nei model39. The tree shows a set of possible nucleotides (states) at each ancestral node based on their inferred likelihood at site 1. For each node, only the most probable state is shown. The initial tree was inferred using the method. The rates among sites were treated as being uniform among sites (Uniform rates option). This analysis involved ten nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + Noncoding. Evolutionary analyses were conducted in MEGA X36 with MUSCLE alignment.
TimeTree construction
Evolutionarily time scale tree construction of different orders of Bivalvia, Mytilidae genus, Dreissena, Mytilus and Perna species by using TimeTree40 (http://www.timetree.org/search/goto_timetree). The TimeTree is a public knowledge-base for information on the evolutionary timescale of life. In the TimeTree server build the time tree of a group of species or custom list41–51.
OBIS map construction
Geographical distribution of selected Bivalvia species map constructed by using OBIS 2.0 server (https://mapper.obis.org/). Ocean biogeographical information system (OBIS) is a global open-access data and information clearing-house on marine biodiversity for science, conservation and sustainable development.
Supplementary information
Acknowledgements
We thank the Department of Zoology, University of Calicut for providing the infrastructural facility. The first author is thankful to Seena, S., Neethu, C.B., Rahila, K., Gayathri, M., Renu, V.V, Anusree, A.S., and Lakshmi S. Nair. Research scholar of Toxicology and Biochemistry division, Department of Zoology, University of Calicut and also Dr. Souji, S.M.
Author contributions
Conceived and designed the experiments: P.P.A. Analyzation and interpretation of the data: P.P.A. and Y.S.V. Wrote the main manuscript: P.P.A. Photographic plate preparation: Y.S.V. Drawing of selected bivalves: Y.S.V. All authors reviewed the manuscript.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
P. P. Anand, Email: anandpp633@gmail.com
Y. Shibu Vardhanan, Email: svardhanan@gmail.com.
Supplementary information
is available for this paper at 10.1038/s41598-020-59169-y.
References
- 1.DeMartini, D. G., Errico, J. M., Sjoestroem, S., Fenster, A. & Waite, H. A cohort of new adhesive proteins identified from transcriptomic analysis of mussel foot glands. J.R.Soc. Interface. 14, 20170151, 10.1098.rsif.2017.0151 (2017). [DOI] [PMC free article] [PubMed]
- 2.Forooshani PK, Lee BP. Recent approaches in designing bio-adhesive materials inspired by mussel adhesive protein. Poly. Chem. 2016;55:9–33. doi: 10.1002/pola.28368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harper, E. M., Taylor, J. D. & Crame, A. (eds) The evolutionary biology of the Bivalvia. Geological society, London, Special Publication. 177, 1–9 (2000).
- 4.Hennebert E, Maldonado B, Ladurner P, Flammang P, Santos R. Experimental strategies for the identification and characterization of adhesive proteins in animals: a review. Interface Focus. The Royal society. 2014;5:20140064. doi: 10.1098/rsfs.2014.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maier G. P., Rapp M. V., Waite J. H., Israelachvili J. N., Butler A. Adaptive synergy between catechol and lysine promotes wet adhesion by surface salt displacement. Science. 2015;349(6248):628–632. doi: 10.1126/science.aab0556. [DOI] [PubMed] [Google Scholar]
- 6.Yu Jing, Wei Wei, Danner Eric, Ashley Rebekah K, Israelachvili Jacob N, Waite J Herbert. Mussel protein adhesion depends on interprotein thiol-mediated redox modulation. Nature Chemical Biology. 2011;7(9):588–590. doi: 10.1038/nchembio.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao H, Waite JH. Linking adhesive and structural proteins in the attachment plaque of Mytilus californianus. J. Biol.Chem. 2006;281(36):26150–26158. doi: 10.1074/jbc.M604357200. [DOI] [PubMed] [Google Scholar]
- 8.Waite JH. The phylogeny and chemical diversity of quinone-tanned glues and varnishes. Comp. Physiol. Biochem. B. 1990;97:19–29. doi: 10.1016/0305-0491(90)90172-P). [DOI] [PubMed] [Google Scholar]
- 9.Olivieri MP, Baier RE, Loomis RE. Surface properties of mussel adhesive protein component films. Biomaterials. 1992;13(14):1000–1008. doi: 10.1016/0142-9612(92)90150-M. [DOI] [PubMed] [Google Scholar]
- 10.Wu S, Zhang Y. Muster: improving protein sequence profile-profile alignments by usingmultiple sources of structure information. Proteins. 2008;72(2):547–556. doi: 10.1002/prot.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleywegt GJ, Jones TA. Phi/Psi-chology: Ramachandran revisited. Structure. 1996;4:1395–1400. doi: 10.1016/S0969-2126(96)00147-5. [DOI] [PubMed] [Google Scholar]
- 12.Laskowski RA, Chistyakov VV, Thornton JM. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucl. Acid. Res. 2005;33:266–268. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrone L, et al. Mussel adhesion is dictated by time-regulated secretion and molecular conformation of mussel adhesive proteins. Nat. Commu. 2015;6:8737. doi: 10.1038/ncomms9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rzepecki LM, Hansen KM, Waite JH. Characterization of a cysteine-rich polyphenolicprotein family from the blue mussel. Biol. Bull. 1992;183:123–137. doi: 10.2307/1542413. [DOI] [PubMed] [Google Scholar]
- 15.Sevier CS, Kaiser C. A. Formation and transfer of disulphides bonds in living cells. Nat. Rev. Mol. Biol. 2002;3(11):836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- 16.Kober L, Zehe C, Bode J. Optimized signal peptides for the development of high expression CHO cell lines. Biotechnol. Bioeng. 2013;110(4):1164–73. doi: 10.1002/bit.24776. [DOI] [PubMed] [Google Scholar]
- 17.Cozzetto, D., Minneci, F., Currant, H. & Jones, D. T. FFPred 3: features-based function predictionfor all gene ontology domains. Nat. Scin. Reprt. 6:31865, 10.38/srep31865 (2016). [DOI] [PMC free article] [PubMed]
- 18.Levine ZA, et al. Surface force measurements and simulations of mussel-derived peptide adhesives on wet organic surfaces. PNAS. 2016;113(16):4332–4337. doi: 10.1073/pnas.1603065113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson NH, Waite JH. Mussel designed protective coatings for complaint substrates. J. Dent .Res. 2008;87(8):701–709. doi: 10.1177/154405910808700808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stager M, Cerasale DJ, Dor R, Winker DW, Cherviron ZA. Signatures of natural selection in the mitochondrial genomes of Tachycineta swallows and their implications for latitudinal patterns of the ‘pace of life’. Gene. 2014;546:104–111. doi: 10.1016/j.gene.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 21.Lesser MP. Oxidative stress in marine environments: biochemistry and physiological ecology. Annu. Rev. Physiol. 2006;68:253–278. doi: 10.1146/annurev.physiol.68.040104.110001. [DOI] [PubMed] [Google Scholar]
- 22.Kowaltowski AJ, Vercesi AE. Mitochondrial damage induced by conditions of oxidative stress. Free Radic. Biol. Med. 1999;26:463–471. doi: 10.1016/S0891-5849(98)00216-0. [DOI] [PubMed] [Google Scholar]
- 23.Smith, A. M. & Callow, J. A. Biological adhesives. (ed. Smith). Berlin, Germany: (Springer 2006).
- 24.Gaita’n-Espitia J, Quintero-Galvis J, Mesas A, D’Eli’a G. Mitogenome of southern hemisphere blue mussels (Bivalvia:Pteriomorphia): insights into the evolutionary characteristics of the Mytilus edulis complex. Scin. Reprt. 2016;6:26853. doi: 10.1038/srep26853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benedict CV, Waite JHJH. Location and analysis of byssal structural proteins of Mytilus edulis. J. Morpho. 1986;2:171–181. doi: 10.1002/jmor.1051890207. [DOI] [PubMed] [Google Scholar]
- 26.Wood AR, Apte S, MacAvoy ES, Gardner JPA. A molecular phylogeny of the marine mussel genus Perna (Bivalvia: Mytilidae) based on nuclear (ITS1&2) and mitochondrial (CO1) DNA sequences. Mol. Phyl. Evol. 2007;44(2):685–698. doi: 10.1016/j.ympev.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 27.Anand PP. Computational modelling of human sarcomeric telethonin protein and predicting the functional effect of missense single nucleotide polymorphism. Curnt.Scin. 2019;117(4):638–648. doi: 10.18520/cs/v117/i4/638-648. [DOI] [Google Scholar]
- 28.Laskowaski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 29.Reynolds CR, Islam SA, Sternberg MJE. EzMol: A web server wizard for the rapid visualization and image production of protein and nucleic acid structures. J. Mol. Biol. 2018;430(15):2244–2248. doi: 10.1016/j.jmb.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kall L, Krogh A, Sonnhammer ELL. Advantages of combined transmembrane topology and signal peptide prediction the Phobius web server. Nucleic Acid Res. 2007;35:429–432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petersen TN, Brunak S, Von Heijne G, Nelsen H. SignaIP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 32.Kumari, S., Malik, M., Florival, C., Manalai, P. & Sonje, S. An assessment of five (PANSS, SAPS, SANS, NSA-16, CGI-SCH) commonly used symptoms rating scales in schizophrenia and comparison to newer scales (CAINS, BNSS). J.Addict Res Ther. 8,3, 14.4172/2155-6105.1000324 (2017). [DOI] [PMC free article] [PubMed]
- 33.Rees DJ, Hanifi A, Obille A, Alexander R, Sone ED. Fingerprinting of proteins that mediate Quagga mussel adhesion using a De Novo assembled foot transcriptome. Nat. Scin. Reprt. 2019;9:6305. doi: 10.1038/s41596019-41975-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willard L, et al. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acid Res. 2003;31(13):3316–3319. doi: 10.1093/nar/gkg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuscó Pol, Filion Guillaume J. Zerone: a ChIP-seq discretizer for multiple replicates with built-in quality control. Bioinformatics. 2016;32(19):2896–2902. doi: 10.1093/bioinformatics/btw336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evoln. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Bioinformatics. 1992;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 38.Nei, M. & Kumar, S. Molecular Evolution and Phylogenetics. (Oxford University Press, New York, 2000).
- 39.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 40.Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 2015;32(4):835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bayern, V., Janek., Grunwald., Ingo (Eds). Biological adhesives systems: from nature to technical and medical application. Vienna, Australia (Springer 2010).
- 42.Gasteiger, E. et al. Protein identification and analysis tools on the ExPasy server, The proteomics protocols Handbook. (In) John M. Walker (ed.) 571–607. (Humana Press, 2005).
- 43.Gray M. W. Mitochondrial Evolution. Cold Spring Harbor Perspectives in Biology. 2012;4(9):a011403–a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guerette PA, et al. Accelerating the design of biomimetic materials by integrating RNA-seq with proteomics and materials science. Nat. Biotechnol. 2013;31:908–915. doi: 10.1038/nbt.2671. [DOI] [PubMed] [Google Scholar]
- 45.Huber, M. Compendium of Bivalves. A full color guide to 3300 of the world’s marine bivalves. A status on Bivalvia after 250 years of Research. Conch Books, Hackenheim, 90pp + CD, ISBN: 978-939767-28-2. (2010)
- 46.Konstantin A, Lorenza B, Jurgen K, Torsten S. The Swiss-Model workspace: a web-based environment for protein structure and homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 47.Silverman HG, Roberto FF. Understanding marine mussel adhesion. Mar. Biotechnol. 2007;9:661–681. doi: 10.1007/s10126-007-9053-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Souji, S.M. Molluscan fauna of southeast coast of India with special reference to bivalvia. PhD Thesis. University of Kerala. (2018).
- 49.Von Heijne G. Signal sequences: The limits of variation. J. Mol. Biol. 1985;184(1):99–105. doi: 10.1016/0022-2386(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 50.Waite JH. Adhesion in byssally attached bivalves. Bio. Rev. 1983;58:209–231. doi: 10.1111/j.1469-185X.1983.tb00387.x. [DOI] [Google Scholar]
- 51.Wilker JJ. Redox and adhesion on the rocks. Nat. Chem. Biol. 2011;7:588–590. doi: 10.1038/nchembio.639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.