Abstract
Streptomyces are Gram-positive bacteria of significant industrial importance due to their ability to produce a wide range of antibiotics and bioactive secondary metabolites. Recent advances in genome mining have revealed that Streptomyces genomes possess a large number of unexplored silent secondary metabolite biosynthetic gene clusters (smBGCs). This indicates that Streptomyces genomes continue to be an invaluable source for new drug discovery. Here, we present high-quality genome sequences of 22 Streptomyces species and eight different Streptomyces venezuelae strains assembled by a hybrid strategy exploiting both long-read and short-read genome sequencing methods. The assembled genomes have more than 97.4% gene space completeness and total lengths ranging from 6.7 to 10.1 Mbp. Their annotation identified 7,000 protein coding genes, 20 rRNAs, and 68 tRNAs on average. In silico prediction of smBGCs identified a total of 922 clusters, including many clusters whose products are unknown. We anticipate that the availability of these genomes will accelerate discovery of novel secondary metabolites from Streptomyces and elucidate complex smBGC regulation.
Subject terms: Genomic analysis, Systems biology, Antibiotics
| Measurement(s) | DNA • genome • sequence_assembly • sequence feature annotation |
| Technology Type(s) | DNA sequencing • sequence assembly process • sequence annotation |
| Factor Type(s) | strain |
| Sample Characteristic - Organism | Streptomyces |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.11791323
Background & Summary
With the rapid emergence of antibiotic microbial resistance (AMR) to all major classes of antibiotics and the decline in number of potential candidates for new antibiotics, there is a pressing need for the discovery of novel antibacterial compounds1. Streptomyces, soil dwelling gram-positive bacteria, continue to be promising microorganisms for the production of clinically important secondary metabolites, including not only antibiotics, but also antiviral, antifungal, and antiparasitic agents, and antitumorals and immunosuppressant compounds2. Streptomyces are distinguished by their complex life cycle and high G + C content (often over 70%) in their linear genomes3,4. Traditionally, drug discovery from Streptomyces has been based on bioactivity screening followed by mass spectrometry and NMR-based molecular identification5. However, recent advances in genomics-based approaches revealed that most of the secondary metabolite biosynthetic gene clusters (smBGCs) of streptomycetes are inactive under laboratory conditions, suggesting that the ability of streptomycetes to produce secondary metabolites has been under-estimated5,6. Each Streptomyces species has the genetic potential to produce more than 30 secondary metabolites on average, which are diverse and differ between species7,8. Considering Streptomyces is the largest genus of actinobacteria with approximately 900 species characterized so far, streptomycetes are a valuable resource for the discovery of novel secondary metabolites9.
SmBGCs, especially polyketide and non-ribosomal peptide synthetase types, are often composed of extraordinarily long genes (>5 kb) encoding multi-modular enzymes with repetitive domain structures. Therefore, accurate gene annotations based on high quality genome sequences are essential for the precise identification of smBGCs10. Gene annotation with the high quality genome of S. clavuligerus revealed that 30% out of a total of 7,163 protein coding genes were incorrectly annotated in the previous draft genome of S. clavuligerus containing ambiguous and inaccurate nucleotides, indicating the importance of high quality genome sequences11. In addition, high quality genome sequences are essential for multi-omics analysis, which facilitates the understanding of the complex regulation on smBGCs and rational engineering for increasing secondary metabolites production11,12.
Among the 1,614 streptomycetes genomes that have been deposited in the NCBI Assembly database to date (as of 9th December 2019), only 189 and 35 assemblies were designated as complete genome level and chromosome level, respectively. More than 86% of assemblies were draft-quality genome sequences, which contain fragmented multiple contigs or ambiguous sequences4,13–15. One of the main obstacles to obtaining high quality genomic information of streptomycetes is the low fidelity of sequencing techniques when dealing with high G w C genomes and frequently repetitive sequences such as terminal inverted repeats13. In addition, since streptomycetes have linear chromosome, it is difficult to confirm the completeness of the assembled chromosome.
In this study, we present the high-quality genome sequences of 30 streptomycetes, increasing the total number of reported complete Streptomyces genome by about 10%. The target streptomycetes were 22 Streptomyces type strains and eight different Streptomyces venezuelae strains, most of which are currently used as industrial strains for producing various bioactive compounds. We applied hybrid assembly strategy with long-read (PacBio) and short-read (Illumina) sequencing techniques to obtain complete genome sequences. PacBio sequencing provides long reads of several kb in length which allows the readthrough of regions with low complexity, enabling the assembly of repetitive regions, which are difficult to assemble by using Illumina sequencing reads, even with the high coverage data16. However, Illumina sequencing provides reads with a lower error rate compared to the PacBio sequencing, and assembled contigs based on the Illumina sequencing reads are not simply a subset of the contigs from PacBio sequencing reads13,17. Therefore, reconciling PacBio and Illumina sequencing methods enables one to generate more complete genomes by overcoming the shortcomings of each method. During the genome assembly using reads from PacBio (0.46~5.18 Gbp) and Illumina (0.5~3.0 Gbp) sequencing, we constructed 6.7 to 10.1 Mbp of streptomycetes genomes, most of which consist of single chromosomes with 72% G + C contents on average. Inaccurate sequences in the assembled genome were corrected using Illumina sequencing reads. The complete streptomycetes genomes have more than 97.4% gene space completeness and on average 7,000 protein coding genes, 20 rRNAs, and 68 tRNAs were annotated. Finally, based on the complete genome sequences and annotations, we predicted a total of 922 smBGCs. The complete genome sequences and newly determined smBGCs in this study should prove to be a fundamental resource for understanding the genetic basis of streptomycetes and for discovering novel secondary metabolites.
Methods
Genomic DNA (gDNA) extraction
Total 30 streptomycetes were purchased from Korean Collection for Type Cultures (KCTC, Korea). A stock of streptomycetes were inoculated to 50 mL of liquid culture medium with 0.16 g mL−1 of glass beads (3 ± 0.3 mm diameter) in 250 mL baffled flask and grown at 30 °C in a 200 rpm orbital shaker. Each streptomycetes was grown in one of four different culture medium, R5(–) medium (25 mM TES (pH 7.2), 103 g L−1 sucrose, 1% (w/v) glucose, 5 g L−1 yeast extract, 10.12 g L−1 MgCl2∙6H2O, 0.25 g L−1 K2SO4, 0.1 g L−1 casamino acids, 0.08 g L−1 ZnCl2, 0.4 mg L−1 FeCl3, 0.02 mg L−1 CuCl2∙2H2O, 0.02 mg L−1 MnCl2∙4H2O, 0.02 mg L−1 Na2B4O7∙10H2O, and 0.02 mg L−1 (NH4)6Mo7O24∙4H2O), 1 × sporulation medium (3.33 g L−1 glucose, 1 g L−1 yeast extract, 1 g L−1 beef extract, 2 g L−1 tryptose, and 0.006 g L−1 FeSO4∙7H2O), YEME medium (340 g L−1 sucrose, 10 g L−1 glucose, 3 g L−1 yeast extract, 5 g L−1 bacto peptone, and 3 g L−1 oxoid malt extract), and MYM medium (4 g L−1 maltose, 4 g L−1 yeast extract, 10 g L−1 malt extract). For gDNA extraction, 25 mL cultured cells were harvested at the exponential growth phase and washed twice with same volume of 10 mM EDTA, followed by the lysozyme (10 mg mL−1) treatment at 37 °C for 45 min. gDNA was extracted using a Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA) according to the manufacturer’s instruction. Quality and quantity of extracted gDNA samples were evaluated using 1% agarose gel electrophoresis and Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA), respectively.
Short-read (Illumina) genome sequencing
For construction of short-read genome sequencing library, 2.5 μg of gDNA was sheared to approximately 350 bp by a Covaris instrument (Covaris Inc., Woburn, MA, USA) with the following conditions; Power 175, Duty factor 20%, C. burst 200, Time 23 s, 8 times. The library was constructed using a TruSeq DNA PCR-Free LT kit (Illumina Inc., San Diego, CA, USA) following manufacturer’s instruction. Briefly, the fragmented DNA samples were cleaned and end-repaired, followed by the adaptor ligation and bead-based size selection ranging from 400 to 500 bp. Quantity of final libraries was measured using Qubit® dsDNA HS Assay Kit (Thermo Fisher Scientific) and the library size was determined using Agilent 2200 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Among the constructed sequencing libraries, 29 libraries were sequenced with the HiSeq. 2500 (Illumina Inc.) as 100 bp single-end reads and remaining one library for S. tsukubaensis was sequenced with the Miseq v.2 (Illumina Inc.) with 50 bp single-read recipe. Finally, 0.46 to 5.18 Gbp of raw sequence data were obtained and the read qualities were examined by creating sequencing QC reports function of CLC genomic workbench version 6.5.1 (CLC bio, Denmark) (Online-only Table 1 and Fig. 1a).
Online-only Table 1.
Summary of PacBio and Illumina genome sequencing data for 30 streptomycetes.
| No. | Species | Strain | Platform | Raw reads (No.) | Mean raw reads length (bp) | Clean reads (No.) | Mean clean reads length (bp) | SRA accession number |
|---|---|---|---|---|---|---|---|---|
| 1 | Streptomyces clavuligerus | ATCC27064 | Illumina | 9,853,205 | 100 | 9,852,872 | 100 | SRR9192366 |
| pacBio | 150,292 | 6,572 | 60,834 | 15,473 | SRR9290551 | |||
| 2 | Streptomyces tsukubaensis | NRRL18488 | Illumina | 9,181,843 | 50 | 9,176,299 | 50 | SRR9192343 |
| pacBio | 150,292 | 8,163 | 93,481 | 12,715 | SRR9290550 | |||
| 3 | Streptomyces galilaeus | ATCC14969 | Illumina | 15,603,988 | 100 | 15,603,366 | 100 | SRR9192357 |
| pacBio | 150,292 | 5,458 | 95,691 | 8,339 | SRR9290549 | |||
| 4 | Streptomyces nitrosporeus | ATCC12769 | Illumina | 15,858,551 | 100 | 15,857,973 | 100 | SRR9192358 |
| pacBio | 150,292 | 5,007 | 101,572 | 7,222 | SRR9290548 | |||
| 5 | Streptomyces subrutilus | ATCC27467 | Illumina | 23,157,475 | 100 | 23,157,337 | 100 | SRR9192359 |
| pacBio | 150,292 | 8,757 | 92,569 | 13,591 | SRR9290547 | |||
| 6 | Streptomyces viridosporus T7A | ATCC39115 | Illumina | 24,919,393 | 100 | 24,918,458 | 100 | SRR9192360 |
| pacBio | 150,292 | 7,910 | 92,139 | 12,571 | SRR9290546 | |||
| 7 | Streptomyces kanamyceticus | ATCC12853 | Illumina | 14,877,606 | 100 | 14,877,044 | 100 | SRR9192361 |
| pacBio | 150,292 | 5,044 | 59,627 | 12,291 | SRR9290545 | |||
| 8 | Streptomyces aureofaciens | ATCC11989 | Illumina | 19,155,832 | 100 | 19,155,718 | 100 | SRR9192362 |
| pacBio | 150,292 | 5,890 | 88,462 | 9,681 | SRR9290544 | |||
| 9 | Streptomyces prasinus | ATCC13879 | Illumina | 18,655,418 | 100 | 18,654,718 | 100 | SRR9192388 |
| pacBio | 150,292 | 5,560 | 84,107 | 9,288 | SRR9290553 | |||
| 10 | Streptomyces fradiae | ATCC10745 | Illumina | 9,446,513 | 100 | 9,445,832 | 100 | SRR9192354 |
| pacBio | 150,292 | 6,843 | 94,568 | 10,538 | SRR9290561 | |||
| 11 | Streptomyces alboniger | ATCC12461 | Illumina | 16,425,356 | 100 | 16,425,241 | 100 | SRR9192355 |
| pacBio | 300,584 | 3,638 | 159,270 | 6,351 | SRR9290560 | |||
| 12 | Streptomyces coeruleorubidus | ATCC13740 | Illumina | 17,546,981 | 100 | 17,546,866 | 100 | SRR9192339 |
| pacBio | 300,584 | 2,836 | 121,377 | 6,164 | SRR9290563 | |||
| 13 | Streptomyces cinereoruber | ATCC19740 | Illumina | 20,182,754 | 100 | 20,181,890 | 100 | SRR9192340 |
| pacBio | 150,292 | 3,319 | 52,279 | 9,266 | SRR9290562 | |||
| 14 | Streptomyces nodosus | ATCC14899 | Illumina | 14,034,927 | 100 | 14,034,843 | 100 | SRR9192341 |
| pacBio | 150,292 | 8,387 | 94,719 | 12,692 | SRR9290557 | |||
| 15 | Streptomyces vinaceus | ATCC27476 | Illumina | 6,612,160 | 100 | 6,610,219 | 100 | SRR9192335 |
| pacBio | 150,292 | 4,984 | 75,928 | 9,432 | SRR9290559 | |||
| 16 | Streptomyces platensis | ATCC23948 | Illumina | 14,819,357 | 100 | 14,819,243 | 100 | SRR9192336 |
| pacBio | 150,292 | 4,869 | 72,573 | 9,619 | SRR9290558 | |||
| 17 | Streptomyces spectabilis | ATCC27465 | Illumina | 18,717,018 | 100 | 18,716,280 | 100 | SRR9192337 |
| pacBio | 300,584 | 10,049 | 202,966 | 14,047 | SRR9290555 | |||
| 18 | Streptomyces chartreusis | ATCC14922 | Illumina | 16,988,966 | 100 | 16,988,850 | 100 | SRR9192338 |
| pacBio | 150,292 | 8,813 | 88,869 | 14,106 | SRR9290554 | |||
| 19 | Stretpomyces rimosus | ATCC10970 | Illumina | 25,109,758 | 100 | 25,108,764 | 100 | SRR9192333 |
| pacBio | 300,584 | 7,066 | 164,891 | 12,342 | SRR9290564 | |||
| 20 | Streptomyces albofaciens | ATCC23873 | Illumina | 19,360,703 | 100 | 19,360,564 | 100 | SRR9192364 |
| pacBio | 150,292 | 11,091 | 110,521 | 14,419 | SRR9290552 | |||
| 21 | Streptomyces filamentosus | ATCC23958 | Illumina | 21,181,460 | 100 | 21,180,615 | 100 | SRR9192342 |
| pacBio | 150,292 | 4,805 | 70,677 | 8,178 | SRR9290556 | |||
| 22 | Streptomyces venezuelae ATCC 10712 | ATCC 10712 | Illumina | 18,667,122 | 100 | 18,664,663 | 100 | SRR9192374 |
| pacBio | 150,292 | 9,101 | 87,860 | 15,328 | SRR9290565 | |||
| 23 | Streptomyces venezuelae ATCC 21113 | ATCC 21113 | Illumina | 21,561,533 | 100 | 21,559,491 | 100 | SRR9192334 |
| pacBio | 150,292 | 10,148 | 104,695 | 14,180 | SRR9290566 | |||
| 24 | Streptomyces venezuelae ATCC 10595 | ATCC 10595 | Illumina | 16,798,923 | 100 | 16,797,236 | 100 | SRR9192369 |
| pacBio | 150,292 | 7,829 | 92,681 | 12,197 | SRR9290567 | |||
| 25 | Streptomyces venezuelae ATCC 15068 | ATCC 15068 | Illumina | 15,310,620 | 100 | 15,308,905 | 100 | SRR9192368 |
| pacBio | 150,292 | 10,754 | 107,412 | 14,515 | SRR9290568 | |||
| 26 | Streptomyces venezuelae ATCC 14583 | ATCC 14583 | Illumina | 19,423,668 | 100 | 19,421,332 | 100 | SRR9192371 |
| pacBio | 150,292 | 10,888 | 106,813 | 14,647 | SRR9290569 | |||
| 27 | Streptomyces venezuelae ATCC 14584 | ATCC 14584 | Illumina | 15,447,783 | 100 | 15,446,240 | 100 | SRR9192370 |
| pacBio | 150,292 | 8,844 | 100,523 | 12,580 | SRR9290540 | |||
| 28 | Streptomyces venezuelae ATCC 14585 | ATCC 14585 | Illumina | 51,795,644 | 100 | 51,791,331 | 100 | SRR9192373 |
| pacBio | 150,292 | 8,275 | 96,243 | 12,388 | SRR9290541 | |||
| 29 | Streptomyces venezuelae ATCC 21782 | ATCC 21782 | Illumina | 19,569,337 | 100 | 19,567,069 | 100 | SRR9192372 |
| pacBio | 150,292 | 6,539 | 70,655 | 13,089 | SRR9290542 | |||
| 30 | Streptomyces venezuelae ATCC 21018 | ATCC 21018 | Illumina | 16,469,406 | 100 | 16,467,790 | 100 | SRR9192375 |
| pacBio | 150,292 | 10,754 | 107,412 | 14,515 | SRR9290543 |
Fig. 1.
Quality of the genome sequencing data. (a) Distribution of Illumina reads quality based on Phred score. (b) Read quality distribution of PacBio reads. Black line indicates total number of bases in the reads which have greater read quality than the corresponding read quality value on x-axis.
Long-read (PacBio) genome sequencing
A total of 5 μg gDNA was used as input for PacBio genome sequencing library preparation. The sequencing library was constructed with the PacBio SMRTbellTM Template Prep Kit (Pacific Biosciences, Menlo Park, CA, USA) following manufacturer’s instructions. Fragments smaller than 20 kbp were removed using the Blue Pippin Size selection system (Sage Science, Beverly, MA, USA) and the constructed libraries were validated using Agilent 2100 Bioanalyzer (Agilent Technologies). Final SMRTbell libraries were sequenced using one or two SMRT cells with P6-C4-chemistry (DNA Sequencing Reagent 4.0) on the PacBio RS II sequencing platform (Pacific Biosciences). Approximately, 0.5 to 3.0 Gbp of raw sequence data were generated (Online-only Table 1).
Genome assembly
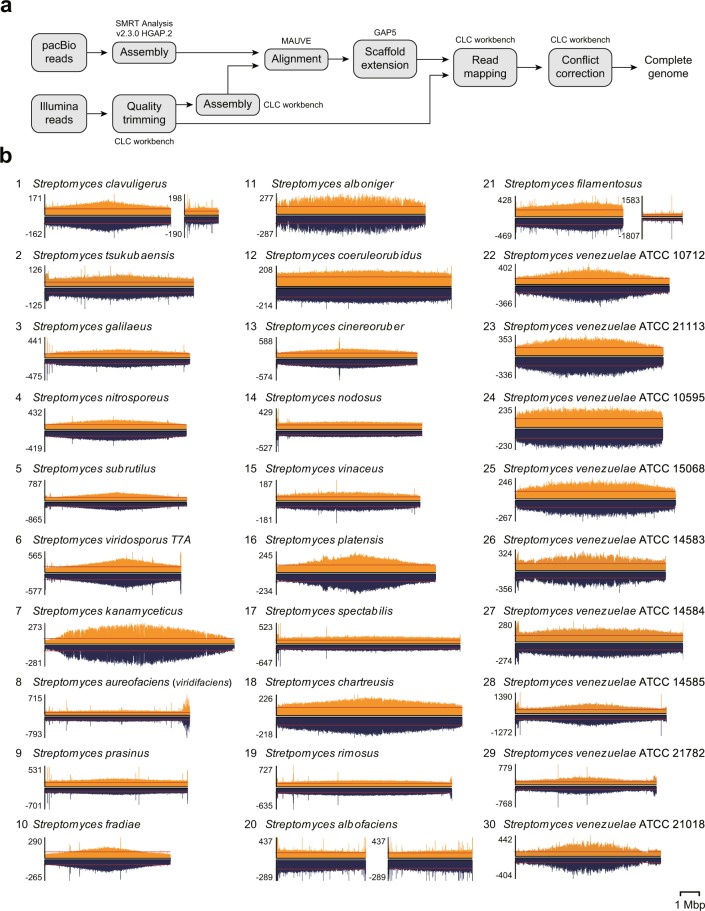
Among the raw PacBio sequencing reads, only the reads with a read quality value greater than 0.75 and a length longer than 50 bp were filtered (Fig. 1b). Post filtered reads were assembled by the hierarchical genome assembly process workflow (HGAP, Version 2.3), including consensus polishing with Quiver18. For each assembled contig, error correction was performed based on their estimated genome size and average coverage. Raw reads from the Illumina sequencing were quality trimmed using CLC genomic workbench version 6.5.1 (ambiguous limit 2 and quality limit 0.05) and assembled using de novo assembly function of CLC genomic workbench version 6.5.1 with default parameters. To expand the assembled contigs, all of assembled PacBio and Illumina contigs were aligned using MAUVE 2.4.019 and linked using GAP5 program (Staden package)20.
Genome correction
Quality trimmed Illumina sequencing reads were mapped to the assembled genome using CLC genomic workbench version 6.5.1 (mismatch cost 2, insertion cost 3, deletion cost 3, length fraction 0.9, and similarity fraction 0.9). Conflicts showing more than 80% frequency for Illumina reads were corrected as Illumina sequence (Table 1). In addition, percentage of mapped Illumina reads on to the assembled genome represents degree of completeness (Table 1 and Fig. 2b). Completeness of gene space was estimated using the BUSCO v3 (Table 2)21.
Table 1.
The statistics of genome assembly and correction.
| No. | Species | Final scaffolds (No.) | Scaffold length before correction (bp) | Mapped Illumina reads (%) | Conflict positions (No.) | Added bases (No.) | Deleted bases (No.) | Scaffold length after correction (bp) | G + C contets (%) | Assembly accession number |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Streptomyces clavuligerus | 2 | 6,748,589 and 1,795,496 | 71.16 and 14.03 | 7 | 4 | 3 | 6,748,591 and 1,795,495 | 72.5 | GCA_005519465.1 |
| 2 | Streptomyces tsukubaensis | 1 | 7,963,727 | 95.13 | 15 | 15 | 0 | 7,963,742 | 71.9 | GCA_003932715.1 |
| 3 | Streptomyces galilaeus | 1 | 7,756,176 | 90.56 | 51 | 34 | 16 | 7,756,194 | 71.4 | GCA_008704575.1 |
| 4 | Streptomyces nitrosporeus | 1 | 7,581,543 | 93.50 | 51 | 35 | 16 | 7,581,562 | 72.2 | GCA_008704555.1 |
| 5 | Streptomyces subrutilus | 1 | 7,604,705 | 96.41 | 286 | 269 | 0 | 7,604,974 | 73.4 | GCA_008704535.1 |
| 6 | Streptomyces viridosporus T7A | 1 | 7,280,447 | 90.44 | 90 | 89 | 0 | 7,280,536 | 72.6 | GCA_008704515.1 |
| 7 | Streptomyces kanamyceticus | 1 | 10,133,525 | 99.09 | 376 | 375 | 3 | 10,133,897 | 71.0 | GCA_008704495.1 |
| 8 | Streptomyces aureofaciens | 1 | 7,757,873 | 84.86 | 16 | 9 | 5 | 7,757,877 | 72.6 | GCA_008704475.1 |
| 9 | Streptomyces prasinus | 1 | 7,646,576 | 89.70 | 1,025 | 1,021 | 5 | 7,647,592 | 72.0 | GCA_008704445.1 |
| 10 | Streptomyces fradiae | 1 | 6,725,574 | 97.63 | 5 | 5 | 0 | 6,725,579 | 74.7 | GCA_008704425.1 |
| 11 | Streptomyces alboniger | 1 | 7,962,594 | 99.12 | 193 | 193 | 1 | 7,962,786 | 71.2 | GCA_008704395.1 |
| 12 | Streptomyces coeruleorubidus | 1 | 9,334,399 | 99.67 | 1,297 | 1,299 | 0 | 9,335,698 | 71.1 | GCA_008705135.1 |
| 13 | Streptomyces cinereoruber | 1 | 7,516,474 | 99.74 | 178 | 178 | 0 | 7,516,652 | 72.9 | GCA_009299385.1 |
| 14 | Streptomyces nodosus | 1 | 7,772,564 | 99.51 | 26 | 25 | 2 | 7,772,587 | 70.9 | GCA_008704995.1 |
| 15 | Streptomyces vinaceus | 1 | 7,673,329 | 92.46 | 180 | 180 | 0 | 7,673,509 | 72.3 | GCA_008704935.1 |
| 16 | Streptomyces platensis | 1 | 8,500,673 | 99.75 | 354 | 352 | 13 | 8,501,012 | 71.1 | GCA_008704855.1 |
| 17 | Streptomyces spectabilis | 1 | 9,806,222 | 95.30 | 934 | 938 | 0 | 9,807,160 | 72.4 | GCA_008704795.1 |
| 18 | Streptomyces chartreusis | 1 | 9,911,637 | 98.42 | 461 | 461 | 0 | 9,912,098 | 71.0 | GCA_008704715.1 |
| 19 | Stretpomyces rimosus | 1 | 9,361,132 | 96.22 | 22 | 22 | 0 | 9,361,154 | 72.0 | GCA_008704655.1 |
| 20 | Streptomyces albofaciens | 2 | 4,757,761 and 4,494,336 | 53.36 and 45.53 | 504 | 501 | 3 | 4,757,978 and 4,494,617 | 72.3 | GCA_008634025.1 |
| 21 | Streptomyces filamentosus | 2 | 5,742,252 and 2,129,928 | 75.22 and 24.28 | 3,218 | 3,228 | 1 | 5,744,022 and 2,131,385 | 73.6 | GCA_008634015.1 |
| 22 | Streptomyces venezuelae ATCC 10712 | 1 | 8,223,439 | 99.84 | 96 | 81 | 15 | 8,223,505 | 72.5 | GCA_008639165.1 |
| 23 | Streptomyces venezuelae ATCC 21113 | 1 | 7,893,622 | 99.85 | 173 | 181 | 0 | 7,893,803 | 72.5 | GCA_008639045.1 |
| 24 | Streptomyces venezuelae ATCC 10595 | 1 | 7,871,449 | 95.50 | 35 | 34 | 3 | 7,871,480 | 72.5 | GCA_008705255.1 |
| 25 | Streptomyces venezuelae ATCC 15068 | 1 | 8,557,615 | 99.71 | 587 | 587 | 0 | 8,558,202 | 71.9 | GCA_008642375.1 |
| 26 | Streptomyces venezuelae ATCC 14583 | 1 | 8,018,461 | 87.17 | 29 | 27 | 4 | 8,018,484 | 71.3 | GCA_008642355.1 |
| 27 | Streptomyces venezuelae ATCC 14584 | 1 | 8,941,823 | 99.00 | 255 | 255 | 0 | 8,942,078 | 71.2 | GCA_008642315.1 |
| 28 | Streptomyces venezuelae ATCC 14585 | 1 | 8,048,139 | 82.34 | 64 | 41 | 26 | 8,048,154 | 71.3 | GCA_008642335.1 |
| 29 | Streptomyces venezuelae ATCC 21782 | 1 | 7,525,235 | 90.50 | 87 | 87 | 0 | 7,525,322 | 71.9 | GCA_008642295.1 |
| 30 | Streptomyces venezuelae ATCC 21018 | 1 | 7,746,214 | 91.61 | 59 | 57 | 4 | 7,746,267 | 72.1 | GCA_008642275.1 |
Fig. 2.
Genome assembly of 30 streptomycetes. (a) Strategy for genome assembly and corrections. (b) Profile of Illumina reads mapped on assembled genomes. Data were visualized using SignalMap (Roche NimbleGen, Inc.). Red line indicates the average Illumina read coverage of all genomic positions.
Table 2.
Gene space completeness of completed genomes.
| No. | Species | Complete and single-copy | Complete and duplicated | Fragmented | Missing | Total | Gene space completeness (%) |
|---|---|---|---|---|---|---|---|
| 1 | Streptomyces clavuligerus | 343 | 0 | 0 | 9 | 352 | 97.4 |
| 2 | Streptomyces tsukubaensis | 350 | 0 | 0 | 2 | 352 | 99.4 |
| 3 | Streptomyces galilaeus | 351 | 0 | 0 | 1 | 352 | 99.7 |
| 4 | Streptomyces nitrosporeus | 352 | 0 | 0 | 0 | 352 | 100.0 |
| 5 | Streptomyces subrutilus | 349 | 0 | 0 | 3 | 352 | 99.1 |
| 6 | Streptomyces viridosporus T7A | 351 | 0 | 0 | 1 | 352 | 99.7 |
| 7 | Streptomyces kanamyceticus | 352 | 0 | 0 | 0 | 352 | 100.0 |
| 8 | Streptomyces aureofaciens | 350 | 0 | 0 | 2 | 352 | 99.4 |
| 9 | Streptomyces prasinus | 350 | 0 | 0 | 2 | 352 | 99.4 |
| 10 | Streptomyces fradiae | 351 | 0 | 0 | 1 | 352 | 99.7 |
| 11 | Streptomyces alboniger | 351 | 0 | 0 | 1 | 352 | 99.7 |
| 12 | Streptomyces coeruleorubidus | 351 | 0 | 0 | 1 | 352 | 99.7 |
| 13 | Streptomyces cinereoruber | 351 | 0 | 0 | 1 | 352 | 99.7 |
| 14 | Streptomyces nodosus | 350 | 0 | 1 | 1 | 352 | 99.4 |
| 15 | Streptomyces vinaceus | 349 | 0 | 1 | 2 | 352 | 99.1 |
| 16 | Streptomyces platensis | 351 | 0 | 0 | 1 | 352 | 99.7 |
| 17 | Streptomyces spectabilis | 350 | 0 | 1 | 1 | 352 | 99.4 |
| 18 | Streptomyces chartreusis | 351 | 0 | 0 | 1 | 352 | 99.7 |
| 19 | Stretpomyces rimosus | 351 | 0 | 0 | 1 | 352 | 99.7 |
| 20 | Streptomyces albofaciens | 346 | 4 | 0 | 2 | 352 | 99.4 |
| 21 | Streptomyces filamentosus | 351 | 0 | 0 | 1 | 352 | 99.7 |
| 22 | Streptomyces venezuelae ATCC 10712 | 352 | 0 | 0 | 0 | 352 | 100.0 |
| 23 | Streptomyces venezuelae ATCC 21113 | 352 | 0 | 0 | 0 | 352 | 100.0 |
| 24 | Streptomyces venezuelae ATCC 10595 | 352 | 0 | 0 | 0 | 352 | 100.0 |
| 25 | Streptomyces venezuelae ATCC 15068 | 351 | 0 | 0 | 1 | 352 | 99.7 |
| 26 | Streptomyces venezuelae ATCC 14583 | 351 | 0 | 0 | 1 | 352 | 99.7 |
| 27 | Streptomyces venezuelae ATCC 14584 | 351 | 0 | 0 | 1 | 352 | 99.7 |
| 28 | Streptomyces venezuelae ATCC 14585 | 351 | 0 | 0 | 1 | 352 | 99.7 |
| 29 | Streptomyces venezuelae ATCC 21782 | 349 | 0 | 0 | 3 | 352 | 99.1 |
| 30 | Streptomyces venezuelae ATCC 21018 | 350 | 0 | 0 | 2 | 352 | 99.4 |
Genome annotation and secondary metabolite biosynthetic gene cluster prediction
The complete genome sequences of streptomycetes were submitted to the NCBI GenBank database and annotated by the latest updated version of NCBI Prokaryotic Genome Annotation Pipeline (PGAP)22. Using the GenBank formatted files of each genomes as input, secondary metabolite biosynthetic gene clusters were predicted by antiSMASH 4.023.
Data Records
Raw reads from short-read (Illumina) and long-read (PacBio) sequencing were deposited in the NCBI Sequence Read Archive (SRA) (Online-only Table 1)24,25. 30 complete genome sequences were deposited in GenBank via the NCBI’s submission portal (Table 3)26–55. Detailed information on the predicted 922 smBGCs in 30 streptomycetes genomes has been deposited in FigShare56.
Table 3.
Summary of genome annotation.
| No. | Species | CDS (No.) | 16s rRNA (No.) | tRNA (No.) | Genome accession number | BioProject accession number |
|---|---|---|---|---|---|---|
| 1 | Streptomyces clavuligerus | 6,880 | 18 | 66 | CP027858 | PRJNA414136 |
| 2 | Streptomyces tsukubaensis | 6,376 | 18 | 66 | CP020700 | PRJNA382016 |
| 3 | Streptomyces galilaeus | 6,725 | 18 | 76 | CP023703 | PRJNA412292 |
| 4 | Streptomyces nitrosporeus | 6,364 | 18 | 74 | CP023702 | PRJNA412292 |
| 5 | Streptomyces subrutilus | 6,431 | 21 | 68 | CP023701 | PRJNA412292 |
| 6 | Streptomyces viridosporus T7A | 6,211 | 18 | 70 | CP023700 | PRJNA412292 |
| 7 | Streptomyces kanamyceticus | 8,384 | 18 | 66 | CP023699 | PRJNA412292 |
| 8 | Streptomyces aureofaciens | 6,453 | 33 | 71 | CP023698 | PRJNA412292 |
| 9 | Streptomyces prasinus | 6,263 | 18 | 68 | CP023697 | PRJNA412292 |
| 10 | Streptomyces fradiae | 5,465 | 18 | 65 | CP023696 | PRJNA412292 |
| 11 | Streptomyces alboniger | 6,613 | 18 | 67 | CP023695 | PRJNA412292 |
| 12 | Streptomyces coeruleorubidus | 8,058 | 18 | 67 | CP023694 | PRJNA412292 |
| 13 | Streptomyces cinereoruber | 6,392 | 18 | 69 | CP023693 | PRJNA412292 |
| 14 | Streptomyces nodosus | 6,491 | 18 | 68 | CP023747 | PRJNA412292 |
| 15 | Streptomyces vinaceus | 6,603 | 21 | 68 | CP023692 | PRJNA412292 |
| 16 | Streptomyces platensis | 7,032 | 21 | 67 | CP023691 | PRJNA412292 |
| 17 | Streptomyces spectabilis | 8,212 | 18 | 65 | CP023690 | PRJNA412292 |
| 18 | Streptomyces chartreusis | 8,396 | 18 | 71 | CP023689 | PRJNA412292 |
| 19 | Stretpomyces rimosus | 7,756 | 21 | 68 | CP023688 | PRJNA412292 |
| 20 | Streptomyces albofaciens | 7,520 | 21 | 67 | PDCM00000000 | PRJNA412292 |
| 21 | Streptomyces filamentosus | 6,832 | 24 | 70 | PDCL00000000 | PRJNA412292 |
| 22 | Streptomyces venezuelae ATCC 10712 | 7,377 | 21 | 67 | CP029197 | PRJNA454547 |
| 23 | Streptomyces venezuelae ATCC 21113 | 6,987 | 21 | 67 | CP029196 | PRJNA454547 |
| 24 | Streptomyces venezuelae ATCC 10595 | 6,942 | 21 | 67 | CP029195 | PRJNA454547 |
| 25 | Streptomyces venezuelae ATCC 15068 | 7,700 | 21 | 69 | CP029194 | PRJNA454547 |
| 26 | Streptomyces venezuelae ATCC 14583 | 7,154 | 18 | 66 | CP029193 | PRJNA454547 |
| 27 | Streptomyces venezuelae ATCC 14584 | 7,832 | 18 | 65 | CP029192 | PRJNA454547 |
| 28 | Streptomyces venezuelae ATCC 14585 | 7,096 | 18 | 66 | CP029191 | PRJNA454547 |
| 29 | Streptomyces venezuelae ATCC 21782 | 6,655 | 18 | 69 | CP029190 | PRJNA454547 |
| 30 | Streptomyces venezuelae ATCC 21018 | 6,769 | 21 | 71 | CP029189 | PRJNA454547 |
Technical Validation
Streptomyces have drawn considerable attention because of their ability to produce various clinically important secondary metabolites. Total 30 streptomycetes genomes were sequenced by using both PacBio and Illumina sequencing methods to elucidate their biosynthetic potential. After cleaning the reads, on average 98,380 PacBio reads with 11,725 bp length and 18,223,235 Illumina reads with 100 bp length (50 bp for S. tsukubaensis) were generated (Fig. 1a,b and Online-only Table 1). Through the assembly of reads from two sequencing platforms using HGAP, CLC workbench, MAUVE, and GAP5 programs, single linear scaffolds ranging from 6.7 to 10.1 Mbp in length with 72% G + C contents were obtained for 27 streptomycetes, whereas two scaffolds were finally constructed for three remaining streptomycetes, S. clavuligerus (6.7 and 1.8 Mbp), S. albofaciens (4.8 and 4.5 Mbp), and S. filamentosus (5.7 and 2.1 Mbp) (Table 1). S. clavuligerus has been reported to have a large linear plasmid with a length of 1.8 Mbp, so the genome was correctly assembled into a single chromosome, while the S. albofaciens and S. filamentosus genomes appear to be assembled into two divided scaffolds11,57. To increase the accuracy of the assembled genome sequences, Illumina sequences showing more than 80% coverage at the conflict sites were taken as the corrected ones (Table 1). Approximately, 96.32% of Illumina sequencing reads were successfully mapped to the corresponding genomes (Table 1 and Fig. 2b). The completeness of the genomes were assessed using the BUSCO approach with a total of 352 orthologue groups from the Actinobacteria Dataset21. Results showed that 29 genomes have more than 99.1% gene space completeness and the S. clavuligerus genome has 97.4% gene space completeness (Table 2). Following NCBI PGAP, 30 genomes were annotated with 7,000 protein coding genes, 20 rRNAs, and 68 tRNAs on average (Table 3). Finally, based on the annotation, a total of 922 smBGCs were predicted in 30 streptomycetes genomes (Fig. 3). Detailed information, such as genomic positions, types, and putative products of each smBGC are publicly available in Figshare56.
Fig. 3.
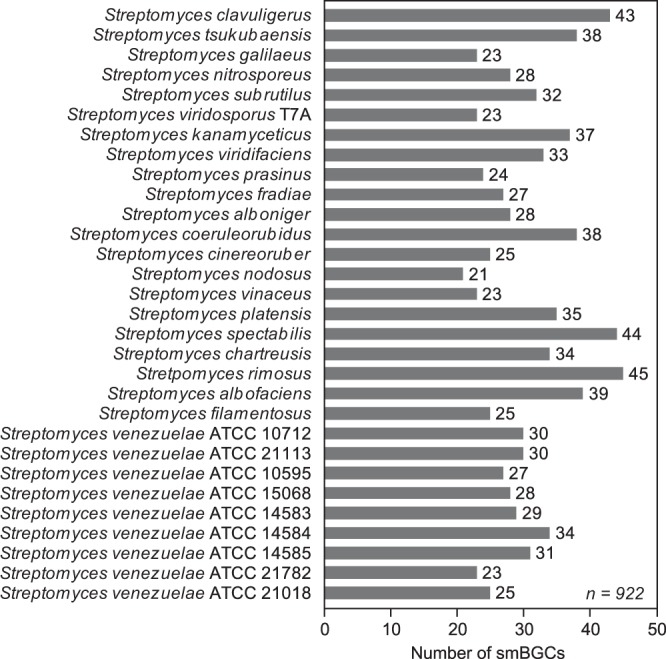
Secondary metabolite biosynthetic gene clusters in 30 complete streptomycetes genomes.
Acknowledgements
This work was supported by a grant from the Novo Nordisk Foundation (grant number NNF10CC1016517). This work was also supported by the Bio & Medical Technology Development Program (2018M3A9F3079664 to B.-K.C.) through the National Research Foundation (NRF) funded by the Ministry of Science and ICT (MSIT).
Online-only Table
Author contributions
B.-K.C. conceived and supervised the study. N.L. and B.-K.C. designed the experiments. N.L., W.K., S.H. and Y.L. performed the experiments. N.L., W.K., S.H., Y.L., S.C., B.P. and B.-K.C. analyzed the data. N.L., W.K., S.C., B.P. and B.-K.C. wrote the manuscript.
Code availability
The version and parameter of all bioinformatics tools used in this work are described in the Methods section.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Namil Lee and Woori Kim.
References
- 1.Genilloud O. The re-emerging role of microbial natural products in antibiotic discovery. Antonie Van Leeuwenhoek. 2014;106:173–188. doi: 10.1007/s10482-014-0204-6. [DOI] [PubMed] [Google Scholar]
- 2.Jones SE, Elliot MA. Streptomyces exploration: Competition, volatile communication and new bacterial behaviours. Trends Microbiol. 2017;25:522–531. doi: 10.1016/j.tim.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Hopwood DA. Soil to genomics: the Streptomyces chromosome. Annu Rev Genet. 2006;40:1–23. doi: 10.1146/annurev.genet.40.110405.090639. [DOI] [PubMed] [Google Scholar]
- 4.Lee N, et al. Synthetic biology tools for novel secondary metabolite discovery in Streptomyces. J Microbiol Biotechnol. 2019;29:667–686. doi: 10.4014/jmb.1904.04015. [DOI] [PubMed] [Google Scholar]
- 5.Ziemert N, Alanjary M, Weber T. The evolution of genome mining in microbes - a review. Nat Prod Rep. 2016;33:988–1005. doi: 10.1039/C6NP00025H. [DOI] [PubMed] [Google Scholar]
- 6.Rebets Y, Brotz E, Tokovenko B, Luzhetskyy A. Actinomycetes biosynthetic potential: how to bridge in silico and in vivo? J Ind Microbiol Biotechnol. 2014;41:387–402. doi: 10.1007/s10295-013-1352-9. [DOI] [PubMed] [Google Scholar]
- 7.Bentley SD, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 8.Omura S, et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reva O, Tummler B. Think big–giant genes in bacteria. Environ Microbiol. 2008;10:768–777. doi: 10.1111/j.1462-2920.2007.01500.x. [DOI] [PubMed] [Google Scholar]
- 11.Hwang S, et al. Primary transcriptome and translatome analysis determines transcriptional and translational regulatory elements encoded in the Streptomyces clavuligerus genome. Nucleic Acids Res. 2019;47:6114–6129. doi: 10.1093/nar/gkz471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Zhang C, Liu C, Ju J, Ma J. Genome sequencing of Streptomyces atratus SCSIOZH16 and activation production of nocardamine via metabolic engineering. Front Microbiol. 2018;9:1269. doi: 10.3389/fmicb.2018.01269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison J, Studholme DJ. Recently published Streptomyces genome sequences. Microb Biotechnol. 2014;7:373–380. doi: 10.1111/1751-7915.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barreiro C, et al. Draft genome of Streptomyces tsukubaensis NRRL 18488, the producer of the clinically important immunosuppressant tacrolimus (FK506) J Bacteriol. 2012;194:3756–3757. doi: 10.1128/JB.00692-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song JY, et al. Draft genome sequence of Streptomyces clavuligerus NRRL 3585, a producer of diverse secondary metabolites. J Bacteriol. 2010;192:6317–6318. doi: 10.1128/JB.00859-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koren S, et al. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 2013;14:R101. doi: 10.1186/gb-2013-14-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ardui S, Ameur A, Vermeesch JR, Hestand MS. Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res. 2018;46:2159–2168. doi: 10.1093/nar/gky066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 19.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonfield JK, Whitwham A. Gap5–editing the billion fragment sequence assembly. Bioinformatics. 2010;26:1699–1703. doi: 10.1093/bioinformatics/btq268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 22.Haft DH, et al. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018;46:D851–D860. doi: 10.1093/nar/gkx1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blin K, et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45:W36–W41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leinonen R, Sugawara H, Shumway M. & International Nucleotide Sequence Database, C. The sequence read archive. Nucleic Acids Res. 2011;39:D19–21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.2019. NCBI Sequence Read Archive. SRP200324
- 26.2018. GenBank. CP020700
- 27.2018. GenBank. CP023688
- 28.2018. GenBank. CP023689
- 29.2018. GenBank. CP023690
- 30.2018. GenBank. CP023691
- 31.2018. GenBank. CP023692
- 32.2018. GenBank. CP023693
- 33.2018. GenBank. CP023694
- 34.2018. GenBank. CP023695
- 35.2018. GenBank. CP023696
- 36.2018. GenBank. CP023697
- 37.2018. GenBank. CP023698
- 38.2018. GenBank. CP023699
- 39.2018. GenBank. CP023700
- 40.2018. GenBank. CP023701
- 41.2018. GenBank. CP023702
- 42.2018. GenBank. CP023703
- 43.2018. GenBank. CP023747
- 44.2018. GenBank. CP029189
- 45.2018. GenBank. CP029190
- 46.2018. GenBank. CP029191
- 47.2018. GenBank. CP029192
- 48.2018. GenBank. CP029193
- 49.2018. GenBank. CP029194
- 50.2018. GenBank. CP029195
- 51.2018. GenBank. CP029196
- 52.2018. GenBank. CP029197
- 53.2018. GenBank. PDCL00000000
- 54.2018. GenBank. PDCM00000000
- 55.2019. GenBank. CP027858
- 56.Lee N, 2020. Thirty complete Streptomyces genome sequences for mining novel secondary metabolite biosynthetic gene clusters. figshare. [DOI] [PMC free article] [PubMed]
- 57.Medema MH, et al. The sequence of a 1.8-Mb bacterial linear plasmid reveals a rich evolutionary reservoir of secondary metabolic pathways. Genome Biol Evol. 2010;2:212–224. doi: 10.1093/gbe/evq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2019. NCBI Sequence Read Archive. SRP200324
- 2018. GenBank. CP020700
- 2018. GenBank. CP023688
- 2018. GenBank. CP023689
- 2018. GenBank. CP023690
- 2018. GenBank. CP023691
- 2018. GenBank. CP023692
- 2018. GenBank. CP023693
- 2018. GenBank. CP023694
- 2018. GenBank. CP023695
- 2018. GenBank. CP023696
- 2018. GenBank. CP023697
- 2018. GenBank. CP023698
- 2018. GenBank. CP023699
- 2018. GenBank. CP023700
- 2018. GenBank. CP023701
- 2018. GenBank. CP023702
- 2018. GenBank. CP023703
- 2018. GenBank. CP023747
- 2018. GenBank. CP029189
- 2018. GenBank. CP029190
- 2018. GenBank. CP029191
- 2018. GenBank. CP029192
- 2018. GenBank. CP029193
- 2018. GenBank. CP029194
- 2018. GenBank. CP029195
- 2018. GenBank. CP029196
- 2018. GenBank. CP029197
- 2018. GenBank. PDCL00000000
- 2018. GenBank. PDCM00000000
- 2019. GenBank. CP027858
- Lee N, 2020. Thirty complete Streptomyces genome sequences for mining novel secondary metabolite biosynthetic gene clusters. figshare. [DOI] [PMC free article] [PubMed]
Data Availability Statement
The version and parameter of all bioinformatics tools used in this work are described in the Methods section.