Abstract
Variation in the microbiome among individual organisms may play a critical role in the relative susceptibility of those organisms to infection, disease, and death. However, predicting microbiome function is difficult because of spatial and temporal variation in microbial diversity, and taxonomic diversity is not predictive of microbiome functional diversity. Addressing this issue may be particularly important when addressing pandemic diseases, such as the global amphibian die-off associated with Bd. Some of the most important factors in probiotic development for disease treatment are whether bacteria with desired function can be found on native amphibians in the local environment. To address this issue, we isolated, sequenced, and assayed the cutaneous bacterial communities of Plethodon cinereus along a gradient of land use change. Our results suggest that cutaneous community composition, but not overall diversity, change with changes in land use, but this does not correspond to significant change in Bd-inhibitory function. We found that Bd-inhibition is a functionally redundant trait, but that level of inhibition varies over phylogenetic, spatial, and temporal scales. This research provides further evidence for the importance of continued examination of amphibian microbial communities across environmental gradients, including biotic and abiotic interactions, when considering disease dynamics.
Keywords: Batrachochytrium dendrobatidis, microbiome, disease, urbanization, functional redundancy, spatial variation
Introduction
The diversity of microbes on an individual has been implicated in the health and resilience of organisms in many taxa, from plants (Mendes et al., 2013) to animals (Yildirim et al., 2010; Sanchez et al., 2012; Bahrndorff et al., 2016), including humans (Cho and Blaser, 2012; The Human Microbiome Project Consortium et al., 2012b). For instance, the presence of infection or disease is driven by the diversity of, and interactions between, pathogenic and non-pathogenic microbiota and the host’s immune system (Blaser, 2006; Benson et al., 2009, 2010; Blaser and Falkow, 2009; Maldonado-Contreras et al., 2011; Feng et al., 2017). Yet, the taxonomic diversity of microbes on an organism is an imperfect predictor of functional diversity (Gilbert et al., 2010; Delgado-Baquerizo et al., 2016; Widder et al., 2016). Predicting community function is further confounded because microbiomes vary extensively over space and time. Hence, predicting the role of microbiota in regulating infection and disease requires a better understanding of spatial and temporal variation in both taxonomic and functional diversity (The Human Microbiome Project Consortium et al., 2012a; Lloyd-Price et al., 2017; Kolde et al., 2018).
Bacterial communities may exhibit substantial biogeographic variation at both local (Costello et al., 2009; Bakker et al., 2014; Zhang et al., 2014) and continental scales (Fierer and Jackson, 2006; Meyer et al., 2018). Further, genetic divergence among populations of bacteria is common, due to the limited ability of bacteria to cross geographic barriers, their short generation times, and high population densities (Martiny et al., 2006; Taylor et al., 2006; Kuehne et al., 2007; Krause and Whitaker, 2015). So, while some genera may appear to be cosmopolitan (Fenchel et al., 1997), morphological and functional differences can arise from geographic divergence, creating distinct ecotypes or strains (Cohan, 2002; Bissett et al., 2010; Booth et al., 2016).
An emerging approach to study how microbial communities respond to environmental differences at both local and regional spatial scales is analyzing the effects of land use change. Habitat destruction, intensification of agriculture, and urbanization all alter edaphic properties, with correlated effects on microbial community composition (Zhao and Guo, 2010; Suleiman et al., 2013; Hawkes and Keitt, 2015; Plassart et al., 2019). In particular, urbanization may markedly alter the physicochemical properties of soil, leading to changes in the abundance and taxa of soil microbes (Zhao and Guo, 2010; Xu et al., 2014; Reese et al., 2016). Studies of the effects of land use change, and spatial variation in microbiota more generally, have rapidly increased in prevalence due to improvements in sequencing technologies (Caporaso et al., 2012; Lal and Seshasayee, 2014; Thompson et al., 2017; Averill et al., 2019); of critical importance now is understanding how these differences relate to community function (Robinson et al., 2010; Belzer and de Vos, 2012; Durrant and Bhatt, 2019).
Studies of genes linked to key environmental processes have related disturbances to changes in both microbial taxonomic and functional diversity (Bissett et al., 2011; Paula et al., 2014; Plassart et al., 2019), yet microbiomes usually are classified solely using the 16S rRNA gene. As others note, this gene alone is not a reliable indicator of the functional ability of a microbiome (Mollet et al., 1997; Fukushima et al., 2002; Hilario et al., 2004; Janda and Abbott, 2007; Becker et al., 2015). This weakness partly is because genomic diversity is high within microbial species, and adaptation to the local habitat can change the gene pool, creating sequence-discrete populations (Philippot et al., 2010; Smillie et al., 2011; Mell and Redfield, 2014; Shapiro and Polz, 2014; Becker et al., 2015; Gómez et al., 2016; Garcia et al., 2018; Ellegaard and Engel, 2019). Thus, 16S data may underestimate functional ability of microbiomes, due to strain-level adaptations to local conditions, or overestimate functional ability, due to functional redundancy across taxa (Delgado-Baquerizo et al., 2016; Mori et al., 2016; Vieira-Silva et al., 2016). As a result, understanding the complex interactions that shape microbial community assembly and structure requires complementing taxonomic analyses with assessments of functional differences within species and across environments (Burke et al., 2011; Jiang et al., 2019).
The current global amphibian decline is a critical system for assessing associations between microbial diversity and disease within a conservation context (Wake and Vredenburg, 2008; Ceballos et al., 2015; Bower et al., 2017). The decline is driven by the combined effects of habitat degradation and the fungal disease, Batrachochytrium dendrobatidis (Bd) (Collins and Storfer, 2003; Stuart et al., 2004). Amphibian infection by Bd, however, is blocked by some species of cutaneous bacteria, such as Janthinobacterium lividum, which produce antifungal metabolites and compete for space and resources with each other and the fungus (Longcore et al., 1999; Becker and Harris, 2010; Familiar López et al., 2017; Bates et al., 2018). However, it is still unknown in a variety of systems if these functional defense mechanisms, which many times are assumed to remain consistent with taxonomic identity, actually differ over space and time (Daskin et al., 2014; Jia and Whalen, 2019).
To address this issue, we examined bacterial diversity and function of the salamander microbiome across a 64 km gradient of land use change in the New York metropolitan area (NY, USA). We hypothesized that both composition and diversity of the cutaneous community would differ with urbanization: urban communities comprised of mainly cosmopolitan species, exurban communities of locally unique species, and suburban a mix of both making it the most diverse. In addition, we hypothesized that Bd-inhibitory ability would be a widespread and redundant trait but would increase with urbanization. This is because bacteria regularly come in contact with and compete with other fungi, not only Bd. While Bd prevalence has been suggested to be lower in urban areas (Becker and Zamudio, 2011), large urban areas such as New York are known centers for urban invaders and major points of entry for amphibians and Bd in the wildlife pet trade. Additionally, New York has the optimal climate for Bd persistence even if hosts are unavailable during certain seasons (Schloegel et al., 2009). Given that urban bacteria exist in areas with adverse environmental conditions and an increase in invaders, higher Bd-resistance could be an indirect benefit of their general response to these stressors. Lastly, we also predicted inhibition to vary among isolates of the same species based on location and season due to limitations in dispersal and potential local adaptations. The results of this study further uncover inhibitory bacteria in novel environments, such as urban areas, and aid in the discovery of potential native probiotics for other at-risk amphibian species in this species-rich region.
Methods
Study Species
A terrestrial species of salamander, the eastern redback salamander (Plethodon cinereus), was sampled at each of the nine study sites (n = 65 across all sites). We chose eastern redback salamanders for this study because they are locally abundant and are one of the most commonly used amphibians for studies on Bd-resistance in the United States. Chytridiomycosis has not been demonstrated to affect P. cinereus in nature, but chytridiomycosis symptoms can be induced in artificially infected salamanders by removing or augmenting their microbiome (Harris et al., 2009a,b; Becker and Harris, 2010). Thus, because this salamander can be infected by Bd but is not susceptible to it, studying its microbiome may provide the key to finding anti-fungal bacteria to use on vulnerable individuals. Permission to sample P. cinereus was obtained through the New York State Department of Environmental Conservation (permit no. 1159), the New York City Department of Parks and Recreation (permits 2016-2018), and the New York State Office of Parks, Recreation, and Historic Preservation (permit no. 2016-MP-008) and the Fordham University IACUC #JL-17-01.
Study Sites
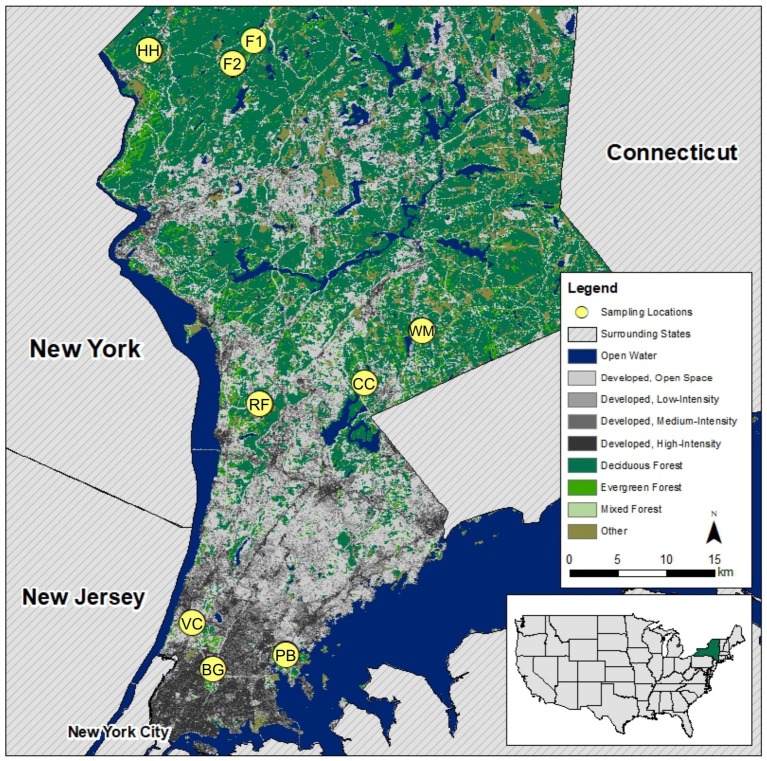
This research took place in parks and preserves in New York City and the two counties north of the city (Westchester and Putnam, NY, USA). We sampled from a total of nine study sites (Figure 1), three sites in each of the following categories: urban (Van Cortlandt Park, the New York Botanical Garden, and Pelham Bay Park), suburban (Rockefeller State Park Preserve, Fordham University’s Louis Calder Center, and Westmoreland Sanctuary), and exurban (two sites within Clarence Fahnestock State Park and one within Hudson Highlands State Park Preserve). Study site classification was based upon each site’s distance from Central Park in New York City, human population density, and GIS landcover analysis of percent-developed land within 5 miles of each study site (urban: >80%, suburban: 20–80%, exurban: <20%). All sites were located in mixed deciduous forests in New York State east of the Hudson River.
Figure 1.
Map of sampling locations along urban-to-exurban gradient showing developed land cover (from low to high intensity) in gray and forest in green adapted from the National Land Cover Database (NLCD) (2011) United States land cover dataset. Site code used throughout the manuscript is shown within the marker to identify each sampling location. Map in bottom-right corner locates New York State within the USA. Sites were classified as urban, suburban, or exurban based upon each site’s distance from Central Park in New York City, human population density, and GIS landcover analysis of percent-developed land within 5 miles of each study site.
Sampling
Samples were collected in Fall 2016 (September through October) and Spring 2017 (April through May), except for 10 samples taken from salamanders at Fahnestock and Hudson Highlands State Parks in Spring 2018. Salamanders were captured and handled using sterile gloves, which were changed between each individual. A non-invasive sampling method of bacterial swabbing was used based on Boyle et al. (2004). In brief, each salamander was rinsed twice with 25 ml of sterile water and swabbed using a sterile cotton swab on their ventral, right, and left sides (Harris et al., 2006). Swabs were immediately streaked onto LB agar (5 g NaCl, 5 g tryptone, 2.5 g yeast extract, 7.5 g agar, 200 μl 2.5 N NaOH, and 500 ml H2O) and were incubated at ambient temperature for 72 h. We chose to use LB agar to provide a high nutrient alternative to R2A, which is commonly used in amphibian microbiome studies as the nutrient content of salamander skin is still undetermined. Instead, we focused on swabbing a higher number of individuals across our sites to maximize culture diversity regardless of media (Medina et al., 2017b).
Identification of Bacterial Isolates
For each individual, we isolated bacteria based on morphotype (colony shape, elevation, margin, surface, and color) to pure cultures on LB agar. DNA from these cultures was extracted using the Qiagen DNeasy Blood and Tissue kit (Germantown, MD, USA) following manufacturer’s protocols, with the addition prior to purification of lysozyme incubation for Gram-positive bacteria. DNA was amplified with the 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACNVGGGTWTCTAAT-3′) primer pair, which targets the V4 region of the 16S rRNA gene, in 25 μl reactions: 12.5 μl GoTaq Green PCR Master Mix (2X), 1 μl forward primer, 1 μl reverse primer, 2 μl DNA, and 8.5 μl nuclease-free water. PCR protocols included: 95°C for 5 min, followed by 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min for 30 cycles, and 72°C for 5 min. This primer pair was chosen as it targets a small region commonly sequenced by high-throughput 16S rRNA sequencing, allowing our results to be easily compared to results from other studies. Amplicon lengths were assessed using gel electrophoresis (1% agarose), and samples yielding amplicons of ~250 bp were then sequenced using Sanger sequencing at Macrogen (New York, NY, USA).
Forward and reverse amplicon sequences were aligned to obtain a consensus sequence for each isolate in Geneious R11.1 (Biomatters Inc., Auckland 1010, New Zealand). Fourteen sequences were discarded due to poor read quality. The remaining 204 sequences were then run in the EZBioCloud database, a curated and integrated database of all 16S rRNA bacterial sequences from NCBI, to assign taxonomy (Yoon et al., 2017). OTU identification was assigned based on the closest reference sequence match with a taxonomic classification if sequence similarity was >97%. In instances where sequence similarity was >97% match to two or more OTUs, samples were classified based on highest percent query cover and completion of read. Additionally, 16S rRNA gene trees for isolates within each level of urbanization were built with MrBayes v. 3.2.6 (Huelsenbeck and Ronquist, 2001). Our runs consisted of four simultaneous Markov chains, each with 1,000,000 generations, a subsampling frequency of 200, and a burn-in fraction of 0.15. Trees were then visualized and adapted in FigTree v. 1.4.3 (Rambaut, 2012).
Characterization of Anti-Bd Ability
All bacterial isolates (including multiple isolates of the same OTU) were challenged with Bd using the 96-well cell-free supernatant (CFS) method developed by Bell et al. (2013). Samples of Bd strain JEL423, a hypervirulent strain from the global panzootic lineage BdGPL (Farrer et al., 2011; DiRenzo et al., 2014), were obtained from Dr. Joyce Longcore (University of Maine). Each isolate was diluted to 1:104 cfu and plated on LB agar plates until colonies formed, after which 1 cm2 of agar with colonies was excised and placed in 7 ml of 1% tryptone broth and allowed to grow at 23°C for 48 h. To obtain Bd used in each microcosm, 1 ml of Bd culture was plated on 1% tryptone plates and grown at 23°C for 4 days, after which 25 ml of 1% tryptone broth was inoculated with decanted zoospores. This Bd liquid culture was then grown at 23°C for 48 h.
Each isolate was co-cultured with 2 × 106 zoospores Bd (100 μl of each) in 800 μl of 1% tryptone, a minimal-nutrient media in which Bd often is grown (Longcore et al., 1999; Piotrowski et al., 2004), for 72 h at 23°C. Cultures were checked for turbidity at OD600 to confirm that they were in late log to early stationary phase and then centrifuged for 5 min at 10,000g. Cultures that did not reach these phases (<5% of isolates) were incubated for an additional 24 h. The supernatant was then filtered through a sterile, 0.22-μm cellulose acetate syringe filter (VWR, Radnor, PA, USA) to obtain CFS. CFS was then assayed with Bd (50 μl CFS: 50 μl Bd-zoospore suspension) in triplicate in a 96-well plate for 10 days at 23°C. The Bd-zoospore suspension consisted of growing Bd on a 1% tryptone agar plate for 4 days, flooding the plate with 3 ml 1% tryptone, and then filtering this suspension through a 20-μm filter. Zoospore concentration was estimated at 2 × 106 zoospores per ml using an INCYTO C-Chip™ hemocytometer (SKC Inc., Covington, GA, USA). Both positive (50 μl Bd CFS: 50 μl Bd-zoospore suspension) and negative controls (50 μl Bd CFS: 50 μl heat-killed Bd-zoospore suspension) were included to control for potential variation in Bd zoospore concentrations, Bd-produced metabolites, and Bd growth when alone. Optical density was measured at 0, 4, 7, and 10 days using an Infinite 200 Pro microplate reader (Tecan Trading AG, Männedorf, Switzerland).
To calculate Bd inhibition, absorbance readings were transformed using the equation: ln[OD/(1-OD)] following Becker et al. (2015). A linear regression was run to determine the growth rate of Bd in the presence of CFS from each isolate. The average slope of each triplicate was then divided by the average growth rate of the positive control, and this value was subtracted from one. This calculated value represented percent Bd inhibition, and bacterial OTUs were then classified by taking the mean percentage inhibition of multiple isolates of the same OTU.
Statistical Analysis
Alpha diversity was calculated using the Shannon index due to its reduced sensitivity to sampling depth differences, as compared to Chao-1 (Haegeman et al., 2013; Preheim et al., 2013). Chi-square test of independence was used to account for the relationship between year and site in our sampling. Beta diversity between sites was measured by generating a Bray-Curtis distance matrix on OTU-level presence-absence data (Legendre and Legendre, 1998) and conducting a permutational multivariate analysis of variance (PERMANOVA) with each site nested by level of urbanization. These data were then used for ordination by nonmetric multidimensional scaling (NMDS) with 1,000 randomized runs. Pairwise analyses were conducted using the package pairwiseAdonis (Martinez, 2019). All analyses were conducted in vegan v. 2.5-2 in R 3.6.0 (Oksanen et al., 2013).
We examined the distribution of isolate inhibition using Hartigan’s dip test (Hartigan and Hartigan, 1985) with the diptest package (Maechler and Ringach, 2013) to test for multimodality. The distribution was then visualized using the EM algorithm in the mixtools package (Benaglia et al., 2009), and the mean and standard deviation of each mode was calculated. We compared each isolate’s percent inhibition with each plate’s Bd control using a Mann-Whitney U-test to classify Bd-inhibitory ability. Differences among bacterial OTUs, genera, phyla, site, and level of urbanization were tested with a Kruskal-Wallis test followed by Dunn post-hoc test. Differences in level of inhibition between the Spring and Fall seasons were tested with a Mann-Whitney U-test.
Results
Community Diversity and Composition
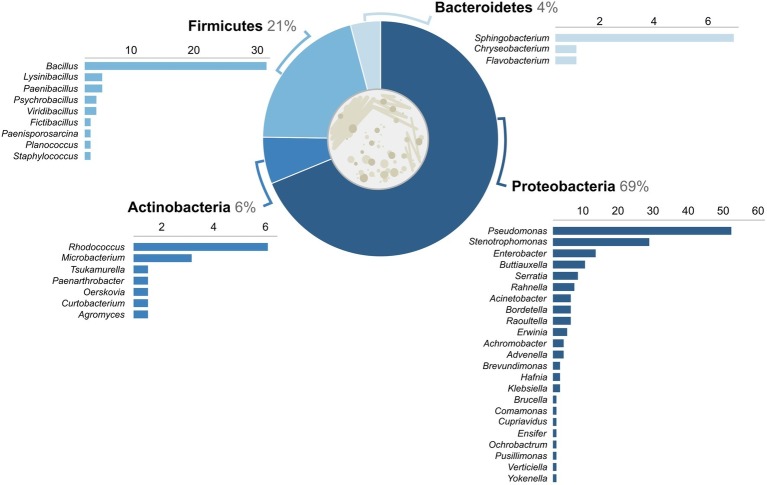
One of our primary objectives was to characterize the diversity of bacteria on salamanders from a large urban area in the northeastern USA. From 69 salamanders, a total of 218 isolates were cultured, 204 of which could be identified as OTUs (Table 1). Four phyla were identified: Proteobacteria (69%), Firmicutes (21%), Actinobacteria (6%), and Bacteroidetes (4%) (Figure 2). Out of 42 total genera, the three most common genera were Pseudomonas (43 isolates), Bacillus (28 isolates), and Stenotrophomonas (26 isolates). The majority of other genera were represented by less than 10 isolates each. While no one bacterial OTU was found at all nine sites, Stenotrophomonas rhizophila was found across all three levels of urbanization and at seven out of nine sites.
Table 1.
Summary of salamanders sampled across nine sites in three levels of urbanization.
| Level urbanization | Site | No. Indls sampled (n = 69) | No. isolates (n = 218) | No. Genera; Most common | No. OTUs; Most common | Diversity (H) | |
|---|---|---|---|---|---|---|---|
| Urban | Van Cortlandt Park (VC) |
7 | 28 | 12; Pseudomonas (PS) |
19; B. gaemokensis (BAGA) |
2.82 | Total: 3.44 |
| New York Botanical Garden (BG) |
8 | 20 | 11; Bacillus (BA) |
16; P. helmanticensis (PSHE) |
2.69 | ||
| Pelham Bay Park (PB) |
9 | 29 | 13; Bacillus (BA) |
21; S. rhizophila (STRH) B. wiedmannii (BAWI) |
2.95 | ||
| Suburban | Rockefeller State Park Preserve (RF) |
11 | 29 | 12; Pseudomonas (PS) |
17; E. cloacae (ENCL) B. wiedmannii (BAWI) A. modestus (AIMO) |
2.65 | Total: 3.48 |
| Louis Calder Center (CC) |
5 | 16 | 9; Pseudomonas (PS) |
13; S. myotis (SEMY) |
2.48 | ||
| Westmoreland Sanctuary (WM) |
10 | 27 | 15; Stenotrophomonas (ST) |
22; E. cloacae (ENCL) |
3.02 | ||
| Exurban | Fahnestock State Park 1 (F1) |
13 | 42 | 15; Pseudomonas (PS) |
24; R. inusitata (RAIN) |
3.01 | Total: 3.29 |
| Fahnestock State Park 2 (F2) |
4 | 16 | 6; Buttiauxella (BU) |
6; B. gaviniae (BUGA) |
1.68 | ||
| Hudson Highlands (HH) |
2 | 11 | 9; Bacillus (BA) Pseudomonas (PS) |
10; B. gaemokensis (BAGA) |
2.27 | ||
Site codes and OTU abbreviations (genus and species) are in parentheses. Diversity is shown for each site and for level of urbanization and was calculated using the Shannon Index.
Figure 2.
Percent of bacterial isolates (n = 204) across the gradient by phylum. Number of isolates identified within genera is displayed below each phylum.
OTU richness and abundance varied by site (Table 1) and a chi-square test of independence showed that both year and site were correlated with regard to OTU richness (X2 = 137.4, p < 0.05). While there was no trend of richness with land use type (p = 0.73), only 11 genera were found at all three levels of urbanization. These include: Pseudomonas, Bacillus, Stenotrophomonas, Sphingobacterium, Serratia, Rhodococcus, Microbacterium, Lysinibacillus, Enterobacter, Buttiauxella, and Bordetella. The remaining 31 genera were unique to certain levels of urbanization, with 24 genera (42% of total genera) unique to a single site. Interestingly, the most common OTU at each site often belonged to a different genus than the genus found to be most common at that site (Table 1).
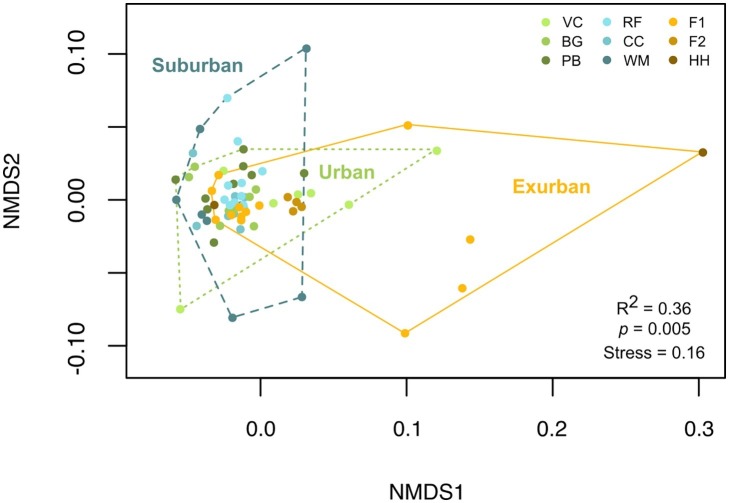
While urban sites had the greatest number of individual isolates compared to suburban and exurban sites, suburban sites had the largest overall Shannon’s diversity score, 3.48 (p = 0.56, Table 1). However, high OTU richness did not correlate to phylogenetic diversity (Figure 3), as many of the isolates were closely related and came from a small number of genera. An NMDS of beta diversity indicated that urban, suburban, and exurban bacterial communities grouped by level of urbanization (R2 = 0.36, p = 0.005; Figure 4). Urban and suburban communities overlapped to the left of the plot, but urban communities distributed along the first axis while suburban communities distributed along the second axis. Exurban sites separated to the right of the ordination plot, with some overlap with urban communities and a pairwise PERMANOVA revealed a significant difference with suburban communities (R2 = 0.15, p = 0.003).
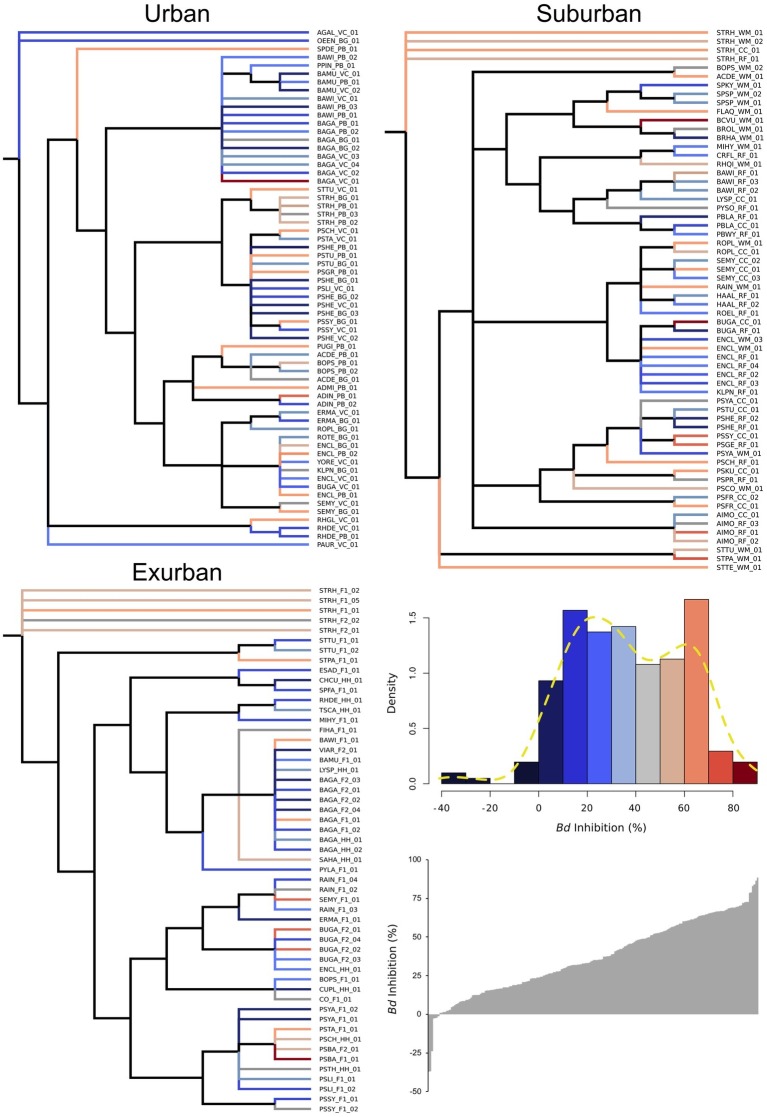
Figure 3.
Salamander skin bacterial OTUs (n = 204) at each level of urbanization displayed as phylogenies. OTUs are identified by the first two letters of the genera and species names (from the 16S rRNA sequence) followed by the site code and isolate number. Color of each isolate’s branch on phylogenetic tree corresponds to Bd-inhibitory ability. Bd-inhibitory ability is divided into intervals of 10% and color is denoted as displayed in the density histogram. Cooler colors indicate low Bd-inhibition, whereas warmer colors indicate high Bd-inhibition. Dashed yellow line in the density histogram shows a bimodal distribution of Bd-inhibition (p = 0.030).
Figure 4.
NMDS of beta diversity by individual salamander (n = 69) blocked by level of urbanization. Beta diversity was measured by calculating a Bray-Curtis dissimilarity matrix of OTU-level presence-absence culture data. Color denotes sampling location. PERMANOVA of beta diversity by level of urbanization indicates a significant difference between suburban and exurban skin bacterial communities (R2 = 0.375, p = 0.005).
Functional Redundancy
To assess whether Bd-inhibitory ability was multimodally distributed in our sample of salamanders, we used Hartigan’s dip test. We observed that Bd-inhibitory ability was bimodal (p = 0.030), exhibiting at least two groups of isolates: the first with a mean of 25.9% (SD = 18.5%), and the second with a mean of 63.9% Bd-inhibition (SD = 9.5%) relative to the control (Figure 3). Isolates within 1.5 SD of the second mean (all isolates above 49.6% Bd inhibition) were considered inhibitory—32% of all isolates. Isolates above the mean of the first mode but below 1.5 SD of the second mean (i.e., isolates between 25.9 and 49.6% Bd inhibition) were considered mildly inhibitory—30% of isolates, while isolates below this group but within 1.5 SD of the mean of the first mode were considered to have no effect on Bd-growth—34% of isolates. Lastly, the seven isolates (3%) that fell below this threshold were considered facilitative: increased Bd-growth relative to the control.
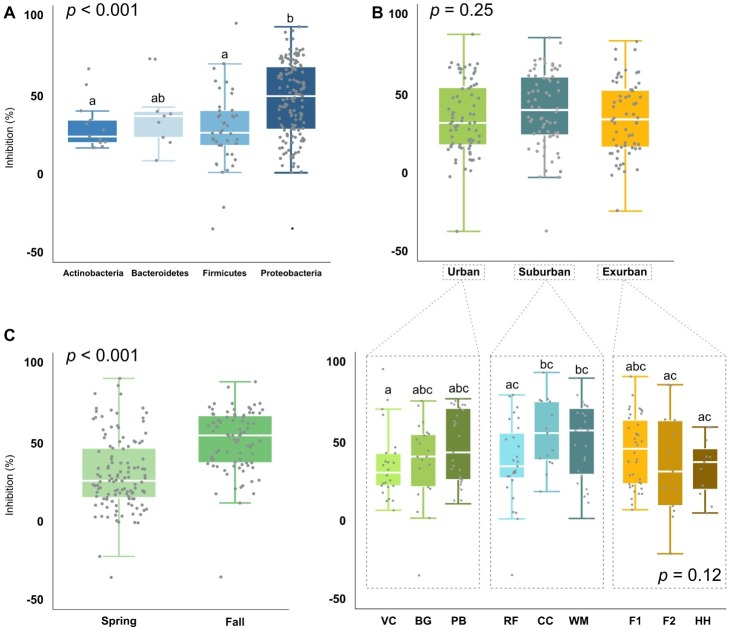
To assess whether Bd-inhibitory function was phylogenetically unique vs. redundant across those groups, we analyzed isolate Bd inhibition by phylum, genus, and OTU. When analyzed by phylum, Proteobacteria were on average 68% more inhibitory than Actinobacteria (p = 0.004) and Firmicutes (p < 0.001) and 26% more inhibitory than Bacteroidetes (X2 = 22.1, Figure 5A). At the genus and OTU level, isolates exhibited a large range of inhibitory ability, with few distinct phylogenetic patterns (Figure 3). In some extreme cases, isolates within the same genus, such as Bacillus and Pseudomonas (identified as “BA” and “PS” on the branch tips of the phylogenetic trees), were identified as inhibitory, while others were facilitative to Bd growth (p < 0.05). When we compared a subset of the most common bacterial genera (n ≥ 5 isolates), we found differences between genera (p = 0.012), driven by two genera: Bacillus (non-inhibitory, mean = 25%) and Stenotrophomonas (inhibitory, mean = 54%) (Table 2).
Figure 5.
Bd-inhibition of isolates (n = 204) by (A) phylum (p < 0.001), (B) level of urbanization (p = 0.252) and site [p = 0.117, but significant (p < 0.05) pairwise site effects], and (C) season (p < 0.001).
Table 2.
Anti-Bd activity of bacterial genera with greater than five isolates.
| Inhibition of Bd (%) | |||||
|---|---|---|---|---|---|
| Genus | No. inhibitory isolates/total | Mean | SD | Range | Mean by level of urbanization |
| Pseudomonas†,§ | 16/43 | 37.17 | 25.6 | −2.7 to 83.9 |
U = 29.6 S = 43.2 E = 40.9 |
| Bacillus§ | 4/28 | 25.0 | 24.1 | −37.1 to 88.5 |
U = 23.1 S = 39.9 E = 23.5 |
| Stenotrophomonas† | 19/26 | 53.8 | 11.8 | 18.2–72.6 |
U = 51.7 S = 60.4 E = 50.3 |
| Enterobacter§ | 4/12 | 34.9 | 22.4 | 10.9–69.1 |
U = 52.1 S = 26.1 E = 27.7 |
| Buttiauxella† | 4/8 | 47.8 | 32.0 | 9.8–86.4 |
U = 43.6 S = 48.1 E = 49.8 |
| Serratia† | 3/8 | 48.1 | 18.4 | 24.2–72.5 |
U = 50.0 S = 42.9 E = 53.0 |
| Sphingobacterium§ | 1/7 | 32.7 | 16.7 | 15.7–66.8 |
U = 47.4 S = 28.9 E = 23.7 |
| Rhodococcus§ | 2/5 | 30.7 | 23.2 | 12.5–60.6 |
U = 29.9 S = 51.0 E = 12.5 |
| Rahnella | 2/5 | 42.7 | 22.8 | 16.5–66.8 |
S = 66.8 E = 36.7 |
| Acinetobacter | 2/5 | 34.0 | 42.4 | −36.9 to 72.7 | S = 34.0 |
| Bordetella | 2/5 | 44.2 | 14.8 | 24.3–61.9 |
U = 46.5 S = 51.8 E = 24.3 |
| Raoultella | 2/5 | 41.9 | 17.5 | 20.8–66.1 |
U = 35.2 S = 46.4 |
Number of isolates determined to be greater than 49.6% Bd-inhibitory are given out of total number of isolates within each genus. Mean level of Bd-inhibition displayed by genus and level of urbanization (U = urban, S = suburban, E = exurban).
Denotes significant difference (p < 0.05) from Bacillus (non-inhibitory).
Denotes significant difference from Stenotrophomonas (inhibitory).
Spatial and Temporal Variation Within Genera and Species
We further analyzed Bd-inhibitory function by level of urbanization, site, and season. Overall, suburban sites appeared to have the largest proportion of inhibitory isolates with 41%, followed by exurban (34%) and urban (30%). Similarly, individual salamanders also varied in the number of Bd inhibitory isolates cultured from their skin. Depending on the site, 17–50% of isolates per individual were >49.5% inhibitory, with salamanders from suburban sites having on average 15% more inhibitory isolates than individuals from urban or exurban sites. However, the effect of urbanization was not statistically significant (X2 = 2.8, p = 0.25, Figure 5B)—so, while there appeared to be fairly large relative differences in the proportion of inhibitory isolates among levels of urbanization, with suburban communities 10–60% more inhibitory than urban and exurban communities (X2 = 2.5, p = 0.28), there was a lot of variation within genera even at the site level.
In addition, Pseudomonas and Bacillus, which make up a large proportion (35% of all isolates) of the cultured bacterial community, exhibited substantial variation in Bd inhibition, with differences by both site (p < 0.05) and season (Pseudomonas: p < 0.001, Bacillus: p = 0.022). In Bacillus, differences were even observed among isolates at the individual site level, such as B. gaemokensis between VC and F2 (p = 0.021), and B. wiedmannii between PB and F2 (p = 0.021). In contrast, Stenotrophomonas was consistently inhibitory, exhibiting no clear variation in inhibition by level of urbanization, site, season, or OTU. When analyzed by season, bacteria isolated in the fall season were 72% more inhibitory than bacteria in the spring (W = 2291.5, p < 0.001; Figure 5C). This same trend still held when a subset of the bacteria, which were found at the same site in both seasons, were compared (W = 437, p = 0.002). When we compared Bd inhibition by year, we found no significant difference between both the 2017 and 2018 spring seasons (X2 = 43.5, p = 0.40).
Discussion
Studies on the distribution of bacteria and the variability of their function are limited (Kueneman et al., 2014; Becker et al., 2015; Bletz et al., 2017; Jiménez and Sommer, 2017; Kruger, 2019) but are increasingly relevant to the amphibian microbiome as changes in microbial community structure have been hypothesized to result in changes in anti-fungal function. In line with these studies, our results suggest that predicting microbiome function across space and time is difficult due to strain-level differences and functional redundancy across taxa. This knowledge is foundational to understanding the dynamic nature of host-associated microbiomes and their function against pathogens, especially given the urgent need to conserve and protect vulnerable taxa in environments around the globe.
A key constraint to understand the distribution and variability of the amphibian microbiome is the limited number of studies in certain geographic areas, as indicated by the studies compiled in the Antifungal Isolates Database (Woodhams et al., 2015). These areas notably include the Northeastern United States, which has comparatively high salamander diversity and urban land use (~11%; Nickerson et al., 2011). Thus, we chose to sample Plethodon cinereus along a gradient of land use in the New York metropolitan area. We acknowledge that our sampling design and site selection may result in overlapping effects of land use and site microenvironment. However, this study design ultimately enabled us to better explore how environmental changes over even small geographic distances in these regions can influence variability of host microbial community diversity, composition, and function.
Community Diversity and Composition
Contrary to our hypothesis, Shannon’s diversity was similar regardless of site or land use. This pattern is in contrast to other studies on the effects of urbanization on plant and animal biodiversity, which suggest that native species richness tends to be reduced in areas with extreme urbanization (Shochat et al., 2006; Faeth et al., 2011; Laforest-Lapointe et al., 2017; Yan et al., 2019). Additionally, the presence of three cosmopolitan genera (Pseudomonas, Bacillus, and Stenotrophomonas) across the entire gradient, representing nearly half (48%) of all isolates, and our observation that urban sites had the highest OTU richness, contrasts with the idea that urbanization generally drives homogenization of taxa in these communities at least for bacteria (McKinney, 2006). However, we did find that site and year were correlated with regard to richness likely due to poor sampling conditions at Fahnestock and Hudson Highlands in 2017 leading us to also sample only those sites in Spring 2018. Despite this, we found no significant difference in Bd inhibition with year regardless of site.
While our use of culturing to examine overall taxonomic diversity is known to miss a proportion of taxa, it allowed us to assess diversity of many actives, and likely functionally relevant, bacteria on salamanders at these locations. However, we do acknowledge that the results of culturing alone are sensitive to the growth media chosen and does not enable us to distinguish bacterial relative abundances as they occur on salamander skin. Our results suggest that while diversity does not differ with urbanization, taxonomic composition does. This pattern has been seen repeatedly in other studies on microbial communities, which suggest that urbanization likely impacts relative abundance of taxa in local communities rather than overall diversity (Cousins et al., 2003; Hall et al., 2009; Xu et al., 2014; Reese et al., 2016; Epp Schmidt et al., 2017; Lehtimäki et al., 2017).
In our comparisons of beta diversity, composition of bacterial communities grouped by level of urbanization. Composition differed at the site level as well. This is likely because 31 genera were unique to certain levels of urbanization with 24 genera (42% of total genera) unique to a single site. Even though three cosmopolitan genera dominated most communities, sites differed in which OTU within these genera were present. This variation we see in OTU-specific composition could partially be explained by dispersal limitation or neutral processes, at least across small geographic distances.
Our finding that composition varies with land use is in line with our hypotheses: we saw the most variation between individual salamander bacterial communities at exurban sites (Figure 4), and more similarity at urban and suburban sites. Further, when we compare the abundance of the 11 genera found at all three levels of urbanization, urban sites had on average 5–12% more cosmopolitan isolates (a total of 78% of urban isolates) than suburban and exurban sites. In these cases, regional variation is likely driven by local environmental conditions associated with urbanization (i.e., changes in nutrient concentrations, habitat fragmentation, heavy metal deposition, etc.) that overwhelm stochastic factors to select for particularly resilient specialist OTUs as well as cosmopolitan generalists.
This larger role of microenvironment versus stochastic factors has repeatedly been seen in soil communities, where soils with similar environmental characteristics have similar microbial communities regardless of geographic distance (Fierer and Jackson, 2006; Lauber et al., 2009; Xu et al., 2014; Lladó et al., 2018; Plassart et al., 2019). Interestingly, we see this same trend in the bacterial communities of the soil-dwelling salamanders sampled in this study. If skin conditions were the primary selective mechanism, we would see no difference in bacterial composition across our gradient. As others have demonstrated, the soil and water environments in which amphibians live in likely function as species pools providing colonists to amphibian hosts (Loudon et al., 2016), and all of the cosmopolitan genera we found on our salamanders are common soil bacteria known to associate with plants or animals. Thus, our results indicate some combination of contemporary environmental variation and lingering historical effects driving distinct microbial biogeography on these salamanders (Martiny et al., 2006; Kraemer and Kassen, 2015). This study serves as an initial investigation into the overall effects of land use associated environmental changes on microbial communities, but further investigation is needed to determine if and how direct land-use associated effects on the soil bacterial reservoir indirectly alter amphibian cutaneous microbiomes.
It is worth noting that we did not find Janthinobacterium lividum, a well-known Bd-inhibitory bacteria (Brucker et al., 2008), on any of the Plethodon cinereus salamanders sampled despite knowledge that strains have been cultured from aquatic and soil environments in the Hudson Valley of New York (O’Brien et al., 2018). Instead, Stenotrophomonas, an antifungal plant mutualist (Wolf et al., 2002; Ryan et al., 2009), was found on salamanders at nearly all sites across our sampling gradient. This genus has a number of favorable characteristics for use as a potential native probiotic in degraded habitats: resistance to heavy metals (including lead), high hydrolytic potential, high bacterial growth rate, and the ability to grow in low-nutrient environments (Ryan et al., 2009). All of these have allowed Stenotrophomonas sp. to colonize and compete in many different biotypes adding to its value as potential anti-Bd isolate for amphibians in our region.
Functional Redundancy
Despite the compositional differences we saw in bacterial taxonomy across sites, overall Bd-inhibitory ability was similar among urban, suburban, and exurban bacterial communities but varied among individuals. Each site possessed at least one bacterial OTU capable of high Bd-inhibition. Given this, Bd-inhibition seems to be a functionally redundant characteristic across the gradient, despite differences in the environment and OTU composition. This pattern is not surprising given that naturally occurring bacteria regularly interact and compete with other microbial taxa, including fungi, for space and resources. Whether inhibitory metabolites are Bd-specific, fungal-specific, or generally anti-microbial is still undetermined for the majority of bacterial strains isolated from amphibian skin in this and other studies and we encourage continued efforts to characterize the amphibian metabolome to identify the mechanisms responsible for anti-Bd function (Woodhams et al., 2014; Belden et al., 2015; Walke et al., 2015).
Despite functional redundancy of Bd-inhibition across the gradient, it does not appear to be a trait associated with phylogenetic relatedness among bacteria (Figure 3). We found variation in inhibition across isolates at the phylum, genus, and OTU levels. For example, in comparing isolates from two different individuals with bacterial communities composed of the same genera, we found Bacillus, Pseudomonas, and Sphingobacterium to vary from 8, 9, and 67% Bd-inhibition at PB (urban) to 61, 44, and 16% at F1 (exurban). Given this, it might be better for researchers to focus on maintaining functional diversity rather than taxonomic diversity in relation to the conservation of amphibian microbiomes. Overall function of the community considers distribution, abundance, and the products of fluctuating interactions between species in the community (Holling, 2001; Alberti, 2005). As shown in this study, taxonomic differences are poor predictors of community functional differences and presence of particular isolates on amphibians in separate locations does not equate to similarities in function.
Spatial and Temporal Variation Within Genera and Species
Contrary to our hypothesis, suburban sites associated with medium levels of environmental change appeared to have the largest proportion of inhibitory isolates with 41%, followed by exurban (34%) and urban (30%) sites. In addition, only 32% of all isolates were inhibitory, which suggests that the majority of the salamander microbiomes cultured in this study show little to no effect on Bd growth in vitro. While inhibition did not significantly differ by level of urbanization, there were site-level differences (Figure 5B). Significant differences among our sites were driven by one suburban site, CC, which consisted of isolates that were on average more inhibitory than all other sites. Differences in Bd inhibition among sites could be due to environmental characteristics, absence of particular isolates, changes in overall community composition, or isolate-level trait differences. Often times, these local factors are not reported or considered in other studies using the Anti-fungal Isolates Database to determine community function from high throughput 16S rRNA sequencing. As mentioned before and reiterated by our results, the 16S rRNA gene alone is not a reliable indicator of Bd-inhibitory function. Thus, averaging the results of globally distributed functional assays likely blurs the fine-scale differences in microbiome function in different regions and can lead researchers to over- or underestimate microbiome function when not complemented by functional assays from local isolates (Mollet et al., 1997; Fukushima et al., 2002; Hilario et al., 2004; Janda and Abbott, 2007; Becker et al., 2015).
We found a high level of anti-Bd functional variation observed across exurban salamander bacterial communities, which was not in line with our initial hypotheses. Our exurban sampling locations (F1, F2, and HH) consist of well-connected hardwood forests and streamside habitats where salamander dispersal is far less limited than in both our urban and suburban sites. However, habitat connectedness or salamander dispersal may not matter at the microbial level. The variation in exurban isolate’s Bd-inhibitory function despite high habitat connectivity is not unexpected given that bacteria can engage in horizontal gene transfer and clonal evolution. Studies have shown that multiple clone-types can exist and swap in dominance based on abiotic and biotic community factors (Fenchel, 2003; Devevey et al., 2015; Wright and Vetsigian, 2016). We also observed isolate-level trait differences in comparisons of isolates across sites from the two most dominant genera, Pseudomonas and Bacillus (Figure 3; Table 2). Less than half of the number of isolates from these genera was inhibitory in urban sites as compared to exurban and suburban sites. These results agree with the findings of Becker et al. (2015), in that there can be substantial functional variation within phylogenetically similar bacterial isolates and provide further evidence for the use of native bacteria in developing potential probiotics.
Differences in an individual bacterial isolate’s interactions with the environment and other microbes are also known to alter bacterial metabolite production (Daskin et al., 2014; Wolz et al., 2017; Medina et al., 2017a; Bird et al., 2018). Thus, another consideration is whether Bd is present and when is it most active, as changes in microbial community composition have been linked with the onset of Bd infection (Pullen et al., 2010; Longo et al., 2015; Clare et al., 2016; Varela et al., 2018). Specifically, Bd is more prevalent during seasons of moderate temperatures (between 17 and 25°C) and high precipitation. In our study, we observed significantly larger Bd-inhibition in the fall as opposed to the spring, which is different than that found in previous studies (Figure 5C; Pullen et al., 2010, Varela et al., 2018).
Despite marginal difference in precipitation, Spring in the New York region is generally cooler than Fall (April/May mean = 15.2°C, September/October mean = 18.2°C according to the National Weather Service’s New York City records over the last 10 years). Based on these temperatures, Spring may be too cold for Bd growth, whereas Fall is within Bd’s optimal temperature range increasing bacterial anti-Bd metabolite response (~17–23°C; Piotrowski et al., 2004). Additionally, amphibians have been shown to lower intensity of Bd infections through over-wintering (Savage et al., 2011; Longo et al., 2015) or raising their own body temperature (Woodhams et al., 2003), which could have confounding effects on their entire microbiome. These temperature differences both in the environment and on the host could also impact which bacteria are present, their physiologies, and their function. Thus, we believe these results warrant further research into seasonal patterns of Bd-inhibition at a range of sites.
Conclusion
Our results indicate omitting functional assays on locally isolated bacteria can lead researchers to over- or underestimate microbiome function with potentially drastic implications for probiotic development or bioaugmentation. While our study of community variation along a relatively small (<65 km) urban land use gradient complements other regional high-throughput taxonomic analyses, further investigation is needed to determine if and how direct land-use associated effects on the soil bacterial reservoir indirectly alter amphibian cutaneous microbiomes. Additionally, our results highlight the need for research into metabolite composition and the genetic mechanisms responsible for their production. These analyses should continue to be performed in both pristine and altered environments to account for the continued habitat degradation occurring alongside the amphibian disease epidemic. In sum, our results provide further evidence for examining the whole microbiome, including micro-differences in species-species interactions and environmental variability, for understanding how it provides disease protection.
Data Availability Statement
The sequencing data generated for this study can be found in GenBank using accession numbers MN582766-MN582943.
Ethics Statement
The animal study was reviewed and approved by Fordham University, Institutional Animal Care and Use Committee Protocol #JL-17-01.
Author Contributions
EB and JL conceived and designed the experiments and wrote the paper. EB and EC performed the experiments and analyzed the data. EB, EC, and JL contributed materials and reagents and reviewed drafts of the paper.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the following organizations for access to their sites for sampling: New York City Department of Parks & Recreation, the Office of Parks and Historic Preservation including the managers of Rockefeller, Fahnestock and Hudson Highlands State Parks, the New York Botanical Gardens, the Westmoreland Sanctuary, and the Louis Calder Center. We thank Steve Kutos, Kiara Taveras, Julia Piccirillo-Stosser, and Sabrina Piccirillo-Stosser for field support.
Footnotes
Funding. This research was done with the support of the Louis Calder Center graduate student research fund and the Henry Luce Foundation which supported EB through a Clare Boothe Luce fellowship and research fund.
References
- Alberti M. (2005). The effects of urban patterns on ecosystem function. Int. Reg. Sci. Rev. 28, 168–192. 10.1177/0160017605275160 [DOI] [Google Scholar]
- Averill C., Cates L. L., Dietze M. C., Bhatnagar J. M. (2019). Spatial vs. temporal controls over soil fungal community similarity at continental and global scales. ISME J. 13, 2082–2093. 10.1038/s41396-019-0420-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrndorff S., Alemu T., Alemneh T., Lund Nielsen J. (2016). The microbiome of animals: implications for conservation biology. Research article. Available at: https://www.hindawi.com/journals/ijg/2016/5304028/abs/ [DOI] [PMC free article] [PubMed]
- Bakker M. G., Schlatter D. C., Otto-Hanson L., Kinkel L. L. (2014). Diffuse symbioses: roles of plant–plant, plant–microbe and microbe–microbe interactions in structuring the soil microbiome. Mol. Ecol. 23, 1571–1583. 10.1111/mec.12571, PMID: [DOI] [PubMed] [Google Scholar]
- Bates K. A., Clare F. C., O’Hanlon S., Bosch J., Brookes L., Hopkins K., et al. (2018). Amphibian chytridiomycosis outbreak dynamics are linked with host skin bacterial community structure. Nat. Commun. 9:693. 10.1038/s41467-018-02967-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M. H., Harris R. N. (2010). Cutaneous bacteria of the Redback salamander prevent morbidity associated with a lethal disease. PLoS One 5, 1–6. 10.1371/journal.pone.0010957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M. H., Walke J. B., Murrill L., Woodhams D. C., Reinert L. K., Rollins-Smith L. A., et al. (2015). Phylogenetic distribution of symbiotic bacteria from Panamanian amphibians that inhibit growth of the lethal fungal pathogen Batrachochytrium dendrobatidis. Mol. Ecol. 24, 1628–1641. 10.1111/mec.13135, PMID: [DOI] [PubMed] [Google Scholar]
- Becker C. G., Zamudio K. R. (2011). Tropical amphibian populations experience higher disease risk in natural habitats. Proc. Natl. Acad. Sci. USA 108, 9893–9898. 10.1073/pnas.1014497108, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden L. K., Hughey M. C., Rebollar E. A., Umile T. P., Loftus S. C., Burzynski E. A., et al. (2015). Panamanian frog species host unique skin bacterial communities. Front. Microbiol. 6:1171. 10.3389/fmicb.2015.01171, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. C., Alford R. A., Garland S., Padilla G., Thomas A. D. (2013). Screening bacterial metabolites for inhibitory effects against Batrachochytrium dendrobatidis using a spectrophotometric assay. Dis. Aquat. Org. 103, 77–85. 10.3354/dao02560, PMID: [DOI] [PubMed] [Google Scholar]
- Belzer C., de Vos W. M. (2012). Microbes inside—from diversity to function: the case of Akkermansia. ISME J. 6, 1449–1458. 10.1038/ismej.2012.6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaglia T., Chauveau D., Hunter D., Young D. (2009). Mixtools: an R package for analyzing finite mixture models. J. Stat. Softw. 32, 1–29. Available at: https://hal.archives-ouvertes.fr/hal-00384896 [Google Scholar]
- Benson A. K., Kelly S. A., Legge R., Ma F., Low S. J., Kim J., et al. (2010). Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. USA 107, 18933–18938. 10.1073/pnas.1007028107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson A., Pifer R., Behrendt C. L., Hooper L. V., Yarovinsky F. (2009). Gut commensal bacteria direct a protective immune response against toxoplasma gondii. Cell Host Microbe 6, 187–196. 10.1016/j.chom.2009.06.005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. K., Prado-Irwin S. R., Vredenburg V. T., Zink A. G. (2018). Skin microbiomes of California terrestrial salamanders are influenced by habitat more than host phylogeny. Front. Microbiol. 9:442. 10.3389/fmicb.2018.00442, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissett A., Richardson A. E., Baker G., Thrall P. H. (2011). Long-term land use effects on soil microbial community structure and function. Appl. Soil Ecol. 51, 66–78. 10.1016/j.apsoil.2011.08.010 [DOI] [Google Scholar]
- Bissett A., Richardson A. E., Baker G., Wakelin S., Thrall P. H. (2010). Life history determines biogeographical patterns of soil bacterial communities over multiple spatial scales. Mol. Ecol. 19, 4315–4327. 10.1111/j.1365-294X.2010.04804.x, PMID: [DOI] [PubMed] [Google Scholar]
- Blaser M. J. (2006). Who are we? Indigenous microbes and the ecology of human diseases. EMBO Rep. 7, 956–960. 10.1038/sj.embor.7400812, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser M. J., Falkow S. (2009). What are the consequences of the disappearing human microbiota? Nat. Rev. Microbiol. 7, 887–894. 10.1038/nrmicro2245, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bletz M. C., Perl R. G. B., Bobowski B. T., Japke L. M., Tebbe C. C., Dohrmann A. B., et al. (2017). Amphibian skin microbiota exhibits temporal variation in community structure but stability of predicted Bd-inhibitory function. ISME J. 11, 1521–1534. 10.1038/ismej.2017.41, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth A., Mariscal C., Doolittle W. F. (2016). The modern synthesis in the light of microbial genomics. Annu. Rev. Microbiol. 70, 279–297. 10.1146/annurev-micro-102215-095456, PMID: [DOI] [PubMed] [Google Scholar]
- Bower D. S., Lips K. R., Schwarzkopf L., Georges A., Clulow S. (2017). Amphibians on the brink. Science 357, 454–455. 10.1126/science.aao0500, PMID: [DOI] [PubMed] [Google Scholar]
- Boyle D., Boyle D., Olsen V., Morgan J., Hyatt A. (2004). Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis. Aquat. Org. 60, 141–148. 10.3354/dao060141, PMID: [DOI] [PubMed] [Google Scholar]
- Brucker R. M., Harris R. N., Schwantes C. R., Gallaher T. N., Flaherty D. C., Lam B. A., et al. (2008). Amphibian chemical defense: antifungal metabolites of the Microsymbiont Janthinobacterium lividum on the salamander Plethodon cinereus. J. Chem. Ecol. 34, 1422–1429. 10.1007/s10886-008-9555-7, PMID: [DOI] [PubMed] [Google Scholar]
- Burke C., Steinberg P., Rusch D., Kjelleberg S., Thomas T. (2011). Bacterial community assembly based on functional genes rather than species. Proc. Natl. Acad. Sci. USA 108, 14288–14293. 10.1073/pnas.1101591108, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. 10.1038/ismej.2012.8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos G., Ehrlich P. R., Barnosky A. D., García A., Pringle R. M., Palmer T. M. (2015). Accelerated modern human-induced species losses: entering the sixth mass extinction. Sci. Adv. 1:e1400253. 10.1126/sciadv.1400253, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I., Blaser M. J. (2012). The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270. 10.1038/nrg3182, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare F. C., Halder J. B., Daniel O., Bielby J., Semenov M. A., Jombart T., et al. (2016). Climate forcing of an emerging pathogenic fungus across a montane multi-host community. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, pii: 20150454. 10.1098/rstb.2015.0454, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan F. M. (2002). What are bacterial species? Annu. Rev. Microbiol. 56, 457–487. 10.1146/annurev.micro.56.012302.160634, PMID: [DOI] [PubMed] [Google Scholar]
- Collins J. P., Storfer A. (2003). Global amphibian declines: sorting the hypotheses. Divers. Distrib. 9, 89–98. 10.1046/j.1472-4642.2003.00012.x [DOI] [Google Scholar]
- Costello E. K., Lauber C. L., Hamady M., Fierer N., Gordon J. I., Knight R. (2009). Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697. 10.1126/science.1177486, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins J. R., Hope D., Gries C., Stutz J. C. (2003). Preliminary assessment of arbuscular mycorrhizal fungal diversity and community structure in an urban ecosystem. Mycorrhiza 13, 319–326. 10.1007/s00572-003-0239-4, PMID: [DOI] [PubMed] [Google Scholar]
- Daskin J. H., Bell S. C., Schwarzkopf L., Alford R. A. (2014). Cool temperatures reduce antifungal activity of symbiotic bacteria of threatened amphibians – implications for disease management and patterns of decline. PLoS One 9:e100378. 10.1371/journal.pone.0100378, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Baquerizo M., Giaramida L., Reich P. B., Khachane A. N., Hamonts K., Edwards C., et al. (2016). Lack of functional redundancy in the relationship between microbial diversity and ecosystem functioning. J. Ecol. 104, 936–946. 10.1111/1365-2745.12585 [DOI] [Google Scholar]
- Devevey G., Dang T., Graves C. J., Murray S., Brisson D. (2015). First arrived takes all: inhibitory priority effects dominate competition between co-infecting Borrelia burgdorferi strains. BMC Microbiol. 15:1. 10.1186/s12866-015-0381-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiRenzo G. V., Langhammer P. F., Zamudio K. R., Lips K. R. (2014). Fungal infection intensity and zoospore output of Atelopus zeteki, a potential acute Chytrid Supershedder. PLoS One 9:e93356. 10.1371/journal.pone.0093356, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant M. G., Bhatt A. S. (2019). Microbiome genome structure drives function. Nat. Microbiol. 4, 912–913. 10.1038/s41564-019-0473-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegaard K. M., Engel P. (2019). Genomic diversity landscape of the honey bee gut microbiota. Nat. Commun. 10:446. 10.1038/s41467-019-08303-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp Schmidt D. J., Pouyat R., Szlavecz K., Setälä H., Kotze D. J., Yesilonis I., et al. (2017). Urbanization erodes ectomycorrhizal fungal diversity and may cause microbial communities to converge. Nat. Ecol. Evol. 1:0123. 10.1038/s41559-017-0123, PMID: [DOI] [PubMed] [Google Scholar]
- Faeth S. H., Bang C., Saari S. (2011). Urban biodiversity: patterns and mechanisms. Ann. N. Y. Acad. Sci. 1223, 69–81. 10.1111/j.1749-6632.2010.05925.x, PMID: [DOI] [PubMed] [Google Scholar]
- Familiar López M., Rebollar E. A., Harris R. N., Vredenburg V. T., Hero J.-M. (2017). Temporal variation of the skin bacterial community and Batrachochytrium dendrobatidis infection in the terrestrial cryptic frog Philoria loveridgei. Front. Microbiol. 8:2535. 10.3389/fmicb.2017.02535, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer R. A., Weinert L. A., Bielby J., Garner T. W. J., Balloux F., Clare F., et al. (2011). Multiple emergences of genetically diverse amphibian-infecting chytrids include a globalized hypervirulent recombinant lineage. Proc. Natl. Acad. Sci. USA 108, 18732–18736. 10.1073/pnas.1111915108, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenchel T. (2003). Biogeography for bacteria. Science 301, 925–926. 10.1126/science.1089242, PMID: [DOI] [PubMed] [Google Scholar]
- Fenchel T., Esteban G. F., Finlay B. J. (1997). Local versus global diversity of microorganisms: cryptic diversity of ciliated protozoa. Oikos 80, 220–225. 10.2307/3546589 [DOI] [Google Scholar]
- Feng K., Zhang Z., Cai W., Liu W., Xu M., Yin H., et al. (2017). Biodiversity and species competition regulate the resilience of microbial biofilm community. Mol. Ecol. 26, 6170–6182. 10.1111/mec.14356, PMID: [DOI] [PubMed] [Google Scholar]
- Fierer N., Jackson R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 103, 626–631. 10.1073/pnas.0507535103, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima M., Kakinuma K., Kawaguchi R. (2002). Phylogenetic analysis of salmonella, Shigella, and Escherichia coli strains on the basis of the gyrB gene sequence. J. Clin. Microbiol. 40, 2779–2785. 10.1128/JCM.40.8.2779-2785.2002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia S. L., Stevens S. L. R., Crary B., Martinez-Garcia M., Stepanauskas R., Woyke T., et al. (2018). Contrasting patterns of genome-level diversity across distinct co-occurring bacterial populations. ISME J. 12, 742–755. 10.1038/s41396-017-0001-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J. A., Field D., Swift P., Thomas S., Cummings D., Temperton B., et al. (2010). The taxonomic and functional diversity of microbes at a temperate coastal site: a ‘multi-Omic’ study of seasonal and Diel temporal variation. PLoS One 5:e15545. 10.1371/journal.pone.0015545, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez P., Paterson S., Meester L. D., Liu X., Lenzi L., Sharma M. D., et al. (2016). Local adaptation of a bacterium is as important as its presence in structuring a natural microbial community. Nat. Commun. 7:12453. 10.1038/ncomms12453, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman B., Hamelin J., Moriarty J., Neal P., Dushoff J., Weitz J. S. (2013). Robust estimation of microbial diversity in theory and in practice. ISME J. 7, 1092–1101. 10.1038/ismej.2013.10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S. J., Ahmed B., Ortiz P., Davies R., Sponseller R. A., Grimm N. B. (2009). Urbanization alters soil microbial functioning in the Sonoran Desert. Ecosystems 12, 654–671. 10.1007/s10021-009-9249-1 [DOI] [Google Scholar]
- Harris R. N., Brucker R. M., Walke J. B., Becker M. H., Schwantes C. R., Flaherty D. C., et al. (2009a). Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J. 3, 818–824. 10.1038/ismej.2009.27 [DOI] [PubMed] [Google Scholar]
- Harris R., James T., Lauer A., Simon M., Patel A. (2006). Amphibian pathogen Batrachochytrium dendrobatidis is inhibited by the cutaneous bacteria of amphibian species. EcoHealth 3, 53–56. 10.1007/s10393-005-0009-1 [DOI] [Google Scholar]
- Harris R. N., Lauer A., Simon M. A., Banning J. L., Alford R. A. (2009b). Addition of antifungal skin bacteria to salamanders ameliorates the effects of chytridiomycosis. Dis. Aquat. Org. 83, 11–16. 10.3354/dao02004 [DOI] [PubMed] [Google Scholar]
- Hartigan J. A., Hartigan P. M. (1985). The dip test of unimodality. Ann. Stat. 13, 70–84. 10.1214/aos/1176346577 [DOI] [Google Scholar]
- Hawkes C. V., Keitt T. H. (2015). Resilience vs. historical contingency in microbial responses to environmental change. Ecol. Lett. 18, 612–625. 10.1111/ele.12451, PMID: [DOI] [PubMed] [Google Scholar]
- Hilario E., Buckley T. R., Young J. M. (2004). Improved resolution on the phylogenetic relationships among pseudomonas by the combined analysis of atpD, carA, recA and 16S rDNA. Antonie Van Leeuwenhoek 86, 51–64. 10.1023/B:ANTO.0000024910.57117.16, PMID: [DOI] [PubMed] [Google Scholar]
- Holling C. S. (2001). Understanding the complexity of economic, ecological, and social systems. Ecosystems 4, 390–405. 10.1007/s10021-001-0101-5 [DOI] [Google Scholar]
- Huelsenbeck J. P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. 10.1093/bioinformatics/17.8.754, PMID: [DOI] [PubMed] [Google Scholar]
- Janda J. M., Abbott S. L. (2007). 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45, 2761–2764. 10.1128/JCM.01228-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Whalen J. K. (2019). A new perspective on functional redundancy and phylogenetic niche conservatism in soil microbial communities. Pedosphere 30, 18–24. 10.1016/s1002-0160(19)60826-x [DOI] [Google Scholar]
- Jiang X., Li X., Yang L., Liu C., Wang Q., Chi W., et al. (2019). How microbes shape their communities? A microbial community model based on functional genes. Genomics Proteomics Bioinformatics 17, 91–105. 10.1016/j.gpb.2018.09.003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez R. R., Sommer S. (2017). The amphibian microbiome: natural range of variation, pathogenic dysbiosis, and role in conservation. Biodivers. Conserv. 26, 763–786. 10.1007/s10531-016-1272-x [DOI] [Google Scholar]
- Kolde R., Franzosa E. A., Rahnavard G., Hall A. B., Vlamakis H., Stevens C., et al. (2018). Host genetic variation and its microbiome interactions within the human microbiome project. Genome Med. 10:6. 10.1186/s13073-018-0515-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer S. A., Kassen R. (2015). Patterns of local adaptation in space and time among soil bacteria. Am. Nat. 185, 317–331. 10.1086/679585, PMID: [DOI] [PubMed] [Google Scholar]
- Krause D. J., Whitaker R. J. (2015). Inferring speciation processes from patterns of natural variation in microbial genomes. Syst. Biol. 64, 926–935. 10.1093/sysbio/syv050, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger A. (2019). Functional redundancy of Batrachochytrium dendrobatidis inhibition in bacterial communities isolated from Lithobates clamitans skin. Microb. Ecol. 79, 231–240. 10.1007/s00248-019-01387-7 [DOI] [PubMed] [Google Scholar]
- Kuehne H. A., Murphy H. A., Francis C. A., Sniegowski P. D. (2007). Allopatric divergence, secondary contact, and genetic isolation in wild yeast populations. Curr. Biol. 17, 407–411. 10.1016/j.cub.2006.12.047, PMID: [DOI] [PubMed] [Google Scholar]
- Kueneman J. G., Parfrey L. W., Woodhams D. C., Archer H. M., Knight R., McKenzie V. J. (2014). The amphibian skin-associated microbiome across species, space and life history stages. Mol. Ecol. 23, 1238–1250. 10.1111/mec.12510, PMID: [DOI] [PubMed] [Google Scholar]
- Laforest-Lapointe I., Messier C., Kembel S. W. (2017). Tree leaf bacterial community structure and diversity differ along a gradient of urban intensity. mSystems 2:e00087-17. 10.1128/msystems.00087-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A., Seshasayee A. S. N. (2014). “The impact of next-generation sequencing technology on bacterial genomics” in A systems theoretic approach to systems and synthetic biology II: Analysis and design of cellular systems. eds. Kulkarni V. V., Stan G-B., Raman K. (Dordrecht, Netherlands: Springer; ), 31–58. [Google Scholar]
- Lauber C. L., Hamady M., Knight R., Fierer N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120. 10.1128/AEM.00335-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P., Legendre L. (1998). Numerical ecology, volume 24, (developments in environmental modelling). Oxford, UK: Elsevier. [Google Scholar]
- Lehtimäki J., Karkman A., Laatikainen T., Paalanen L., von Hertzen L., Haahtela T., et al. (2017). Patterns in the skin microbiota differ in children and teenagers between rural and urban environments. Sci. Rep. 7:45651. 10.1038/srep45651, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lladó S., López-Mondéjar R., Baldrian P. (2018). Drivers of microbial community structure in forest soils. Appl. Microbiol. Biotechnol. 102, 4331–4338. 10.1007/s00253-018-8950-4 [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J., Mahurkar A., Rahnavard G., Crabtree J., Orvis J., Hall A. B., et al. (2017). Strains, functions and dynamics in the expanded human microbiome project. Nature 550, 61–66. 10.1038/nature23889, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longcore J. E., Pessier A. P., Nichols D. K. (1999). Batrachochytrium dendrobatidis gen. et sp. nov., a chytrid pathogenic to amphibians. Mycologia 91, 219–227. [Google Scholar]
- Longo A. V., Savage A. E., Hewson I., Zamudio K. R. (2015). Seasonal and ontogenetic variation of skin microbial communities and relationships to natural disease dynamics in declining amphibians. R. Soc. Open Sci. 2:140377. 10.1098/rsos.140377, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudon A. H., Venkataraman A., VanTreuren W., Woodhams D., Parfrey L. W., McKenzie V., et al. (2016). Vertebrate hosts as islands: dynamics of selection, immigration, loss, persistence and potential function of bacteria on salamander skin. Front. Microbiol. 7:333. 10.3389/fmicb.2016.00333, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maechler M., Ringach D. (2013). Diptest: Hartigan’s dip test statistic for Unimodality. R package, Version 0.75-7. Available at: https://CRAN.R-project.org/package=diptest [Google Scholar]
- Maldonado-Contreras A., Goldfarb K. C., Godoy-Vitorino F., Karaoz U., Contreras M., Blaser M. J., et al. (2011). Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 5, 574–579. 10.1038/ismej.2010.149, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P. (2019). pairwiseAdonis: Pairwise multilevel comparison using adonis. R package version 0.3.
- Martiny J. B. H., Bohannan B. J. M., Brown J. H., Colwell R. K., Fuhrman J. A., Green J. L., et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112. 10.1038/nrmicro1341 [DOI] [PubMed] [Google Scholar]
- McKinney M. L. (2006). Urbanization as a major cause of biotic homogenization. Urbanization 127, 247–260. 10.1016/j.biocon.2005.09.005 [DOI] [Google Scholar]
- Medina D., Hughey M. C., Becker M. H., Walke J. B., Umile T. P., Burzynski E. A., et al. (2017a). c. Microb. Ecol. 74, 227–238. 10.1007/s00248-017-0933-y [DOI] [PubMed] [Google Scholar]
- Medina D., Walke J. B., Gajewski Z., Becker M. H., Swartwout M. C., Belden L. K. (2017b). Culture media and individual hosts affect the recovery of culturable bacterial diversity from amphibian skin. Front. Microbiol. 8:1574. 10.3389/fmicb.2017.01574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mell J. C., Redfield R. J. (2014). Natural competence and the evolution of DNA uptake specificity. J. Bacteriol. 196, 1471–1483. 10.1128/JB.01293-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R., Garbeva P., Raaijmakers J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. 10.1111/1574-6976.12028, PMID: [DOI] [PubMed] [Google Scholar]
- Meyer K. M., Memiaghe H., Korte L., Kenfack D., Alonso A., Bohannan B. J. M. (2018). Why do microbes exhibit weak biogeographic patterns? ISME J. 12, 1404–1413. 10.1038/s41396-018-0103-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet C., Drancourt M., Raoult D. (1997). rpoB sequence analysis as a novel basis for bacterial identification. Mol. Microbiol. 26, 1005–1011. [DOI] [PubMed] [Google Scholar]
- Mori A. S., Isbell F., Fujii S., Makoto K., Matsuoka S., Osono T. (2016). Low multifunctional redundancy of soil fungal diversity at multiple scales. Ecol. Lett. 19, 249–259. 10.1111/ele.12560, PMID: [DOI] [PubMed] [Google Scholar]
- National Land Cover Database (NLCD) (2011). Multi-Resolution Land Characteristics Consortium (MRLC). Available at: https://data.nal.usda.gov/dataset/national-land-cover-database-2011-nlcd-2011 (Accessed February 07, 2019). [Google Scholar]
- Nickerson C., Ebel R., Borchers A., Carriazo F. (2011). Major uses of land in the United States, 2007, EIB-89. Washington, DC: United States Department of Agriculture, Economic Research Service. [Google Scholar]
- O’Brien K., Perron G. G., Jude B. A. (2018). Draft genome sequence of a red-pigmented Janthinobacterium sp. native to the Hudson Valley watershed. Genome Announc. 6:e01429-17. 10.1128/genomea.01429-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., O’hara R., et al. (2013). Package ‘vegan.’ Community ecology package, version 2
- Paula F. S., Rodrigues J. L. M., Zhou J., Wu L., Mueller R. C., Mirza B. S., et al. (2014). Land use change alters functional gene diversity, composition and abundance in Amazon forest soil microbial communities. Mol. Ecol. 23, 2988–2999. 10.1111/mec.12786, PMID: [DOI] [PubMed] [Google Scholar]
- Philippot L., Andersson S. G. E., Battin T. J., Prosser J. I., Schimel J. P., Whitman W. B., et al. (2010). The ecological coherence of high bacterial taxonomic ranks. Nat. Rev. Microbiol. 8, 523–529. 10.1038/nrmicro2367, PMID: [DOI] [PubMed] [Google Scholar]
- Piotrowski J. S., Annis S. L., Longcore J. E. (2004). Physiology of Batrachochytrium dendrobatidis, a chytrid pathogen of amphibians. Mycologia 96, 9–15. 10.2307/3761981 [DOI] [PubMed] [Google Scholar]
- Plassart P., Prévost-Bouré N. C., Uroz S., Dequiedt S., Stone D., Creamer R., et al. (2019). Soil parameters, land use, and geographical distance drive soil bacterial communities along a European transect. Sci. Rep. 9:605. 10.1038/s41598-018-36867-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preheim S. P., Perrotta A. R., Friedman J., Smilie C., Brito I., Smith M. B., et al. (2013). “Chapter eighteen - computational methods for high-throughput comparative analyses of natural microbial communities” in Methods in enzymology. ed. DeLong E. F. (Cambridge, MA: Academic Press; ), 353–370. [DOI] [PubMed] [Google Scholar]
- Pullen K. D., Best A. M., Ware J. L. (2010). Amphibian pathogen Batrachochytrium dendrobatidis prevalence is correlated with season and not urbanization in Central Virginia. Dis. Aquat. Org. 91, 9–16. 10.3354/dao02249, PMID: [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2012). FigTree v.1.4.3. http://tree.bio.ed.ac.uk/software/figtree/ (Accessed October 12, 2018).
- Reese A. T., Savage A., Youngsteadt E., McGuire K. L., Koling A., Watkins O., et al. (2016). Urban stress is associated with variation in microbial species composition[mdash]but not richness[mdash]in Manhattan. ISME J. 10, 751–760. 10.1038/ismej.2015.152, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C. J., Bohannan B. J. M., Young V. B. (2010). From structure to function: the ecology of host-associated microbial communities. Microbiol. Mol. Biol. Rev. 74, 453–476. 10.1128/MMBR.00014-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan R. P., Monchy S., Cardinale M., Taghavi S., Crossman L., Avison M. B., et al. (2009). The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7, 514–525. 10.1038/nrmicro2163, PMID: [DOI] [PubMed] [Google Scholar]
- Sanchez L. M., Wong W. R., Riener R. M., Schulze C. J., Linington R. G. (2012). Examining the fish microbiome: vertebrate-derived bacteria as an environmental niche for the discovery of unique marine natural products. PLoS One 7:e35398. 10.1371/journal.pone.0035398, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage A. E., Sredl M. J., Zamudio K. R. (2011). Disease dynamics vary spatially and temporally in a North American amphibian. Biol. Conserv. 144, 1910–1915. 10.1016/j.biocon.2011.03.018 [DOI] [Google Scholar]
- Schloegel L. M., Picco A. M., Kilpatrick A. M., Davies A. J., Hyatt A. D., Daszak P. (2009). Magnitude of the US trade in amphibians and presence of Batrachochytrium dendrobatidis and ranavirus infection in imported North American bullfrogs (Rana catesbeiana). Biol. Conserv. 142, 1420–1426. 10.1016/j.biocon.2009.02.007 [DOI] [Google Scholar]
- Shapiro B. J., Polz M. F. (2014). Ordering microbial diversity into ecologically and genetically cohesive units. Trends Microbiol. 22, 235–247. 10.1016/j.tim.2014.02.006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shochat E., Warren P. S., Faeth S. H., McIntyre N. E., Hope D. (2006). From patterns to emerging processes in mechanistic urban ecology. Trends Ecol. Evol. 21, 186–191. 10.1016/j.tree.2005.11.019, PMID: [DOI] [PubMed] [Google Scholar]
- Smillie C. S., Smith M. B., Friedman J., Cordero O. X., David L. A., Alm E. J. (2011). Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480, 241–244. 10.1038/nature10571, PMID: [DOI] [PubMed] [Google Scholar]
- Stuart S. N., Chanson J. S., Cox N. A., Young B. E., Rodrigues A. S. L., Fischman D. L., et al. (2004). Status and trends of amphibian declines and extinctions worldwide. Science 306, 1783–1786. 10.1126/science.1103538, PMID: [DOI] [PubMed] [Google Scholar]
- Suleiman A. K. A., Manoeli L., Boldo J. T., Pereira M. G., Roesch L. F. W. (2013). Shifts in soil bacterial community after eight years of land-use change. Syst. Appl. Microbiol. 36, 137–144. 10.1016/j.syapm.2012.10.007, PMID: [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Turner E., Townsend J. P., Dettman J. R., Jacobson D. (2006). Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom fungi. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1947–1963. 10.1098/rstb.2006.1923, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Human Microbiome Project Consortium. Huttenhower C., Gevers D., Knight R., Abubucker S., Badger J. H., et al. (2012a). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Human Microbiome Project Consortium. Methé B. A., Nelson K. E., Pop M., Creasy H. H., Giglio M. G., et al. (2012b). A framework for human microbiome research. Nature 486, 215–221. 10.1038/nature11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L. R., Sanders J. G., McDonald D., Amir A., Ladau J., Locey K. J., et al. (2017). A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463. 10.1038/nature24621, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela B. J., Lesbarrères D., Ibáñez R., Green D. M. (2018). Environmental and host effects on skin bacterial community composition in Panamanian frogs. Front. Microbiol. 9:298. 10.3389/fmicb.2018.00298, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira-Silva S., Falony G., Darzi Y., Lima-Mendez G., Garcia Yunta R., Okuda S., et al. (2016). Species–function relationships shape ecological properties of the human gut microbiome. Nat. Microbiol. 1:16088. 10.1038/nmicrobiol.2016.88 [DOI] [PubMed] [Google Scholar]
- Wake D. B., Vredenburg V. T. (2008). Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc. Natl. Acad. Sci. USA 105, 11466–11473. 10.1073/pnas.0801921105, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walke J. B., Becker M. H., Loftus S. C., House L. L., Teotonio T. L., Minbiole K. P. C., et al. (2015). Community structure and function of amphibian skin microbes: an experiment with bullfrogs exposed to a Chytrid fungus. PLoS One 10:e0139848. 10.1371/journal.pone.0139848, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widder S., Allen R. J., Pfeiffer T., Curtis T. P., Wiuf C., Sloan W. T., et al. (2016). Challenges in microbial ecology: building predictive understanding of community function and dynamics. ISME J. 10, 2557–2568. 10.1038/ismej.2016.45, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf A., Fritze A., Hagemann M., Berg G. (2002). Stenotrophomonas rhizophila sp. nov., a novel plant-associated bacterium with antifungal properties. Int. J. Syst. Evol. Microbiol. 52, 1937–1944. 10.1099/00207713-52-6-1937, PMID: [DOI] [PubMed] [Google Scholar]
- Wolz M., Carly R., Yarwood S. A., Grant E. H. C., Fleischer R. C., Lips K. R. (2017). Effects of host species and environment on the skin microbiome of Plethodontid salamanders. J. Anim. Ecol. 87, 341–353. 10.1111/1365-2656.12726 [DOI] [PubMed] [Google Scholar]
- Woodhams D. C., Alford R. A., Antwis R. E., Archer H., Becker M. H., Belden L. K., et al. (2015). Antifungal isolates database of amphibian skin-associated bacteria and function against emerging fungal pathogens. Ecology 96:595. 10.1890/14-1837.1 [DOI] [Google Scholar]
- Woodhams D., Alford R., Marantelli G. (2003). Emerging disease of amphibians cured by elevated body temperature. Dis. Aquat. Org. 55, 65–67. 10.3354/dao055065, PMID: [DOI] [PubMed] [Google Scholar]
- Woodhams D. C., Brandt H., Baumgartner S., Kielgast J., Küpfer E., Tobler U., et al. (2014). Interacting symbionts and immunity in the amphibian skin mucosome predict disease risk and probiotic effectiveness. PLoS One 9:e96375. 10.1371/journal.pone.0096375, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E. S., Vetsigian K. H. (2016). Inhibitory interactions promote frequent bistability among competing bacteria. Nat. Commun. 7:11274. 10.1038/ncomms11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.-J., Li S., Su J.-Q., Gibson V., Li H., Zhu Y.-G. (2014). Does urbanization shape bacterial community composition in urban park soils? A case study in 16 representative Chinese cities based on the pyrosequencing method. FEMS Microbiol. Ecol. 87, 182–192. 10.1111/1574-6941.12215, PMID: [DOI] [PubMed] [Google Scholar]
- Yan Y., Klinkhamer P. G. L., van Veen J. A., Kuramae E. E. (2019). Environmental filtering: a case of bacterial community assembly in soil. Soil Biol. Biochem. 136:107531. 10.1016/j.soilbio.2019.107531 [DOI] [Google Scholar]
- Yildirim S., Yeoman C. J., Sipos M., Torralba M., Wilson B. A., Goldberg T. L., et al. (2010). Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS One 5:e13963. 10.1371/journal.pone.0013963, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S.-H., Ha S.-M., Kwon S., Lim J., Kim Y., Seo H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. 10.1099/ijsem.0.001755, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Geng J., Tang X., Fan H., Xu J., Wen X., et al. (2014). Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISME J. 8, 881–893. 10.1038/ismej.2013.185, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Guo H. G. (2010). Effects of urbanization on the quantity changes of microbes in urban-to-rural gradient forest soil. Agric. Sci. Technol. 11, 118–122. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequencing data generated for this study can be found in GenBank using accession numbers MN582766-MN582943.