Abstract
An increasing number of candidate genes related to abiotic stress tolerance are being discovered and proposed to improve the existing cultivars of the high oil-bearing crop sesame (Sesamum indicum L.). However, the in planta functional validation of these genes is remarkably lacking. In this study, we cloned a novel sesame R2-R3 MYB gene SiMYB75 which is strongly induced by drought, sodium chloride (NaCl), abscisic acid (ABA) and mannitol. SiMYB75 is expressed in various sesame tissues, especially in root and its protein is predicted to be located in the nucleus. Ectopic over-expression of SiMYB75 in Arabidopsis notably promoted root growth and improved plant tolerance to drought, NaCl and mannitol treatments. Furthermore, SiMYB75 over-expressing lines accumulated higher content of ABA than wild-type plants under stresses and also increased sensitivity to ABA. Physiological analyses revealed that SiMYB75 confers abiotic stress tolerance by promoting stomatal closure to reduce water loss; inducing a strong reactive oxygen species scavenging activity to alleviate cell damage and apoptosis; and also, up-regulating the expression levels of various stress-marker genes in the ABA-dependent pathways. Our data suggested that SiMYB75 positively modulates drought, salt and osmotic stresses responses through ABA-mediated pathways. Thus, SiMYB75 could be a promising candidate gene for the improvement of abiotic stress tolerance in crop species including sesame.
Keywords: Abiotic stress, abscisic acid, MYB transcription factors, root growth, Sesamum indicum, SiMYB75
In this study, the gene SiMYB75 was cloned from sesame (Sesamum indicum L.) and was found to be strongly induced by drought, sodium chloride (NaCl), abscisic acid (ABA) and mannitol. SiMYB75 displayed a root preferential expression and the ectopic over-expression in Arabidopsis thaliana demonstrated that it promotes root growth and is involved in drought and salinity tolerance by modulating the expression of ABA-related genes and strongly reducing reactive oxygen species production in cells. SiMYB75 could be a promising candidate gene for the improvement of abiotic stress tolerance in crop species including sesame..
Introduction
Unfavourable environmental conditions, including extreme temperatures, flooding, drought and high salinity limit crop growth and productivity resulting in considerable yield loss worldwide. Hence, improving crop tolerance to environmental stresses is of a paramount significance for global food security. Biotechnological manipulation of genes with high potential to impart cellular processes of stress tolerance is among the strategies for improving crop tolerance (Seok et al. 2017).
To protect themselves from abiotic stresses, plants translate environmental inputs into internal signals through hormones, second messengers which activate transcription factors (Wilkinsonet and Davies 2002; Shinozaki and Yamaguchi-Shinozaki 2007). In general, plants respond to abiotic stresses through abscisic acid (ABA)-dependent pathway and ABA-independent pathway. ABA is a broad-spectrum phytohormone that coordinates various stress signal transduction pathways during abiotic stress responses in plants (Agarwal and Jha 2010). Transcription factors (TFs) are the key mediators of these stress signal transduction pathways by regulating the expression of downstream target genes (Shinozaki et al. 2003). Several classes of TFs such as Apetala 2/ethylene-responsive element binding factor (AP2/ERF), NAM/ATAF1/CUC2 (NAC), WRKY, basic leucine zipper (bZIP), basic helix-loop-helix (bHLH), C2H2 zinc fingers (ZFs) and Myeloblastosis (MYB) were identified as involved in abiotic stress responsive pathways in plants (Abe et al. 1997; Nakashima et al. 2007; Kodaira et al. 2011; Jisha et al. 2015; Tu et al. 2016;Zhao et al. 2018).
MYBs are a vital gene family with a large number of members which modulate various biological processes in plants such as shoot growth, root formation, organ development, metabolism, hormone signal transduction and response to biotic and abiotic stresses (Jin and Martin 1999; Dubos et al. 2010; Ambawat et al. 2013; Roy 2016). They have attracted tremendous investigations in various plant species mainly regarding their involvement in plant abiotic stresses tolerance. The MYB domain is composed of one to four imperfect tandem repeats (R) and each repeat adopted a helix-turn-helix (HTH) conformation with 51–53 amino acid residues (Stracke et al. 2001). MYB proteins are classified into four major subfamilies based on the number of imperfect adjacent repeats in the MYB domain: R1-MYB or MYB-related (1R), R2R3-type MYB (2R), R1R2R3-type MYB (3R) and 4R-type MYB or atypical MYB (4R). Among them, the R2R3-MYB subfamily is the most abundant in plants (Stracke et al. 2001). In the model plant species Arabidopsis thaliana which contains 198 MYB genes, the functional characterization of the AtMYB96 gene showed that its over-expression increases cuticular wax biosynthesis under drought conditions as a tolerance mechanism (Lee et al. 2014). Moreover, several other genes including AtMYB44, AtMYB73, AtMYB20, AtMYB2 and AtMYB15 were reported to confer drought and salt tolerance through ABA-mediated pathways (Jung et al. 2008; Ding et al. 2009; Cui et al. 2013; Kim et al. 2013). Ectopic over-expression of the rice gene OsMYB3R-2 confers resistance to multiple abiotic stresses in Arabidopsis (Dai et al. 2007). Similarly, Yin et al. (2017) characterized the rice gene OsMYBR1 and found that transgenic Arabidopsis plants up-regulate many stress-related genes and strongly accumulate osmoprotectants, leading to improved drought tolerance. Recent studies of Butt et al. (2017) and Li et al. (2017b) revealed that two novel genes PbrMYB21 from Pyrus betulaefolia and GaMYB85 from Gossypium arboreum control the polyamine levels and stomata density as key mechanisms for drought tolerance.
Sesame (Sesamum indicum L.) is a traditional oilseed crop highly valued for its health-promoting oil (Anilakumar et al. 2010). With the growing interest in this crop, the global production and the area down to sesame are rapidly increasing. However, sesame is yet to become a major crop in the world because of the weak productivity, the lack of improved varieties with tolerance to biotic and abiotic stresses (Dossa et al. 2017a). Sesame is mainly grown in arid and semi-arid areas and is often challenged by severe drought (Langham 2007). Although sesame is rated as a relatively drought tolerant crop, the plant growth and yield are critically affected under prolonged stress (Sun et al. 2010;Dossa et al. 2017b). Similarly, adverse effects of salt stress, especially NaCl, on sesame seed germination, seedling growth and yield were reported (Koca et al. 2007; Yahya 2010; Bazrafshan and Ehsanzadeh 2014; Bekele et al. 2017; Li et al. 2018). Gene mining for improvement for abiotic stress tolerance in sesame, particularly, towards drought and salt stress tolerance represents the trending issue in the current sesame research (Dossa et al. 2017a). An increasing number of abiotic stress tolerance candidate genes are being discovered in sesame (Dossa et al. 2016a, b, c; Wang et al. 2016; Dossa et al. 2017c; Li et al. 2017b; Mmadi et al. 2017;Li et al. 2018; Wang et al. 2018b; You et al. 2018; Zhang et al. 2018; Dossa et al. 2019a), but their functional validation is notably lacking (Chowdhury et al. 2017; Dossa et al. 2019b). Sesame resilience to the genetic manipulation is still significantly enough to justify the use of a heterologous system such as Arabidopsis thaliana.
In a previous report, Mmadi et al. (2017) identified at the genome-wide level, 287 MYB encoding genes, which were predicted to be involved in various biological pathways in sesame. They identified 28 MYB genes strongly affected by drought and waterlogging stresses. In this study, we cloned one of these genes (SiMYB75) and characterized its function in mediating drought and salt stress tolerance. SiMYB75 was induced by drought, salt, ABA and osmotic stresses in sesame. Its ectopic over-expression in Arabidopsis promoted root growth, increased the endogenous ABA level, induced ABA sensitivity phenotype and enhanced tolerance to drought and salt stresses. Our results indicate that SiMYB75 functions through the ABA-dependent pathways and is a potential gene for the genetic improvement of sesame abiotic stress tolerance.
Methods
Bioinformatics analysis of SiMYB75
The whole gene, promoter, coding and protein sequences of SiMYB75 (SIN_1015311) were obtained from Sinbase (http://ocri-genomics.org/Sinbase/; (Wang et al. 2014)). The Pfam 26.0 (http://pfam.xfam.org/) database was exploited to identify the putative MYB conserved domains in the SiMYB75 protein. The theoretical Molecular weight and Isoelectric point of the SiMYB75 protein were predicted using the Compute pI/Mw tool (www.web.expasy.org/compute_pi). The promoter region (1 Kb) of SiMYB75 and AtMYB71 were analysed using the PLACE online tool (Higo et al. 1998). Next, using BLASTp search with an e-value ≤ 1E-10 and amino acid sequence >100 residues, we identified the homologues of SiMYB75 protein in closely related species, viz. potato, grape, oleaster, jatropha, rice and in the model plant species Arabidopsis. The amino acid sequences were aligned with Clustal W in the MEGA 7.0 software (Kumar et al. 2016) and the result was used to construct a Neighbor Joining phylogenetic tree with 1000 bootstrap replicates.
Sesame materials and stress treatments
The sesame modern cultivar Zhongzhi No. 13 was obtained from the Sesame Germplasm Resource Preserving Center, of the Oil Crops Research Institute-Chinese Academy of Agricultural Sciences (Wuhan, China). The seeds were sterilized with 3 % sodium hypochlorite for 7 min and washed three times using sterile water. For the drought experiment, the seeds were sown in pots containing loam soil mixed with 10 % vermiculite and plants were regularly watered. After 2 weeks, the seedlings were submitted to a water stress for 7 days. Concerning the salt and osmotic stress treatments, seedlings were hydroponically grown in a box containing half-strength Hoagland solution for 2 weeks. Then, seedlings were transferred into a new nutrient solution containing 200 mM sodium chloride (NaCl) for 48 h (salt stress treatment) and a nutrient solution containing 2 % PEG6000 for 5 days (osmotic stress treatment). For abscisic acid (ABA) treatment, 1 week-old seedlings initially grown in a box containing half-strength Hoagland solution were transferred into a nutrient solution containing 100 µM ABA for 12 h. The whole experiment was conducted in a greenhouse with the temperature and relative humidity kept at 35 °C and 60 %, respectively, and under long-day conditions (16 h day/8 h night). Root samples of stressed and control plants were collected simultaneously at the end of each stress treatment.
qRT–PCR expression profiling of SiMYB75 under various abiotic stress treatments in sesame
RNAs from root of stressed and control sesame plants were extracted using the EASYspin Plus kit (Aidlab Biotechnologies, China) according to the manufacturer’s instructions. The RNA was treated with DNaseI and reverse transcribed with oligo (dT23) primer using the FastQuant RT kit (Tiangen Biotech, China). The SiMYB75 specific primer pairs (Table S1) were designed using the Primer5.0 software (Lalitha 2000). The quantitative reverse transcriptase polymerase chain reaction analysis of SiMYB75 was performed as described by Dossa et al. (2018) using the ChamQ SYBR qPCR Master Mix (Vazyme Biotec, China) on a Light Cycler 480 II (Roche, Switzerland). The relative expression level of SiMYB75 was normalized to the expression level of the sesame Actin 7 gene (SIN_1006268) based on the 2-∆∆Ct method (Livak and Schmittgen 2001). This analysis was carried out in three independent biological replicates and three technical replicates of each biological replicate.
Tissue expression analysis of SiMYB75 in sesame
The expression pattern of SiMYB75 in various tissues was analysed using the transcriptome sequencing data generated in our group from root, stem tip, leaf and seed of cv. Zhongzhi No. 13 under normal growth conditions (Wei et al. 2011).
Vector construction and Arabidopsis genetic transformation
To functionally characterize the gene SiMYB75 in Arabidopsis thaliana, we cloned its protein coding region by PCR from sesame root cDNA (F- GCTTTCGCGAGCTCGGTACCATGTCTTGGGGAGTAAT GGG; R-CGACTCTAGAGGATCCTCAATAGAAAGTTGCAGCTA), and inserted the into a pCAMBIA 1301s vector (which is a modified form of the pCAMBIA1301 vector) between KPnI (5’-end) and BamHI (3’-end) sites, driven by the CaMV 35S promoter (see Supporting Information—Fig. S1). The plasmid containing the 35S::SiMYB75 construct was transformed first into Agrobacterium tumefaciens strain LBA4404 and then into Arabidopsis ecotype Col-0 cv. Columbia by the floral dipping method (Clough and Bent 1998). Transgenic seeds were screened by sowing on MS medium containing 1 % agar, and 1 % sucrose and 50 μg.ml-1 hygromicin. All the putative T1 transgenic plants and wild type (WT) plants were screened by PCR with genomic DNA from leaves as described by Dossa et al. (2018) (see Supporting Information—Fig. S1). Furthermore, RT–PCR and β-glucuronidase (GUS) staining were used to confirm the integration of the construct (Wang et al. 2009). Three independent T3 transgenic homozygous lines were used for the stress treatments, gene expression assay and phenotypic analyses.
Sub-cellular localization of SiMYB75 protein
To examine the sub-cellular localization of SiMYB75 protein, the full sequence was loaded in the TargetP 1.1 server with default parameters (Emanuelsson et al. 2000) and the WoLF PSORT web site (Horton et al. 2007).
In addition, the full-length ORF was PCR amplified and the fragment was ligated into the vector pBWA(V)HS-GLosgfp. Then, the construct pBWA(V)HS-SiMYB75-GLosgfp and the nucleus marker vector (pBWA(V)HS-Nucleus-mKate) were co-transformed into Arabidopsis protoplasts under PEG mixture (40 % (w/v) PEG 4000, 2 M mannitol and 0.1 M CaCl2). An empty vector, pBWA(V)HS-GFP, was used as a control (Ma et al. 2019). After incubation at room temperature for 16–24 h, the expression of plasmid was observed under a laser scanning confocal microscope.
Evaluation of transgenic lines exposed to ABA, osmotic, salt and drought stresses
First, to analyse the response of the transgenic plants to salt and osmotic stresses, about 30 seeds of WT and three T3 lines (#L1, #L2 and #L3) were surface sterilized and plated on solid Murashige and Skoog (MS) medium containing 0/100 mM mannitol and 0/100 mM NaCl. The seeds were stratified for 2 days in the dark at 4 °C and then transferred to growth chamber at 22 °C under long-day conditions (16 h light/8 h dark). The percentage of germinated seeds with green cotyledons was recorded after 7 days (Manuka et al. 2018). Next, 10-days-old over-expressing SiMYB75 seedlings and WT plants were transferred into solid MS medium supplemented with 0/150 mM mannitol, 0/150 mM NaCl and 0/10 µM ABA. Plates were placed vertically and after 10 days, seedling root length was recorded. In addition, 10-days-old seedlings (transgenic lines and WT) were transferred into pot (two plants per pot) and grown in normal conditions for 15 days. Then, one-third of the pots were subjected to dehydration stress for 17 days. Another one-third of the pots were watered with 200 mM NaCl solution every 2 days for a week. The remaining plants were kept in a normal growth condition throughout the whole experiment. At the end of each stress application, the number of dead/survived plants and rosette biomass dry weight were recorded and pictures were captured to show visible phenotypes. We estimated the relative rosette biomass as the ratio of the records under stress and control conditions. To evaluate the effect of the transgene on the plant growth, daily pictures of the plants were taken starting from 25 days after sowing (beginning of the stress) to 45 days after sowing (after stress treatments). The rosette diameter was estimated; the leaf number and the bolting time were identified. The experiment was repeated once more with four replicates in each experiment for statistical analysis. Leaf samples were collected for physiological, biochemical and gene expression analysis.
Gene expression analysis in Arabidopsis
The qRT–PCR was performed on RNA extracted from leaf samples as described by Dossa et al. (2018) using the Arabidopsis gene Actin 2 (AT3G18780) as the internal control. Specific primer pairs of SiMYB75, four antioxidant genes AtSOD, AtPOD, AtAPX, AtCAT and 11 stress marker genes including 11 stress marker genes including AtRD22, AtDREB2C, AtABA3, AtWRKY28, AtNCED3, AtEREBP, RD29B, ATCBF2, HSP17.4B, ATHSP70–14 and AtNACO19 were designed (see Supporting Information—Table S1). Samples in the control condition (non-stress) were used as reference and data are presented as relative transcript level based on the 2-∆∆Ct method (Livak and Schmittgen 2001).
Biochemical analysis
Leaf from salt, drought and control treated plants (five individual plants from each line/treatment) was used for assessing the ABA content. ABA was measured using the high performance liquid chromatography–tandem mass spectrometry (HPLC–MS/MS) method as described by Liu et al. (2014). Similarly, for the detection of superoxide (O2-), leaves were transferred in nitro blue tetrazolium (NBT) (1 mg.ml-1) for 5 h. The leaves were boiled in absolute ethanol for 10 min and then pictures were captured for record. Localization of O2- was visualized as blue coloration. Leaf tissue was homogenized in 1 mL 80 % methanol, and then centrifuged at 12 000×g for 10 min at room temperature. The ability of the extract to scavenge hydrogen peroxide (H2O2) was determined according to the method of Ruch et al. (1989). Ten microlitres of the extract was mixed with 600 μL of the reaction solution (10 mM of hydrogen peroxide in phosphate buffer, pH = 4). After 10 min of incubation at room temperature, the absorbance was measured at 240 nm. Blank is the reaction solution without hydrogen peroxide. Ascorbic acid was used as a standard. The scavenging effect was expressed in % and calculated:
Measurements of protective enzyme activities in leaf samples, including ascorbate peroxidase (APX, EC 1.11.1.11), peroxidase (POD, EC 1.11.1.7), total superoxide dismutase (SOD, EC 1.15.1.1), catalase (CAT, EC 1.11.1.6) as well as the leaf chlorophyll content and the concentration of malondialdehyde (MDA) were determined according to descriptions of Dossa et al. (2017c).
Water loss and stomata closure assays
To examine the possible role of SiMYB75 in drought stress tolerance, we further conducted water loss experiments using detached leaves (Wang et al. 2018a). Four similar shaped and sized leaves were excised from each 4-week-old transgenic and WT lines, and the fresh weight was immediately determined. All leaves were placed on a laboratory bench at room temperature for 210 min. Every 30 min, leaf weight was recorded in the same order that leaves were detached. For the stomata closure assay, the leaves were immersed in a solution containing 10 mM KCl, and 10 mM MES (pH = 6.15) under white light for 3 h. Subsequently, 20 μM ABA was added into the solution for 1 h. In addition, we measured the stomatal movements in leaves detached from drought and salt treated plants. The pictures were taken using a compound microscope with magnification ×40 (OLYMPUS DP72, Japan). We measured more than 100 stomata of each line using the IMAGEJ 1.8.0 software (Broken Symmetry Software). Each experiment was performed in triplicates.
Cell membrane stability assay
The ion leakage technique was carried out basically as described by Cabello and Chan (2012) on detached leaves at the end of drought and salt treatments. Briefly, detached leaves (~100 mg) were placed in 20 mL of deionized water in two test tubes. One tube was incubated in a water bath at a constant temperature of 40 ºC for 30 min, and its conductivity (C1) was measured with a conductivity metre (Orion Star A322). The second tube was placed in a boiling water bath (100 ºC) for 10 min, cooled, and conductivity was recorded (C2). Ion leakage was expressed as a percentage using the formula: [1-(C1/C2)] × 100.
Statistical analyses
All the data were analysed with the R software (www.r-project.org). One-way analysis of variance was performed by comparing each transgenic line to the wild type plants. This was followed by Tukey HSD test for mean comparison. The error bars were calculated with data from a single experiment.
Results and Discussion
SiMYB75 is an R2R3 MYB type gene and is induced by various environmental stresses
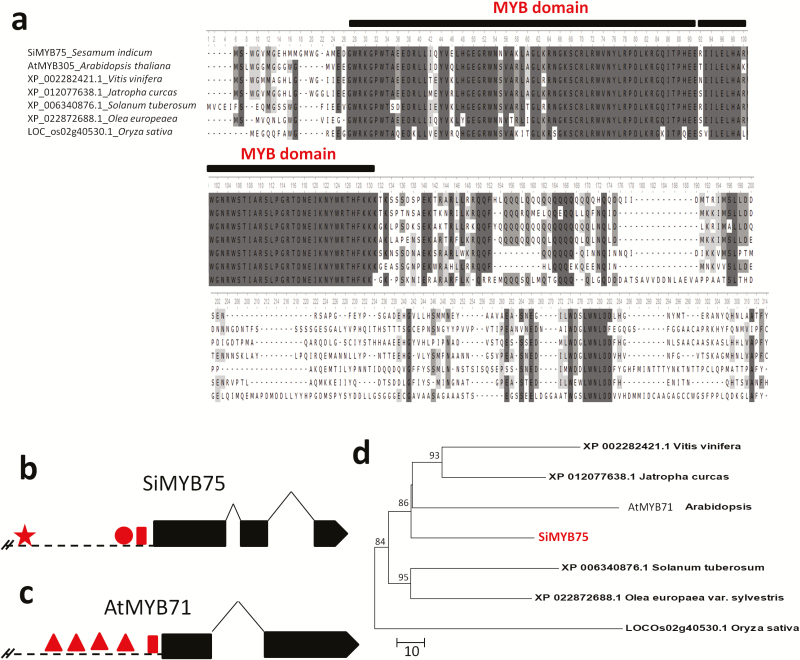
In this study, we retrieved the gene SIN_1015311 (SiMYB75) and found that it contains 2 MYB domains (from 23 to 70 and from 76 to 121 amino acids), indicating an R2R3 MYB type (Fig. 1a). Gene structure analysis showed that SiMYB75 comprises 3 exons and 2 introns with a total length of 1400 bp (Fig. 1b). The gene is located on the linkage group 3 and encodes an ORF of 256 amino acids with a predicted molecular mass of 29.8 kD and an isoelectric point of 7.92. Prediction of the sub-cellular localization of SiMYB75 using the TargetP 1.1 server and the WoLF PSORT web site indicated that SiMYB75 protein targets the nucleus, similarly as its homolog AtMYB71 (At3g24310) in Arabidopsis. Furthermore, using the pBWA(V)HS-SiMYB75-GFP construct transformed into Arabidopsis protoplasts, SiMYB75the GFP signals were co-localized with nuclear marker signals which confirmed that SiMYB75 was localized in the nucleus in accordance with the results predicted by the software (see Supporting Information—Fig. S2). Promoter sequence analysis of SiMYB75 revealed a TATA box (TATAAA) at the position -142 bp. Furthermore, we identified a CRT/DRE element (ACCGAC) (Dubouzet et al. 2003) at the position -243 bp and an ABRE element (ACGTGGC) (Nakashima et al. 2006) at the position -914 bp (Fig. 1b). The discovery of a putative functional promoter and presence of stress-related cis-acting elements further indicate that SiMYB75 may serve as an intact gene and function in sesame abiotic stress responses (Ren et al. 2016). Similarly, by analysing the promoter region of the homologous gene AtMYB71 in Arabidopsis, we observed four stress response elements (STRE, AGGGG), implying that AtMYB71 may be also involved in stress responses (Fig. 1c). The construction of the phylogenetic tree based on the alignment of the complete protein sequences of SiMYB75 and its homologues from some related plant species showed that SiMYB75 is more close to its homologues from Arabidopsis, jatropha and grape (Fig. 1a and d).
Figure 1.
Sesamum indicum SiMYB75 amino acid sequence analysis. (a) Alignment of deduced protein sequence of SiMYB75 (NCBI protein ID: XP_011073726.1) with those of related proteins in other species including Arabidopsis thaliana (National Center for Biotechnology Information (NCBI) protein ID: NP_189074.1), Solanum tuberosum (NCBI protein ID: XP_006340876.1), Japtropha curcas (NCBI protein ID: XP_012077638.1), Vitis vinifera (NCBI protein ID: XP_002282421.1), Olea europaea var. sylvestris (NCBI protein ID: XP_022872688.1), Oryza sativa (NCBI protein ID: XP_015624171.1). Numbers above indicate the amino acid position and the positions of the two MYB domains are shown; (b) Gene structure of SiMYB75. Exons and introns are represented by black boxes and lines, respectively. Dashed lines represent 1 Kb promoter region with the position of ABRE motif (red star), the CRT/DRE motif (red circle) and the TATA box (red rectangle); (c) Gene structure of AtMYB71. Exons and introns are represented by black boxes and lines, respectively. Dashed lines represent 1 Kb promoter region with the position of STRE motifs (red triangle) and the TATA box (red rectangle); (d) A phylogenetic tree was constructed in the MEGA7 software with the seven protein sequences using the neighbour-joining method with 1000 bootstrap replicates.
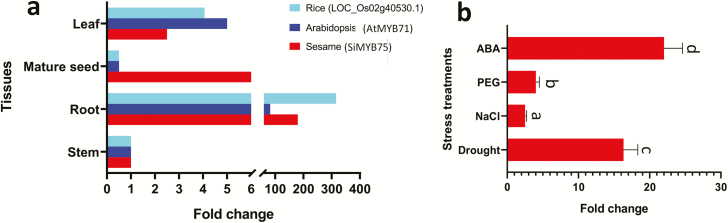
SiMYB75 was expressed in various sesame tissues but was more vigorous in the root, implying that SiMYB75 may play an important role in root growth and development (Fig. 2a). The homologues of SiMYB75 in Arabidopsis (AtMYB71) and in rice (LOC_Os02g40530.1) were also found strikingly expressed in the root tissue, indicating a conserved spatial expression pattern of this gene in various species (Fig. 2a) (Schmid et al. 2005; Wang et al. 2015). Moreover, we investigated the expression patterns of SiMYB75 under different abiotic stresses, including drought, salt, ABA and osmotic stresses, in the sesame root. As expected, we found that SiMYB75 was strongly induced by all applied stresses, showing that SiMYB75 may act as a regulator of drought, salt, ABA and osmotic stresses tolerance in sesame. It is worth mentioning that ABA and drought stresses induced significantly higher expression levels of SiMYB75 as compared to the osmotic and salt treatments (P < 0.05) (Fig. 2b). Also, osmotic stress higher induced the SiMYB75 as compared to salt stress. In agreement with our results, AtMYB71 was found more up-regulated in Arabidopsis root under 300 mM mannitol than 150 mM NaCl treatments (Kilian et al. 2007). But it was not strongly induced by other abiotic stress treatments such as wounding, UV-B, heat and genotoxic (Kilian et al. 2007). Nonetheless, the concentration and duration of osmotic and salt stress treatments were fixed in this study and the possibility that higher expression of SiMYB75 may be observed at other concentrations or time points cannot be excluded.
Figure 2.
SiMYB75 expression profiling in tissues and various abiotic stresses. (a) Expression in root, seed, leaf and stem of Sesame (www.sesame-bioinfo.org/cgi-bin/Sinbase2.0/search_gene.cgi), Arabidopsis (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi?ncbi_accn=822 019&modeInput = Absolute&dataSource=Developmental_Map) and Rice (www.ncbi.nlm.nih.gov/gene/4 330 001). Data expressed as fold change using the stem expression level as the reference; (b) Transcript fold change of SiMYB75 under sodium chloride (NaCl) (200 mM for 48 h), polyethylene glycol (PEG) 6000 (2 % for 5 days), abscisic acid (ABA) (100 µM for 12 h) and drought (water withholding for 7 days) treatments compared with the control treatment in the sesame root. Data shown are average and SD of triplicate quantitative reverse transcriptase polymerase chain reactions. The sesame Actin 7 gene was used as the internal control. Different letters above bars represent a significant difference between treatments at P ≤ 0.05.
Over-expression of SiMYB75 increases tolerance to NaCl, osmotic, drought and confers ABA sensitivity in transgenic Arabidopsis
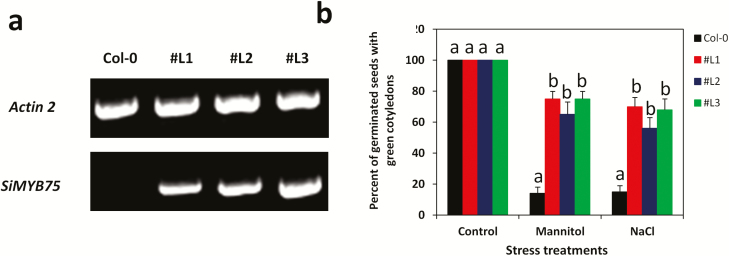
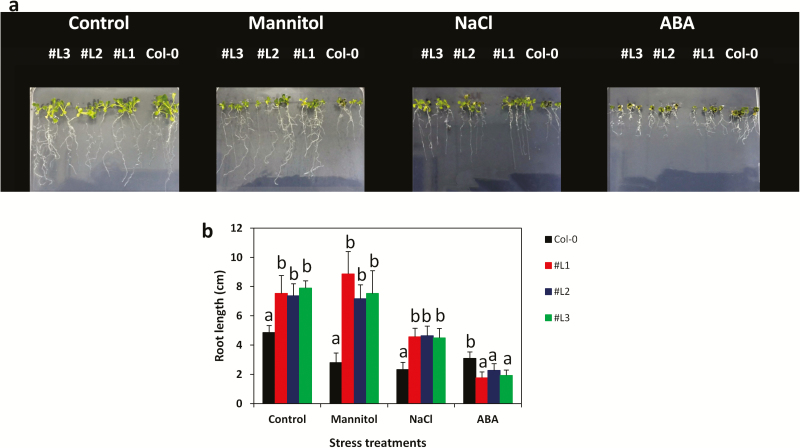
To study the function of SiMYB75, we developed several transgenic Arabidopsis lines and selected three independent and homozygous T3 lines (#L1, #L2 and #L3) highly expressing the transgene for functional characterizations. This was confirmed through RT–PCR (Fig. 3a). The three over-expressing Arabidopsis lines and the wild-type (WT) plants were subjected to NaCl, mannitol and drought treatments. In the control condition (MS medium), SiMYB75 over-expressing lines and WT plants germinated well and have green cotyledons (100 %). Under 100 mM mannitol and 100 mM NaCl treatments, the percentage of germinated seeds with green cotyledons was reduced in all lines (Fig. 3b). However, the transgenic plants displayed significantly higher percentages of germinated seeds with green cotyledons than the WT plants (P ≤ 0.001), indicating that the over-expression of SiMYB75 imparts osmotic and salt tolerance in Arabidopsis at the germination stage. Since SiMYB75 was found highly expressed in the sesame root, we further analysed the root growth of the transgenic Arabidopsis plants under normal and stress conditions (Fig. 4a). In the normal MS medium, the SiMYB75 over-expressing lines exhibited significantly longer roots than those of WT plants (P ≤ 0.01) (Fig. 4b). The root length was not affected by mannitol application in the transgenic plants while it was significantly decreased in the WT plants (P ≤ 0.001) (Fig. 4b). This result suggests that SiMYB75 promotes root growth maintenance under osmotic stress, which seems to be a tolerance mechanism. The MYB proteins have been described as key components involved in root growth regulation, particularly under abiotic stresses (Baldoni et al. 2015). Fang et al. (2017) by investigating a novel R2R3 MYB gene PtrSS1 from poplar found that the transgenic lines increased the lateral root number to resist under salt stress conditions. Similar to our findings, the over-expression of the gene AtMYB60 increased root growth and conferred tolerance to short-term drought stress in Arabidopsis (Oh et al. 2011). Moreover, using a phylogenetic approach, Mmadi et al. (2017) classified the gene SiMYB75 in the subgroup C3 together with the gene AtMYB59, which was defined as a root development related gene (Mu et al. 2009). Under salt treatment, the root growth was reduced in all plants when compared to the growth in the normal MS medium, though, the over-expressing lines showed significantly higher root length than the WT plants (P ≤ 0.001). This highlights the higher ability of the transgenic lines to cope with the NaCl stress (Fig. 4b). Conversely, we observed an increased ABA sensitivity of the transgenic lines.
Figure 3.
Identification of positive Arabidopsis over-expressing lines. (a) Reverse transcriptase polymerase chain reaction analysis of SIMYB75 in the wild type (Col-0) and transgenic lines (#L1, #L2 and #L3).The Actin 2 gene was used as internal control. (b) Evaluation of Arabidopsis SiMYB75 over-expressing lines (#L1, #L2 and #L3) and wild type plants (Col-0) under mannitol (0/100 mM) and sodium chloride (NaCl) (0/100 mM) treatments at the germination stage. Percentage of germinated seeds with green cotyledons recorded after 7 days. For each experiment, about 30 plants/ lines were used. Data represent means ± SD of five measurements from one experiment and the experiment was repeated once more and similar results were obtained. Different letters above bar mean significant difference between each transgenic line and Col-0 at P ≤ 0.05.
Figure 4.
Root growth of Arabidopsis SiMYB75 over-expressing lines (#L1, #L2 and #L3) and wild type (Col-0) seedlings under various abiotic stress treatments. (a) Phenotypes of the transgenic and Col-0 plants under normal Murashige and Skoog (MS) medium and MS medium supplemented with 150 mM mannitol, 150 mM sodium chloride (NaCl) and 10 µM abscisic acid (ABA); (b) Root length was recorded after 10 days. For each experiment, 16 seedlings/lines were used. Data represent means ± SD from one experiment and the experiment was repeated once more and similar results were obtained. Different letters above bar mean significant difference between each transgenic line and Col-0 at P ≤ 0.05.
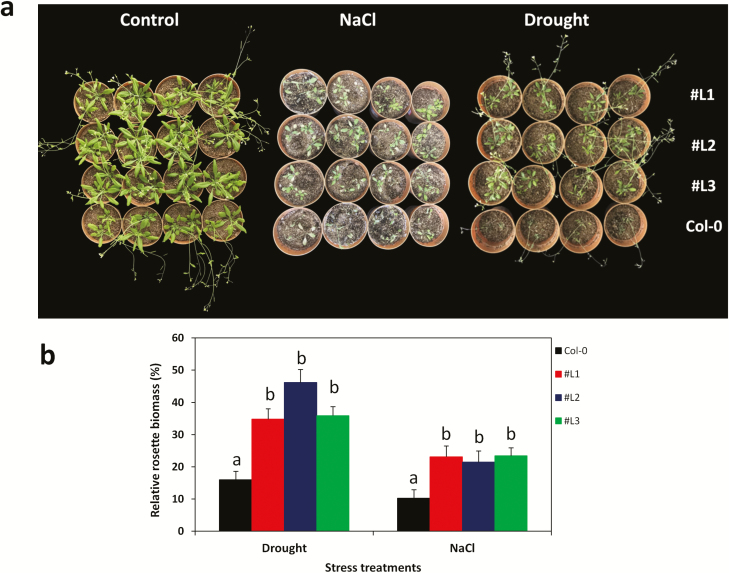
Next, we examined the tolerance of the over-expressing SiMYB75 lines at the seedling stage either under water deprivation (drought) or high NaCl concentration (200 mM) (Fig. 5a). The transgene did not have significant effect on plant size, bolting time and dry matter under normal growth conditions (see Supporting Information—Fig. S3). However under drought and salt stress conditions, the growth of both WT and transgenic plants was inhibited. All plants bolted later, and had lower leaf numbers, rosette diameters and dry weights when compared with the controls. Nonetheless, much less severe effects were observed for the transgenic lines than WT plants (see Supporting Information—Fig. S3). As shown in Supporting Information—Table S2, nearly all the transgenic plants survived 17 days drought stress while most of WT plants died. Similarly, the transgenic lines had higher survival rates than the WT plants under salt stress (see Supporting Information—Table S2). In addition, the transgenic lines were able to maintain a significantly higher biomass production under stress than the WT plants, as shown by the relative rosette biomass under drought (P ≤ 0.001) and salt (P ≤ 0.001) (Fig. 5b).
Figure 5.
Drought and salt stress tolerance assay at the seedling stage of Arabidopsis SiMYB75 over-expressing lines (#L1, #L2 and #L3) and wild type plants (Col-0). (a) Phenotypes of the transgenic and Col-0 plants in control, sodium chloride (NaCl) (200 mM for 7 days) and drought (17 days water withholding) treatments; (b) relative rosette biomass estimated as the ration of the dry weight in the stress and control treatments. For each experiment, 8 plants/lines were used. Data represent means ± SD from one experiment and the experiment was repeated once more and similar results were obtained. Different letters above bar mean significant difference between each transgenic line and Col-0 at P ≤ 0.05.
Overall, the Arabidopsis transgenic experiment confirmed that SiMYB75 is a positive modulator of osmotic, drought and salt tolerance. SiMYB75 seems to be more effective under osmotic/drought stress than salt stress. In fact, plants exposed to salt and drought stresses both suffer from osmotic stress. Additionally, salt stress provokes a strong ion toxicity resulting from a fast ion uptake by the plants (Jakab et al. 2005). Therefore, it is probable that SiMYB75 primarily mediates cellular osmotic adjustment but is not involved in ion toxicity prevention. This finding correlates well with the observed levels of SiMYB75 induction under drought, osmotic and salt stresses in sesame (Fig. 2b).
Physiological and biochemical adaptations under drought and NaCl stresses in SiMYB75 over-expressing Arabidopsis lines
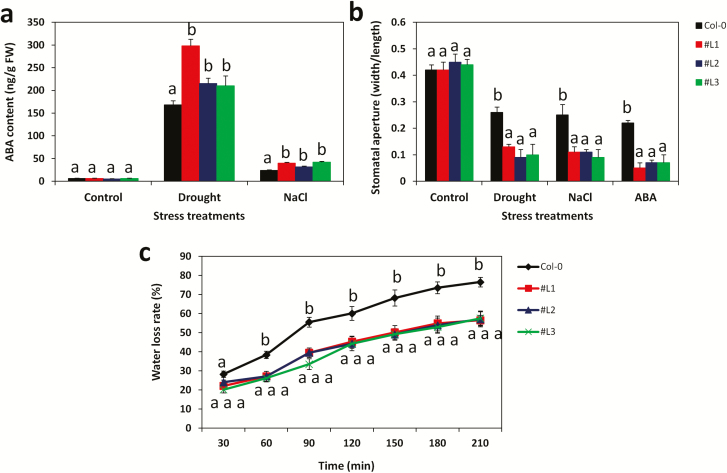
Under drought and salinity stress conditions, ABA is usually generated in many biological systems (Mittler 2002). We quantified the endogenous ABA content under normal, drought and salt conditions in leaves of the WT and the transgenic plants. As presented in Fig. 6a, SiMYB75 over-expressing lines accumulated significantly higher content of ABA under stress treatments than the WT plants (P ≤ 0.001), with a higher accumulation in drought stress than in salt stress treatments. Most stress-responsive MYBs, including ZmMYB48, OsMYB2, SbMYB8, AtMYB44, AtMYB73, AtMYB20, AtMYB2 and AtMYB15 (Abe et al. 2003; Jung et al. 2008; Ding et al. 2009; Yang et al. 2012; Cui et al. 2013; Kim et al. 2013; Yuan et al. 2015; Wang et al. 2017), were reported to regulate plant response to abiotic stress through the ABA-dependent pathways. In this study, we noticed that SiMYB75 over-expressing lines were sensitive to the exogenous ABA application (Fig. 4a) and they strongly accumulated the endogenous ABA under abiotic stress treatments. Therefore, we deduce that SiMYB75 also functions through the ABA-mediated signalling pathways.
Figure 6.
Physiological and biochemical changes in the SiMYB75 over-expressing lines (#L1, #L2 and #L3) compared to wild type plants (Col-0) under drought and salt treatments. (a) abscisic acid (ABA) content in leaves of transgenic and Col-0 plants in control, sodium chloride (NaCl) (200 mM for 7 days) and drought (17 days water withholding) treatments; (b) stomata aperture (width/length) assay of transgenic and Col-0 plants in control, drought, salt and 20 µM ABA treatments. Bright-field pictures were taken at ×40 magnification using an OLYMPUS DP72 microscope and over 100 stomata per line/treatment were analysed using the IMAGEJ software; (c) Water loss rates in the leaves detached from transgenic and Col-0 plants under normal conditions during a 210-min period; For each experiment, four plants/lines were used. Data represent means ± SD from one experiment and the experiment was repeated once more and similar results were obtained. Different letters above bar mean significant difference between each transgenic line and Col-0 at P ≤ 0.05.
Increased biosynthesis and accumulation of ABA limit transpirational water loss through adjustment of the stomatal aperture in plants (Farooq et al. 2009). Our stomatal closure (width/maximum length) assay revealed that, in contrast to the WT plants, the transgenic lines almost closed the stomata under ABA, drought and salt treatments (Fig. 6b). In addition, the water loss experiment performed on detached leaves showed that during a 210-min period, the WT plants lost rapidly and more intracellular water than the transgenic lines (Fig. 6c). Since, the stomatal closure is an important gateway for plants to decrease water loss, we infer that the low ability of WT plants to regulate the stomata closure under stress has led to an increased water loss and probably to cell damage and death. Our results corroborate well the conclusions from previous studies of Ren et al. (2016) and Wang et al. (2018) who showed that over-expression of Maize ZmMYB48 and Wheat TaCIPK27, two genes functioning in the ABA-dependent pathways, increased the endogenous ABA levels under drought stress, promoted the stomatal closure and reduced the intracellular water loss in transgenic Arabidopsis, resulting in an enhanced drought tolerance.
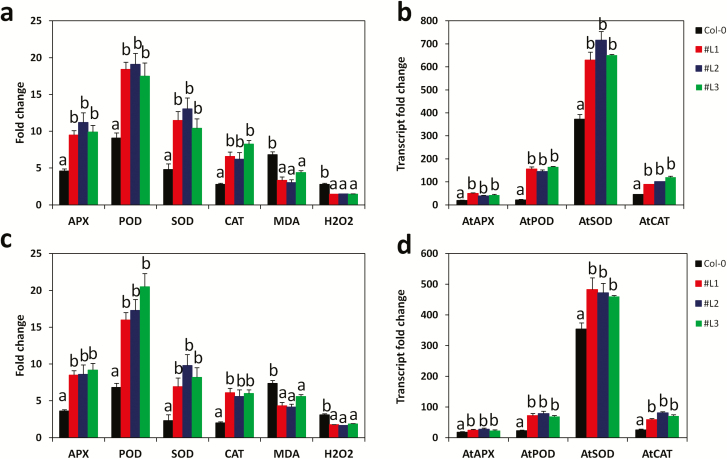
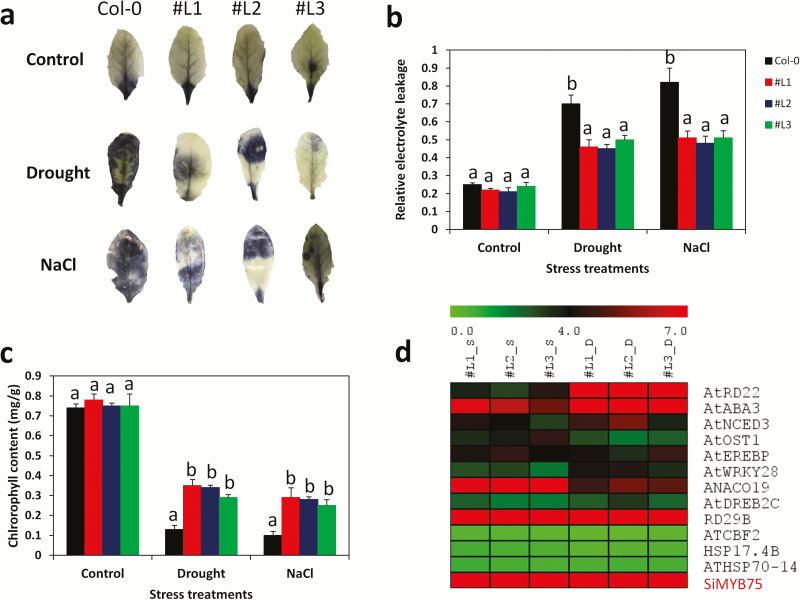
In plants, drought and salinity stresses cause oxidative damage via the production of reactive oxygen species (ROS), such as H2O2 and superoxide (Mittler 2002; Xiong and Zhu 2002). High ROS-scavenging activities decrease the over-accumulation of ROS in plants, thereby inhibiting the onset of programmed cell death (Mittler 2002; Farooq et al. 2009). To investigate the ROS-scavenging activities in the transgenic lines and WT plants, the levels of the enzymes SOD, POD, CAT and APX and their corresponding transcripts AtSOD, AtPOD, AtCAT and AtAPX (Gao et al. 2012) were evaluated under drought (Fig. 7a and b) and salt (Fig. 7c and d). For the entire assayed enzymes, we observed an increase in their activities under stress compared to the control conditions. Similar observations were noted for their corresponding transcript levels. However, these inductions were significantly higher in the transgenic lines than in WT plants. These results denote that the over-expressing lines have an elevated ROS scavenging ability as a potent mechanism underlying their strong tolerances to drought and salt stresses. The molecule malondialdehyde (MDA) has been associated with lipid peroxidation via an increased generation of ROS, and thus its quantification has been suggested as a general indicator for stress tolerance (Dossa et al. 2017c). We, therefore, analysed the MDA and H2O2 contents in leaf tissues under stress conditions in relation to the control treatment (Fig. 7a and c). Our results showed that the stress-induced accumulations of MDA and H2O2 in cells of WT plants were significantly higher than in cells of the transgenic lines (P ≤ 0.001). This implies that WT plants suffered more from stresses than the transgenic lines. Fig. 8a showed that there was no difference between the WT plants and the transgenic lines regarding the superoxide production under control condition. However, under drought and salt stress treatments, WT plants produced more superoxide than transgenic lines further providing a solid support to our previous results. Next, we examined the relative electrolyte leakage and the chlorophyll content in leaves of stressed and control plants (Fig. 8b and c). Both drought and salt increase the cell ion leakage while decrease the chlorophyll contents in all plants. By comparing the WT plants and the transgenic lines, we found that the transgenic lines suffered lower cell membrane damage than the WT counterparts. On the other hand, the chlorophyll content was significantly higher in the transgenic lines compared to the WT plants (P ≤ 0.001)
Figure 7.
Analysis of the reactive oxygen species (ROS)-scavenging machinery. Enzymes activity (ascorbate peroxidase (APX), superoxide dismutase (SOD), peroxidase (POD), catalase (CAT)) and ROS level indicators (malondialdehyde (MDA), hydrogen peroxide (H2O2)) in the SiMYB75 over-expressing lines (#L1, #L2 and #L3) compared to wild-type plants (Col-0) under (a) drought and (c) salt treatments. Transcript fold change (FC) of four antioxidant genes AtAPX, AtPOD, AtCAT and AtSOD under (b) drought and (d) salt treatments. For each experiment, four plants/lines were used. Data represent means ± SD from one experiment and the experiment was repeated once more and similar results were obtained. Different letters above bar mean significant difference between each transgenic line and Col-0 at P ≤ 0.05.
Figure 8.
The transgenic lines (#L1, #L2 and #L3) suffered less than wild-type plants (Col-0) from drought and salt. (a) Nitro blue tetrazolium staining of leaves detached from plants under control, drought and sodium chloride (NaCl) treatments. (b) Electrolyte leakage. (c) Measurement of chlorophyll content. For each experiment, four plants/lines were used. Data represent means ± SD from one experiment and the experiment was repeated once more and similar results were obtained. Different letters above bar mean significant difference between each transgenic line and Col-0 at P ≤ 0.05. (d) Heatmap displaying fold changes (FC) of abiotic stress marker genes and abscisic acid (ABA) signalling and biosynthesis genes in the transgenic lines compared to Col-0 plants (FC transgenic line/ FC wild-type plants) under drought and NaCl treatments based on quantitative reverse transcriptase polymerase chain reaction. The Actin 2 gene was used as internal control. Data shown are average of triplicate reactions. S and D represent salt and drought treatments, respectively.
In summary, our physiological and biochemical investigations established that over-expression of SiMYB75 increases ABA content under stress conditions, which leads to stomatal closure and reduced water loss in the leaves. In addition, SiMYB75 acts through the ROS signalling pathway by increasing the activity of antioxidant enzymes and probably other non-enzymatic components to reduce ROS production and alleviate cell damage and apoptosis.
Over-expression of SiMYB75 increases the expression of abiotic stress-responsive genes in the transgenic plants
Under adverse conditions, transcription factors can induce or repress the activity of the RNA polymerase, thus regulating target gene expression in plants (Saibo et al. 2009). Since, SiMYB75 over-expressing lines displayed improved tolerance to drought, salt and osmotic stresses through ABA-mediated pathways in transgenic Arabidopsis plants, we hypothesized that SiMYB75 modulates the expression levels of downstream abiotic-stress-marker genes in Arabidopsis. To test this hypothesis, we performed qRT–PCR analysis of stress-marker genes related to salinity, cold, heat, drought and genes related to ABA biosynthesis, e.g. AtRD22, AtABA3, AtNCED3, AtOST1, AtEREBP, AtWRKY28, ANAC019, RD29B, ATCBF2, HSP17.4B, ATHSP70–14 and AtDREB2C (Swindell et al. 2007; Manuka et al. 2018; Park et al. 2018), under control, drought and salt treatments. The results revealed that drought and salt treatments increased the expression levels of all tested stress marker genes in WT and transgenic lines. However, by comparing the fold changes (FC) of the expression levels of those genes in the transgenic lines with the WT plants (FC transgenic lines/FC WT plants), we observed that all the tested stress marker genes were up-regulated in the transgenic lines under both stress treatments except ATCBF2, HSP17.4B and ATHSP70–14 (Fig. 8d).
Previous studies demonstrated that the expression levels of the genes AtNCED3 and AtABA3 involved in ABA synthesis increased under abiotic stresses such as drought and salinity (Yamaguchi-Shinozaki et al. 2006; Nakashima et al. 2009; Xie et al. 2014). Our results are in accordance with these reports. It implies that the elevated concentration of ABA in SiMYB75 over-expressing lines (Fig. 6a) is favoured by the increased expression levels of ABA synthesis genes. RD29B which is the main ABA-dependent pathway marker gene (Msanne et al. 2011) was found strongly up-regulated in the transgenic lines as compared to the wild-type plants, further supporting the premise that SiMYB75 over expression acts in the ABA-dependent pathway. According to Yunta et al. (2011), an increase in ABA content stimulates within minutes the regulators such as the OPEN STOMATA 1 (OST1) gene, which modulates the stomatal closure. Therefore, we interpreted that the over-expression of SiMYB75 further triggered the expression level of AtOST1 in the transgenic lines under stress treatments resulting in a rapid stomatal closure (Fig. 6b). The RESPONSIVE TO DEHYDRATATION 22 (RD22) gene is used as a reliable ABA early response marker in plants (Yamaguchi-Shinozaki et al. 1993; Matus et al. 2014). The Arabidopsis gene AtRD22 is up-regulated by moisture stress, salinity and exogenously applied ABA (Yamaguchi-Shinozaki et al. 1993). Wang et al. (2012) also demonstrated that the soybean GmRD22 gene could directly improve salt stress tolerance when over-expressed in both Arabidopsis and rice. Similarly, the strong up-regulation of several stress-induced transcription factors such as AtEREBP, AtWRKY28, ANAC019 and AtDREB2C were shown to be a tolerance mechanism upon exposure to abiotic stresses in Arabidopsis (Tran et al. 2004; Babitha et al. 2013; Liu et al. 2013; Manuka et al. 2018; Wang et al. 2018). Interestingly, the three genes (ATCBF2, HSP17.4B and ATHSP70–14) that did not exhibit significant difference between transgenic lines and WT plants, have been shown to regulate different abiotic stresses such as heat and cold and are not induced by ABA (Swindell et al. 2007; Park et al. 2018). In that sense, our results validate the formulated hypothesis referring to the participation of SiMYB75 in the ABA-dependent pathways and indicate that the enhanced ability to cope with drought, salt and osmotic stresses of the SiMYB75-over-expressing lines is due in part to the increased expression of these abiotic stress-marker genes.
Conclusions
In this report, we cloned and characterized a novel R2R3-MYB gene SiMYB75 from sesame. We demonstrated that SiMYB75 works through the ABA-mediated pathways and positively modulates drought, salt and osmotic stresses responses. These results increase our understanding of the roles of sesame MYB transcription factors in response to abiotic stresses. SiMYB75 is, therefore, a promising candidate gene for sesame crop improvement. However, the difficulties related to the sesame genetic transformation hinder the development of genetically modified cultivars with improved performances in field. Nonetheless, to harness the potential of SIMYB75, we will study the natural variation in its promoter and genic regions in association with drought and salt stress responses in a large panel of sesame (Dossa et al. 2019a). This will help identify superior haplotypes and develop specific molecular markers for breeding applications.
Supporting Information
The following additional information is available in the online version of this article—
Table S1. List of the primers used for qRT-PCR experiments in sesame and Arabidopsis
Table S2. Number of survived and dead plants aftr 17 days drought and 7 days under 200 mM NaCl treatments. For each experiment, 8 plants/ lines were used.
Fig. S1. Map of the vector used for the transformation of Arabidopsis and PCR results of the T1 transgenic plants. M represents 2 Kb DNA Ladder.
Fig. S2. Subcellular localization of the SiMYB75 protein in Arabidopsis protoplasts. A, fluorescence signal in target protein co-localizations with a Nucleus marker; B, fluorescence signal of the GFP empty vector.
Fig. S3. Growth parameters of transgenic SiMYB75 lines and its Wild Type (WT) under control, drought and salinity conditions, including leaf number, rosette diameter, day to bolting, rosette dry weight at 25 days after sowing (DAS) and at 45 DAS. Data represent means ± SD of five measurements from one experiment and the experiment was repeated once more and similar results were obtained. Different letters above bar mean significant difference between each transgenic line and Col-0 at P ≤ 0.05.
Acknowledgements
This research was funded by the China Agriculture research System, grant number (CARS-14), the Agricultural Science and Technology Innovation Project of Agricultural Sciences, grant number (CAAS-ASTIP-2013-OCRI), the National Natural Science Foundation of China, grant number (31500223) and the Fundamental Research Funds for Central Non-profit Scientific Institution, grant number (1610172018007).
Dossa K, Mmadi MA, Zhou R, Liu A, Yang Y, Diouf D, You J and Zhang X. 2019. Ectopic expression of the sesame MYB transcription factor SiMYB305 promotes root growth and modulates ABA-mediated tolerance to drought and salt stresses in Arabidopsis. AoB PLANTS 11: plz081; doi: 10.1093/aobpla/plz081
Data
All data generated or analysed during this study are included in this published article and its supplementary information files.
Contributions by the Authors
K.D., M.A.M., D.D. and X.Z. conceived, designed and supervised the experiment; K.D. and M.A.M. wrote the manuscript; K.D., M.A.M., A.L. and R.Z. performed the experiment; Y.J. and Y.Y. provided support in lab experiment and data analysis. K.D. analysed the data. All authors read and approved the manuscript.
Conflict of Interest Statement
None declared.
Literature Cited
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2003. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. The Plant Cell 15:63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. 1997. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. The Plant Cell 9:1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal PK, Jha B. 2010. Transcription factors in plants and ABA dependent and independent abiotic stress signaling. Biologia Plantarum 54:201–212. [Google Scholar]
- Ambawat S, Sharma P, Yadav NR, Yadav RC. 2013. MYB transcription factor genes as regulators for plant responses: an overview. Physiology and Molecular Biology of Plants: An International Journal of Functional Plant Biology 19:307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anilakumar KR, Pal A, Khanum F, Bawas AS. 2010. Nutritional medicinal and industrial uses of sesame (Sesamum indicum L.) seeds. Agriculturae Conspectus Scientificus 75:159–168. [Google Scholar]
- Babitha KC, Ramu SV, Pruthvi V, Mahesh P, Nataraja KN, Udayakumar M. 2013. Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Research 22:327–341. [DOI] [PubMed] [Google Scholar]
- Baldoni E, Genga A, Cominelli E. 2015. Plant MYB transcription factors: their role in drought response mechanisms. International Journal of Molecular Sciences 16:15811–15851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazrafshan AH, Ehsanzadeh P. 2014. Growth photosynthesis and ion balance of sesame (Sesamum indicum L.) genotypes in response to NaCl concentration in hydroponic solutions. Photosynthetica 52:134–147. [Google Scholar]
- Bekele A, Besufekad Y, Adugna S, Yinur D. 2017. Screening of selected accessions of Ethiopian sesame (Sesamum indicum L) for salt tolerance. Biocatalysis and Agricultural Biotechnology 9:82–94 [Google Scholar]
- Butt HI, Yang Z, Gong Q, Chen E, Wang X, Zhao G, Ge X, Zhang X, Li F. 2017. GaMYB85, an R2R3 MYB gene, in transgenic Arabidopsis plays an important role in drought tolerance. BMC Plant Biology 17:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello JV, Chan RL. 2012. The homologous homeodomain-leucine zipper transcription factors HaHB1 and AtHB13 confer tolerance to drought and salinity stresses via the induction of proteins that stabilize membranes. Plant Biotechnology Journal 10:815–825. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Basu A, Kundu S. 2017. Overexpression of a new osmotin-like protein gene (SindOLP) confers tolerance against biotic and abiotic stresses in sesame. Frontiers in Plant Science 8:410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal: for Cell and Molecular Biology 16:735–743. [DOI] [PubMed] [Google Scholar]
- Cui MH, Yoo KS, Hyoung S, Nguyen HT, Kim YY, Kim HJ, Ok SH, Yoo SD, Shin JS. 2013. An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Letters 587:1773–1778. [DOI] [PubMed] [Google Scholar]
- Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K. 2007. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiology 143:1739–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Li S, An X, Liu X, Qin H, Wang D. 2009. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. Journal of Genetics and Genomics = Yi Chuan Xue Bao 36:17–29. [DOI] [PubMed] [Google Scholar]
- Dossa K, Diouf D, Cissé N. 2016a. Genome-wide investigation of Hsf genes in sesame reveals their segmental duplication expansion and their active role in drought stress response. Frontiers in Plant Science 7:1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossa K, Niang M, Assogbadjo AE, Cisse N, Diouf D. 2016b. Whole genome homology-based identification of candidate genes for drought resistance in (Sesamum indicum L). African Journal of Biotechnology 15:1464–1475. [Google Scholar]
- Dossa K, Wei X, Li D, Fonceka D, Zhang Y, Wang L, Yu J, Boshou L, Diouf D, Cissé N, Zhang X. 2016c. Insight into the AP2/ERF transcription factor superfamily in sesame and expression profiling of DREB subfamily under drought stress. BMC Plant Biology 16:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossa K, Diouf D, Wang L, Wei X, Zhang Y, Niang M, Fonceka D, Yu J, Mmadi MA, Yehouessi LW, Liao B, Zhang X, Cisse N. 2017a. The emerging oilseed crop sesamum indicum fenters the “Omics” Era. Frontiers in Plant Science 8:1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossa K, Li D, Wang L, Zheng X, Liu A, Yu J, Wei X, Zhou R, Fonceka D, Diouf D, Liao B, Cissé N, Zhang X. 2017b. Transcriptomic, biochemical and physio-anatomical investigations shed more light on responses to drought stress in two contrasting sesame genotypes. Scientific Reports 7:8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossa K, Yehouessi LW, Likeng-Li-Ngue BC, Diouf D, Liao B, Zhang X, Cissé N, Bell JM. 2017c. Comprehensive screening of some west and central African sesame genotypes for drought resistance probing by agromorphological physiological biochemical and seed quality traits. Agronomy 7:83. [Google Scholar]
- Dossa K, Mmadi MA, Zhou R, Zhou Q, Yang M, Cisse N, Diouf D, Wang L, Zhang X. 2018. The contrasting response to drought and waterlogging is underpinned by divergent DNA methylation programs associated with transcript accumulation in sesame. Plant Science 277:207–217. [DOI] [PubMed] [Google Scholar]
- Dossa K, Li D, Zhou R, Yu J, Wang L, Zhang Y, You J, Liu A, Mmadi MA, Fonceka D, Diouf D, Cissé N, Wei X, Zhang X. 2019a. The genetic basis of drought tolerance in the high oil crop Sesamum indicum. Plant Biotechnology Journal 17:1788–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossa K, Mmadi MA, Zhou R, Zhang T, Su R, Zhang Y, Wang L, You J, Zhang X. 2019b. Depicting the core transcriptome modulating multiple abiotic stresses responses in sesame (Sesamum indicum L.). International Journal of Molecular Sciences 20:3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. 2010. MYB transcription factors in Arabidopsis. Trends in Plant Science 15:573–581. [DOI] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2003. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. The Plant Journal: for Cell and Molecular Biology 33:751–763. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of Molecular Biology 300:1005–1016. [DOI] [PubMed] [Google Scholar]
- Fang Q, Jiang T, Xu L, Liu H, Mao H, Wang X, Jiao B, Duan Y, Wang Q, Dong Q, Yang L, Tian G, Zhang C, Zhou Y, Liu X, Wang H, Fan D, Wang B, Luo K. 2017. A salt-stress-regulator from the Poplar R2R3 MYB family integrates the regulation of lateral root emergence and ABA signaling to mediate salt stress tolerance in Arabidopsis. Plant Physiology and Biochemistry: PPB 114:100–110. [DOI] [PubMed] [Google Scholar]
- Farooq M, Wahid A, Lee DJ, Ito O, Siddique KHM. 2009. Advances in drought resistance of rice. Critical Reviews in Plant Sciences 28:199–217. [Google Scholar]
- Gao H, Song A, Zhu X, Chen F, Jiang J, Chen Y, Sun Y, Shan H, Gu C, Li P, Chen S. 2012. The heterologous expression in Arabidopsis of a chrysanthemum Cys2/His2 zinc finger protein gene confers salinity and drought tolerance. Planta 235:979–993. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Higo H. 1998. PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Research 26:358–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. 2007. WoLF PSORT: protein localization predictor. Nucleic Acids Research 35:W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab G, Ton J, Flors V, Zimmerli L, Métraux JP, Mauch-Mani B. 2005. Enhancing Arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiology 139:267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C. 1999. Multifunctionality and diversity within the plant MYB-gene family. Plant Molecular Biology 41:577–585. [DOI] [PubMed] [Google Scholar]
- Jisha V, Dampanaboina L, Vadassery J, Mithöfer A, Kappara S, Ramanan R. 2015. Overexpression of an AP2/ERF type transcription factor OsEREBP1 confers biotic and abiotic stress tolerance in rice. Plos One 10:e0127831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi YD, Cheong JJ. 2008. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiology 146:623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. 2007. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. The Plant Journal: for Cell and Molecular Biology 50:347–363. [DOI] [PubMed] [Google Scholar]
- Kim JH, Nguyen NH, Jeong CY, Nguyen NT, Hong SW, Lee H. 2013. Loss of the R2R3 MYB, AtMyb73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. Journal of Plant Physiology 170:1461–1465. [DOI] [PubMed] [Google Scholar]
- Koca H, Bor M, Ozdemir F, Turkan I. 2007. The effect of salt stress on lipid peroxidation antioxidative enzymes and proline content of sesame cultivars. Environmental and Experimental Botany 60:344–351. [Google Scholar]
- Kodaira KS, Qin F, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. 2011. Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiology 157:742–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalitha S. 2000. Primer premier 5. Biotechnol Softw Internet Rep 1:270–272. [Google Scholar]
- Langham DR. 2007. “ Phenology of sesame.” In: Janick J and Whipkey A eds. Issues in new crops and new uses. Alexandria, VA:ASHS Press, 144–182. [Google Scholar]
- Lee SB, Kim H, Kim RJ, Suh MC. 2014. Overexpression of Arabidopsis MYB96 confers drought resistance in Camelina sativa via cuticular wax accumulation. Plant Cell Reports 33:1535–1546. [DOI] [PubMed] [Google Scholar]
- Li D, Dossa K, Zhang Y, Wei X, Wang L, Zhang Y, Liu A, Zhou R, Zhang X. 2018. GWAS Uncovers uncovers differential genetic bases for drought and salt tolerances in sesame at the germination stage. Genes 9:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu P, Yu J, Wang L, Dossa K, Zhang Y, Zhou R, Wei X, Zhang X. 2017a. Genome-wide analysis of WRKY gene family in the sesame genome and identification of the WRKY genes involved in responses to abiotic stresses. BMC Plant Biology 17:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Xing C, Yao Z, Huang X. 2017b. PbrMYB21, a novel MYB protein of Pyrus betulaefolia, functions in drought tolerance and modulates polyamine levels by regulating arginine decarboxylase gene. Plant Biotechnology Journal 15:1186–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JF, Ding J, Yuan BF, Feng YQ. 2014. Magnetic solid phase extraction coupled with in situ derivatization for the highly sensitive determination of acidic phytohormones in rice leaves by UPLC-MS/MS. The Analyst 139:5605–5613. [DOI] [PubMed] [Google Scholar]
- Liu Y, Ji X, Zheng L, Nie X, Wang Y. 2013. Microarray analysis of transcriptional responses to abscisic acid and salt stress in Arabidopsis thaliana. International Journal of Molecular Sciences 14:9979–9998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif.) 25:402–408. [DOI] [PubMed] [Google Scholar]
- Ma G, Zou Q, Shi X, Tian D, Sheng Q. 2019. Ectopic expression of the AaFUL1 gene identified in Anthurium andraeanum affected floral organ development and seed fertility in tobacco. Gene 696:197–205. [DOI] [PubMed] [Google Scholar]
- Manuka R, Saddhe AA, Kumar K. 2018. Expression of OsWNK9 in Arabidopsis conferred tolerance to salt and drought stress. Plant Science: An International Journal of Experimental Plant Biology 270:58–71. [DOI] [PubMed] [Google Scholar]
- Matus JT, Aquea F, Espinoza C, Vega A, Cavallini E, Dal Santo S, Cañón P, Rodríguez-Hoces de la Guardia A, Serrano J, Tornielli GB, Arce-Johnson P. 2014. Inspection of the grapevine BURP superfamily highlights an expansion of RD22 genes with distinctive expression features in berry development and ABA-mediated stress responses. Plos One 9:e110372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. 2002. Oxidative stress, antioxidants and stress tolerance. Trends in Plant Science 7:405–410. [DOI] [PubMed] [Google Scholar]
- Mmadi MA, Dossa K, Wang L, Zhou R, Wang Y, Cisse N, Sy MO, Zhang X. 2017. Functional characterization of the versatile MYB gene family uncovered their important roles in plant development and responses to drought and waterlogging in sesame. Genes 8:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Msanne J, Lin J, Stone JM, Awada T. 2011. Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234:97–107. [DOI] [PubMed] [Google Scholar]
- Mu RL, Cao YR, Liu YF, Lei G, Zou HF, Liao Y, Wang HW, Zhang WK, Ma B, Du JZ, Yuan M, Zhang JS, Chen SY. 2009. An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Research 19:1291–1304. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2006. Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Molecular Biology 60:51–68. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K. 2009. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiology 149:88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K. 2007. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. The Plant Journal: for Cell and Molecular Biology 51:617–630. [DOI] [PubMed] [Google Scholar]
- Oh JE, Kwon Y, Kim JH, Noh H, Hong SW, Lee H. 2011. A dual role for MYB60 in stomatal regulation and root growth of Arabidopsis thaliana under drought stress. Plant Molecular Biology 77:91–103. [DOI] [PubMed] [Google Scholar]
- Park S, Gilmour SJ, Grumet R, Thomashow MF. 2018. CBF-dependent and CBF-independent regulatory pathways contribute to the differences in freezing tolerance and cold-regulated gene expression of two Arabidopsis ecotypes locally adapted to sites in Sweden and Italy. Plos One 13:e0207723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Lyle C, Jiang GL, Penumala A. 2016. Soybean salt tolerance 1 (GmST1) reduces ROS production, enhances ABA sensitivity, and abiotic stress tolerance in Arabidopsis thaliana. Frontiers in Plant Science 7:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S. 2016. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signaling & Behavior 11:e1117723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch RJ, Cheng SJ, Klaunig JE. 1989. Prevention of cytotoxicity and inhibition of intercellular communication by antioxidant catechins isolated from Chinese green tea. Carcinogenesis 10:1003–1008. [DOI] [PubMed] [Google Scholar]
- Saibo NJ, Lourenço T, Oliveira MM. 2009. Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Annals of Botany 103:609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. 2005. A gene expression map of Arabidopsis thaliana development. Nature Genetics 37:501–506. [DOI] [PubMed] [Google Scholar]
- Seok HY, Woo DH, Nguyen LV, Tran HT, Tarte VN, Mehdi SM, Lee SY, Moon YH. 2017. Arabidopsis AtNAP functions as a negative regulator via repression of AREB1 in salt stress response. Planta 245:329–341. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. 2003. Regulatory network of gene expression in the drought and cold stress responses. Current Opinion in Plant Biology 6:410–417. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. 2007. Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany 58:221–227. [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. 2001. The R2R3-MYB gene family in Arabidopsis thaliana. Current Opinion in Plant Biology 4:447–456. [DOI] [PubMed] [Google Scholar]
- Sun J, Rao Y, Mei L, Yan T, Yan X, Zhou H. 2010. Effects of drought stress on sesame growth and yield characteristics and comprehensive evaluation of drought tolerance. Chinese Journal of Oil Crop Sciences 32:525–533. [Google Scholar]
- Swindell WR, Huebner M, Weber AP. 2007. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2004. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. The Plant Cell 16:2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu M, Wang X, Huang L, Guo R, Zhang H, Cai J, Wang X. 2016. Expression of a grape bZIP transcription factor VqbZIP39 in transgenic Arabidopsis thaliana confers tolerance of multiple abiotic stresses. Plant Cell Tissue and Organ Culture 125:537. [Google Scholar]
- Wang Y, Li T, John SJ, Chen M, Chang J, Yang G, He G. 2018a. A CBL-interacting protein kinase TaCIPK27 confers drought tolerance and exogenous ABA sensitivity in transgenic Arabidopsis. Plant Physiology and Biochemistry: PPB 123:103–113. [DOI] [PubMed] [Google Scholar]
- Wang L, Li D, Zhang Y, Gao Y, Yu J, Wei X, Zhang X. 2016. Tolerant and susceptible sesame genotypes reveal waterlogging stress response patterns. Plos One 11:e0149912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Niu QW, Wu HW, Liu J, Ye J, Yu N, Chua NH. 2015. Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. The Plant Journal: for Cell and Molecular Biology 84:404–416. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang Q, Liu M, Bo C, Wang X, Ma Q, Cheng B, Cai R. 2017. Over-expression of a maize MYB48 gene confers drought tolerance in transgenic Arabidopsis plants. Journal of Plant Biology 60:612–621. [Google Scholar]
- Wang L, Yu J, Li D, Zhang X. 2014. Sinbase: an integrated database to study genomics genetics and comparative genomics in Sesamum indicum. Plant and Cell Physiology 56:e2. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Zhou R, Dossa K, Yu J, Li D, Liu A, Mmadi MA, Zhang X, You J. 2018b. Identification and characterization of the bZIP transcription factor family and its expression in response to abiotic stresses in sesame. Plos One 13:e0200850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhou L, Fu Y, Cheung MY, Wong FL, Phang TH, Sun Z, Lam HM. 2012. Expression of an apoplast-localized BURP-domain protein from soybean (GMRD22) enhances tolerance towards abiotic stress. Plant, Cell & Environment 35:1932–1947. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhu Y, Wang L, Liu X, Liu Y, Phillips J, Deng X. 2009. A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter. Planta 230:1155–1166. [DOI] [PubMed] [Google Scholar]
- Wei W, Qi X, Wang L, Zhang Y, Hua W, Li D, Lv H, Zhang X. 2011. Characterization of the sesame (Sesamum indicum L.) global transcriptome using Illumina paired-end sequencing and development of EST-SSR markers. BMC Genomics 12:451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson S, Davies WJ. 2002. ABA-based chemical signalling: the co-ordination of responses to stress in plants. Plant, Cell & Environment 25:195–210. [DOI] [PubMed] [Google Scholar]
- Xie M, Wu D, Duan G, Wang L, He R, Li X, Tang D, Zhao X, Liu X. 2014. AtWNK9 is regulated by ABA and dehydration and is involved in drought tolerance in Arabidopsis. Plant Physiology and Biochemistry: PPB 77:73–83. [DOI] [PubMed] [Google Scholar]
- Xiong L, Zhu JK. 2002. Molecular and genetic aspects of plant responses to osmotic stress. Plant, Cell & Environment 25:131–139. [DOI] [PubMed] [Google Scholar]
- Yahya A. 2010. Selectivity and partitioning of potassium and sodium in sesame. J Plant Nutr 33:670–683. [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 1993. The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Molecular & General Genetics: MGG 238:17–25. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 2006. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology 57:781–803. [DOI] [PubMed] [Google Scholar]
- Yang A, Dai X, Zhang WH. 2012. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. Journal of Experimental Botany 63:2541–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Cui Y, Wang M, Xia X. 2017. Overexpression of a novel MYB-related transcription factor, OsMYBR1, confers improved drought tolerance and decreased ABA sensitivity in rice. Biochemical and Biophysical Research Communications 490:1355–1361. [DOI] [PubMed] [Google Scholar]
- You J, Wang Y, Zhang Y, Dossa K, Li D, Zhou R, Wang L, Zhang X. 2018. Genome-wide identification and expression analyses of genes involved in raffinose accumulation in sesame. Scientific Reports 8:4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Qi L, Yang J, Wu C, Liu Y, Huang LA. 2015. Scutellaria baicalensis R2R3-MYB gene SbMYB8 regulates flavonoid biosynthesis and improves drought stress tolerance in transgenic tobacco. Plant Cell Tissue and Organ Culture 120:961− 972. [Google Scholar]
- Yunta C, Martínez-Ripoll M, Zhu JK, Albert A. 2011. The structure of Arabidopsis thaliana OST1 provides insights into the kinase regulation mechanism in response to osmotic stress. Journal of Molecular Biology 414:135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li D, Wang Y, Zhou R, Wang L, Zhang Y, Yu J, Gong H, You J, Zhang X. 2018. Genome-wide identification and comprehensive analysis of the NAC transcription factor family in Sesamum indicum. Plos One 13:e0199262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhang X, Guo R, Wang Y, Guo C, Li Z, Chen Z, Gao H, Wan X. 2018. Over-expression of a grape WRKY transcription factor gene VlWRKY48 in Arabidopsis thaliana increases disease resistance and drought stress tolerance. Plant Cell Tissue and Organ Culture 132:359–370. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.