Abstract
Colon cancer (CC), one of the major causes of tumor-associated death, is often presented with a heterogenic pool of cells with unique differentiation patterns. This study explored the functions that LINC00460 displayed in CC by regulating microRNA-433-3p (miR-433-3p) and Annexin A2 (ANXA2). LINC00460 expression was either silenced or overexpressed in HCT-116 and LOVO cells to explore the functional roles of LINC00460 in CC. The relationship between miR-433-3p and LINC00460/ANXA2 was analyzed using dual-luciferase reporter assay, RNA-pull down, and RNA immunoprecipitation (RIP) assays. Cell proliferation, metastasis, invasion, and apoptosis were examined in vitro, and tumorigenicity was evaluated in vivo following LINC00460 silencing. Additionally, the regulatory mechanisms were investigated using LINC00460 and ANXA2 gain- or loss-of-function experiments. We found that LINC00460 was expressed highly in CC. Downregulation of LINC00460 inhibited cell invasion and proliferation in vitro and restrained tumor growth in vivo. Moreover, LINC00460 was able to specifically bind to miR-433-3p to increase the expression of ANXA2. Furthermore, LINC00460 downregulated the E-cadherin expression and upregulated the vimentin and N-cadherin expression by upregulating ANXA2, therefore inducing epithelial-mesenchymal transition. These findings suggested that LINC00460 might function as an oncogenic long non-coding RNA (lncRNA) in CC development and could be explored as a potential biomarker and therapeutic target for CC.
Keywords: long non-coding RNA, LINC00460, competing endogenous RNA, microRNA-433-3p, Annexin A2, colon cancer, cell invasion, cell proliferation, cell metastasis, xenograft tumor in nude mice
Introduction
Colon cancer (CC) is regarded as one of the common tumors across the world, which takes half a million lives every year.1 It has been revealed that more than 90% of the mortality in CC is associated with distant metastasis and invasion and that epithelial-mesenchymal transition (EMT) has a significant impact on primary metastatic development in CC.2 CC progression is a gradual process that evolves from normal cell transformation induced by increasing multiple epigenetic and genetic changes to tumor at initial stage and at last deteriorates into malignancy.3 When it comes to risk factors of CC, smoking, cholecystectomy, hormone treatment, usage of nonsteroidal anti-inflammatory drugs, waist girth, and diabetes were all accountable.4 Although the survival rates of CC patients in stage III can be improved by surgical procedures in combination with adjuvant chemotherapy, the efficacy that current adjuvant treatments have on CC patients in various stages is still unsatisfactory. Moreover, CC patients in stage II frequently suffer from different risks, such as worse differentiation and T4 tumors.5 Increasing evidence has proved that long non-coding RNAs (lncRNAs) represent a novel therapeutic direction for tumor treatment.6 Based on this, we decided that the influence of lncRNAs on CC formation and progression needed to be explored.
lncRNAs, a class of non-protein coding transcripts, are associated with the progression of multiple cancers, including CC.7 lncRNAs are involved in tumor formation, proliferation, invasion, and migration, as well as apoptosis through the regulation of diverse signaling pathways.8 For instance, LINC00460 has been found to correlate to nasopharyngeal carcinoma progression by regulation of the miR-149-5p/IL-6 (interleukin-6) signaling pathway.9 Moreover, a previous study reported that LINC00460 is overexpressed in colorectal cancer.10 In a biological prediction website (https://cm.jefferson.edu/rna22/Interactive/), LINC00460 has been predicted to bind to microRNA-433-3p (miR-433-3p). miR-433-3p has been proved to correlate to tumor formation and progression, contributing to restrained cell proliferation and promoting chemosensitivity through regulation of the cyclic AMP (cAMP) response element-binding protein in human glioma.11 Furthermore, an online website (https://cm.jefferson.edu/rna22/Interactive/) has predicted binding sites between miR-433-3p and the 3′ UTR of Annexin A2 (ANXA2). Moreover, ANXA2 is associated with tumor cell adhesion, invasion, and metastasis, indicating that ANXA2 could function as a diagnostic marker for CC.12 Moreover, co-expression of ANXA2, SOD2, and HOXA13 results in poor therapeutic efficacy for patients with esophageal squamous cell cancer.13 Taken together, we hypothesized a close relationship between LINC00460, miR-433-3p, and ANXA2 during the development of CC. Therefore, our study aimed to investigate the mechanisms of LINC00460 in EMT of CC cells via the mediation of ANXA2 by binding to miR-433-3p.
Results
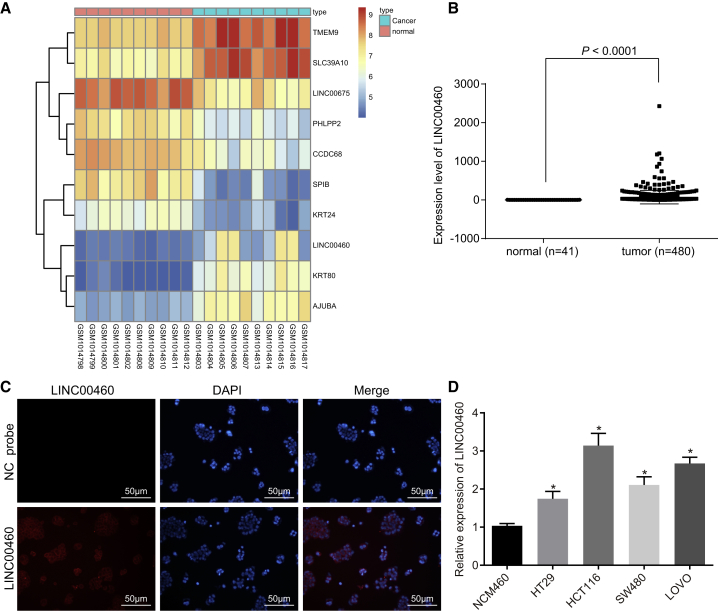
LINC00460 Is Highly Expressed in CC
Bioinformatics analysis was conducted to study the LINC00460 expression in CC. Data analysis of the GSE41328 dataset revealed that LINC00460 was highly expressed in CC (Figure 1A). It was further shown by The Cancer Genome Atlas (TCGA) database that LINC00460 was expressed higher in CC tissues than in normal tissues (Figure 1B). The results from RNA-fluorescence in situ hybridization (FISH) detection revealed that LINC00460 was mainly located in the cytoplasm (Figure 1C). The expression of LINC00460 in four cell lines was determined by qRT-PCR and the results (Figure 1D) showed that compared with human normal colon epithelial cells, the expression of LINC00460 in four CC cell lines was significantly increased. Therefore, we performed loss- and gain-of-function with HCT-116 and LOVO cell lines to elucidate the biological functions of LINC00460 in CC cells.
Figure 1.
LINC00460 Is Highly Expressed in CC Cells
(A) The heatmap of CC microarray GEO: GSE41328. (B) LINC00460 expression in CC and normal tissues in TCGA database. (C) Location of LINC00460 detected by RNA-FISH assay; gray represents LINC00460 expression, and red represents nuclei stained by DAPI (×200). (D) The expression of LINC00460 in four CC cell lines and normal cells (the comparison among multiple groups was analyzed by one-way analysis of variance with Tukey’s post hoc test); *p < 0.05 versus human normal colon mucosal epithelial cell NCM-460. NC, negative control; miR-433-3p, microRNA-433-3p; ceRNA, competing endogenous RNA; CC, colon cancer; RNA-FISH, RNA-fluorescence in situ hybridization; lncRNA, long non-coding RNA; TCGA, The Cancer Genome Atlas; DAPI, 4’,6-diamidino-2-phenylindole.
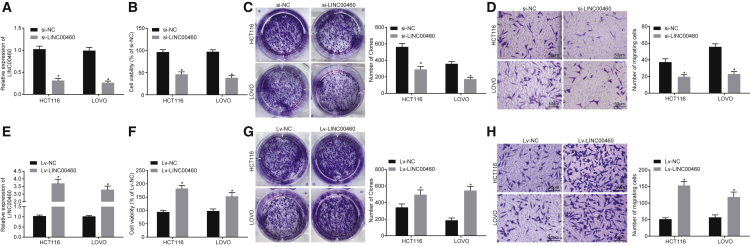
Knock Down of LINC00460 Suppresses the Proliferation and Invasion of HCT-116 Cells
Whether LINC00460 affects the viability and invasion of CC cells was determined through loss-of-function experiments. The knockdown efficiency of LINC00460 was verified by qRT-PCR. HCT-116 and LOVO cells transfected with small interfering RNA (siRNA) targeting LINC00460 (si-LINC00460) showed a significant decrease in the expression of LINC00460 (Figure 2A). Cell Counting Kit-8 (CCK-8), colony formation, and Transwell assays were used to evaluate cell proliferation, colony formation, and invasion capabilities of CC cells. The results showed that knock down of LINC00460 inhibited cell proliferation and colony formation (Figures 2B and 2C), while impairing the cell invasion ability (Figure 2D). In addition, HCT-116 and LOVO cells were transfected with the LINC00460 overexpression plasmids, and as a result, the expression of LINC00460 was significantly increased (Figure 2E). The proliferation, colony formation, and invasion abilities of HCT-116 and LOVO cells were enhanced following LINC00460 overexpression (Figures 2F–2H). Overall, the results demonstrated that the silencing of LINC00460 can inhibit the proliferation, colony formation, and invasion abilities of HCT-116 and LOVO cells.
Figure 2.
LINC00460 Promotes the Proliferation and Invasion Abilities of CC Cells In Vitro (×200)
(A) The transfection efficiency of si-LINC00460 in HCT-116 and LOVO cells was determined by qRT-PCR. (B) The viability of HCT-116 and LOVO cells after transfection of si-LINC00460 or si-NC was measured by CCK-8. (C) Colony formation analysis of HCT-116 and LOVO cells after transfection with si-LINC00460 or si-NC. (D) The invasion ability of HCT-116 and LOVO cells after transfection with si-LINC00460 or si-NC was evaluated by Transwell assay (×200). (E) The transfection efficiency of LINC00460 overexpression plasmid in HCT-116 and LOVO cells determined by qRT-PCR. (F) The viability of HCT-116 and LOVO cells after LINC00460 overexpression assessed by CCK-8. (G) Colony formation of HCT-116 and LOVO cells after LINC00460 overexpression. (H) Invasion ability of HV-116 and LOVO cells after LINC00460 overexpression assessed by Transwell assay (×200). The independent parallel experiments were repeated three times; **p < 0.01 versus the si-NC or Lv-NC groups; comparisons between two groups were analyzed using two-tailed independent t test. CC, colon cancer; NC, negative control; CCK-8, cell counting kit-8; miR-433-3p, microRNA-433-3p.
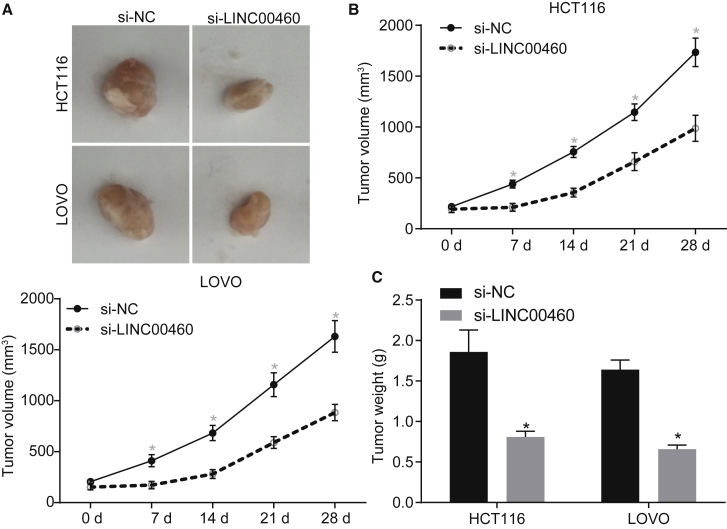
Downregulation of LINC00460 Restrains Carcinogenicity of HCT-116 Cells In Vivo
To evaluate the effects of LINC00460 on tumor growth in nude mice, we subcutaneously injected HCT-116 and LOVO cells transfected with si-NC or si-LINC00460 into nude mice. The results from the tumor xenografts in nude mice showed that the growth rate of subcutaneous tumors was significantly slowed down by LINC00460 knockdown (Figure 3A) and that the volume and weight of subcutaneous tumors of the nude mice injected with si-LINC00460-transfected cells were both reduced compared to the nude mice injected with si-NC-transfected cells (p < 0.05) (Figures 3B and 3C). Overall, the functional data demonstrated that silencing of LINC00460 blocked tumor growth in CC cells.
Figure 3.
LINC00460 Promotes the Oncogenicity of HCT-116 and LOVO Cells
(A) Representative images of subcutaneous tumors in nude mice after silencing of LINC00460. (B) Tumor growth curve after silencing of LINC00460. (C) Tumor weight in nude mice after silencing of LINC00460. **p < 0.05 versus the si-NC group; tumor volume at different time points were compared using repeated-measures ANOVA; comparison of tumor weight between two groups was analyzed by two-tailed independent t test. NC, negative control.
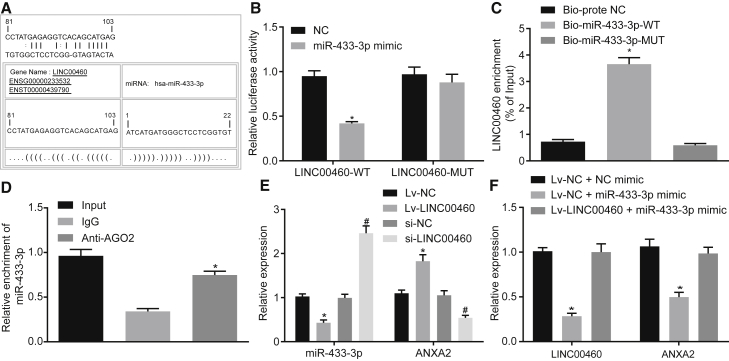
LINC00460 Binds to miR-433-3p to Upregulate the Expression of ANXA2 in CC Cells
The LINC00460-miRNA interaction was analyzed using bioinformatics method. An online analysis software revealed the presence of specific binding regions between the LINC00460 gene sequence and the miR-433-3p sequence (Figure 4A). The dual-luciferase reporter assay was performed to verify the relationship between LINC00460 and miR-433-3p. The results showed that the luciferase activity of the cells co-transfected with wild-type (WT)-LINC00460/miR-433-3p mimic was reduced compared to that of the cells co-transfected with WT-LINC00460/NC (p < 0.05); however, the luciferase activity of the cells co-transfected with mutant (MUT)-LINC00460/miR-433-3p mimic was not significantly different (p > 0.05), indicating that LINC00460 could specifically bind to the miR-433-3p (Figure 4B). Furthermore, both Bio-miR-433-3p-WT and LINC00460 were enriched in the pull-down fraction of cells using the RNA pull-down assay (p < 0.05); however, there was no significant difference when the cells were transfected with Bio-miR-433-3p-MUT (p > 0.05) (Figure 4C).
Figure 4.
LINC00460 Binds to miR-433-3p to Increase the Expression of ANXA2
(A) The predicted binding sites of miR-433-3p on LINC00460. (B) The luciferase activity detection of LINC00460-WT/LINC00460-MUT after miR-433-3p mimic transfection. (C) The enrichment of LINC00460 detected by RNA pull-down assay. (D) The enrichment of LINC00460 in HCT-116 cells and miR-433-3p in IP complexes detected using anti-AGO2 RIP method, *p < 0.01 versus anti-IgG. (E) miR-433-3p and ANXA2 expression after LINC00460 silencing or overexpression determined by qRT-PCR. (F) LINC00460 and ANXA2 expression after mimic alone or with LINC00460 overexpression determined by qRT-PCR; *p < 0.05 versus the NC, Bio-probe-NC, control, empty vector, or Lv-NC + NC mimic groups; # p < 0.05 versus the control, si-NC, or Lv-NC + miR-433-3p mimic groups. Comparison between two groups was analyzed using two-tailed independent t test; the comparison among multiple groups was analyzed by one-way analysis of variance with Tukey’s post hoc test. miR-433-3p, microRNA-433-3p; ANXA2, Annexin A2; UTR, untranslated region; RIP, RNA immunoprecipitation; miR-433-3p, microRNA-433-3p; ANXA2, annexin A2; NC, negative control.
The RNA-induced silencing complex is formed by a miRNA ribonucleic acid-protein complex present in the anti-AGO2 co-immunoprecipitation system. Therefore, the anti-AGO2 co-immunoprecipitation system contains miRNA and its interacting RNA components. RNA immunoprecipitation (RIP) experiments were performed using anti-AGO2 in HCT-116 cell lysate, and it was shown that LINC00460 and miR-433-3p were preferentially enriched in AGO2-containing miRNA ribonucleoprotein complexes (miRNPs) compared to anti-immunoglobulin G (IgG) immunoprecipitation system (Figure 4D).
The results from qRT-PCR further showed that in the cells transfected with the LINC00460 overexpression plasmid, the expression of miR-433-3p was decreased and the expression of ANXA2 mRNA was increased (p < 0.05). Compared with the cells transfected with si-NC, the expression of miR-433-3p was increased in the cells transfected with si-LINC00460, and the expression of ANXA2 mRNA was decreased (p < 0.05; Figure 4E). LINC00460 and ANXA2 mRNA expression was significantly decreased in cells after transfection with the miR-433-3p mimic, while the LINC00460 overexpression showed the opposite effects (Figure 4F). It was suggested that the upregulation of LINC00460 inhibited the expression of miR-433-3p and increased the expression of ANXA2.
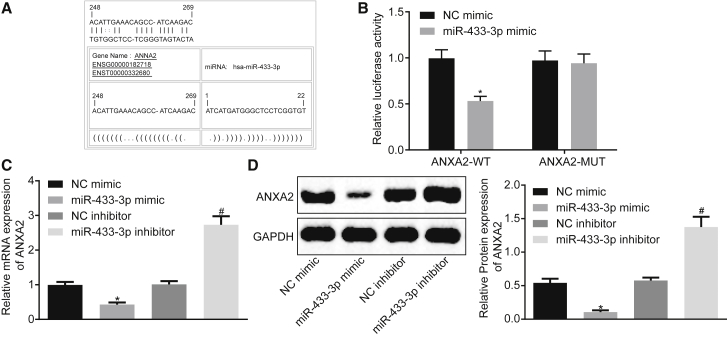
miR-433-3p Targets ANXA2
By using online software analysis, the relationship between ANXA2 and miR-433-3p was predicted. The results revealed the presence of a specific binding region between the ANXA2 and the miR-433-3p sequences, suggesting that ANXA2 was likely to be targeted by miR-433-3p (Figure 5A). The dual-luciferase reporter assay verified that ANXA2 was a target of miR-433-3p (Figure 5B). The luciferase activity of ANXA2-WT in the cells transfected with miR-433-3p mimic was decreased (p < 0.05), while the luciferase activity of the ANXA2-MUT was not significantly different (p > 0.05), suggesting that miR-433-3p could specifically bind to ANXA2. The effects of miR-433-3p on the expression of ANXA2 were determined by qRT-PCR and western blot assays. The results showed that the overexpression of miR-433-3p significantly inhibited the expression of ANXA2, and the inhibition of miR-433-3p significantly upregulated the expression of ANXA2 (Figures 5C and 5D). The above results suggested that miR-433-3p directly interacted with ANXA2 in CC cells.
Figure 5.
miR-433-3p Targets ANXA2
(A) Predicted binding sites of miR-433-3p on ANXA2-3′ UTR. (B) Luciferase activity detection of ANXA2-WT/ANXA2-MUT after miR-433-3p mimic transfection. (C) The mRNA expression of ANXA2 after miR-433-3p mimic or miR-433-3p inhibitor transfection determined by qRT-PCR. (D) The protein expression of ANXA2 after miR-433-3p mimic or inhibitor transfection determined by western blot assay. *p < 0.05 versus the NC mimic group; # p < 0.05 versus the NC inhibitor group. Comparisons between two groups were analyzed using two-tailed independent t test; the comparison among multiple groups was analyzed by one-way analysis of variance with Tukey’s post hoc test. miR-433-3p, microRNA-433-3p; ANXA2, Annexin A2; UTR, untranslated region; NC, negative control.
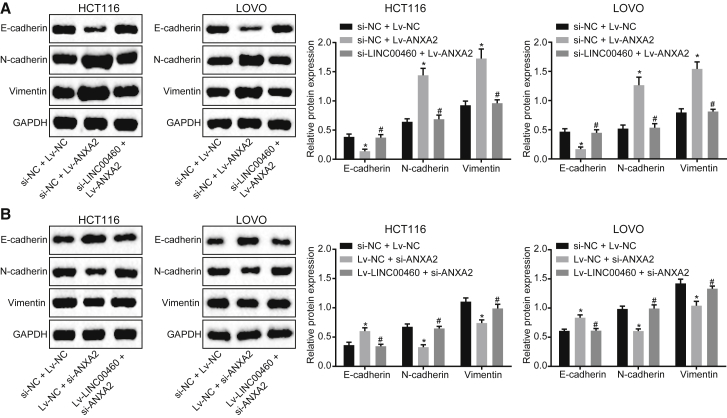
The LINC00460/miR-433-3p/ANXA2 Axis Regulates EMT in CC
EMT is one of the main reasons for the enhanced migration and invasion ability of epithelial-derived malignant cells.14 In order to evaluate the effects of LINC00460 on EMT, the expression of the epithelial markers E-cadherin and N-cadherin, as well as the interstitial marker vimentin, was measured by western blot assay. As shown in the results, knock down of LINC00460 reversed the inhibitory effect of ANXA2 on the expression of E-cadherin and the promoting effect on that of N-cadherin and vimentin (Figure 6A). Accordingly, overexpression of LINC00460 abolished the promotion of E-cadherin and the inhibition of N-cadherin and vimentin induced by silencing of ANXA2 (Figure 6B). The results suggested that the LINC00460/miR-433-3p/ANXA2 axis mediated the development and EMT of CC.
Figure 6.
LINC00460 Induces the EMT in HCT-116 and LOVO Cells by Increasing ANXA2
(A) The protein expression of N-cadherin, E-cadherin, vimentin in HCT-116, and LOVO cells after ANXA2 overexpression alone or with LINC00460 silencing, as measured by western blot assay. (B) The protein expression of N-cadherin, E-cadherin, vimentin in HCT-116, and LOVO cells in response to ANXA2 silencing alone or with LINC00460 overexpression, measured by western blot assay; **p < 0.01. The comparisons between multiple groups were performed using one-way analysis of variance with Tukey’s post hoc test. The experiments were performed in triplicate. miR-433-3p, microRNA-433-3p; ANXA2, Annexin A2; NC, negative control.
Discussion
CC, the third most frequent tumor worldwide, is characterized by distant metastasis and invasion, causing one-tenth of all deaths induced by tumors in humans.2 Although some CC screening tests, such as colonoscopy and fecal occult-blood determination, have been used for prognostic diagnosis, the efficacy remains unsatisfactory and CC patients still face relatively higher mortality.12 In a former study, lncRNAs have been observed to correlate to various types of tumor progression, including CC.8 In this study, the effects of LINC00460 on cell proliferation, invasion, and EMT were explored. The data obtained from our study showed that downregulated LINC00460 repressed cell proliferation, invasion, and tumor xenografts of CC. More importantly, we have proved that LINC00460 played a stimulative role in CC by downregulating miR-433-3p and upregulating the expression of ANXA2.
Initially, we demonstrated that LINC00460 was highly expressed and miR-433-3p was downregulated in CC. A recent study indicated that LINC00460 showed significantly high expression in non-small lung cancer (NSCLC), inducing an unfavorable prognosis for NSCLC patients.10 Similarly, Li et al.10 revealed an upregulation of LINC00460 in colorectal cancer. Moreover, based on the target prediction websites, miR-433-3p was found to interact with LINC00460 in our results. Previously, miR-433-3p was demonstrated to exhibit decreased expression levels in glioma cell lines and tissues.11 Furthermore, we also found that LINC00460 could function as a competing endogenous RNA (ceRNA) to suppress miR-433-3p and enhance ANXA2. ceRNAs, one of the most recently discovered classes of RNAs, play important roles in gene modulation regulated by miRNAs and share miRNA recognition elements, thus achieving an effect of mutual regulation.15 ANXA2 is a tumor-induced molecule and its overexpression has been linked to colorectal cancer.16 Wu et al.17 have also reported that ANXA2 expression is increased in metastatic cancers and correlated with the phenotype of metastasis and drug resistance. Moreover, ANXA2 has been demonstrated to be negatively targeted by miR-101.18 In the present study, ANXA2 was identified to be a putative target gene of miR-433-3p, matching with the evidence obtained from the online analysis. A recent study has suggested that the lncRNA-MUF can bind ANXA2 and act as a ceRNA to sponge miR-34a, while its silencing contributed to the inhibition of EMT and tumor cell formation in hepatocellular carcinoma.19 Another research group indicated that LINC00460 sponged miR-149-5p to promote nasopharyngeal carcinoma tumorigenesis.9 Therefore, the findings mentioned above are in accordance with our result showing that the suppressed LINC00460 functions as ceRNA to promote miR-433-3p and repress ANXA2.
Furthermore, we have found that LINC00460 silencing and miR-433-3p overexpression could suppress CC cell proliferation and invasion together with EMT, which agrees with the results obtained from other studies. Shi et al.20 indicated that the upregulation of miR-433-3p contributed to the suppression of esophageal squamous cell carcinoma cell proliferation, migration, and invasion. Also, a previous study reported similar findings, showing that downregulation of LINC00460 inhibited cell proliferation and invasion, as well as migration in NSCLC cells.21 Moreover, enhanced ANXA2 expression was linked to lymph node metastasis, distal metastasis, advanced tumor-node-metastasis stage, and E-cadherin expression in patients with gastric adenocarcinoma.22 Specifically, repression of miR-206 expression in prostate cancer cells upregulated the protein expression of ANXA2 to regulate E-cadherin, N-cadherin and vimentin expression and promoting cell invasion in vitro.23 Consistently, we monitored a positive correlation between ANXA2 and E-cadherin expression and an inverse correlation between ANXA2 and N-cadherin and vimentin expression. Furthermore, our rescue experiments validated our findings, showing that LINC00460 could reverse the effects of si-ANXA2 on these EMT markers.
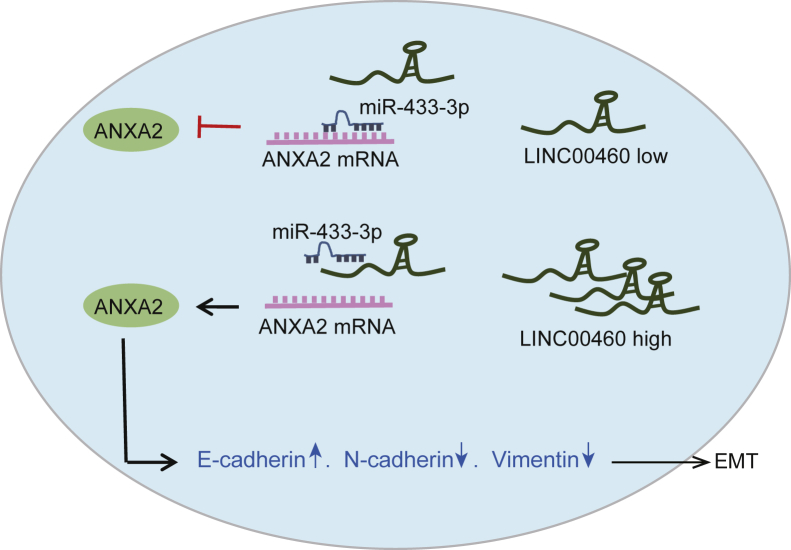
Taken together, the results obtained from the present study allowed us to draw the conclusion that LINC00460 silencing suppressed the proliferation, invasion, and tumorigenesis in CC cells by downregulating ANXA2 via miR-433-3p (Figure 7). LINC00460 showed high expression levels in CC cells and functioned as a ceRNA to sponge miR-433-3p to upregulate ANXA2. These findings elucidated the molecular mechanism involving LINC00460 in CC tumorigenesis. However, due to the limited sample size and experimental conditions, further studies are still required to define the detailed mechanisms by which LINC00460 and miR-433-3p interact and influence CC cells. Additionally, since LINC00460 and miR-433-3p may have multiple target genes,24, 25, 26 we only probed the regulation of LINC00460C/miR-433-3p/ANXA2 axis in CC, and other possible regulatory pathways need to be explored.
Figure 7.
A Schematic Diagram of LINC00460 Implicated in the Progression of CC
LINC00460 serves as a ceRNA of miR-433-3p to increase the expression of ANXA2, thus contributing to the induction of EMT in CC. miR-433-3p, microRNA-433-3p; ANXA2, Annexin A2; EMT, epithelial-mesenchymal transition.
Materials and Methods
Ethics Statement
All of the experimental animals for medical research were used following the animal care guidelines from the Animal Experiment Committee in Taizhou First People’s Hospital. The animal utilization protocols were approved by the Animal Experiment Committee.
Bioinformatics Analysis
The RNA sequencing expression data (GEO: GSE41328) and the annotation probe files were downloaded from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) which analyzed 5 CC tumor samples and 5 normal samples (control).27 The Affy package for the R programming language was used for background correction and normalization.28 Then, the linear model-Empirical Bayes method in the Limma installation package with the traditional t test was used to non-specifically filter the expression spectral data to select the differentially expressed lncRNAs.29 At the same time, TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) database was used to obtain the gene expression data from CC samples. The R programing language was used for statistical analysis. Differential analysis was performed for the transcriptome profiling data with the package edgeR for R.30 False-positive discovery (FDR) correction was applied on p value with package multitest. Lastly, the differentially expressed genes (DEGs) were screened out with the screening threshold of FDR < 0.05 and |log2 (fold change)| > 2.
Cell Culture
Human CC cell lines HT-29 (from a 44-year-old Caucasian woman, negative for CD4, but with the existence of galactose ceramide), HCT-116 (from a male patient and has a mutation in codon 13 of the ras proto-oncogene), SW480 (from a 50-year-old Caucasian male; positive for c-myc, K-ras, H-ras, N-ras, myb, sis, and fos), LOVO (initiated in 1971 from a fragment of a metastatic tumor nodule in the left supraclavicular region of a 56-year-old Caucasian male), and human normal colon mucosal epithelial cell NCM-460 were obtained from the Shanghai Institute for Cell Biology (Chinese Academy of Sciences, Shanghai, China).30,31 Human CC cell lines were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (GIBCO Company, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS; Kete Biotech, Yancheng, Jiangsu, China) in an incubator with 5% CO2 at 37°C.
Cell Grouping and Transfection
According to the known LINC00460 and miR-433-3p sequences from the National Center for Biotechnology Information, the empty vector plasmid, LINC00460 overexpression plasmid, LINC00460 siRNA (si-LINC00460) negative control (NC) plasmid, si-LINC00460 plasmid, miR-433-3p mimic NC plasmid, miR-433-3p mimic plasmid, miR-433-3p inhibitor NC plasmid, and miR-433-3p inhibitor plasmids were constructed by Shanghai Sangon Biological Engineering Technology & Services (Shanghai, China).
The cells at passage three were treated with trypsin and seeded into a 24-well plate to grow into a monolayer. Afterward, the cells were transfected with the empty vector (Lv-NC), LINC00460 overexpression plasmid, NC siRNA (si-NC), or si-LINC00460.
24 h before transfection, the cells were plated into a 6-well plate. When cells grew to be 30%∼50% confluent, 50 nM siRNAs or miRNAs were transfected into cells using Lipofectamine 2000 (11668-019, Invitrogen, New York, CA, USA) following the manufacturer’s instructions. Afterward, the cells were incubated for 6∼8 h under the condition of 37°C with 5% CO2 and then further cultured in the complete medium for 24∼48 h for subsequent experiments.
Dual-Luciferase Reporter Assay
The targeting relationship between LINC00460 and miR-433-3p, as well as ANXA2 and miR-433-3p, was analyzed online using a biological prediction website (https://cm.jefferson.edu/rna22/Interactive/) and verified by the dual-luciferase reporter assay. The target and mutation sequences were designed and chemically synthesized based on the predicted binding sequences of the LINC00460 WT and the ANXA2 3′ UTR with miR-433-3p. The XhoI and NotI restriction sites were used to flank the target sequence, which was cloned into the PUC57 vector first and then sub-cloned into the psiCHECK-2 vector. Cells at a density of 2 × 105 cells/well were transfected with the luciferase reporter and/or miR-433-3p. The cells were harvested 48 h after transfection, and the luciferase activities were determined using the Genecopoeia’s dual-luciferase detection kit (D0010, Beijing Solarbio Life Sciences, Beijing, China) on a Promega’s Glomax20/20 luminometer (E5311, Zhongmei Biotech, Xi’an Shaanxi, China).
lncRNA Subcellular Location and RNA-FISH Assay
The lncRNA subcellular website (http://lncatlas.crg.eu/) was used to predict the localization of LINC00460 in HCT-116 cells. The subcellular localization of LINC00460 was verified by a FISH Kit (Hoffmann-La Roche, Basel, Switzerland). After transfection, the HCT-116 cells from each group were washed two times with cold phosphate-buffered solution (PBS) and fixed with 4% paraformaldehyde. The hybrid solution containing digoxigenin-labeled LINC00460 probes (Sigma, St. Louis, MO, USA) was added into the cell culture plate with the antagonistic LINC00460 probe as NC. The nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI) (Sigma, St. Louis, MO, USA) for 10 min at room temperature. The fluorescent images were visualized and recorded under a confocal laser scan microscope (FV1000, Olympus, Tokyo, Japan).
RIP
The binding of LINC00460 to the AGO2 protein was detected using a RIP kit (Merck Millipore, Billerica, MA, USA). After cell lysis with the RIP lysis buffer (P0013B, Beyotime, Shanghai, China), a portion of the lysate was incubated with the RIP buffer containing 50 μL of magnetic beads, which were conjugated with 5 μg of anti-AGO2 (ab32381, 1:50, Abcam, Cambridge, UK) and IgG (ab109489, 1:100, Abcam, Cambridge, UK). Among the antibodies, IgG was considered as the NC. Proteinase K buffer was then added to the samples. Finally, the target RNA was extracted and purified for further study by qRT-PCR.
RNA Pull-Down
HCT-116 cells were transfected with 50 nM WT biotinylated miR-433-3p and 50 nM MUT biotinylated miR-433-3p. After 48 h of transfection, the cells were incubated with the specific cell lysis buffer (Ambion, Austin, TX, USA) for 10 min. A total of 50 mL of cell lysate was collected for later use. The remaining volume was incubated with the M-280 streptomycin and magnetic beads (Sigma, St. Louis, MO, USA) pre-coated with RNase-free and yeast tRNA (Sigma, St. Louis, MO, USA) at 4°C for 3 h. Subsequently, the sample was washed two times with cold lysate, three times with low salt buffer, and one time with high salt buffer. The antagonistic miR-433-3p probe was used as NC. The total RNA was extracted with Trizol, and the LINC00460 expression was detected by qRT-PCR.
qRT-PCR
The total RNA of CC cells was extracted using a Trizol extraction kit. The LINC00460, miR-433-3p, and ANXA2 primers (Table 1) were designed and synthesized by Invitrogen (Carlsbad, CA, USA). An Open Array MicroRNA Real-time PCR Master Mix kit was used for the qRT-PCR assay on an ABI7300 system (ABI Company, Oyster Bay, NY, USA). The level of miR-433-3p was normalized to the housekeeping gene U6, while the expression of ANXA2 and LINC00460 was normalized to housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The 2-ΔΔCT method was used to calculate the relative mRNA transcription level.
Table 1.
Gene Primers for qRT-PCR
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| LINC00460 | 5′-GCATGCACACTTCTCGGCTA-3′ | 5′-GAATGCGTCTTCTTTCCCACG-3′ |
| miR-433-3p | 5′-GGAGAAGTACGGTGAGCCTGT-3′ | 5′-GAACACCGAGGAGCCCATCAT-3′ |
| ANXA2 | 5′-CACGGCCCAGGTTATCTTGT-3′ | 5′-TGCTGCGGTTGGTCAAAATG-3′ |
| U6 | 5′-CATGCTTGTAGCTGCCCCAT-3′ | 5′-GAGAGTACTGGGTGTCCGTTT-3′ |
| GAPDH | 5′-TATCATGGAATCCACCGGTGTCC-3′ | 5′-TGACCTTGCCCACAACCTTA-3′ |
miR-433-3p, microRNA-433-3p; ANXA2, Annexin A2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Western Blot Analysis
The total protein was extracted using a RIP Assay Kit (R0010, Beijing Solarbio Life Sciences, Beijing, China). The protein concentration was measured using a bicinchoninic acid Kit (G3522-1, GBCBIO Technologies, Guangzhou, Guangdong, China). Based on the different concentrations, the proteins were quantified, separated by polyacrylamide gel electrophoresis, transferred onto a nitrocellulose membrane using a wet transfer method, and blocked with 5% bovine serum albumin (BSA) at room temperature for 1 h. Then diluted primary antibodies rabbit polyclonal antibodies to vimentin (1:1,000, Abcam, Cambridge, UK), N-cadherin (1:1,000, Abcam, Cambridge, UK), and E-cadherin (1:1,000, ab76055, Abcam, Cambridge, UK) were added for incubation overnight at 4°C. After that, the membrane was incubated with horseradish peroxidase-labeled goat-anti-rabbit IgG antibody (1:5,000, Beijing Zhongshan Biotechnology, Beijing, China), and reacted with enhanced chemiluminescence solution (ECL808-25, Biomiga, San Diego, USA) at the room temperature for 1 min. The anti-GAPDH (1:1,000, ab8245, Abcam, Cambridge, UK) antibody served as the internal reference. The relative levels of proteins were calculated by the ratio of the gray value between the target band and the internal reference band.
Cell Viability Assessment
After transfection for 48 h, the cells in the logarithmic growth phase were dispersed into a cell suspension at a density of 2 × 104 cells/mL in Dulbecco’s modified eagle’s medium containing 10% FBS. The cells were seeded into 96-well culture plates with 100 μL per well and 8 parallel wells set. Following incubation at 37°C with 5% CO2 for 24 h, 10 μL of CCK-8 (Sigma-Aldrich, St. Louis, MO, USA) reagent was added and incubated for 2 h. The optical density (OD) of each well was recorded at 570 nm using a microplate reader (NYW-96M, Beijing NuoYawei Instrument, Beijing, China). The blank transfection was set as normalization 100% survival.
Clone Formation Assay
After detachment with 0.25% trypsin, the cells were seeded into a 6-well plate at a density of 500 cells/well and incubated at 37°C with 5% CO2 for 12 days to form cell colonies. The colonies with more than 50 cells were counted under the microscope.
Transwell Assay
After 48 h of transfection, the cells were starved in serum-free medium for 24 h and resuspended with serum-free Opti-MEMI medium (31985008, Nanjing SenBeiJia Biological Technology, Nanjing, Jiangsu, China) containing 10 g/L of BSA to adjust the density to 3 × 104 cells/mL. A total of 1 × 105 cells in 200 μL of RPMI 1640 were added into the apical chamber coated with diluted Matrigel (1:8, 40111ES08, Shanghai Yeasen Biological Technology, Shanghai, China), while 600 μL of RPMI 1640 medium containing 20% FBS was added into the basolateral chamber. After 24 h of routine culture, the invaded cells were fixed with 4% polyformaldehyde for 15 min and stained by 0.5% crystal violet solution (prepared by methanol) for 15 min. After the cells on the upper surface of the filter were removed, five randomly selected microscopic fields of fixed cells per filter (×200) were imaged under an inverted microscope (XDS-800D, Caikon Optical Instrument, Shanghai, China). The number of cells across the membrane was counted.
Tumor Xenograft in Nude Mice
HCT-116 and LOVO cells were resuspended with serum-free RPMI 1640 medium (GIBCO Company, Grand Island, NY, USA) and the cell density was adjusted to 1 × 107 cells/mL. A total of 48, 4-week-old, specific-pathogen-free male BALB/c nude mice (weight 16–18 g; SLAC Laboratory Animal, Shanghai, China) were selected. The HCT-116 and LOVO cells transfected with si-NC or si-LINC00460 were injected subcutaneously into BALB/c nude mice. The nude mice were routinely sterilized after the ether anesthetization and later intraperitoneally and subcutaneously injected with 100 μL of tumor cell suspension. The mice were euthanized 4 weeks later and the tumor xenograft and lymph node metastatic tumors from the abdominal cavity were surgically resected.
Statistical Analysis
The SPSS 21 statistical software (IBM, Armonk, NY, USA) was used to analyze all data. The data were presented as mean ± standard deviation. The comparisons between two groups were analyzed using two-tailed independent t test. The comparisons among multiple groups were analyzed using one-way analysis of variance (ANOVA) with Tukey’s post hoc test. The data at different time points were compared using repeated-measures ANOVA. A p < 0.05 indicates a statistically significant difference.
Author Contributions
W.H. and H.Y. designed the study. F.L. and W.W. collated the data, carried out data analyses, and produced the initial draft of the manuscript. M.Z. contributed to drafting the manuscript. R.D. revised it critically for important intellectual content. All authors have read and approved the final submitted manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
There are no acknowledgments.
References
- 1.Wu G., Zhu Y.Z., Zhang J.C. Sox4 up-regulates Cyr61 expression in colon cancer cells. Cell. Physiol. Biochem. 2014;34:405–412. doi: 10.1159/000363009. [DOI] [PubMed] [Google Scholar]
- 2.Liu C.C., Cai D.L., Sun F., Wu Z.H., Yue B., Zhao S.L., Wu X.S., Zhang M., Zhu X.W., Peng Z.H., Yan D.W. FERMT1 mediates epithelial-mesenchymal transition to promote colon cancer metastasis via modulation of β-catenin transcriptional activity. Oncogene. 2017;36:1779–1792. doi: 10.1038/onc.2016.339. [DOI] [PubMed] [Google Scholar]
- 3.Oberg A.L., French A.J., Sarver A.L., Subramanian S., Morlan B.W., Riska S.M., Borralho P.M., Cunningham J.M., Boardman L.A., Wang L. miRNA expression in colon polyps provides evidence for a multihit model of colon cancer. PLoS ONE. 2011;6:e20465. doi: 10.1371/journal.pone.0020465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartz A., He T., Ross J.J. Risk factors for colon cancer in 150,912 postmenopausal women. Cancer Causes Control. 2012;23:1599–1605. doi: 10.1007/s10552-012-0037-4. [DOI] [PubMed] [Google Scholar]
- 5.Oue N., Anami K., Schetter A.J., Moehler M., Okayama H., Khan M.A., Bowman E.D., Mueller A., Schad A., Shimomura M. High miR-21 expression from FFPE tissues is associated with poor survival and response to adjuvant chemotherapy in colon cancer. Int. J. Cancer. 2014;134:1926–1934. doi: 10.1002/ijc.28522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao W., Liu J.N., Liu Z., Wang X., Han Z.G., Ji T., Chen W.T., Zou X. A three-lncRNA signature derived from the Atlas of ncRNA in cancer (TANRIC) database predicts the survival of patients with head and neck squamous cell carcinoma. Oral Oncol. 2017;65:94–101. doi: 10.1016/j.oraloncology.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 7.Li T., Mo X., Fu L., Xiao B., Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7:8601–8612. doi: 10.18632/oncotarget.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Q., Meng W.Y., Jie Y., Zhao H. LncRNA MALAT1 induces colon cancer development by regulating miR-129-5p/HMGB1 axis. J. Cell. Physiol. 2018;233:6750–6757. doi: 10.1002/jcp.26383. [DOI] [PubMed] [Google Scholar]
- 9.Kong Y.G., Cui M., Chen S.M., Xu Y., Xu Y., Tao Z.Z. LncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene. 2018;639:77–84. doi: 10.1016/j.gene.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Li K., Sun D., Gou Q., Ke X., Gong Y., Zuo Y., Zhou J.K., Guo C., Xia Z., Liu L. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018;420:80–90. doi: 10.1016/j.canlet.2018.01.060. [DOI] [PubMed] [Google Scholar]
- 11.Sun S., Wang X., Xu X., Di H., Du J., Xu B., Wang Q., Wang J. MiR-433-3p suppresses cell growth and enhances chemosensitivity by targeting CREB in human glioma. Oncotarget. 2017;8:5057–5068. doi: 10.18632/oncotarget.13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurluler E., Guner O.S., Tumay L.V., Turkel Kucukmetin N., Hizli B., Zorluoglu A. Serum annexin A2 levels in patients with colon cancer in comparison to healthy controls and in relation to tumor pathology. Med. Sci. Monit. 2014;20:1801–1807. doi: 10.12659/MSM.892319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma R.L., Shen L.Y., Chen K.N. Coexpression of ANXA2, SOD2 and HOXA13 predicts poor prognosis of esophageal squamous cell carcinoma. Oncol. Rep. 2014;31:2157–2164. doi: 10.3892/or.2014.3088. [DOI] [PubMed] [Google Scholar]
- 14.Lee J.M., Dedhar S., Kalluri R., Thompson E.W. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J. Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kartha R.V., Subramanian S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front. Genet. 2014;5:8. doi: 10.3389/fgene.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang T., Peng H., Wang J., Yang J., Nice E.C., Xie K., Huang C. Prognostic and diagnostic significance of annexin A2 in colorectal cancer. Colorectal Dis. 2013;15:e373–e381. doi: 10.1111/codi.12207. [DOI] [PubMed] [Google Scholar]
- 17.Wu B., Zhang F., Yu M., Zhao P., Ji W., Zhang H., Han J., Niu R. Up-regulation of Anxa2 gene promotes proliferation and invasion of breast cancer MCF-7 cells. Cell Prolif. 2012;45:189–198. doi: 10.1111/j.1365-2184.2012.00820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bao J., Xu Y., Wang Q., Zhang J., Li Z., Li D., Li J. miR-101 alleviates chemoresistance of gastric cancer cells by targeting ANXA2. Biomed. Pharmacother. 2017;92:1030–1037. doi: 10.1016/j.biopha.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Yan X., Zhang D., Wu W., Wu S., Qian J., Hao Y., Yan F., Zhu P., Wu J., Huang G. Mesenchymal Stem Cells Promote Hepatocarcinogenesis via lncRNA-MUF Interaction with ANXA2 and miR-34a. Cancer Res. 2017;77:6704–6716. doi: 10.1158/0008-5472.CAN-17-1915. [DOI] [PubMed] [Google Scholar]
- 20.Shi Q., Wang Y., Mu Y., Wang X., Fan Q. MiR-433-3p Inhibits Proliferation and Invasion of Esophageal Squamous Cell Carcinoma by Targeting GRB2. Cell. Physiol. Biochem. 2018;46:2187–2196. doi: 10.1159/000489548. [DOI] [PubMed] [Google Scholar]
- 21.Yue Q.Y., Zhang Y. Effects of Linc00460 on cell migration and invasion through regulating epithelial-mesenchymal transition (EMT) in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2018;22:1003–1010. doi: 10.26355/eurrev_201802_14382. [DOI] [PubMed] [Google Scholar]
- 22.Han Y., Ye J., Dong Y., Xu Z., Du Q. Expression and significance of annexin A2 in patients with gastric adenocarcinoma and the association with E-cadherin. Exp. Ther. Med. 2015;10:549–554. doi: 10.3892/etm.2015.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang N., Wang L., Liu J., Liu L., Huang J., Chen X., Luo Z. MicroRNA-206 regulates the epithelial-mesenchymal transition and inhibits the invasion and metastasis of prostate cancer cells by targeting Annexin A2. Oncol. Lett. 2018;15:8295–8302. doi: 10.3892/ol.2018.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang A., Xu M., Mo Y.Y. Role of the lncRNA-p53 regulatory network in cancer. J. Mol. Cell Biol. 2014;6:181–191. doi: 10.1093/jmcb/mju013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhan A., Soleimani M., Mandal S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritchie W. microRNA Target Prediction. Methods Mol. Biol. 2017;1513:193–200. doi: 10.1007/978-1-4939-6539-7_13. [DOI] [PubMed] [Google Scholar]
- 27.Lin G., He X., Ji H., Shi L., Davis R.W., Zhong S. Reproducibility Probability Score--incorporating measurement variability across laboratories for gene selection. Nat. Biotechnol. 2006;24:1476–1477. doi: 10.1038/nbt1206-1476. [DOI] [PubMed] [Google Scholar]
- 28.Fujita A., Sato J.R., Rodrigues Lde.O., Ferreira C.E., Sogayar M.C. Evaluating different methods of microarray data normalization. BMC Bioinformatics. 2006;7:469. doi: 10.1186/1471-2105-7-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 30.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukui R., Tanabe E., Kitayoshi M., Yoshikawa K., Fukushima N., Tsujiuchi T. Negative regulation of cell motile and invasive activities by lysophosphatidic acid receptor-3 in colon cancer HCT116 cells. Tumour Biol. 2012;33:1899–1905. doi: 10.1007/s13277-012-0450-z. [DOI] [PubMed] [Google Scholar]