Abstract
Stem cell-based therapy is one of the most attractive approaches to ischemic heart diseases, such as myocardial infarction (MI). We evaluated the cardio-protective effects of the human umbilical cord blood-derived mesenchymal stem cells (hUCB-MSCs) stably expressing lymphoid enhancer-binding factor 1 (LEF1; LEF1/hUCB-MSCs) in a rat model of MI. LEF1 overexpression in hUCB-MSCs promoted cell-proliferation and anti-apoptotic effects in hypoxic conditions. For the application of its therapeutic effects in vivo, the LEF1 gene was introduced into an adeno-associated virus integration site 1 (AAVS1) locus, known as a safe harbor site on chromosome 19 by CRISPR/Cas9-mediated gene integration in hUCB-MSCs. Transplantation of LEF1/hUCB-MSCs onto the infarction region in the rat model significantly improved overall survival. The cardio-protective effect of LEF1/hUCB-MSCs was proven by echocardiogram parameters, including greatly improved left-ventricle ejection fraction (EF) and fractional shortening (FS). Moreover, histology and immunohistochemistry successfully presented reduced MI region and fibrosis by LEF1/hUCB-MSCs. We found that these overall positive effects of LEF1/hUCB-MSCs are attributed by increased proliferation and survival of stem cells in oxidative stress conditions and by the secretion of various growth factors by LEF1. In conclusion, this study suggests that the stem cell-based therapy, conjugated with genome editing of transcription factor LEF1, which promotes cell survival, could be an effective therapeutic strategy for cardiovascular disease.
Keywords: LEF1, hUCB-MSCs, CRISPR/Cas9, myocardial infarction, cardio-protection, growth factors
Introduction
Myocardial infarction (MI) is the most common coronary heart disease, which in turn, is the leading cause of morbidity and mortality worldwide.1,2 The main cause of the disease is an obstruction of the coronary artery that leads to a massive loss of cardiomyocytes, resulting in myocardial dysfunction and heart failure.3 The primary therapy should be the restoration of lost cardiomyocytes, but there is a clear limitation to the regenerative capacity of the adult mammalian heart, meaning that additional therapeutic approaches are mandatory.
Stem cell therapy has received constant attention as a potential strategy for the regeneration of infarcted hearts, and human umbilical cord blood-derived mesenchymal stem cells (hUCB-MSCs) are regarded as a promising candidate for this therapy due to its unique properties, such as multiple lineage potential, lack of teratoma formation, easy expansion, and low immunogenicity.4 Various trials have treated the injured myocardium using diverse MSC populations, including hUCB-MSCs. Some of these studies showed restoration of damaged cardiomyocytes, enhancement of cardiac function, and to certain degrees, reduced infarct size.5, 6, 7 Although many studies have attempted to utilize hUCB-MSCs and provided positive outcomes in preclinical trials, there are still some hurdles to be surmounted, such as low graft and survival rates in the hostile microenvironment of the infarcted region with its insufficient supply of oxygen and nutrients. Therefore, to enhance hUCB-MSCs’ functionality, additional strategies, such as cell patch and genome editing of the hUCB-MSCs, are required to improve cell-survival rate and paracrine effects. There have been a few trials with different target genes that obtained the expected outcomes and addressed these issues. Our recent study reported that hUCB-MSCs overexpressing the vascular endothelial growth factor (VEGF) gene controlled by doxycycline (Dox) induction successfully improved cardiac function via the increase of angiogenesis in MI.8 VEGF is an angiogenic factor that can promote endothelial cell survival and can be suggested as a therapeutic reagent.9 Although methods were varied, introduction of islet (ISL) and hepatocyte growth factor (HGF) also enhanced therapeutic effects of stem cells.10,11 However, there is still room to improve the functionality of hUCB-MSCs via not only endocrine and paracrine growth factors but also autocrine effects. It is ultimately necessary to survey as many therapeutic target genes as possible.
We indeed focused on the activation of canonical wingless/integrase-1 (Wnt) signaling pathways that have been known to contribute to mouse embryonic stem cell (ESC) self-renewal and help maintain the undifferentiated status of ESCs through modulation of Oct4 and Nanog.12, 13, 14 Lymphoid enhancer-binding factor-1 (LEF1), a 48-kD nuclear protein, is a crucial transcription factor for proliferation and survival of B and T cells.15,16 The involvement of LEF1 in canonical Wnt signaling pathways has been studied, and activation of the pathways via the overexpression of LEF1 has been shown to contribute to mouse ESC self-renewal.13 Furthermore, it has also been reported that the gene regulates the follicle morphogenesis and proliferation of neural progenitor cells.17,18
Recent studies have focused on the functions of LEF1 related to stem cells and cardiogenesis. High expression of LEF1 between the mesoderm and cardiac progenitor cell stage has been identified in various studies, and it has been suggested that the gene plays an important role between these stages.19,20 Moreover, direct and temporal contribution of LEF1 in mouse heart maturation has also been reported. It has been demonstrated that the gene is mainly expressed in MSCs in the valvular region during the murine heart development,21 but its function in mouse and human MSCs (hMSCs) needs additional study. Despite this positive potential in cell proliferation, survival, and cardiac differentiation, the cardio-protective effects from MI in stem cell therapy have not been demonstrated yet.
In parallel, diverse gene introduction systems, such as viruses, zinc-finger nucleases, transcription activator-like effector nucleases (TALENs), and CRISPR and CRISPR-associated 9 (Cas9), have been developed.22, 23, 24, 25 Since 2012, when the CRISPR system was initially demonstrated, it has been improved to reduce off-target mutations25 and has been widely adopted as the simplest and most efficient gene-integration systems in various cells and organisms, including human, rat, and mouse.26, 27, 28
Taken together, the objective of this study is to examine the therapeutic efficacy of the hUCB-MSCs stably expressing the LEF1 by CRISPR/CAS9-mediated gene integration (LEF1/hUCB-MSCs) in MI. We first investigated autocrine effects of overexpressing LEF1 in hUCB-MSCs on cell proliferation and survival of hUCB-MSCs in both normal and oxidative stress conditions and then survival-promoting effects of the cells in the MI region in vivo. Moreover, paracrine effects via stimulation of growth factor and cytokine secretion by LEF1 were also analyzed to explain the recovery of cardiac function in the MI animal model by LEF1.
Results
LEF1 Promotes hUCB-MSC Proliferation
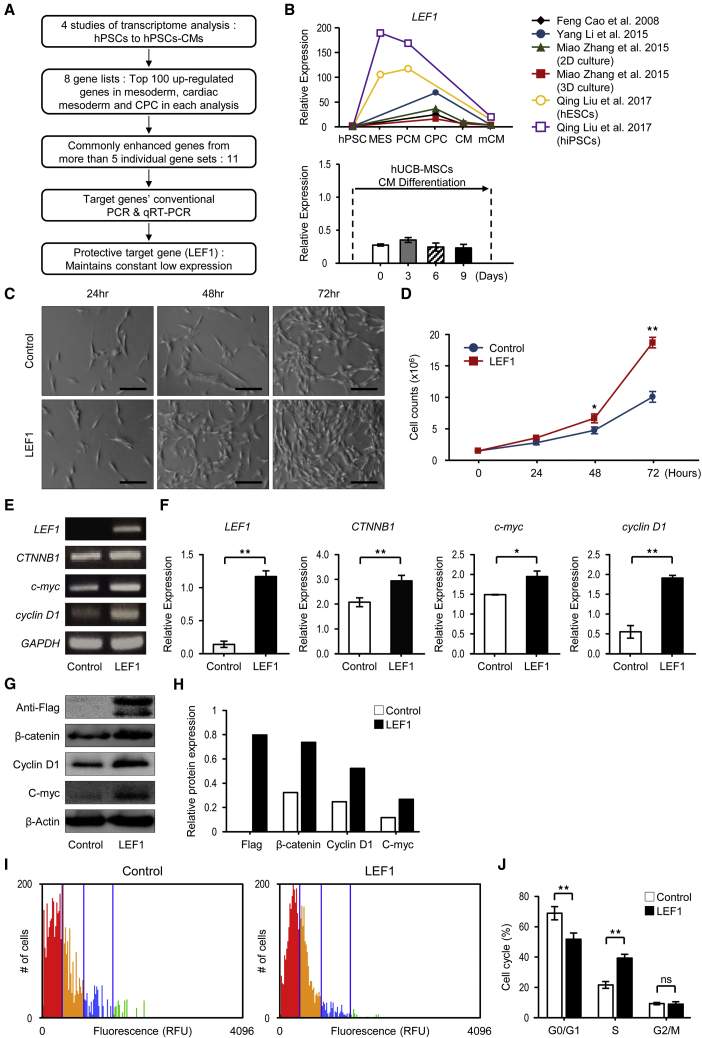
To find the target gene for hUCB-MSC therapy, we examined multiple transcriptome analyses from 4 different studies that were associated with cardiomyocyte differentiation from pluripotent cells, since the genes that exhibit cell-specific expression from mesoderm to cardiac progenitor cell stage promote self-renewal to maintain the stage or differentiation to cardiac progenitor cells.29, 30, 31 We focused on genes that are enriched in these stages. From 8 different analyses, we first examined the 100 most strongly upregulated genes in these stages in comparison to human pluripotent stem cells and then listed them individually in supplemental data (Table S1). From these gene lists, the 11 genes that were commonly enriched in 5 or more analyses were selected (Figure S1A). Among these, 36% (4 out of 11) of the genes are known to function in cardiac differentiation, e.g., PLXNA2, TBX3, and BMP4, and in MSC proliferation, e.g., LIX1, and were excluded from further target selection, finally leaving only 7 target genes.
The 7 target genes selected from the in silico literature surveys were subjected to conventional PCR (Figure S1B) and qRT-PCR to examine the gene expression in hUCB-MSCs and expression patterns during the cardiomyocyte differentiation. Various patterns were observed (Figure S2), but the aim of this study was to enhance the protection efficacy of hUCB-MSCs by insertion of target genes, so we focused on the genes that maintained a constant low expression (LEF1, ZEB2). However, the effects of LEF1 on cell proliferation, as well as the direct and temporal contributions to heart development, have been reported.13,21,32 Consequently, all additional studies proceeded with LEF1 (Figures 1A and 1B). Since low-proliferative capacity is one of the limits in the use of differentiated stem cells,33 we first examined if introducing LEF1 would enhance hUCB-MSC proliferation. The hUCB-MSCs transfected with LEF1 showed significantly increased numbers of cells, 72 h after incubation, without any aberrant changes in morphology (Figures 1C and 1D). The activation of canonical Wnt/β-catenin signaling by LEF1 transfection was confirmed by RT-PCR of β-catenin (CTNNB1) and c-Myc, as well as LEF1 itself. LEF1 significantly increased the expression of CTNNB1 and c-Myc. Cyclin D1 expression was also dramatically increased, representing stimulated cell cycles by LEF1 (Figures 1E and 1F). This result was confirmed in protein levels, as western blot and densitometry analysis clearly showed that Wnt/β-catenin signaling and cyclin D1 were upregulated by LEF1 (Figures 1G and 1H). We then presented the evidence for the proliferative fate of hUCB-MSCs enhanced in LEF1 transfection. Cell-cycle analysis using propidium iodide (PI) staining, combined with an automated fluorescence cell counter, showed cell fates shifted up from G0/G1 to S phases in the LEF1-transfected hUCB-MSC population. Approximately 40% cell-cycle enhancement was observed within the hUCB-MSCs transfected with LEF1 (Figures 1I and 1J).
Figure 1.
Selection Process of Therapeutic Target Gene LEF1 and Cell-Proliferation Effects of LEF1 in hUCB-MSCs
(A) Schematic depiction of the overall workflow. (B) Comparison of LEF1 gene expression with in silico literature surveys and qRT-PCR data. (C) Representative images from phase-contrast microscopy at 24, 48, and 72 h after LEF1 transfection of hUCB-MSCs. Scale bars, 100 μm. Control (Ctrl): no DNA; LEF1: LEF1:pDC3.1. (D) hUCB-MSCs (1.5 × 105 cells), treated with no DNA or LEF1:pDC3.1, were seeded, and the increased number of cells was counted at the 24-, 48-, and 72-h time points. *p < 0.05 and **p < 0.01. (E) Conventional PCR for the Wnt pathway and cell-cycle-related genes. (F) Significant difference in gene-expression levels was confirmed by real-time PCR. Relative expression to GAPDH was calculated by the ΔΔ CT method. **p < 0.01. (G) Western blot analysis confirmed the increased expression in protein level under LEF1 overexpression. (H) Densitometry showed relative protein expression to β-actin level. (I) Cell-cycle analysis was performed by automated fluorescence cell counting in hUCB-MSCs differentially treated with no DNA or LEF1:pDC3.1. Colors indicate different stages; red: G0/G1 phase; yellow: S phase; blue: G2/M phase. (J) The histogram for the cell-cycle distribution after transfection of LEF1 and scrambled control DNA. **p < 0.01 compared to control.
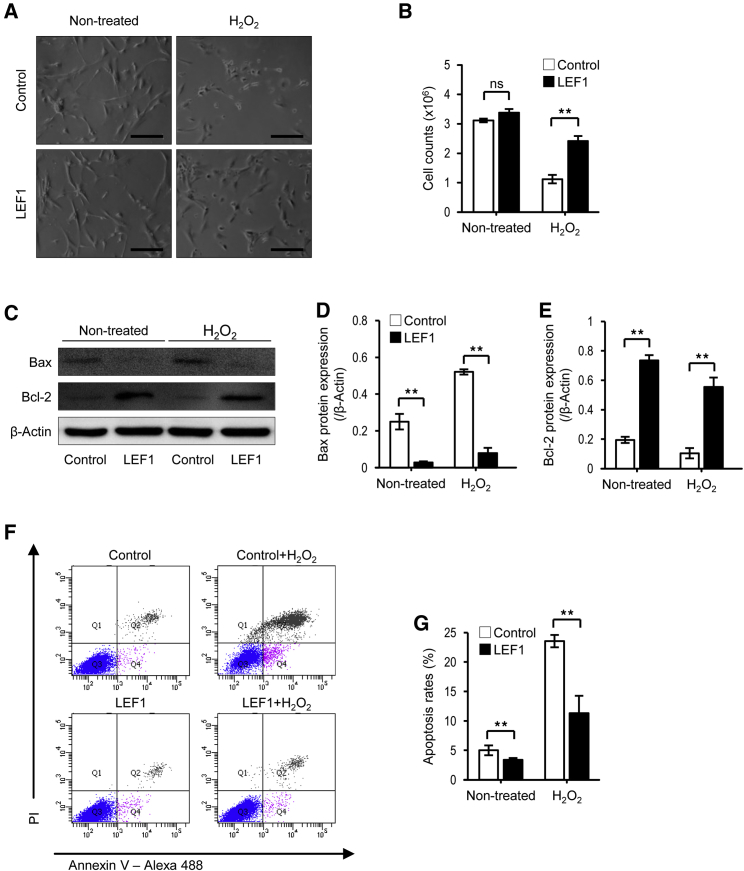
LEF1 Prevents hUCB-MSCs from Hydrogen Peroxide-Induced Apoptosis
A major issue in stem cell therapy for ischemic heart diseases, including MI, is the low survival of transplanted cells in the ischemic region. It is reported that most hMSCs implanted onto ischemic hearts died within 4 days after transplantation.34 Therefore, we examined the protective function of LEF1 from the hydrogen peroxide-induced cell death of hUCB-MSCs in vitro. Apoptosis was induced by 500 μM H2O2 treatment for 48 h in hUCB-MSCs, whereas hUCB-MSCs expressing LEF1 still survived at a significantly higher number of cells (Figure 2A). Slightly more cells were counted in LEF1-transfected hUCB-MSCs than in the control hUCB-MSCs under normal conditions. Yet, the number of cells remained drastically higher in the LEF1-expressing hUCB-MSC group (80% survival) than the control hUCB-MSC group (30% survival) under severe oxidative stress conditions induced by H2O2 (Figure 2B). Western blot analysis for Bax and Bcl-2 proteins revealed that LEF1 blocked proapoptotic Bax but induced anti-apoptotic Bcl-1, even without oxidative stress, thus presenting a reduced level of Bax but an increased level of Bcl-2 in both normal and severe oxidative stress conditions (Figures 2C–2E). These results demonstrate that the cells had a protective effect upon H2O2 treatment. We next demonstrated that LEF1 could protect hUCB-MSCs from apoptosis under severe oxidative stress using flow cytometry with Annexin V/PI labeling. As shown in Figures 2F and 2G, a drastic increase in the apoptotic cells was observed in control hUCB-MSCs upon H2O2 treatment (∼25%). However, a remarkable reduction in the apoptotic cell ratio was shown in LEF1-transfected hUCB-MSCs under both normal (∼4%) and oxidative stress conditions (∼10%). Altogether, these results indicate that the LEF1 plays an important role in anti-apoptosis of hUCB-MSCs under oxidative stress.
Figure 2.
LEF1 Overexpression Protects hUCB-MSCs from Oxidative Stress-Induced Apoptosis
(A) Microscopy demonstrated that oxidative stress-caused cell death was attenuated by LEF1 overexpression. Scale bars, 100 μm. (B) Quantification of reduced cell death in LEF1 overexpression under oxidative stress condition. **p < 0.01. (C) Drastic increase in Bcl-2 and decrease in Bax expression in LEF1-overexpressing hUCB-MSCs were observed by western blot. (D and E) Image analysis confirmed the significant changes of Bax (D) and Bcl-2 (E) expression in LEF1-overexpressing hUCB-MSCs. (F) Fluorescence-activated cell sorting (FACS) analysis using PI and Annexin V successfully presented increased apoptotic cell populations (Q2) under H2O2 treatment in hUCB-MSCs (top right), but less cells were dead in LEF1-expressing hUCB-MSCs (bottom right). (G) Reduced apoptosis triggered by H2O2 in LEF1 transfection. **p < 0.01.
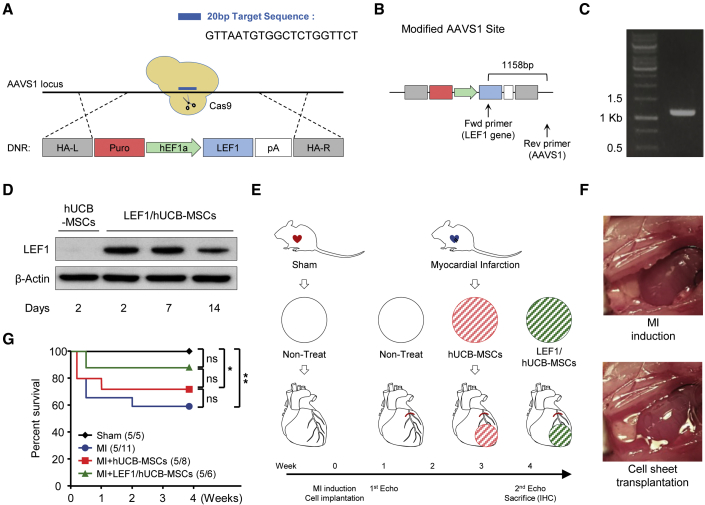
LEF1/hUCB-MSC Transplantation Improves Cardiac Dysfunction after Myocardial Infarction
A number of studies have demonstrated that stem cell therapy is effective in myocardial regeneration via enhancement of angiogenesis in the region of MI.33 Various types of stem cells, different treatments or gene editing, and transplantation methods have been developed and tested to improve therapeutic efficacy.35 In the present study, we first constructed LEF1/hUCB-MSCs that steadily express LEF1 using the CRISPR/Cas9 system, followed by homologous recombination on the AAVS1 genomic safe harbor site (Figure 3A). Successful integration of LEF1 on AAVS1 was confirmed by genomic PCR spanning from LEF1 to the AAVS1 site (Figures 3B and 3C). LEF1 protein was stably expressed by LEF1/hUCB-MSCs until 14 days after transfection (Figure 3D). To determine the cardioprotective potential of LEF1/hUCB-MSCs in ischemic heart disease, we conducted an experiment using transplantation of LEF1/hUCB-MSCs and control hUCB-MSCs in a post-MI rat model. This experiment included a group of sham rats (surgery without MI) and MI rats (nontreat with MI) as controls (Figure 3E). The cell transplantation of each group was administered as a cell patch using the UpCell system (Figure 3F).
Figure 3.
Experimental Strategy of the Therapeutic hUCB-MSC Transplantation System
(A) Schematic diagram of the LEF1 gene-integration procedure by CRISPR/Cas9-mediated knockin to the AAVS1 site of hUCB-MSCs. 20-nt-length single-guide RNA (sgRNA) target sequence on the AAVS1 locus; LEF1 introduction cassette flanked by homologous arm (HA) left (HA-L) and right (HA-R) were indicated. (B) The architecture of donor DNA in the AAVS1 locus. Arrows: primers designed for the detection of successful homologous recombination. (C) Confirmation of the correct integration of the LEF1 cassette into the AAVS1 locus using PCR (1,158 expected size). (D) Western blot analysis showing stable expression of LEF1 protein from LEF1/hUCB-MSCs for 2 weeks. (E) Schematic illustration of the therapeutic procedure with cell-sheet transplantation of hUCB-MSCs and LEF1/hUCB-MSCs in an MI model. Of note, the cell-sheet transplantation was performed with induction surgery of MI on day 0, and echocardiography measurements were performed prior to MI surgery, as well as 1 week and 4 weeks after transplantation. The four groups are Sham: surgery without MI; MI: MI alone; MI + hUCB-MSCs: MI treated with hUCB-MSCs; and MI + LEF1/hUCB-MSCs: MI treated with LEF1/hUCB-MSCs. (F) Representative images depicting before and after MI induction and stem cell transplantation using the UpCell system. (G) Survival curves of the experimental groups. The survival rate was significantly enhanced in the LEF1/hUCB-MSC group (n = 5–11). *p < 0.05; **p < 0.01; ns, not significant.
A total of 20 rats, 5 rats in each group, was initially designed and subjected to MI surgery. The rats were randomized to the following groups: operation + non-MI, MI + nontreat, MI + hUCB-MSCs, MI + LEF1/hUCB-MSCs. Each cell sheet was transplanted 30 min after visual inspection of the infarction. However, 6 out of 11 rats with MI died within 2 weeks after the operation (45.5% survival). Three out of 8 (62.5% survival) died in the MI + hUCB-MSC group and 1 out of 6 in the MI + LEF1/hUCB-MSC group (83.3% survival) (Figure 3G).
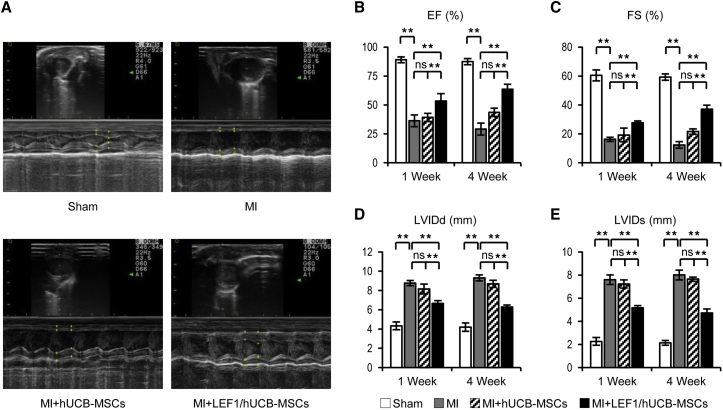
To measure the combined therapy in MI, we performed echocardiography at 1 week and 4 weeks after MI surgery (Figure 4A). A successful induction of MI in the rat model was confirmed by echocardiography showing drastic reduction of left-ventricular ejection fraction (EF) (sham: 89.01 ± 2.56% versus MI: 36.33 ± 5.11%) and fraction shortening (FS) (sham: 60.48 ± 3.82% versus MI: 16.24 ± 1.41%) when comparing the sham and MI group at 1 week postsurgery (Figures 4B and 4C). Damage of the heart muscle was also measured by an increase of the left ventricle (LV) inner diameter at diastole (LVIDd) (sham: 4.33 ± 0.41 mm versus MI: 8.77 ± 0.27 mm) and LV inner diameter at systole (LVIDs) (sham: 2.26 ± 0.33 mm versus MI: 7.60 ± 0.41 mm) (Figures 4D and 4E). To evaluate functional improvement of hUCB-MSCs and LEF1 overexpression in hUCB-MSCs on the MI, we also measured these 4 values at 4 weeks postsurgery (Figures 4B–4E). The MI + hUCB-MSC group tended to have some protective effects when compared with MI, but there was no significant difference between the values taken 1 and 4 weeks postsurgery: EF (43.56 ± 3.62%), FS (21.6 ± 1.88%), LVIDd (8.68 ± 0.37 mm), and LVIDs (7.64 ± 0.18 mm). Of note, in the comparison among the three MI groups (MI alone, MI + hUCB-MSCs, and MI + LEF1/hUCB-MSCs), LEF1 expressing hUCB-MSCs significantly improved in all 4 functional values in 4 weeks: EF (63.53 ± 4.34%), FS (37.11 ± 2.78%), LVIDd (6.29 ± 0.19 mm), and LVIDs (4.72 ± 0.34 mm). These results demonstrate the protective effects of LEF1/hUCB-MSCs in MI when compared with the MI-alone group and with the MI + hUCB-MSC group as well: EF (29.18 ± 5.13%), FS (12.36 ± 2.21%), LVIDd (9.30 ± 0.31 mm), and LVIDs (7.71 ± 0.42 mm).
Figure 4.
Transplantation of LEF1/hUCB-MSCs Recovered Cardiac Function in MI Rat
(A) Representative images of echocardiography showed successively improved cardiac function in MI, MI + hUCB-MSCs, and MI + LEF1/hUCB-MSCs at 4 weeks postsurgery. (B–E) Four values representing cardiac function (EF in B; FS in C; LVIDd in D; and LVIDs in E) were measured and compared in histograms. Significant improvements were detected in all MI + LEF1/hUCB-MSC groups.*p < 0.05, **p < 0.01; ns, not significant. Error bar: standard error. White bars, Sham; gray bars, MI; striped bars, MI + hUCB-MSCs; black bars, MI + LEF1/UCB-MSCs.
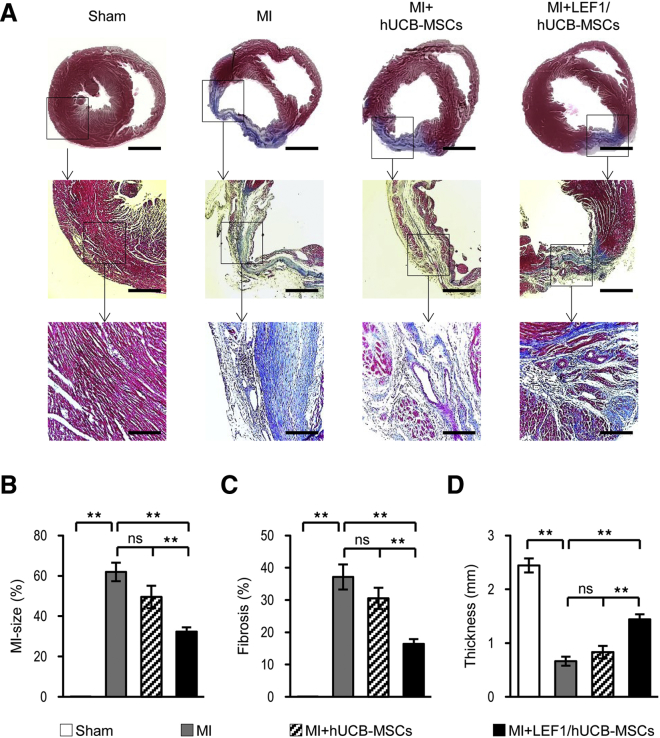
LEF1/hUCB-MSCs Show Reduced MI Size and Fibrosis and Protect the Left-Ventricular Wall from Thinning
Transplantation of LEF1/hUCB-MSCs reduced MI size and fibrosis and protected the left-ventricular wall from thinning (Figure 5). The improvement of cardiac function was observed from a histological analysis. Masson’s trichrome staining on the rat heart, 4 weeks after sham surgery, presented heart muscle in red (Sham in Figure 5A). The other three hearts with induced MI produced different ranges of regions stained with blue, which represents fibrosis in the LV. A very thin, blue-stained wall fiber was detected in the MI-induced control model. No live heart muscle was stained in the MI region. Instead, a little thicker LV was noted in the hUCB-MSC-treated MI group, but large fibrosis was still stained. Some live heart cells stained in red could be seen in the region of fibrosis. This may indicate that hUCB-MSC itself has a minor protective effect (MI + hUCB-MSCs in Figure 5A). In particular though, a clear improvement in the anti-fibrosis effect was detected in the group treated with LEF1/hUCB-MSCs (MI + LEF/hUCB-MSCs in Figure 5A). The region of fibrosis stained blue was greatly reduced and the number of heart muscle cells remaining (stained with red) in the fibrotic region were more readily observed in the magnified image (Figure 5A). For the quantitation of efficacy in cardiac function, three factors, MI-size, fibrosis, and wall thickness, were measured by analyzing the stained areas. MI-size and fibrosis were dramatically decreased in the LEF1/hUCB-MSC group when compared with both the MI and MI + hUCB-MSC groups (Figures 5B and 5C). Furthermore, the decrease of LV wall thickness resulting from MI was significantly protected from the LEF1/hUCB-MSC group, whereas the LV wall thickness tends to appear drastically reduced in the hUCB-MSC group, which was not significantly different from the MI group (Figure 5D). These results suggest that LEF1 abundantly expressed in hUCB-MSCs not only has an autocrine effect (enhancing the survival of hUCB-MSCs) but also somehow has paracrine effects, possibly via growth factors and cytokines, which can protect and regenerate damaged rat heart cells in the region of MI.
Figure 5.
The LEF1/hUCB-MSC Transplantation Greatly Reduced MI Size and Fibrosis and Restored the LV Wall Thickness
(A) Masson’s trichrome staining showed MI regions at 4 weeks after surgery. Scale bars: top row, 1 mm; middle row, 400 μm; bottom row, 200 μm. The red-stained region means the viable myocardium, and the blue-stained region means the fibrotic area. The large, fibrous region stained in blue is found in MI alone compared to MI + hUCB-MSCs and MI + LEF1/hUCB-MSCs. Serial magnification distinctively presented damaged heart in MI and enhanced heart-protective efficacy of MI + LEF1/hUCB-MSCs. (B–D) Quantification of infarct size (B), fibrosis region (C), and the wall thickness (D) of LV in each group (n = 5). *p < 0.05, **p < 0.01; ns, not significant.
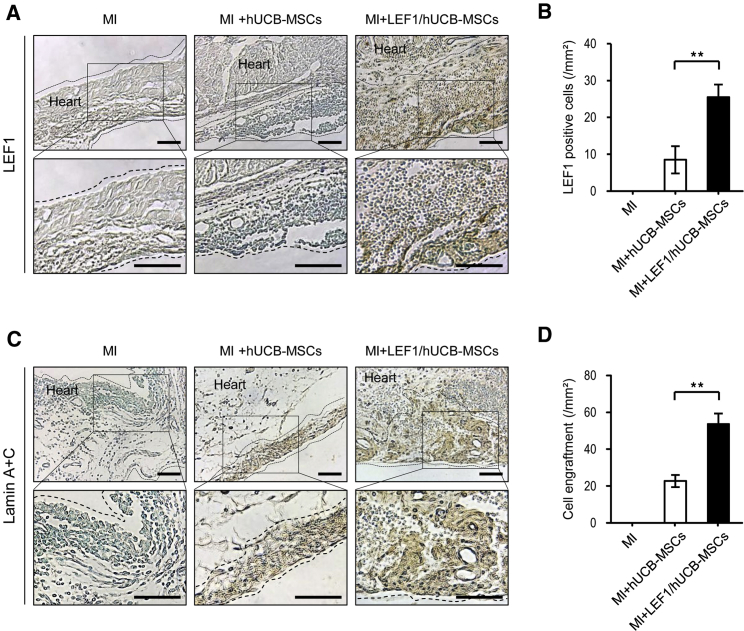
LEF1 Expression in hUCB-MSCs Helps to Prolong Survival of Implanted Stem Cells in MI
LEF1 expression was confirmed by immunohistochemical staining in MI-induced rat heart tissues. Fibrous heart structure was found in the MI-only group with few cells stained with methyl green (MI in Figure 6A). There were some cells that survived in the patch of hUCB-MSCs, but no LEF1 expression was detected in the MI + hUCB-MSC group. LEF1 expression was stained only in the tissues from the cell patch of LEF1/hUCB-MSCs attached on the MI region (MI + LEF1/hUCB-MSCs in Figure 6A). This suggests that there was no endogenous expression of LEF1 in the patched hUCB-MSCs. In addition, substantially more cells were counted in the LEF1/hUCB-MSC group than the hUCB-MSC group, 4 weeks after surgery. This might contribute to the thicker LV in the MI region with LEF1/hUCB-MSCs. Quantification of the LEF1 immunohistochemistry (IHC) data successfully displayed prolonged survival of LEF1/hUCB-MSCs (Figure 6B). To distinguish implanted human stem cells from rat heart cells, IHC with anti-lamin A + C antibody was performed in the MI region. We took a picture from the edge of the MI region to include rat heart cells as a control. No lamin A + C-positive cells were detected from rat heart cells in the MI group (Figure 6C). Only a thin layer of human cells was stained with lamin A + C-positive cells in the hUCB-MSC groups, whereas a large number of lamin A + C-positive, implanted human stem cells were detected in the MI + LEF1/hUCB-MSC groups. Strikingly, rat heart myocardium still remained thicker at 4 weeks after MI induction following LEF1/hUCB-MSC treatment (Figure 6D). These results suggest that LEF1 expression in hUCB-MSCs has positive effects, not only on the survival of stem cells but also on protecting rat myocardial cells from MI via paracrine effects.
Figure 6.
Immunohistochemical Staining Confirmed the Cells Surviving and Expressing LEF1 from the Engrafted MI + LEF1/hUCB-MSCs
(A) LEF1 was detected only from MI + LEF1/hUCB-MSCs. No rat heart cells and hUCB-MSCs expressed LEF1. Scale bars, 100 μm. (B) LEF1-positive cells in the heart tissue were quantitatively measured (n = 5). (C) No lamin signal was detected in the MI-alone group. A thin layer was found in the group of MI + hUCB-MSCs, and a thicker layer was stained from MI + LEF1/hUCB-MSC-engrafted MI. Scale bars, 100 μm. (D) Human cell engraftment was quantitatively measured (n = 5). **p < 0.01.
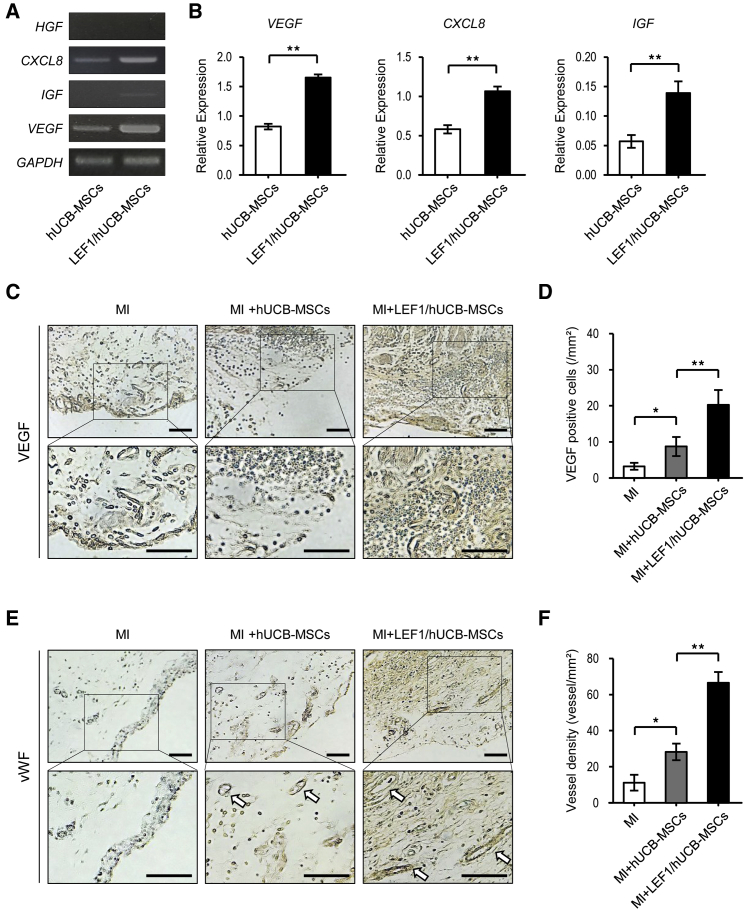
LEF1 Expression in hUCB-MSCs Triggers the Production of Various Growth Factors that Protect the Heart and Promote VEGF Production and Angiogenesis in MI
Growth factors and cytokines, such as VEGF, insulin-like growth factor (IGF), HGF, interleukin-1β (IL-1β), and IL-6, have been investigated as therapeutic targets in various cell types and diseases.8,36, 37, 38 Recently, these molecules, secreted by MSCs, have been examined in repair and regeneration therapy.35 We thus investigated the paracrine effect of LEF1/hUCB-MSCs via enhanced secretion of growth factors and cytokines. Expression levels of three growth factors (HGF, IGF, and VEGF) and the cytokine (IL-8) were compared between control hUCB-MSCs and LEF1/hUCB-MSCs. As expected, HGF, IGF, VEGF, and IL-8 were increased in LEF1/hUCB-MSCs (Figure 7A). Quantitative RT-PCR showed an approximate 2-fold increase in the expression of VEGF, IL-8, and IGF in LEF1/hUCB-MSCs (Figure 7B). VEGF protein expression was confirmed by IHC staining (Figure 7C). Rat heart at the edge of the MI region was faintly stained due to crossreactivity of the VEGF antibody between human and rat. A strong VEGF signal was detected in the surrounding regions of LEF1/hUCB-MSCs (Figure 7D). Since VEGF is known to assist in angiogenesis, we measured the vascular genesis in the MI regions of the MI, MI + hUCB-MSC, and MI + LEF1/hUCB-MSC groups using von Willebrand factor (vWF) IHC staining. Of these, the LEF1/hUCB-MSC groups showed higher vessel density than the other two groups (Figures 7E and 7F). These results determined that LEF1 integration in hUCB-MSCs triggers the expression of diverse growth factors and enhances vessel formation, resulting in protective effects on myocardial cells in the rat MI model.
Figure 7.
LEF1 Triggered Therapeutic Gene (VEGF, IL-8, IGF) Expressions and Enhanced Angiogenesis in MI + LEF1/hUCB-MSCs
(A) Expression of growth factors HGF, CXCL8, IGF, and VEGF was measured by conventional PCR. (B) Increased mRNA expression of growth factors in LEF1/hUCB-MSCs compared with hUCB-MSCs. Real-time PCR demonstrated that the relative gene expressions of VEGF, CXCL8, and IGF to GAPDH were increased in LEF1/hUCB-MSCs. **p < 0.01. (C) Immunohistochemical staining for VEGF in three MI groups: MI alone, MI + hUCB-MSCs, and LEF1/MI + hUCB-MSCs. Anti-VEGF antibody faintly stained rat heart, but a stronger signal was detected in LEF1/hUCB-MSCs. Scale bars, 100 μm. (D) VEGF protein expression was measured in infarcted heart among different groups (n = 5). (E) Immunohistochemical staining for vWF in three MI groups: MI alone, MI + hUCB-MSCs, and LEF1/MI + hUCB-MSCs. The blood vessels are stained in brown (arrows). More vessels were observed in MI + LEF1/hUCB-MSCs. Scale bars, 100 μm. (F) Higher vascular densities were detected in the MI + LEF1/hUCB-MSC group than in the MI-alone and MI + hUCB-MSC groups (n = 5). *p < 0.05 and **p < 0.01.
Discussion
Stem cell-based therapies have emerged as a promising treatment for heart regeneration after infarction.33 Among them, hUCB-MSCs have been recognized as good candidates, due to trophic activities, such as multiple lineage potential, lack of teratoma formation, easy expansion, and low immunogenicity.39 In cardiovascular disease, MSC-based therapies have been attempted in a lot of preclinical research and have shown sufficient potential. However, the low cell-survival rates and the engraftment failure of implanted cells in the infarcted region are the main obstacles in this field that still remain. Therefore, enhancement of stem cell proliferation and its survival under the harsh conditions of the ischemic area is necessary to utilize MSCs for clinical application.
In the current study, we thus focused on enhancing stem cell activities in cell proliferation and its survival in the harsh conditions of the ischemic region. LEF1 has been known to play a role in regulating cell proliferation via Wnt/β-catenin signaling.12,32,40 Huang and Qin13 recently reported that LEF1 plays a crucial role in sustaining self-renewal in mouse ESCs as well. Therefore, we investigated the effects of LEF1 expression in hUCB-MSCs in normal and harsh conditions, such as oxidative stress in vitro. Introduction of LEF1 to hUCB-MSCs resulted in the overexpression of LEF1-stimulated stem cells’ cell cycle and proliferation via the canonical Wnt pathway in normal conditions (Figure 1). In addition, the understanding of the cell response to oxidative stress in vitro is important because the cells’ microenvironment after implantation is hypoxic, which can lead to apoptosis.41 This study clearly showed that LEF1 expression protected hUCB-MSCs from oxidative stress conditions by increasing Bcl-2 expression. This was confirmed through flow cytometry, showing a reduced number of apoptotic cells induced by H2O2 (Figure 2).
We then generated therapeutic hUCB-MSCs that stably express LEF1 through CRISPR/Cas9-mediated genome editing (LEF1/hUCB-MSCs) in order to examine whether the induction of the LEF1 gene in hUCB-MSCs affects the cell engraftment, survival, and tolerance in hypoxic conditions. The CRISPR/Cas9 gene integration system was employed on the AAVS1 locus to overcome side effects, such as tumorigenesis, or unpredictable integration of the transgene, which could be induced by the viral approach.28 Steady expression of LEF1 was detected until 2 weeks in LEF1/hUCB-MSCs. As expected from the in vitro study, the LEF1/hUCB-MSC group showed strong, positive effects in the in vivo MI model. Echocardiography and histological staining analysis clearly showed evidence that LEF1/hUCB-MSCs have a protective effect in the MI region. EF, FS, LVIDd, and LVIDs, which represent left-ventricular cardiac functions, were greatly improved in LEF1/hUCB-MSCs compared with MI alone and hUCB-MSC treatment.
In addition, the protective effect of LEF1/hUCB-MSCs was measured by MI-size, fibrosis, and wall thickness using Masson’s trichrome staining. MI and fibrosis were formed approximately 52% less in LEF1/hUCB-MSCs compared with the MI control group. This is a huge improvement compared with the hUCB-MSC group, showing only a 21% reduction. Furthermore, wall-thickness loss in MI was suppressed by LEF1/hUCB-MSCs, whereas it was not significantly different between MI alone and the MI + hUCB-MSC group. Most cardiac muscle cells were replaced by fibrosis in MI alone, but heart muscle structure was still retained in the MI region treated with LEF1/hUCB-MSCs. This result may suggest two major mechanisms to explain these enhanced therapeutic effects. One is a paracrine effect, and the other one is direct transdifferentiation. After transplantation, the engrafted MSCs could secrete the therapeutic factors that regenerate the damaged cardiac tissue and cause neovascularization via paracrine effects and also prevent the cell loss of cardiomyocytes by direct transdifferentiation or inhibition of fibrosis. However, according to previous studies, there is little evidence that the induction of the LEF1 gene in stem cells directly affected the differentiation of cardiomyocytes or the decrease of fibrosis. In contrast, it has been reported that LEF1 expression secretes many therapeutic factors associated with cell cycling, proliferation, and survival.12,13,32,42 This mechanism that enhances positive effects in cell-cycle, proliferation, and cell survival under oxidative stress was confirmed by this in vitro study (Figures 1 and 2). We also tested paracrine effects of LEF1 expression in hUCB-MSCs. As we showed in Figure 7, increased VEGF and IL-8 expressions were detected in LEF1/hUCB-MSCs in vitro and were confirmed by IHC in a rat MI model with transplanted LEF1/hUCB-MSCs. These results indicated that LEF1/hUCB-MSCs enhanced the secretion of various growth factors associated with microenvironmental neovascularization, proliferation, and immune responses, resulting in protective effects on hearts damaged by MI.
Although we presented several positive aspects of genome editing to lead LEF1 expression in hUCB-MSCs, there are also several studies regarding the aberration of LEF1 gene expression in various cases of tumorigenesis and cancer cell proliferation, migration, and invasion.43,44 Particularly, LEF1 expression has been reported in cancer cell types, such as some leukemias, lymphoma, squamous cell carcinoma, and colorectal cancer.45,46 Lack of teratoma formation has been known in MSCs; however, the tumorigenic effect of LEF1 in hUCB-MSCs needs be tested in regard to long-term expression and survival.
In conclusion, we provide the evidence that LEF1 promotes hUCB-MSC proliferation and attenuates the apoptosis from oxidative stress. The hUCB-MSCs in which the LEF1 gene was integrated by the CRISPR/Cas9 system displayed enhanced cell survival and improved cardio-protective effects in an animal model of MI. These results suggest that the introduction of LEF1 could be a novel strategy in stem cell therapy after MI.
Materials and Methods
Isolation and Culture of hUCB-MSCs
hUCB-MSCs were isolated, as previously described,47 and by following the procedure approved by the Borame Hospital institutional review board (IRB) and Seoul National University (IRB No. 0603/001-002-07C1). In brief, UCB samples were harvested from term and preterm deliveries at the time of birth with the mother’s informed consent (Seoul City Borame Hospital Cord Blood Bank). Separation of the MSCs from UCB was performed using Ficoll-Paque PLUS (Amersham Bioscience, Uppsala, Sweden). The cells were suspended in DMEM (Gibco, Grand Island, NY, USA) with 20% fetal bovine serum (FBS; Gibco), 100 IU/mL penicillin, 100 mg/mL streptomycin, 2 mM L-glutamine, and 1 mM sodium pyruvate. After 24 h, the cells were washed twice in PBS and cultured in DMEM with 10% FBS.
Transfection and Cell-Proliferation Assays
To introduce the expression vector, transfection was performed using Lipofectamine 3000 by following the manufacturer’s manual (Thermo Fisher Scientific, Waltham, MA, USA). The hUCB-MSCs and LEF1-introduced hUCB-MSCs were subjected to a cell-proliferation assay. In brief, 150,000 cells were seeded in 60 mm plates on day 0 and counted after 24 h, 48 h, and 72 h by an automated cell counter (Arthur; NanoEnTek, Seoul, Korea). Experiments were run in triplicate.
RNA Isolation and cDNA Synthesis
The total RNA was extracted by using the TRIzol reagent (Invitrogen, Carlsbad, USA), and the quality was measured by the ratio of 260/280 using the Nanodrop Epoch Microplate Spectrophotometer (BioTek Instruments, VT, USA). Reverse transcription was performed on 2 μg of RNA using the Omniscript RT kit, following the manufacturer’s guideline (QIAGEN), TRIzol reagent (Invitrogen, Carlsbad, USA).
Conventional and Real-Time PCR Analyses
Conventional PCR was performed on 1 μL of cDNA using GoTaq polymerase in the following conditions: denaturation at 95°C for 2 min, 35 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, followed by final extension at 72°C for 5 min. Real-time PCR was performed in the same reaction conditions except for the introduction of SYBR Green (Bio-Rad CFX Manager, CA, USA), and the relative expressions were calculated by the delta-delta comparative threshold (ΔΔ CT) method. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as an internal control.
Western Blot Analysis
After treatment with radioimmunoprecipitation assay (RIPA) buffer for 30 min, approximately 20 to 30 μg of cell lysates was used for western blot analysis, as previously reported.48 Proteins were detected with the following antibodies: anti-LEF1 (1: 1,000; Cell Signaling Technology; C12A5), anti-c-Myc (1:1,000; Cell Signaling Technology; D84C12), anti-cyclin D1 (1: 1,000; Cell Signaling Technology; 92G2), anti-Bcl-2 (1:1,000; Abcam; ab692), anti-Bax (1:1,000; Santa Cruz Biotechnology; SC-493), and anti-β-actin (1:2,000; Santa Cruz Biotechnology; SC-32233).
Cell-Cycle Analysis
The cells in each group were collected after 48 h of transfection. After washing with cold PBS three times, the cell pellets were resuspended in PBS with the cell density of 1 × 105 cells/mL. After treatment with 400 μL of PI (Sigma-Aldrich, St. Louis, MO, USA), the samples were incubated for 30 min in the darkness at 4°C. The cell cycle was detected using red fluorescence with an automated fluorescence cell counter (Arthur; NanoEnTek, Seoul, Korea).
Flow Cytometry
After cell transfection, the cells in each group were digested with ethylene diamine tetraacetic acid (EDTA) and then collected in a flow tube. After 3 PBS washes, each sample was subjected to the Annexin-V fluorescein isothiocyanate (FITC) cell apoptosis kit (Sigma-Aldrich, St. Louis, MO, USA), following the kit instructions. The Annexin-V-FITC/PI dye was added to the 1 × 106 cell suspension. After a 15-min incubation at room temperature, the 525-nm and 620-nm band-pass filter was used for FITC and PI fluorescence detection, and the excitation wavelength of 488 nm was used for the detection of cell apoptosis.
Donor Construct Design and CRISPR/Cas9-Mediated Gene Editing
The transgene insertion system using CRISPR/Cas9 into the AAVS1 locus was derived from OriGene Technologies (https://www.origene.com/products/gene-expression/crispr-cas9) and consists of pCas-Guide-AAVS1 (#GE 100023) and pAAVS1-EF1a-Puro-DNR (#GE 100046). The donor vector contains 550 bp left, right homology arms matched to the AAVS1 locus in each side, followed by the EF1α (EF1a) promoter for the constitutive expression of the target gene.
To insert the LEF1 gene in the donor vector, the LEF1 plasmid (#RC 208663) was purchased from OriGene (Rockville, MD, USA). The donor vector and LEF1 plasmid were digested with SgfI/MluI (NEB, Ipswich, MA, USA). This fragment was synthesized de novo and ligated into the donor vector to generate pAAVS1-EF1a-LEF1-DNR. To generate the LEF1/hUCB-MSCs, hUCB-MSCs were cotransfected with pCas-Guide-AAVS1 and pAAVS1-EF1a-LEF1-DNR through electroporation with the Neon Transfection System (Thermo Fisher Scientific).
MI Rat Model and Cell-Sheet Transplantation
The MI rat models were surgically induced, as previously described.8 Briefly, male Sprague-Dawley rats, weighing 260–300 g (Orient Bio, Seongnam, Korea), were used as the MI model. All animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC; SNU-190307-2; Seoul National University, Korea). The rats were divided into four groups: rats that received LEF1/hUCB-MSCs with MI, hUCB-MSCs with MI, nontreated with MI, and sham (no-MI), and each group consisted of at least 5 rats.
For the transplantation of stem cells, the UpCell system was employed (Thermo Fisher Scientific, Waltham, MA). Cell sheets of LEF1/hUCB-MSCs or hUCB-MSCs were detached from the plates by incubating at 20°C for 20 min. After transplantation, the supporting membrane was removed, and the cell sheets showed stable attachment. To prevent the rat’s immune rejection of the human cells, all rats received cyclosporine, as previously described.47
Masson’s Trichrome Staining and Histological Analysis
After extraction of all mice hearts, they were washed with PBS, fixed, embedded, and sectioned into 5 μm sections. Masson’s trichrome staining was performed according to instructions of the trichrome stain kit (Sigma-Aldrich, St. Louis, MO, USA). The infarct size, fibrosis, and scar thickness were quantified with ImageJ software (NIH, Bethesda, MD, USA).
Immunohistochemical Staining
Immunohistochemical staining was performed using a standard protocol. The heart sections were incubated overnight with the following primary antibodies: anti-lamin A + C (1:200; Abcam; ab108922), anti-LEF1 (1:200; Cell Signaling Technology; C12A5), anti-VEGF (1:200; Abcam; ab69479), and anti-vWF (1:200; Abcam; ab6994), and then they were subsequently exposed to biotinylated a secondary antibody and streptavidin peroxidase complex using the Histostain-Plus kit (Sigma-Aldrich, St. Louis, MO, USA). Then, the sections were stained with, 3′-diaminobenzidine tetra hydrochloride (Liquid DAB substrate kit; Abcam).
Functional Assessment of the Infarcted Myocardium
The measurement of cardiac function was performed, as previously reported.8 Briefly, it was assessed by transthoracic echocardiography prior to MI surgery (normal baseline) and both 1 and 4 weeks after MI for every experimental group.
Statistical Analysis
Data are expressed as the mean ± SEM. For statistical analysis of multiple groups, 1-way ANOVA was performed, followed by a Bonferroni post hoc test. For verification of the therapeutic effects in MI, 2-way ANOVA was carried out, followed by a Bonferroni post hoc test. A p value less than 0.05 was considered statistically significant.
Author Contributions
J.-Y.C. conceived and developed the entire study. H.-M.C. mainly analyzed data. K.-H.L. wrote the first draft of the manuscript. Y.-m.S. performed rat surgery for MI induction. T.-J.S. assisted all experiments. P.-D.R. supported scientific discussion. M.-C.C. performed echocardiography. K.-S.K. provided hUCB-MSCs. All of the authors discussed the results and contributed to the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
All supporting materials were included in Supplemental Information. This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) of Korea, funded by the Ministry of Science and ICT (2016M3A9B6026771 and 2012M3A9C6049716).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.01.007.
Supplemental Information
References
- 1.GBD 2016 Causes of Death Collaborators Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1151–1210. doi: 10.1016/S0140-6736(17)32152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jessup M., Brozena S. Heart failure. N. Engl. J. Med. 2003;348:2007–2018. doi: 10.1056/NEJMra021498. [DOI] [PubMed] [Google Scholar]
- 3.Neri M., Riezzo I., Pascale N., Pomara C., Turillazzi E. Ischemia/Reperfusion Injury following Acute Myocardial Infarction: A Critical Issue for Clinicians and Forensic Pathologists. Mediators Inflamm. 2017;2017:7018393. doi: 10.1155/2017/7018393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagamura-Inoue T., He H. Umbilical cord-derived mesenchymal stem cells: Their advantages and potential clinical utility. World J. Stem Cells. 2014;6:195–202. doi: 10.4252/wjsc.v6.i2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amado L.C., Saliaris A.P., Schuleri K.H., St John M., Xie J.S., Cattaneo S., Durand D.J., Fitton T., Kuang J.Q., Stewart G. Cardiac repair with intramyocardial injection of allogeneic mesenchymal stem cells after myocardial infarction. Proc. Natl. Acad. Sci. USA. 2005;102:11474–11479. doi: 10.1073/pnas.0504388102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Y.L., Zhao Q., Qin X., Shen L., Cheng L., Ge J., Phillips M.I. Paracrine action enhances the effects of autologous mesenchymal stem cell transplantation on vascular regeneration in rat model of myocardial infarction. Ann. Thorac. Surg. 2005;80:229–236. doi: 10.1016/j.athoracsur.2005.02.072. discussion 236–237. [DOI] [PubMed] [Google Scholar]
- 7.Cai M., Shen R., Song L., Lu M., Wang J., Zhao S., Tang Y., Meng X., Li Z., He Z.X. Bone Marrow Mesenchymal Stem Cells (BM-MSCs) Improve Heart Function in Swine Myocardial Infarction Model through Paracrine Effects. Sci. Rep. 2016;6:28250. doi: 10.1038/srep28250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho H.M., Kim P.H., Chang H.K., Shen Y.M., Bonsra K., Kang B.J., Yum S.Y., Kim J.H., Lee S.Y., Choi M.C. Targeted Genome Engineering to Control VEGF Expression in Human Umbilical Cord Blood-Derived Mesenchymal Stem Cells: Potential Implications for the Treatment of Myocardial Infarction. Stem Cells Transl. Med. 2017;6:1040–1051. doi: 10.1002/sctm.16-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrara N., Gerber H.P., LeCouter J. The biology of VEGF and its receptors. Nat. Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 10.Xiang Q., Liao Y., Chao H., Huang W., Liu J., Chen H., Hong D., Zou Z., Xiang A.P., Li W. ISL1 overexpression enhances the survival of transplanted human mesenchymal stem cells in a murine myocardial infarction model. Stem Cell Res. Ther. 2018;9:51. doi: 10.1186/s13287-018-0803-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao L., Liu X., Zhang Y., Liang X., Ding Y., Xu Y., Fang Z., Zhang F. Enhanced cell survival and paracrine effects of mesenchymal stem cells overexpressing hepatocyte growth factor promote cardioprotection in myocardial infarction. Exp. Cell Res. 2016;344:30–39. doi: 10.1016/j.yexcr.2016.03.024. [DOI] [PubMed] [Google Scholar]
- 12.Li J., Li J., Chen B. Oct4 was a novel target of Wnt signaling pathway. Mol. Cell. Biochem. 2012;362:233–240. doi: 10.1007/s11010-011-1148-z. [DOI] [PubMed] [Google Scholar]
- 13.Huang C., Qin D. Role of Lef1 in sustaining self-renewal in mouse embryonic stem cells. J. Genet. Genomics. 2010;37:441–449. doi: 10.1016/S1673-8527(09)60063-1. [DOI] [PubMed] [Google Scholar]
- 14.Kim C.G., Chung I.Y., Lim Y., Lee Y.H., Shin S.Y. A Tcf/Lef element within the enhancer region of the human NANOG gene plays a role in promoter activation. Biochem. Biophys. Res. Commun. 2011;410:637–642. doi: 10.1016/j.bbrc.2011.06.044. [DOI] [PubMed] [Google Scholar]
- 15.Okamura R.M., Sigvardsson M., Galceran J., Verbeek S., Clevers H., Grosschedl R. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8:11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- 16.Staal F.J., Luis T.C., Tiemessen M.M. WNT signalling in the immune system: WNT is spreading its wings. Nat. Rev. Immunol. 2008;8:581–593. doi: 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- 17.Armenteros T., Andreu Z., Hortigüela R., Lie D.C., Mira H. BMP and WNT signalling cooperate through LEF1 in the neuronal specification of adult hippocampal neural stem and progenitor cells. Sci. Rep. 2018;8:9241. doi: 10.1038/s41598-018-27581-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Yu J., Shi C., Huang Y., Wang Y., Yang T., Yang J. Lef1 contributes to the differentiation of bulge stem cells by nuclear translocation and cross-talk with the Notch signaling pathway. Int. J. Med. Sci. 2013;10:738–746. doi: 10.7150/ijms.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y., Lin B., Yang L. Comparative transcriptomic analysis of multiple cardiovascular fates from embryonic stem cells predicts novel regulators in human cardiogenesis. Sci. Rep. 2015;5:9758. doi: 10.1038/srep09758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Q., Jiang C., Xu J., Zhao M.T., Van Bortle K., Cheng X., Wang G., Chang H.Y., Wu J.C., Snyder M.P. Genome-Wide Temporal Profiling of Transcriptome and Open Chromatin of Early Cardiomyocyte Differentiation Derived From hiPSCs and hESCs. Circ. Res. 2017;121:376–391. doi: 10.1161/CIRCRESAHA.116.310456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye B., Li L., Xu H., Chen Y., Li F. Opposing roles of TCF7/LEF1 and TCF7L2 in cyclin D2 and Bmp4 expression and cardiomyocyte cell cycle control during late heart development. Lab. Invest. 2019;99:807–818. doi: 10.1038/s41374-019-0204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaj T., Gersbach C.A., Barbas C.F., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joung J.K., Sander J.D. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de la Fuente-Núñez C., Lu T.K. CRISPR-Cas9 technology: applications in genome engineering, development of sequence-specific antimicrobials, and future prospects. Integr. Biol. 2017;9:109–122. doi: 10.1039/c6ib00140h. [DOI] [PubMed] [Google Scholar]
- 29.Tateno H., Hiemori K., Hirayasu K., Sougawa N., Fukuda M., Warashina M., Amano M., Funakoshi T., Sadamura Y., Miyagawa S. Development of a practical sandwich assay to detect human pluripotent stem cells using cell culture media. Regen Ther. 2017;6:1–8. doi: 10.1016/j.reth.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M., Raval K.K., Zhang J., Kamp T.J., Palecek S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Witman N., Sahara M. Cardiac Progenitor Cells in Basic Biology and Regenerative Medicine. Stem Cells Int. 2018;2018:8283648. doi: 10.1155/2018/8283648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reya T., O’Riordan M., Okamura R., Devaney E., Willert K., Nusse R., Grosschedl R. Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity. 2000;13:15–24. doi: 10.1016/s1074-7613(00)00004-2. [DOI] [PubMed] [Google Scholar]
- 33.Segers V.F., Lee R.T. Stem-cell therapy for cardiac disease. Nature. 2008;451:937–942. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- 34.Toma C., Pittenger M.F., Cahill K.S., Byrne B.J., Kessler P.D. Human mesenchymal stem cells differentiate to a cardiomyocyte phenotype in the adult murine heart. Circulation. 2002;105:93–98. doi: 10.1161/hc0102.101442. [DOI] [PubMed] [Google Scholar]
- 35.Tompkins B.A., Natsumeda M., Balkan W., Hare J.M. What Is the Future of Cell-Based Therapy for Acute Myocardial Infarction. Circ. Res. 2017;120:252–255. doi: 10.1161/CIRCRESAHA.116.310340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Enoki C., Otani H., Sato D., Okada T., Hattori R., Imamura H. Enhanced mesenchymal cell engraftment by IGF-1 improves left ventricular function in rats undergoing myocardial infarction. Int. J. Cardiol. 2010;138:9–18. doi: 10.1016/j.ijcard.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 37.Chang H.K., Kim P.H., Kim D.W., Cho H.M., Jeong M.J., Kim D.H., Joung Y.K., Lim K.S., Kim H.B., Lim H.C. Coronary stents with inducible VEGF/HGF-secreting UCB-MSCs reduced restenosis and increased re-endothelialization in a swine model. Exp. Mol. Med. 2018;50:114. doi: 10.1038/s12276-018-0143-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pannitteri G., Marino B., Campa P.P., Martucci R., Testa U., Peschle C. Interleukins 6 and 8 as mediators of acute phase response in acute myocardial infarction. Am. J. Cardiol. 1997;80:622–625. doi: 10.1016/s0002-9149(97)00434-7. [DOI] [PubMed] [Google Scholar]
- 39.Lee C.Y., Kim R., Ham O., Lee J., Kim P., Lee S., Oh S., Lee H., Lee M., Kim J., Chang W. Therapeutic Potential of Stem Cells Strategy for Cardiovascular Diseases. Stem Cells Int. 2016;2016:4285938. doi: 10.1155/2016/4285938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aloysius A., DasGupta R., Dhawan J. The transcription factor Lef1 switches partners from β-catenin to Smad3 during muscle stem cell quiescence. Sci. Signal. 2018;11:eaan3000. doi: 10.1126/scisignal.aan3000. [DOI] [PubMed] [Google Scholar]
- 41.Denu R.A., Hematti P. Effects of Oxidative Stress on Mesenchymal Stem Cell Biology. Oxid. Med. Cell. Longev. 2016;2016:2989076. doi: 10.1155/2016/2989076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim K., Pang K.M., Evans M., Hay E.D. Overexpression of beta-catenin induces apoptosis independent of its transactivation function with LEF-1 or the involvement of major G1 cell cycle regulators. Mol. Biol. Cell. 2000;11:3509–3523. doi: 10.1091/mbc.11.10.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Y., Li C., Huang L., Niu S., Lu Q., Gong D., Huang S., Yuan Y., Chen H. Prognostic value of association of OCT4 with LEF1 expression in esophageal squamous cell carcinoma and their impact on epithelial-mesenchymal transition, invasion, and migration. Cancer Med. 2018;7:3977–3987. doi: 10.1002/cam4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamb R., Ablett M.P., Spence K., Landberg G., Sims A.H., Clarke R.B. Wnt pathway activity in breast cancer sub-types and stem-like cells. PLoS ONE. 2013;8:e67811. doi: 10.1371/journal.pone.0067811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shtutman M., Zhurinsky J., Simcha I., Albanese C., D’Amico M., Pestell R., Ben-Ze’ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Espada J., Calvo M.B., Díaz-Prado S., Medina V. Wnt signalling and cancer stem cells. Clin. Transl. Oncol. 2009;11:411–427. doi: 10.1007/s12094-009-0380-4. [DOI] [PubMed] [Google Scholar]
- 47.Kang B.J., Kim H., Lee S.K., Kim J., Shen Y., Jung S., Kang K.S., Im S.G., Lee S.Y., Choi M. Umbilical-cord-blood-derived mesenchymal stem cells seeded onto fibronectin-immobilized polycaprolactone nanofiber improve cardiac function. Acta Biomater. 2014;10:3007–3017. doi: 10.1016/j.actbio.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 48.Lee J.H., Kim B.G., Ahn J.M., Park H.J., Park S.K., Yoo J.S., Yates J.R., 3rd, Cho J.Y. Role of PI3K on the regulation of BMP2-induced beta-Catenin activation in human bone marrow stem cells. Bone. 2010;46:1522–1532. doi: 10.1016/j.bone.2010.02.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.