Abstract
Strigolactones (SLs) are plant-derived signaling molecules that stimulate the hyphal branching of arbuscular mycorrhizal fungi (AMF), and consequently promote symbiotic interaction between the fungus and the plant. Currently, our knowledge on the molecular mechanism of SL transport is restricted to the Solanaceae family. In the Solanaceae family, SL translocation toward the rhizosphere occurs through the exodermis via hypodermal passage cells and involves a member of the G subfamily, of the ATP-binding cassette (ABC) membrane transporters. Most Fabaceae species, including those that are agriculturally important, have a different root anatomy compared to most angiosperm plants (i.e., lacking an exodermis). Thus, we have investigated how SL transport occurs in the model legume Medicago truncatula. Here, we show that overexpression of a SL transporter from petunia (PaPDR1) enhances AMF colonization rates in M. truncatula. This result demonstrates the importance of ABCG proteins for the translocation of orobanchol-type molecules to facilitate arbuscular mycorrhiza, regardless of root anatomy and phylogenetic relationships. Moreover, our research has led to the identification of Medicago ABCG59, a close homologue of Petunia PDR1, that exhibits root specific expression and is up-regulated by phosphate starvation as well as in the presence of rac-GR24, a synthetic SL. Its promoter is active in cortical cells, root tips, and the meristematic zone of nodules. The mtabcg59 loss-of-function mutant displayed a reduced level of mycorrhization compared to the WT plants but had no impact on the number of nodules after Sinorhizobium meliloti inoculation. The reduced mycorrhization indicates that less SLs are secreted by the mutant plants, which is in line with the observation that mtabcg59 exudates exhibit a reduced stimulatory effect on the germination of the parasitic plant Phelipanche ramosa compared to the corresponding wild type.
Keywords: ABC transporters, arbuscular mycorrhiza, exodermis, Medicago truncatula, strigolactones, symbioses
Introduction
Arbuscular mycorrhiza (AM) represents an ancient and widespread beneficial association, established between most terrestrial plants and filamentous fungi from the subphylum Glomeromycotina (Parniske, 2008; Spatafora et al., 2016). Arbuscular mycorrhizal fungi (AMF) improve plant nutrient acquisition, especially phosphate, depletion of which is a major factor limiting plant growth (Karandashov and Bucher, 2005). In return, plants supply the fungi with sugars (Doidy et al., 2012) and lipids (Jiang et al., 2017; Luginbuehl et al., 2017; Rich et al., 2017). This nutrient transfer between symbionts occurs via highly branched, fungus-derived structures called arbuscules, formed inside root cortical cells (Parniske, 2008; Luginbuehl and Oldroyd, 2017).
The establishment of AM is preceded by a signal exchange between partners that mediates reciprocal recognition, prior to the establishment of direct contact. One of the breakthroughs in the field of AM research was the discovery of strigolactones (SLs) as host-derived precolonization signals (Akiyama et al., 2005). SLs, released into the rhizosphere upon phosphate deficiency, stimulate fungal mitochondrial metabolism, and promote hyphal branching (Akiyama et al., 2005; Besserer et al., 2006). These signaling molecules also contribute to the transcriptional reprogramming of AMF (Tsuzuki et al., 2016) and induce the extrusion of chitin oligosaccharides, the so-called “myc factors”, responsible for the activation of nuclear Ca2+ spikes in the host root epidermis (Genre et al., 2013). In the rhizosphere, SLs are active at very low concentrations and are chemically instable. Thus, it has been postulated that SLs form a steep concentration gradient in the soil, indicating the proximity of a plant host and guide fungal hyphae to the root (Akiyama and Hayashi, 2006; Boyer et al., 2012; Nadal and Paszkowski, 2013; Oldroyd, 2013). Moreover, elevated concentrations of SLs near to the root surface may affect the initiation of hyphopodium formation. It has recently been shown that in rice this process is severely attenuated in the SL biosynthetic mutants (Kobae et al., 2018).
Following physical contact and fungal hyphopodium formation on the epidermis, internal root colonization begins. Having penetrated the epidermal cells, the hyphae encounters the hypodermis, the outermost cortical layer. In most angiosperm plants, cell walls within the hypodermis might be modified and form an apoplastic barrier called an exodermis, comprising cells with Casparian bands and suberin lamellae, as well as unsuberized ones. The suberized cells become impenetrable to fungi, therefore deeper cortex colonization occurs only through the suberin-free cells called hypodermal passage cells (HPCs) (Sharda and Koide, 2008). Interestingly, Petunia hybrida pleiotropic drug resistance 1 (PhPDR1), the first characterized SL exporter, is specifically expressed in HPCs. Loss-of-function mutations of this gene negatively affects the formation of arbuscular mycorrhiza (Kretzschmar et al., 2012). An orthologue of PhPDR1 from Petunia axillaris exhibits a cell-type-specific polar localization in HPCs. PaPDR1 is localized in the outer-lateral plasma membrane of HPCs, in accordance with its proposed function in releasing SL toward the rhizosphere and in creating SL gradients guiding AM fungi to preferred access sites (Sasse et al., 2015). PhPDR1 and PaPDR1 are members of the ancient and ubiquitous ATP-binding cassette (ABC) protein family, and belong to the ABCG subfamily, which is the most abundant in plants and appears to have a great impact on plant adaptations to terrestrial life (Kang et al., 2011; Hwang et al., 2016).
Currently, our knowledge about the molecular mechanisms of SL transport is restricted to the Solanaceae family (Kretzschmar et al., 2012; Xie et al., 2015). This is a surprising constraint as SLs influence other types of plant-rhizosphere interactions, both negative and positive. Their presence in the rhizosphere promotes the germination of seeds of parasitic plants of the genera Striga and Orobanche (Matusova et al., 2005). Recently, a role of SLs in the legume - rhizobium symbiosis (LRS) has also been described. While the function of SL in AM symbiosis is well established, their role in LRS is still emerging. It has been shown, that mutations in the SL biosynthetic genes (CCD7 and CDD8) lead to a decrease in the number of nodules in Pisum sativum (pea) and Lotus japonicus (Foo and Davies, 2011; Liu et al., 2013). Further analysis in pea revealed that SLs promote the formation of infection threads by influencing the bacterial partner (McAdam et al., 2017). The question about SL transporters seems to be especially intriguing in the case of the leguminous plants (Fabaceae), since it was reported that unlike most other angiosperm plants, they do not form an exodermis (Hose et al., 2001; Seago and Fernando, 2013). Moreover, Fabaceae such as Medicago sativa (alfalfa) and Glycine max (soybean) are crops exhibiting an important impact on agriculture and they have evolved several strategies to survive in low nutrient soils (Stagnari et al., 2017). Nitrogen and phosphorus are limiting nutrients in most natural soils. A large part of phosphorus in the soil is not available to plants due to its tendency to interact with cations and it is often present as the sparingly soluble rock phosphate. This non-renewable resource is being mined at an increasing rate to meet the demand for artificial fertilizers (Cordell et al., 2009). In the case of phosphorus, in many ecosystems and, in particular in acidic soils, the response of plants to sparingly available phosphate results mainly in the association with mycorrhizal fungi (Kluber et al., 2012). In this regard it is also worth considering how the root anatomical traits in leguminous plants may influence SL dispersal as well as affect SL coordinated symbiotic processes.
In this study, by functional overexpression of the petunia’s PaPDR1 in Medicago truncatula, we have shown that SL transport mechanism, based on ABCG transporters, is conserved between species. Additionally, by analyzing transcriptomic and phylogenetic data, we have identified a full-size ATP-binding cassette (ABC) transporter, namely MtABCG59. This protein is important for establishing mycorrhizal symbiosis in Medicago by acting as an SL exporter localized in the root cortex.
Materials and Methods
Plant Materials, Treatments, and Growth Conditions
Medicago truncatula seeds were acquired from A. Kondorosi of CNRS, Gif-sur-Yvette, France. Seeds of the mtabcg59 Tnt1 retrotransposon insertion mutant lines (NF15758 and NF12356) were obtained from the Noble Research Institute. The presence of Tnt1 was confirmed using gene-specific primers and primers annealing to the Tnt1 border (Supplementary Table 1).
The Medicago seeds were scarified with 96% sulfuric acid (10 min), stratified (4°C in the dark for 3 days), and were then germinated on solid, half-strength Murashige and Skoog medium (M5524, Sigma-Aldrich). For all experiments plants were grown in a greenhouse (one plant per 0.5 L pot) or growth chamber (in vitro experiments) under a 16 h light/8 h dark (24/22°C) regime, at 50%–60% relative humidity.
For the mycorrhization trials the Medicago plants (R-108 and mtabcg59 mutants) were grown for three weeks (one plant per 0.5 L pot) in clay granules (Oil Dri US-Special, Damolin) with mycorrhizal inoculum (2 g per pot) (Swiss Collection of Arbuscular Mycorrhizal Fungi (SAF)). Clay granules were supplemented once a week with half-strength Hoagland solution. Afterwards the mycorrhizal roots were collected for quantification of AM colonization or AM-marker genes (MtBCP1 and MtPT4) expression analyses. For the experiments five and four biological replicates were performed respectively, 3 independent plants were pooled per replicate.
For the rac-GR24 treatments, seven‐day‐old M. truncatula seedlings were transferred onto fresh half-strength MS medium supplemented with 10 µM rac-GR24 (Chiralix) dissolved in acetone or the equal volume of acetone (mock). Roots were collected 24 h after transfer and immediately frozen. Roots from three plants were pooled for each condition and four independent replicates were performed. The collected material was used for qRT-PCR analyses.
For the phosphorous starvation experiments seven‐day‐old M. truncatula seedlings were planted in a mixture of perlite and vermiculite (2:5). Plants were watered twice a week with water and custom prepared fertilizer solution optimized for legumes (Supplementary Table 2) or fertilizer without KH2PO4 and NH4H2PO4. Roots were collected four weeks after they were planted and immediately frozen. Roots from two plants were pooled for each condition and three independent replicates were performed. The collected material was used for qRT-PCR analyses.
Quantification of Mycorrhization
To determine mycorrhizal colonization, a modified gridline intersect method (Mcgonigle et al., 1990) was applied. Mycorrhizal roots were washed in tap water and boiled for 10 min in 10% KOH. Then the roots were rinsed with ddH2O, boiled for 10 min in a solution containing 5% black ink and 5% acetic acid. After washing and de-staining the roots were stored in 5% acetic acid at 4°C. The roots were then spread evenly on a petri dish with 4 mm2 grid and the percentage of positive events (roots with intra-radical structures) were calculated from the total amount of segments analyzed. Five independent biological experiments (i.e., 5 pools of 3 plants each) were performed.
Phelipanche ramosa Germination Assay
Rhizotron chambers for assessing the germination of P. ramosa seeds were assembled from square petri dishes. Three and one holes were respectively carved on the bottom and on the top of the petri dish to allow for water uptake and plant growth. Ten days after germination on half-strength MS agarose medium, the WT and mtabcg59 Medicago seedlings were moved onto a round wet filter paper placed in the rhizotron. Then, 150 seeds per filter paper were positioned on the roots and off the roots (seeds placed 1 to 3 mm aside). Another wet filter paper was positioned on the top, creating a paper/roots + seeds/paper sandwich. The rhizotron chambers were then filled up with clay (Oil-Dri), sealed with Micropore Tape (3M) and positioned in a try with water on its bottom to keep the rhizotron moist but not water-logged. Five days later, the rhizotron chambers were opened and germination of P. ramosa was quantified. At this stage of development, no significant differences were present for the biomass and surface of seedling roots between the two genotypes (R-108 and mtabcg59). No germination was detected without the presence of a plant root. Around 4,000 micro-pilar openings and the eventual presence of radicle were quantified with the help of a binocular microscope.
Nodulation Assay
For in vitro nodulation experiments M. truncatula seedlings (R-108, mtabcg59) were grown vertically on the surface of the modified solid Fahraeus (-N) medium, pH 6.5. Sinorhizobium meliloti strain 1021 was grown overnight at 28°C in liquid Bergensen’s Modified Medium (BMM) containing streptomycin (500 µg/L), and then diluted with sterile BMM to an OD600 = 0.1. Six-day-old M. truncatula seedling roots were flood-inoculated with 200 µl of bacterial culture spread along the root. Nodule numbers were counted 14 and 21 days after inoculation. There were five replicates (N = 5) with nine plants (n = 9) each, for each genotype.
Quantitative RT-PCR Analyses
Total RNA of the collected samples were extracted using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). The cDNA was then synthesized with an Omniscript Reverse Transcription (RT) Kit (Qiagen) or Moloney Murine Leukemia Virus Reverse Transcriptase Reverse Transcriptase (M-MLV RT) (Promega, Madison, Wisconsin, USA). Quantitative PCR analyses for MtABCG43, MtABCG44, MtABCG59, MtCCD7, and MtCCD8 were performed in a CFX Connect Real-Time System machine (BioRad, Hercules, CA, USA) using iTaq Universal SYBR Green supermix (BioRad) with at least three biological replicates each with three technical repeats. Quantitative PCR analyses for the AM-marker genes (MtBCP1 and MtPT4) were performed in a 7500 Fast Real-Time PCR System (Applied Biosystems, Waltham, Massachusetts, USA) using SYBR Green PCR Master Mix (Applied Biosystems) with four biological replicates each with two technical repeats. Expression levels were normalized to Mtactin and calculated with the ΔΔCt method. For primer sequences, see Supplementary Table 1.
Genetic Constructs and Plant Transformation
A genomic DNA fragment (1,752 bp) corresponding to the promoter region of MtABCG59 was amplified with KOD Hot Start DNA Polymerase (Novagen, Madison, Wisconsin, USA), and cloned into the following vectors: (i) pPR97, carrying the β-glucuronidase (gusA) reporter gene (Szabados et al., 1995), by restriction-ligation using the restriction sites for EcoRI and HindIII; and (ii) pPLV04_v2, carrying a GFP reporter gene tagged with a nuclear localization signal (NLS), by ligation-independent cloning (De Rybel et al., 2011). The genomic DNA fragment (7,500 bp) of MtABCG59 was used for subcellular localization and was amplified with KOD Hot Start DNA Polymerase (Novagen) and cloned into the Gateway entry vectors pENTR4 (Invitrogen, Carlsbed, CA, USA) by restriction-ligation, using restriction sites for NotI and SalI. The resulting entry clone was used for recombination with a destination vector pMDC43 (Curtis and Grossniklaus, 2003) using Gateway LR Clonase II Enzyme Mix (Invitrogen). The gPaPDR1 cloning procedure was described in (Kretzschmar et al., 2012). For primers, see Supplementary Table 1.
Plant Transformation
Transgenic roots carrying ProMtABCG59:GUS or ProMtABCG59:NLS-GFP constructs were obtained from M. truncatula after inoculation of a severed radicle with Agrobacterium rhizogenes Arqua1 (at least 30 composite plants for each construct). Stably transformed M. truncatula plants carrying ProMtABCG59:GUS (6 independent F0 lines) or Pro35S:GFP-PaPDR1 (3 independent F0 lines) were obtained by Agrobacterium tumefaciens AGL1-mediated transformation using leaf explants and regeneration via somatic embryogenesis. The M. truncatula transformation was performed according to the protocol described in the Medicago handbook (http://www.noble.org/medicagohandbook). Arabidopsis protoplasts that transiently expressed Pro35S:GFP-MtABCG59 or free GFP were obtained using PEG-mediated direct gene transfer, as described previously (Smolarkiewicz et al., 2014).
Microscopic Observation and Staining Procedures
Transgenic hairy roots and stably transformed M. truncatula (plants of the F1 generation) carrying GUS reporter constructs were stained using 5-bromo-4-chloro-3-indolyl-β-D-glucuronide, as previously described (Gallagher, 1992) and visualized by light microscopy.
Microscopic observations of the transgenic roots carrying the ProMtABCG59:NLS-GFP reporter construct and Arabidopsis protoplasts transiently expressing Pro35S:GFP-MtABCG59 or free GFP were performed using laser scanning confocal microscopy (Leica TCS SP5).
For suberin staining, 6-week-old M. truncatula and Nicotiana tabacum roots (10 of each species) were incubated in 0.01% solution of Fluorol Yellow 088 (Santa Cruz Biotechnology, Dallas, Texas, USA) at 70°C for 30 min. Free-hand root sections (from the regions located at least 3 cm above the root tip) were observed under UV-light with fluorescence microscopy (Leica DMI 4000B, Wetzlar, Germany).
For arbuscules visualization, mycorrhizal roots were fixed in 50% ethanol for 3 h, cleared in 20% (w/v) KOH for 3 days at room temperature (21°C–23°C), followed by 0.1 M HCl for 1 h. Then roots were washed in PBS and incubated in WGA-AlexaFluor 488 staining solution (1 µg/ml in PBS and 0.01% v/v Tween 20) in the dark for 16 h. The roots were imaged with laser scanning confocal microscopy (Leica TCS SP5).
Results
Medicago truncatula Root Anatomy
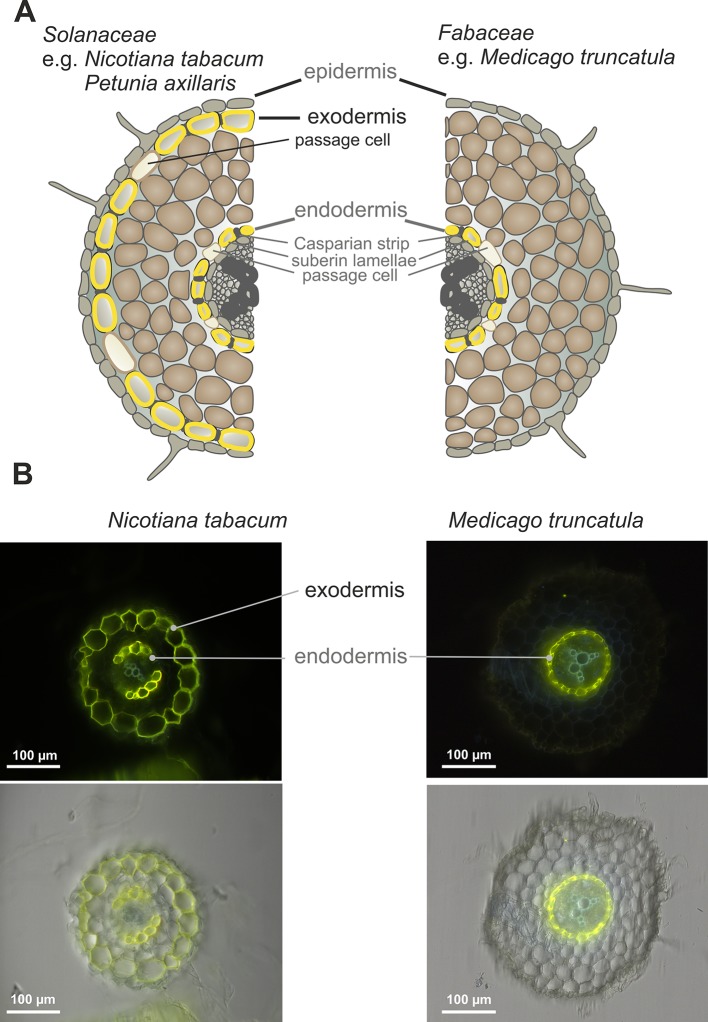
It has been shown that the secretion of SLs in petunia, belonging to the Solanaceae family, occurs through the hypodermal passage cells (HPCs), located within the exodermis (Kretzschmar et al., 2012). Notably, most Fabaceae species have a different root anatomy and do not form an exodermis (Perumalla et al., 1990; Seago and Fernando, 2013). This is also true for the model legume Medicago truncatula, as shown by the Fluorol Yellow 088 fluorescent staining of the suberin lamella in cross-sections of its roots. In contrast to a plant such as Nicotiana tabacum, whose roots possess two layers, an exo- and endodermis, which are stained by this dye, Medicago possesses only one layer corresponding to the endodermis (Figure 1). The lack of exodermis in Medicago roots has been observed regardless of the plant age or the region of the root examined (Supplementary Figure 1).
Figure 1.
Root anatomical structure. (A) Scheme of Nicotiana tabacum (Solanaceae) and Medicago truncatula (Fabaceae) root layers. (B) Deposition of suberin lamellae in roots of 6-week-old N. tabacum and M. truncatula (representative images from 10 roots). The cross-sections of the roots comes from the regions located 3 cm above the root tip. Fluoral yellow 088 fluorescence (upper panel), fluorescence images overlayed with corresponding bright-field images (lower panel).
In petunia, SLs have been shown to be translocated and secreted into the rhizosphere by the membrane transporter PDR1, which belongs to the full-size ABCG (PDR) subfamily of ABC transporters (Kretzschmar et al., 2012). Interestingly, overexpression of the PaPDR1 (the only up to date cloned SL transporter), in Medicago enhanced AMF colonization rates, and did not affect the arbuscule structures in Medicago (Supplementary Figure 2). This observation underlines the importance of ABCG proteins for the translocation of orobanchol-type molecules to facilitate AM symbiotic interactions, despite the differences in the anatomical features of the roots between the plant families. The M. truncatula transgenic plants overexpressing PaPDR1 also displayed several SL-related phenotypes in the shoot, demonstrating the importance of SL redistribution in Medicago aboveground organs by this transporter. PaPDR1 OE plants possessed: (i) modified leaf shape with deeper margin serrations, (ii) enhanced internode elongation, and iii) petioles of a reduced length (Supplementary Figure 3).
Identification of a Potential Strigolactone Transporter in Medicago truncatula
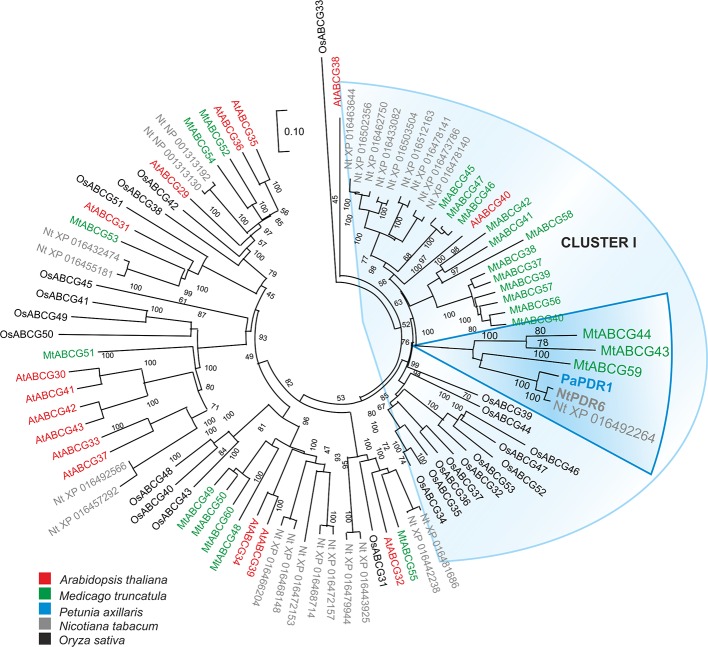
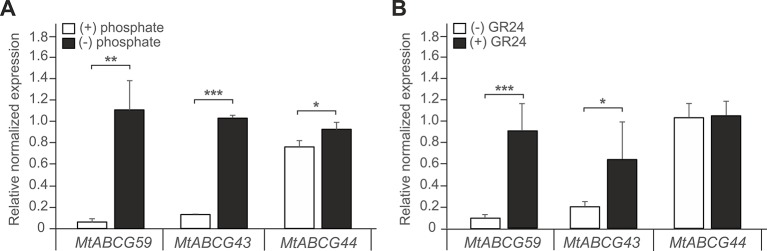
We have previously identified several ABCG transporters in the M. truncatula genome (Banasiak and Jasinski, 2014). The phylogenetic analysis showed that three ABCG proteins: MtABCG59 (MtPDR23), MtABCG44 (MtPDR8), and MtABCG43 (MtPDR7), clustered with the previously characterized SL transporter PaPDR1 from P. axillaris (Figure 2, Supplementary Table 3). They share 74%, 71%, and 68% amino acid sequence identity with PaPDR1, respectively. Since nutrient starvation stimulates AM formation as well as biosynthesis and secretion of SLs in Petunia (Liu et al., 2018b), we have tested the response of those three genes to phosphorous deficiency. The highest level of gene induction was observed for MtABCG59 and MtABCG43 exhibiting 18 and 17-fold changes, respectively (Figure 3A). Furthermore, exogenous applications of a synthetic SLs analogue – rac-GR24 onto the roots, increased the mRNA accumulation of MtABCG59 (9-fold) and again at a lower extent MtABCG43 (3-fold change) (Figure 3B). Previously it was reported that the expression of several ABCG transporters could be substrate inducible (Jasinski et al., 2001; Biala et al., 2017; Pawela et al., 2019). The MtABCG44 was only slightly induced (1.2-fold change) under phosphate limiting condition, and rac-GR24 had no impact on its expression (Figure 3). Based on this expression profile, and phylogenetic relationships, we selected MtABCG59 for further and more comprehensive functional analyses.
Figure 2.
Phylogenetic analysis of full-size ABCG (Pleiotropic drug resistance - PDR) proteins in various plants. Neighbor-joining phylogenetic tree (bootstraps: 1000) was conducted using MEGA X software (Kumar et al., 2018) based on the amino acid sequences generated after multiple sequence alignments with MUSCLE. The highlighted PaPDR1 and NtPDR6, as well as their homologues from Medicago truncatula belong to Cluster I (Crouzet et al., 2013).
Figure 3.
Expression profiles of Medicago truncatula ABCG59, ABCG43, and ABCG44. (A, B) Quantitative Real-Time PCR expression analysis of MtABCG59, MtABCG43, and MtABCG44 in roots under phosphate-limiting conditions (A) or treated with the synthetic strigolactone analogue rac-GR24 (B). The transcript levels were normalized to the actin gene from Medicago truncatula. Data represent the mean ± SD of three and four independent biological experiments, respectively, and three technical repeats. Significant differences from the control plants determined by Student's t-test are indicated as follows: *P < 0.05, **P < 0.01, ***P < 0.001.
Gene Expression Pattern of MtABCG59 in M. truncatula and Subcellular Localization of the Corresponding Protein
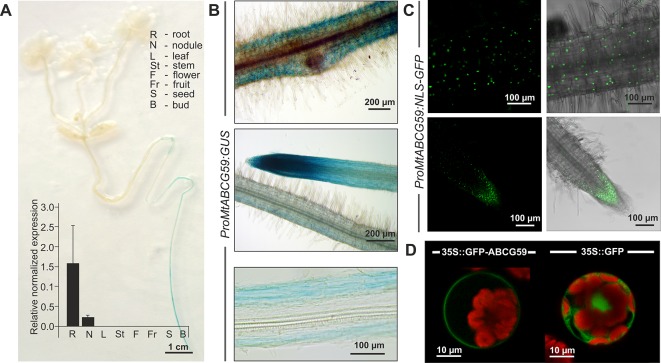
The qPCR analyses revealed that MtABCG59 mRNA accumulates in roots and root-derived nodules (Figure 4A). To further explore the spatial expression patterns of MtABCG59, we generated M. truncatula transgenic plants expressing the β-glucuronidase (GUS) reporter gene under the control of the native MtABCG59 promoter (ProMtABCG59:GUS). Histochemical staining of the 2-week-old transgenic seedlings, as well as qPCR, showed that MtABCG59 expression is root specific (Figure 4A). Moreover, microscopic observations of transgenic Medicago hairy roots expressing ProMtABCG59:GUS and ProMtABCG59 fused with a nuclear-localized version of green fluorescent protein (ProMtABCG59:NLS-GFP) revealed that the root tip, where SLs are biosynthesized (van Zeijl et al., 2015a), and the cortical cells, are the main sites of MtABCG59 promoter activity (Figures 4B, C).
Figure 4.
Expression pattern and subcellular localization of MtABCG59. (A) GUS staining of 2-week-old transgenic Medicago truncatula expressing ProMtABCG59:GUS and quantitative PCR expression analysis of MtABCG59 in different M. truncatula organs (three-month-old plants) revealed the MtABCG59 transcript accumulation exclusively in the roots and nodules. The transcript levels were normalized to the actin gene from M. truncatula. Data represent the mean ± SD of three independent biological experiments and three technical repeats. (B) X-Gluc staining of ProMtABCG59:GUS reporter line in root cortex (upper panel) and in root meristem (middle panel), X-Gluc staining of root cross-sections (bottom panel). (C) Fluorescence signal of ProMtABCG59:NLS-GFP reporter line in root cortex (upper panel) and in root meristem (bottom panel). (D) Arabidopsis mesophyll protoplast expressing the fusion gene Pro35S:GFP-MtABCG59. The GFP signal was observed in the plasma membrane (left panel). Control Arabidopsis protoplast expressing free cytoplasmic GFP (right panel). The red color represents chlorophyll autofluorescence.
To determine the subcellular localization of MtABCG59, we fused GFP to the C-terminal end of MtABCG59 and transiently expressed the Pro35S:GFP-MtABCG59 recombinant gene in Arabidopsis leaf mesophyll protoplasts. Visualization of GFP fluorescence was performed using confocal microscopy. The GFP signal from the fusion protein was detected in the plasma membrane. In control protoplasts, transformed with Pro35S:GFP, the fluorescence signal was observed in the cytoplasm (Figure 4D).
Mycorrhizal Phenotype of mtabcg59 Mutant Lines
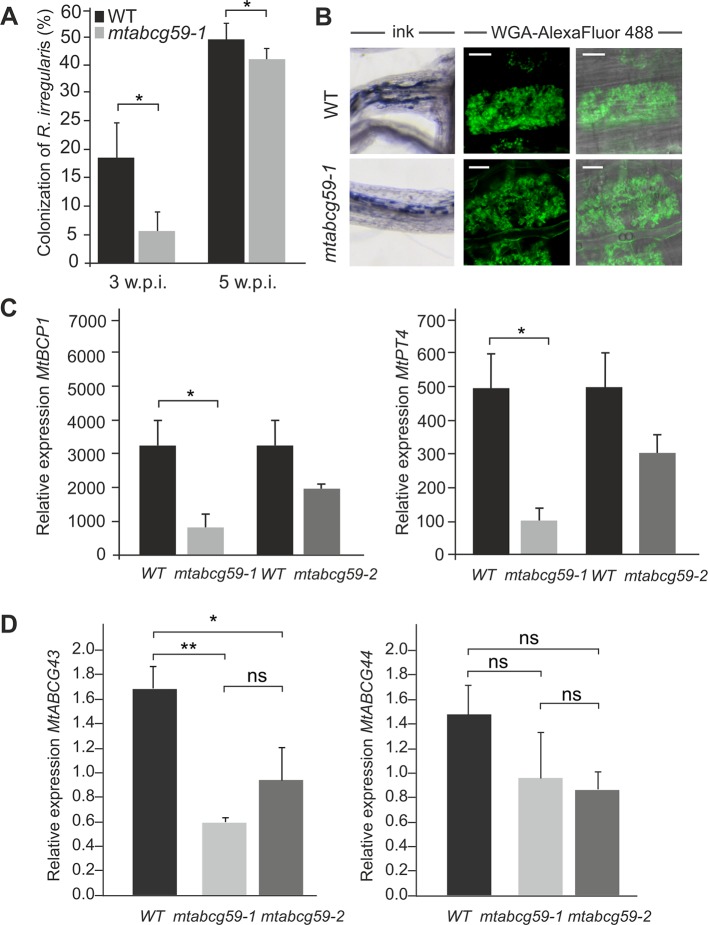
To test the hypothesis that MtABCG59 can influence the AM symbiosis we identified the tobacco retrotransposon (Tnt1) inserted line (NF15758) for MtABCG59 and assessed with the grid-line intersect method (see M&M) the colonization rate of AMF in the roots derived from the WT and mtabcg59-1. The percent of root length colonization by Rhizophagus irregularis was estimated 3 and 5 weeks after inoculation with the fungal spores. The experiments showed that the mtabcg59-1 mutant exhibits 70% and 11% lower levels of colonization than the WT, respectively (Figure 5A). The arbuscule structures appeared unaffected in mtabcg59-1, suggesting that MtABCG59 is required for initiation of AM, rather than proper development of arbuscules (Figure 5B). The reduction of AM formation in mtabcg59-1 as well as in the second mutant line (mtabcg59-2, NF12356) 3wpi was further demonstrated with the usage of AM-related marker genes MtBCP1 and MtPT4 (Figure 5C). MtBCP1, encoding a blue copper protein, is expressed in the epidermal and cortical cells containing fungal structures and in adjacent noncolonized cortical cells (Hohnjec et al., 2005; Hogekamp and Kuster, 2013; Zhang et al., 2015), while MtPT4 encodes an arbuscule-specific phosphate transporter (Javot et al., 2007; Hogekamp and Kuster, 2013). Real-time PCR analyses revealed that AM marker genes had lower expression levels in the mtabcg59-1 and mtabcg59-2, reflecting the lower mycorrhization rate in these plants compared to the WT (Figure 5C).
Figure 5.
Phenotypic characterization of the mtabcg59 mutant in mycorrhization contexts. (A) Mycorrhizal colonization of Medicago truncatula control (WT) and mtabcg59-1 mutant roots 3 and 5 weeks after inoculation with Rhizophagus irregularis. The percentage of the root length colonized by the mycorrhizal fungi is shown. Data represent the means ± SE of five independent biological experiments (i.e. 5 pools of 3 plants each). Significant differences from the control plants were determined by Student's t-test and are indicated by: *P < 0.05. (B) Arbuscules formed in the WT and mtabcg59-1 mutant were morphologically similar. Ink-staining of fungal structures (left panel), WGA-AlexaFluor 488 staining of arbuscules (right panel), scale bar, 10 µm. (C) Transcript accumulation of AM-related marker genes MtBCP1 (left panel) and MtPT4 (right panel) in mtabcg59 mutants and control (WT) roots measured by quantitative real-time PCR. The data represents the mean ± SD of four independent biological experiments and two technical repeats. Significant differences from the control plants were determined by Student's t-test and are indicated by: * P < 0.05. (D) Relative expression of MtABCG43 and MtABCG44 in WT and mtabcg59 mutant plants. Values represent the mean of three biological replicates ± SD. Significant differences in genes expression between WT and mtabcg59 mutants were determined by an ANOVA test and Tukey's multiple comparison test, and are as follows: ns, not significant; *P < 0.05, **P < 0.01.
Taking into the consideration the possible functional redundancy, we have investigated if MtABCG59 paralogues, namely MtABCG43 and MtABCG44 might take over the function of MtACG59. Interestingly, we have observed that upon phosphate deficient condition MtABCG43 is downregulated in both mtabcg59 mutants background. Concomitantly, the expression of MtABCG44 did not show significant differences (Figure 5D).
Root Exudates From mtabcg59 Exhibit a Reduced Ability to Promote Germination of Phelipanche ramosa Seeds
To investigate whether MtABCG59 is involved in SL secretion, mtabcg59-1 root exudates were assessed for their ability to stimulate parasitic weed Phelipanche ramosa seed germination. Compared to the WT, a significant decrease in germination capacity was observed for P. ramosa seeds exposed to root exudates from mtabcg59-1. Interestingly, this difference was present only with seeds placed 1 to 3 mm aside, no difference in germination rates were observed with seeds placed directly on the root surface (Figure 6, Supplementary Figure 4).
Figure 6.
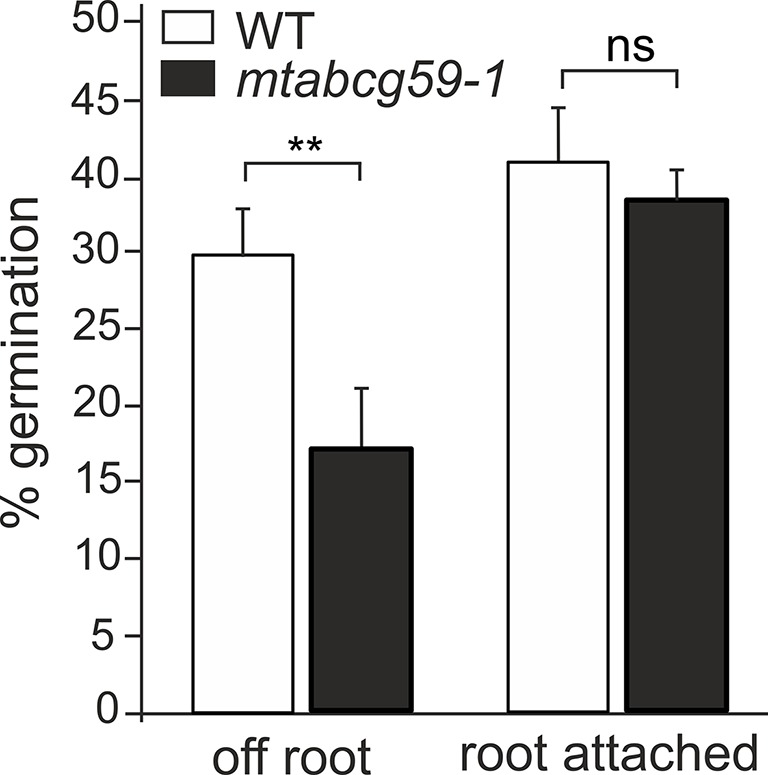
Germination rates of Phelipanche ramosa seeds exposed to WT and mtabcg59-1 exudates. Root attached (seeds placed directly in root surface); off root (seeds placed 1 to 3 mm aside). The data represent the mean ± SE of six to seven independent biological experiments (approx. 150 seeds screened for each biological replicate). Significant differences from the control plants were determined by Student's t-test and are indicated by: ns, not significant; **P < 0.005.
Lack of MtABCG59 Disturbs Strigolactone Biosynthesis
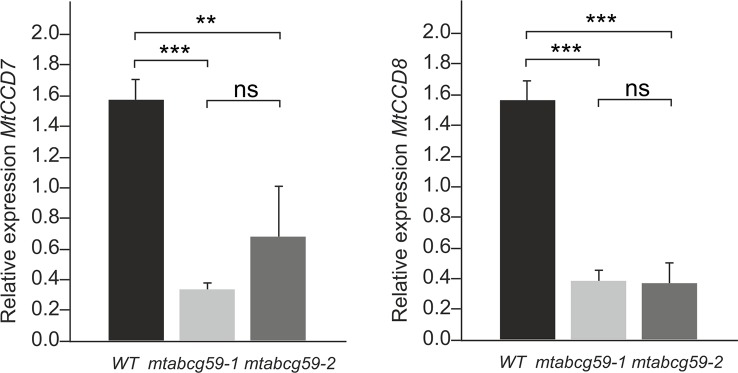
The presence of MtABCG59 transcripts in root tips suggests that MtABCG59 might be involved in the export of SL from the site of biosynthesis. Since SL content can influence the transcript level of SL biosynthetic genes, CCD7 and CCD8 (Foo et al., 2005; Hayward et al., 2009) we have examined their expression in WT and mtabcg59 mutant lines. We have noticed a decrease in mRNA accumulation of CCD7 and CCD8 in both mtabcg59 mutants background. The observed effect might be due to excessive accumulation of SL in the cells that can trigger a negative feedback regulation mechanism in SL biosynthesis (Figure 7).
Figure 7.
MtABCG59 affects the SL-biosynthetic pathway. Relative expression of SL-biosynthesis gene (MtCCD7 and MtCCD8) in WT and mtabcg59 mutant plants. Values represent the mean of three biological replicates ± SD. Significant differences in genes expression between WT and mtabcg59 mutants were determined by an ANOVA test and Tukey's multiple comparison test, and are as follows: ns, not significant; **P < 0.01, ***P < 0.001.
Lack of MtABCG59 Does Not Affect Nodulation
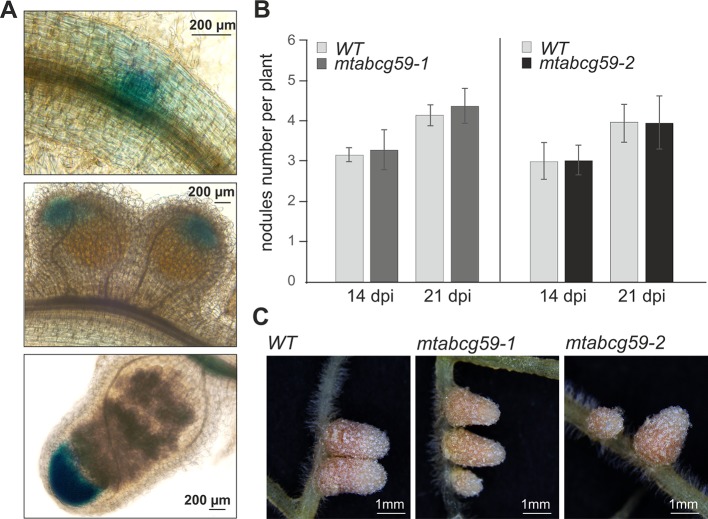
Recent studies have found that SLs positively affect interactions between legumes and nitrogen-fixing bacteria (McAdam et al., 2017). Moreover, we have shown that the expression pattern of MtABCG59 in nodule primordia as well as in the meristematic zone of developing and mature nodules (Figure 8A) is similar to that reported for genes encoding SL biosynthetic enzymes (van Zeijl et al., 2015a). Despite this, mtabcg59 plants inoculated with Sinorhizobium meliloti had a comparable number of root nodules to the WT, and did not differ morphologically (Figures 8B, C), indicating that MtABCG59 is either not required for nodulation or that a redundant, yet unknown protein compensates its loss of function.
Figure 8.
Phenotypic characterization of mtabcg59 in nodulation context. (A) Expression of ProMtABCG59:GUS in nodules. (B) Average nodule number per plant in WT and mtabcg59 plants. Six-day-old seedlings were inoculated with Sinorhizobium meliloti and grown on modified Fahraeus (-N) medium. At 14- and 21-days post-inoculation (dpi), the nodules were quantified. The data represent the mean ± SD of (N = 5, n = 9), per line. (C) WT and mtabcg59 nodules morphology.
Discussion
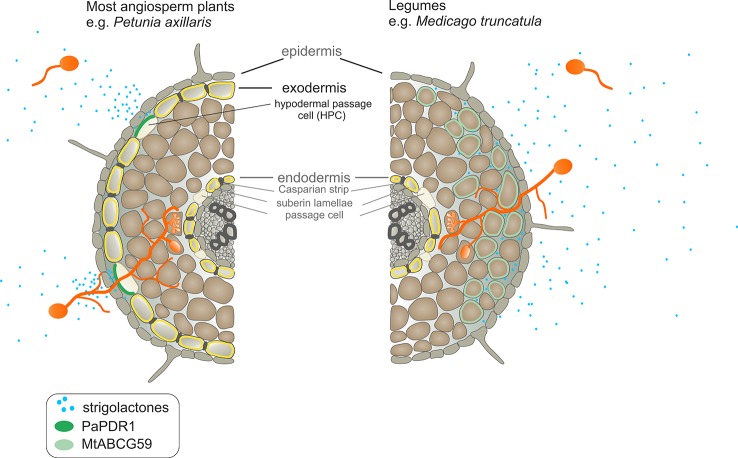
The only SL transporter characterized in detail is the PaPDR1 from Petunia axillaris, and it operates in a highly cell specific manner, being active inter alia in the hypodermal passage cells (HPCs) located within exoderemis (Sasse et al., 2015). We have shown that the roots of the model legume plant Medicago truncatula, related to the important forage crop Medicago sativa, do not possess an exodermis and HPCs (Figure 1). Overexpression of PaPDR1 in Medicago revealed that despite these anatomical differences, this transporter is functional in Medicago and its presence promotes AM. The increased mycorrhization in Medicago overexpressing PaPDR1, indicates that SL transport mechanisms based on ABCG transporters, exhibit similar properties between species. Notably, canonical orobanchol-type molecules are the most abundant SLs exuded into the rhizosphere by both petunia and M. truncatula (Kretzschmar et al., 2012; Tokunaga et al., 2015). Thus, the PaPDR1 OE strategy appears to be useful for ameliorating mycorrhization rate in Fabaceae. It could improve phosphate acquisition and plant biomass production as it was reported for petunia (Liu et al., 2018a; Liu et al., 2018b).
It has been demonstrated that AMF hyphal branching can be initiated by the SL present in the root exudates of legume plants (Akiyama et al., 2005; Steinkellner et al., 2007). However, transporters responsible for SL release into the rhizosphere from this plant family have not been discovered. To identify a putative SL transporter in M. truncatula, we combined the phylogenetic information with expression profile analyses. Among the tested genes, MtABCG59, besides sharing the highest level of sequence identity with the previously described SL exporter PaPDR1 from petunia (Figure 2), it had the highest induction rates in roots in phosphate limited conditions and after rac-GR24 treatment (Figure 3). We have shown that in Medicago roots, the expression of the putative SL transporter MtABCG59 occurs mainly in the outer cortical cells (Figure 4B). The observed expression pattern is compatible with the proposed function of MtABCG59 in SL secretion from the roots toward the soil to induce hyphal branching of AM fungi and guide them to host roots. As MtABCG59 is also expressed in Medicago colonized roots (Supplementary Figure 5), it cannot be excluded that it also participates in the SL translocation to the apoplast, guiding intraradical hyphal branching within the root cortex. Additionally, the presence of MtABCG59 transcripts in root tips suggests that MtABCG59 might be involved in the export of SL from the site of biosynthesis towards the upper root, as has been previously revealed for PaPDR1 (Sasse et al., 2015). This hypothesis can be supported by the observation that SL-biosynthesis genes MtCCD7 and MtCCD8 are downregulated in mtabcg59 mutant lines compared to WT (Figure 7, Supplementary Figure 6). Previously, it was proposed that excessive accumulation of SL in the cells can trigger a negative feedback regulation mechanism in SL biosynthesis and affect mRNA accumulation of CCD7 and CCD8 (Foo et al., 2005; Hayward et al., 2009; Liu et al., 2018b).
Phenotypic characterization of the mtabcg59-1 mutant showed that its loss of function negatively influences AM formation, especially, at the early stage of interaction. Previously, it was proposed that altered presymbiotic AM fungal growth could result in delayed colonization, which is apparent at early time points (Montero et al., 2019). That is also what we observed for mtabcg59 mutant (Figure 5). Likewise, as in other reported SL-related mutants (e.g., biosynthetic, perception, and transport) (Kretzschmar et al., 2012; Lanfranco et al., 2018), the arbuscular structure in colonized root cells was not altered in mtabcg59 plants (Figure 5). This observation supports the notion that SLs influence the fungal morphology mainly in the rhizosphere but do not affect its intercellular accommodation and development. SLs ex planta stimulate the seed germination of parasitic weeds such as witchweeds (Striga spp.) and broomrapes (Orobanche and Phelipanche spp.) (Cook et al., 1966; Yoneyama et al., 2018). To verify whether the decrease of AM formation in mtabcg59-1 is indeed caused by a reduction in the secretion of SLs, we applied a parasitic seed germination bioassay. This approach allows for the indirect but sensitive determination of the relative amounts of SLs exuded by roots (Guillotin et al., 2017). The SL levels secreted by the mtabcg59-1 were assessed by the germination of Phelipanche ramosa seeds that exhibit a broad host range, including M. truncatula (Fernandez-Aparicio et al., 2009). Our experiments revealed that root exudates derived from mtabcg59-1 possess lower germination-inducing activity than exudates from the WT. Interestingly, the effect observed concerned only seeds which were not directly in touch with the roots. Further studies are needed to test whether this lack of difference in seed germination in seeds that are in touch with the root is due to a second, less active transporter or due to diffusion of the hydrophobic SLs. In Medicago apart from MtABCG59, expression of MtABCG43 is induced by phosphorous deficiency and rac-GR24 (Figure 3). Further investigation is required to demonstrate that MtABCG43 participates in SL secretion, however, functional redundancy is possible. Nevertheless, since the expression of MtABCG43 decreased in mtabcg59 mutants compared to WT (Figure 5D), the diffusion of SL through the exodermis-free cortex appears to be more likely. In the case of mtabcg59 mutant, the concentration of diffusion-derived SL on the root surface is still sufficient for parasitic seed germination. Taken together, our findings suggest that MtABCG59 is involved in SL extrusion to the soil to maintain an SL concentration sufficiently high in the proximity of the roots, allowing to induce hyphal branching and guiding the fungi towards the host root (Figure 9).
Figure 9.
Comparison of strigolactone (SL) exudation into the soil and SL-dependent guidance of arbuscular mycorrhizal fungi to the host root between Petunia axillaris and Medicago truncatula. In P. axillaris, SLs are secreted into the rhizosphere by PaPDR1 that is localized in the hypodermal passage cells (HPCs) within the exodermis. A steep concentration gradient of SLs, created by PaPDR1, guides arbuscular mycorrhizal fungi (AMF) to access penetration sites – i.e. unsuberized HPCs. In the M. truncatula, due to a lack of exodermis, SLs could passively enter the rhizosphere because they do not encounter the apoplastic diffusion barrier. However, we suggest that to achieve the full extent of AMF colonization, the active export of SLs, mediated by MtABCG59, is required. Its action ensures sufficiently high concentrations of SLs around the roots to attract AM fungi, which together with the characteristic root anatomical traits in M. truncatula, enables the colonization of the whole root surface.
In addition to arbuscular mycorrhiza, SLs are implicated in the legume-rhizobium symbiosis (LRS) (Lopez-Raez et al., 2017). A characteristic feature of this type of interaction is the formation of root nodules, which are the place of nitrogen fixation and nutrient exchange between partners. Phenotypic analyses of SL biosynthetic mutants of Pisum sativum, Lotus japonicus, and Glycine max demonstrated the positive role of SLs in LRS establishment (Foo and Davies, 2011; Liu et al., 2013; Rehman et al., 2018). In the case of M. truncatula, exogenous application of rac-GR24 had either a positive or negative effect on this process, depending on the SL concentrations used (De Cuyper et al., 2015). We could not detect a significant difference in the root nodule structures or the number of root nodules between the mtabcg59 and WT despite the fact that MtABCG59 is expressed in the nodule meristem (Figure 8) and co-expressed with the SL biosynthetic genes (MtD27, MtCCD7, and MtCDD8) (van Zeijl et al., 2015a). Although SLs are biosynthesized in mature nodules, their role in this organ remains currently unknown. Pea SL-deficient mutants produced less nodules, but their size and ability to fix atmospheric nitrogen were comparable to those of WT plants. Therefore, it was postulated that SLs affect the LRS mainly in the early stages of the interaction. Recently, it has been shown that SLs promote infection thread formation influencing the bacterial partner (McAdam et al., 2017). Nevertheless, the function of the SL transporter in the nodule, like in the root, could be the removal of SLs from the site of biosynthesis to avoid SL accumulation in the meristem and/or deliver them to the place of perception. It cannot be excluded that a redundant transporter can take over the SL transport under LRS. Analysis of publicly available transcriptomic data showed that in Medicago expression of another PaPDR1 homologue, MtABCG43, is induced after Nod factor treatment (van Zeijl et al., 2015b; Jardinaud et al., 2016) and its mRNA was found to accumulate at the highest level in nodules (Supplementary Figure 7). It is also possible that in M. truncatula SLs do not play an essential role in determining the nodule number. This assumption can be supported by the observation that the silencing of SL biosynthetic gene MtD27 did not affect the efficiency of nodulation in Medicago (van Zeijl et al., 2015a).
SL acts not only as a signaling molecule in the rhizosphere but also as a phytohormone that adjusts the overall plant architecture to nutrient availability. They control inter alia internode elongation (de Saint Germain et al., 2013) and shoot branching by inhibiting axillary bud outgrowth (Gomez-Roldan et al., 2008; Umehara et al., 2008) as well as increasing root surface, and thus, influencing the lateral root and root hair development (Kapulnik et al., 2011; Ruyter-Spira et al., 2011). It is worth noting that PhPDR1 and NtPDR6, in addition to the roots, are expressed in aerial tissues, especially in nodes. Their mutation and/or silencing enhance shoot branching, giving plants a bushy phenotype (Kretzschmar et al., 2012; Xie et al., 2015). Notably, the PaPDR-overexpressing Medicago showed several SL-related phenotypes in the shoot, such as deeper leaf margin serrations and increased internode length. Analogous phenotypes were previously reported for Medicago after the exogenous application of rac-GR24 at the primary shoot apex. Conversely, SL deficient (ccd7, ccd8) and insensitive (d14) mutants displayed shallower serrations as well as reduced shoot elongation than the WT (Lauressergues et al., 2015). We did not detect expression of MtABCG59 in aboveground tissues (Figure 4A) or observe alterations in the shoot architecture of the mtabcg59 mutant (Supplementary Figure 8). Hence, we propose that in Medicago one or more additional SL transporter(s) can operate to control aerial developmental traits. One of the candidates might be MtABCG44 (71% amino acid identity to MtABCG59) that exhibits a broad expression pattern (Supplementary Figure 7).
Conclusion
In conclusion, in the present study we have identified and characterized MtABCG59, an ABC class G transporter involved in SL export towards the soil that positively affects arbuscular mycorrhiza formation in Medicago truncatula. We propose that its primary function is releasing SLs into the rhizosphere to attract AMF during the presymbiotic stage of interactions. The root specific expression of MtABCG59 and the lack of mtabcg59 aboveground phenotypes suggests that in M. truncatula, SL distribution can be mediated by more than one transporter. Moreover, with the overexpression of the petunia SL transporter PaPDR1 in M. truncatula, we have demonstrated the importance of ABCG proteins for the translocation of orobanchol-type molecules, regardless of root anatomy and phylogenetic relationships.
Data Availability Statement
Sequence data from this article can be found in the GenBank database under the following accession numbers: MTR_3g107870 for MtABCG59, MTR_1g011640 for MtABCG43, MTR_1g011650 for MtABCG44, MTR_3g095530 for Mtactin, AFA43815 for PaPDR1, MTR_1g028600 for MtPT4, JQ292812 for PaPDR1.
Author Contributions
JB and LB conceived and designed the study. JB, LB, and NS performed the experiments. JB and EM secured funding for the project. JB, MJ, LB, and EM analyzed and interpreted the data. JB wrote the manuscript (initial draft preparation). MJ, LB, and EM wrote the manuscript (critical editing and review of the manuscript).
Funding
National Science Centre grant number: SONATA 2015/17/D/NZ3/03625. Swiss National Foundation grants number 31003A-152831 and 31003A-169546.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank I. Femiak and J. Matuszek for excellent technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00018/full#supplementary-material
Abbreviations
AM, arbuscular mycorrhiza; HPC, hypodermal passage cell; LRS, legume-rhizobium symbiosis; SL, strigolactone.
References
- Akiyama K., Hayashi H. (2006). Strigolactones: chemical signals for fungal symbionts and parasitic weeds in plant roots. Ann. Bot. 97 (6), 925–931. 10.1093/aob/mcl063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K., Matsuzaki K., Hayashi H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435 (7043), 824–827. 10.1038/nature03608 [DOI] [PubMed] [Google Scholar]
- Banasiak J., Jasinski M. (2014). Defence, Symbiosis and ABCG Transporters. Plant ABC Transporters 22, 163–184. 10.1007/978-3-319-06511-3_9 [DOI] [Google Scholar]
- Besserer A., Puech-Pages V., Kiefer P., Gomez-Roldan V., Jauneau A., Roy S., et al. (2006). Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PloS Biol. 4 (7), e226. 10.1371/journal.pbio.0040226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biala W., Banasiak J., Jarzyniak K., Pawela A., Jasinski M. (2017). Medicago truncatula ABCG10 is a transporter of 4-coumarate and liquiritigenin in the medicarpin biosynthetic pathway. J. Exp. Bot. 68 (12), 3231–3241. 10.1093/jxb/erx059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer F. D., de Saint Germain A., Pillot J. P., Pouvreau J. B., Chen V. X., Ramos S., et al. (2012). Structure-activity relationship studies of strigolactone-related molecules for branching inhibition in garden pea: molecule design for shoot branching. Plant Physiol. 159 (4), 1524–1544. 10.1104/pp.112.195826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook C. E., Whichard L. P., Turner B., Wall M. E., Egley G. H. (1966). Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154 (3753), 1189–1190. 10.1126/science.154.3753.1189 [DOI] [PubMed] [Google Scholar]
- Cordell D., Drangert J. O., White S. (2009). The story of phosphorus: global food security and food for thought. Global Environ. Change-Hum. Policy Dimensions 19 (2), 292–305. 10.1016/j.gloenvcha.2008.10.009 [DOI] [Google Scholar]
- Crouzet J., Roland J., Peeters E., Trombik T., Ducos E., Nader J., et al. (2013). NtPDR1, a plasma membrane ABC transporter from Nicotiana tabacum, is involved in diterpene transport. Plant Mol. Biol. 82 (1-2), 181–192. 10.1007/s11103-013-0053-0 [DOI] [PubMed] [Google Scholar]
- Curtis M. D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 (2), 462–469. 10.1104/pp.103.027979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cuyper C., Fromentin J., Yocgo R. E., De Keyser A., Guillotin B., Kunert K., et al. (2015). From lateral root density to nodule number, the strigolactone analogue GR24 shapes the root architecture of Medicago truncatula. J. Exp. Bot. 66 (13), 4091. 10.1093/jxb/erv227 [DOI] [PubMed] [Google Scholar]
- De Rybel B., van den Berg W., Lokerse A., Liao C. Y., van Mourik H., Moller B., et al. (2011). A versatile set of ligation-independent cloning vectors for functional studies in plants. Plant Physiol. 156 (3), 1292–1299. 10.1104/pp.111.177337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A., Ligerot Y., Dun E. A., Pillot J. P., Ross J. J., Beveridge C. A., et al. (2013). Strigolactones stimulate internode elongation independently of gibberellins. Plant Physiol. 163 (2), 1012–1025. 10.1104/pp.113.220541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doidy J., Grace E., Kuhn C., Simon-Plas F., Casieri L., Wipf D. (2012). Sugar transporters in plants and in their interactions with fungi. Trends Plant Sci. 17 (7), 413–422. 10.1016/j.tplants.2012.03.009 [DOI] [PubMed] [Google Scholar]
- Fernandez-Aparicio M., Flores F., Rubiales D. (2009). Recognition of root exudates by seeds of broomrape (Orobanche and Phelipanche) species. Ann. Bot. 103 (3), 423–431. 10.1093/aob/mcn236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E., Davies N. W. (2011). Strigolactones promote nodulation in pea. Planta 234 (5), 1073–1081. 10.1007/s00425-011-1516-7 [DOI] [PubMed] [Google Scholar]
- Foo E., Buillier E., Goussot M., Foucher F., Rameau C., Beveridge C. A. (2005). The branching gene RAMOSUS1 mediates interactions among two novel signals 464 and auxin in pea. Plant Cell 17 (2), 464–474. 10.1105/tpc.104.026716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher S. R. (1992). GUS protocols: using the GUS gene as a reporter of gene expression (San Diego: Academic Press; ). [Google Scholar]
- Genre A., Chabaud M., Balzergue C., Puech-Pages V., Novero M., Rey T., et al. (2013). Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 198 (1), 190–202. 10.1111/nph.12146 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V., Fermas S., Brewer P. B., Puech-Pages V., Dun E. A., Pillot J. P., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455 (7210), 189–194. 10.1038/nature07271 [DOI] [PubMed] [Google Scholar]
- Guillotin B., Etemadi M., Audran C., Bouzayen M., Becard G., Combier J. P. (2017). Sl-IAA27 regulates strigolactone biosynthesis and mycorrhization in tomato (var. MicroTom). New Phytol. 213 (3), 1124–1132. 10.1111/nph.14246 [DOI] [PubMed] [Google Scholar]
- Hayward A., Stirnberg P., Beveridge C., Leyser O. (2009). Interactions between Auxin and Strigolactone in Shoot Branching Control. Plant Physiol. 151 (1), 400–412. 10.1104/pp.109.137646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogekamp C., Kuster H. (2013). A roadmap of cell-type specific gene expression during sequential stages of the arbuscular mycorrhiza symbiosis. BMC Genomics 14, 306. 10.1186/1471-2164-14-306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohnjec N., Vieweg M. F., Puhler A., Becker A., Kuster H. (2005). Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol. 137 (4), 1283–1301. 10.1104/pp.104.056572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hose E., Clarkson D. T., Steudle E., Schreiber L., Hartung W. (2001). The exodermis: a variable apoplastic barrier. J. Exp. Bot. 52 (365), 2245–2264. 10.1093/jexbot/52.365.2245 [DOI] [PubMed] [Google Scholar]
- Hwang J. U., Song W. Y., Hong D., Ko D., Yamaoka Y., Jang S., et al. (2016). Plant ABC transporters enable many unique aspects of a terrestrial plant’s lifestyle. Mol. Plant 9 (3), 338–355. 10.1016/j.molp.2016.02.003 [DOI] [PubMed] [Google Scholar]
- Jardinaud M. F., Boivin S., Rodde N., Catrice O., Kisiala A., Lepage A., et al. (2016). A laser dissection-rnaseq analysis highlights the activation of cytokinin pathways by nod factors in the medicago truncatula root epidermis. Plant Physiol. 171 (3), 2256–2276. 10.1104/pp.16.00711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski M., Stukkens Y., Degand H., Purnelle B., Marchand-Brynaert J., Boutry M. (2001). A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. Plant Cell 13 (5), 1095–1107. 10.1105/tpc.13.5.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H., Penmetsa R. V., Terzaghi N., Cook D. R., Harrison M. J. (2007). A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 104 (5), 1720–1725. 10.1073/pnas.0608136104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Wang W., Xie Q., Liu N., Liu L., Wang D., et al. (2017). Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356 (6343), 1172–1175. 10.1126/science.aam9970 [DOI] [PubMed] [Google Scholar]
- Kang J., Park J., Choi H., Burla B., Kretzschmar T., Lee Y., et al. (2011). Plant ABC Transporters. Arabidopsis Book 9, e0153. 10.1199/tab.0153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik Y., Resnick N., Mayzlish-Gati E., Kaplan Y., Wininger S., Hershenhorn J., et al. (2011). Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis. J. Exp. Bot. 62 (8), 2915–2924. 10.1093/jxb/erq464 [DOI] [PubMed] [Google Scholar]
- Karandashov V., Bucher M. (2005). Symbiotic phosphate transport in arbuscular mycorrhizas. Trends Plant Sci. 10 (1), 22–29. 10.1016/j.tplants.2004.12.003 [DOI] [PubMed] [Google Scholar]
- Kluber L. A., Carrino-Kyker S. R., Coyle K. P., DeForest J. L., Hewins C. R., Shaw A. N., et al. (2012). Mycorrhizal response to experimental pH and P manipulation in acidic hardwood forests. PloS One 7 (11), e48946. 10.1371/journal.pone.0048946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobae Y., Kameoka H., Sugimura Y., Saito K., Ohtomo R., Fujiwara T., et al. (2018). Strigolactone biosynthesis genes of rice are required for the punctual entry of arbuscular mycorrhizal fungi into the roots. Plant Cell Physiol. 59 (3), 544–553. 10.1093/pcp/pcy001 [DOI] [PubMed] [Google Scholar]
- Kretzschmar T., Kohlen W., Sasse J., Borghi L., Schlegel M., Bachelier J. B., et al. (2012). A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483 (7389), 341–344. 10.1038/nature10873 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfranco L., Fiorilli V., Venice F., Bonfante P. (2018). Strigolactones cross the kingdoms: plants, fungi, and bacteria in the arbuscular mycorrhizal symbiosis. J. Exp. Bot. 69 (9), 2175–2188. 10.1093/jxb/erx432 [DOI] [PubMed] [Google Scholar]
- Lauressergues D., Andre O., Peng J., Wen J., Chen R., Ratet P., et al. (2015). Strigolactones contribute to shoot elongation and to the formation of leaf margin serrations in Medicago truncatula R108. J. Exp. Bot. 66 (5), 1237–1244. 10.1093/jxb/eru471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Novero M., Charnikhova T., Ferrandino A., Schubert A., Ruyter-Spira C., et al. (2013). Carotenoid cleavage dioxygenase 7 modulates plant growth, reproduction, senescence, and determinate nodulation in the model legume Lotus japonicus. J. Exp. Bot. 64 (7), 1967–1981. 10.1093/jxb/ert056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Bollier D., Gubeli C., Peter N., Arnold P., Egli M., et al. (2018. a). Simulated microgravity and the antagonistic influence of strigolactone on plant nutrient uptake in low nutrient conditions. NPJ Microgravity 4, 20. 10.1038/s41526-018-0054-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Pfeifer J., de Brito Francisco R., Emonet A., Stirnemann M., Gubeli C., et al. (2018. b). Changes in the allocation of endogenous strigolactone improve plant biomass production on phosphate-poor soils. New Phytol. 217 (2), 784–798. 10.1111/nph.14847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Raez J. A., Shirasu K., Foo E. (2017). Strigolactones in plant interactions with beneficial and detrimental organisms: the Yin and Yang. Trends Plant Sci. 22 (6), 527–537. 10.1016/j.tplants.2017.03.011 [DOI] [PubMed] [Google Scholar]
- Luginbuehl L. H., Oldroyd G. E. D. (2017). Understanding the arbuscule at the heart of endomycorrhizal symbioses in plants. Curr. Biol. 27 (17), R952–R963. 10.1016/j.cub.2017.06.042 [DOI] [PubMed] [Google Scholar]
- Luginbuehl L. H., Menard G. N., Kurup S., Van Erp H., Radhakrishnan G. V., Breakspear A., et al. (2017). Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356 (6343), 1175–1178. 10.1126/science.aan0081 [DOI] [PubMed] [Google Scholar]
- Matusova R., Rani K., Verstappen F. W., Franssen M. C., Beale M. H., Bouwmeester H. J. (2005). The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiol. 139 (2), 920–934. 10.1104/pp.105.061382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam E. L., Hugill C., Fort S., Samain E., Cottaz S., Davies N. W., et al. (2017). Determining the site of action of strigolactones during nodulation. Plant Physiol. 175 (1), 529–542. 10.1104/pp.17.00741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcgonigle T. P., Miller M. H., Evans D. G., Fairchild G. L., Swan J. A. (1990). A new method which gives an objective-measure of colonization of roots by vesicular arbuscular mycorrhizal fungi. New Phytol. 115 (3), 495–501. 10.1111/j.1469-8137.1990.tb00476.x [DOI] [PubMed] [Google Scholar]
- Montero H., Choi J., Paszkowski U. (2019). Arbuscular mycorrhizal phenotyping: the dos and don’ts. New Phytol. 221 (3), 1182–1186. 10.1111/nph.15489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal M., Paszkowski U. (2013). Polyphony in the rhizosphere: presymbiotic communication in arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 16 (4), 473–479. 10.1016/j.pbi.2013.06.005 [DOI] [PubMed] [Google Scholar]
- Oldroyd G. E. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11 (4), 252–263. 10.1038/nrmicro2990 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6 (10), 763–775. 10.1038/nrmicro1987 [DOI] [PubMed] [Google Scholar]
- Pawela A., Banasiak J., Biala W., Martinoia E., Jasinski M. (2019). MtABCG20 is an ABA exporter influencing root morphology and seed germination of Medicago truncatula. Plant J. 98 (3), 511–523. 10.1111/tpj.14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumalla C. J., Peterson C. A., Enstone D. E. (1990). A survey of angiosperm species to detect hypodermal casparian bands.1. Roots with a uniseriate hypodermis and epidermis. Bot. J. Linn. Soc 103 (2), 93–112. 10.1111/j.1095-8339.1990.tb00176.x [DOI] [Google Scholar]
- Rehman N. U., Ali M., Ahmad M. Z., Liang G., Zhao J. (2018). Strigolactones promote rhizobia interaction and increase nodulation in soybean (Glycine max). Microb. Pathog. 114, 420–430. 10.1016/j.micpath.2017.11.049 [DOI] [PubMed] [Google Scholar]
- Rich M. K., Nouri E., Courty P. E., Reinhardt D. (2017). Diet of arbuscular mycorrhizal fungi: bread and butter? Trends Plant Sci. 22 (8), 652–660. 10.1016/j.tplants.2017.05.008 [DOI] [PubMed] [Google Scholar]
- Ruyter-Spira C., Kohlen W., Charnikhova T., van Zeijl A., van Bezouwen L., de Ruijter N., et al. (2011). Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol. 155 (2), 721–734. 10.1104/pp.110.166645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse J., Simon S., Gubeli C., Liu G. W., Cheng X., Friml J., et al. (2015). Asymmetric localizations of the ABC transporter PaPDR1 trace paths of directional strigolactone transport. Curr. Biol. 25 (5), 647–655. 10.1016/j.cub.2015.01.015 [DOI] [PubMed] [Google Scholar]
- Seago J. L., Jr., Fernando D. D. (2013). Anatomical aspects of angiosperm root evolution. Ann. Bot. 112 (2), 223–238. 10.1093/aob/mcs266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharda J. N., Koide R. T. (2008). Can hypodermal passage cell distribution limit root penetration by mycorrhizal fungi? New Phytol. 180 (3), 696–701. 10.1111/j.1469-8137.2008.02600.x [DOI] [PubMed] [Google Scholar]
- Smolarkiewicz M., Skrzypczak T., Michalak M., Lesniewicz K., Walker J. R., Ingram G., et al. (2014). Gamma-secretase subunits associate in intracellular membrane compartments in Arabidopsis thaliana. J. Exp. Bot. 65 (12), 3015–3027. 10.1093/jxb/eru147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora J. W., Chang Y., Benny G. L., Lazarus K., Smith M. E., Berbee M. L., et al. (2016). A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108 (5), 1028–1046. 10.3852/16-042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagnari F., Maggio A., Galieni A., Pisante M. (2017). Multiple benefits of legumes for agriculture sustainability: an overview. Chem. Biol. Technol. Agric. 4. 10.1186/s40538-016-0085-1. UNSP 2. [DOI] [Google Scholar]
- Steinkellner S., Lendzemo V., Langer I., Schweiger P., Khaosaad T., Toussaint J. P., et al. (2007). Flavonoids and strigolactones in root exudates as signals in symbiotic and pathogenic plant-fungus interactions. Molecules 12 (7), 1290–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados L., Charrier B., Kondorosi A., Debruijn F. J., Ratet P. (1995). New plant promoter and enhancer testing vectors. Mol. Breed. 1 (4), 419–423. 10.1007/Bf01248419 [DOI] [Google Scholar]
- Tokunaga T., Hayashi H., Akiyama K. (2015). Medicaol, a strigolactone identified as a putative didehydro-orobanchol isomer, from Medicago truncatula. Phytochemistry 111, 91–97. 10.1016/j.phytochem.2014.12.024 [DOI] [PubMed] [Google Scholar]
- Tsuzuki S., Handa Y., Takeda N., Kawaguchi M. (2016). Strigolactone-induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus rhizophagus irregularis. Mol. Plant Microbe Interact. 29 (4), 277–286. 10.1094/MPMI-10-15-0234-R [DOI] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., et al. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455 (7210), 195–200. 10.1038/nature07272 [DOI] [PubMed] [Google Scholar]
- van Zeijl A., Liu W., Xiao T. T., Kohlen W., Yang W. C., Bisseling T., et al. (2015. a). The strigolactone biosynthesis gene DWARF27 is co-opted in rhizobium symbiosis. BMC Plant Biol. 15, 260. 10.1186/s12870-015-0651-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zeijl A., Op den Camp R. H., Deinum E. E., Charnikhova T., Franssen H., Op den Camp H. J., et al. (2015. b). Rhizobium lipo-chitooligosaccharide signaling triggers accumulation of cytokinins in medicago truncatula roots. Mol. Plant 8 (8), 1213–1226. 10.1016/j.molp.2015.03.010 [DOI] [PubMed] [Google Scholar]
- Xie X., Wang G., Yang L., Cheng T., Gao J., Wu Y., et al. (2015). Cloning and characterization of a novel Nicotiana tabacum ABC transporter involved in shoot branching. Physiol. Plant 153 (2), 299–306. 10.1111/ppl.12267 [DOI] [PubMed] [Google Scholar]
- Yoneyama K., Xie X., Yoneyama K., Kisugi T., Nomura T., Nakatani Y., et al. (2018). Which are the major players, canonical or non-canonical strigolactones? J. Exp. Bot. 69 (9), 2231–2239. 10.1093/jxb/ery090 [DOI] [PubMed] [Google Scholar]
- Zhang X., Pumplin N., Ivanov S., Harrison M. J. (2015). EXO70I is required for development of a sub-domain of the periarbuscular membrane during arbuscular mycorrhizal symbiosis. Curr. Biol. 25 (16), 2189–2195. 10.1016/j.cub.2015.06.075 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data from this article can be found in the GenBank database under the following accession numbers: MTR_3g107870 for MtABCG59, MTR_1g011640 for MtABCG43, MTR_1g011650 for MtABCG44, MTR_3g095530 for Mtactin, AFA43815 for PaPDR1, MTR_1g028600 for MtPT4, JQ292812 for PaPDR1.