Abstract
Chlamydia spp. are a group of obligate intracellular pathogens causing a number of diseases in animals and humans. Avian chlamydiosis (AC), caused by Chlamydia psittaci (C. psittaci) as well as new emerging C. avium, C. gallinacea and C. ibidis, have been described in nearly 500 avian species worldwidely. The Crested Ibis (Nipponia nippon) is a world endangered avian species with limited population and vulnerable for various infections. To get a better understanding of the prevalence of Chlamydia spp. in the endangered Crested Ibis, faecal samples were collected and analysed. The results confirmed that 20.20% (20/99) of the faecal samples were positive for Chlamydiaceae and were identified as C. ibidis with co-existence of C. psittaci in one of the 20 positive samples. In addition, ompA sequence of C. psittaci obtained in this study was classified into the provisional genotype Matt116, while that of C. ibidis showed high genetic diversity, sharing only 77% identity with C. ibidis reference strain 10-1398/6. We report for the first time the presence of C. ibidis and C. psittaci in the Crested Ibis, which may indicate a potential threat to the endangered birds and should be aware of the future protection practice.
Key words: Chlamydia ibidis, Chlamydia psittaci, Crested Ibis (Nipponia nippon)
Introduction
Chlamydia spp. are a group of obligate intracellular bacteria that cause a number of important diseases in animals and humans. Avian chlamydiosis (AC), caused by Chlamydia psittaci (C. psittaci), has been shown to occur in nearly 500 avian species worldwidely, including domestic, companion and wild birds [1]. Recently, new emerging avian chlamydial pathogens, such as C. avium, C. gallinacea, C. ibidis (Candidatus taxon) and C. buteonis have been described [2–4]. Birds infection with Chlamydia spp. usually show respiratory, ocular and enteric symptoms occasionally with fatal outcome, but asymptomatic, latent infections are also common. The clinical signs vary greatly in severity and depend on the species and age of the bird, as well as the causative strain involved [1]. Shedding of Chlamydia spp. through respiratory and intestinal routes occurs intermittently by infected birds, which represent a reservoir of infection [5]. Notably, Chlamydia species hosted by birds may present risks of zoonotic infections to humans. Several reports have shown the prevalence and diversity of Chlamydia spp. in domestic poultry and pet birds in China. A nationwide survey of Chlamydia spp. in domestic birds including chickens, ducks, geese and pigeons has shown that C. gallinacea and C. psittaci presented in all four avian species investigated, and C. gallinacea is the endemic chlamydial species in chickens, whereas C. psittaci dominates only in pigeons [6]. Investigation and genotyping of Chlamydia spp. in pigeons in Northern China showed that the total prevalence was 20.4% (215/963) and was identified as genotype B of C. psittaci [7]. Recently, an outbreak of Chlamydia-related egg production declines in domestic duck farms has been reported due to the infection of C. psittaci (genotype A) in Southeast of China [8]. In addition, this pathogen has also been detected and isolated occasionally from pet birds such as parrots [9]. These reports suggest domestic and companion birds might be a neglected reservoir of chlamydial pathogens which may threaten the poultry production and present risks to public health in China. However, there is no data about the prevalence of Chlamydia spp. in wild birds, which may also be reservoirs for poultry and human infections.
The Crested Ibis (Nipponia nippon) is a rare and precious species in the world and is red-listed as ‘Endangered’ by the International Union for Conservation of Nature. This bird was thought to be extinct in the wild until the last seven individuals were rediscovered in China in 1981 [10]. Since then, great efforts have been made on the protection of Crested Ibises. The total number of Crested Ibis has now increased to over 3000. Most of the Crested Ibis inhabit in areas of Qinling Mountains in Shaanxi province, China, while a certain number of the birds are in Sado Island in Japan, which have been introduced from China [11]. However, recovery of the Crested Ibis population is still difficult due to small population size, low genetic diversity as well as habitat loss, winter starvation and persecution [12, 13]. In addition, diseases and undetermined fitness also pose a threat to the Crested Ibis [14]. The wide distribution and broad-host infection of avian chlamydial pathogens may also exhibit a potential healthy risk to the endangered Crested Ibis. To get a better understanding of the prevalence of Chlamydia spp. in Crested Ibis, we analysed faecal samples of Crested Ibis collected in a breeding centre in Shaanxi, China.
Materials and methods
The faecal samples were collected from the Crested Ibis in Shaanxi Rare Wildlife Rescue and Breeding Research Center, Louguantai, a Qinling Mountain area in Xi'an, Shaanxi province, China. There were 206 of the captive-bred Crested Ibis living in this area during the sampling time. The living area (about 3600 m2) of Crested Ibis was segregated by nets to avoid the escape of the birds and also to keep away other animals which may threat them. In this Center, the living area was divided into 44 cages. The birds were fed and given water by specialized breeders of the Center every day. When the birds were routinely fed, a clean plastic sheet was put on the floor nearby the feeding site in each cage. The plastic sheet was removed after the Crested Ibis finished their meal and get away. Only fresh faecal samples were collected from the sheet. Faecal samples were collected only once from each cage. A total of 99 faecal samples were random collected from this population. A formal ethical approval is not required for this kind of study and sampling was performed by a specialized breeder of the Crested Ibis during routine feeding activities. At the time of sampling, all the birds showed no evident signs of disease. The faeces were transported on ice to the laboratory and stored at −80 °C until use.
DNA extraction from faeces was performed using the commercial QIAamp DNA Stool Mini Kit (Qiagen) following the manufacturer's protocol. Extracted DNA from faeces was initially screened by using a Chlamydiaceae-specific real-time polymerase chain reaction (PCR) targeting the 23S rRNA gene fragment [15]. A plasmid containing a fragment of 23S rRNA gene of C. abortus was used as a positive control. All Chlamydiaceae-positive DNA were retested with genus-specific real-time PCR assays to identify avian Chlamydia species including C. psittaci [16], C. avium [17], C. gallinacea [17] as well as C. abortus [16], which was also detected in birds [18]. For the detection of C. ibidis, a pair of primers and probe were designed according to the ompA sequences (GenBank access no: MN106768) obtained from this study. The oligo sequences were synthesized as a forward primer (5′-TCTTTGGGAATGTGGTTGG-3′), reverse (5′-GGTATCCTTCTCCGTCCCAG-3′) and the probe (5′-FAM-CCGCAGCGCAA-TTCAAGTGCACAMRA-3′). Positive DNA samples were further confirmed by amplification and sequencing of specific fragments of ompA, 16S rRNA and 16S rRNA-23S rRNA intergenic spacer together with 23S rRNA domain I (IGS-23S rRNA) as described previously [19–21]. PCR amplicons were sent to an external company (Shanghai Sangon Biotech, China) for sequencing. The obtained sequences were deposited in the GenBank database.
The data were analysed using MEGA 5.05 software [22]. Amplicons were subjected to BLAST analysis against the GenBank database (NCBI) to identify related entries and aligned with a panel of Chlamydia reference strains. To assess the phylogenetic relationship between Chlamydia spp. and the tested samples, phylogenetic trees for 16S rRNA (1358 bp) and IGS-23S rRNA (990 bp) as well as ompA (940 bp) were constructed by the neighbour-joining method with 1000 replicates' bootstrap using the Maximum Composite Likelihood model with MEGA 5.05.
Results and discussion
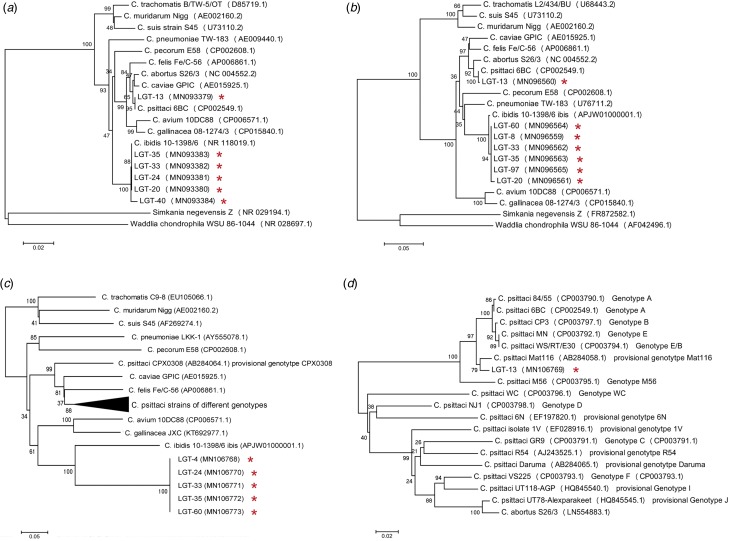
The results of real-time PCR assay showed 20 of the 99 collected faecal samples were positive for Chlamydiaceae giving a positive rate of 20.20%. One of the 20 positive DNA samples was identified as C. psittaci by the species-specific real-time PCR, while no C. avium, C. gallinacea or C. abortus was detected. To further identification and confirmation of the Chlamydia species in the faeces of Crested Ibis, all Chlamydiaceae-positive DNA were subjected to amplification and sequencing for the specific fragments of 16S rRNA, IGS-23S rRNA and ompA genes. Out of the 20 positive samples, six amplicons for 16S rRNA (MN093379-MN093384), seven for IGS-23S rRNA (MN096559-MN096565) and six for ompA (MN106768-MN106773) were successfully acquired and sequenced. BLAST analysis against the GenBank database led to the identification of C. ibidis and confirmation of C. psittaci existence in the faeces of Crested Ibis. Based on the ompA sequences of C. ibidis obtained in this study, a pair of primers and probe for real-time PCR detection were designed and the Chlamydiaceae-positive DNA was retested. The results showed that all the 20 positive DNA samples were identified as C. ibidis. Furthermore, dendrograms were constructed on the basis of the obtained 16S rRNA, IGS-23S rRNA and ompA gene fragments aligning with Chlamydia species reference sequences available in GenBank and altogether showed a similar topology. All sequences obtained in this study were assigned to either C. psittaci or C. ibidis within the Chlamydiaceae family (Fig. 1). These results suggested that the Crested Ibis was a host of this novel Candidatus Chlamydia species with C. psittaci co-existence occasionally.
Fig. 1.
Phylogenetic tree based on 16S rRNA gene fragment (1356 bp) (a), 16S-23S intergenic spacer and full length of 23S rRNA domain I fragment (923 bp) (b) and ompA gene fragment (720 bp) of all Chlamydiaceae members (c) and C. psittaci strains of different ompA genotypes (d). Representative sequences of established Chlamydiaceae species as well as the sequences obtained from this study (GenBank accession number was marked by red asterisks) were included. S. negevensis strain Z and Waddlia chondrophila strain WSU 86-1044 were used as an outgroup. Based on these alignments, phylogenetic trees were constructed by the neighbour-joining method using the Maximum Composite Likelihood model with MEGA 5.05.
The ompA gene, coding the major outer membrane protein (MOMP), is one of the sources of diversity among chlamydial genomes in some species including C. psittaci [23, 24]. Genotyping using ompA sequencing was employed commonly to detection and identification of C. psittaci strains. Based on the sequences of ompA, nine classical genotypes were described in C. psittaci (A to F, E/B, M56 and WC), along with a number of provisional genotypes (CPX0308, YP84, R54, Matt116, 6N, 1V, I and J) which were untypable so far [23, 24]. The ompA sequence of C. psittaci we obtained in this study was identical to a parrot isolated strain Matt116 (GenBank access: AB284058.1), which was classified into the provisional genotype Matt116 (Fig. 1d) [24]. It was evident that C. psittaci strains of each genotype were related to a certain range of hosts and pathogenicity [25]. Considering that the strain Matt116 was isolated among psittacine birds during an AC oubreak, C. psittaci strains classified into this genotype might be of high virulence. Therefore, the presence of this pathogen should be aware, especially in the endangered Crested Ibis, even though it was identified only in one sample.
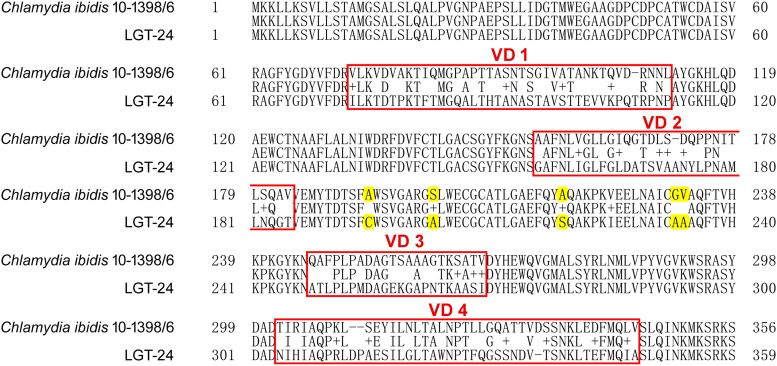
In this study, the data provided evidence of the presence of C. ibidis in the faecal samples of Crested Ibis. To further characterization of the prevalent strains, we analysed the obtained ompA sequences (MN106768, MN106770–MN106773). The results showed that these sequences matched totally, while shared only 77% identity with that of the C. ibidis reference strain 10-1398/6 (data not shown). As described above, ompA sequences variation was common among strains in a chlamydial species, which represented by the variations of amino acid sequences of MOMP coded by ompA genes [26]. To determine whether the nucleotide mismatches were random or with regularity between the obtained and reference sequences, the putative amino acid sequences of MOMP coded by ompA genes were also analysed. It presented four regions of amino acid sequences variations among the obtained and reference sequences, namely VD1–VD4, with a few amino acid substitutions between VD2 and VD3 (Fig. 2). These data were coincidence with the previous works that the amino acid sequences of MOMPs were interspersed by four short variable domains (VD1–VD4) whose sequences vary among different strains of genotypes or serotypes within the species of C. trachomatis or C. pecorum [27, 28]. In addition, when aligning the MOMP sequences of C. trachomatis strains belonging to different biovars (trachoma biovar, genital tract infection biovar and lymphogranuloma venereum biovar), it also presented four variation regions, which were almost the same at positions and in sizes as that shown in Figure 2 (data not shown). These data suggested that C. ibidis was a species of high genetic diversity. It also implied that the C. ibidis strains circulated in Crested Ibis presented a novel ompA genotype that differed from the reference strain isolated previously.
Fig. 2.
Alignment of the two amino acid sequences of MOMP from C. ibidis reference strain (10-1398/6) and a field prevalent strain (LGT-24) in the Crested ibis in China.
C. ibidis was first identified in feral African Sacred Ibises (Threskiornis aethiopicus) in western France and was confirmed as a new candidatus species of the family Chlamydiaceae [3]. However, this Chlamydia species has not yet been detected in any other birds or elsewhere since its discovery. In this study, we provided evidence for the presence of C. ibidis in the Crested Ibis. It seemed that C. ibidis was a species of highly genetic diversity, and the genetic diversity might be related to the hosts and/or geographic origin of the strains. Although no evidence showed the pathogenicity of C. ibidis in either feral African Sacred Ibises or Crested Ibises yet, due to the less isolated strains and few known infected hosts, intensive investigate is still needed to uncover the prevalence, transmission and pathogenicity of this new emerging Chlamydia species. However, C. psittaci, the major caused agent of AC, is a highly pathogenic species with various genotypes, knowing for its wide distribution and high prevalence in birds. The C. psittaci strains of different genotypes present relative host specificity, and most of them have been identified in cases of zoonotic transmission [25]. The harbouring and shedding of C. psittaci by the Crested Ibis may pose a potential risk to the whole population and as a source of environmental contamination.
Finally, there are some limitations to this study. The faecal samples tested in this work were collected randomly from the living area of the Crested Ibis. It is possible that some of the faecal samples may be from the same bird, and therefore the chlamydial prevalence of 20% may be overestimated in this population. On the other hand, the faecal sample could not be linked to a specific individual so that it remained unclear about the intrinsic characteristic related to the prevalence of this chlamydial species in this bird. Nevertheless, it was still evident that this endangered avian species harboured C. psittaci and C. ibidis. This study not only identified new a chlamydial host but could also help to gain deeper insights into the evolution of Chlamydiaceae. In addition, the presence of C. psittaci and C. ibidis in Crested Ibis might indicate a potential threat to the endangered birds, which should be aware in the future protection practice.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (31502081), Graduated Student Innovation Projects in Northwest Minzu University from Fundamental Research Funds for the Central Universities (Yxm2019150) and National Key R&D program of China (2017YFD0500905).
Conflict of interest
The authors declare that there is no conflict of interest.
References
- 1.Kaleta EF and Taday EMA (2003) Avian host range of Chlamydophila Spp. based on isolation, antigen detection and serology. Avian Pathology 32, 435–462. [DOI] [PubMed] [Google Scholar]
- 2.Sachse K et al. (2015) Emendation of the family Chlamydiaceae: proposal of a single genus, Chlamydia, to include all currently recognized species. Systematic and Applied Microbiology 38, 99–103. [DOI] [PubMed] [Google Scholar]
- 3.Vorimore F et al. (2013) Isolation of a New Chlamydia species from the Feral Sacred Ibis (Threskiornis aethiopicus): Chlamydia ibidis. Plos One 8, e74823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laroucau K et al. (2019) Chlamydia buteonis, a new Chlamydia species isolated from a red-shouldered hawk. Systematic and Applied Microbiology 42, 125997. [DOI] [PubMed] [Google Scholar]
- 5.Harkinezhad T, Geens T and Vanrompay D (2009) Chlamydophila psittaci infections in birds: a review with emphasis on zoonotic consequences. Veterinary Microbiology 135, 68–77. [DOI] [PubMed] [Google Scholar]
- 6.Guo W et al. (2016) Chlamydia gallinacea, not C. psittaci, is the endemic chlamydial species in chicken (Gallus Gallus). Scientific Reports 6, 19638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X et al. (2018) Epidemiological investigation and genotype of chlamydia exposure in Pigeons in Three Provinces in Northern China. Vector-borne and Zoonotic Diseases 18, 181–184. [DOI] [PubMed] [Google Scholar]
- 8.Lin W et al. (2019) A parrot-type Chlamydia psittaci strain is in association with egg production drop in laying ducks. Transboundary and Emerging Diseases 66, 2002–2010. [DOI] [PubMed] [Google Scholar]
- 9.Wang C et al. (2019) Isolation and characterization of avian Chlamydia psittaci from Symptomatic Pet Birds in Southern Hunan, China. Avian Diseases 63, 31–37. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y (1981) Recovery of Japanese Crested Ibis in Qin-ling range. Acta Zoologica Sinica 27, 273–273, (in Chinese). [Google Scholar]
- 11.Urano K et al. (2013) Genetic diversity and structure in the Sado captive population of the Japanese crested ibis. Zoological Science 30, 432–438. [DOI] [PubMed] [Google Scholar]
- 12.Zhang B, Fang SG and Xi YM (2006) Major histocompatibility complex variation in the Endangered Crested Ibis Nipponia Nippon and implications for reintroduction. Biochemical Genetics 44, 110–120. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y et al. (2015) Decline of traditional rice farming constrains the recovery of the endangered Asian crested ibis (Nipponia Nippon). Ambio 44, 803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yongmei X et al. (2007) Prevalence of a septicemia disease in the crested ibis (Nipponia Nippon) in China. Avian Diseases 51, 614–617. [DOI] [PubMed] [Google Scholar]
- 15.Ehricht R et al. (2006) Optimized DNA microarray assay allows detection and genotyping of single PCR-amplifiable target copies. Molecular & Cellular Probes 20, 60–63. [DOI] [PubMed] [Google Scholar]
- 16.Pantchev A et al. (2010) Detection of all Chlamydophila and Chlamydia spp. of veterinary interest using species-specific real-time PCR assays. Comparative Immunology, Microbiology and Infectious Diseases 33, 473–484. [DOI] [PubMed] [Google Scholar]
- 17.Aleksandar Z et al. (2013) A real-time PCR assay for the detection of atypical strains of Chlamydiaceae from pigeons. Plos One 8, e58741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szymańska-Czerwińska M et al. (2017) Dissemination and genetic diversity of chlamydial agents in Polish wildfowl: isolation and molecular characterisation of avian Chlamydia abortus strains. Plos One 12, e0174599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett KD, Bush RM and Andersen AA (1999) Emended description of the order Chlamydiales, proposal of Parachlamydiaceae Fam. Nov. and Simkaniaceae Fam. Nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. International Journal of Systematic Bacteriology 49, 415–440. [DOI] [PubMed] [Google Scholar]
- 20.Pudjiatmoko et al. (1997) Phylogenetic analysis of the genus Chlamydia Based on 16S rRNA gene sequences. International Journal of Systematic Bacteriology 47, 425–431. [DOI] [PubMed] [Google Scholar]
- 21.Denamur E et al. (1991) Restriction pattern of the major outer-membrane protein gene provides evidence for a homogeneous invasive group among ruminant isolates of Chlamydia psittaci. Journal of General Microbiology 137, 2525–2530. [DOI] [PubMed] [Google Scholar]
- 22.Tamura K et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madani SA and Peighambari SM (2013) PCR-based diagnosis, molecular characterization and detection of atypical strains of avian Chlamydia psittaci in companion and wild birds. Avian Pathology 42, 38–44. [DOI] [PubMed] [Google Scholar]
- 24.Sachse K et al. (2009) DNA microarray-based genotyping of Chlamydophila psittaci strains from culture and clinical samples. Veterinary Microbiology 135, 22–30. [DOI] [PubMed] [Google Scholar]
- 25.Vanrompay et al. (2007) Chlamydophila psittaci transmission from pet birds to humans. Emerging Infectious Diseases 13, 1108–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan Y et al. (1989) Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 Chlamydia trachomatis serovars. Infection & Immunity 57, 1040–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins DP et al. (2012) Within-population diversity of koala Chlamydophila pecorum at ompA VD1-VD3 and the ORF663 hypothetical gene. Veterinary Microbiology 156, 353–358. [DOI] [PubMed] [Google Scholar]
- 28.Hulin V et al. (2015) Assessment of Chlamydia psittaci shedding and environmental contamination as potential sources of worker exposure throughout the mule duck breeding process. Applied and Environmental Microbiology 82, 1504–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]