Abstract
Most cancers develop with one of two types of genomic instability, namely, chromosomal instability (CIN) or microsatellite instability (MSI). Both are induced by replication stress-associated DNA double-strand breaks (DSBs). The type of genomic instability that arises is dependent on the choice of DNA repair pathway. Specifically, MSI is induced via a PolQ-dependent repair pathway called microhomology-mediated end joining (MMEJ) in a mismatch repair (MMR)-deficient background. However, it is unclear how the MMR status determines the choice of DSB repair pathway. Here, we show that replication stress-associated DSBs initially targeted by the homologous recombination (HR) system were subsequently hijacked by PolQ-dependent MMEJ in MMR-deficient cells, but persisted as HR intermediates in MMR-proficient cells. PolQ interacting with MMR factors was effectively loaded onto damaged chromatin in an MMR-deficient background, in which merged MRE11/γH2AX foci also effectively formed. Thus, the choice of DNA repair pathway according to the MMR status determines whether CIN or MSI is induced.
Keywords: Biological sciences, Cell biology, Genetics, DNA repair, Molecular biology, Genomic instability, Mismatch repair, Microhomology-mediated end joining, DNA polymerase theta
Biological sciences; Cell biology; Genetics; DNA repair; Molecular biology; Genomic instability; DNA repair; Mismatch repair; Microhomology-mediated end joining; DNA polymerase theta
1. Introduction
Most cancer cells develop with genomic instability, which can be separated into two types, namely, chromosomal instability (CIN) and microsatellite instability (MSI) [1]. While CIN is induced in most cases, MSI occurs in an MMR-deficient background, which can arise due to mutations in MSH2, MSH6, MLH1, and PMS2 [2,3]. CIN encompasses a wide variety of chromosomal abnormalities, including chromosomal deletions and translocations, ploidy abnormalities, loss of heterozygosity, and gene amplifications [4, 5]. By contrast, MSI is defined as changes in the lengths of microsatellite fragments that contain short repetitive sequences (1–6 bases) [6]. Importantly, such genomic destabilization is associated with mutation induction and clonal evolution of cells with abrogated defense systems, such as the ARF/p53 module [7]. Given that most cancers develop with genomic instability, mutagenesis associated with genomic destabilization is probably the major risk factor for step-wise cancer development.
Cancer risk elevates in association with age. Possibly related with this, DNA double-strand breaks (DSBs) risking CIN and MSI are widely accumulated in association with age in vivo and cultivation passages in vitro where cells are generally senesced [8]. Such cells are specifically defective in repairing replication stress-associated DSBs but not DSBs directly caused by radiation [9]. Such cellular senescence is further associated with aging and aging-associated disorders [10, 11]. Both CIN and MSI inductions are triggered by replication stress-associated DNA double-strand breaks (DSBs) [7, 12, 13, 14, 15], which are generally targeted by homologous recombination (HR) factors [16, 17].
CIN arises when the DSBs are not effectively repaired, in which persisted DSBs are often carried over into G2–M phases, leading to cytokinesis failure and tetraploidy (CIN) [15]. Probably associating with this, cells with CIN are often initially developed with tetraploidy [18, 19], such as mouse embryonic fibroblast cells (MEFs) just acquired immortality and cells at pre-cancerous stages [20, 21]. By contrast, identical DSBs in MMR-deficient cells are erroneously repaired by microhomology-mediated end joining (MMEJ) and this induces MSI, i.e., specific insertions/deletions at microsatellite loci that are potential micro-homologous sites [22]. Since MSI is induced with eliminating DSBs, CIN is suppressed during the MSI induction [7]. Thus, CIN and MSI are induced when those DSBs are persisted or erroneously repaired by MMEJ. Those persisted DSB accumulation is associated with cellular senescence [23].
Even in MMR-deficient cells, MSI is blocked upon suppression of MMEJ by PolQ knockdown (KD) and PARP inhibition [7]. Under these conditions, cells are similar to MMR-proficient cells. Specifically, replication stress-associated DSBs persist, leading to CIN induction (tetraploidization) [7]. This implies that MMEJ specifically occurs in an MMR-deficient background to induce MSI and suppress CIN, posing the question of how MMR deficiency is associated with MMEJ induction. This may be due to differences in the interactions of repair factors according to the MMR status. In support of this hypothesis, MMR factors constitutively interact with multiple repair factors by forming a large repair factor complex called the BASC, which includes BRCA1 and MRE11 [24].
This study demonstrated that replication stress-associated DSBs were initially targeted by the HR system and subsequently hijacked by a PolQ-mediated repair pathway in MMR-deficient cells, but persisted as HR intermediates in MMR-proficient cells. PolQ physiologically interacting with MMR factors was more efficiently loaded onto damaged chromatin in MMR-deficient cells than in MMR-proficient cells. This helps to explain how CIN and MSI are differentially induced according to the MMR status.
2. Materials and methods
2.1. Cell culture
Wild-type (WT) and MMR-deficient (Msh2−/−) mouse embryos were prepared from Msh2+/- mice [25]. Each embryo was separately minced by sterile razors, and was subsequently washed with PBS and cultivated as MEFs at passage 1 (P1) [26]. MEFs cultured using a Std-3T3 passage protocol [26] after the determination their genotypes [25]. HCT116 and HeLa cells, as well as HCT116 cells expressing PolQ-targeting shRNA and the negative control (NC), were also used [7]. All cells were cultured in Dulbecco's Modified Eagle's Medium (Wako) supplemented with 10% (v/v) fetal calf serum (Gibco). Cell lysates were fractionated as previously reported [27].
2.2. Antibodies
Antibodies against the following proteins were obtained from the indicated suppliers: β-actin (AC-74, Sigma), Flag (F1804, Sigma; 8142, Cell Signaling), H3 (MABI0301, MBL), γH2AX (JBW301, Upstate; 9718, Cell Signaling), 53BP1 (Merck, PC712), MLH1 (3515, Cell Signaling; [28]), MRE11 (ab33125, Abcam), MSH2 ([28]; A300-452A, Bethyl), PCNA (ab29, Abcam), Rad50 (ab87918, ab119708 Abcam), Rad51 (8875, Cell Signaling), Rad51-pS14 [29], Rad51-pT13pS14 [29], and non-immune IgG (sc-2025, Santa Cruz).
2.3. Cell biological experiments
DNA damage was induced by treatment with 0.2 mM hydroxyurea (HU, Sigma). Western blotting was performed using antibodies indicated above after blotting onto PVDF membrane [30]. Proteins were transferred to PVDF membranes for 2 or 6 h for detection of Flag-PolQ. Cells for immunofluorescence were prepared using primary and secondary antibodies indicated below after 4 % paraformaldehyde fixation, permeabilization with 0.1 % Triton X-100/PBS, and blocking (2 % Goat serum in PBS containing 0.3 % Tritin X-100 [30]. Immunofluorescence was performed using a confocal laser microscope (Olympus FV10i). Immunoprecipitation was performed using antibodies indicated above and protein A magnetic beads (Tamagawa-Seiki) in the presence of a complete protease inhibitor cocktail (Roche) Benzonase Nuclease (Millipore) [27]. Lipofectamine 3000 (Life Technologies) was used to transfect the Flag-tagged PolQ expression vector. Lipofectamine RNAiMax (Life Technologies) was used to transfect siRNAs. The sequences of the top-strand siRNAs targeting PolQ were GCUUCAGUGAUGACUAUCUAGUAAA (siPolQ#1) and CAUUCGGGUCUUGGCGGCAACUUCU (siPolQ#2).
3. Results
3.1. Persistence of HR intermediates in MMR-proficient cells
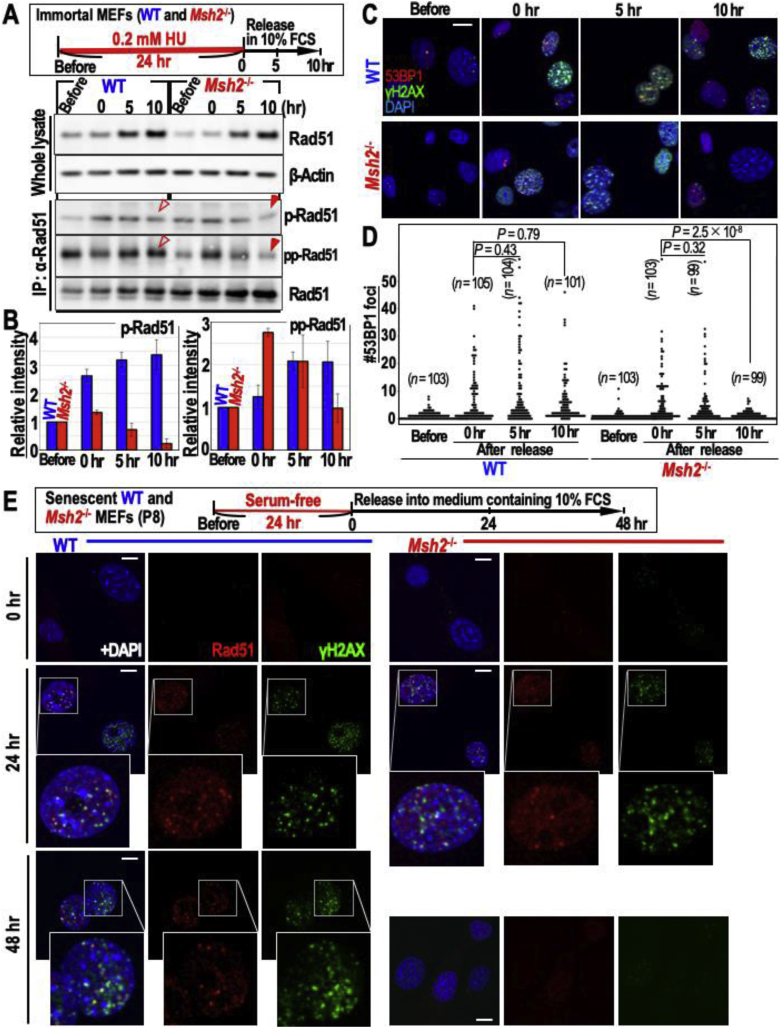
To investigate the repair factors targeting replication stress-associated DSBs that trigger CIN and MSI, we first studied Rad51 statuses because such DSBs are usually repaired by HR and CIN is induced when these DSBs persist and are carried over into M phase [31]. Therefore, we specifically monitored phosphorylation of Rad51 at serine (S) 14 (p-Rad51) and di-phosphorylation of Rad51 at threonine 13 and S14 (pp-Rad51) because the levels of these forms are elevated during HR in G2–M phases [29]. Rad51 was upregulated and phosphorylated when WT and Msh2−/− MEFs were treated with HU, which causes replication stress (Figure 1A and B), indicating that HR is the primary repair pathway targeting these DSBs. While p- and pp-Rad51 signals remained high in WT MEFs, they were reduced in Msh2−/− MEFs at 10 h after release (Figure 1A; see red arrowheads). These are associated with DSB-accumulation statuses that are monitored by γH2AX/53BP1 foci: while those foci levels are high until 10 h after release in WT MEFs, those in Msh2−/− MEFs are reduced (Figure 1C and D). These results suggest that persistent DSBs in WT MEFs are HR intermediates and hence are associated with a risk of damage carryover into G2–M phases, consistent with the previous finding that CIN is specifically induced in WT MEFs when those cells are senesced and become defective in repairing replication stress-associated DSBs [7]. The pp-Rad51 signal was higher in WT MEFs than in Msh2−/− MEFs even before HU treatment. This probably reflects the background risk of genomic destabilization because the genomic rearrangement rate is generally higher in MMR-deficient cancer cells than in MMR-proficient cancer cells even during normal proliferation [32].
Figure 1.
Replication stress-associated DSBs are persisted under intermediates of HR in MMR-proficient cells. (A–D) WT and Msh2−/− MEFs were treated with HU, as shown in the workflow (A). HR statuses were determined by the intensities of p- and pp-Rad51 signals (A,B). MMR-deficiency dependent decay of p- and pp-Rad51 signals are pointed by red arrowheads (A). Source images of blots are shown in Supplementary Fig. 1. p- and pp-Rad51 signals normalized with β-Actin signal are shown (B). DSB statuses were determined by the merged foci of γH2AX and 53BP1 (C,D). The representative images are shown (C). The numbers of 53BP1 foci in each cell were plotted (n numbers and P values are indicated in graph) (D). Two-tailed Welch's t-test was used for statistical analysis. (E) WT and Msh2−/− MEFs were exogenously growth-accelerated, as shown in the workflow. HR and DSB statuses were determined by the merged foci of Rad51 and γH2AX. Representative cells are zoomed. Error bars represent ±SD. Scale bars, 10 μm.
We next sought to confirm that HR is the major repair pathway that initially targets replication stress-associated DSBs in both MMR-proficient and -deficient cells and that HR intermediates are the major form of persistent DSBs in MMR-proficient cells. To this end, we investigated merged γH2AX/Rad51 foci in cells treated as shown in the workflow to induce replication stress (Figure 1E). As expected, merged γH2AX/Rad51 foci initially arose in both WT and Msh2−/− MEFs. However, these foci were continuously detected for 48 h after release in WT MEFs, but not in Msh2−/− MEFs. Thus, we conclude that replication stress-associated DSBs are primarily targeted by HR factors in both MMR-proficient and -deficient cells, but persist as HR intermediates specifically in MMR-proficient cells.
3.2. PolQ-dependent reduction of DSBs in MMR-deficient cells
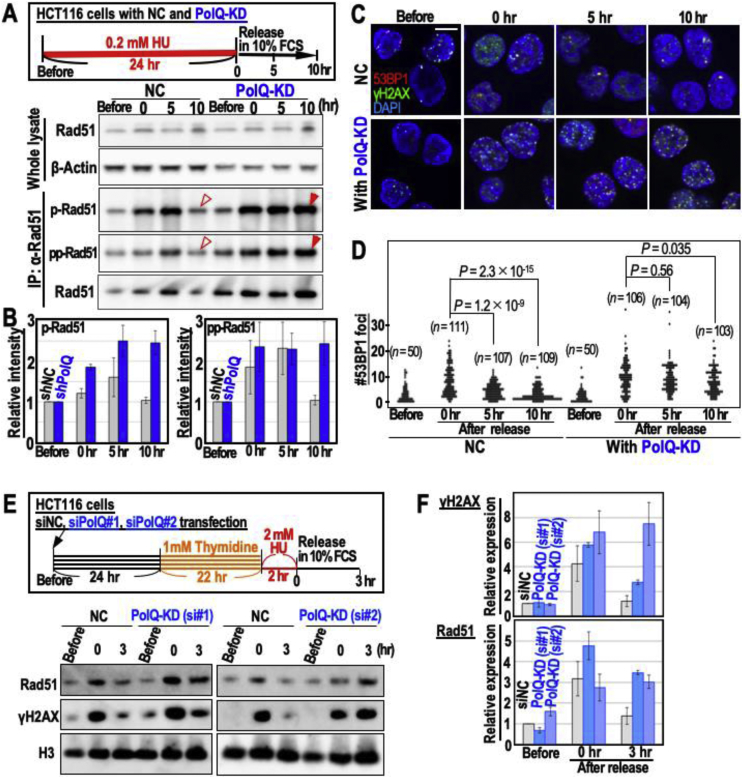
Replication stress-associated DSBs in MMR-deficient cells are repaired by MMEJ with inducing MSI [7]. To determine the MMEJ dependence in the termination of HR and the resulting repair, PolQ mediating MMEJ was knocked down in MMR-deficient HCT116 cells and the statuses of p- and pp-Rad51 signals were monitored after cells were treated as in the workflow (Figure 2A and B). As expected, while these signals, which arose after 0–5 h, were reduced at 10 h after release in HCT116 cells (NC), those under PolQ-KD were continuously detected (Figure 2A and B). This is consistently associated with γH2AX/53BP1-focus levels: while γH2AX/53BP1 foci were reduced by 10 h after the release in HCT116 cells (NC), those under PolQ-KD were continuously detected (Figure 2C and D). Identical results are also shown by western signals of γH2AX and Rad51 (Figure 2E and F). Together, these data suggest that the PolQ-dependent DSB repair pathway, i.e., MMEJ is specifically induced in MMR-deficient cells and hijacks HR intermediates, while such intermediates persist in MMR-proficient cells. Although such DSBs are persisted under PolQ-KD even in MMR-deficient HCT116 cells, this is consistent with a previous study, because such cells reveal reversal of the genomic instability phenotype of these cells such that CIN is induced and MSI is suppressed [7].
Figure 2.
Replication stress-associated DSBs are repaired through a PolQ-mediated repair pathway with terminating HR in MMR-proficient cells. (A–D) HCT116 cells were treated with HU, as shown in the workflow (A). The statuses of HR were determined by the intensities of p-Rad51 and pp-Rad51 signals (A,B). Signals observed PolQ dependent decay are pointed by red arrowheads (A). Source images of blots are shown in Supplementary Fig. 2. p- and pp-Rad51 signals normalized with β-Actin signal are shown (B). DSB statuses were determined by the merged foci of γH2AX and 53BP1 (C,D). Representative images are shown (C). The numbers of 53BP1 foci in each cell were plotted (n numbers and P values are indicated in graph) (D). Two-tailed Welch's t-test was used for statistical analysis. (E,F) Rad51 accumulation status were determined together with the damage status that was determined by γH2AX signal after cells were treated as in the workflow (E). Source images of blots are shown in Supplementary Fig. 3. Rad51 and γH2AX signals normalized with histone H3 signal are shown (F). Error bars represent ±SD.
3.3. Effective loading of PolQ and MRE11 onto chromatin in MMR-deficient cells
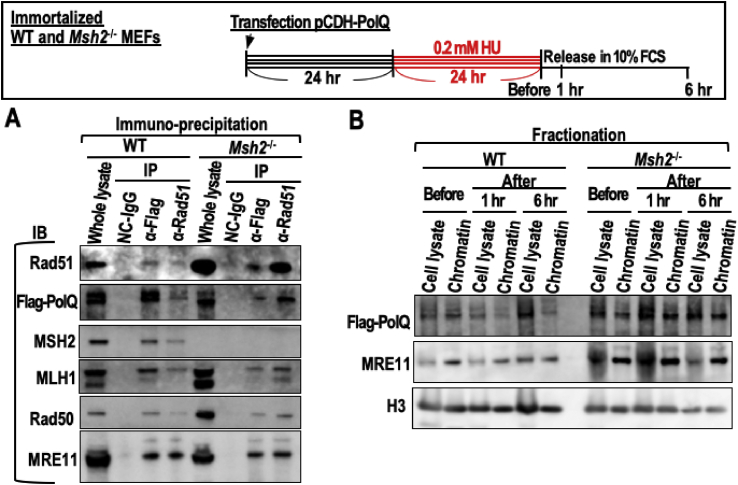
A remaining question is how the MMR status determines the choice of DNA repair pathway i.e., induction of MMEJ in an MMR-deficient background but not in an MMR-proficient background. One possible explanation is that complex formation by repair factors is altered, which affects damage recognition. In fact, MMR factors widely interact with multiple DSB repair factors present in the BASC, a super complex of BRCA1-associated proteins including Rad50 and MRE11 [24]. To compare complex formation between MMR-proficient and -deficient cells, we performed immunoprecipitation (IP) using WT and Msh2−/− MEFs transfected with the Flag-tagged PolQ expression vector and treated with HU, as shown in the workflow. As expected, MSH2 and MLH1 co-immunoprecipitated with Rad51 and Flag-PolQ, with the exception of MSH2 in Msh2−/− MEFs (Figure 3A), suggesting that they physiologically interact. The co-IP efficiencies of Rad51 and Flag-PolQ were higher in Msh2−/− MEFs than in WT MEFs, indicating that complex formation of DNA repair factors differs between these cells (Figure 3A). MRE11 and Rad50 also co-immunoprecipitated with Rad51 more effectively in Msh2−/− MEFs than in WT MEFs (Figure 3A).
Figure 3.
Damaged chromatin loading of PolQ and MRE11 are more effective under MMR deficient background compared to those of WT cells. (A,B) WT and Msh2−/− MEFs were treated with HU as shown in the workflow, and then IP (A) and chromatin fractionation (B) was performed. IP was performed with cells at 1 h after release from HU using an anti-Rad51 or -Flag antibody. Source images of blots are shown in Supplementary Fig. 4. Their reciprocal associations were detected by immunoblotting (IB). H3 was detected as a loading control (B). Source images of blots are shown in Supplementary Fig. 5.
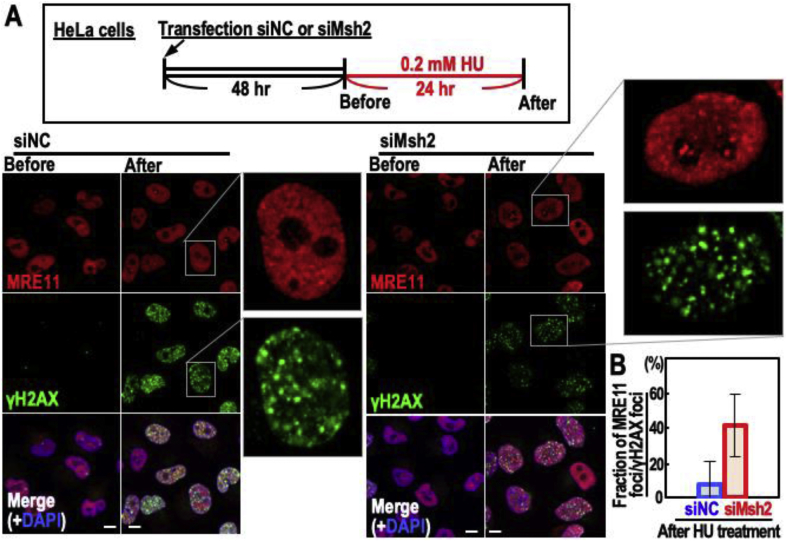
We next tested whether the MMR status affects the loading of DSB repair factors onto damaged chromatin. WT and Msh2−/− MEFs were treated as shown in the workflow, cell lysates were fractionated, and loading of Flag-PolQ and MRE11 onto damaged chromatin was investigated (Figure 3B). Intriguingly, Flag-PolQ and MRE11 signals were higher on chromatin isolated from Msh2−/− MEFs than on chromatin isolated from WT MEFs (Figure 3B), indicating that loading of repair factors including PolQ and MRE11 onto damaged chromatin differs according to the MMR status. In addition to the efficient loading of MRE11 onto damaged chromatin in Msh2−/− MEFs (Figure 3B), merged MRE11/γH2AX foci formed more efficiently in MSH2-depleted HeLa cells than in NC HeLa cells (Figure 4). These results imply that recruitment of repair factors to replication stress-associated DSB loci differs according to the MMR status, which helps to explain how CIN and MSI are differentially induced.
Figure 4.
MRE11 foci merged with γH2AX are more effectively formed under MMR deficient background compared to those of WT cells. (A,B) HeLa cells knocked down for MSH2 and the control were treated with HU as shown in the top box, and γH2AX and MRE11 foci were assessed by immunofluorescence microscopy (A). Representative cells are zoomed. Foci numbers were counted (B). Error bars indicate SD. Scale bars, 10 μm.
4. Discussion
The current study illustrates that replication stress-associated DSBs persist as HR intermediates in MMR-proficient cells and there is a risk of carryover of these DSBs into G2–M phases, leading to CIN induction. By contrast, although DSBs are also initially targeted by HR factors in MMR-deficient cells, they are subsequently hijacked by a PolQ-mediated repair pathway called MMEJ. These findings are consistent with a recent study showing that replication stress-associated DSBs induce MSI in MMR-deficient cells and CIN in MMR-proficient cells [7]. Induction of both MSI and CIN can lead to clonal evolution of cells with abrogated defense systems.
CIN and MSI are differentially induced according to the MMR status owing to the choice of DNA repair pathway. Although the underlying mechanism has not been fully elucidated, this study revealed that it involves differences in complex formation and loading of associated repair factors onto damaged chromatin. PolQ and MRE11 physiologically interacting with MMR factors were more efficiently loaded onto damaged chromatin in an MMR-deficient background (Figure 3B). Merged MRE11/γH2AX foci also formed more effectively in an MMR-defective background (Figure 4). The cross co-IP efficiencies of Flag-PolQ and Rad51 were higher in MMR-deficient MEFs than in MMR-proficient MEFs (Figure 3A). Such MMR factor dependence of complex formation and associated damage recognition probably underlies how CIN and MSI are differentially induced.
MMR proteins interact with a wide variety of repair factors, including MRE11, Rad50, and BRCA1 [24]; however, the significance of such complex formation remained unclear for a long time. Based on the current study together with a previous report of replication stress-triggered MSI induction [7], these interactions are probably associated with the choice of DNA repair pathway, i.e., suppression of MMEJ in an MMR-proficient background. One important question is how MMR functions when normal cells are subjected to replication stress. This is probably associated with genome stability maintenance and suppression of MMEJ. Replication stress-associated DSBs can be repaired in normally proliferating cells through HR. Therefore, it is probably important to block erroneous repair by MMEJ in order to prevent induction of insertions/deletions in microsatellite loci and to maintain genome stability. In fact, the risk of clonal evolution of cells with mutated cancer-driver genes is higher with MSI induction than with CIN induction [7]. However, this MMR-dependent barrier is ineffective after cells become senescent because replication stress-associated DSBs are not actively repaired by HR during senescence [9], and the risk of CIN induction increases to avoid clonal evolution of cells with mutated cancer-driver genes.
Declarations
Author contribution statement
H. Fujimori, M. Hyodo, Y. Minakawa and Y. Atsumi: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data.
A. Shimizu: Conceived and designed the experiments; Performed the experiments.
Y. Matsuno and Y. Murakami: Analyzed and interpreted the data.
K. Yoshioka: Analyzed and interpreted the data; Wrote the paper.
Y. Nakatsu and T. Tsuzuki: Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by was supported by the MEXT/JSPS KAKENHI (20770136) and the National Cancer Center Research and Development Fund (23-C-10).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Lengauer C., Kinzler K., Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 2.Schofield M.J., Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 3.Whitelaw N.C., Whitelaw E. Transgenerational epigenetic inheritance in health and disease. Curr. Opin. Genet. Dev. 2008;18:273–279. doi: 10.1016/j.gde.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Rajagopalan H., Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 5.Korbel J.O., Campbell P.J. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 6.Woerner S.M., Kloor M., von Knebel Doeberitz M., Gebert J.F. Microsatellite instability in the development of DNA mismatch repair deficient tumors. Cancer Biomark. 2006;2:69–86. doi: 10.3233/cbm-2006-21-208. [DOI] [PubMed] [Google Scholar]
- 7.Matsuno Y., Atsumi Y., Shimizu A., Katayama K., Fujimori H., Hyodo M., Minakawa Y., Nakatsu Y., Kaneko S., Hamamoto R., Shimamura T., Miyano S., Tsuzuki T., Hanaoka F., Yoshioka K. Replication stress triggers microsatellite destabilization and hypermutation leading to clonal expansion in vitro. Nat. Commun. 2019;10:3925. doi: 10.1038/s41467-019-11760-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sedelnikova O.A., Horikawa I., Zimonjic D.B., Popescu N.C., Bonner W.M., Barrett J.C. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat. Cell Biol. 2004;6:168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- 9.Minakawa Y., Atsumi Y., Shinohara A., Murakami Y., Yoshioka K. Gamma-irradiated quiescent cells repair directly induced double-strand breaks but accumulate persistent double-strand breaks during subsequent DNA replication. Genes Cells. 2016;21:789–797. doi: 10.1111/gtc.12381. [DOI] [PubMed] [Google Scholar]
- 10.Baker D.J., Wijshake T., Tchkonia T., Lebrasseur N.K., Childs B.G., Van De Sluis B., Kirkland J.L., Van Deursen J.M. Clearance of p16 Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simboeck E., Di Croce L. P16INK4a in cellular senescence. Aging (Albany. NY) 2013;5:590–591. doi: 10.18632/aging.100592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorgoulis V.G., Vassiliou L.-V.F., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., DiTullio R.A., Jr., Kastrinakis N.G., Levy B., Kletsas D., Yoneta A., Herlyn M., Kittas C., Halazonetis T.D. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 13.Bartkova J., Hořejší Z., Koed K., Krämer A., Tort F., Zleger K., Guldberg P., Sehested M., Nesland J.M., Lukas C., Orntoft T., Lukas J., Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 14.Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L.V.F., Kolettas E., Niforou K., Zoumpourlis V.C., Takaoka M., Nakagawa H., Tort F., Fugger K., Johansson F., Sehested M., Andersen C.L., Dyrskjot L., Ørntoft T., Lukas J., Kittas C., Helleday T., Halazonetis T.D., Bartek J., Gorgoulis V.G. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 15.Ichijima Y., Yoshioka K., Yoshioka Y., Shinohe K., Fujimori H., Unno J., Takagi M., Goto H., Inagaki M., Mizutani S., Teraoka H. DNA lesions induced by replication stress trigger mitotic aberration and tetraploidy development. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petermann E., Orta M.L., Issaeva N., Schultz N., Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsvetkova A., Ozerov I.V., Pustovalova M., Grekhova A., Eremin P., Vorobyeva N., Eremin I., Pulin A., Zorin V., Kopnin P., Leonov S., Zhavoronkov A., Klokov D., Osipov A.N. γH2AX, 53BP1 and Rad51 protein foci changes in mesenchymal stem cells during prolonged X-ray irradiation. Oncotarget. 2017;8:64317–64329. doi: 10.18632/oncotarget.19203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atsumi Y., Fujimori H., Fukuda H., Inase A., Shinohe K., Yoshioka Y., Shikanai M., Ichijima Y., Unno J., Mizutani S., Tsuchiya N., Hippo Y., Nakagama H., Masutani M., Teraoka H., Yoshioka K. Onset of quiescence following p53 mediated down-regulation of H2AX in normal cells. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osawa T., Atsumi Y., Sugihara E., Saya H., Kanno M., Tashiro F., Masutani M., Yoshioka K. Arf and p53 act as guardians of a quiescent cellular state by protecting against immortalization of cells with stable genomes. Biochem. Biophys. Res. Commun. 2013;432:34–39. doi: 10.1016/j.bbrc.2013.01.091. [DOI] [PubMed] [Google Scholar]
- 20.Vitale I., Galluzzi L., Senovilla L., Criollo A., Jemaà M., Castedo M., Kroemer G. Illicit survival of cancer cells during polyploidization and depolyploidization. Cell Death Differ. 2011;18:1403–1413. doi: 10.1038/cdd.2010.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshioka K., Atsumi Y., Fukuda H., Masutani M., Teraoka H. The quiescent cellular state is Arf/p53-dependent and associated with H2AX downregulation and genome stability. Int. J. Mol. Sci. 2012;13:6492–6506. doi: 10.3390/ijms13056492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshioka K., Matsuno Y., Hyodo M., Fujimori H. Genomic-destabilization-associated mutagenesis and clonal evolution of cells with mutations in tumor-suppressor genes. Cancers. 2019;11:1643. doi: 10.3390/cancers11111643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodier F., Muñoz D.P., Teachenor R., Chu V., Le O., Bhaumik D., Coppé J.P., Campeau E., Beauséjour C.M., Kim S.H., Davalos A.R., Campisi J., DNA-SCARS Distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J. Cell Sci. 2011;124:68–81. doi: 10.1242/jcs.071340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Cortez D., Yazdi P., Neff N., Elledge S.J., Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 25.Egashira A., Yamauchi K., Yoshiyama K., Kawate H., Katsuki M., Sekiguchi M., Sugimachi K., Maki H., Tsuzuki T. Mutational specificity of mice defective in the MTH1 and/or the MSH2 genes. DNA Repair. 2002;1:881–893. doi: 10.1016/s1568-7864(02)00113-1. [DOI] [PubMed] [Google Scholar]
- 26.Todaro G., Green H., Torado G.J., Green M.D. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atsumi Y., Minakawa Y., Ono M., Dobashi S., Shinohe K., Shinohara A., Takeda S., Takagi M., Takamatsu N., Nakagama H., Teraoka H., Yoshioka K. ATM and SIRT6/SNF2H mediate transient H2AX stabilization when DSBs form by blocking HUWE1 to allow efficient γH2AX foci formation. Cell Rep. 2015;13:2728–2740. doi: 10.1016/j.celrep.2015.11.054. [DOI] [PubMed] [Google Scholar]
- 28.Yoshioka K., Yoshioka Y., Hsieh P. ATR kinase activation mediated by MutSα and MutLα in response to cytotoxic O6-methylguanine adducts. Mol. Cell. 2006;22:501–510. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yata K., Lloyd J., Maslen S., Bleuyard J.-Y., Skehel M., Smerdon S.J., Esashi F. Plk1 and CK2 act in concert to regulate Rad51 during DNA double strand break repair. Mol. Cell. 2012;45:371–383. doi: 10.1016/j.molcel.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atsumi Y., Inase A., Osawa T., Sugihara E., Sakasai R., Fujimori H., Teraoka H., Saya H., Kanno M., Tashiro F., Nakagama H., Masutani M., Yoshioka K. The arf/p53 protein module, which induces apoptosis, down-regulates histone H2AX to allow normal cells to survive in the presence of anti-cancer drugs. J. Biol. Chem. 2013;288:13269–13277. doi: 10.1074/jbc.M112.402560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minakawa Y., Shimizu A., Matsuno Y., Yoshioka K. Genomic destabilization triggered by replication stress during senescence. Cancers. 2017;9:159. [Google Scholar]
- 32.Lengauer C., Kinzler K.W., Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.