Highlights
-
•
BMSC-Exos promote malignant tumorigenesis and metastasis in OS cells.
-
•
BMSC-Exos promotes oncogenic autophagy in OS cells.
-
•
BMSC-Exos-mediated autophagy contributes to the BMSC-Exos-induced malignant tumorigenesis metastasis in OS cells.
Keywords: Exosomes, hBMSCs, Autophagy, Osteosarcoma
Abstract
Osteosarcoma (OS) is a malignant bone tumor that frequently occurs in adolescents. It has a high rate of pulmonary metastasis and mortality. Previous studies have demonstrated that human bone marrow mesenchymal stem cells (hBMSCs) can promote the malignant progression in various tumors, including OS. Also, it is recognized that exosomes derived from hBMSCs (hBMSC-Exos) mediate cell-to-cell communication and exhibit similar effects on the development of various tumors. However, the role of hBMSC-Exos in the development of OS is still unclear and the underlying mechanism needs to be elucidated. Our results show that hBMSC-derived exosomes promote OS cell proliferation, migration, and invasion. Meanwhile, silencing autophagy-related gene 5 (ATG5) in OS cells abolishes the pro-tumor effects of hBMSC-Exos in vitro and in vivo. Our present study demonstrates that hBMSC-Exos promotes tumorigenesis and metastasis by promoting oncogenic autophagy in OS.
1. Introduction
Osteosarcoma (OS) has been implicated as the most prevalent malignant tumor of the bone tumors and occurs often in children and adolescents [1]. Pulmonary metastasis is the leading cause of death in patients with OS [2]. Despite the great advancement in strategies in surgery, chemotherapy, and radiotherapy, the 5-year survival rate of OS patients with metastasis is extremely low [2,3]. Henceforth, understanding the mechanism underlying the development of OS and improving the prognosis of OS patients with metastasis is imperative.
Recent studies have demonstrated that mesenchymal stem cells (MSCs) are locally adjacent to the tumor tissues and may interact with tumors directly, contributing to the formation of the tumor microenvironment and cancer progression by producing growth factors or promoting tumor vascularization [4,5]. A previous study has demonstrated that BMSCs may promote the malignant development of OS cells [6]. In addition, both mouse and human BMSCs can promote OS development through genetic modifications or spontaneous transformation [7,8]. Exosomes play an important role in cell–cell communications since they can be released by one cell and captured by a neighboring cell [9,10]. A large body of MSCs research has focused on MSCs-derived exosomes (MSCs-Exos) and has shown that MSCs-Exos exert bio-functions similar to those of MSCs [11,12]. Also, it has been reported that MSCs-Exos can promote tumor growth and metastasis in various tumors [13,14].
In the present study, we showed that hBMSC-Exos can promote cell proliferation, migration, and invasion in OS in vitro and in vivo. Moreover, hBMSC-Exos can also promote autophagy, and hBMSC-Exo-mediated autophagy contributes to hBMSC-Exo-induced promotion of malignant tumorigenesis in OS.
2. Materials and methods
2.1. Cell culture
Human bone marrow mesenchymal stem cells (hBMSCs, HUXMA-01001) were obtained from Cyagen Biosciences Inc. (CA, US) and cultured in Dulbecco's modified Eagle's medium/high glucose (DMEM, Hyclone, UT, USA) with 10% fetal bovine serum (FBS, Gibco Laboratory, Grand Island, NY, USA) and 1% penicillin/streptomycin (Gibco, Carlsbad, CA) at 37 °C and 5% CO2.
The human OS cell lines HOS (CRL-1543) and MG63 (CRL-1427) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). For each experiment, OS cells were cultured in DMEM/high glucose media supplemented with 10% FBS and 1% penicillin/streptomycin.
2.2. Exosome isolation and identification
Exosome isolation and identification were performed as we previously described [15,16]. Exosomes were obtained after a series of centrifugations and were either stored at −80 °C or used immediately for downstream experiments.
The morphology of the exosomes was observed under a transmission electron microscope (Tecnai 12; Philips, Best, The Netherlands). The diameters of the exosomes were analyzed by a Nanosight LM10 System (Nanosight Ltd., Navato, CA). Western blot was used to determine specific exosome surface markers, such as CD63 and CD81.
2.3. Exosome uptake by OS cells
Briefly, 4 mg/mL Dil solution (Molecular Probes, Eugene, OR, USA) was added to PBS containing exosomes, which were then incubated. The uptake of Dil-labeled exosomes by OS cells was observed by confocal microscopy.
2.4. siRNA transfection
Small interfering RNA (siRNA) targeting human autophagy-related gene 5 (ATG5) (siATG5) with its scrambled control siRNA (siNC) were purchased from GenePharma (Shanghai, China) for the transfections of HOS and MG63 cells. Lipofectamine 3000 reagent was used for the transfections, according to the manufacturer's instructions. RT-PCR and western blot analyses were then used to confirm the ATG5 expression levels.
2.5. Cell Counting Kit-8 assay
A Cell Counting Kit-8 (CCK-8) (Dojindo, Kumamoto, Japan) was used for the cell proliferation assay. In brief, OS cells were seeded in a 96-well plate at a density of 2000 cells/100 μL of medium/well and co-cultured with PBS or exosomes at a concentration of 100 μg/mL for 24 h. To this, 10 μL of CCK-8 solution (10 mL; 1:10 dilution) in fresh culture medium was added every 24 h and incubated for 2 h at 37 °C. The optical density (OD) at a wavelength of 450 nm was determined using a microplate reader (ELx800, Bio-Tek, USA).
2.6. 5-Ethynyl-2’-deoxyuridine assay
Cell proliferation was also measured using the 5-ethynyl-2’-deoxyuridine (EDU) assay kit (RiboBio, Guangzhou, China) according to the manufacturer's instructions. Briefly, cells were seeded in 24-well plates at a density of 2.0 × 104 cells/well and co-cultured in the presence of PBS or exosomes for 24 h before EDU (50 mM) was added. After this, Apollo staining and Hoechst 33342 were added. Finally, proliferation images were acquired and analyzed by fluorescence microscopy. The proliferation rate was presented as the percentage of EDU-positive cells relative to the total cell numbers.
2.7. Invasion assay
Cell invasion capacity was measured using 24-well BD Matrigel invasion chambers (BD Biosciences), according to the manufacturer's instructions. To avoid potential effects of cell proliferation on migration and invasion, OS cells were pre-treated with 10 μg/mL Mitomycin C for 3 h for migration and scratch assay. Briefly, for the invasion assays, approximately 2 × 104 OS cells co-cultured with PBS or exosomes were seeded on the upper well of the invasion chamber in DMEM medium without FBS, while the lower chamber well contained DMEM supplemented with 10% FBS to activate invasion. After 24 h, the non-invading upper cells were wiped with a cotton swab. Cells that had invaded the lower chamber were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet for 30 min. The invasive cells were counted in three random microscopic views and photographed.
2.8. Wound healing assay
Cells were cultured in 6-well plates and grown to 80–90% confluence. The cells were then scratched in the central area of the confluent culture using a sterile 200 µL pipette tip. The cells were carefully washed three times with PBS and then PBS or exosomes at 100 µg/mL was added. Wound healing was observed at 0 and 24 h after injury.
2.9. 3D spheroid BME cell invasion assay
For the 3D spheroid BME cell invasion assays, OS cells were seeded at a density of 2 × 104 cells/mL in 96-well ultralow adherence plates (#7007, Costar) in the presence of PBS or exosomes. The cells were induced to aggregate into a multicellular spheroid with an estimated density of 2000 cells after 96 h followed by the addition of Matrigel to the wells. The motion of these multicellular spheroid cells was observed after 48 h using fluorescence microscopy.
2.10. Autophagosome detection by GFP-mRFP-LC3
A GFP-mRFP-LC3 (Obio, Shanghai, China) lentivirus was used for autophagosome detection. OS cells were transfected with a GFP-mRFP-LC3 lentivirus according to the manufacturer's instructions. HOS and MG63 cells were fixed in 4% paraformaldehyde and the nuclei were stained with DAPI. The location and quantitation of the autophagosomes were then confirmed using confocal microscopy. The number of yellow spots (which represent the autophagosomes) and the number of red spots (which represent the autophagic lysosomes) were counted.
2.11. Transmission electron microscopy (TEM)
The cells were fixed in precooled 2% glutaraldehyde in 0.1 M cacodylate buffer at 4 °C overnight and then exposed to phosphate buffer containing 1% osmium tetroxide for 1 h. After being dehydrated in different concentrations of acetone, the cells were infiltrated and embedded in Epon. The embedded materials were sectioned and stained with 3% uranyl acetate and lead citrate. Images were then obtained under an electron microscope.
2.12. Western blot
Briefly, proteins were extracted from cells treated with a RIPA lysis and extraction buffer (KeyGen Biotechnology, Nanjing, China). Equal amounts of protein were separated on 10% SDS-PAGE, transferred to PVDF membranes (EMD Millipore Corp., Burlington, MA, USA), and blocked with bovine serum albumin (5%, v/v) followed by incubation overnight at 4 °C with primary antibodies. After washing with TBS-Tween (3 × 10 min), the membranes were incubated for 120 min at room temperature with the secondary antibodies. Reacting bands were visualized using ECL reagent (Thermo Fisher Scientific), and the density of the protein bands was semi-quantified using ImageJ software (National Institutes of Health, Bethesda, MD, USA). Anti- N-cadherin, E-cadherin, vimentin, Beclin1, p62, ATG5, LC3-II, and β-actin were purchased from Abcam (Cambridge, UK).
2.13. Animal experiments
The animal studies were approved by the Institutional Animal Care and Use Committee of Affiliated Hospital of Nanjing University of Chinese Medicine. Four-week-old nude mice (BALB/c nude mice) used for the in vivo tumor growth assays were purchased from the Animal Model Institute of Nanjing University (Nanjing, China). OS cells were treated with PBS or exosomes for 24 h before implantation. The nude mice were randomly divided into three groups (n = 6 per group). OS cells labeled with firefly luciferase (2 × 106 cells in 100 μL PBS) were subcutaneously injected into the nude mice. For tumor metastasis assays, PBS containing 2 × 106 cells were injected into nude mice via the caudal vein. The metastases were imaged using an IVIS200 imaging system (Caliper Life Sciences, Waltham, MA, USA).
2.14. Statistical analysis
All experiments were performed in triplicate, and data are expressed as the mean ± standard deviation. The Student's t-test was used to compare two groups. Statistical analyses were performed using SPSS v. 22.0 (SPSS Inc., Chicago, IL, USA).
3. Results
3.1. Identification of exosomes
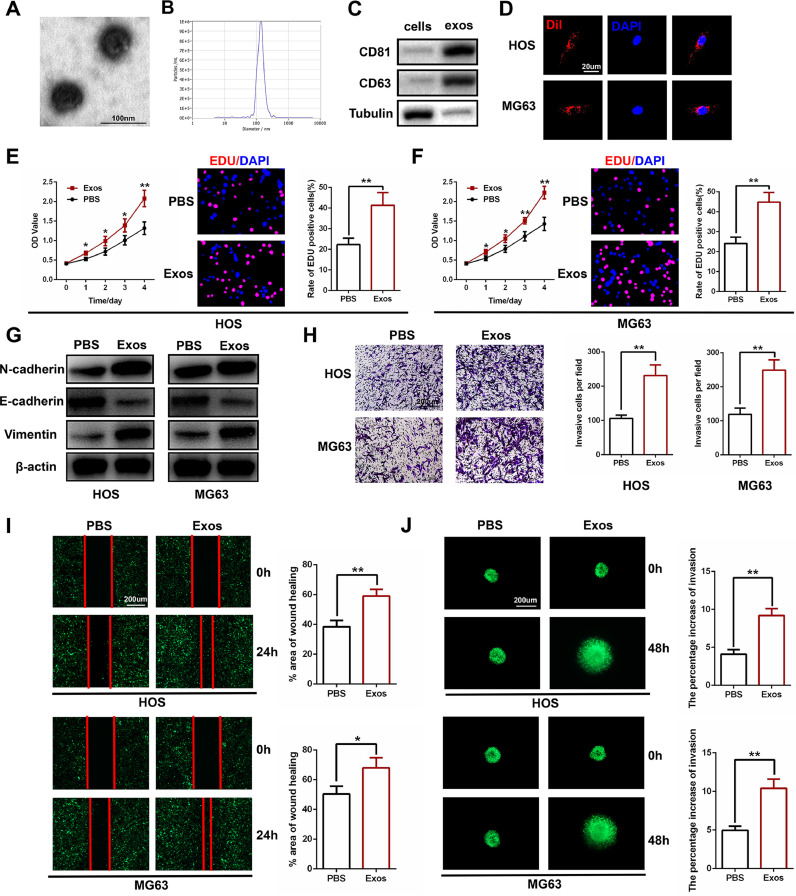
Transmission electron microscopy (TEM), nanoparticle tracking analysis (NTA), and western blots were utilized to identify the exosomes. As shown in Fig. 1A, TEM revealed typically rounded nanoparticles ranging from 50 to 150 nm in size. NTA exhibited a similar size distribution (average 114.9 nm) (Fig. 1B). Western blot revealed the presence of exosome surface markers, including CD63 and CD81 (Fig. 1C). Then, a Dil dye was used to label the exosomes that were then co-cultured with target OS cells for 24 h. Confocal microscopy was used to capture the Dil-labeled exosome uptake by the OS cells (Fig. 1D).
Fig. 1.
Human bone marrow mesenchymal stem cell-derived exosomes (HBMSC-Exos) promote tumor proliferation, migration, and invasion in osteosarcoma (OS) cells. (A) Morphology of hBMSC-Exos under transmission electron microscopy (TEM). (B) NTA analysis of hBMSC-Exos revealed that hBMSC-Exos ranges from 50 to 150 nm. (C) Western blot analysis of exosomal proteins including CD63 and CD81 (n = 3). (D) Uptake of the red fluorescence dye Dil-labeled hBMSC-Exos by HOS and MG63 cells. (E and F) Proliferation of HOS and MG63 cells after administering PBS or hBMSC-Exos as measured by CCK-8 assay and EDU staining (n = 3). (G) Representative images of western blot analysis of invasion-related protein levels in HOS and MG63 cells after administering PBS and hBMSC-Exos (n = 3). (H) Effects of hBMSC-Exos on invasion using the transwell invasion assay in HOS and MG63 cells. Quantification of the transwell invasion assay is shown (n = 3). (I) Wound healing assay was performed in HOS and MG63 cells and cell migration was measured 24 h after scratching (n = 3). (J) Representative images of the 3D spheroid BME cell invasion assay in HOS and MG63 cells after administering PBS and hBMSC-Exos (n = 3). *P < 0.05, ⁎⁎P < 0.01, ⁎⁎⁎P < 0.001.
3.2. HBMSC-Exos promotes OS cell proliferation, migration, and invasion in vitro
HOS and MG63 cells were used for all of the in vitro experiments. We cultured OS cells with PBS and exosomes at a concentration of 100 µg/mL. The proliferation of HOS and MG63 cells was examined using CCK-8 and EDU assays. As shown in Fig. 1E and F, hBMSC-Exos promoted cell proliferation in OS cells compared to the PBS control group.
In the presence of hBMSC-Exos, the expression levels of mesenchymal markers, including N-cadherin and vimentin, were increased, but the expression of the epithelial cell marker E-cadherin was decreased as shown by western blot analysis (Fig. 1G). A transwell invasion assay was also conducted to investigate the impact of hBMSC-Exos on the cell invasion ability of OS cells. As shown in Fig. 1H, administering hBMSC-Exos significantly promoted OS cell invasiveness when compared to the PBS control group. A wound healing assay was then performed, and the results demonstrate that hBMSC-Exos markedly promoted the migration of OS cells into the scratch-wounded area (Fig. 1I). These results were also confirmed using 3D spheroid BME cell invasion assays (Fig. 1J). Taken together, these results demonstrate that hBMSC-Exos can enhance the proliferation, migration, and invasion of OS cells.
3.3. HBMSC-derived exosomes promote autophagy in OS cells
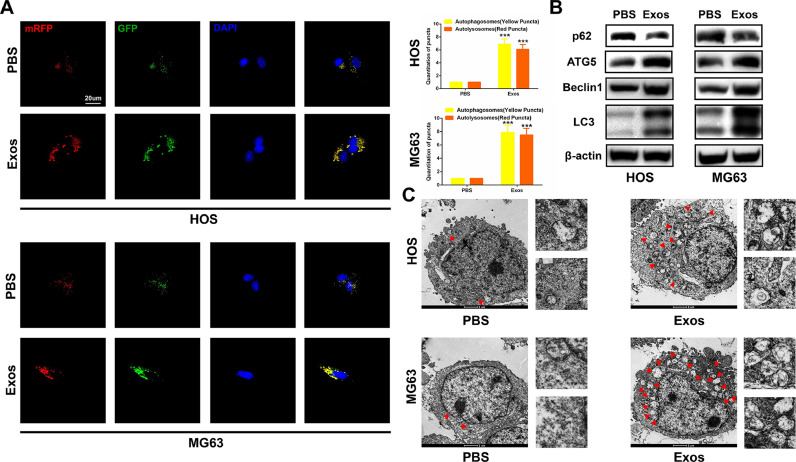
Numerous studies have proven that autophagy has a double-edged effect on the development of tumors. Therefore, we determined the level of autophagy when hBMSC-Exos was administered. Firstly, we transfected OS cell lines with GFP-mRFP-LC3 and observed the formation of autophagy flux under confocal microscopy. GFP-mRFP-LC3 dot accumulation was more obvious in the presence of hBMSC-Exos than in the PBS control group in HOS and MG63 cells (Fig. 2A). Next, western blot analysis indicated an increased level of ATG5, Beclin1, and LC3-II, but a decreased level of p62 when hBMSC-Exos was administered (Fig. 2B). We detected autophagosomes using TEM and found that exposure to hBMSC-Exos had favorable effects in promoting autophagy (Fig. 2C). Collectively, these results suggest that hBMSC-Exos can promote autophagy in OS cells.
Fig. 2.
HBMSC-Exos promotes autophagy in OS cells. (A) HOS and MG63 cells transfected with GFP-mRFP-LC3 lentivirus were cultured with PBS and hBMSC-Exos. Yellow and red puncta were observed and counted under confocal microscopy (n = 3). (B) Western blot analysis of autophagy-related protein levels, including Beclin1, p62, ATG5, and LC3-II in HOS and MG63 cells (n = 3). (C) TEM was used to detect the autophagic microstructure of HOS and MG63 cells after administering PBS and hBMSC-Exos. Red arrows indicate a cellular autophagosome with a double-layer structure or an autolysosome generated by the fusion of an autophagosome and lysosome. *P < 0.05, ⁎⁎P < 0.01, ⁎⁎⁎P < 0.001.
3.4. HBMSC-Exo-mediated autophagy contributes to HBMSC-Exo-induced promotion of tumor proliferation, migration, and invasion
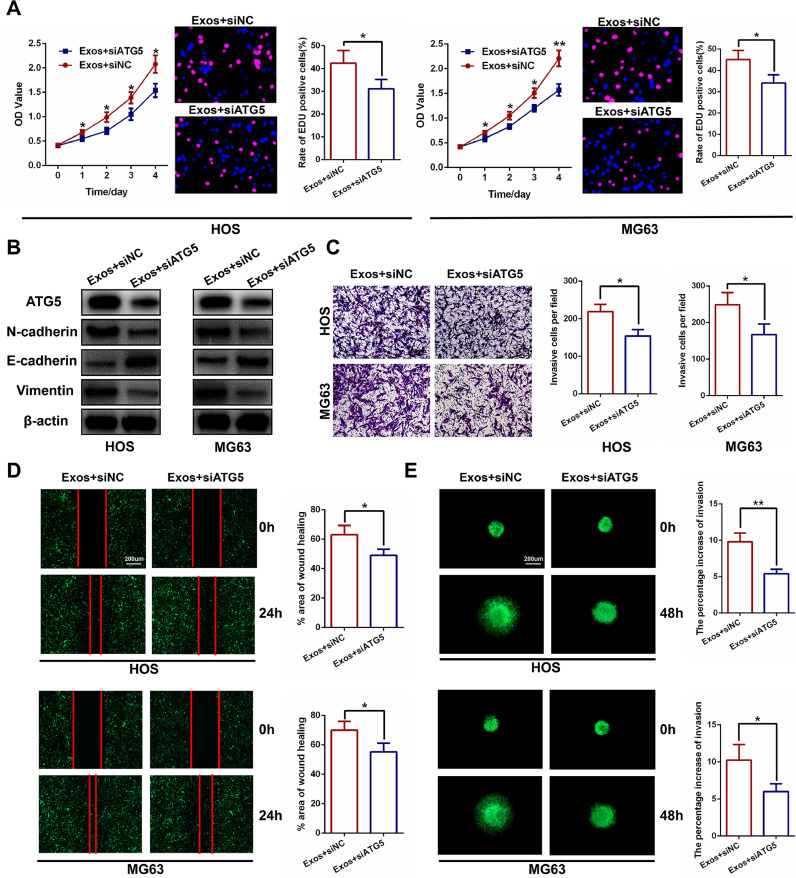
To investigate whether autophagy leads to the progression of OS, we abrogated autophagy by downregulating the expression of ATG5, a crucial factor related to autophagosome elongation. We confirmed the transfection efficiency of siATG5 in HOS and MG63 cells using RT-PCR and western blot analyses (Fig. S1A and B). The CCK-8 assay and EDU assay were performed and demonstrated that silencing ATG5 abolished the effects on OS proliferation caused by hBMSC-Exos (Fig. 3A). Western blot analysis (Fig. 3B), the transwell invasion assay (Fig. 3C), wound healing assay (Fig. 3D), and 3D spheroid BME cell invasion assay (Fig. 3E) were also conducted and proved that silencing ATG5 abolished the effects on OS migration and invasion caused by hBMSC-Exos. These data show that hBMSC-Exo-mediated autophagy contributes to hBMSC-Exo-induced promotion of tumor proliferation, migration, and invasion.
Fig. 3.
HBMSC-Exo-mediated autophagy contributes to the hBMSC-Exo-induced promotion of tumor proliferation, migration, and invasion. (A) Silencing ATG5 significantly inhibited cell proliferation measured by CCK-8 and EDU assays in HOS and MG63 cells (n = 3). (B) Silencing ATG5 significantly decreased invasion-related protein levels as shown by western blot analysis (n = 3). (C–E) Silencing of ATG5 significantly inhibited the invasion and migration capacity of HOS and MG63 cells following the addition of hBMSCs-Exos, as shown by the transwell invasion assay, wound healing assay, and 3D spheroid BME cell invasion assay (n = 3).
3.5. HBMSC-derived exosomes accelerate xenograft tumor growth and pulmonary metastasis in vivo
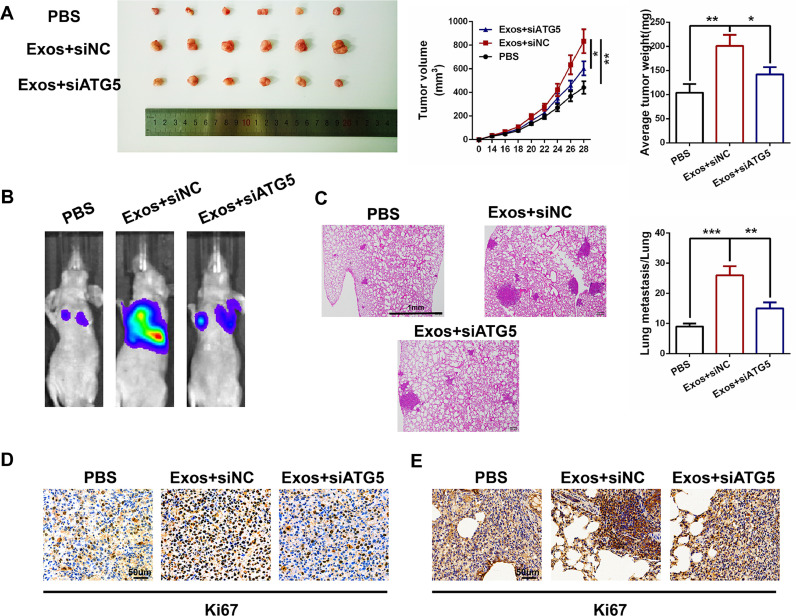
Nude mice were subcutaneously injected with MG63 cells pre-treated with PBS or Exos+siNC or Exos+siATG5 to determine the effects of hBMSC-Exos on tumor progression in vivo. The ATG5 expression levels were confirmed in tumors in different groups in Fig. S2A. As shown in Fig. 4A, the volume and weight of the tumors increased with exposure to hBMSC-Exos, compared to the PBS control group. However, the volume and weight of the tumors decreased when ATG5 was silenced in the MG63 cells. To further confirm the function of hBMSC-Exos in tumor metastasis in vivo, MG63 cells were injected into nude mice via the tail vein and the expression levels of ATG5 in lungs in different groups were shown in Fig. S2B. After 6 weeks, lung metastasis was significantly promoted in the Exos+siNC group compared to the control PBS group while significantly inhibited in the Exos+siATG5 group compared to the Exos+siNC group (Fig. 4B). The mice were then euthanized and the lung tissues were examined by H&E staining, which showed consistent results (Fig. 4C). Immunohistochemistry (IHC) using antibody for specific detection of the Ki67 proliferation marker in tumor and lung sections confirmed the above results (Fig. 4D and E). Taken together, these results demonstrate that hBMSC-Exos accelerates xenograft tumor growth and pulmonary metastasis in vivo and that autophagy plays a vital role in OS progression and metastasis.
Fig. 4.
HBMSC-Exos accelerates xenograft tumor growth and pulmonary metastasis in vivo. (A) MG63 cells treated with hBMSC-Exos markedly promoted tumor growth while silencing ATG5 in MG63 cells significantly inhibited tumor growth. Tumor volume and weight were calculated; n = 6 mice/group. (B) Representative images of pulmonary metastases in the three different groups were obtained by the IVIS Imaging System; n = 6 mice/group. (C) Representative H&E-stained lung sections from mice in the three different groups. (D and E) IHC staining of proliferation marker (Ki67) in tumor and lung sections in the three different groups.
4. Discussion
Osteosarcoma is the most common malignant bone tumor, occurring mainly in adolescents. Regardless of the many treatments that have been developed, there have been limited effects in enhancing the prognosis of OS [17], [18], [19]. Thus, focus should be given to exploring the underlying mechanism in the progression of OS.
Considering that BMSCs are derived from bone marrow tissue, which is abundant in areas close to the osteosarcoma, it is very important to explore their effects in OS development. The accumulating evidence has demonstrated that MSCs participate in the formation of the cancer microenvironment and contribute to the progression of various kinds of tumors, including OS [5], [6], [7], [8]. Recently, studies have concluded that the potential role of MSCs in tumor progression is mediated through secreted paracrine factors [20,21]. As a type of paracrine factor, exosomes can modulate cell-to-cell communication by delivering bio-messages and may participate in the development of tumors.
Previous studies have demonstrated that MSCs-Exos promotes tumor cell proliferation and metastasis in several kinds of cancers, including breast cancer, nasopharyngeal carcinoma, and gastric cancer [21], [22], [23]. However, until now, no study has reported the role of exosomes in the development of OS. In our present study, our results showed that hBMSC-Exos exhibits a pro-tumor role in the progression of OS.
Accumulating evidence has indicated that autophagy might have a double-edged role in the progression of malignant tumors by either promoting cancer cell survival or leading to cancer cell death [24,25]. In order to examine the relationship between hBMSC-Exos and autophagy, western blot analysis, TEM, and the GFP-mRFP-LC3 dot accumulation assays were performed. The results show that administering hBMSC-Exos promoted autophagy in OS cells. Furthermore, in order to examine the underlying relationship between autophagy and OS development, we silenced ATG5, which is recognized to have an essential role in autophagosome formation. In the present study, we found that silencing ATG5 in OS cells impaired OS cell malignancy induced by administering hBMSC-Exos in vitro and in vivo. Thus, our results indicate that hBMSC-Exos induces OS progression by promoting oncogenic autophagy in OS cells. However, in addition to its classical role in the formation of autophagosomes, ATG5 is also known to exhibit autophagy-independent bio-functions including cell death, cell immunity, pathogen control and so on [26], [27], [28]. Thus, we cannot rule out that other ATG5-dependent non-autophagic effects may be involved in these processes. Besides, according to our present study, the results which siATG5 did not totally abolish the exosome-mediated tumorigenesis indicated that other potential underlying mechanisms may participate in. Therefore, the precise mechanism of hBMSC-Exos-induced tumorigenesis in OS cells should be explored in future studies.
It is recognized that exosomes participate in the transport of biochemicals, such as cytokines, mRNAs, non-coding RNAs, and proteins and, as a result, play an essential role in intercellular communication through the transfer of genetic material [29]. Several studies have demonstrated that exosomal miRNAs might participate in the development of tumors [30,31]. Meanwhile, hypoxic microenvironments also participate in the biological process in tumor development, and some recent studies have identified some non-coding RNAs under hypoxia [32,33]. In fact, we have identified several exosomal non-coding RNAs in the development of OS under normoxia and hypoxia. We have also explored the relationship between exosomal non-coding RNAs and autophagy in OS cells (data unpublished), which could further enhance our insight into the progression of OS. However, the specific mechanism of hBMSC-Exos-induced autophagy promoting tumorigenesis in OS cells is still unclear and requires further investigation.
Taken together, our findings demonstrate that exosomes derived from bone marrow mesenchymal stem cells can promote cell proliferation, migration, and invasion in OS cells. Furthermore, our results suggest that autophagy contributes to hBMSC-Exo-mediated promotion of tumorigenesis and metastasis in OS both in vitro and in vivo. In conclusion, we have demonstrated that in the progression of OS, hBMSC-Exos promotes tumorigenesis and metastasis by promoting oncogenic autophagy.
CRediT authorship contribution statement
Yao Huang: Conceptualization, Methodology, Investigation, Resources. Wei Liu: Conceptualization, Software, Validation, Writing - original draft. Bing He: Software, Formal analysis. Lei Wang: Validation, Data curation. Fucheng Zhang: Resources. Hao Shu: Validation. Luning Sun: Writing - review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgements
This work was sponsored by the Natural Science Foundation of Jiangsu Province (Grant no. BK20191505).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jbo.2020.100280.
Appendix. Supplementary materials
References
- 1.Berner K., Johannesen T.B., Berner A., Haugland H.K., Bjerkehagen B., Bohler P.J., Bruland O.S. Time-trends on incidence and survival in a nationwide and unselected cohort of patients with skeletal osteosarcoma. Acta Oncol. 2015;54:25–33. doi: 10.3109/0284186X.2014.923934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aljubran A.H., Griffin A., Pintilie M., Blackstein M. Osteosarcoma in adolescents and adults: survival analysis with and without lung metastases. Ann. Oncol. 2009;20:1136–1141. doi: 10.1093/annonc/mdn731. [DOI] [PubMed] [Google Scholar]
- 3.Gill J., Ahluwalia M.K., Geller D., Gorlick R. New targets and approaches in osteosarcoma. Pharmacol. Ther. 2013;137:89–99. doi: 10.1016/j.pharmthera.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Kucerova L., Altanerova V., Matuskova M., Tyciakova S., Altaner C. Adipose tissue-derived human mesenchymal stem cells mediated prodrug cancer gene therapy. Cancer Res. 2007;67:6304–6313. doi: 10.1158/0008-5472.CAN-06-4024. [DOI] [PubMed] [Google Scholar]
- 5.Yu J.M., Jun E.S., Bae Y.C., Jung J.S. Mesenchymal stem cells derived from human adipose tissues favor tumor cell growth in vivo. Stem Cells Dev. 2008;17:463–473. doi: 10.1089/scd.2007.0181. [DOI] [PubMed] [Google Scholar]
- 6.Yu F.X., Hu W.J., He B., Zheng Y.H., Zhang Q.Y., Chen L. Bone marrow mesenchymal stem cells promote osteosarcoma cell proliferation and invasion. World J. Surg. Oncol. 2015;13:52. doi: 10.1186/s12957-015-0465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao W., Mohseny A.B., Hogendoorn P.C., Cleton-Jansen A.M. Mesenchymal stem cell transformation and sarcoma genesis. Clin. Sarcoma Res. 2013;3:10. doi: 10.1186/2045-3329-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tirode F., Laud-Duval K., Prieur A., Delorme B., Charbord P., Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11:421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 9.Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathieu M., Martin-Jaular L., Lavieu G., Thery C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 11.Nawaz M., Fatima F., Vallabhaneni K.C., Penfornis P., Valadi H., Ekstrom K., Kholia S., Whitt J.D., Fernandes J.D., Pochampally R., Squire J.A., Camussi G. Extracellular vesicles: evolving factors in stem cell biology. Stem Cells Int. 2016 doi: 10.1155/2016/1073140. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan X., Gong Y.Z., Wu P., Liao D.F., Zheng X.L. Mesenchymal stem cell-derived microparticles: a promising therapeutic strategy. Int. J. Mol. Sci. 2014;15:14348–14363. doi: 10.3390/ijms150814348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kidd S., Spaeth E., Dembinski J.L., Dietrich M., Watson K., Klopp A., Battula V.L., Weil M., Andreeff M., Marini F.C. Direct evidence of mesenchymal stem cell tropism for tumor and wounding microenvironments using in vivo bioluminescent imaging. Stem Cells. 2009;27:2614–2623. doi: 10.1002/stem.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu W., Xu W., Jiang R., Qian H., Chen M., Hu J., Cao W., Han C., Chen Y. Mesenchymal stem cells derived from bone marrow favor tumor cell growth in vivo. Exp. Mol. Pathol. 2006;80:267–274. doi: 10.1016/j.yexmp.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Rong Y., Liu W., Wang J., Fan J., Luo Y., Li L., Kong F., Chen J., Tang P., Cai W. Neural stem cell-derived small extracellular vesicles attenuate apoptosis and neuroinflammation after traumatic spinal cord injury by activating autophagy. Cell Death Dis. 2019;10:340. doi: 10.1038/s41419-019-1571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W., Wang Y., Gong F., Rong Y., Luo Y., Tang P., Zhou Z., Zhou Z., Xu T., Jiang T., Yang S., Yin G., Chen J., Fan J., Cai W. Exosomes derived from bone mesenchymal stem cells repair traumatic spinal cord injury by suppressing the activation of A1 neurotoxic reactive astrocytes. J. Neurotrauma. 2019;36:469–484. doi: 10.1089/neu.2018.5835. [DOI] [PubMed] [Google Scholar]
- 17.Luetke A., Meyers P.A., Lewis I., Juergens H. Osteosarcoma treatment – where do we stand? A state of the art review. Cancer Treat. Rev. 2014;40:523–532. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Liu W., Zhou Z., Zhang Q., Rong Y., Li L., Luo Y., Wang J., Yin G., Lv C., Cai W. Overexpression of miR-1258 inhibits cell proliferation by targeting AKT3 in osteosarcoma. Biochem. Biophys. Res. Commun. 2019;510:479–486. doi: 10.1016/j.bbrc.2019.01.139. [DOI] [PubMed] [Google Scholar]
- 19.Luo Y., Liu W., Tang P., Jiang D., Gu C., Huang Y., Gong F., Rong Y., Qian D., Chen J., Zhou Z., Zhao S., Wang J., Xu T., Wei Y., Yin G., Fan J., Cai W. miR-624-5p promoted tumorigenesis and metastasis by suppressing hippo signaling through targeting PTPRB in osteosarcoma cells. J. Exp. Clin. Cancer Res. 2019;38:488. doi: 10.1186/s13046-019-1491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu W., Huang L., Li Y., Zhang X., Gu J., Yan Y., Xu X., Wang M., Qian H., Xu W. Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth in vivo. Cancer Lett. 2012;315:28–37. doi: 10.1016/j.canlet.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Ono M., Kosaka N., Tominaga N., Yoshioka Y., Takeshita F., Takahashi R.U., Yoshida M., Tsuda H., Tamura K., Ochiya T. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci. Signal. 2014;7 doi: 10.1126/scisignal.2005231. ra63. [DOI] [PubMed] [Google Scholar]
- 22.Gu H., Ji R., Zhang X., Wang M., Zhu W., Qian H., Chen Y., Jiang P., Xu W. Exosomes derived from human mesenchymal stem cells promote gastric cancer cell growth and migration via the activation of the Akt pathway. Mol. Med. Rep. 2016;14:3452–3458. doi: 10.3892/mmr.2016.5625. [DOI] [PubMed] [Google Scholar]
- 23.Shi S., Zhang Q., Xia Y., You B., Shan Y., Bao L., Li L., You Y., Gu Z. Mesenchymal stem cell-derived exosomes facilitate nasopharyngeal carcinoma progression. Am. J. Cancer Res. 2016;6:459–472. [PMC free article] [PubMed] [Google Scholar]
- 24.Singh S.S., Vats S., Chia A.Y., Tan T.Z., Deng S., Ong M.S., Arfuso F., Yap C.T., Goh B.C., Sethi G., Huang R.Y., Shen H.M., Manjithaya R., Kumar A.P. Dual role of autophagy in hallmarks of cancer. Oncogene. 2018;37:1142–1158. doi: 10.1038/s41388-017-0046-6. [DOI] [PubMed] [Google Scholar]
- 25.White E. The role for autophagy in cancer. J. Clin. Investig. 2015;125:42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Codogno P., Meijer A.J. Atg5: more than an autophagy factor. Nat. Cell Biol. 2006;8:1045–1047. doi: 10.1038/ncb1006-1045. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Z.J., Fux B., Goodwin M., Dunay I.R., Strong D., Miller B.C., Cadwell K., Delgado M.A., Ponpuak M., Green K.G., Schmidt R.E., Mizushima N., Deretic V., Sibley L.D., Virgin H.W. Autophagosome-independent Essential Function for the Autophagy Protein Atg5 in Cellular Immunity to Intracellular Pathogens. Cell Host Microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galluzzi L., Green D.R. Autophagy-independent functions of the autophagy machinery. Cell. 2019;177:1682–1699. doi: 10.1016/j.cell.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu W., Li L., Rong Y., Qian D., Chen J., Zhou Z., Luo Y., Jiang D., Cheng L., Zhao S., Kong F., Wang J., Zhou Z., Xu T., Gong F., Huang Y., Gu C., Zhao X., Bai J., Wang F., Zhao W., Zhang L., Li X., Yin G., Fan J., Cai W. Hypoxic mesenchymal stem cell-derived exosomes promote bone fracture healing by the transfer of miR-126. Acta Biomater. 2019 doi: 10.1016/j.actbio.2019.12.020. [DOI] [PubMed] [Google Scholar]
- 30.Che Y., Shi X., Shi Y., Jiang X., Ai Q., Shi Y., Gong F., Jiang W. Exosomes derived from miR-143-overexpressing MSCs inhibit cell migration and invasion in human prostate cancer by downregulating TFF3. Mol. Ther. Nucl. Acids. 2019;18:232–244. doi: 10.1016/j.omtn.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Gong L., Bao Q., Hu C., Wang J., Zhou Q., Wei L., Tong L., Zhang W., Shen Y. Exosomal miR-675 from metastatic osteosarcoma promotes cell migration and invasion by targeting CALN1. Biochem. Biophys. Res. Commun. 2018;500:170–176. doi: 10.1016/j.bbrc.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 32.Mao Y., Wang Y., Dong L., Zhang Y., Zhang Y., Wang C., Zhang Q., Yang S., Cao L., Zhang X., Li X., Fu Z. Hypoxic exosomes facilitate angiogenesis and metastasis in esophageal squamous cell carcinoma through altering the phenotype and transcriptome of endothelial cells. J. Exp. Clin. Cancer Res. 2019;38:389. doi: 10.1186/s13046-019-1384-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X., Sai B., Wang F., Wang L., Wang Y., Zheng L., Li G., Tang J., Xiang J. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol. Cancer. 2019;18:40. doi: 10.1186/s12943-019-0959-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.