Abstract.
We are developing automated analysis of corneal-endothelial-cell-layer, specular microscopic images so as to determine quantitative biomarkers indicative of corneal health following corneal transplantation. Especially on these images of varying quality, commercial automated image analysis systems can give inaccurate results, and manual methods are very labor intensive. We have developed a method to automatically segment endothelial cells with a process that included image flattening, U-Net deep learning, and postprocessing to create individual cell segmentations. We used 130 corneal endothelial cell images following one type of corneal transplantation (Descemet stripping automated endothelial keratoplasty) with expert-reader annotated cell borders. We obtained very good pixelwise segmentation performance (e.g., Dice , , across 10 folds). The automated method segmented cells left unmarked by analysts and sometimes segmented cells differently than analysts (e.g., one cell was split or two cells were merged). A clinically informative visual analysis of the held-out test set showed that 92% of cells within manually labeled regions were acceptably segmented and that, as compared to manual segmentation, automation added 21% more correctly segmented cells. We speculate that automation could reduce 15 to 30 min of manual segmentation to 3 to 5 min of manual review and editing.
Keywords: deep learning, endothelial cell segmentation, cornea
1. Introduction
We are developing methods for quantitative analysis of corneal endothelial cell (EC) images as a means of assessing cornea health, particularly following a corneal transplant, one of the most common transplant surgeries (47,683 in the United States in 2018).1 Detection and management of cornea allograft rejection remains a significant problem, resulting in corneal endothelial damage and subsequent graft failure.1 In normal eyes, a single layer of hexagonally arranged ECs contains fluid-coupled ion channels that regulate fluid in the cornea and help maintain a clear cornea. An abnormal EC layer will have fewer ECs, lose its hexagonality, and contain cells with abnormal intensity patterns, leading to various manual analyses of images to assess the EC layer. A dysfunctional endothelium leads to cornea swelling and blurring of vision and in some cases ultimately leads to failure of the transplant.2,3 Our group has led two major studies evaluating the effect of various factors on the success and failure of corneal transplants, which included extensive manual analyses of the EC layer. The cornea donor study was a 10-year study that found there was no association between donor age and graft failure 10 years postpenetrating keratoplasty.4–10 The second study, the cornea preservation time study (CPTS), concluded that at 3 years, corneal transplants saw greater EC loss when the donor corneas were preserved for longer time (8 to 14 days) compared to corneas preserved for a shorter time (0 to 7 days).11–15 Our Cornea Image Analysis Reading Center (hereafter Reading Center) was responsible for manual image analysis of over 1900 EC images for these studies, as well as numerous other studies. Another report suggested that morphologic changes in the endothelium detected by specular microscopy image examination of the endothelium at standardized time intervals in DMEK patients showed that morphological characteristics of ECs were indicative of a future graft rejection.16
While corneal transplant specular microscopic EC images aid in cornea health assessment, they are challenging to manually annotate and analyze consistently for a variety of reasons. They can have poor image quality due to corneal thickening, illumination gradients, poor cell border definition, pleomorphism (cell shape variation), and polymegathism (cell size variation). It is important to annotate or segment the dark regions or cell borders that encompass full ECs as this will enable measurement of clinical morphometrics, such as endothelial cell density (ECD), coefficient of variation (CV), and percent hexagonality (HEX). These morphometrics aid in the indirect assessment of the corneal endothelium health.17
There have been previous reports of automatic segmentation methods, with advantages and disadvantages. Some approaches include watershed algorithms,18–20 genetic algorithms,17 and region-contour and mosaic recognition algorithms.21 However, some of these segmentation processes still require manual editing because they overestimate cell borders.22 Another limitation of such segmentation methods is that they can fail in the case of poor image quality. Poor quality images with low contrast or illumination shading due to specular microscopy optics and light scattering within the cornea hinder the ability of a traditional processing algorithm to learn adequately from these images.23,24 Previously, U-Net has shown promising results of cell segmentation via delineation of the cell borders.6 U-Net has been used to segment neuronal structures in electron microscopy, images that are visually similar to EC images (i.e., both contain dark cell border regions between brighter cell regions), leading us to believe that this approach would be applicable. Recent work by Fabijańska has demonstrated further proof regarding the high performance and accuracy of a U-Net-based learning approach in specular EC images.22 However, this study was conducted on a small set of 30 images, 15 training images and 15 test images, taken by a specular microscope. To the best of our knowledge, these images included varying cell densities of nondiseased ECs.25 Daniel et al. reported to have experimented with training the U-Net architecture on thousands of patches generated from over 300 EC images with a variety of diseases and cell densities. The training labels utilized for this network were generated from cell centroid markings instead of manual cell border tracing, and only the central and contiguous cells from the automatic segmentations were evaluated for comparison with the training labels.26 Finally, Vigueras-Guillén et al. trained a dense U-Net architecture to automatically segment corneal EC images, in particular those from glaucomatous eyes. They added a dense block into their structure but also trained on a dataset with less than 50 images.27
In this report, we applied a fully convolutional neural (FCN) network learning system to segment 130 specular-microscopic, EC images following Descemet stripping automated endothelial keratoplasty (DSAEK) acquired in the CPTS. This is the first report of automated analysis of low-quality images obtained following cornea transplantation. Images from the CPTS were obtained from 40 clinical sites with 70 surgeons examining a total of 1330 eyes over a 4-year duration using two different types of specular microscopes (Konan Medical, Irvine California), (Tomey Inc., Phoenix, Arizona), and a confocal microscope (Nidek Inc., San Jose, California).15 We utilized the U-Net convolution neural network for deep learning and semantic segmentation. Other contributions included new processing approaches suitable for low-quality images (e.g., adaptive Otsu and pruning). We quantitatively assessed results by standard methods (Dice coefficient and Jaccard index). In addition, as an automated method can segment cells not analyzed by operators and segment cells differently from an analyst, we analyzed results visually to determine the number and types of edits that an operator might make on segmented images.
2. Image Processing and Analysis
Our processing methods created a binary output image consisting of black cell border pixels and white “other” pixels, which include both EC regions and regions in the image where no borders could be identified due to poor image quality or an absence of contiguous cells. It should be noted that we define a cell border as a collection of curve segments that enclose a bright cell. The method included image preprocessing to reduce brightness variation in EC images; deep learning, FCN network semantic segmentation (U-Net) to generate probability outputs; and postprocessing to create binary, single-pixel-wide cell borders.
2.1. Image Preprocessing
We corrected the left to right varying brightness in EC images due to the specular microscope technique28 (Fig. 1). We used three different techniques for estimating the bias field. First, we generated a low-pass background image using a Gaussian blur and divided the original image by the background image to create a flattened image. We used a normalized Gaussian filter with standard deviation and a kernel foot print (). We set the 99th percentile of the flattened uint8 image’s intensity to the maximum intensity (255), and then normalized the rest of the image’s intensity so its distribution expanded the (0 to 255) intensity range. This method will be referred to as the division method. For the second preprocessing method, we again generated a low-pass background image via the same Gaussian blur previously mentioned, and then subtracted this background image from the original image. Again, we normalized the flattened image using the same method as before. This method will be referred to as the subtraction method. Third, we used a normalization technique specifically designed to enhance EC images, as described by Piorkowski et al.29 In this method, we normalized the brightness along the vertical and horizontal directions by adding the difference between the average image brightness and the average brightness in the corresponding row/column at each pixel. New pixel values are given as
| (1) |
where is the new pixel value, is the original pixel value, is the average brightness in the image, is the height of the image, and is the width of the image. This will be referred to as the localization method.
Fig. 1.
(a)–(f) Examples of specular microscopy images with varying ECD, duration post-transplant, and contrast from left to right across the image area.
2.2. Deep Learning Architecture
We used U-Net, a popular neural network architecture proposed by Ronneberger et al.30 that has been shown to work better than the conventional sliding window-based convolutional neural network approach for some image segmentation problems. U-Net, as compared to previous neural networks, has many advantages. Due to its decoding (downsampling), encoding (upsampling) architecture, and skip connections, the network is able to recover full spatial resolution in its decoding layers, allowing one to train such deep fully convolutional networks for semantic segmentation. The U-Net architecture contains 16 layers and has a receptive field size of (93, 93).31 The output of U-Net will be an array of probabilities (0 to 1), where values closer to 0 correspond to cell border and values closer to 1 correspond to cell or other. The U-Net network architecture used in this work is shown in Fig. 2. Optimization, training, and testing are described later.
Fig. 2.
U-Net architecture. Numbers on top and the left bottom corner of each tensor indicate number of channels/filters and its height and width, respectively. The network consists of multiple encoding and decoding steps with three skip connections between the layers (white arrows). Arrows are color-coded as follows: blue: convolution with (3, 3) filter size, ReLU activation; green: upconvolution with a (2, 2) kernel using max pooling indices from the encoding layers; red: max pooling with a (2, 2) kernel; and pink: (1, 1) convolution with sigmoid activation.
2.3. Binarization and Postprocessing
Postprocessing converted probability outputs from U-Net into binary images with black, single-pixel width cell borders and white other pixels, consisting of cells and regions where cells could not be identified due to low brightness or absence of cells. This allows for comparison to our cell border ground-truth labels. The binarization and postprocessing pipeline are shown in Fig. 3.
Fig. 3.
Adaptive thresholding (top or T) and adaptive Otsu (bottom or B) were used to convert (a) probability outputs from the U-Net algorithm to binarized, single-pixel width cell borders, which can be compared to ground truth labels. For adaptive threshold, steps are (b-T) binarization, (c-T) thinning to obtain single-pixel wide lines, (d-T) fill and open to remove extraneous branches, (e-T) multiplication with c-T to obtain the cells borders that fully enclose cells, (f-T) and finally pruning to remove small spurious segments. Image (f-T) was compared to the ground truth label image (g) for training and testing. Adaptive Otsu was processed the same way (bottom).
First, the output two-dimensional probability arrays from the network were scaled up from values (0 to 1) to create (0 to 255) grayscale images with dark cell borders and bright cells. An example image is shown in Fig. 3(a). We tested two binarization methods: adaptive Otsu thresholding and a simple sliding window local thresholding approach initially proposed by Sauvola and Pietikäinen32 Before applying adaptive Otsu, images were inverted to give bright borders. Adaptive Otsu generated a binary image with thick white borders and black other pixels consisting of cells and indeterminate regions [Fig. 3(b)-B]. For the sliding window local thresholding approach, the adaptive threshold was calculated within a [] sliding window at each pixel location using
| (2) |
where is the adaptive threshold value, is the mean intensity in the pixel neighborhood, is the standard deviation in the same neighborhood, is the difference between the maximum and minimum standard deviation across the image, and is a tunable parameter of the thresholding algorithm. If the value of the center pixel exceeds , then the pixel will be assigned a value of 255; if the center pixel value is less than , it will be reassigned as 0. The binarized image was inverted so that the end product was white cell borders with black cells and surrounding area [Fig. 3(b)-T].
Following both binarization methods, we performed a morphological thinning operation to narrow the now white cell borders to single-pixel width [Figs. 3(c)-T and 3(c)-B]. A flood-fill morphological operation delineated all borders and encapsulated cells white, keeping the outside surrounding area black. This process left small extraneous, unenclosed branches that needed to be removed. We performed a morphological area opening operation that identified and removed four-connected pixels or any connected components . The result of this step [Figs. 3(d)-T and 3(d)-B] was multiplied by the result directly after the morphological thinning step and generated the images in Figs. 3(e)-Tand 3(e)-B. The product image underwent a final pruning morphological operation to remove spurs, or any stray lines in the image before being inverted once more. The result was a binary image with single-pixel width black borders that were closed and fully encapsulated white cells, with white surrounding area [Figs. 3(f)-T and 3(f)-B].
2.4. Performance Metrics: Dice Coefficient and Jaccard Index
We computed two conventional performance metrics (Dice coefficient and Jaccard index) to determine the accuracy of automatic segmentation of cells as compared to the original manual labeling. Dice coefficient and Jaccard index were calculated on a cell-by-cell basis as
| (3) |
| (4) |
In the equations above, is a manually obtained cell region and its border pixels (the ground truth), and is the same for the corresponding automatic segmentation. To enable a cellwise analysis, each cell from the ground truth image and the automatic segmentation image was numerically labeled via connected components. For each ground truth cell, we identified the nearest, corresponding cell from the automated cell segmentation image, and calculated the corresponding Dice coefficient and Jaccard index.
2.5. Visual Analysis: Questions and Rating Scale
For a more clinically informative evaluation, which can identify cases where editing of automated results might be desired, we conducted a cell-by-cell visual analysis with an expert analyst and an ophthalmologist on held out images. Two regions with different concerns were identified. First, within the manually segmented cell region, we were concerned with merging or splitting of cells by the automated method. Second, within regions containing newly identified cells from the automated algorithm not identified in the ground truth image, we were concerned with identifying appropriately segmented cells. Two questions were asked of readers.
-
1.
When automated and manual cell detections differ (e.g., when automated method identifies two cells and manual identifies one cell or vice versa), which method correctly segmented the cell: manual, automatic, or an equivocal case. The number of discrepancies per image when answering this question was recorded as well.
-
2.
For each newly identified cell, indicate whether it has been automatically segmented accurately, inaccurately, or it is an equivocal case.
3. Experimental Methods
3.1. Labeled Dataset
EC images were collected retrospectively from the Reading Center along with their corresponding corner analysis performed in HAI CAS/EB cell analysis system software (HAI Laboratories, Lexington, Massachusetts).5 A subset of 130 images were randomly collected from the CPTS clinical research study on 1330 eyes of which 1251 were successful grafts and 79 experienced failures. Patient images were collected at 6- to 48-month time points postsurgery. None of the subset images used in this analysis experienced graft failure. All images were of size (446, 304) pixels with a pixel area of from Konan specular microscopes (Konan Medical, Irvine, California). Images were acquired over a 4-year period following DSAEK from different sites in the 40 sites in the study. Images contained between 8 and 130 cells per image with EC densities ranging between 600 and . All images were deidentified and handled in a method approved by the University Hospitals Cleveland Medical Center Institutional Review Board.
All images were manually analyzed using the standard operating procedure in the Reading Center. We used images without any processing steps to remove the varying brightness across the images. Trained readers in the Reading Center used the HAI corners method to segment the cell borders of the raw EC images.5 Cell labeling conventions and analyses are firmly established within the Reading Center, which has performed EC analysis utilizing a variety of analyses tools on over 300,000 EC images from 44 studies over the past 8 years. The HAI CAS/EB software (HAI Laboratories, Lexington, Massachusetts) allows readers to mark corners for each cell and then generates cell borders connecting the corners. In addition, common morphometrics, such as ECD, CV, and % of hexagonal cells (HEX), are computed in this software. Sample images with the manual cell border segmentations overlaid in green is shown in Fig. 4. We noticed that sometimes the manual annotated cell borders would occasionally appear as double borders (arrows in Fig. 4). To correct this, the borders were dilated slightly before thinning, producing single-pixel width borders for the labels.
Fig. 4.
Example EC image and its manual analysis. (a) This is a good quality sample EC image with cells of various shapes and sizes. Note the consistently obtained brightness variation from left to right. (b) The cell-border segmentation overlay (in green) was created by an analyst using the HAI CAS software. In this good quality image, segmentations were obtained throughout the image. Note that cells cut-off at the boundaries of the image are not labeled. (c) This is the binary segmentation map used as a ground-truth label image. Red arrows indicate example double borders generated by the HAI-CAS software in (b) that were replaced with single borders in (c) following processing (see text).
3.2. Classifier Training and Testing
The dataset of 130 images was split into a training/validation set and a held-out test set with 100 and 30 images, respectively. A 10-fold cross-validation approach was applied on the first set of 100 images, with 90 images in the training set and 10 images in the validation set. The validation set was used when training the neural network. Training was stopped when performance in the validation set determined by the metric validation loss did not improve.
Details of training follow. Images were initially padded in a symmetric fashion to ensure that convolutions were valid at the edges. The neural networks were trained using weighted binary cross entropy as the loss function. Class imbalance was accounted for by weighting the loss function by the inverse of the observed class proportions. In this manner, samples of a class that occur at a lower frequency (such as the cell border class in our case) will have a larger weight in the computation of the loss function. The network was optimized using the Adam optimizer with an initial learning rate of 1.0e-4. Finally, data augmentation was performed to allow the network to achieve good generalization performance. Briefly, the augmentations used were in the range of translations in height and width across the image, zoom, shear, and random horizontal flips. The network was trained for 200 epochs.
Software for image preprocessing and binarizing the network predictions were implemented using MATLAB R2018a. U-Net was implemented using the Keras API (with Tensorflow as backend), the Python programming language. Neural network training was performed using two NVIDIA Tesla P100 graphics processing unit cards with 11 GB RAM on each card.
The model with the lowest validation-loss-value during the training phase was applied to the held-out test set. Performance metrics (Dice coefficient and Jaccard index) were obtained on the held-out test sets. Experiments were conducted to evaluate preprocessing and postprocessing steps.
4. Results
We now report algorithm optimizations. Three different types of preprocessing methods (division, subtraction, and localization) were applied to flatten and normalize EC images. An example EC image processed by the three methods is shown in Fig. 5. All three methods enhanced the dark regions of the original image, with division yielding the most evenly bright image. We separately trained and tested the U-Net algorithm on the raw images and the three types of preprocessed images giving probability outputs in Fig. 6 for the test image from Fig. 5. In areas near the edges of the image, U-Net tends to give gray cell borders, indicating less confidence in the border result. As compared to other methods, division and subtraction tend to have better delineation of cell borders at edges of the image. Postprocessing generated final segmentation results (Fig. 7). All the automated results are very similar to those from manual labeling, except that the automated methods in all images, including this example, segmented more cells than were manually annotated [e.g., compare Figs. 7(b)–7(e) with Fig. 7(a)]. Each automated result has both poor (red circles) and good (green circles) cell border segmentations as compared to manual observation of the original input image [Fig. 6(a)]. We prefer division preprocessing, because in this, and other images, it gave new cell annotations and had the fewest number of obvious errors among the preprocessing methods. In addition, we determined that the adaptive Otsu thresholding binarization method was better than the sliding window local thresholding because it generated less erroneous splitting of cells (not shown).
Fig. 5.
Preprocessing shading correction methods applied to a representative EC image. (a) The original EC image is corrected by (b) the localization, (c) division, and (d) subtraction methods (see text). Note that the correction methods all significantly flattened the image.
Fig. 6.
Deep learning probability maps of an example test image following four types of preprocessing. (a) An original test image was processed with deep learning following (b) no preprocessing, (c) localization preprocessing, (d) division preprocessing, and (e) subtraction preprocessing. Note that in the center of the probability outputs, the borders appear more black due to the high confidence of the trained algorithm in these regions; however, toward the edges and some blurry areas of the original images, the borders appear more gray because the algorithm is less confident here (indicated by red arrows).
Fig. 7.
Final segmentation results of the EC image shown in Fig. 5 with four preprocessing methods. (a) Ground truth label, (b) final segmentation result after no preprocessing, (c) after localization preprocessing, (d) after division preprocessing, and (e) after subtraction preprocessing. The green circles indicate correct cell segmentation, whereas red circles indicate incorrect cell segmentation. Blue arrows indicate regions of newly segmented cells not previously annotated.
Using standard performance metrics, we analyzed segmentations on the held-out dataset. There were 1876 and 2362 cells from the manual and automated methods, respectively. Quantitative comparisons were only possible within the manually annotated regions, where manual and automated cell counts were 1876 and 1798, respectively, only a 4% difference. Appropriately high Dice coefficient and Jaccard index values were obtained (Table 1). From the cell-by-cell analysis, we believe the splitting/merging of cells had a low effect on the value of the quantitative metrics. Much of the “error” had to do with inconsequential differences at cell borders because our method gave curved cell border segments while the manual method consisted of straight-line approximations. Within manually annotated regions, 92% of automatically segmented cells had a Dice coefficient , indicating successful segmentation of a very large proportion of cells. Within manually annotated regions, division preprocessing performance was only very slightly better than the other two preprocessing methods (not shown).
Table 1.
Dice coefficient and Jaccard index metrics of segmentation results from the held-out test set images following the division preprocessing.
| Cell basis | ||||
|---|---|---|---|---|
| Metric | Minimum | Average | Maximum | Std. dev. |
| Dice coefficient | 0.00 | 0.87 | 0.98 | 0.17 |
| Jaccard coefficient | 0.00 | 0.80 | 0.96 | 0.18 |
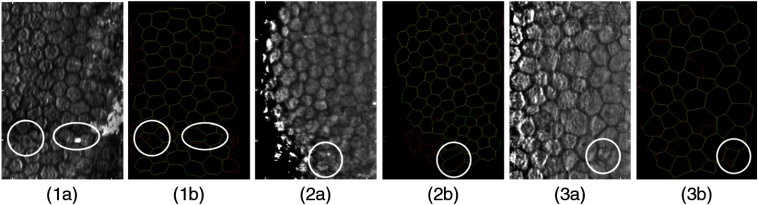
In addition, we analyzed the held-out dataset with more clinically informative, expert visual evaluations, within both the previously manually annotated regions as well as regions newly segmented by the automated method. In Fig. 8, there are three examples of images where the automatic segmentation split or merged cells, as compared to the manual annotations. It is important to note that many of these discrepancies occur because of false dark lines within cells that appear to be borders, faint borders that are indistinguishable from their neighboring cell regions, and cells in dark regions of the image with low-image quality. Figure 9 shows examples where the automated process identified and segmented new cells outside the region of manually annotated cells. These new cells were usually found in dark regions of the image that were enhanced via preprocessing. Results of visual analysis within manually annotated regions are shown in Figs. 10(a) and 10(b). Overall, only 2.3% of automatically segmented cells were deemed inaccurate. Most images (73%) contained only 0 to 2 discrepancies as compared to manual labeling. In regions newly segmented by the automated method, 564 cells were identified. Of these, nearly 70% (390) were deemed acceptably segmented, giving an additional 20% of new cells to be analyzed, as compared to original manual labeling. It is noted that experts could have been biased by previous conceptions of cells to be analyzed, possibly leading to actual cells being labeled as inaccurate. In Sec. 5, we interpret these data to suggest the amount of editing required in an automatically segmented image.
Fig. 8.
Three examples of EC images and corresponding segmentations highlighting cells with differences between manual (green lines) and automatic (red lines) segmentations. Yellow lines represent the overlap of the two segmentation methods. (a) EC images in the held-out test set (b) overlays of the manual and automatic segmentation results. Cases 1(a) and 1(b) show two examples in white circles where the automated method first split a single cell and then merged two cells into one cell. The reviewers agreed the manual annotations were more accurate than the automated segmentations. Cases 2(a) and 2(b) illustrate an example where the automated method merged two cells into one cell, and the reviewers concluded the case equivocal because it was unclear which method actually segmented the cell correctly. Cases 3(a) and 3(b) show an example where the automated method split one of the manually annotated cells into two cells. The reviewers considered these cases to have been segmented more accurately by the automated method than the manual annotation.
Fig. 9.
Three examples of EC images showing the automatic segmentation method identifying and segmenting cells outside of the sample area of the ground truth annotations. (a) Sample EC image from the independent held-out test set; (b) ground truth (green) and automatic (red) segmentations overlay showing cells outside of the ground truth sample area segmented by the U-Net algorithm.
Fig. 10.
Visual analysis of automated cells in the held-out test set consisting of 30 images. Within the manually analyzed regions, experts identified that only 52 of 1876 cells (3%) required reconsideration. A histogram of numbers of discrepancies is shown in (a), where 22 images (73%) contained 0, 1, or 2 potential discrepancies. Notably, among discrepancies, the automated result was deemed better or equivocal 16% of the time, giving only 44 (2.3%) of cells where experts deemed it desirable to change the automated result. In the regions newly segmented by the automated method, 564 new cells (an increase of 30%) were identified, with the image histogram shown in (c). Nearly 70% or 390 of newly segmented cells were deemed acceptable (accurate or equivocal in the pie chart d).
5. Discussion
Our algorithm successfully automatically segmented cell borders of challenging post-DSAEK EC images. As described previously, the CPTS study dataset used in this report includes a variety of EC images, which vary in image quality, time after surgery, ECD, and illumination/contrast. Ground truth annotations were from experts within our very established Reading Center. In the held-out test set, conventional metrics against previously manually annotated cells indicate successful segmentations with an average cell-by-cell Dice coefficient of . For the most part, discrepancy was at cell borders where the automated method gave rounded borders and the manual method was constrained to straight line approximations. The Dice coefficient exceeded 0.80 among these cells 92% of the time, suggesting a very large proportion of acceptable cells segmentations. This was confirmed with the visual analysis of cells within the previously manually annotated cell region. Only 2.3% of the automatically segmented cells were deemed unacceptable, and 73% of images had only 0 to 2 errors. Within the newly segmented regions, we obtained 390 acceptably segmented cells, or roughly an additional 20% as compared to the 1876 manually labeled cells.
We developed and optimized the preprocessing portion to enhance image quality and postprocessing pipeline to convert deep learning probability outputs into segmented images. Notably, final segmentation results were little affected by the preprocessing method within the manually annotated region, where all methods, including no preprocessing gave a bright region with good cell contrast. However, outside this region, the division and subtraction methods gave superior brightness and contrast as compared to other methods. We chose the division process because it accurately segmented more new cells outside of the manually annotated region. On the postprocessing side, adaptive Otsu thresholding proved to be superior to sliding window. It more accurately preserved correct cell borders and omitted less confident/inaccurate curve segments.
Since we intend to create software that includes the possibility of manual editing, we now interpret the visual analysis with this in mind. In the held-out test set, the automated method segmented about 30% more cells than the manual method (9% and 21%, needing and not needing editing, respectively). In general, it is quite easy from examination of the original input image to identify segmentation regions to eliminate. Such regions could be erased quite quickly using a proper interactive tool. More difficult are those cells where one would like to split or merge results. Fortunately, these are a small proportion of the total, suggesting that editing could be done quite quickly once they are recognized. A visual raster scan of a segmentation overlay with toggle viewing can pick up these instances in 1 to 2 min. We think that editing could be done in about 3 to 5 min per image, much less than the 15 to 30 min now required for a full manual segmentation.
In summary, deep learning automated segmentation of cell borders is successful in even challenging EC images following corneal transplantation. Accuracy of fully automated analysis is very good, but, if deemed necessary, results could be edited fairly quickly to give results comparable to fully manual annotations. Methods reported here will be utilized as a corner stone for future calculations of clinical morphometrics to help physicians assess the healthiness of corneal transplants.
Acknowledgments
This project was supported by the National Eye Institute through Grant No. NIH R21EY029498-01. The grants were obtained via collaboration between Case Western Reserve University, University Hospitals Eye Institute, and Cornea Image Analysis Reading Center. The content of this report was solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. There are no conflicts of interest. This work made use of the High-Performance Computing Resource in the Core Facility for Advanced Research Computing at Case Western Reserve University. The veracity guarantor, Chaitanya Kolluru, affirms to the best of his knowledge that all aspects of this paper are accurate. This research was conducted in space renovated using funds from an NIH construction grant (C06 RR12463) awarded to Case Western Reserve University.
Biography
Biographies of the authors are not available.
Disclosures
No conflicts of interest, financial or otherwise, are declared by the authors.
Contributor Information
Naomi Joseph, Email: nmj14@case.edu.
Chaitanya Kolluru, Email: cxk340@case.edu.
Beth A. M. Benetz, Email: beth.benetz@case.edu.
Harry J. Menegay, Email: Harry. Menegay@case.edu.
Jonathan H. Lass, Email: jonathan.lass@uhhospitals.org.
David L. Wilson, Email: david.wilson@case.edu.
References
- 1.Eye Bank Association of America, “Statistical report,” https://restoresight.org/what-we-do/publications/statistical-report/ (2018).
- 2.Waring G. O., et al. , “The corneal endothelium. Normal and pathologic structure and function,” Ophthalmology 89, 531–590 (1982). 10.1016/S0161-6420(82)34746-6 [DOI] [PubMed] [Google Scholar]
- 3.Edelhauser H. F., “The resiliency of the corneal endothelium to refractive and intraocular surgery,” Cornea 19, 263–273 (2000). 10.1097/00003226-200005000-00002 [DOI] [PubMed] [Google Scholar]
- 4.Piorkowski A., et al. , “Influence of applied corneal endothelium image segmentation techniques on the clinical parameters,” Comput. Med. Imaging Graphics 55, 13–27 (2017). 10.1016/j.compmedimag.2016.07.010 [DOI] [PubMed] [Google Scholar]
- 5.Benetz B. A., et al. , “Specular microscopy,” in Cornea: Fundamentals, Diagnosis, Management, Mannis M., Holland E., Eds., pp. 160–179, Elsevier, New York: (2016). [Google Scholar]
- 6.Lass J. H., et al. , “Endothelial cell density to predict endothelial graft failure after penetrating keratoplasty,” Arch Ophthalmol. 128, 63–69 (2010). 10.1001/archophthalmol.2010.128.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benetz B. A., et al. , “Endothelial morphometric measures to predict endothelial graft failure after penetrating keratoplasty,” JAMA Ophthalmol. 131, 601–608 (2013). 10.1001/jamaophthalmol.2013.1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugar A., et al. , “Factors predictive of corneal graft survival in the cornea donor study,” JAMA Ophthalmol. 133, 246–254 (2015). 10.1001/jamaophthalmol.2015.3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lass J. H., et al. , “Donor age and factors related to endothelial cell loss ten years after penetrating keratoplasty: specular microscopy ancillary study,” Ophthalmology 120, 2428–2435 (2013). 10.1016/j.ophtha.2013.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornea Donor Study Investigator Group et al. , “Donor age and corneal endothelial cell loss 5 years after successful corneal transplantation. Specular microscopy ancillary study results,” Ophthalmology 115, 627–632.e8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lass J. H., et al. , “Corneal endothelial cell loss 3 years after successful Descemet stripping automated endothelial keratoplasty in the cornea preservation time study: a randomized clinical trial,” JAMA Ophthalmol. 135, 1394–1400 (2017). 10.1001/jamaophthalmol.2017.4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stulting R. D., et al. , “Factors associated with graft rejection in the cornea preservation time study,” Am. J. Ophthalmol. 196, 197–207 (2018). 10.1016/j.ajo.2018.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel S. V., et al. , “Postoperative endothelial cell density is associated with late endothelial graft failure after Descemet stripping automated endothelial keratoplasty,” Ophthalmology 126, 1076–1083 (2019). 10.1016/j.ophtha.2019.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lass J. H., et al. , “Donor, recipient, and operative factors associated with increased endothelial cell loss in the cornea preservation time study,” JAMA Ophthalmol. 137, 185–193 (2019). 10.1001/jamaophthalmol.2018.5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lass J. H., et al. , “Cornea preservation time study: methods and potential impact on the cornea donor pool in the United States,” Cornea 34, 601–608 (2015). 10.1097/ICO.0000000000000417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monnereau C., et al. , “Endothelial cell changes as an indicator for upcoming allograft rejection following Descemet membrane endothelial keratoplasty,” Am. J. Ophthalmol. 158, 485–495 (2014). 10.1016/j.ajo.2014.05.030 [DOI] [PubMed] [Google Scholar]
- 17.Scarpa F., Ruggeri A., “Automated morphometric description of human corneal endothelium from in-vivo specular and confocal microscopy,” in 38th Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. (EMBC), pp. 1296–1299 (2016). 10.1109/EMBC.2016.7590944 [DOI] [PubMed] [Google Scholar]
- 18.Selig B., et al. , “Fully automatic evaluation of the corneal endothelium from in vivo confocal microscopy,” BMC Med. Imaging 15, 13 (2015). 10.1186/s12880-015-0054-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gavet Y., Pinoli J.-C., “Visual perception based automatic recognition of cell mosaics in human corneal endothelium microscopy images,” Image Anal. Stereol. 27, 53–61 (2011). 10.5566/ias.v27.p53-61 [DOI] [Google Scholar]
- 20.Vincent L. M., Masters B. R., “Morphological image processing and network analysis of cornea endothelial cell images,” Proc. SPIE 1769, 212–227 (1992). 10.1117/12.60644 [DOI] [Google Scholar]
- 21.Poletti E., Ruggeri A., “Segmentation of corneal endothelial cells contour through classification of individual component signatures,” in XIII Mediterranean Conf. Med. and Biol. Eng. and Comput. 2013, Springer, Cham, pp. 411–414 (2014). [Google Scholar]
- 22.Fabijańska A., “Segmentation of corneal endothelium images using a U-Net-based convolutional neural network,” Artif. Intell. Med. 88, 1–13 (2018). 10.1016/j.artmed.2018.04.004 [DOI] [PubMed] [Google Scholar]
- 23.Likar B., et al. , “Retrospective shading correction based on entropy minimization,” J. Microsc. 197, 285–295 (2000). 10.1046/j.1365-2818.2000.00669.x [DOI] [PubMed] [Google Scholar]
- 24.Piórkowski A., Gronkowska-Serafin J., “Selected issues of corneal endothelial image segmentation,” J. Med. Inf. Technol. 17 (2011). [Google Scholar]
- 25.Gavet Y., Pinoli J.-C., “A geometric dissimilarity criterion between Jordan spatial mosaics. Theoretical aspects and application to segmentation evaluation,” J. Math. Imaging Vision 42, 25–49 (2012). 10.1007/s10851-011-0272-4 [DOI] [Google Scholar]
- 26.Daniel M. C., et al. , “Automated segmentation of the corneal endothelium in a large set of ‘real-world’ specular microscopy images using the U-Net architecture,” Sci. Rep. 9, 4752 (2019). 10.1038/s41598-019-41034-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vigueras-Guillén J. P., et al. , “Fully convolutional architecture versus sliding-window CNN for corneal endothelium cell segmentation,” BMC Biomed. Eng. 1, 4 (2019). 10.1186/s42490-019-0003-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarey B. E., Edelhauser H. F., Lynn M. J., “Review of corneal endothelial specular microscopy for FDA clinical trials of refractive procedures, surgical devices and new intraocular drugs and solutions,” Cornea 27, 1–16 (2008). 10.1097/ICO.0b013e31815892da [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piorkowski A., et al. , “Selected aspects of corneal endothelial segmentation quality,” J. Med. Inf. Technol. 24, 155–163 (2015). [Google Scholar]
- 30.Ronneberger O., Fischer P., Brox T., “U-net: convolutional networks for biomedical image segmentation,” Lect. Notes Comput. Sci. 9351, 234–241 (2015). 10.1007/978-3-319-24574-4 [DOI] [Google Scholar]
- 31.Drozdzal M., et al. , “The importance of skip connections in biomedical image segmentation,” in Deep Learning and Data Labeling for Medical Applications, pp. 179–187, Springer, Cham: (2016). [Google Scholar]
- 32.Sauvola J., Pietikäinen M., “Adaptive document image binarization,” Pattern Recognit., 33, 225–236 (2000). 10.1016/S0031-3203(99)00055-2 [DOI] [Google Scholar]