Abstract
Mass spectrometry (MS) offers high levels of specificity and sensitivity in clinical applications, and we have previously been able to demonstrate that matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS is capable of distinguishing two-component cell mixtures at low limits of detection. Ovarian cancer is notoriously difficult to detect due to the lack of diagnostic techniques available to the medical community. By sampling a local microenvironment, such as the vaginal canal and cervix, a MS based method is presented for monitoring disease progression from proximal samples to the diseased tissue. A murine xenograft model of high grade serous ovarian carcinoma (HGSOC) was used for this study and vaginal lavages were obtained from mice on a weekly basis throughout disease progression and subjected to our MALDI-TOF MS workflow followed by statistical analyses. Proteins in the 4–20 kDa region of the mass spectrum yielded a fingerprint that we could consistently measure over time that correlated with disease progression. These fingerprints were found to be largely stable across all mice with the protein fingerprint converging towards the end point of the study. MALDI-TOF MS serves as a unique analytical technique for measuring a sampled vaginal microenvironment in a specific and sensitive manner for the detection of HGSOC in a murine model.
Keywords: MALDI-TOF MS, ovarian cancer, proteins, fingerprints, vaginal, microenvironment, murine model, detection, lavage, disease progression
Introduction
Ovarian cancer is a severe gynecological disease that is currently the fifth leading cause of cancer deaths among women in the United States, with an estimated 22,530 newly diagnosed cases and 13,980 deaths expected in 2019.1 This disease has a five-year survival rate of 93% if diagnosed when tumor growth is limited to reproductive organs such as the ovaries or fallopian tubes.2,3 However, the majority of diagnoses occur during the later stages of the disease when tumors have already metastasized, resulting in a five-year survival rate of less than 30%. Around 90% of these diagnoses are of epithelial origin, of which high-grade serous ovarian cancer (HGSOC) is the most common subtype.3–5 These late-stage diagnoses can be attributed to the limited diagnostic methods available for implementation in routine women’s health exams. Current diagnostic methods, such as transvaginal ultrasounds and CA-125 biomarker tests, are generally used only after patients present with symptoms as they are more invasive procedures than what is performed in a typical health screening. These methods also suffer from inaccuracy and a lack of specificity, which limits their use in diagnosing ovarian cancer in its early stages.6,7
In a routine health screening, women receive a Pap smear to determine cervical health but recent data suggests that these samples may be repurposed for ovarian cancer detection.8 This, along with other studies, shows that DNA from ovarian cancer cells can be detected in clinical samples, such as Pap smears, which represents a concentrated sample from the local microenvironment in the female reproductive system.8–10 This clinical evidence would suggest that cells and/or cellular fragments from ovarian tumors reside outside of the proximal environment in areas such as the cervix or vagina. Further confirmation of the Pap smear linked to HGSOC detection was recently detailed in a case report, in which adenocarcinoma cells derived from HGSOC precursor lesions were observed in a cervical smear.11 This case study indicates that developing a technology or measurement using Pap smears may allow for the detection of HGSOC.
While reliable early diagnostic markers for ovarian cancer have remained elusive, research has moved closer towards characterizing the proteome of a healthy female reproductive system using MS.12 For instance, the iKnife and MassSpec Pen are both innovative devices that couple MS signatures in lipids to the detection of cancerous tissue in a surgical setting.13,14 Work has recently been done using mass spectrometry (MS) to investigate Pap smears under the assumption that cells, cellular debris or secreted proteins from the female genital tract would be present.12 Bottom-up proteomics using a LTQ Orbitrap mass spectrometer was performed on liquid Pap smear samples from women considered to be healthy to create a ‘Normal’ Pap Test Core Proteome.12 This analytical method is highly sensitive but it does require significant sample preparation such as sample cleanup to remove salts and insoluble particulates, protein digestion, and lengthy liquid chromatography experiments leading to a time investment, as well as a high degree of expertise needed to operate a high mass resolution tandem mass spectrometer (LC-MS/MS). We chose to investigate the use of matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS since this is considered a versatile analytical technique used in a variety of applications ranging from the characterization of microbial species to biomarker detection using protein signatures.15–17 For example, the MALDI-TOF Biotyper system is currently FDA approved for use in clinical diagnostic laboratories for microbial typing.18 MALDI-TOF MS has also been used to characterize mammalian cells based on spectral fingerprints, which in turn, allows for their identification.19–21 Perhaps in combination with genomic sequencing, the ability to directly detect proteins, particularly those that change in abundance over time from a local source, may improve strategies that can be leveraged for detection.
As previous studies have highlighted that ovarian cancer cells and associated cellular debris have the ability to travel through the female reproductive system to the cervix, we felt it would be feasible to detect these changes to the reproductive environment using MALDI-TOF MS based on whole-cell fingerprinting with protein signatures.8,10,21 As a proof of principle for this concept, we used a murine model of HGSOC. In this study, we used vaginal lavages (analogous to Pap smears) in a murine xenograft model to detect the increasing burden of ovarian cancer based on protein fingerprints in the 4–20 kDa mass range obtained using MALDI-TOF MS (Figure 1). Statistical analysis of the protein signatures found candidate proteins that both increase and decrease over time as disease progresses highlighting the importance of sampling a local microenvironment proximal to disease origin. By sourcing a local microenvironment, we hoped to achieve greater sensitivity and specificity while also exploring the ability of small proteins to lend themselves as possible biomarkers of disease progression.
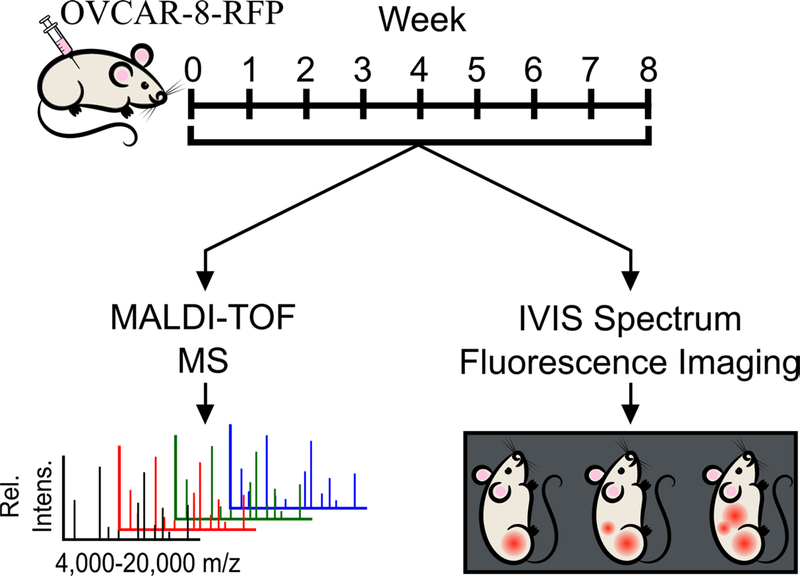
Figure 1.
Workflow of murine xenograft study. Female athymic mice were IP injected with OVCAR-8-RFP and tumors were allowed to develop over an eight-week period. Each week, mice were given vaginal lavages and were imaged using an IVIS imaging system to track tumor burden. Vaginal lavages were analyzed using MALDI-TOF MS to obtain mass spectra for statistical analyses.
Materials and Methods
OVCAR-8, a human-derived ovarian cancer cell line, expressing red fluorescent protein (RFP) was a gift from M. Sharon Stack at the University of Notre Dame.22,23 Additional information regarding the Materials and Methods section can be found in the Supporting Information.
Fluorescence-activated cell sorting (FACS)
Cells obtained from murine vaginal lavages of healthy mice were obtained and counted using a Cellometer K2 cell counter (Nexcelom, Lawrence, MA) to obtain quantitative values for observed leukocytes and cornified epithelial cells. Standard solutions containing 1×106 cells for each condition were prepared in PBS. Additionally, OVCAR-8-RFP cells were trypsinized, resuspended in medium and subsequently counted using a Cellometer K2 cell counter. Spiked lavage samples containing 0%, 0.1%, 1%, 10% and 100% OVCAR-8-RFP cells were prepared in PBS with a final volume of 500 μL. Flow cytometry was used to measure the percentage of OVCAR-8-RFP (Cytoflex, Brea, CA). Samples comprised of 100% murine lavage cells (negative control) and 100% OVCAR-8-RFP cells (positive control) were used to set up the gates.
In Vivo Murine Xenograft Study
5×106 OVCAR-8-RFP cells suspended in PBS were injected intraperitoneally into NCr nu/nu athymic female mice (N=5) aged 10 to 12 weeks (Taconic, Rensselaer, NY). Tumor burden was monitored on a weekly basis using a Xenogen IVIS® Spectrum In Vivo Imaging System (PerkinElmer, Waltham, MA). Mice were also given weekly vaginal lavages using 200 μL of sterile PBS throughout the study to collect cells from the local microenvironment of the reproductive organs. Cells sourced from lavages were counted and spun down to remove PBS as it has been shown to suppress MALDI-TOF ionization.21 This initial centrifugation step also removes mucous and mucous-associated proteins, such as mucin. Cells are then normalized to 10,000 cells/μL using DI water and stored at −80°C following collection. Upon addition of DI water, cells undergo osmotic stress and lyse, which removes the need for a wash step as all remaining proteins will be in solution. This lysing step prior to analysis also allows for proteins or other similarly sized molecules to be detected via MALDI-TOF MS. After two months of tumor progression, all animals were humanely sacrificed followed by collection of tumors and reproductive organs.
MALDI-TOF MS
Frozen lavage suspensions were thawed on ice and diluted to 5,000 cells/μL using deionized water. Equal volumes of lavage samples and a 20 mg/mL sinapic acid matrix solution (Sigma Aldrich, St. Louis, MO) in 30/70 ACN/H2O with 1% TFA were mixed together for a final concentration of 2,500 cells/μL. Samples were placed on ice for ten minutes and 1.5 μL of each sample was spotted onto a 384- well ground steel MALDI target plate with 24 technical replicates per lavage with Protein Standard I (Bruker Daltonics, Billerica, MA) used as a calibrant. This procedure was previously optimized and described in depth in Petukhova et. al.21 An Autoflex Speed LRF MALDI-TOF mass spectrometer (Bruker Daltonics, Billerica, MA) was used to acquire mass spectra of murine vaginal lavages samples in positive linear mode with a mass range of 4 to 20 kDa using a laser power of 75%, a laser width of 3 (medium) and a gain of 18.1x. Protein spectra was acquired using AutoXecute, an automated run function of FlexControl v. 3.4 software (Bruker Daltonics, Billerica, MA). 4,000 laser shots were accumulated in fifty shot increments for each sample using the random walk option. All mass spectra were externally calibrated using Protein Standard I (Bruker Daltonics, Billerica, MA), which had been spotted directly adjacent to each sample spot in a square pattern.
Spectra were pre-processed using FlexAnalysis v. 3.4 (Bruker Daltonics, Billerica, MA) with baseline subtraction (TopHat algorithm) and smoothing (SavitzkyGolay algorithm, width of m/z 5 over five cycles). Peak picking was performed using a signal-to-noise threshold of four following processing. Mass spectra were converted to a mzML format and further pre-processed using MALDIquant, an open-source R package, (https://cran.r-project.org/web/packages/MALDIquant/MALDIquant.pdf) using a standard workflow consisting of steps such as baseline correction and spectra alignment; resulting in a feature matrix comprised of intensity and m/z values shown in (Figure S1). Spectral data are publicly available online (ftp://MSV000083628@massive.ucsd.edu).
Statistical Analysis
In order to search for features that were differentially expressed throughout the longitudinal sampling points in every mouse, a Wilcoxon rank-sum test was applied, using the abundances of each feature, to assess whether the abundance differences between all pairs of days within one mouse were statistically significant. The Wilcoxon rank-sum test was chosen because it makes very few assumptions about the distribution of the data, e.g. normality in the case of t-tests. A Bonferroni-correction was applied to the p-value of each test to account for multiple hypothesis testing and a threshold of 0.01 was applied on the corrected p-value for each test. Source code is available online (https://github.com/mwang87/ometa_mousemaldi).
Results and Discussion
In Vivo Imaging to Monitor Tumor Progression in OVCAR-8 Xenograft Model
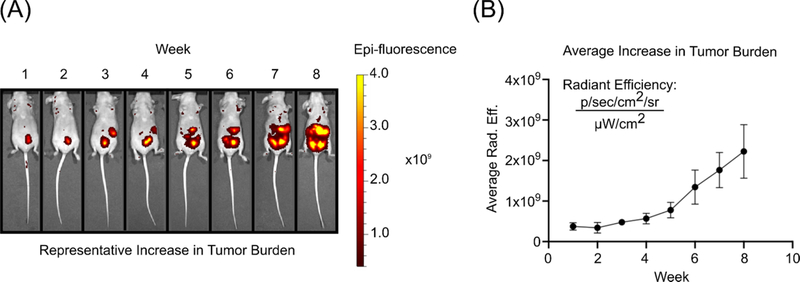
The presentation of HGSOC in vivo in our xenograft model was brought about by the use of OVCAR-8, a human-derived ovarian carcinoma cell line. OVCAR-8 displays a p53 mutation which is found in up to 96% of HGSOC tumors and is responsible for increased tumorigenic and metastatic potential as evidenced in previous animal studies.24–26 Following the IP injection of OVCAR-8-RFP cells, all mice were imaged every seven days and their respective fluorescence increased in intensity and tumors colonized the peritoneum over time (Figure 2 and S2). At the conclusion of the longitudinal study, all five of the mice had significant metastatic tumor burden and four out of the five presented with ascites, a buildup of fluid in the abdominal cavity, which is a symptom present in over a third of ovarian cancer patients.27
Figure 2.
(A) In vivo fluorescence imaging of OVCAR-8-RFP tumors in athymic nude mice over an eight-week period (B) Average radiant efficiency over time for all mice indicating an increase in tumor burden
Vaginal Lavages from a HGSOC Model Yields Distinct Differences from Other Disease Models
Based on our previous work with two-component mixtures with different cell lines, it was anticipated that a vaginal lavage taken over time would be useful for monitoring HGSOC.21 In order to verify that these protein fingerprints are unique to disease state and not a result of any peritoneal disease, mice with non-alcoholic steatohepatitis (NASH), which results in severe liver damage, were age-matched to those in our OVCAR-8 xenograft model in its seventh week and vaginal lavages were collected. Upon comparison of average spectra for each condition, it was observed that the protein signatures were distinct between the two peritoneal diseases (Figure S4). Therefore, our MALDI-TOF MS methodology yields distinct, disease specific protein fingerprints from the vaginal microenvironment. Additionally, while our study consisted of vaginal lavages on a weekly basis, we wanted to ensure that significant changes to the protein fingerprint are truly due to the progression of HGSOC and not inflammation of the vaginal cavity. Repetitive vaginal lavages were performed on healthy athymic nude mice over a five day period and protein signatures were not significantly different from day-to-day (Figure S5). It is also important to note that the murine estrous cycle spans a period of four to five days so slight changes could be attributed to this phenomenon.
Use of MALDI-TOF for Analysis of Vaginal Lavage Samples
MALDI-TOF MS had several advantages for this study such as solvent-free, rapid data acquisition (milliseconds), minimal sample requirement and the availability of this technology in clinical laboratories.15–18 It also allows rapid collection of a protein fingerprint over a large mass range, providing intact protein masses. Similar to how the Biotyper system (Bruker Daltonics) works for microbial identifications, the collection of a protein fingerprint allows for the comparison of different spectral patterns, in terms of the appearance and absence of features, to look for matches based on characteristic peaks.18 This is advantageous to our study as we have the ability to statistically analyze and compare protein fingerprints collected at various time points throughout the study to look for changes in protein peaks during tumor progression. Our lab had previously identified the small protein mass range (4–20 kDa) as having a higher number of characteristic peaks as well as the most distinctive when comparing cell types, particularly OVCAR and murine oviductal cell lines.21 Therefore, protein fingerprints from this mass range were collected for murine vaginal lavages from weeks one through seven of the xenograft study, as they were most likely to yield the richest spectral data. It is also important to note that each mouse in the study acted as their own biological control prior to the IP injection with OVCAR-8-RFP. This not only reduced the overall number of animals necessary to complete the study but also facilitated as few biological variables as possible when comparing protein fingerprints from their respective lavages.
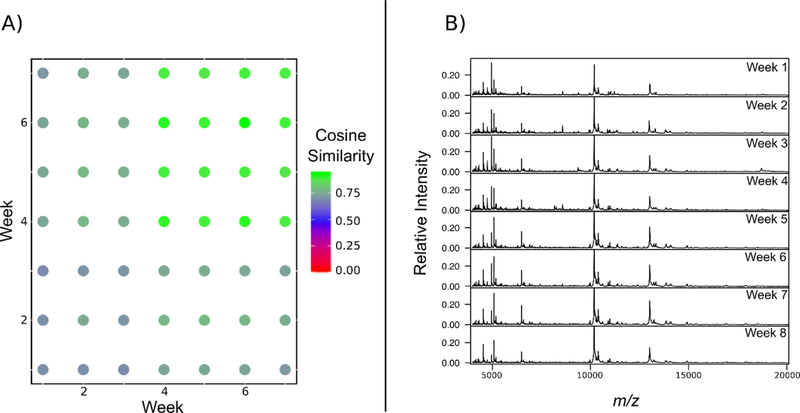
In order to begin to visualize the data, the cosine similarity of the murine vaginal lavages in the form of a dot product matrix was used (Figure 3A). In this instance, cosine similarity is a measurement of how similar two groups of spectra are from each other with 0 being completely dissimilar and 1 being an exact match.28 This allows for an easily accessible view of how the profiles may differ over time when compared to specific weeks in the study. Additionally, representative consensus spectra from each time point of all five mice was generated (Figure 3B), with consensus spectra for each individual mouse in the study is found in the Supporting Information (Figures S6–10). At the beginning of the study, each biological replicate (N=5) had a relatively unique profile when compared to others at or around the same time point with lower cosine score of around 0.7 being observed. However, one of the striking results was that at the end of the study during the seventh week, the protein fingerprints from the vaginal lavages are largely conserved, which would indicate that all five mice have similar protein profiles once the tumor burden is representative of late stage ovarian cancer. This would imply that there are unique processes and/or cellular responses associated with the progression of HGSOC that can be captured in a spectral protein fingerprint. All detected peaks that were found to be differentially expressed across time points can be found in the Supporting Information (Table S1).
Figure 3.
(A) Dot matrix plot comparing spectral similarity between time points. High cosine scores (green) indicates that the global profiles are highly similar between intersecting time point (i.e. weeks six and seven) while low cosine scores (red) indicates less similarity in features between the spectra of the respective time points. (B) Consensus spectra of vaginal lavages from all mice (N=5) throughout tumor progression for a total of eight weeks (n=24 for each mouse at each time point).
This late stage similarity prompted us to search for the presence and absence of protein features across all mice across paired time points (i.e. week one versus week seven). We found that multiple proteins began to dominate the mass spectra throughout disease progression and that a larger number of proteins either appeared and become absent from the protein fingerprint over time. This led to the investigation of specific regions of the spectrum and the verification that specific peaks became either significantly up- or downregulated over time, which would suggest that tumor progression does, in fact, induce change in specific processes in our murine model system. Several research groups have studied the proteome of cervical-vaginal fluid (CVF) obtained from women found to be in good health and while some of the proteins that were identified fell within the mass range detailed in this study, such as thioredoxin and profilin-1, there is not much known about the reproductive proteome between m/z 4,000–20,000.12,29–33 However, some studies have shown that proteins falling into our targeted mass range can be detected via proteomic analysis of CVF, inclusive of proteins responsible for cell proliferation and immune response.30,31 Some have speculated that this mass range contains an abundance of histones or ribosomal proteins based on previous whole-cell fingerprinting research and we hope to expand the knowledge of this area in the future through the identification of detected proteins using MS.19
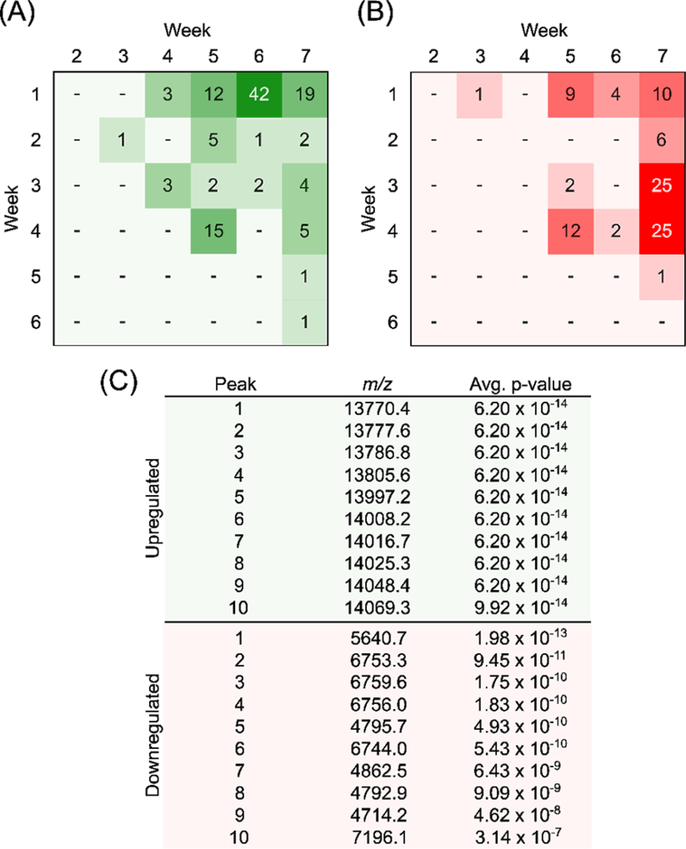
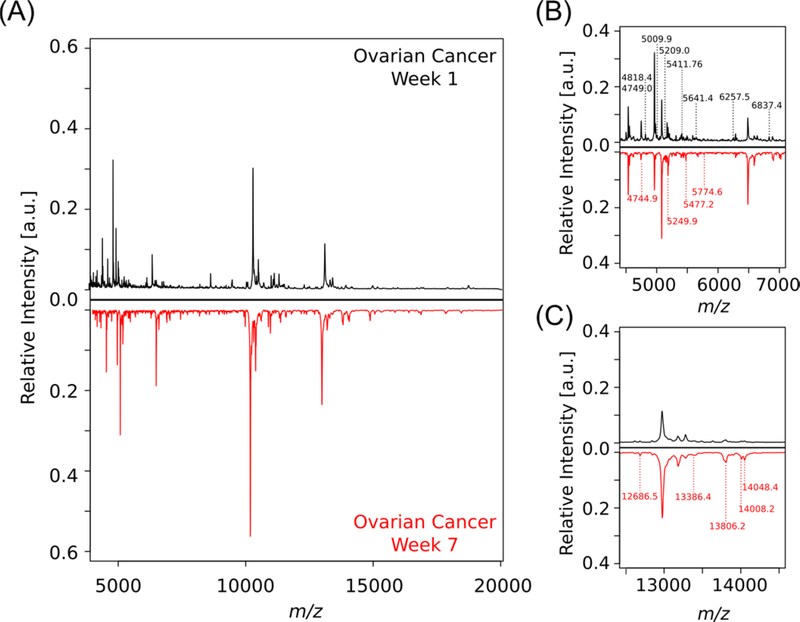
The Wilcoxon-rank sum test was used for the normalized intensity values from each observed peak and ranks them in terms of value for comparison. This statistical test makes minimal assumptions about the data, which allows it to be used in cases of non-normal distributions. Following analysis, specific features, shared between all mice, were found to be significantly change in intensity throughout the time course of the study (Figure 4). The determination of the test statistic in each comparison (week to week) allowed for the calculation of a p-value for significance. This analysis allowed for the identification of 118 upregulated and 97 downregulated spectral features when comparing data from different time points, implying that there were multiple significant changes over time in our mouse model. In particular, there are 19 m/z values and 10 m/z values that are significantly up- and downregulated, respectively, in all mice (p <0.001). Interestingly, the significant features between the initial and final time points of the study are segregated into different subsets of the mass range (Figure 5). The majority of the signals that appear over time are approximately twice the mass as those observed to be downregulated. This may point to a dysregulation in protease activity and requires further validation on the identity of these proteins to further test the possible mechanism by which these proteins accumulate in the disease state.
Figure 4.
(A) Pivot table showing number of significantly upregulated signals observed across time points across all five mice. (B) Pivot table showing number of significantly downregulated signals observed across time points across all five mice. (C) Top ten m/z values according to average p-values when comparing weeks one and seven.
Figure 5.
Mirror plot of average protein fingerprints of murine lavages comparing weeks one and seven in the ovarian cancer murine model (N=5, n=24). (A) Full spectra (m/z 4,000–20,000) mirror plot. In regions with significant differences peaks have been labelled in B and C. Peaks with no significant differences have been left unlabelled. (B) Spectral features are in this region (m/z 4,500–7,000) differ in terms of peaks and intensity with between time points peaks labelled in week 1 are those that are downregulated overtime, whereas peaks shown in week 7 have been upregulated overtime. (C) Spectral features in this region (m/z 12,500–14,500) present a second mass region in which labeled peaks are indicative of those unique to a specific time point which have been upregulated overtime.
Origin of the proteins detected do not appear to be escaped cells
Given our ability to detect unique signatures using MALDI-TOF MS, the next question we sought to examine was whether we were detecting signatures from escaped cells. Deep sequencing technologies have demonstrated the ability to amplify ovarian-cancer derived cells in vaginal fluids, vide supra. As part of the study design we sought to utilize the fluorescence of the OVCAR-8 RFP cells in vaginal lavages to correlate protein signatures to escaped cell populations. However, we were not able to detect any fluorescent proteins during the lavage cell counting. This finding was confirmed using FACS in a controlled cell spiking study; some fluorescence quenching occurs in controlled mixtures and yields robust fluorescence when the composition of RFP labelled cells is greater than 1% of the otherwise healthy lavage background (Figure S3). This is inline with our previous whole cell MALDI-TOF MS proof of principle study in which detection of multiple distinguishing features occurs with increasing percentages of cancer cells (5–10%) mixed with healthy cells and yields only a limited number of unique features when 1% of cancer cells are in an otherwise healthy heterogenous two-component mixture.21
Taken together, we believe that it is possible that while a small proportion of tumorigenic cells escape into the vaginal microenvironment, it is unlikely that the signatures we are directly detecting arises from these escaped cells. In our MALDI-TOF MS method, cells are lysed prior to analysis, and our detection of proteins is from all cell types and cellular debris within lavage samples instead of whole intact cells, making the source of these proteins ambiguous. For instance, we cannot differentiate between cellular origination from tumor cells versus those found in the murine reproductive system. Moreover, it could also be possible that we are observing the cellular response to cancer. In which case, this represents an exciting avenue for further study as these protein signatures does not rely on our ability to detect escaped cells within the lavage, as we do not have the ability to amplify our MS signals, as is done in deep sequencing studies. This could potentially provide an excellent opportunity to multi-plex sequencing and amplification that comes from tumor cells with changes in the vaginal microenvironment. We have also further demonstrated that the response is unique at least to ovarian cancer as the NASH murine fingerprint in aged, sexed matched mice were distinct, which represents an entirely different peritoneal disease. The observance of statistically significant changes in feature intensity, regardless of whether the proteins are from ovarian cancer tumor cells or not, is of importance for this study as we are not focused on the appearance of tumor cells in the murine lavage but rather changes in the overall system.
While the spectral fingerprint obtained from the murine vaginal lavages is a promising start to better understand the cellular processes that occur throughout the progression of ovarian cancer, it is of note that while MALDI-TOF mass spectrometers allow for the observance of intact protein masses, they do not have the mass resolving power in linear mode to provide an exact mass for the detected spectral features. Bottom-up proteomics, performed on high-resolution instruments such as hybrid quadrupole-orbitraps, will be required to definitively identify these proteins via analysis of peptide fragments. Bottom-up proteomics has its own limitations such as potentially not having enough cleavage sites for the digestion enzyme due to the small size of the proteins. However, using a combination of both MS methods to obtain an intact mass along with tandem fragmentation will allow us to identify these proteins in the future. The identification of these proteins could allow us to differentiate the species origin of the proteins (murine versus human) which may continue to shed light on our understanding of cellular escape versus host response as it relates to our ability to directly detect proteins using MALDI-TOF MS from vaginal fluids.
Conclusion
Based on statistical analysis with the Wilcoxon rank-sum test, we have been able to identify intact m/z values of small proteins obtained from murine vaginal lavages. We conclude that the protein fingerprint region obtained from our MALDI-TOF MS technique can be used to detect the presence of ovarian cancer. By sourcing samples from a local microenvironment, such as the vaginal cavity or cervix, we have shown that significant changes in protein signatures can be observed over time in the progression of ovarian cancer in a murine model. Our next step will be to further investigate and identify these proteins of interest using tandem MS to gain a better understanding of their role in disease progression in our in vivo model. A deeper understanding of these signature proteins and the processes they are involved in may further enhance our ability to design a diagnostic test.
Supplementary Material
Figure S1. Pre-processing workflow using the MALDIquant package in R
Figure S2. In vivo fluorescence imaging of OVCAR-8-RFP tumors in athymic mice
Figure S3. Fluorescent Cell Spiking Study
Figure S4. Mirror plot comparing protein fingerprints from week seven in a HGSOC and NASH model
Figure S5. Mass spectra of repeat murine lavages from healthy mice
Figure S6. Consensus spectra of vaginal lavages from Mouse 901
Figure S7. Consensus spectra of vaginal lavages from Mouse 902
Figure S8. Consensus spectra of vaginal lavages from Mouse 903
Figure S9. Consensus spectra of vaginal lavages from Mouse 904
Figure S10. Consensus spectra of vaginal lavages from Mouse 905
Table S1. Up-and downregulated spectral features differentially expressed across various time points throughout the study
Acknowledgements
This research was supported by the National Center for Advancing Translational Sciences, National Institute of Health, through grant UL1TR002003 (JEB & LMS), UG3 ES029073 (JEB), R01 CA240301 (JEB), F30 CA217079 (ANY), the Research Corporation for Science Advancement Scialog® award #26222 (LMS), UIC Provost’s Graduate Research Award (MMG), and University of Illinois at Chicago Start-up Funds (LMS). The authors would also like to acknowledge Daniel D. Lantvit, a research specialist in the Burdette lab, for his expertise and assistance with initiating our in vivo work. We would also like to acknowledge Dr. Natalia Nieto at UIC for allowing us to collect samples during her animal study concerning NASH.
Footnotes
Dr. Mingxun Wang is a consultant for Prometheus Laboratories Inc., a diagnostics company. However, Dr. Wang’s involvement with Prometheus Laboratories Inc. is not related to the material in the submitted work.
Supporting Information
The following supporting information is available free of charge at ACS website http://pubs.acs.org:
References
- (1).Siegel RL; Miller KD; Jemal A Cancer Statistics, 2019. CA Cancer J. Clin. 2019, 69 (1), 7–34. [DOI] [PubMed] [Google Scholar]
- (2).Noone AM; Howlader N; Krapcho M; Miller D; Brest A; Yu M; Ruhl J; Tatalovich Z; Mariotto A; Lewis DR; et al. SEER Cancer Statistics Review, 1975–2015, National Cancer Institute; Bethesda, MD, https://seer.cancer.gov/csr/1975_2015/, based on November 2017 SEER data submission, posted to the SEER web site, April 2018 [Google Scholar]
- (3).Torre LA; Trabert B; DeSantis CE; Miller KD; Samimi G; Runowicz CD; Gaudet MM; Jemal A; Siegel RL Ovarian Cancer Statistics, 2018. CA Cancer J. Clin. 2018, 68 (4), 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Klinkebiel D; Zhang W; Akers SN; Odunsi K; Karpf AR DNA Methylome Analyses Implicate Fallopian Tube Epithelia as the Origin for High-Grade Serous Ovarian Cancer. Mol. Cancer Res. 2016, 14 (9), 787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Labidi-Galy SI; Papp E; Hallberg D; Niknafs N; Adleff V; Noe M; Bhattacharya R; Novak M; Jones S; Phallen J; et al. High Grade Serous Ovarian Carcinomas Originate in the Fallopian Tube. Nat. Commun. 2017, 8 (1), 1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).van Nagell JR, Jr, DePriest PD; Reedy MB; Gallion HH; Ueland FR; Pavlik EJ; Kryscio RJ The Efficacy of Transvaginal Sonographic Screening in Asymptomatic Women at Risk for Ovarian Cancer. Gynecol. Oncol. 2000, 77 (3), 350–356. [DOI] [PubMed] [Google Scholar]
- (7).Buys SS; Partridge E; Black A; Johnson CC; Lamerato L; Isaacs C; Reding DJ; Greenlee RT; Yokochi LA; Kessel B; et al. Effect of Screening on Ovarian Cancer Mortality: The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Randomized Controlled Trial. JAMA 2011, 305 (22), 2295–2303. [DOI] [PubMed] [Google Scholar]
- (8).Kinde I; Bettegowda C; Wang Y; Wu J; Agrawal N; Shih I-M; Kurman R; Dao F; Levine DA; Giuntoli R; et al. Evaluation of DNA from the Papanicolaou Test to Detect Ovarian and Endometrial Cancers. Sci. Transl. Med 2013, 5 (167), 167ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Bakkum-Gamez JN; Wentzensen N; Maurer MJ; Hawthorne KM; Voss JS; Kroneman TN; Famuyide AO; Clayton AC; Halling KC; Kerr SE; et al. Detection of Endometrial Cancer via Molecular Analysis of DNA Collected with Vaginal Tampons. Gynecol. Oncol. 2015, 137 (1), 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Erickson BK; Kinde I; Dobbin ZC; Wang Y; Martin JY; Alvarez RD; Conner MG; Huh WK; Roden RBS; Kinzler KW; et al. Detection of Somatic TP53 Mutations in Tampons of Patients with High-Grade Serous Ovarian Cancer. Obstet. Gynecol. 2014, 124 (5), 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Shintaku M; Taniguchi H; Yamamoto Y; Kono F; Sumitomo M Detection of Tumor Cells of Serous Tubal Intraepithelial Carcinoma (STIC) in Cervical Smears and Rapid Development of the Ovarian Involvement: A Case Report. Diagn. Cytopathol 2018, 46 (11), 945–949. [DOI] [PubMed] [Google Scholar]
- (12).Boylan KL; Afiuni-Zadeh S; Geller MA; Hickey K; Griffin TJ; Pambuccian SE; Skubitz AP A Feasibility Study to Identify Proteins in the Residual Pap Test Fluid of Women with Normal Cytology by Mass Spectrometry-Based Proteomics. Clin. Proteomics 2014, 11 (1), 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Balog J; Sasi-Szabó L; Kinross J; Lewis MR; Muirhead LJ; Veselkov K; Mirnezami R; Dezső B; Damjanovich L; Darzi A; et al. Intraoperative Tissue Identification Using Rapid Evaporative Ionization Mass Spectrometry. Sci. Transl. Med. 2013, 5 (194), 194ra93. [DOI] [PubMed] [Google Scholar]
- (14).Zhang J; Rector J; Lin JQ; Young JH; Sans M; Katta N; Giese N; Yu W; Nagi C; Suliburk J; et al. Nondestructive Tissue Analysis for Ex Vivo and in Vivo Cancer Diagnosis Using a Handheld Mass Spectrometry System. Sci. Transl. Med 2017, 9 (406). 10.1126/scitranslmed.aan3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Fenselau C; Demirev PA Characterization of Intact Microorganisms by MALDI Mass Spectrometry. Mass Spectrom. Rev. 2001, 20 (4), 157–171. [DOI] [PubMed] [Google Scholar]
- (16).Croxatto A; Prod’hom G; Greub G Applications of MALDI-TOF Mass Spectrometry in Clinical Diagnostic Microbiology. FEMS Microbiol. Rev. 2012, 36 (2), 380–407. [DOI] [PubMed] [Google Scholar]
- (17).Rodrigo MAM; Zitka O; Krizkova S; Moulick A; Adam V; Kizek R MALDI-TOF MS as Evolving Cancer Diagnostic Tool: A Review. J. Pharm. Biomed. Anal. 2014, 95, 245–255. [DOI] [PubMed] [Google Scholar]
- (18).Maier T; Kostrzewa M Spectrophotometric Identification of Microbe Subspecies. 20110012016:A1, January 20, 2011. [Google Scholar]
- (19).Munteanu B; Hopf C Emergence of Whole-Cell MALDI-MS Biotyping for High-Throughput Bioanalysis of Mammalian Cells? Bioanalysis 2013, 5 (8), 885–893. [DOI] [PubMed] [Google Scholar]
- (20).Munteanu B; Hopf C Whole/Intact Cell MALDI MS Biotyping in Mammalian Cell Analysis In Advances in MALDI and Laser-Induced Soft Ionization Mass Spectrometry; Cramer R Ed.; Springer International Publishing: Cham, 2016; pp 249–262. [Google Scholar]
- (21).Petukhova VZ; Young AN; Wang J; Wang M; Ladanyi A; Kothari R; Burdette JE; Sanchez LM Whole Cell MALDI Fingerprinting Is a Robust Tool for Differential Profiling of Two-Component Mammalian Cell Mixtures. J. Am. Soc. Mass Spectrom. 2019, 30 (2), 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Russo A; Czarnecki AA; Dean M; Modi DA; Lantvit DD; Hardy L; Baligod S; Davis DA; Wei J-J; Burdette JE PTEN Loss in the Fallopian Tube Induces Hyperplasia and Ovarian Tumor Formation. Oncogene 2018, 37 (15), 1976–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Hardy LR; Pergande MR; Esparza K; Heath KN; Önyüksel H; Cologna SM; Burdette JE Proteomic Analysis Reveals a Role for PAX8 in Peritoneal Colonization of High Grade Serous Ovarian Cancer That Can Be Targeted with Micelle Encapsulated Thiostrepton. Oncogene 2019, 38 (32), 6003–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Domcke S; Sinha R; Levine DA; Sander C; Schultz N Evaluating Cell Lines as Tumour Models by Comparison of Genomic Profiles. Nat. Commun 2013, 4, 2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Olive KP; Tuveson DA; Ruhe ZC; Yin B; Willis NA; Bronson RT; Crowley D; Jacks T Mutant p53 Gain of Function in Two Mouse Models of Li-Fraumeni Syndrome. Cell 2004, 119 (6), 847–860. [DOI] [PubMed] [Google Scholar]
- (26).Quartuccio SM; Karthikeyan S; Eddie SL; Lantvit DD; Ó hAinmhire E; Modi DA; Wei J-J; Burdette JE Mutant p53 Expression in Fallopian Tube Epithelium Drives Cell Migration. Int. J. Cancer 2015, 137 (7), 1528–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Ahmed N; Stenvers KL Getting to Know Ovarian Cancer Ascites: Opportunities for Targeted Therapy-Based Translational Research. Front. Oncol. 2013, 3, 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Wan KX; Vidavsky I; Gross ML Comparing Similar Spectra: From Similarity Index to Spectral Contrast Angle. J. Am. Soc. Mass Spectrom. 2002, 13 (1), 85–88. [DOI] [PubMed] [Google Scholar]
- (29).Dasari S; Pereira L; Reddy AP; Michaels J-EA; Lu X; Jacob T; Thomas A; Rodland M; Roberts CT; Gravett MG; et al. Comprehensive Proteomic Analysis of Human Cervical−Vaginal Fluid. Journal of Proteome Research. 2007, pp 1258–1268. 10.1021/pr0605419. [DOI] [PubMed] [Google Scholar]
- (30).Tang L-J; De Seta F; Odreman F; Venge P; Piva C; Guaschino S; Garcia RC Proteomic Analysis of Human Cervical-Vaginal Fluids. J. Proteome Res. 2007, 6 (7), 2874–2883. [DOI] [PubMed] [Google Scholar]
- (31).Shaw JLV; Smith CR; Diamandis EP Proteomic Analysis of Human Cervico-Vaginal Fluid. J. Proteome Res. 2007, 6 (7), 2859–2865. [DOI] [PubMed] [Google Scholar]
- (32).Panicker G; Ye Y; Wang D; Unger ER Characterization of the Human Cervical Mucous Proteome. Clin. Proteomics 2010, 6 (1–2), 18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Soleilhavoup C; Riou C; Tsikis G; Labas V; Harichaux G; Kohnke P; Reynaud K; de Graaf SP; Gerard N; Druart X Proteomes of the Female Genital Tract During the Oestrous Cycle. Mol. Cell. Proteomics 2016, 15 (1), 93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Pre-processing workflow using the MALDIquant package in R
Figure S2. In vivo fluorescence imaging of OVCAR-8-RFP tumors in athymic mice
Figure S3. Fluorescent Cell Spiking Study
Figure S4. Mirror plot comparing protein fingerprints from week seven in a HGSOC and NASH model
Figure S5. Mass spectra of repeat murine lavages from healthy mice
Figure S6. Consensus spectra of vaginal lavages from Mouse 901
Figure S7. Consensus spectra of vaginal lavages from Mouse 902
Figure S8. Consensus spectra of vaginal lavages from Mouse 903
Figure S9. Consensus spectra of vaginal lavages from Mouse 904
Figure S10. Consensus spectra of vaginal lavages from Mouse 905
Table S1. Up-and downregulated spectral features differentially expressed across various time points throughout the study