Abstract
Flowers with more petals are of more ornamental value. It is well known that AGAMOUS (AG) is the core member of the C-class gene which plays an essential role in double flower formation and identification of stamens and carpels in Arabidopsis thaliana. We searched C-class genes in the genome of the carnation, and found two AG orthologs (DcaAGa, DcaAGb). Phylogenetic analysis showed that the two genes were closely related to the euAG subclade. Then we searched the genomes of other Caryophyllales plants (Beta vulgaris, Spinacia oleracea, Chenopodium quinoa) for C-class genes, and found that their C-class genes all belonged to the euAG subclade. Semi-quantitative PCR (sq-PCR) analysis indicated that the expression of DcaAG genes in the single flower phenotype was higher than that in the double flower phenotype. Quantitative real-time RT-PCR (qRT-PCR) analysis showed that the expressions of DcaAG genes in the flower bud were significantly different from those in the root, stem, and leaf between the single and double flower phenotype carnations, and that DcaAG genes were specifically expressed in the stamen and carpel of carnation. Moreover, the expression of other floral organ identity genes (AP1 and AP2, PI and AP3, SEP1 and SEP3 corresponding to the A-, B-, and E-class of genes, respectively) showed no significant difference in all floral organs between the single and double flower phenotype carnations, suggesting that C-class (DcaAG) genes might play an important role in the double flower phenotype in carnation. Petal loss or decrease, precocious flowering, silique shortening, and seed sterility were observed in 35S::DcaAGa and 35S::DcaAGb transgenic Arabidopsis plants. All these results show that DcaAG genes might affect the petal number negatively and have a specific function in stamen and carpel development in carnation.
Keywords: carnation, Caryophyllales, AGAMOUS, bioinformatics analysis, expression analysis, genetic transformation
1. Introduction
Flowers are important sexual organs of angiosperms, and the flower development process has been a research hotspot in ornamental plants for a long time. The molecular mechanism of the flower development in Arabidopsis thaliana [1] and Antirrhinum majus [2] is extremely clear, but little is known about the regulation network of flower development in many non-model plants. The ABC model explains the genetic regulatory mechanism of floral organs in A. majus and Arabidopsis [3]. Class A genes control the formation of the first whorl floral organ, class A and B genes work together to control petal formation, class B and C genes control stamen formation, and class C genes regulate the formation of carpel [4,5]. On the basis of the original ABC model, the ABCDE model is proposed with the additional class D genes function for ovule development and the class E genes function for all floral organs’ development [4,5,6].
The AG subfamily experienced many duplication events, but two of the duplication events are representative during the evolutionary process [7]. In the early evolution stage of core dicotyledonous plants, AG and PLENA (PLE) were parallel branches derived from a common ancestor after they experienced the gene duplication event. This duplication event resulted in the formation of the euAG subseries (FARINELLI (FAR) and AG) and the PLE subseries (SHATTERPROOF1/2 (SHP1/2) and PLE) [7,8,9,10]. Another earlier duplication event occurred before the emergence of angiosperms, which resulted in the emergence of the C-class gene (AG) and D-class gene (AGL11) [7,10,11]. The mutations of AG and PLE led to consistent phenotypic features, the stamens converted into petals, together with the replacement of carpels by sepals [12,13]. In A. majus, although the PLE lineage gene (PLE) and euAG lineage gene (FAR) exhibited the partial redundant function of C-class genes [12,14], the function of PLE and FAR are slightly different. PLE functions to identify stamen and carpel, and to terminate floral meristem, while FAR plays an important role in male fertility [14]. In Cyclamen persicum, the repressive expression of CpAG1 resulted in the conversion of stamens into petals, but the inhibited expression of CpAG2 only caused incomplete formation of stamens and carpels. Although CpAG1 and CpAG2 had similar protein sequence, but they had different roles in whorl 3 and whorl 4 [15]. In Phalaenopsis aphrodita, two AG genes PhalAG1 and PhalAG2 belong to the C- and D-lineages, respectively. The expressions of PhalAG1 and PhalAG2 were much higher in flower buds than that in vegetative organs, and they played a redundant role in flower development [16]. In Arabidopsis, AG belonged to the C-class genes of the MADS-box family, played an essential role in the formation of floral meristem and sexual organs during flower development [17]. Previous studies reported that in C-class gene mutants of Arabidopsis, stamens are homologously transformed into petals, and AG inhibited A-class genes’ activity in petals and carpels, and that AG was able to identify the third and fourth whorls [18,19,20]. Moreover, some floral organ identity genes also play an important role in flower development. APETALA1 (AP1) and APETALA2 (AP2) are A-class genes in Arabidopsis, AP1 functions to identify sepals and petals and ap1 mutant phenotype shows the formation of leaf-like sepals [21]. With the exception of AP2, all A-, B-, C-, D-, and E- class genes are MADS-box genes. AP2 is necessary for the specification of the first and second whorls [17,22]. APETALA3 (AP3) and PISTILLATA (PI) represent B class genes, which play a major role in specifying petal and stamen identities in Arabidopsis [23,24]. SEPALLATA1/2/3 (SEP1/2/3) are E-class genes, play a major role in the specification of all floral organs and floral determinacy [6,25,26]. Plants with a triple mutant of sep1/2/3 show the phenomenon of all floral organs changing to sepals [26].
As one of the famous cut flowers, the carnation is widely used in daily life throughout the world. Recently, double flower trait have attracted researcher’s attention due to its improvement for ornamental value of many species such as carnation, rose, and lily. Many studies have been performed to clarify the molecular mechanism associated with double flower formation, such as Arabidopsis [22], Eschscholzia californica [27], and Prunus lannesiana [28]. In carnation, D85 locus controlled flower phenotype and two SSR markers (CES0212 and CES1982) tightly linked to this locus were identified [29]. Despite the preliminary localization of the double flower phenotype and the genomic information in the carnation, suitable candidate genes cannot be screened at present due to the lack of chromosomal information in the genome [30].
In our study, firstly, we obtained two C-class genes in carnation, and found that the two C-class genes (DcaAGa and DcaAGb) belong to the euAG subclade. In order to determine whether the C-class genes in Caryophyllales all belong to the euAG subclade, C-class genes in B. vulgaris, S. oleracea, C. quinoa were searched and analyzed, and they all belong to the euAG subclade consistent with previous studies [31,32]. Subsequently, two C-class genes, DcaAGa and DcaAGb from two flower phenotype carnations were cloned and analyzed. The expressions of two DcaAG genes were analyzed using qRT-PCR and semi-quantitative PCR between single and double flower phenotype carnations, and the functions of these two genes were analyzed through overexpression of DcaAGa and DcaAGb in Arabidopsis. The expression analysis of other floral organ identity genes was performed in floral organs between single and double flower phenotype carnations by using qRT-PCR. Our data suggests that the two DcaAG genes might play a major role in affecting petal number, and that they play a specific role in regulating stamen and carpel development.
2. Results
2.1. Isolation and Sequence Analysis of the DcaAG Genes from Single and Double Flower Phenotype Carnations
Single flower phenotype carnation (‘Da hong’) which was collected in south China in 2015, has five petals (Figure 1, Table 1). Double flower phenotype carnation (‘Master’), a common cultivar, is wildly used in the cut flower market, has more than 40 petals (Figure 1, Table 1). Two DcaAG genes were identified from the carnation genome [30] and cloned from these two flower phenotype carnations, named DcaAGa and DcaAGb, respectively. The CDS sequences of DcaAGa and DcaAGb between two flower phenotype carnations were the same, respectively. The DcaAGa encoded a 251 amino acid protein, while the DcaAGb encoded a 249 amino acid protein (Figure 2), both of which contained the MADS domain (20–81), a short I region (82–105), a not highly conserved K domain (106–199), and the C-terminal region (200–251) with AG motifs I and II (Figure 2).
Figure 1.
Characteristics of single (‘Da hong’) and double (‘Master’) flower phenotype carnations. (A–D) represent flower, sepals, petals, and carpel of ‘Da hong’, respectively. (E–H) represent flower, sepals, petals, and carpel of ‘Master’, respectively. ‘Da hong’ and ‘Master’ with petals of 5 and more than 40, respectively. Bar = 1 cm.
Table 1.
Information of the experimental materials.
| Flower Phenotype | Name of Material | Petal Number | Color |
|---|---|---|---|
| Single | ‘Da-Hong’ | 5 | Red |
| Double | ‘Master’ | >40 | Dark red |
Figure 2.
Protein sequence comparison of DcaAGa, DcaAGb, and the AG-related MADS domain proteins in Arabidopsis thaliana and Antirrhinum majus. The MADS domain, K domain, and I domain are marked with an arrow box. The AG motifs I and II in the C terminal region are boxed, highly conserved for AG-like protein.
Bioinformatics analysis revealed that the molecular weights (MW) of DcaAGa and DcaAGb were 28.73 and 28.49 kDa, respectively (Table 2), and the isoelectric points (pI) of DcaAGa and DcaAGb were 9.43 and 9.28, respectively (Table 2). Prediction of subcellular localization revealed that both DcaAGs were predicted to be located in the nucleus (Table 2).
Table 2.
Protein properties of DcaAGa and DcaAGb.
| Gene | Accession ID | AA Length | Molecular Weight (kDa) | Isoelectric Point | Subcellular Localization | Signal Peptide |
|---|---|---|---|---|---|---|
| DcaAGa | Dca35398.1 | 251 | 28.72751 | 9.43 | nucl | NO |
| DcaAGb | Dca50159.1 | 249 | 28.48724 | 9.28 | nucl | NO |
AA: Amino acid; Predication of subcellular localization was analyzed by Plant-mPloc.
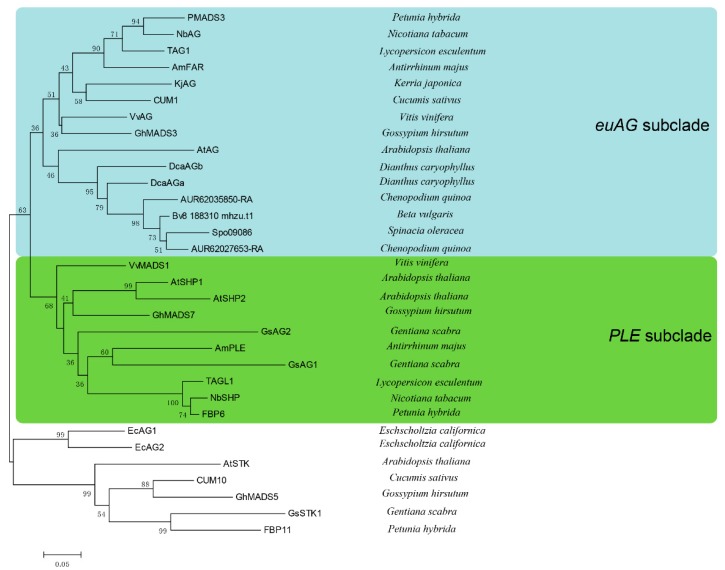
Phylogenetic analysis showed that DcaAGa and DcaAGb all fall into the euAG branch (Figure 3). Interestingly, our result was consistent with the previous study findings that AG homologs from Caryophyllales belonged to only the euAG subclade rather than the PLE subclade [7,31,32]. We identified one, one, and two C-class genes from B. vulgaris, S. oleracea and C. quinoa, respectively, and found that they all belonged to the euAG subclade (Figure 3).
Figure 3.
Phylogenetic tree showing the relationship between DcaAG genes and major C/D-class MADS-box proteins of other plants. DcaAG genes and C-class genes in Caryophyllales plants (Beta vulgaris, Spinacia oleracea, and Chenopodium quinoa) divide into euAG subclade rather than PLE subclade. MEGA 5.0 is used to construct phylogenetic tree, the maximum evolution method is used to infer evolutionary relationships. The phylogenetic tree is estimated by setting 2000 bootstrap replicates. The numbers below the branches refer to bootstrap value.
2.2. Expression Analysis of DcaAG Genes in Different Tissues and Floral Organs of Carnations
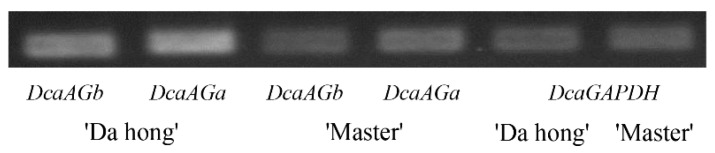
Semi-quantitative PCR analysis indicated that the expression levels of DcaAG genes were lower in the double flower phenotype carnation than that in the single flower phenotype carnation, and that the expression patterns of DcaAGa and DcaAGb in these two flower phenotype carnations were basically consistent, while the expression of the former was slightly higher than that of the latter (Figure 4).
Figure 4.
Semi-quantitative PCR (sq-PCR) analysis of two DcaAG genes in single and double flower phenotype carnations. The expression of both DcaAG genes in the double flower phenotype carnation were lower than that in the single flower phenotype carnation.
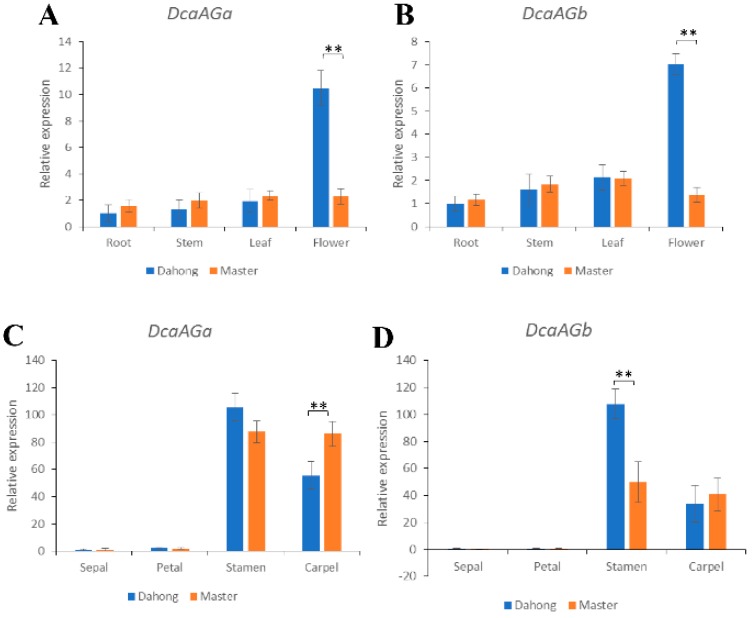
To examine the expression of DcaAG genes between the single and double flower phenotype in different tissues, qRT-PCR analysis was performed. Significant differential expression was detected in the flower bud (Figure 5). The expression levels of DcaAG genes in the root and stem of the single flower phenotype carnation were slightly lower than those of the double flower phenotype carnation (Figure 5). On the contrary, the expression levels of DcaAG genes in the flower bud of the single flower phenotype carnation plant were much higher than that of the double flower phenotype carnation (Figure 5), which indicated that DcaAG genes might play a major role in the formation of differential characteristics between single and double flower phenotype carnations.
Figure 5.
Quantitative PCR analysis of two DcaAG genes at different tissues (A,B) and floral organs (C,D) in single and double flower phenotype carnations. In different tissues, the expression of DcaAGa (A) and DcaAGb (B) were obviously different in the flower bud, and no significant difference was detected in the root, stem and leaf between two flower phenotype carnations. In floral organs, the expression of DcaAGa (C) and DcaAGb (D) were exclusively expressed in the stamen and carpel, and low expression levels were detected in the sepal and petal. DcaAGa (C) and DcaAGb (D) showed significant differential expression in the carpel and stamen between two flower phenotype carnations, respectively. ** indicates p < 0.01 by student’s t-test.
The expression patterns of DcaAG genes between the two flower phenotypes in different floral organs (carpel, stamen, petal, and sepal) were analyzed by qRT-PCR. DcaAG genes were exclusively expressed in the carpel and stamen, while extremely low expression levels were detected in the sepal and petal (Figure 5). The expression levels of DcaAG genes were higher in the stamen of the single flower phenotype carnation than that of the double flower phenotype carnation (Figure 5). On the contrary, the expression levels of DcaAG genes were lower in the carpel of the single flower phenotype carnation than that of the double flower phenotype carnation (Figure 5).
2.3. Expression Pattern Analysis of A-, B-, and E-Class Genes in Single and Double Flower Phenotype Carnations
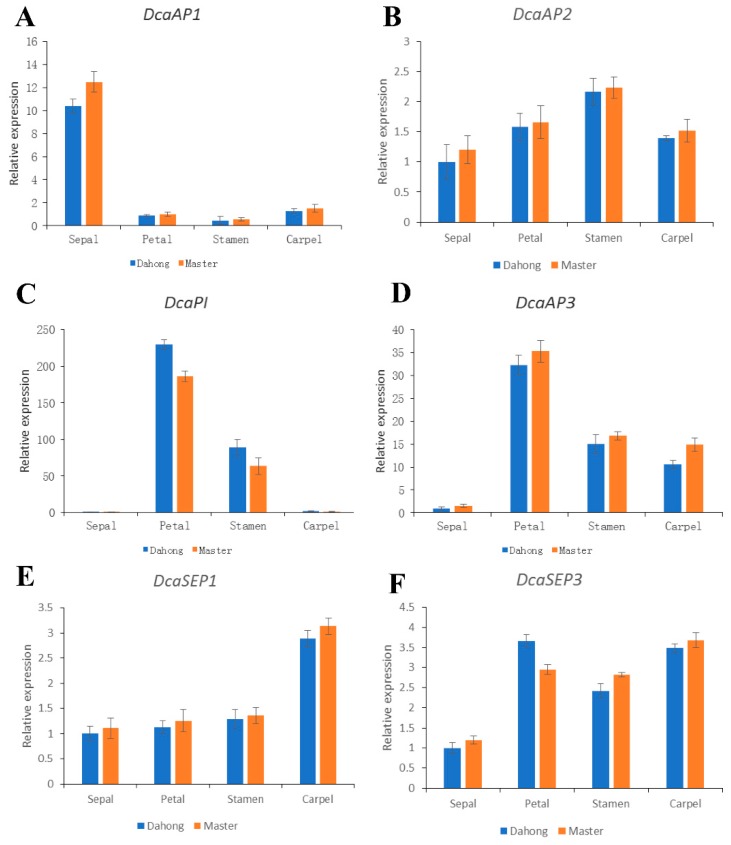
In order to investigate the expression levels of the floral organ identity genes between single and double flower phenotype carnations quantitative RT-RCR was performed to analyze the expression pattern of DcaAP1, DcaAP2, DcaAP3, DcaPI, DcaSEP1, and DcaSEP3. DcaAP1 had especially higher expression in the sepal than that in the petal, stamen, and carpel, and expressed slightly lower in all floral organs of the single flower phenotype carnation compared to that of the double flower phenotype carnation (Figure 6A). DcaAP2 expressed in all floral organs stably and was slightly lower in all floral organs of the single flower phenotype carnation than that of the double flower phenotype carnation (Figure 6B). DcaPI and DcaAP3 both had higher expression in the second and third whorls than that in the first and fourth whorls, and there was no significant difference in expression between single and double flower phenotype carnations (Figure 6C,D). The expression pattern of DcaSEP1 and DcaSEP3 were relatively smooth in all floral organs between single and double flower phenotype carnations (Figure 6E,F). The huge difference of expression in C-class genes and no significant difference of expression in A-, B-, and E-class genes between single and double flower phenotype carnations indicated that the C-class genes may result in the differential characteristics between single and double flower phenotype carnations. A previous study on Kerria japonica revealed that repressing the expression of Df-KjAG could affect the expression of other floral organ identity genes, such as KjAGL2, KjAGL9, KjAP1, KjAP2, KjAP3, and KjPI in the double flower K.japonica [33], which is different from the situation in carnation.
Figure 6.
Quantitative PCR analysis of floral organ identity genes at different floral organs in single and double flower phenotype carnations. DcaAP1 (A) had higher expression in the sepal than that in petal, stamen, and carpel. DcaAP2 (B) expressed in all floral organs stably. DcaPI (C) and DcaAP3 (D) had higher expression in the second and third whorls than that in the first and fourth whorls. DcaSEP1 (E) and DcaSEP3 (F) expressed relatively smoothly in all floral organs between single and double flower phenotype carnations. The expression of DcaAP1 (A), DcaAP2 (B), DcaPI (C), DcaAP3 (D), DcaSEP1 (E), and DcaSEP3 (F) had no significant difference in sepal, petal, stamen and carpel between two flower phenotype carnations. Student’s t-test was used to determine significant differences in expression.
2.4. Phenotypes of DcaAG Genes Overexpression in Arabidopsis
We investigated the function of DcaAG genes in flower initiation and development by overexpressing DcaAG genes in Arabidopsis. Twenty independent 35S::DcaAGa and 33 independent 35S::DcaAGb transgenic Arabidopsis plants were obtained. The expression levels of DcaAGa and DcaAGb in the transgenic Arabidopsis lines were tested by semi-quantitative PCR. Both DcaAG genes were found to express in transgenic plants (Figure S1). The 15 independent 35S::DcaAGa and 14 independent 35S::DcaAGb transgenic plants showed severe phenotypic alteration (Figure 7), namely, their petals became significantly shorter and even almost disappeared, and a large number of deformed carpels were observed (Figure 7C–F). All the transgenic plants displayed abnormal growth of narrow and curly leaves in the early developmental stage (Figure 7I–J). These transgenic plants flowered earlier than the wild type plants and the positive control group (Figure 7K,L). All the transgenic plants showed a significant decrease in trichome on the leaf surface margin and vein (Figure 7O–P). The seeds of 35S::DcaAGa and 35S::DcaAGb transgenic lines were sterile or with low activity, and their siliques were shorter, wrinkled, and convex near the top (Figure 7Q). These results indicated that overexpression of DcaAG genes in Arabidopsis lead to a series of effects on Arabidopsis development, especially on flower development.
Figure 7.
Phenotypic analysis of transgenic plants by overexpressing DcaAG genes in Arabidopsis. The flower of the wild-type Arabidopsis (A) and positive control group (B) (Arabidopsis transformed with empty vector). The petals of 35::DcaAGa (C,D) and 35::DcaAGb (E,F) transgenic plants is decreased or almost disappears compared to the wild type plant and positive control group. Phenotype of leaves in the early developmental stage of wild type plants (G), positive control group (H), 35::DcaAGa (I) and 35::DcaAGb (J) transgenic plants, the transgenic plants display abnormal growth of narrow and curly leaves. 35::DcaAGa (K) and 35::DcaAGb (L) transgenic plants flowering earlier than that of the wild type plant and positive control group. Trichome on the leaf surface margin and vein in 35::DcaAGa (O) and 35::DcaAGb (P) transgenic plants are decreased compared to wild type plant (M) and positive control group (N). The siliques are shorter, wrinkled with convex near the top in transgenic plants, compared to wild type plants (Q). se: sepal, pe: petal, st: stamen, ca: carpel. Bar = 5 mm in A–J, and Q. Bar = 1 cm in K and L. Bar = 1 mm in M–P.
2.5. Expression Analysis in Transgenic Arabidopsis
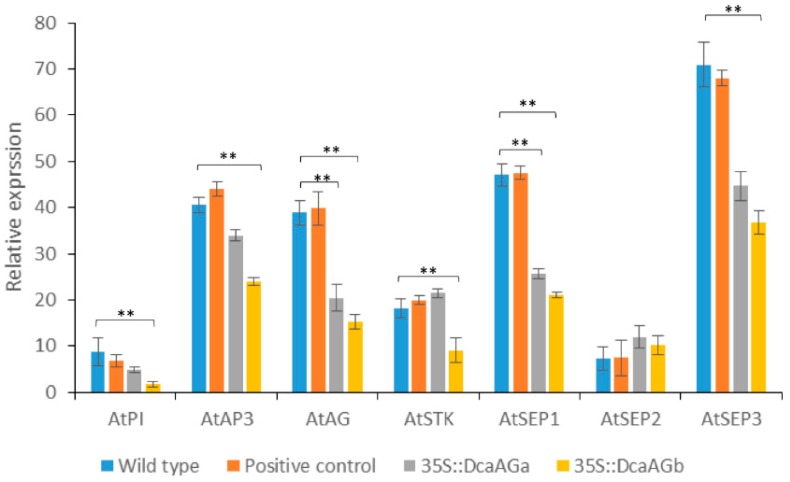
The expression levels of floral organ identity genes in transgenic Arabidopsis were detected by qRT-PCR analysis. The results showed that the expressions of AtPI, AtAP3, AtAG, AtSEP1, and AtSEP3 were down-regulated in the 35S::DcaAGa and 35S::DcaAGb transgenic T1 lines, while the expression of AtSEP2 was up-regulated, compared with the wild type plants and positive control group (Figure 8). The expression of AtSTK was slightly higher in the 35S::DcaAGa transgenic plant than that in the wild type plants and positive control group, and was down-regulated in 35S::DcaAGb transgenic plant (Figure 8). It indicated that the overexpression of DcaAG genes had an effect on genes related to flower development in Arabidopsis.
Figure 8.
Quantitative PCR analysis of AtPI, AtAP3, AtAG, AtSTK, AtSEP1, AtSEP2, and AtSEP3 in wild type, positive control group, 35::DcaAGa and 35::DcaAGb transgenic Arabidopsis. The expression of AtPI, AtAP3, AtAG, AtSTK, AtSEP1, and AtSEP3 in transgenic Arabidopsis show a significant difference, compared to the wild type and positive control group plant. ** indicates p < 0.01 by student’s t-test. Positive control represents the plants are transformed with pCAMBIA2300s vector.
3. Discussion
C-class genes have been identified from the genome of the carnation and cloned from single and double flower phenotype carnations. No difference was observed in the CDS sequence of DcaAG genes between the two flower phenotype carnations. The amino acid sequence consistency of DcaAGa and DcaAGb achieves 89%, with a consistency of 98% at the N terminal and M domain, 79% in the I domain, 82% in the K domain, and 88% in the C terminal. Moreover, the CDS sequences of DcaAGa and DcaAGb exhibit a similarity as high as 71% to that of AG in Arabidopsis, while 69% and 66% similarity to that of FAR and PLE in A. majus, respectively. This suggests that the domains of DcaAG genes are relatively conservative during plant evolution.
In Arabidopsis, AG is one of the key genes for double flower formation. SHP was not physically present in the meristem or primordia cells at the investigated time, indicating that AG is the only C function factor in the process of floral primordia development [34,35]. The change in expression pattern is reported to lead to subfunctionalization of C function genes [36,37]. Semi-quantitative PCR analysis shows that the expression levels of DcaAG genes are lower in the double flower phenotype carnation than that of the single flower phenotype carnation (Figure 4), suggesting DcaAG genes in the carnation might be a negative regulator of double flower formation, which has been proved in many other species, such as Arabidopsis [38], Rose [39], and Tricyrtis macranthopsis [40]. The expression pattern of DcaAG genes in different tissues reveals a larger difference in the flower bud than that in the root, stem and leaf between two flower phenotype carnations (Figure 5), which indicates that the inhibition of the DcaAG gene might have an important effect on the flower development of the double flower phenotype carnation. This situation in the carnation is also reported in the studies of Rose [41], Ranunculids [18], and Cyclamen [15]. The expression pattern of DcaAG genes in different floral organs of two flower phenotype carnations indicates that the DcaAG genes are both expressed in the third and fourth whorls, but nearly not expressed in the first and second whorls (Figure 5). Conjointly with the expression analysis of DcaAG genes and the floral organ identity genes in floral organs between two flower phenotype carnations, the decreased expression of DcaAG genes in the double flower phenotype carnation corresponds to no significant difference in expression of other floral organ identity genes between the two flower phenotype carnations (Figure 5, Figure 6 ), indicating that C-class (DcaAG) genes might have an important role in double flower formation in the carnation. Moreover, the expression of DcaAGa is slightly higher than that of DcaAGb (Figure 4) and both DcaAG genes have a similar expression pattern in the two flower phenotype carnations (Figure 4 and Figure 5), which is similar to the expression pattern of EScaAG1, EScaAG2 in E. californica [27] and MtAGa, MtAGb in Medicago truncatula [42], suggesting that two DcaAG genes might have a functional redundancy in regulating the flower organs in plants. However, the different expression level between two DcaAG genes might be due to their different regulation roles in exerting C function.
Compared to wild type Arabidopsis, early flowering is observed as a novel phenomenon in 35S::DcaAGa and 35S::DcaAGb transgenic Arabidopsis plants which is rarely reported in other species (Figure 7K,L). The phenotypes of 35S::DcaAGa and 35S::DcaAGb transgenic Arabidopsis plants exhibit petal loss, short silique, and almost seed sterility (Figure 7C–F,Q). These results indicate that overexpression of DcaAG genes in Arabidopsis cause the abnormal development in the third and fourth whorls and inhibit growth of petals in transgenic plants. The phenotypes of 35S::DcaAGa and 35S::DcaAGb transgenic plants are highly similar (Figure 7), which might be attributed to the similar C-class function of two DcaAG genes. AG and FAR are orthologue genes [7,43]. The flower of the FAR mutant plant shows stamen sterility in A. majus, indicating that FAR has an important role in promoting stamen development [44], while AG plays an essential role in stamen and carpel development [23]. Our results suggest that both DcaAGa and DcaAGb in carnation could regulate the stamen and carpel in a different manner to the euAG genes (AG and FAR) [7].
In this research, phylogenetic analysis indicates that both DcaAGa and DcaAGb fall into the euAG subclade (Figure 3). C-class genes in other Caryophyllales plants fall into the same clade as DcaAG genes by phylogenetic analysis (Figure 3). Our findings are in agreement with previous study shows that AG/FAR and SHP/PLE represent two distinct paralogous lineages rather than single genetic orthologs [7,31,32]. The AG/FAR subgroup functions in identifying male and female organs, while the SHP/PLE subgroup plays an important role in the identification of floral organs in A. majus and Arabidopsis [12,17]. According to the phylogenetic analysis, expression analysis of DcaAG genes (Figure 3, Figure 4 and Figure 5) and the phenotype of the 35S::DcaAGa and 35S::DcaAGb transgenic Arabidopsis plants (Figure 7), we speculate that members of the SHP/PLE may disappear in some species of Caryophyllales, and only genes in the AG/FAR subgroup perform C-class function. AG and SHP1/2 are the core C-class MADS-box genes in Arabidopsis, which is unlike many species with two C-class MADS-box genes, such as A. majus (PLE/FAR) [14], tomato (TAG1/TAGL1) [45], and cyclamen (CpAG1/CpAG2) [15]. In cyclamen, CpAG1 and CpAG2 have their own distinct roles in the stamen and carpel, respectively [15]. The situation in carnation is slightly different to cyclamen, the functions in the stamen and carpel of DcaAGa and DcaAGb are similar (Figure 7) which may indicate that DcaAGa and DcaAGb performed C function redundantly in carnation, and DcaAGa exhibits a stronger function than DcaAGb according to the expression pattern of the two DcaAG genes (Figure 4, Figure 5). Overall, our results indicate that C-class genes in the carnation all belong to the euAG subclade, and that DcaAG genes in the carnation play an essential role in regulating the formation of the stamen and carpel, especially in regulating the stamen, silique, and in seed development. Also, these two genes may have an important role in regulating petal number.
4. Materials and Methods
4.1. Plant Materials and RNA Sample Preparation
Two varieties of carnations, ‘Master’ (double flower phenotype) and ‘Da Hong’ (single flower phenotype), with different flower phenotype and biological characteristics (Table 1, Figure 1) were planted in Huazhong Agricultural University, China. Young root, stem, leaf, flower bud, and floral organs (sepal, petal, stamen and carpel) of two flower phenotype carnations were collected and frozen immediately in liquid nitrogen, and then stored at −80 °C for RNA extraction. RNA was extracted in the method described in a previous study [46]. A. thaliana Columbia (Col) ecotype plants were grown under a long-day regime (22 °C, 16/8 h light/dark) with light intensity of 8000 lx and humidity of 75%.
4.2. Search for C-Class Genes in the Genome of Caryophyllales Plants and Molecular Cloning of DcaAG Genes in Single and Double Flower Phenotype Carnations
The protein sequence of AG and SHP1/2 from Arabidopsis, downloaded from the TAIR website (http://www.arabidopsis.org/), were used to search C-class genes in the genome of carnation (http://carnation.kazusa.or.jp/), S. oleracea (http://www.spinachbase.org), C. quinoa (https://www.cbrc.kaust.edu.sa/chenopodiumdb/) and B. vulgaris (http://bvseq.boku.ac.at/index.shtml) by blastp (e-value 1e−6) program. The protein sequences obtained from these databases were aligned by DNAMAN 6.0 software. Those protein sequences without AG conserved domain were filtered. The flower bud’s cDNA of two flower phenotype carnations was used as a template for amplification of the target fragment. The CDS sequences of DcaAGa and DcaAGb in the carnation’s genome and Primer Premier 5.0 were used to design cloning primers. Primers are listed in Supplementary Table S1. PCR program was performed as follows: 4 min at 94 °C for 1 cycle, followed by 35 cycles at 94 °C for 30 s, 55 °C (DcaAGa)/58 °C (DcaAGb) for 30 s, and 72 °C for 1 min, and followed by 10 min at 72 °C for 1 cycle. A fragment of PCR product was collected and cloned into the pMD18-T vector (Takara, Japan, http://www.takara.com.cn), and then connected to Sangon Biotech (Shanghai) for sequencing.
4.3. Bioinformatics and Phylogenetic Analysis
The putative protein sequences were obtained from translation of CDS sequence of DcaAGa and DcaAGb. ProtParam (http://web.expasy.org/protparam/) was used to predicate molecular weight (MW) and isoelectric point (pI) of the amino acid sequences with default parameters. The predication of subcellular localization was analyzed by Plant-mPloc [47].
C-class genes in Caryophyllales plants (carnation, B. vulgaris, S. oleracea and C. quinoa) and the other major C/D-class MADS-box proteins in Arabidopsis, A. majus, Lycopersicon esculentum, Petunia hybrid, Nicotiana tabacum, Eschscholtzia californica, Vitis vinifera, Kerria japonica, Cucumis sativus, Gossypium hirsutum, and Gentiana scabra were used for phylogenetic analysis by using MEGA 5.0 [48]. Full-length protein sequences were aligned by MUSCLE with default parameters. The maximum evolution method with bootstrap of 2000 replicates and the Nearest-Neighbor-Interchange (NNI) algorithm were used to evaluate evolutionary relationships.
The accession number of the proteins used in the phylogenetic analysis are as follows: EcAG1 (DQ088996); EcAG2 (DQ088997); AtAG (X53579); AmFAR (AJ239057); AtSTK (NP_001329612); TAG1 (L26295); PMADS3 (X72912); NbAG (JQ699177); AtSHP1 (M55550); AtSHP2 (M55553); AmPLE (S53900); TAGL1 (AY098735); FBP6 (X68675); FBP11 (CAA57445); NbSHP (JQ699178); VvAG (NM_001281168); VvMADS1 (NP_001268105); KjAG (AMQ23646); CUM1 (AAC08528); CUM10 (AAC08529); GhMADS3 (XP_016711294); GhMADS5 (ABM69043); GhMADS7 (ABM69045); GsAG1 (LC022775); GsAG2 (LC022779); GsSTK1 (LC022768). All the protein sequences were downloaded from the Phytozome website (https://phytozome.jgi.doe.gov/pz/portal.html) and listed in Supplementary Table S2.
4.4. Quantitative PCR and Semi-Quantitative PCR Analysis of DcaAG Gene Expression in Single and Double Flower Phenotype Carnations
The RNA extracted from different tissues (root, stem, leaf, and flower bud), and floral organs (sepal, petal, stamen, and carpel) were used for qRT-PCR analysis to investigate the expression pattern of DcaAGa and DcaAGb in two flower phenotype carnations. Then, the expression patterns of A- (AP1, AP2), B- (PI, AP3), and E-class (SEP1, SEP3) genes in floral organs (sepal, petal, stamen, and carpel) were analyzed by qRT-PCR. A-, B-, and E-class Genes were identified in a previous study [46]. Quantitative real-time PCR analysis was performed as described previously [46]. The experiment with each sample was conducted with three biological replicates. The DcaGAPDH (glyceraldehyde-3-phosphate dehydrogenase) and β-actin were used as housekeeping genes. The comparative CT(2−ΔΔCT) method was used to calculate the relative expression value.
For semi-quantitative PCR analysis, the cycle parameters were 95 °C/4 min for 1 cycle, 95 °C/30 s, 57 °C/30 s, 72 °C/20 s for 26 cycles, and followed by 10 min at 72 °C for 1 cycle. DcaGAPDH was used as reference gene. Primer Premier 5.0 was used to design qRT-PCR and semi-quantitative PCR primers. The sequences of primers are listed in Supplementary Table S3.
4.5. Overexpression of DcaAGa and DcaAGb in Arabidopsis
BamH I and SalI were used to digest pMD18-T vector containing the full-length CDS sequence of two DcaAG genes. The full-length CDS sequence of DcaAG genes were subcloned into binary vector pCAMBIA2300s under the control of cauliflower mosaic virus 35S promoter in sense orientation. The 35S::DcaAG genes were introduced to the Agrobacterium tumefaciens strain GV3101, which were subsequently transformed into Arabidopsis plants by the floral dip procedure. Total RNA in the flower bud of 35S::DcaAGa and 35S::DcaAGb transgenic plants was extracted, and used for semi-quantitative PCR and qRT-PCR analysis. The expression levels of DcaAGa and DcaAGb in the transgenic Arabidopsis lines were tested by semi-quantitative PCR, and AtACT2 was used as reference gene. The PCR program was performed as follows: 95 °C/3 min for 1 cycle, 95 °C/30 s, 58 °C/30 s, 72 °C/20 s for 27 cycles, and followed by 8 min at 72 °C for 1 cycle. The qRT-PCR analysis of genes associated with flower development including PI, AP3, AG, SEEDSTICK (STK), AG, SEP1, SEP2, and SEP3 were performed to observe the expression difference between wild type, positive control and transgenic Arabidopsis plants. The AtACT2 and AtACT11 were used as housekeeping genes. Primer Premier 5.0 was used to design semi-quantitative PCR and qRT-PCR primers. Sequences of primers are listed in Supplementary Table S4.
5. Conclusions
In this research, we identified two C-class genes (DcaAGa and DcaAGb) in carnation and found that they fall intothe euAG subclade rather than the PLE subclade. DcaAGa and DcaAGb are cloned from single and double flower phenotype carnations and the CDS sequence of DcaAGa and DcaAGb show no difference between the two flower phenotype carnations, respectively. The expression of DcaAG genes in different tissues (root, stem, leaf, and flower bud) shows that DcaAG genes have a larger difference in flower bud than that in the root, stem and leaf between single and double flower phenotype carnations. The expression levels of DcaAG genes and other floral organ identity genes (A-class of AP1 and AP2, B-class of AP3 and PI, E-class of SEP1 and SEP3) were analyzed in floral organs (sepal, petal, stamen, and carpel) between two flower phenotype carnations. This revealed that only DcaAG genes show significant differential expression in stamen and carpel. Overexpression of DcaAGa and DcaAGb in Arabidopsis show that DcaAGa and DcaAGb play an important role in stamen and carpel development, regulating the petal number negatively, while having a similar function which indicates a redundant function between them. This study provides a foundation for future study on the double flower trait in the carnation.
Acknowledgments
We would to like to thank all colleagues in our laboratory for providing constructive suggestions and experimental support.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/1/87/s1, Table S1: The sequences of primers used for cloning DcaAG genes, Table S2: Protein sequences used for phylogenetic analysis, Table S3: The sequences of primers used for semi-quantitative PCR and qRT-PCR analysis in carnation. Table S4: The sequences of primers used for semi-quantitative PCR and qRT-PCR in transgenic Arabidopsis. Figure S1: Semi-quantitative PCR analysis of two DcaAG genes in different transgenic Arabidopsis lines.
Author Contributions
X.F. and Q.W. conceived and designed the experiments. Q.W., N.D., X.Z., and S.L. performed the experiments and analyzed the data. X.F. and Q.W. wrote the paper. M.B. and Q.W. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (31872135) and Fundamental Research Funds for the Central Universities (2662018JC036).
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- 1.Irish V.F. The flowering of Arabidopsis flower development. Plant J. Cell Mol. Biol. 2010;61:1014–1028. doi: 10.1111/j.1365-313X.2009.04065.x. [DOI] [PubMed] [Google Scholar]
- 2.Davies B., Cartolano M., Schwarz-Sommer Z. Flower development: The Antirrhinum perspective. Adv. Bot. Res. 2006;44:279–321. [Google Scholar]
- 3.Litt A., Kramer E.M. The ABC model and the diversification of floral organ identity. Semin. Cell Dev. Biol. 2010;21:129–137. doi: 10.1016/j.semcdb.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Coen E.S., Meyerowitz E.M. The War Of the Whorls—Genetic Interactions Controlling Flower Development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 5.Weigel D., Meyerowitz E.M. The Abcs Of Floral Homeotic Genes. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- 6.Honma T., Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409:525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- 7.Kramer E.M., Jaramillo M.A., Di Stilio V.S. Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics. 2004;166:1011–1023. doi: 10.1534/genetics.166.2.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker A. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 2003;29:464–489. doi: 10.1016/S1055-7903(03)00207-0. [DOI] [PubMed] [Google Scholar]
- 9.Litt A., Irish V.F. Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: Implications for the evolution of floral development. Genetics. 2003;165:821–833. doi: 10.1093/genetics/165.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zahn L.M., Leebens-Mack J., DePamphilis C.W., Ma H., Theissen G. To B or Not to B a flower: The role of DEFICIENS and GLOBOSA orthologs in the evolution of the angiosperms. J. Hered. 2005;96:225–240. doi: 10.1093/jhered/esi033. [DOI] [PubMed] [Google Scholar]
- 11.Moyroud E., Minguet E.G., Ott F., Yant L., Pose D., Monniaux M., Blanchet S., Bastien O., Thevenon E., Weigel D., et al. Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell. 2011;23:1293–1306. doi: 10.1105/tpc.111.083329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradley D., Carpenter R., Sommer H., Hartley N., Coen E. Complementary Floral Homeotic Phenotypes Result From Opposite Orientations Of a Transposon at the Plena-Locus Of Antirrhinum. Cell. 1993;72:85–95. doi: 10.1016/0092-8674(93)90052-R. [DOI] [PubMed] [Google Scholar]
- 13.Pinyopich A., Ditta G.S., Savidge B., Liljegren S.J., Baumann E., Wisman E., Yanofsky M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature. 2003;424:85–88. doi: 10.1038/nature01741. [DOI] [PubMed] [Google Scholar]
- 14.Davies B., Motte P., Keck E., Saedler H., Sommer H., Schwarz-Sommer Z. PLENA and FARINELLI: Redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J. 1999;18:4023–4034. doi: 10.1093/emboj/18.14.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka Y., Oshima Y., Yamamura T., Sugiyama M., Mitsuda N., Ohtsubo N., Ohme-Takagi M., Terakawa T. Multi-petal cyclamen flowers produced by AGAMOUS chimeric repressor expression. Sci. Rep. 2013;3:2641. doi: 10.1038/srep02641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song I.J., Nakamura T., Fukuda T., Yokoyama J., Ito T., Ichikawa H., Horikawa Y., Kameya T., Kanno A. Spatiotemporal expression of duplicate AGAMOUS orthologues during floral development in Phalaenopsis. Dev. Genes Evol. 2006;216:301–313. doi: 10.1007/s00427-005-0057-0. [DOI] [PubMed] [Google Scholar]
- 17.Bowman J.L., Smyth D.R., Meyerowitz E.M. Genetic Interactions among Floral Homeotic Genes Of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Galimba K.D., Tolkin T.R., Sullivan A.M., Melzer R., Theissen G., Di Stilio V.S. Loss of deeply conserved C-class floral homeotic gene function and C- and E-class protein interaction in a double-flowered ranunculid mutant. Proc. Natl. Acad. Sci. USA. 2012;109:E2267–E2275. doi: 10.1073/pnas.1203686109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizukami Y., Ma H. Separation Of Ag Function In Floral Meristem Determinacy From That In Reproductive Organ Identity by Expressing Antisense Ag Rna. Plant Mol. Biol. 1995;28:767–784. doi: 10.1007/BF00042064. [DOI] [PubMed] [Google Scholar]
- 20.Wang S.Y., Lee P.F., Lee Y.I., Hsiao Y.Y., Chen Y.Y., Pan Z.J., Liu Z.J., Tsai W.C. Duplicated C-Class MADS-Box Genes Reveal Distinct Roles in Gynostemium Development in Cymbidium ensifolium (Orchidaceae) Plant Cell Physiol. 2011;52:563–577. doi: 10.1093/pcp/pcr015. [DOI] [PubMed] [Google Scholar]
- 21.Pabon-Mora N., Ambrose B.A., Litt A. Poppy APETALA1/FRUITFULL orthologs control flowering time, branching, perianth identity, and fruit development. Plant Physiol. 2012;158:1685–1704. doi: 10.1104/pp.111.192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowman J.L., Smyth D.R., Meyerowitz E.M. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowman J.L., Drews G.N., Meyerowitz E.M. Expression Of the Arabidopsis Floral Homeotic Gene Agamous Is Restricted To Specific Cell-Types Late In Flower Development. Plant Cell. 1991;3:749–758. doi: 10.1105/tpc.3.8.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jack T., Fox G.L., Meyerowitz E.M. Arabidopsis homeotic gene APETALA3 ectopic expression: Transcriptional and posttranscriptional regulation determine floral organ identity. Cell. 1994;76:703–716. doi: 10.1016/0092-8674(94)90509-6. [DOI] [PubMed] [Google Scholar]
- 25.Ditta G., Pinyopich A., Robles P., Pelaz S., Yanofsky M.F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004;14:1935–1940. doi: 10.1016/j.cub.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 26.Pelaz S., Ditta G.S., Baumann E., Wisman E., Yanofsky M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- 27.Yellina A.L., Orashakova S., Lange S., Erdmann R., Leebens-Mack J., Becker A. Floral homeotic C function genes repress specific B function genes in the carpel whorl of the basal eudicot California poppy (Eschscholzia californica) EvoDevo. 2010;1:13. doi: 10.1186/2041-9139-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhixiong L., Dandan Z., Di L., Fenglan L., Hai L. Exon skipping of AGAMOUS homolog PrseAG in developing double flowers of Prunus lannesiana (Rosaceae) Plant Cell Rep. 2013;32:227–237. doi: 10.1007/s00299-012-1357-2. [DOI] [PubMed] [Google Scholar]
- 29.Yagi M., Yamamoto T., Isobe S., Tabata S., Hirakawa H., Yamaguchi H., Tanase K., Onozaki T. Identification of tightly linked SSR markers for flower type in carnation (Dianthus caryophyllus L.) Euphytica. 2014;198:175–183. doi: 10.1007/s10681-014-1090-8. [DOI] [Google Scholar]
- 30.Yagi M., Kosugi S., Hirakawa H., Ohmiya A., Tanase K., Harada T., Kishimoto K., Nakayama M., Ichimura K., Onozaki T., et al. Sequence analysis of the genome of carnation (Dianthus caryophyllus L.) DNA Res. 2014;21:231–241. doi: 10.1093/dnares/dst053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brockington S.F., Rudall P.J., Frohlich M.W., Oppenheimer D.G., Soltis P.S., Soltis D.E. ‘Living stones’ reveal alternative petal identity programs within the core eudicots. Plant J. 2012;69:193–203. doi: 10.1111/j.1365-313X.2011.04797.x. [DOI] [PubMed] [Google Scholar]
- 32.Sather D.N., York A., Pobursky K.J., Golenberg E.M. Sequence evolution and sex-specific expression patterns of the C class floral identity gene, SpAGAMOUS, in dioecious Spinacia oleracea L. Planta. 2005;222:284–292. doi: 10.1007/s00425-005-1544-2. [DOI] [PubMed] [Google Scholar]
- 33.Ma J., Shen X., Liu Z., Zhang D., Liu W., Liang H., Wang Y., He Z., Chen F. Isolation and Characterization of AGAMOUS-Like Genes Associated With Double-Flower Morphogenesis in Kerria japonica (Rosaceae) Front. Plant Sci. 2018;9:959. doi: 10.3389/fpls.2018.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DS O.M., Wuest S.E., Rae L., Raganelli A., Ryan P.T., Kwasniewska K., Das P., Lohan A.J., Loftus B., Graciet E., et al. Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell. 2013;25:2482–2503. doi: 10.1105/tpc.113.113209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ehlers K., Bhide A.S., Tekleyohans D.G., Wittkop B., Snowdon R.J., Becker A. The MADS Box Genes ABS, SHP1, and SHP2 Are Essential for the Coordination of Cell Divisions in Ovule and Seed Coat Development and for Endosperm Formation in Arabidopsis thaliana. PLoS ONE. 2016;11:e0165075. doi: 10.1371/journal.pone.0165075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Airoldi C.A., Davies B. Gene duplication and the evolution of plant MADS-box transcription factors. J. Genet. Genom. 2012;39:157–165. doi: 10.1016/j.jgg.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 37.Dreni L., Kater M.M. MADS reloaded: Evolution of the AGAMOUS subfamily genes. New Phytol. 2014;201:717–732. doi: 10.1111/nph.12555. [DOI] [PubMed] [Google Scholar]
- 38.Drews G.N., Bowman J.L., Meyerowitz E.M. Negative Regulation Of the Arabidopsis Homeotic Gene Agamous by the Apetala2 Product. Cell. 1991;65:991–1002. doi: 10.1016/0092-8674(91)90551-9. [DOI] [PubMed] [Google Scholar]
- 39.Bendahmane M., Dubois A., Raymond O., Bris M.L. Genetics and genomics of flower initiation and development in roses. J. Exp. Bot. 2013;64:847–857. doi: 10.1093/jxb/ers387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharifi A., Oizumi K., Kubota S., Bagheri A., Shafaroudi S.M., Nakano M., Kanno A. Double flower formation in Tricyrtis macranthopsis is related to low expression of AGAMOUS ortholog gene. Sci. Hortic. 2015;193:337–345. doi: 10.1016/j.scienta.2015.06.050. [DOI] [Google Scholar]
- 41.Dubois A., Raymond O., Maene M., Baudino S., Langlade N.B., Boltz V., Vergne P., Bendahmane M. Tinkering with the C-Function: A Molecular Frame for the Selection of Double Flowers in Cultivated Roses. PLoS ONE. 2010;5:e9288. doi: 10.1371/journal.pone.0009288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serwatowska J., Roque E., Gomez-Mena C., Constantin G.D., Wen J., Mysore K.S., Lund O.S., Johansen E., Beltran J.P., Canas L.A. Two euAGAMOUS genes control C-function in Medicago truncatula. PLoS ONE. 2014;9:e103770. doi: 10.1371/journal.pone.0103770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng M., Yanofsky M.F. Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2001;2:186–195. doi: 10.1038/35056041. [DOI] [PubMed] [Google Scholar]
- 44.Causier B., Kieffer M., Davies B. MADS-Box genes reach maturity. Science. 2002;296:275–276. doi: 10.1126/science.1071401. [DOI] [PubMed] [Google Scholar]
- 45.Pan I.L., McQuinn R., Giovannoni J.J., Irish V.F. Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J. Exp. Bot. 2010;61:1795–1806. doi: 10.1093/jxb/erq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X., Wang Q., Yang S., Lin S., Bao M., Bendahmane M., Wu Q., Wang C., Fu X. Identification and Characterization of the MADS-Box Genes and Their Contribution to Flower Organ in Carnation (Dianthus caryophyllus L.) Genes. 2018;9:193. doi: 10.3390/genes9040193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chou K.C., Shen H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE. 2010;5:e11335. doi: 10.1371/journal.pone.0011335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.