Abstract
Fumagillin is a mycotoxin produced, above all, by the saprophytic filamentous fungus Aspergillus fumigatus. This mold is an opportunistic pathogen that can cause invasive aspergillosis, a disease that has high mortality rates linked to it. Its ability to adapt to environmental stresses through the production of secondary metabolites, including several mycotoxins (gliotoxin, fumagillin, pseurotin A, etc.) also seem to play an important role in causing these infections. Since the discovery of the A. fumigatus fumagillin in 1949, many studies have focused on this toxin and in this review we gather all the information currently available. First of all, the structural characteristics of this mycotoxin and the different methods developed for its determination are given in detail. Then, the biosynthetic gene cluster and the metabolic pathway involved in its production and regulation are explained. The activity of fumagillin on its target, the methionine aminopeptidase type 2 (MetAP2) enzyme, and the effects of blocking this enzyme in the host are also described. Finally, the applications that this toxin and its derivatives have in different fields, such as the treatment of cancer and its microsporicidal activity in the treatment of honeybee hive infections with Nosema spp., are reviewed. Therefore, this work offers a complete review of all the information currently related to the fumagillin mycotoxin secreted by A. fumigatus, important because of its role in the fungal infection process but also because it has many other applications, notably in beekeeping, the treatment of infectious diseases, and in oncology.
Keywords: fumagillin, Aspergillus fumigatus, chemical detection, metabolic pathway and regulation, MetAP2 enzyme, cancer treatment, microsporicidal activity, honeybee hive infections
1. Introduction
Aspergillus fumigatus is a saprophytic mold that plays an important ecological role as a decomposer, recycling carbon and nitrogen sources [1,2,3]. It is a ubiquitous fungus with a worldwide distribution, which can be detected in air and soil samples, and even on the International Space Station [4,5,6,7]. This ubiquity is because it is highly adaptive; able to colonize a wide range of environments because of its metabolic diversity, broad stress, and thermal tolerances; and has the ability to spread its conidia easily [4,6,8,9]. In addition, this mold has gone from being considered as just a saprophytic fungus to recognition as one of the most important opportunistic fungal pathogens around, and it is the main causal agent of invasive aspergillosis which has a high mortality rate, between 40% and 90% [2,10,11].
Filamentous fungi produce a remarkable diversity of specialized secondary metabolites (SMs), characterized as bioactive molecules of low molecular weight that are not required for the growth of the organism. Production of these SMs can help fungi in their adaptation to different environmental conditions, improving competitiveness against other microbes or with immune responses during infections [12]. These SMs play diverse ecological roles in fungal defense, communication, and virulence [13], and some of them, owing to their toxic activity, are collectively known as mycotoxins. In recent years, there have been many reviews on the production of theses type of compounds by species from the genus Aspergillus [12,14,15,16,17,18,19,20], and specifically, A. fumigatus has the potential to produce 226 of these compounds [21]. The genes responsible for the synthesis of SMs are commonly associated with biosynthetic gene clusters [16,22,23] and the A. fumigatus genome contains between 26 and 36 putative SMs gene clusters depending on the authors [23,24,25].
Fumagillin is one of these mycotoxins. First isolated from A. fumigatus in 1949 [26], it is encoded inside a supercluster on chromosome eight [27,28]. The target of this mycotoxin is the methionine aminopeptidase (MetAP) type 2 enzyme to which it binds and inactivates irreversibly [29]. As MetAPs are essential for the hydrolyzation of the initial methione (iMet) located in the N-terminal of the new proteins being synthesized [30,31], any imbalance produced by MetAP2 inhibition can affect many proteins, some of them implicated in the correct maintenance of cellular safety.
This activity is the basis of the different effects associated with fumagillin. On the one hand, this toxin showed an antibiotic effect as amoebicidal activity inhibiting the growth of Entamoeba histolytica [32], and shows similar functions during interaction with macrophages [33]. These studies, among others, led Casadevall et al. [34] to hypothesize that fungal virulence can be based on mechanisms developed to defend against ameboid predators. Besides, fumagillin has pharmaceutical potential for the treatment of microsporidiosis [35], as it is the only effective chemical treatment currently available for nosemiasis caused by the parasitic fungi from the Microsporidia phylum on Apis spp. [36]. In fact, it is usually used for the treatment of pests in bee hives [36,37]. However, due to the toxicity of fumagillin, it should be used very carefully and it cannot be used widely. Therefore, less toxic derivatives have been developed to replace fumagillin in some applications. On the other hand, fumagillin has anti-angiogenic activity [29], probably because of its inhibitory activity against the MetAP2 enzyme; consequently, it has valuable pharmaceutical potential and a potential role in the treatment of cancer [30]. Moreover, this toxin is able to inhibit the function of neutrophils [38], inducing cell death in erythrocytes [39] and plays a role in damaging lung epithelial cells which opens the way to fungal invasion [40], perhaps owing to its antiangiogenic properties.
The objective of this review was to collate all current knowledge of this toxin—its chemical characteristics, detection methods, production, metabolic regulation, effects, uses, and its applications in different fields.
2. Fumagillin from a Chemical/Analytical Point of View
2.1. Fumagillin Physichochemical Properties
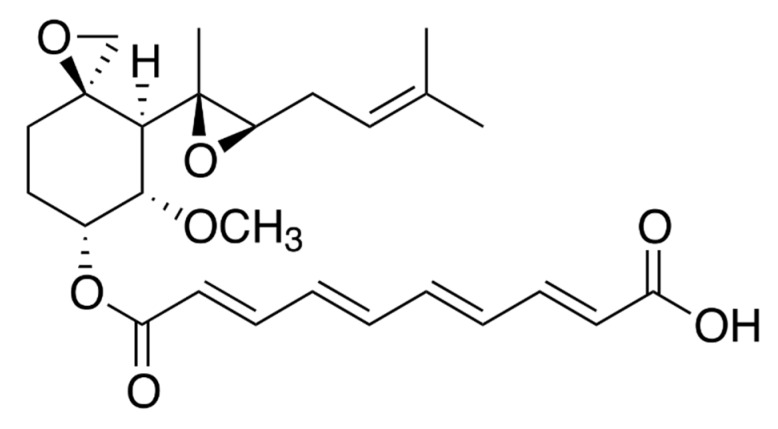
Fumagillin (Figure 1) is a small molecule with a molecular weight of 458.54 g·mol−1. A decatetraenedioic acid connected to a cyclohexane by an ester bond characterizes its chemical structure. The cyclohexane has also a methoxy group, an epoxide, and an aliphatic chain that derives from a terpene and contains another epoxide. These epoxides are in part responsible for the instability of the molecule.
Figure 1.
Chemical structure of fumagillin.
As a relatively non-polar molecule (predicted partition coefficient, logP: 4.05 [41]), fumagillin has poor water solubility (3.3 mg·L−1 [42]) but can be dissolved in organic solvents such as ethanol or DMSO. However, owing to the acidic nature of the carboxylic group (predicted pKa: 4.65 [41]), the polarity of the molecule increases with pH (predicted distribution coefficient, logD: 0.53 at pH 10 [43]). Therefore, the solubility of fumagillin in water is highly dependent on pH.
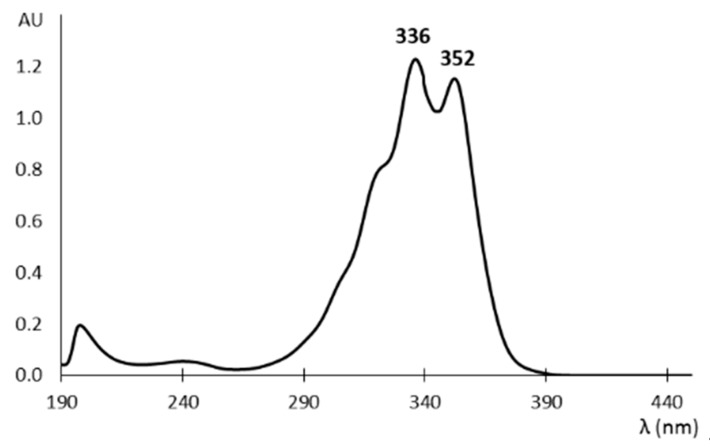
The acidic portion of the molecule is also a good chromophore responsible for the strong absorption of fumagillin in the ultraviolet range, with maxima around 335 and 350 nm (Figure 2) [44].
Figure 2.
Absorption spectrum of fumagillin solution at pH 2.
2.2. Degradation of Fumagillin
In 1954, soon after the discovery of fumagillin [26], a series of three articles on the stability of fumagillin were published: photolytic degradation in alcohol solution [44], photolytic degradation of crystalline fumagillin [45], and thermal degradation in the presence and absence of air [46]. These early papers expressed concerns regarding the degradation of the molecule when exposed to light. In the first publication, Garrett and Eble [44] observed a loss of fumagillin’s activity that they correlated to a decrease in absorptivity at 351 nm. This degradation was attributed to a photolytic degradation caused, primarily, by light below 400 nm. They proposed that the degradation was due to an isomerization process of the tetraenedioic chromophore, and for the first time, coined the term neofumagillin for the degradation products.
Due to the obvious impact of photodegradation on the analytical results, several authors have studied fumagillin stability. Brackett et al. [47] subjected fumagillin to different stress conditions and observed degradation under both strong acidic (1 M HCl) and strong basic (1 M NaOH) conditions. Interestingly, when they studied the impact of UV light exposure, they observed a degradation at 366 nm but not at 254 nm, probably because the molecule does not absorb UV light at that wavelength. They also exposed a fumagillin solution to normal fluorescent laboratory lights and estimated that only around 60% of the analyte remained after six hours of exposure. In a deeper study into the photostability of fumagillin, Kochansky and Nasr [48] exposed 50% ethanol solutions and sugar syrup samples to sunlight and fluorescent light. The former proved to degrade fumagillin faster than the latter and exhibited an initial exponential decay followed by a linear decrease. Dmitrovic and Durden [49] also observed that the degradation of fumagillin in an acetonitrile solution was much faster under sunlight than under a laboratory’s fluorescent light.
The photolytic degradation of fumagillin has also been studied in honey. Assil et al. [50] concluded that after one day of light exposure, only about one-third of the molecule remained. They observed a major degradation product that was analyzed in depth by diode array spectrophotometer, nuclear magnetic resonance (NMR), and infrared spectroscopy (IR). All the results agree with the isomerization process previously proposed by Garret and Eble [44]. In order to reduce the photodegradation of fumagillin the use of amber vials is a useful alternative, as demonstrated by Higes et al. [51].
Although photodegradation is the most important factor related to fumagillin stability, thermal degradation should also be mentioned. For instance, van den Heever et al. [52] paid special attention to this variable in order to study the degradation of fumagillin in bee-hive conditions. They monitored the concentration of fumagillin in honey samples at 21 °C (in the dark and exposed to light) and at 34 °C in darkness (simulated hive conditions). They observed that the fastest degradation occurred at 21 °C under light exposure and more importantly, that the degradation was faster at 34 °C than at 21 °C in the absence of light. The faster degradation at higher temperatures is in accordance with the results obtained by Higes et al. [51] in syrup and in sugar honey patties. In that case, they studied four temperatures (4, 22, 30 and 40 °C) and observed that the degradation rate in syrup increased with temperature, especially in the samples exposed at 40 °C (where no fumagillin was detected after 20 days). On the other hand, fumagillin was more stable in sugar honey patties with losses around 10% after 45 days. Assil et al. [50] also observed that fumagillin in pure honey was moderately stable for 35 days, even at 80 °C. Under the same conditions, water:ethanol solutions were not stable and they attributed this fact to a protective effect of honey, since high sugar concentrations might limit the availability of reactant water.
Considering the degradation processes that fumagillin undergoes, the light induced and thermal decomposition products have been studied for decades [44]. Probably, the most important contributions to the field are the works by Assil et al. [50] using NMR and IR, and by Nozal et al. [53] using mass spectrometry (MS). On the one hand, thermal degradation products have been related to compounds more polar than fumagillin since they elute earlier in reverse phase chromatography. Among these, dihydroxyfumagillin comes from the hydrolysis of the unstable epoxide in the cyclohexane, and fumagillol via a further hydrolysis of the ester. On the other hand, the light induced degradation compounds are slightly more non-polar than the precursor molecule and are related to the aforementioned neofumagillin derived from the isomerization of the decatetraenedioic moiety. Four isomers have been proposed: two cis–trans diastereoisomers and cis–cis and cis–trans isomers (Figure 3).
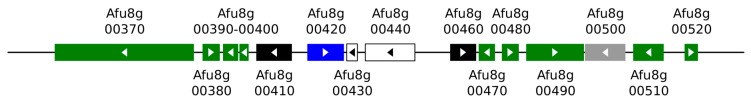
Figure 3.
Arrangement of genes in the fumagillin cluster of A. fumigatus Af293 strain (Afu8g00370-Afu8g00520; ORF names, Source NCBI Database [65]. Green: genes implicated in biosynthetic pathway; blue: regulatory gene; gray and black: other genes related with fumagillin; white: non-related fumagillin genes.
2.3. Analytical Methods for the Determination of Fumagillin
Fumagillin has been widely employed as an antimicrobial agent with honeybees and fish; thus, most analytical methods have been developed for its identification in these matrices. Although these methods are usually only intended for fumagillin’s detection, it has also been analyzed together with bicyclohexylamine [54], a toxic compound used as counter ion in commercial formulations, or together with other residues in honey [55], surface waters [56], and dairy products [57].
Fumagillin has traditionally been analyzed with photometric detection coupled with high performance liquid chromatography (HPLC), using either fixed wavelength instruments or diode array instruments (DAD). More recently, MS based methods have been also developed.
A solid phase extraction (SPE) treatment is commonly applied prior to chromatographic analysis, although this step is not necessary in simple matrices such as fumagillin powder, for which a sample dilution is enough [47]. Direct analysis after sample dilution (10%–20%, w/v) has also been used for honey analysis employing water or water:acetonitrile solutions [47,51], but is not the most common practice. Nowadays, SPE is usually performed after diluting honey with water, typically using polymeric sorbents (Strata-X). After sample loading, the cartridge is cleaned with water: methanol mixtures and fumagillin is eluted with acetonitrile, basified acetonitrile, or acidified acetonitrile depending on the author [49,52,53]. It is remarkable that Kanda et al. [58] proposed a different SPE approach based on the use of weak anion exchange cartridges after QuEChERS extraction.
SPE based sample treatments have also been applied to the analysis of fumagillin in fish tissue, but due to the nature of this matrix, an additional sample treatment step is necessary. Hence, Guyonnet et al. [59] proposed a method which includes a sample extraction with water and acetonitrile prior to SPE. Fekete et al. [60] developed two sample treatments that could be used, depending on the sensitivity requirements. In both of them, acetonitrile was added to tissue and sonicated in order to disrupt the cells. Then, a simple clean-up or an enrichment procedure was proposed using a C18 cartridge in both cases. For the former, the supernatant is passed through the column and directly analyzed. The latter requires a higher amount of tissue and water addition to the supernatant in order to retain fumagillin in the sorbent. Finally, the analyte is eluted with acetonitrile:water (9:1) solution and analyzed by HPLC.
The chromatographic analysis of fumagillin is performed on non-polar stationary phases (C18 or C8), mainly in isocratic mode, although gradient mode has also been applied for some multiresidue or MS methods. Regarding the mobile phase, acetonitrile is the most common organic modifier with the exception of the method developed by Kanda et al. [58], where methanol is employed and also the method proposed by van den Heever et al. [54] where the acetonitrile contains a small amount of methanol (10%). The aqueous mobile phase is usually slightly acidic due to the addition of phosphoric acid [60], formic acid [55], or acetic acid [47,48,50]. Nevertheless, other authors use ammonium formate and formic acid buffers [49,54] or ammonium formate alone to adjust the pH of the mobile phase. The latter was used by Nozal et al. [53] for the determination of fumagillin and is especially interesting because at this pH value (6.5) the carboxylic group is deprotonated. For this reason, the acidic nature of fumagillin was employed by Guyonnet et al. [59] to develop a method based on ion-pairing. A mobile phase at pH 7.8 was used, where tetrabutylammonium acts as a cation to retain fumagillin in a C8 column. To end this section on chromatographic conditions, the comparison of methods proposed by Fekete et al. [61] for the determination of the analyte in fish tissue should be mentioned. This team developed, as far as we know, the only normal phase method for fumagillin using a silica gel column and a complex mobile phase (n-hexane:dichloromethane:dioxane:2-propanol:acetic acid 43:43:9:5:0.1, v/v percent). One of the advantages of the method is that it can be more easily coupled to a liquid-liquid extraction, such as the one the group proposed with dichloromethane:dioxane:2-propanol. Furthermore, it offered complementary selectivity to the normal phase method they developed (which includes bicyclohexylamine as additive in the mobile phase).
The chromatographic separation has traditionally been coupled with photometric detection since fumagillin shows two adsorption maxima in the UV region around 335 and 350 nm (Figure 2). Although the absorption at 335 nm is slightly higher, the 350 nm wavelength has more commonly been used because a higher selectivity is expected [47,50,53,59,60,61]. In the last few years MS based methods have emerged in which fumagillin is analyzed in positive electrospray mode with the exception of the Orbitrap based method developed by Jia et al. [57] in which the [M-H]-ion is used (m/z 457.22318) for the analysis of dairy products. Nozal et al. [53] used a single quadrupole instrument and measured the protonated molecule (m/z 459) in selected ion monitoring mode (SIM) for quantitative analysis. Furthermore, they used the potassium adduct (m/z 497) and a fragment (m/z 427) for confirmation. All the tandem MS (MS/MS) methods for the analysis of fumagillin use the protonated molecule as the precursor ion but different transitions are monitored for quantitative purposes: 459.2 > 177.0 [52], 459.1 > 233.3 [55], 459.2 > 131 [49], or 459 > 427 [56]. In addition to these quantitation transitions 459.1 > 215.3 and 459.2 > 102.8 transitions have been used for the confirmation analysis.
As expected, MS based analytical methods offer lower limits of quantitation (LOQ) than photometric detection. In this sense, Nozal et al. [53] obtained LOQ levels several times lower with MS compared to photometric detection when they applied a LC-DAD-MS method to honey samples. While the LOQ for the DAD detection at 350 nm ranged from 95 to 150 ng·g−1 depending on the origin of the honey, the LOQ for the same honeys ranged from 3 to 10 ng·g−1 using SIM detection. Also, 10 ng·g−1 was the LOQ achieved by van den Heever et al. in honey using a MS/MS method [54] and the one obtained by Dmitrovic et al. [49] was slightly lower. They calculated the LOQ with different MS/MS instruments and obtained values ranging from 1.1 to 1.6 ng·g−1. An order of magnitude lower (0.1 ng·g−1) is the LOQ reported by Kanda et al. [58] obtained with a LC-MS/MS method that included sample treatment with QuEChERS and a SPE. Regarding photometric detection, LOQ values reported by different authors in honey and fish range from 5 to 100 ng·g−1. Indeed, Fekete et al. [60] proposed two different sample treatments in which 5 ng·g−1 LOQ was obtained when enriching the samples and 100 ng·g−1 when applying a simple clean-up procedure.
Finally, it must be highlighted that apart from the chromatographic methods described here an enzyme-linked immunosorbent assay (ELISA) method was proposed by Assil et al. [50] in order to detect fumagillin in honey, reaching levels as low as 20 ng·g−1.
3. Fumagillin from a Biological Point of View
3.1. Fumagillin Biosynthetic Gene Cluster
Genetic information for fumagillin production in A. fumigatus is found in a supercluster of genes that encodes genes related to the production of three bioactive metabolites, among others, fumitremorgin B, pseurotin A, and fumagillin, which are located on the subtelomeric region of chromosome 8 (Afu8g00100-Afu8g00720) [27,28]. The fumagillin biosynthetic gene cluster, which encodes 15 genes inside this supercluster (from Afu8g00370 to Afu8g00520), is named the fma cluster (Figure 3) [28,62]. Unlike other clusters that encode genes for other biosynthetic pathways, an interwoven net between the genes of the pseurotin A and fumagillin clusters has been detected [28]. The nomenclature of the genes involved in this biosynthetic pathway is complex since each research group applied their own terminology. One of them named the genes and enzymes encoded by their enzymatic activity, fma-PKS (gene encoding fumagillin biosynthesis polyketide synthase, Afu8g00370) and when the activity was unknown, by the position in the genome of A. fumigatus Af293 strain, as af490 (Afu8g00490) [62,63,64]. Almost simultaneously, another group named the genes in a progression from fmaA to fmaG using the A. fumigatus A1163 strain [28]. Table 1 shows the different nomenclatures used and the information collected in different databases. In this review, the open reading frame (ORF), gene, and protein names of NCBI Database [65], Aspergillus Genome Database [66], and UniprotKB Database [67] were used, respectively.
Table 1.
Composition of fumagillin biosynthetic cluster.
| Gene Name b | Other Gene Names c | Protein Abbreviated (Complete) Name d | UniProtKB d | Deletion Mutant | Ref. | |
|---|---|---|---|---|---|---|
| Afu8g00370 | fma-PKS |
af370
fmaB |
Fma-PKS (Fumagillin dodecapentaenoate synthase/Fumagillin biosynthesis polyketide synthase) | Q4WAY3 | Fumagillin production abolished Fumagillol production |
[28,62] |
| Afu8g00380 | fma-AT |
af380
fmaC |
Fma-AT (Polyketide transferase af380/Fumagillin biosynthesis acyltransferase) | Q4WAY4 | Fumagillin production abolished Fumagillol production |
[62] |
| Afu8g00390 /00400 | Afu8g00390 |
af390-400
fmaD |
Fma-MT (O-methyltransferase af390-400/Fumagillin biosynthesis methyltransferase) | A0A067Z9B6 | Fumagillin production abolished Demethyl-fumagillin production |
[28,63] |
| Afu8g00410 | metAP | fpaII | MAP2-1/MetAP2-1 (Methionine aminopeptidase 2-1/Peptidase M) | Q4WAY7 | [68] | |
| Afu8g00420 | fumR | fapR | FumR (C6 finger transcription factor fumR/Fumagillin gene cluster regulator) | Q4WAY8 | Fumagillin and pseurotin gene clusters silenced | [28,64] |
| Afu8g00430 | Afu8g00430 | (EthD domain-containing protein) | Q4WAY9 | [62,69] | ||
| Afu8g00440 | Afu8g00440 | psoF | PsoF (Baeyer-Villiger monooxygenase/Dual-functional monooxygenase/methyltransferase psoF/Pseurotin biosynthesis protein F) | Q4WAZ0 | Pseurotin production abolished | [28,62,69,70,71] |
| Afu8g00460 | Afu8g00460 | fpaI | (Methionine aminopeptidase) | Q4WAZ1 | [28,62] | |
| Afu8g00470 | Afu8g00470 |
af470
fmaE |
Fma-ABM (Monooxygenase af470/Fumagillin biosynthesis antibiotic biosynthesis monooxygenase superfamily monooxygenase) | Q4WAZ2 | Fumagillin production abolished Prefumagillin accumulation |
[28,62,63] |
| Afu8g00480 | Afu8g00480 |
af480
fmaF |
Fma-C6H (Dioxygenase af480/Fumagillin biosynthesis cluster C-6 hydroxylase) | Q4WAZ3 | Fumagillin production abolished 6-demethoxy-fumagillin accumulation |
[28,62,63] |
| Afu8g00490 | Afu8g00490 | af490 | Fma-KR (Stereoselective keto-reductase af490/Fumagillin biosynthesis cluster keto-reductase) | Q4WAZ4 | Fumagillin production strongly decreased | [62,63,72] |
| Afu8g00500 | Afu8g00500 | (Acetate-CoA ligase, putative) | Q4WAZ5 | [62,72] | ||
| Afu8g00510 | Afu8g00510 |
af510
fmaG |
Fma-P450 (Multifunctional cytochrome P450 monooxygenase af510/ Fumagillin biosynthesis cluster P450 monooxygenase) | Q4WAZ6 | Fumagillin production abolished, β-trans-bergamotene accumulation |
[28,62,63] |
| Afu8g00520 | fma-TC |
af520
fmaA |
Fma-TC (Fumagillin beta-trans-bergamotene synthase af520/Fumagillin biosynthesis terpene cyclase) | M4VQY9 | Fumagillin production abolished | [28,62] |
Inside the fma cluster, there are other genes related to fumagillin production. Among these, the Afu8g00500 gene encodes a putative acetate-CoA ligase, which may be related to the production of the substrates necessary for the enzyme farnesyl synthase to create farnesyl pyrophosphate. However, this has not been proven.
In addition, the gene regulating the production of this mycotoxin (fumR) is also found inside this cluster. Dhingra et al. [64] and Wiemann et al. [28] detected this gene and named it fumR and fapR, respectively. The deletion of this gene brought about a lack of fumagillin and pseurotin A production [28,64].
On the other hand, the genes Afu8g00430 and Afu8g00440, are not related to fumagillin production but are also in this cluster. They encode an EthD domain-containing protein and PsoF (pseurotin biosynthesis protein F) respectively and seem to be related to the production cluster of pseurotin A [28].
Finally, it is worth mentioning that genes encoding two proteins with MetAP activity also belong to this cluster. These genes, similar to the fumagillin target, may be related to the resistance of the fungus to its own toxin, as explained below. One of them, encoded by the metAP gene, is a MetAP2-1, and the other one encoded by the gene Afu8g00460, is a Putative MetAP type 1 (MetAP1).
3.2. Fumagillin Biosynthetic Pathways
The fumagillin metabolic pathway was elucidated by Dhingra et al. [64], Lin et al. [62,63], and Wiemann et al. [28], based on studies carried out with defective mutants of A. fumigatus, cloning and obtaining the different enzymes in the pathway in Saccharomyces, and performing in vitro studies of its enzymatic activity using intermediaries as substrate. Figure 4 shows the metabolic reactions involved in the pathway.
Figure 4.
Fumagillin biosynthetic pathway.
Briefly, two components are required for the generation of fumagillin, a structure composed of a rearranged highly oxygenated sesquiterpene and a polyketide-derived tetraenoic diacid. They are produced by two different biosynthetic branches [62,63], which are subsequently joined by esterification.
To be specific, one branch of the pathway begins with the conversion of farnesyl pyrophosphate (FPP) to β-trans-bergamotene by the integral membrane-enzyme, Fma-TC, a beta-trans-bergamotene synthase [62]. This intermediate component undergoes three sequential steps of oxidative transformations catalyzed by another membrane enzyme, Fma-P450, a multifunctional cytochrome P450 monooxygenase [63]. In these oxidation steps, β-trans-bergamotene is first hydroxylated at the bridgehead C5 position to yield 5R-hydroxyl-beta-trans-bergamotene; that is subsequently oxidized at the C9 position coupled to cleavage of the cyclobutane C5—C8 bond originating the intermediate component 5-keto-cordycol. Then, the final step of oxidation, an additional epoxidation via this Fma-P450, transforms this compound into 5-keto-demethoxyfumagillol. In Figure 4, these three oxidation steps are shown as a single action. The biosynthetic pathway continues with two sequential transformations, a hydroxylation and subsequent methylation in the C6 position of 5-keto-demethoxyfumagillol to yield 5-keto-fumagillol [63]. Two enzymes participate in these transformations, Fma-C6H, a dioxygenase, and Fma-MT, a polyketide transferase that causes the hydroxylation and methylation, respectively. Lin et al. [63] indicated that Fma-MT was misannotated, as coded by two separated genes, Afu8g00390 and Afu8g00400 in the NCBI Database (https://www.ncbi.nlm.nih.gov/), detecting a single joint transcript for these two genes. The protein database UniProtKB [67] has accepted this proposal and these two genes have now been unified as one single protein. The final step consists of a stereoselective reduction of 5-keto-fumagillol by Fma-KR, a stereoselective keto-reductase to obtain fumagillol.
The second branch of the fumagillin biosynthetic pathway produces the dodecapentaenoyl group via Fma-PKS, a fumagillin dodecapentaenoate synthase. This dodecapentaenoyl group is not detected as a free molecule, and Lin et al. [62] have pointed out that it stays attached to the Fma-PKS enzyme. The next step is due the action of Fma-AT, a polyketide transferase that catalyzes the transfer of the synthesized dodecapentaenoyl group onto the fumagillol produced in the other branch, to produce prefumagillin [62]. Finally, the Fma-ABM, converts prefumagillin to fumagillin via an oxidative cleavage with a monooxygenase [63].
3.3. Regulation of Fumagillin Cluster
Sensing and responding to environmental clues is critical to the lifestyle of filamentous fungi [73]. In fact, many of the environmental signals that regulate SMs production in Aspergillus fungi, including temperature, pH, and carbon or nitrogen sources, also trigger the onset of asexual and sexual development [74]. In spite of this, it is not well understood how environmental variations influence fungi to produce a wide diversity of ecologically important SMs.
During the expression of the genes involved in the synthesis and secretion of SMs, a hierarchical network of master regulators that respond to multiple environmental cues is implied [75] and they are also involved in the complex processes that regulate development in fungi [25]. In fact, to control the transcription of these SMs production-clusters, not only cluster-specific transcription factors, but globally acting transcriptional regulators, are often involved [25].
As pointed out above, the FumR regulates the expression of both the fma and the pseurotin clusters [28,64]. In turn, the global regulator of the fungal secondary metabolism, LaeA, controls the supercluster that includes both clusters [17,27,76,77,78,79]. The relevance of LaeA is worth highlighting as it influences the expression of at least 9.5% of the genome (943 of 9,626 genes in A. fumigatus), and to be specific, controls the expression of 20%–40% of major classes of SM biosynthesis genes [27]. Moreover, it is part of the “velvet system,” which is a light-dependent system made up by the heterotrimeric complex VelB/VeA/LaeA and is responsible for developmental regulation and the secondary metabolism [73,78]. In this complex, the components do not affect the activation of the gene clusters under different conditions equally [64,73,79] and while the LaeA always regulates the genes positively, the VeA sometimes shows a negative regulation.
In addition to these cellular networks, other signaling pathways related to fumagillin expression, are the G-protein-coupled receptors GprC and GprD [80], the developmental regulators FlbB and FlbE [81], the nonredundant phosphopantetheinyl transferase PptA [82], the catalytic subunit of protein phosphatase Z PpzA [83], the developmental regulators BrlA and WetA [25,84], the VeA-dependent regulator of secondary metabolism MtfA [85], the developmental transcription factor StuA [68], and MAT1-1 and MAT1-2 genes of one of the mating-types [86]. All of these signaling networks affect many or even all secondary metabolism clusters, not only the genes involved in fumagillin pathways.
Finally, the host immune response during colonization/infection and the antifungal treatment employed can also activate these secondary metabolism regulators. The activation of the fumagillin cluster during experimental animal infections has been detected, contributing to the invasion and generation of cell damage [40]. Other authors have shown that, in contact with caspofungin, fumagillin [87] and gliotoxin [88] are overexpressed. This implies that the treatment with caspofungin could have adverse effects on host cells during infection. Moreover, taking into account the fact that several fungi can produce echinocandins, such as caspofungin [89], these responses can represent a normal response to environmental signals between fungi [87].
4. Biological Activities of Fumagillin
4.1. Methionine Aminopeptidases as Cellular Targets of the Mycotoxin
The MetAP is a family of intracellular proteolytic enzymes, originally described as cytosolic proteins [90,91], that fulfils an important role during protein post-transcriptional and co-translational modifications (NH2-terminal myristoylation and acetylation among others) [30]. Furthermore, MetAPs are essential for the new proteins because they control the hydrolyzed iMet located in the N-terminal [30,31]. The correct excision of iMet is also necessary to expose a glycine residue where a lipid acid could bind covalently, allowing an efficient association with membranes or other proteins [31]. The maintenance of MetAP activity throughout evolution may have an energy recycling purpose because in lower organisms methionine is the most expensive amino acid to synthesize from an energy efficiency point of view [92].
Broadly speaking, two different types of MetAP enzymes, known as MetAP1 and MetAP2, exist [90,91]. Both types possess distinct substrate specificity but share a similar enzymatic activity. The specificity of MetAPs relies on the second residue of the target protein, and therefore, depending on that amino acid, only one of the isoforms, both, or neither of them, can hydrolyze the N-terminal methionine [93]. Furthermore, while MetAP2 is only responsible for the N-terminal processing of the proteins synthesized de novo, MetAP1 is responsible for processing most of the proteins [94,95]. Structurally, they are quite similar, but the most important difference between them is the helical domain insertion on the surface of MetAP2 [96,97,98]. It is also worth mentioning the fact that both types are not usually expressed simultaneously, Eubacteria expressing only MetAP1, archaebacteria only MetAP2, and eukaryotes both types [99,100,101,102]. An exception is Synechocystis sp., which express MetAP1, 2 and a novel type MetAP3 only found in this cyanobacteria [103].
In spite of the fact that in general, only two types of MetAPs are described, there are many isoforms of each enzyme. The classical examples are the six isoforms of MetAP1 that Arabidopsis thaliana carries in its genome [104]. Furthermore, each type of MetAP can include different sub-families, such as the human MetAP-1D, also called MAP1D, which is a sub-family of type 1 and is overexpressed in colon cancer [105]. This different distribution and the wide variety of MetAPs point out the complexity of this enzyme group.
Although MetAP1 is very interesting, MetAP2 has been studied more owing to its inhibitory effect on angiogenesis and endothelial cell proliferation. To be precise, the function of MetAP2 related to endothelial cell proliferation was discovered serendipitously when Ingber et al. [106] suffered a fungal contamination in some capillary endothelial cell cultures. It was observed that the fungus, identified as A. fumigatus Fresenius, produced a cell rounding gradient zone instead of the normal toxicity produced by other fungal contaminations in cell cultures. Then, they studied the effect of conditioned medium and isolated the active fraction, finally identifying fumagillin as the active compound. This purified fumagillin completely inhibited endothelial cell proliferation, angiogenesis, and tumor-induced neovascularization [106].
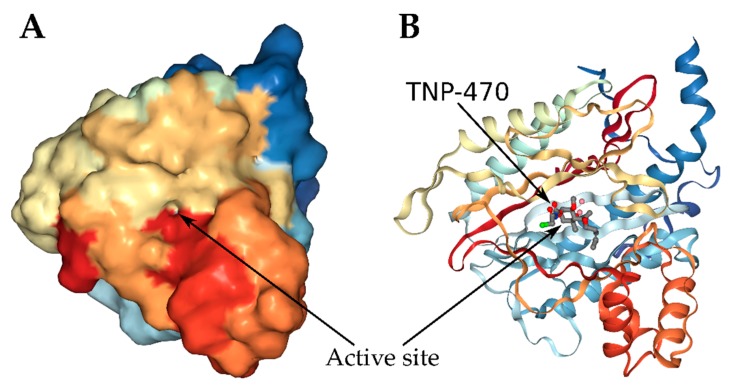
However, it was not until 1997 when Sin et al. [29], using TNP-470, a fumagillin derivate, discovered that the fumagillin-binding protein was the MetAP2, which was irreversibly inactivated by the toxin. For that, they used mutant models of S. cerevisae null for MetAP1 (∆map1) and MetAP2 (∆map2) and concluded that because fumagillin inhibited the growth of the ∆map1 strain, but not the ∆map2 and the wild type strains, the target of the toxin was the metalloprotease MetAP2, which is highly conserved between humans and fungi [29]. One year later, Liu et al. [107] were the first to define the precise connection between the toxin and MetAP2 as a covalent bond formed between the C3 ring epoxide group of fumagillin and the imidazole nitrogen (Nε2) of the histidine 231 located on the active site of the MetAP2 [107] (Figure 5). The exposure to this mycotoxin inhibits irreversibly the aminopeptidase activity of MetAP2 because fumagillin binds at the action site of the enzyme [29,98].
Figure 5.
Human MetAP2 3D image. (A) Frontal view of enzyme with the active site and (B) a view of the MetAP2 chain coupled to TNP-470, a fumagillin derivative in active site. Rainbow colors: blue (N-terminus) to red (C-terminus). TNP-470 view as ball and stick. (PDB ID: 1B6A, NGL Viewer [108] RCSB PDB [109,110]).
It must be recognized that in eukaryotic cells, MetAP2 were previously described as a eukaryotic initiation factor-2-associated protein (p67), which protects the phosphorylation of the eukaryotic initiation factor-2 alpha (eIF2α) [91,101]. Specifically, the N-terminal lysine of the MetAP2 seems to take part in the protection of eIF2α phosphorylation [111]. Furthermore, p67 (MetAP2) can also bind to ERK1 and ERK2, two kinases that have a crucial function in the cell proliferation process [112], inhibiting its phosphorylation [113].
The role of MetAP2 on cell proliferation, and therefore on cancer, has also been widely studied. In fact, several researchers have shown that the proliferation of endothelial cells decreases if MetAP2 is strongly downregulated by an anti-sense oligonucleotide or siRNAs [114,115,116]. Catalano et al. [97], studying mesothelioma cancer cells, discovered that these cells expressed MetAP2 mRNA levels higher than non-cancerous mesothelial cells and that the treatment with fumagillin induces their apoptosis. In contrast, MetAP2 inhibition increased caspase activity, prevented fumagillin-induced apoptosis, but did not affect the telomerase activity of the mesothelioma cancer cells [97]. Overexpression of MetAP2 had been also related with B cells of malignant lymphomas of various subtypes [117], and with cholangiocarcinoma (CAA) cells [116]. In the final case, inactivation of MetAP2 eliminated the proliferation of metastatic CCA [116].
Regarding the MetAPs’ importance to microorganisms, experimental studies demonstrated that MetAP1 is an essential gene in Escherichia coli [118] and Salmonella typhimurium [119]. However, in S. cerevisae, MetAP1 and MetAP2 are only essential when both are silenced [120], affecting the deletion of one of them only to the growth rate [94,120]. In the A. fumigatus genome, some genes encode for MetAP2 enzymes, the housekeeping Afu2g01750, and the Afu8g00410 that is encoded inside the fumagillin biosynthetic cluster. The final gene, which encodes MetAP2-1, is expressed together with this mycotoxin when the cluster is activated. Production of this MetAP2-1 enzyme might protect the fungus from the action of its own toxin because it is insensitive to fumagillin activity and/or because the concentration of the target is considerably increased [62,121]. However, it is not yet clear whether this hypothesis is correct.
4.2. Effects of Fumagillin Activity on Host Cells
As the MetAP activity is essential for cell viability as explained above [92], fumagillin activity affects not only MetAPs proteins but also all the subsequent activities necessary for the well-functioning of proteins related to cell viability and growth [31].
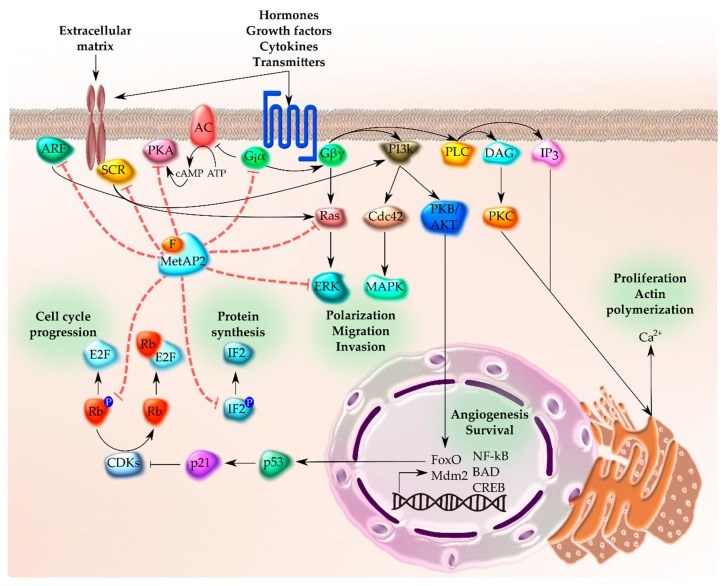
Among the molecules affected by the inhibition of MetAP, the guanine nucleotide-binding proteins (G protein-coupled receptors or GPCRs) are a ubiquitous and well-studied family of proteins that need to be N-terminally processed [122]. Namely, the myristoylation of Gi/o α subunit allows the association with membranes, indispensable for the efficiency of the protein Gi/o but not for Gs [123]. The pathway starts with an extracellular stimulus caused by the binding of hormones, cytokines, neurotransmitters, growth factors, etc., with its membrane GPCRs characterized by seven membrane-spanning regions [124]. Afterwards, these proteins activate intracellular signaling involved in numerous signal transduction pathways related to development, survival, proliferation, invasion, migration, tumorigenesis, etc. [31,122,123,124,125]. Focusing on the activation of Gi/o, its α subunit can inhibit adenylyl cyclase (AC), incrementing ATP levels and inhibiting protein kinase A (PKA) [124]. At the same time, Giα can pass the signal flow to Giβγ, which regulates phospholipase C (PLC), phosphatidylinositol 3 kinase (PI3K), Ras, and K+ channels. PLC stimulates inositol (1,4,5)-trisphosphate (IP3) which mobilizes Ca2+ and protein kinase C (PKC) via diacylglycerol (DAG) [126]. In addition, Ras activates the ERK pathway and calcium dependent proliferation. Furthermore, PI3K promotes the cell division control protein Cdc42, responsible for actin reorganization, necessary for cell adhesion, polarization, migration and invasion [127], and the protein kinase B (PKB/AKT), triggering the activation of numerous anti-apoptotic genes (NF-κB, BAD, CREB, etc.) and inhibiting p53 via the activation of FoxO and Mdm2 (Figure 6) [128]. Besides, PI3K activates the extracellular signal regulated protein kinase (ERK) cascade.
Figure 6.
Schematic diagram depicting MetAP2 implication in different pathways. The scheme represents the normal functioning of cascades (black arrows) and the effect of MetAP2 inhibited by fumagillin (red discontinuous arrows). Fumagillin (F) binds covalently to MetAP2 and provokes the inhibition of G protein, SRC, and the ARF signaling pathway at different levels, and the protection of Rb and IF2 against phosphorylation (P).
Other molecules that must also suffer N-terminal processing, such as Src [129], PKA [130], ARF [131], and calcineurin [132], are also involved in the same processes. Depending on the stimuli, either Gs or Gi/o pathways are activated, and their effects are related to cytoskeletal contraction or relaxation, respectively [124,127]. Together with Cdc42 and G proteins, these signals are able to mediate extracellular matrix cilia formation, phagocytosis, cell adhesion, and cytoskeletal remodeling [133].
Therefore, the activity of fumagillin could affect multiple signaling pathways, so it is easy to understand that there are a large number of different phenotypes and sensitivities produced by fumagillin. For example, an inadequate localization of Gi/o, Src, and Arf proteins produced by the lack of iMet excision means that the cell is unable to produce the transduction signals after the reception of an external stimulus. This would explain the lack of growth in cells exposed to fumagillin when they are treated with insulin-like grow factor (IGF), platelet-derived growth factor (PDGF), or endothelial growth factor (VEGF) [30,134,135]. Moreover, as signal transduction pathways have the objective of rearranging the cytoskeleton via actin polymerization; their alteration may be the reason for failures in the invasion and migration of some cell types [30].
However, the intracellular signaling pathways are not the only metabolic pathways where MetAP2 is involved. For example: thioredoxin (Trx1), cyclophilin A (CypA), Eukaryotic elongation factor (eEF2), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) are in vivo proven MetAP2 substrates [136].
In addition, the cytostatic and cytotoxic effect observed in fumagillin sensitive cell lines [137] could be due to an increase in reactive oxygen species (ROS). This affects the cell redox state, causing the incorrect functioning of signalization pathways and the lack of thioredoxin antioxidant function [136,137]. In support of this theory is the fact that it has been demonstrated that the addition of N-acetylcysteine, a potent antioxidant, to the medium protected human melanoma tumor cell line B16F10 from TNP-470, induced death by avoiding an increase in ROS [138]. Nevertheless, in neutrophils the exposure to fumagillin inhibits the NADH oxide complex formation by blocking the translocation of the cytosolic component p47phox to the membrane, thus, preventing the production of superoxide. In addition, neutrophils treated with fumagillin show less degranulation and reduced levels of actin filaments, something that could contribute to the inhibition of the structural reorganizations necessary for neutrophil activation [38]. Neutrophils are the first line of defense against conidia reaching the pulmonary alveoli, and therefore, fumagillin can reduce their ability to kill the hyphae or phagocyte conidia of A. fumigatus [38].
Another effect attributed to fumagillin is the promotion of eryptosis, which is the suicidal death of erythrocytes by a dysregulation of intracellular calcium concentration [39], which may also contribute to the virulence of A. fumigatus. Calcium is regulated by Ras, G proteins, and calcineurin, all of them targets for MetAP2 [122,132,139]. This mechanism significantly increases intracellular calcium concentration, enhances ceramide abundance, triggers phosphatidylserine exposure, and lowers the volume of erythrocytes.
4.3. Fumagillin and Other Similar Toxins
The fungal metabolism allows the production of a huge variety of toxins, and that is the case for A. fumigatus [1]. In the case of fumagillin, this molecule does not share its effects or structure with other toxins produced by this fungus [140], but there are other organisms that produce similar toxins; for example, Ovalicin, a sesquiterpene produced by Pseudorotium ovalis, structurally similar to fumagillin [141]. In fact, they share the same specific target and effect over cells causing same cytostatic and cytotoxic effects [142]. In spite of their being closely related, the mechanism of action of the two toxins has not yet been clarified.
In relation to cellular effect, other toxins are also involved in similar metabolic pathways. Pertussis toxin (PTX), cholera toxin, Escherichia coli heat-labile enterotoxin (LT), diphtheria toxin, and Pseudomonas exotoxin A are ADP-ribosylating toxins, which do not affect METAP2 activity, but affect the later stages of that signal transduction pathway [143]. Specifically, the A-promoter of PTX catalyzes the ADP-ribosylation of the C-terminal in the α subunits of Gi/o, inhibiting the membrane localization, and therefore, stops signal transduction [144]. Regarding PTX, this also induces an accumulation of cAMP that affects cell polarization, migration, apoptosis, and the response to growth factors [127,143,144]. In the case of the cholera and LT toxins, these enhance AC activity stimulating the Gs pathway, producing the same effect as Gi/o inhibition [145]. Finally, the diphtheria toxin and the Pseudomonas exotoxin A have the same mechanism of action, ADP-ribosylation of EF2 producing the inhibition of protein synthesis [146].
Angiogenesis inhibitors or tumor suppressors, Bevacizumab, Dovitinib, Volociximab, etc., focus on suppressing the growth factors’ signal transduction pathways at the membrane, and at the receptor tyrosine kinases level, blocking cytoskeletal reorganization, or interfering with mediator molecules, such as histamine and nitric oxide [147]. These actions are also in accordance with the theories expressed in this review about fumagillin mechanism of action.
5. Fumagillin Applications
5.1. Angiogenesis and Antitumor Activities
In the early nineties, the antiangiogenic effect of fumagillin was proven. Ingber et al. [106] stated that fumagillin completely inhibits endothelial cell proliferation in the presence of basic fibroblast growth factor (bFGF), and also inhibits tumor suppressor-induced neovascularization in mice [106]. Shortly after, Kusaka et al. [148] proved that fumagillin and its derivative AGM-1470 are antiangiogenic compounds with four different assays. Summarizing their results, locally administered AGM-1470 inhibited the angiogenesis in a chick embryo chorioallantoic membrane assay and in a rat corneal assay. On the other hand, in a rat sponge implantation assay, AGM-1470 inhibited angiogenesis induced by bFGF. Moreover, in a rat blood vessel organ culture assay, both fumagillin and AGM-1470, reduced the formation of blood vessels and the growth of endothelial cells by 90%, although AGM-1470 did so at a lower concentration [148].
As angiogenesis is an important step in tumor and metastasis development, angiogenesis inhibitors have been studied as promising molecules for cancer treatment. In this regard, some fumagillin derivatives have been tested for antitumor effects on tumor cell lines and different animal models, mainly rats and mice, and in only a few human clinical trials. Some of them lowered the growth of human umbilical vein endothelial cells (HUVEC) at low concentrations and caused cell apoptosis at higher concentrations. In an animal model they suppressed tumor growth [149]. Similar effects have also been observed in a CCA cholangiocarcinoma cell line with fumagillin and its derivative TNP-470 [116,150]. In other animal studies with TNP-470, inhibition of tumor growth and lower tumor vascularization were also observed [151]. This compound also prevented an increase in vascular endothelial growth factor (VEGF), hepatocyte growth factor beta (HGF-β), cyclin D, cyclin E, and cyclin-dependent kinase Cdk4 and Cdk3 levels that are present in hepatocarcinogenesis, promoting cell cycle inhibition [152]. On the other hand, the administration of TNP-470 to patients with colon adenocarcinoma reduced liver metastasis in a phase I study [153]. However, the toxicity of these compounds must be taken into account as they can cause serious but reversible side effects, such as encephalopathy, thrombocytopenia, and ataxia [153,154,155].
It has been observed that in murine models of colorectal cancer, fumagillin and its derivative fr-11887 can inhibit colorectal growth and prevent metastasis [156,157]. On the other hand, it has also been demonstrated that the combination of fumagillin derivatives with antitumor molecules like 5-fluorouracil can inhibit the growth of colon adenocarcinoma tumor cell lines [158] or prolong its tumor growth inhibitory effect after the termination of treatment in an experimental murine model [159].
Similar growth inhibitory effects have been also described when TNP-470 was added to pancreatic cancer cell lines cultures. However, although angiogenesis and tumor volume reduction in a murine model of pancreatic tumor after TNP-470 administration was reported, it did not improve animal survival rates [160]. Other studies with TNP-470 treatment reported a reduction in metastatic nodules or tumor dissemination in murine models [161,162,163]. Interestingly, the co-administration of TNP-470 with immune response inducers, such as dendritic cells (DC), has also been studied in a murine model of pancreatic tumors, bringing a reduction in the tumor volume, the density of blood microvessels, and raising the animal survival rate [164]. The role of TNP-470 as an adjuvant in DC tumor vaccines is currently being studied, as it seems that it could stimulate, in vitro and in vivo, the immunogenicity of the DC, and thus, promote polarization of the immune response towards the specific antigen differentiation of TH1 lymphocytes [165].
Some authors have also studied the effects of fumagillin on other tumor processes, in such cases as anaplastic thyroid carcinoma, lymphoma, or neuroblastoma in vitro and in vivo, with promising results [30]. Good results with fumagillin derivatives have been observed, in vitro and in vivo, with prostate cancer models [159,166,167], but in humans no conclusive anti-tumor activity was found [168]. In other types of cancer, such as esophageal cancer, we only have some preliminary and unclear results [169].
5.2. Fumagillin Antibiotic Uses
5.2.1. Treatment of Microsporidiosis
Intestinal microsporidiosis due to Enterocytozoon bieneusi can cause chronic diarrhea, malabsorption, and wasting in immunocompromised patients. It has been demonstrated that fumagillin treatment leads to the clearance of microsporidia and that it could be an effective treatment for chronic E. bieneusi infection in immunocompromised patients; for example, those with AIDS [35,170]. However, some patients developed serious adverse events, such as neutropenia and thrombocytopenia, that completely disappeared after the suspension of treatment. Similar results were observed for the treatment of intestinal microsporidiosis with fumagillin in renal transplant recipients [171]. In this case, the secondary effects were not so significant—some abdominal cramps and severe but reversible thrombocytopenia; a decrease in the blood concentration of the immunosuppressor tacrolimus was also observed. It was also reported that fumagillin was a successful treatment for E. bieneusi microsporidiosis in two patients with an allogeneic hematopoietic stem cell transplant [172].
Other specialized parasitic fungi from the Microsporidia phylum are Nosema apis and Nosema ceranae. These two parasites can infect Apis spp., such as A. mellifera, the European bee (also known as the domestic bee), and A. cerana, the Asiatic bee. These parasites infect the bee digestive tract midgut epithelial cells and are spread by fecal-oral transmission. The infection lowers worker bee life expectancy, inhibits pollen digestion that leads to insufficient nutrition, raises winter mortality, and lowers honey production [173,174]. The effect of fumagillin on N. apis is well known [175,176,177,178]; however, its role against N. ceranae has been questioned. Williams et al. [179] suggests that fumagillin is successful at temporarily reducing N. ceranae, whereas others suggest that fumagillin is more toxic for honeybees than for N. ceranae and those fumagillin low-level residues could lead to hyperproliferation of Nosema spp. [180]. However, other authors demonstrated the effectiveness of fumagillin against this parasite [37]. In any case, fumagillin is the only effective chemical treatment currently available for nosemiasis [36]. Nevertheless, it is licensed for use in beekeeping in the United States and Canada but not in Europe, where its use is restricted to special circumstances because of its toxic effects [36,51,181].
5.2.2. Treatment of Other Parasitosis
Another antiparasitic effect of fumagillin has been described. Both fumagillin and TNP-470, potently blocked in vitro growth of Plasmodium falciparum and Leishmania donavani [182]. Later, it was described that fumagillin and its derivative fumarranol can bind to P. falciparum MetAP2, inhibiting its growth in vitro and in vivo in a murine model [183]. This opens the door to a possible new treatment for malaria produced by both chloroquine-sensitive and chloroquine resistant strains, and maybe for the treatment of other cryptosporidiosis. In fact, Arico-Muendel et al. [32] described the role of several fumagillin derivatives in inhibiting the in vitro growth of Entamoeba histolytica, P. falciparum, and Trypanosoma brucei. On the other hand, Hillmann et al. [184] described an amoebicidal effect of A. fumigatus against the amoeba Dictyostelium discoideum, which is abundant in soil, but the effect was principally attributed to gliotoxin by the authors. However, fumagillin had comparably little influence on the viability of amoeba, with a minimum inhibitory concentration 50 (MIC50) towards D. discoideum 40 times higher than gliotoxin [184].
5.3. Other Applications
The Vpr is a protein that appears on HIV-1 virions. It aids efficient translocation of the proviral DNA into the nucleus and is required for the HIV-1 infection of non-dividing cells, such as macrophages. It is also involved in the activation of viral transcription, induction of cell cycle G2 arrest, and apoptosis of the host cells. For all these reasons, it is a suitable candidate for the development of new treatments based on its inhibition. Some authors have seen that fumagillin can inhibit Vpr-dependent viral gene expression upon the infection of human macrophages [185] and proposed a role of fumagillin as a novel type of AIDS treatment.
On the other hand, it has been described that in early stage development of adipose tissue, adipogenesis is closely associated to angiogenesis [186]. Taking into account the fact that fumagillin is an angiogenesis inhibitor and that weight loss is a common side effect of fumagillin administration in human trials, some authors have studied the role of fumagillin in adipose tissue development. They observed that treatment with fumagillin impaired diet-induced obesity in mice, associated with adipocyte hypotrophy, without any significant effect on adipose tissue angiogenesis [187]. Nevertheless, another study expounded that effects on differentiation of preadipocytes cannot explain the inhibitory effect of fumagillin-like compounds on adipose tissue formation, as they showed only a minor effect on in vivo adipocyte differentiation [188].
6. Conclusions
In conclusion, fumagillin seems to be an important factor in promoting rapid adaptation of the fungus A. fumigatus to different stresses, including contact with the immune system and lung tissue of the host, favoring infections. As that delay in diagnosis is one of the causes of the high mortality rate associated with invasive aspergillosis, the use of the fumagillin as a biomarker could favor an early diagnosis. The fumagillin activity triggers the inactivation of the enzyme MetAP2, which is of paramount importance for the processing of multiple important proteins. Therefore, this mycotoxin causes damage to the host during infective processes and protects the fungus against environmental aggressions, such as predators. This toxic activity has been used widely by humans in different areas. In fact, due to the inhibitory effect of fumagillin over the angiogenesis and cell proliferation, one of the most promising applications is the control of different types of cancer cells. Likewise, the activity that fumagillin presents against some parasites has allowed its use, for example, in treatment of infections by Nosema spp. in bees. Consequently, many analytical methods have been developed for the analysis of fumagillin in several matrices, including honey. The problem that its toxicity brings may be solved through the development of fumagillin derivatives that maintain their activity but with lower levels of toxicity, an endeavour that may generate new alternative treatments for some types of cancers or infections.
Acknowledgments
We thank the member of the Chartered of Linguists (number 022913) for improving the English in the manuscript.
Author Contributions
All authors have equally contributed to the writing, figure designing, and correcting the manuscript. A.R., A.R.-G. and X.G. have also coordinated the manuscript elaboration. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Basque Government, grant number IT1362-19. X.G. and U.P.-C. were funded by the Basque Government and the UPV/EHU, respectively.
Conflicts of Interest
The authors declare no conflicts of interest.
Key Contribution
Fumagillin is a mycotoxin produced during infections caused by A. fumigatus and in the adaptation processes of this species to multiple environmental stresses. In this review; we include detailed information on the fungal mechanisms employed in the production of this mycotoxin and those present in its regulation, how it acts on its target, and how it can be detected, all of which may be useful for diagnosis or the development of new treatments. Its usefulness in different fields is also described; because of its anti-angiogenic activity it is used to attack different types of tumors; at the same time its antibiotic activity is useful against several parasites.
References
- 1.Latgé J.P. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 1999;12:310–350. doi: 10.1128/CMR.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tekaia F., Latgé J.P. Aspergillus fumigatus: Saprophyte or pathogen? Curr. Opin. Microbiol. 2005;8:385–392. doi: 10.1016/j.mib.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 3.Dagenais T.R., Keller N.P. Pathogenesis of Aspergillus fumigatus in Invasive Aspergillosis. Clin. Microbiol. Rev. 2009;22:447–465. doi: 10.1128/CMR.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwon-Chung K.J., Sugui J.A. Aspergillus fumigatus—What makes the species a ubiquitous human fungal pathogen? PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knox B.P., Blachowicz A., Palmer J.M., Romsdahl J., Huttenlocher A., Wang C.C., Keller N.P., Venkateswaran K. Characterization of Aspergillus fumigatus isolates from air and surfaces of the international Space Station. mSphere. 2016;1:e00227-16. doi: 10.1128/mSphere.00227-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paulussen C., Hallsworth J.E., Álvarez-Pérez S., Nierman W.C., Hamill P.G., Blain D., Rediers H., Lievens B. Ecology of aspergillosis: Insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microb. Biotechnol. 2017;10:296–322. doi: 10.1111/1751-7915.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blachowicz A., Chiang A.J., Romsdahl J., Kalkum M., Wang C.C.C., Venkateswaran K. Proteomic characterization of Aspergillus fumigatus isolated from air and surfaces of the International Space Station. Fungal Genet. Biol. 2019;124:39–46. doi: 10.1016/j.fgb.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alshareef F., Robson G.D. Prevalence, persistence, and phenotypic variation of Aspergillus fumigatus in the outdoor environment in Manchester, UK, over a 2-year period. Med. Mycol. 2014;52:367–375. doi: 10.1093/mmy/myu008. [DOI] [PubMed] [Google Scholar]
- 9.Amarsaikhan N., O’Dea E.M., Tsoggerel A., Owegi H., Gillenwater J., Templeton S.P. Isolate-dependent growth, virulence, and cell wall composition in the human pathogen Aspergillus fumigatus. PLoS ONE. 2014;9:100430. doi: 10.1371/journal.pone.0100430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown G.D., Denning D.W., Gow N.A., Levitz S.M., Netea M.G., White T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 11.Gauthier G.M., Keller N.P. Crossover fungal pathogens: The biology and pathogenesis of fungi capable of crossing kingdoms to infect plants and humans. Fungal Genet. Biol. 2013;61:146–157. doi: 10.1016/j.fgb.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 12.Raffa N., Keller N.P. A call to arms: Mustering secondary metabolites for success and survival of an opportunistic pathogen. PLoS Pathog. 2019;15:1007606. doi: 10.1371/journal.ppat.1007606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett J.W., Bentley R. What’s in a name?—Microbial secondary metabolism. Adv. Appl. Microbiol. 1989;34:1–28. doi: 10.1016/S0065-2164(08)70316-2. [DOI] [Google Scholar]
- 14.Arias M., Santiago L., Vidal-García M., Redrado S., Lanuza P., Comas L., Domingo M.P., Rezusta A., Gálvez E.M. Preparations for invasion: Modulation of host lung immunity during pulmonary aspergillosis by gliotoxin and other fungal secondary metabolites. Front. Immunol. 2018;9:2549. doi: 10.3389/fimmu.2018.02549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamei K., Watanabe A. Aspergillus mycotoxins and their effect on the host. Med. Mycol. 2005;43:S95–S99. doi: 10.1080/13693780500051547. [DOI] [PubMed] [Google Scholar]
- 16.Keller N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019;17:167–180. doi: 10.1038/s41579-018-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller N.P., Bok J.W., Chung D., Perrin R.M., Shwab E.K. LaeA, a global regulator of Aspergillus toxins. Med. Mycol. 2006;44:83–85. doi: 10.1080/13693780600835773. [DOI] [PubMed] [Google Scholar]
- 18.Raffa N., Osherov N., Keller N.P. Copper utilization, regulation, and acquisition by Aspergillus fumigatus. Int. J. Mol. Sci. 2019;20:1980. doi: 10.3390/ijms20081980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romsdahl J., Wang C.C.C. Recent advances in the genome mining of Aspergillus secondary metabolites (covering 2012–2018) Med. Chem. Commun. 2019;10:840–866. doi: 10.1039/C9MD00054B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shankar J., Tiwari S., Shishodia S.K., Gangwar M., Hoda S., Thakur R., Vijayaraghavan P. Molecular insights into development and virulence determinants of Aspergilli: A proteomic perspective. Front. Cell. Infect. Microbiol. 2018;8:180. doi: 10.3389/fcimb.2018.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frisvad J.C., Rank C., Nielsen K.F., Larsen T.O. Metabolomics of Aspergillus fumigatus. Med. Mycol. 2009;47:53–71. doi: 10.1080/13693780802307720. [DOI] [PubMed] [Google Scholar]
- 22.Keller N.P., Turner G., Bennett J.W. Fungal secondary metabolism—from biochemistry to genomics. Nat. Rev. Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 23.Bignell E., Cairns T.C., Throckmorton K., Nierman W.C., Keller N.P. Secondary metabolite arsenal of an opportunistic pathogenic fungus. Philos Trans R. Soc. B. 2016;371:20160023. doi: 10.1098/rstb.2016.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lind A.L., Wisecaver J.H., Lameiras C., Wiemann P., Palmer J.M., Keller N.P., Rodrigues F., Goldman G.H., Rokas A. Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species. PLoS Biol. 2017;15 doi: 10.1371/journal.pbio.2003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lind A.L., Lim F.Y., Soukup A.A., Keller N.P., Rokas A. An LaeA- and BrlA-dependent cellular network governs tissue-specific secondary metabolism in the human pathogen Aspergillus fumigatus. mSphere. 2018;3 doi: 10.1128/mSphere.00050-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanson F.R., Eble T.E. An antiphage agent isolated from Aspergillus sp. J. Bacteriol. 1949;58:527–529. doi: 10.1128/jb.58.4.527-529.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perrin R.M., Fedorova N.D., Bok J.W., Cramer R.A., Wortman J.R., Kim H.S., Nierman W.C., Keller N.P. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007;3:50. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiemann P., Guo C., Palmer J.M., Sekonyela R., Wang C.C.C., Keller N.P. Prototype of an intertwined secondary-metabolite supercluster. Proc. Natl. Acad. Sci. USA. 2013;110:17065–17070. doi: 10.1073/pnas.1313258110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sin N., Meng L., Wang M.Q., Wen J.J., Bornmann W.G., Crews C.M. The anti-angiogenic agent fumagillin covalently binds and inhibits the methionine aminopeptidase, MetAP-2. Proc. Natl. Acad. Sci. USA. 1997;94:6099–6103. doi: 10.1073/pnas.94.12.6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mauriz J.L., Martín-Renedo J., García-Palomo A., Tuñón M.J., González-Gallego J. Methionine aminopeptidases as potential targets for treatment of gastrointestinal cancers and other tumours. Curr. Drug Targets. 2010;11:1439–1457. doi: 10.2174/1389450111009011439. [DOI] [PubMed] [Google Scholar]
- 31.Vetro J.A., Dummitt B., Chang Y.H. Methionine-aminopeptidase: Emerging role in angiogenesis. In: Hooper N.M., Lendeckel U., editors. Aminopeptidases in Biology and Disease. Kluwer Academic/Plenum Publishers; New York, NY, USA: 2004. pp. 17–44. [Google Scholar]
- 32.Arico-Muendel C., Centrella P.A., Contonio B.D., Morgan B.A., O’Donovan G., Paradise C.L., Skinner S.R., Sluboski B., Svendsen J.L., White K.F., et al. Antiparasitic activities of novel, orally available fumagillin analogs. Bioorganic Med. Chem. Lett. 2009;19:5128–5131. doi: 10.1016/j.bmcl.2009.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novohradska S., Ferling I., Hillmann F. Exploring Virulence Determinants of Filamentous Fungal Pathogens through Interactions with Soil Amoebae. Front. Cell. Infect. Microbiol. 2017;7:497. doi: 10.3389/fcimb.2017.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casadevall A., Fu M.S., Guimaraes A.J., Albuquerque P. The ‘Amoeboid Predator-Fungal Animal Virulence’ Hypothesis. J. Fungi. 2019;5:10. doi: 10.3390/jof5010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molina J.M., Tourneur M., Sarfati C., Chevret S., de Gouvello A., Gobert J.G., Balkan S., Derouin F. Fumagillin treatment of intestinal microsporidiosis. N. Engl. J. Med. 2002;346:1963–1969. doi: 10.1056/NEJMoa012924. [DOI] [PubMed] [Google Scholar]
- 36.van den Heever J.P., Thompson T.S., Curtis J.M., Ibrahim A., Pernal S.F. Fumagillin: An overview of recent scientific advances and their significance for apiculture. J. Agric. Food Chem. 2014;62:2728–2737. doi: 10.1021/jf4055374. [DOI] [PubMed] [Google Scholar]
- 37.van den Heever J.P., Thompson T.S., Otto S.J.G., Curtis J.M., Ibrahim A., Pernal S.F. Evaluation of Fumagilin-B and other potential alternative chemotherapies against Nosema ceranae-infected honeybees (Apis mellifera) in cage trial assays. Apidologie. 2016;47:617–630. doi: 10.1007/s13592-015-0409-3. [DOI] [Google Scholar]
- 38.Fallon J.P., Reeves E.P., Kavanagh K. Inhibition of neutrophil function following exposure to the Aspergillus fumigatus toxin fumagillin. J. Med. Microbiol. 2010;59:625–633. doi: 10.1099/jmm.0.018192-0. [DOI] [PubMed] [Google Scholar]
- 39.Zbidah M., Lupescu A., Jilani K., Lang F. Stimulation of suicidal erythrocyte death by fumagillin. Basic Clin. Pharmacol. Toxicol. 2013;112:346–351. doi: 10.1111/bcpt.12033. [DOI] [PubMed] [Google Scholar]
- 40.Guruceaga X., Ezpeleta G., Mayayo E., Sueiro-Olivares M., Abad-Diaz-De-Cerio A., Aguirre Urizar J.M., Liu H.G., Wiemann P., Bok J.W., Filler S.G., et al. A possible role for fumagillin in cellular damage during host infection by Aspergillus fumigatus. Virulence. 2018;9:1548–1561. doi: 10.1080/21505594.2018.1526528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MarvinSketch, (version 17.22.0, calculation module developed by ChemAxon, 2017).
- 42.DrugBank. [(accessed on 6 November 2019)]; Available online: https://www.drugbank.ca/about.
- 43.Log D Predictor. [(accessed on 15 October 2019)]; Available online: https://disco.chemaxon.com/apps/demos/logd/
- 44.Garrett E.R., Eble T.E. Studies on the stability of fumagillin. I. Photolytic degradation in alcohol solution. J. Am. Pharm. Assoc. Sci. Ed. 1954;43:385–390. doi: 10.1002/jps.3030430702. [DOI] [PubMed] [Google Scholar]
- 45.Eble T.E., Garrett E.R. Studies on the stability of fumagillin. II. Photolytic degradation of crystalline fumagillin. J. Am. Pharm. Assoc. Sci. Ed. 1954;43:536–538. doi: 10.1002/jps.3030430906. [DOI] [PubMed] [Google Scholar]
- 46.Garrett E.R. Studies on the stability of fumagillin. III. Thermal degradation in the presence and absence of air. J. Am. Pharm. Assoc. Sci. Ed. 1954;43:539–543. doi: 10.1002/jps.3030430907. [DOI] [PubMed] [Google Scholar]
- 47.Brackett J.M., Arguello M.D., Schaar J.C. Determination of fumagillin by high-performance liquid-chromatography. J. Agric. Food Chem. 1988;36:762–764. doi: 10.1021/jf00082a022. [DOI] [Google Scholar]
- 48.Kochansky J., Nasr M. Laboratory studies on the photostability of fumagillin, the active ingredient of Fumidil B. Apidologie. 2004;35:301–310. doi: 10.1051/apido:2004017. [DOI] [Google Scholar]
- 49.Dmitrovic J., Durden D.A. Analysis of fumagillin in honey by LC-MS/MS. J. AOAC Int. 2013;96:687–695. doi: 10.5740/jaoacint.12-174. [DOI] [Google Scholar]
- 50.Assil H.I., Sporns P. ELISA and HPLC methods for analysis of fumagillin and its decomposition products in honey. J. Agric. Food Chem. 1991;39:2206–2213. doi: 10.1021/jf00012a021. [DOI] [Google Scholar]
- 51.Higes M., Nozal M.J., Alvaro A., Barrios L., Meana A., Martin-Hernandez R., Bernal J.L., Bernal J. The stability and effectiveness of fumagillin in controlling Nosema ceranae (Microsporidia) infection in honey bees (Apis mellifera) under laboratory and field conditions. Apidologie. 2011;42:364–377. doi: 10.1007/s13592-011-0003-2. [DOI] [Google Scholar]
- 52.Van den Heever J.P., Thompson T.S., Curtis J.M., Pernal S.F. Stability of dicyclohexylamine and fumagillin in honey. Food Chem. 2015;179:152–158. doi: 10.1016/j.foodchem.2015.01.111. [DOI] [PubMed] [Google Scholar]
- 53.Nozal M.J., Bernal J.L., Martin M.T., Bernal J., Alvaro A., Martin R., Higes M. Trace analysis of fumagillin in honey by liquid chromatography-diode array-electrospray ionization mass spectrometry. J. Chromatogr. A. 2008;1190:224–231. doi: 10.1016/j.chroma.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 54.Van den Heever J.P., Thompson T.S., Curtis J.M., Pernal S.F. Determination of dicyclohexylamine and fumagillin in Honey by LC-MS/MS. Food Anal. Meth. 2015;8:767–777. doi: 10.1007/s12161-014-9956-x. [DOI] [PubMed] [Google Scholar]
- 55.Lopez M.I., Pettis J.S., Smith I.B., Chu P.S. Multiclass determination and confirmation of antibiotic residues in honey using LC-MS/MS. J. Agric. Food Chem. 2008;56:1553–1559. doi: 10.1021/jf073236w. [DOI] [PubMed] [Google Scholar]
- 56.Ivešić M., Krivohlavek A., Žuntar I., Tolić S., Šikić S., Musić V., Pavlić I., Bursik A., Galić N. Monitoring of selected pharmaceuticals in surface waters of Croatia. Environ. Sci. Pollut. Res. 2017;24:23389–23400. doi: 10.1007/s11356-017-9894-4. [DOI] [PubMed] [Google Scholar]
- 57.Jia W., Chua X., Ling Y., Huang J., Chang J. Multi-mycotoxin analysis in dairy products by liquid chromatography coupled to quadrupole orbitrap mass spectrometry. J. Chromatogr. A. 2014;1345:107–114. doi: 10.1016/j.chroma.2014.04.021. [DOI] [PubMed] [Google Scholar]
- 58.Kanda M., Sasamoto T., Takeba K., Hayashi H., Kusano T., Matsushima Y., Nakajima T., Kanai S., Takano I. Rapid Determination of Fumagillin Residues in Honey by Liquid Chromatography-Tandem Mass Spectrometry Using the QuEChERS Method. J. AOAC Int. 2011;94:878–885. [PubMed] [Google Scholar]
- 59.Guyonnet J., Richard M., Hellings P. Determination of fumagillin in muscle-tissue of rainbow-trout using automated ion-pairing liquid-chromatography. J. Chromatogr. B-Biomed. Appl. 1995;666:354–359. doi: 10.1016/0378-4347(94)00577-R. [DOI] [PubMed] [Google Scholar]
- 60.Fekete J., Romvari Z., Szepesi I., Morovjan G. Liquid-chromatographic determination of the antibiotic fumagillin in fish meat samples. J. Chromatogr. A. 1995;712:378–381. doi: 10.1016/0021-9673(95)00567-7. [DOI] [PubMed] [Google Scholar]
- 61.Fekete J., Romvari Z., Gebefugi I., Kettrup A. Comparative study on determination of fumagillin in fish by normal and reversed phase chromatography. Chromatographia. 1998;48:48–52. doi: 10.1007/BF02467515. [DOI] [Google Scholar]
- 62.Lin H.C., Chooi Y.H., Dhingra S., Xu W., Calvo A.M., Tang Y.J. The fumagillin biosynthetic gene cluster in Aspergillus fumigatus encodes a cryptic terpene cyclase involved in the formation of β-trans-bergamotene. J. Am. Chem. Soc. 2013;135:4616–4619. doi: 10.1021/ja312503y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin H.C., Tsunematsu Y., Dhingra S., Xu W., Fukutomi M., Chooi Y.H., Cane D.E., Calvo A.M., Watanabe K., Tang Y. Generation of complexity in fungal terpene biosynthesis, discovery of a multifunctional cytochrome P450 in the fumagillin pathway. J. Am. Chem. Soc. 2014;136:4426–4436. doi: 10.1021/ja500881e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dhingra S., Lind A.L., Lin H.C., Tang Y., Rokas A., Calvo A.M. The fumagillin gene cluster, an example of hundreds of genes under veA control in Aspergillus fumigatus. PLoS ONE. 2013;8:77147. doi: 10.1371/journal.pone.0077147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.NCBI (National Center for Biotechnology Information) [(accessed on 6 November 2019)]; Available online: https://www.ncbi.nlm.nih.gov/
- 66.AspGD (Aspergillus Genome Database) [(accessed on 6 November 2019)]; Available online: http://www.aspgd.org/
- 67.UniProtKB (UniProt Knowledgebase) [(accessed on 6 November 2019)]; Available online: https://www.uniprot.org/
- 68.Gravelat F.N., Doedt T., Chiang L.Y., Liu H., Filler S.G., Patterson T.F., Sheppard D.C. In Vivo analysis of Aspergillus fumigatus developmental gene expression determined by real-time reverse transcription-PCR. Infect. Immun. 2008;76:3632–3639. doi: 10.1128/IAI.01483-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vödisch M., Scherlach K., Winkler R., Hertweck C., Braun H.P., Roth M., Haas H., Werner E.R., Brakhage A.A., Kniemeyer O. Analysis of the Aspergillus fumigatus proteome reveals metabolic changes and the activation of the pseurotin A biosynthesis gene cluster in response to hypoxia. J. Proteome Res. 2011;10:2508–2524. doi: 10.1021/pr1012812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mascotti M.L., Ayub M.J., Dudek H., Sanz M.K., Fraaije M.W. Cloning, overexpression and biocatalytic exploration of a novel Baeyer-Villiger monooxygenase from Aspergillus fumigatus Af293. AMB Express. 2013;3:33. doi: 10.1186/2191-0855-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsunematsu Y., Fukutomi M., Saruwatari T., Noguchi H., Hotta K., Tang Y., Watanabe K. Elucidation of pseurotin biosynthetic pathway points to trans-acting C-methyltransferase: Generation of chemical diversity. Angew. Chem. Int. Ed. 2014;53:8475–8479. doi: 10.1002/anie.201404804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bruns S., Seidler M., Albrecht D., Salvenmoser S., Remme N., Hertweck C., Brakhage A.A., Kniemeyer O., Müller F.M. Functional genomic profiling of Aspergillus fumigatus biofilm reveals enhanced production of the mycotoxin gliotoxin. Proteomics. 2010;10:3097–3107. doi: 10.1002/pmic.201000129. [DOI] [PubMed] [Google Scholar]
- 73.Lind A.L., Smith T.D., Saterlee T., Calvo A.M., Rokas A. Regulation of secondary metabolism by the Velvet complex is temperature-responsive in Aspergillus. G3 Bethesda. 2016;6:4023–4033. doi: 10.1534/g3.116.033084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Calvo A.M., Wilson R.A., Bok J.W., Keller N.P. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 2002;66:447–459. doi: 10.1128/MMBR.66.3.447-459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brakhage A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 76.Bok J.W., Keller N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bok J.W., Balajee S.A., Marr K.A., Andes D., Nielsen K.F., Frisvad J.C., Keller N.P. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot Cell. 2005;4:1574–1582. doi: 10.1128/EC.4.9.1574-1582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bayram O., Krappmann S., Ni M., Bok J.W., Helmstaedt K., Valerius O., Braus-Stromeyer S., Kwon N.J., Keller N.P., Yu J.H., et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- 79.Amaike S., Keller N.P. Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus. Eukaryot Cell. 2009;8:1051–1060. doi: 10.1128/EC.00088-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gehrke A., Heinekamp T., Jacobsen I.D., Brakhage A.A. Heptahelical receptors GprC and GprD of Aspergillus fumigatus are essential regulators of colony growth, hyphal morphogenesis, and virulence. Appl. Environ. Microbiol. 2010;76:3989–3998. doi: 10.1128/AEM.00052-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim S.S., Kim Y.H., Shin K.S. The developmental regulators, FlbB and FlbE, are involved in the virulence of Aspergillus fumigatus. J. Microbiol. Biotechnol. 2013;23:766–770. doi: 10.4014/jmb.1301.01002. [DOI] [PubMed] [Google Scholar]
- 82.Johns A., Scharf D.H., Gsaller F., Schmidt H., Heinekamp T., Straßburger M., Oliver J.D., Birch M., Beckmann N., Dobb K.S., et al. A nonredundant phosphopantetheinyl transferase, PptA, is a novel antifungal target that directs secondary metabolite, siderophore, and lysine biosynthesis in Aspergillus fumigatus and is critical for pathogenicity. MBio. 2017;8 doi: 10.1128/mBio.01504-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Manfiolli A.O., de Castro P.A., Dos Reis T.F., Dolan S., Doyle S., Jones G., Riaño Pachón D.M., Ulaş M., Noble L.M., Mattern D.J., et al. Aspergillus fumigatus protein phosphatase PpzA is involved in iron assimilation, secondary metabolite production, and virulence. Cell. Microbiol. 2017;19 doi: 10.1111/cmi.12770. [DOI] [PubMed] [Google Scholar]
- 84.Wu M.Y., Mead M.E., Lee M.K., Ostrem Loss E.M., Kim S.C., Rokas A., Yu J.H. Systematic dissection of the evolutionarily conserved WetA developmental regulator across a genus of filamentous fungi. mBio. 2018;9 doi: 10.1128/mBio.01130-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lind A.L., Wisecaver J.H., Smith T.D., Feng X., Calvo A.M., Rokas A. Examining the evolution of the regulatory circuit controlling secondary metabolism and development in the fungal genus Aspergillus. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu Y., Blachowicz A., Will C., Szewczyk E., Glenn S., Gensberger-Reigl S., Nowrousian M., Wang C.C.C., Krappmann S. Mating-type factor-specific regulation of the fumagillin/pseurotin secondary metabolite supercluster in Aspergillus fumigatus. Mol. Microbiol. 2018;110:1045–1065. doi: 10.1111/mmi.14136. [DOI] [PubMed] [Google Scholar]
- 87.Conrad T., Kniemeyer O., Henkel S.G., Krüger T., Mattern D.J., Valiante V., Guthke R., Jacobsen I.D., Brakhage A.A., Vlaic S., et al. Module-detection approaches for the integration of multilevel omics data highlight the comprehensive response of Aspergillus fumigatus to caspofungin. BMC Syst. Biol. 2018;1:88. doi: 10.1186/s12918-018-0620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eshwika A., Kelly J., Fallon J.P., Kavanagh K. Exposure of Aspergillus fumigatus to caspofungin results in the release, and de novo biosynthesis, of gliotoxin. Med. Mycol. 2013;51:121–127. doi: 10.3109/13693786.2012.688180. [DOI] [PubMed] [Google Scholar]
- 89.Netzker T., Fischer J., Weber J., Mattern D.J., König C.C., Valiante V., Schroeckh V., Brakhage A.A. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front. Microbiol. 2015;6:299. doi: 10.3389/fmicb.2015.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arfin S.M., Kendall R.L., Hall L., Weaver L.H., Stewart A.E., Matthews B.W., Bradshaw R.A. Eukaryotic methionyl aminopeptidases: Two classes of cobalt-dependent enzymes. Proc. Natl. Acad. Sci. USA. 1995;92:7714–7718. doi: 10.1073/pnas.92.17.7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li X., Chang Y.H. Evidence that the human homologue of a rat initiation factor-2 associated protein (p67) is a methionine aminopeptidase. Biochem. Biophys. Res. Commun. 1996;227:152–159. doi: 10.1006/bbrc.1996.1482. [DOI] [PubMed] [Google Scholar]
- 92.Meinnel T., Mechulam Y., Blanquet S. Methionine as translation start signal: A review of the enzymes of the pathway in Escherichia coli. Biochimie. 1993;75:1061–1075. doi: 10.1016/0300-9084(93)90005-D. [DOI] [PubMed] [Google Scholar]
- 93.Boissel J.P., Kasper T.J., Shah S.C., Malone J.I., Bunn H.F. Amino-terminal processing of proteins: Hemoglobin South Florida, a variant with retention of initiator methionine and N(α)-acetylation. Proc. Natl. Acad. Sci. USA. 1985;82:8448–8452. doi: 10.1073/pnas.82.24.8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chang Y.H., Teichert U., Smith J.A. Molecular cloning, sequencing, deletion, and overexpression of a methionine aminopeptidase gene from Saccharomyces cerevisiae. J. Biol. Chem. 1992;267:8007–8011. [PubMed] [Google Scholar]
- 95.Chen S., Vetro J.A., Chang Y.H. The specificity in vivo of two distinct methionine aminopeptidases in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2002;398:87–93. doi: 10.1006/abbi.2001.2675. [DOI] [PubMed] [Google Scholar]
- 96.Walker K.W., Bradshaw R.A. Yeast Methionine Aminopeptidase, I. Alteration of substrate specificity by site-directed mutagenesis. J. Biol. Chem. 1999;274:13403–13409. doi: 10.1074/jbc.274.19.13403. [DOI] [PubMed] [Google Scholar]
- 97.Catalano A., Romano M., Robuffo I., Strizzi L., Procopio A. Methionine aminopeptidase-2 regulates human mesothelioma cell survival: Role of Bcl-2 expression and telomerase activity. Am. J. Pathol. 2001;159:721–731. doi: 10.1016/S0002-9440(10)61743-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lowther W.T., Matthews B.W. Structure and function of the methionine aminopeptidases. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2000;1477:157–167. doi: 10.1016/S0167-4838(99)00271-X. [DOI] [PubMed] [Google Scholar]
- 99.Endo H., Takenaga K., Kanno T., Satoh H., Mori S. Methionine aminopeptidase 2 is a new target for the metastasis-associated protein, S100A4. J. Biol. Chem. 2002;277:26396–26402. doi: 10.1074/jbc.M202244200. [DOI] [PubMed] [Google Scholar]
- 100.Bradshaw R.A., Brickey W.W., Walker K.W. N-terminal processing: The methionine aminopeptidase and N(α)-acetyl transferase families. Trends Biochem. Sci. 1998;23:263–267. doi: 10.1016/S0968-0004(98)01227-4. [DOI] [PubMed] [Google Scholar]
- 101.Ray M.K., Datta B., Chakraborty A., Chattopadhyay A., Meza-Keuthen S., Gupta N.K. The eukaryotic initiation factor 2-associated 67-kDa polypeptide (p67) plays a critical role in regulation of protein synthesis initiation in animal cells. Proc. Natl. Acad. Sci. USA. 1992;89:539–543. doi: 10.1073/pnas.89.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Datta B., Ray M.K., Chakrabarti D., Wylie D.E., Gupta N.K. Glycosylation of eukaryotic peptide chain initiation factor 2 (eIF-2)-associated 67-kDa polypeptide (p67) and its possible role in the inhibition of eIF-2 kinase-catalyzed phosphorylation of the eIF-2 α-subunit. J. Biol. Chem. 1989;264:20620–20624. [PubMed] [Google Scholar]
- 103.Atanassova A., Sugita M., Sugiura M., Pajpanova T., Ivanov I. Molecular cloning, expression and characterization of three distinctive genes encoding methionine aminopeptidases in cyanobacterium Synechocystis sp. strain PCC6803. Arch. Microbio. 2003;180:185–193. doi: 10.1007/s00203-003-0576-x. [DOI] [PubMed] [Google Scholar]
- 104.Giglione C., Serero A., Pierre M., Boisson B., Meinnel T. Identification of eukaryotic peptide deformylases reveals universality of N-terminal protein processing mechanisms. EMBO J. 2000;19:5916–5929. doi: 10.1093/emboj/19.21.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Leszczyniecka M., Bhatia U., Cueto M., Nirmala N.R., Towbin H., Vattay A., Wang B., Zabludoff S., Phillips P.E. MAP1D, a novel methionine aminopeptidase family member is overexpressed in colon cancer. Oncogene. 2006;25:3471–3478. doi: 10.1038/sj.onc.1209383. [DOI] [PubMed] [Google Scholar]
- 106.Ingber D., Fujita T., Kishimoto S., Kanamaru T., Brem H., Folkman J. Synthetic analogues of fumagillin that inhibit angiogenesis and suppress tumour growth. Nature. 1990;348:555–557. doi: 10.1038/348555a0. [DOI] [PubMed] [Google Scholar]
- 107.Liu S., Widom J., Kemp C.W., Crews C.M., Clardy J. Structure of Human Methionine Aminopeptidase-2 complexed with Fumagillin. Science. 1998;282:1324–1328. doi: 10.1126/science.282.5392.1324. [DOI] [PubMed] [Google Scholar]
- 108.Rose A.S., Bradley A.R., Valasatava Y., Duarte J.M., Prlić A., Rose P.W. NGL viewer: Web-based molecular graphics for large complexes. Bioinformatics. 2018;34:3755–3758. doi: 10.1093/bioinformatics/bty419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.RCSB PDB (RCSB Protein Data Bank) [(accessed on 21 November 2019)]; Available online: http://www.rcsb.org/3d-view/1B6A.
- 110.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Datta R., Choudhury P., Bhattacharya M., Soto Leon F., Zhou Y., Datta B. Protection of translation initiation factor eIF2 phosphorylation correlates with eIF2-associated glycoprotein p67 levels and requires the lysine-rich domain I of p67. Biochimie. 2001;83:919–931. doi: 10.1016/S0300-9084(01)01344-X. [DOI] [PubMed] [Google Scholar]
- 112.Buscà R., Pouysségur J., Lenormand P. ERK1 and ERK2 map kinases: Specific roles or functional redundancy? Front. Cell. Dev. Biol. 2016;4:1–23. doi: 10.3389/fcell.2016.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Datta B., Majumdar A., Datta R., Balusu R. Treatment of cells with the angiogenic inhibitor fumagillin results in increased stability of eukaryotic initiation factor 2-associated glycoprotein, p67, and reduced phosphorylation of extracellular signal-regulated kinases. Biochemistry. 2004;43:14821–14831. doi: 10.1021/bi049172p. [DOI] [PubMed] [Google Scholar]
- 114.Datta R., Choudhury P., Ghosh A., Datta B. A glycosylation site, 60SGTS63, of p67 is required for its ability to regulate the phosphorylation and activity of eukaryotic initiation factor 2α. Biochemistry. 2003;42:5453–5460. doi: 10.1021/bi020699g. [DOI] [PubMed] [Google Scholar]
- 115.Bernier S.G., Taghizadeh N., Thompson C.D., Westlin W.F., Hannig G. Methionine aminopeptidases type I and type II are essential to control cell proliferation. J. Cell. Biochem. 2005;95:1191–1203. doi: 10.1002/jcb.20493. [DOI] [PubMed] [Google Scholar]
- 116.Sawanyawisuth K., Wongkham C., Pairojkul C., Saeseow O.T., Riggins G.J., Araki N., Wongkham S. Methionine aminopeptidase 2 over-expressed in cholangiocarcinoma: Potential for drug target. Acta Oncol. 2007;46:378–385. doi: 10.1080/02841860600871061. [DOI] [PubMed] [Google Scholar]
- 117.Kanno T., Endo H., Takeuchi K., Moristhita Y., Fukayama M., Mori S. High expression of methionine aminopeptidase type 2 in germinal center B cells and their neoplastic counterparts. Lab. Investig. 2002;82:893–901. doi: 10.1097/01.LAB.0000020419.25365.C4. [DOI] [PubMed] [Google Scholar]
- 118.Chang S.Y.P., McGary E.C., Chang S. Methionine aminopeptidase gene of Escherichia coli is essential for cell growth. J. Bacteriol. 1989;171:4071–4072. doi: 10.1128/jb.171.7.4071-4072.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Miller C.G., Strauch K.L., Kukral A.M., Miller J.L., Wingfield P.T., Mazzei G.J., Werlen R.C., Graber P., Movva N.R. N-terminal methionine-specific peptidase in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA. 1987;84:2718–2722. doi: 10.1073/pnas.84.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li X., Chang Y.H. Amino-terminal protein processing in Saccharomyces cerevisiae is an essential function that requires two distinct methionine aminopeptidases. Proc. Natl. Acad. Sci. USA. 1995;92:12357–12361. doi: 10.1073/pnas.92.26.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Keller N.P. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat. Chem. Biol. 2015;11:671–677. doi: 10.1038/nchembio.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Buss J.E., Mumby S.M., Casey P.J., Gilman A.G., Sefton B.M. Myristoylated alpha subunits of guanine nucleotide-binding regulatory proteins. Proc. Natl. Acad. Sci. USA. 1987;84:7493–7497. doi: 10.1073/pnas.84.21.7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang Z., Wensel T.G. N-Myristoylation of the rod outer segment G protein, transducin, in cultured retinas. J. Biol. Chem. 1992;267:23197–23201. [PubMed] [Google Scholar]
- 124.Neves S.R., Ram P.T., Iyengar R. G protein pathways. Science. 2002;296:1636–1639. doi: 10.1126/science.1071550. [DOI] [PubMed] [Google Scholar]
- 125.Van Keulen S.C., Rothlisberger U. Effect of N-terminal myristoylation on the active conformation of Gαi1-GTP. Biochemistry. 2017;56:271–280. doi: 10.1021/acs.biochem.6b00388. [DOI] [PubMed] [Google Scholar]
- 126.Gresset A., Sondek J., Harden T.K. The phospholipase C isozymes and their regulation. Subcellular Biochemistry. In: Balla T., Wymann M., York J., editors. Phosphoinositides I: Enzymes of Synthesis and Degradation. Volume 58. Springer; Dordrecht, The Netherlands: 2012. pp. 61–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xu J., Wang F., Van Keymeulen A., Herzmark P., Straight A., Kelly K., Takuwa Y., Sugimoto N., Mitchison T., Bourne H.R. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell. 2003;114:201–214. doi: 10.1016/S0092-8674(03)00555-5. [DOI] [PubMed] [Google Scholar]
- 128.Manning B.D., Toker A. AKT/PKB signaling: Navigating the network. Cell. 2018;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Roskoski R., Jr. Src protein–tyrosine kinase structure and regulation. Biochem. Biophys. Res. Commun. 2004;324:1155–1164. doi: 10.1016/j.bbrc.2004.09.171. [DOI] [PubMed] [Google Scholar]
- 130.Tillo S.E., Xiong W.H., Thakahashi M., Miao S., Andrade A.L., Fortin D.A., Yang G., Qin M., Smoody B.F., Stork P.J.S., et al. Liberated PKA catalytic subunits associate with the membrane via myristoylation to preferentially phosphorylate membrane substrates. Cell Rep. 2017;19:617–629. doi: 10.1016/j.celrep.2017.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Behnia R., Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. doi: 10.1038/nature04397. [DOI] [PubMed] [Google Scholar]
- 132.Aitken A., Cohen P., Santikarn S., Williams D.H., Calder A.G., Smith A., Kleet C.B. Identification of the NHZ-terminal blocking group of calcineurin B as myristic acid. FEBS Lett. 1982;150:314–318. doi: 10.1016/0014-5793(82)80759-X. [DOI] [PubMed] [Google Scholar]
- 133.D’Souza-Schorey C., Chavrier P. ARF proteins: Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 134.Mahajan-Thakur S., Bien-Möller S., Marx S., Schroeder H., Rauch B.H. Sphingosine 1-phosphate (S1P) signaling in glioblastoma multiforme—A systematic review. Int. J. Mol. Sci. 2017;18:2448. doi: 10.3390/ijms18112448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Erpel T., Courtneidge S.A. Src family protein tyrosine kinases and cellular signal transduction pathways. Curr. Opin. Cell Biol. 1995;7:176–182. doi: 10.1016/0955-0674(95)80025-5. [DOI] [PubMed] [Google Scholar]
- 136.Warder S.E., Tucker L.A., Mcloughlin S.M., Strelitzer T.J., Meuth J.L., Zhang Q., Sheppard G.S., Richardson P.L., Lesniewski R., Davidsen S.K., et al. Discovery, identification, and characterization of candidate pharmacodynamic markers of methionine aminopeptidase-2 inhibition. J. Proteome Res. 2008;7:4807–4820. doi: 10.1021/pr800388p. [DOI] [PubMed] [Google Scholar]
- 137.Frottin F., Bienvenut W.V., Bignon J., Jacquet E., Vaca Jacome A.S., Van Dorsselaer A., Cianferani S., Carapito C., Meinnel T., Giglione C., et al. MetAP1 and MetAP2 drive cell selectivity for a potent anti-cancer agent in synergy, by controlling glutathione redox state. Oncotarget. 2016;7:63306–63323. doi: 10.18632/oncotarget.11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Okrój M., Kamysz W., Slominska E.M., Mysliwski A., Bigda J. A novel mechanism of action of the fumagillin analog, TNP-470, in the B16F10 murine melanoma cell line. Anticancer Drugs. 2005;16:817–823. doi: 10.1097/01.cad.0000172835.60142.a5. [DOI] [PubMed] [Google Scholar]
- 139.Liu Y., Kahn R.A., Prestegard J.H. Structure and Membrane Interaction of Myristoylated ARF1. Structure. 2009;17:79–87. doi: 10.1016/j.str.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Abad A., Fernández-Molina J.V., Bikandi J., Ramírez A., Margareto J., Sendino J., Hernando F.L., Pontón J., Garaizar J., Rementeria A. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev. Iberoam. Micol. 2010;27:155–182. doi: 10.1016/j.riam.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 141.Sigg H.P., Weber H.P. Isolierung und strukturaufklarung von ovalicin. Helv. Chim. Acta. 1968;51:1395–1408. doi: 10.1002/hlca.19680510624. [DOI] [PubMed] [Google Scholar]
- 142.Griffith E.C., Su Z., Turk B.E., Chen S., Chang Y.H., Wu Z., Biemann K., Liu J.O. Methionine aminopeptidase (type 2) is the common target for angiogenesis inhibitors AGM-1470 and ovalicin. Chem. Biol. 1997;4:461–471. doi: 10.1016/S1074-5521(97)90198-8. [DOI] [PubMed] [Google Scholar]
- 143.Mangmool S., Kurose H. Gi/o protein-dependent and -independent actions of pertussis toxin (PTX) Toxins. 2011;3:884–899. doi: 10.3390/toxins3070884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Locht C., Coutte L., Mielcarek N. The ins and outs of pertussis toxin. FEBS J. 2011;278:4668–4682. doi: 10.1111/j.1742-4658.2011.08237.x. [DOI] [PubMed] [Google Scholar]
- 145.Sanchez J., Holmgren J. Cholera toxin structure, gene regulation and pathophysiological and immunological aspects. Cell. Mol. Life Sci. 2008;65:1347–1360. doi: 10.1007/s00018-008-7496-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Collier R.J. Understanding the mode of action of diphtheria toxin: A perspective on progress during the 20th century. Toxicon. 2001;39:1793–1803. doi: 10.1016/S0041-0101(01)00165-9. [DOI] [PubMed] [Google Scholar]
- 147.El-Kenawi A.E., El-Remessy A.B. Angiogenesis inhibitors in cancer therapy: Mechanistic perspective on classification and treatment rationales. Br. J. Pharmacol. 2013;170:712–729. doi: 10.1111/bph.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kusaka M., Sudo K., Fujita T., Marui S., Itoh F., Ingber D., Folkman J. Potent anti-angiogenic action of AGM-1470: Comparison to the fumagillin parent. Biochem. Biophys. Res. Commun. 1991;174:1070–1076. doi: 10.1016/0006-291X(91)91529-L. [DOI] [PubMed] [Google Scholar]
- 149.Chun E., Han C.K., Yoon J.H., Sim T.B., Kim Y.K., Lee K.Y. Novel inhibitors targeted to methionine aminopeptidase 2 (MetAP2) strongly inhibit the growth of cancers in xenografted nude model. Int. J. Cancer. 2005;114:124–130. doi: 10.1002/ijc.20687. [DOI] [PubMed] [Google Scholar]
- 150.Kidoikhammouan S., Seubwai W., Silsirivanit A., Wongkham S., Sawanyawisuth K., Wongkham C. Blocking of methionine aminopeptidase-2 by TNP-470 induces apoptosis and increases chemosensitivity of cholangiocarcinoma. J. Cancer Res. Ther. 2019;15:148–152. doi: 10.4103/jcrt.JCRT_250_17. [DOI] [PubMed] [Google Scholar]
- 151.Niwano M., Arii S., Mori A., Ishigami S., Harada T., Mise M., Furutani M., Fujioka M., Imamura M. Inhibition of tumor growth and microvascular angiogenesis by the potent angiogenesis inhibitor, TNP-470, in rats. Surg. Today. 1998;28:915–922. doi: 10.1007/s005950050252. [DOI] [PubMed] [Google Scholar]
- 152.Mauriz J.L., Gonzalez P., Durán M.C., Molpeceres V., Culebras J.M., Gonzalez-Gallego J. Cell-cycle inhibition by TNP-470 in an in vivo model of hepatocarcinoma is mediated by a p53 and p21WAF1/CIP1 mechanism. Transl. Res. 2007;149:46–53. doi: 10.1016/j.trsl.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 153.Bhargava P., Marshall J.L., Rizvi N., Dahut W., Yoe J., Figuera M., Phipps K., Ong V.S., Kato A., Hawkins M.J. A Phase I and pharmacokinetic study of TNP-470 administered weekly to patients with advanced cancer. Clin. Cancer Res. 1999;5:1989–1995. [PubMed] [Google Scholar]
- 154.Audemard A., Le Bellec M.L., Carluer L., Dargère S., Verdon R., Castrale C., Lobbedez T., Hurault de Ligny B. Fumagillin-induced aseptic meningoencephalitis in a kidney transplant recipient with microsporidiosis. Transpl. Infect. Dis. 2012;14:147–149. doi: 10.1111/tid.12010. [DOI] [PubMed] [Google Scholar]
- 155.Molina J.M., Goguel J., Sarfati C., Chastang C., Desportes-Livage I., Michiels J.F., Maslo C., Katlama C., Cotte L., Leport C., et al. Potential efficiency of fumagillin in intestinal microsporidiosis due to Enterocytozoon bieneusi in patients with HIV Infection: Results of a drug screening study. AIDS. 1997;11:1603–1610. doi: 10.1097/00002030-199713000-00009. [DOI] [PubMed] [Google Scholar]
- 156.Hou L., Mori D., Takase Y., Meihua P., Kai K., Tokunaga O. Fumagillin inhibits colorectal cancer growth and metastasis in mice: In vivo and in vitro study of anti-angiogenesis. Pathol. Int. 2009;59:448–461. doi: 10.1111/j.1440-1827.2009.02393.x. [DOI] [PubMed] [Google Scholar]
- 157.Tanaka S., Arii S. Current status and perspective of antiangiogenic therapy for cancer: Hepatocellular carcinoma. Int. J. Clin. Oncol. 2006;11:82–89. doi: 10.1007/s10147-006-0566-5. [DOI] [PubMed] [Google Scholar]
- 158.Ogawa H., Sato Y., Kondo M., Takahashi N., Oshima T., Sasaki F., Une Y., Nishihira J., Todo S. Combined treatment with TNP-470 and 5-fluorouracil effectively inhibits growth of murine colon cancer cells in vitro and liver metastasis in vivo. Oncol. Rep. 2000;7:467–472. doi: 10.3892/or.7.3.467. [DOI] [PubMed] [Google Scholar]
- 159.Wang J., Sheppard G.S., Lou P., Kawai M., BaMaung N., Erickson S.A., Tucker-Garcia L., Park C., Bouska J., Wang Y.C., et al. Tumor suppression by a rationally designed reversible inhibitor of methionine aminopeptidase-2. Cancer Res. 2003;63:7861–7869. [PubMed] [Google Scholar]
- 160.Hotz H.G., Reber J.A., Hotz B., Sanghavi P.C., Yu T., Foitzik T., Buhr H.J., Hines O.J. Angiogenesis inhibitor TNP-470 reduces human pancreatic cancer growth. J. Gastrointest. Surg. 2001;5:131–138. doi: 10.1016/S1091-255X(01)80024-X. [DOI] [PubMed] [Google Scholar]
- 161.Kawarada Y., Ishikura H., Kishimoto T., Saito K., Takahashi T., Kato H., Yoshiki T. Inhibitory effects of the antiangiogenic agent TNP-470 on establishment and growth of hematogenous metastasis of human pancreatic carcinoma in SCID beige mice in vivo. Pancreas. 1997;15:251–257. doi: 10.1097/00006676-199710000-00006. [DOI] [PubMed] [Google Scholar]
- 162.Shishido T., Yasoshima T., Denno R., Mukaiya M., Sato N., Hirata K. Inhibition of liver metastasis of human pancreatic carcinoma by angiogenesis inhibitor TNP-470 in combination with cisplatin. Jpn. J. Cancer Res. 1998;89:963–969. doi: 10.1111/j.1349-7006.1998.tb00655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kato H., Ishikura H., Kawarada Y., Furuya M., Kondo S., Kato H., Yoshiki T. Anti-angiogenic treatment for peritoneal dissemination of pancreas adenocarcinoma: A study using TNP-470. Jpn. J. Cancer Res. 2001;92:67–73. doi: 10.1111/j.1349-7006.2001.tb01049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Miyazaki J., Tsuzuki Y., Matsuzaki K., Hokari R., Okada Y., Kawaguchi A., Nagao S., Itoh K., Miura S. Combination therapy with tumor-lysate pulsed dendritic cells and antiangiogenic drug TNP-470 for mouse pancreatic cancer. Int. J. Cancer. 2005;117:499–505. doi: 10.1002/ijc.21202. [DOI] [PubMed] [Google Scholar]
- 165.Ho D.H., Wong R.H. TNP-470 skews DC differentiation to Th1-stimulatory phenotypes and can serve as a novel adjuvant in a cancer vaccine. Blood Adv. 2018;2:1664–1679. doi: 10.1182/bloodadvances.2017013433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yamaoka M., Yamamoto T., Masaki T., Ikeyama S., Sudo K., Fujita T. Angiogenesis inhibitor TNP-470 (AGM-1470) potently inhibits the tumor growth of hormone-independent human breast and prostate carcinoma cell lines. Cancer Res. 2003;53:5233–5236. [PubMed] [Google Scholar]
- 167.Tucker L.A., Zhang Q., Sheppard G.S., Jiang F., McKeegan E., Lesniewski R., Davidsen S.K., Bell R.L., Wang J. Ectopic expression of methionine aminopeptidase-2 causes cell transformation and stimulates proliferation. Oncogene. 2008;27:3967–3976. doi: 10.1038/onc.2008.14. [DOI] [PubMed] [Google Scholar]
- 168.Logothetis C.J., Wu K.K., Finn L.D., Figg W., Ghaddar H., Gutterman J.U. Phase I trial of the angiogenesis inhibitor TNP-470 for progressive androgen-independent prostate cancer. Clin. Cancer Res. 2001;7:1198–1203. [PubMed] [Google Scholar]
- 169.Bo H., Ghazizadeh M., Shimizu H., Kurihara Y., Egawa S., Moriyama Y., Tajiri T., Kawanami O. Effect of ionizing irradiation on human esophageal cancer cell lines by cDNA microarray gene expression analysis. J. Nippon Med. Sch. 2004;71:172–180. doi: 10.1272/jnms.71.172. [DOI] [PubMed] [Google Scholar]
- 170.Conteas C.N., Berlin O.G., Ash L.R., Pruthi J.S. Therapy for human gastrointestinal microsporidiosis. Am. J. Trop. Med. Hyg. 2000;63:121–127. doi: 10.4269/ajtmh.2000.63.121. [DOI] [PubMed] [Google Scholar]
- 171.Champion L., Durrbach A., Lang P., Delahousse M., Chauvet C., Sarfati C., Glotz D., Molina J.M. Fumagillin for treatment of intestinal microsporidiosis in renal transplant recipients. Am. J. Transpl. 2010;10:1925–1930. doi: 10.1111/j.1600-6143.2010.03166.x. [DOI] [PubMed] [Google Scholar]
- 172.Bukreyeva I., Angoulvant A., Bendib I., Gagnard J.C., Bourhis J.H., Dargère S., Bonhomme J., Thellier M., Gachot B., Wyplosz B. Enterocytozoon bieneusi microsporidiosis in stem cell transplant recipients treated with fumagillin. Emerg. Infect. Dis. 2017;23:1039–1041. doi: 10.3201/eid2306.161825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bailey L., Ball B.V. Honey Bee Pathology. 2nd ed. Academic Press; London, UK: 1991. pp. 1–298. [Google Scholar]
- 174.Fries I. Nosema apis—A parasite in the honey bee colony. Bee World. 1993;74:5–19. doi: 10.1080/0005772X.1993.11099149. [DOI] [Google Scholar]
- 175.Bailey L. Effect of fumagillin upon Nosema apis (Zander) Nature. 1953;171:212–213. doi: 10.1038/171212a0. [DOI] [PubMed] [Google Scholar]
- 176.Katznelson H., Jamieson C.A. Control of Nosema disease of honeybees with fumagillin. Science. 1952;115:70–71. doi: 10.1126/science.115.2977.70. [DOI] [PubMed] [Google Scholar]
- 177.Hartwig A., Przełȩcka A. Nucleic acids in the intestine of Apis mellifera infected with Nosema apis and treated with fumagillin DCH: Cytochemical and autoradiographic studies. J. Invertebr. Pathol. 1971;18:331–336. doi: 10.1016/0022-2011(71)90034-6. [DOI] [PubMed] [Google Scholar]
- 178.Webster T.C. Fumagillin affects Nosema apis and honey bees (Hymenopterai Apidae) J. Econ. Entomol. 1994;87:601–604. doi: 10.1093/jee/87.3.601. [DOI] [Google Scholar]
- 179.Williams G.R., Sampson M.A., Shutler D., Rogers R.E.L. Does fumagillin control the recently detected invasive parasite Nosema ceranae in Western honey bees (Apis mellifera)? J. Invertebr. Pathol. 2008;99:342–344. doi: 10.1016/j.jip.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 180.Huang W.F., Solter L.F., Yau P.M., Imai B.S. Nosema ceranae escapes fumagillin control in honey bees. PLoS Pathol. 2013;9 doi: 10.1371/journal.ppat.1003185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhang Y., Li X.Q., Li H.M., Zhang Q.H., Gao Y., Li X.J. Antibiotic residues in honey: A review on analytical methods by liquid chromatography tandem mass spectrometry. Trends Anal. Chem. 2019;110:344–356. doi: 10.1016/j.trac.2018.11.015. [DOI] [Google Scholar]
- 182.Zhang P., Nicholson D.E., Bujnicki J.M., Su X., Brendle J.J., Ferdig M., Kyle D.E., Milhous W.K., Chiang P.K. Angiogenesis inhibitors specific for methionine aminopeptidase 2 as drugs for malaria and leishmaniasis. J. Biomed. Sci. 2002;9:34–40. doi: 10.1007/BF02256576. [DOI] [PubMed] [Google Scholar]
- 183.Chen X., Xie S., Bhat S., Kumar N., Shapiro T.A., Liu J.O. Fumagillin and fumarranol interact with P. falciparum methionine aminopeptidase 2 and inhibit malaria parasite growth in vitro and in vivo. Chem. Biol. 2009;16:193–202. doi: 10.1016/j.chembiol.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 184.Hillmann F., Novohradská S., Mattern D.J., Forberger T., Heinekamp T., Westermann M., Winckler T., Brakhage A.A. Virulence determinants of the human pathogenic fungus Aspergillus fumigatus protect against soil amoeba predation. Environ. Microbiol. 2015;17:2858–2869. doi: 10.1111/1462-2920.12808. [DOI] [PubMed] [Google Scholar]
- 185.Watanabe N., Nishihara Y., Yamaguchi T., Koito A., Miyoshi H., Kakeya H., Osada H. Fumagillin suppresses HIV-1 infection of macrophages through the inhibition of Vpr activity. FEBS Lett. 2006;580:2598–2602. doi: 10.1016/j.febslet.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 186.Cao Y. Angiogenesis modulates adipogenesis and obesity. J. Clin. Investig. 2007;117:2362–2368. doi: 10.1172/JCI32239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lijnen H.R., Frederix L., Van Hoef B. Fumagillin reduces adipose tissue formation in murine models of nutritionally induced obesity. Obesity. 2010;18:2241–2246. doi: 10.1038/oby.2009.503. [DOI] [PubMed] [Google Scholar]
- 188.Scroyen I., Christiaens V., Lijnen H.R. Effect of fumagillin on adipocyte differentiation and adipogenesis. Biochim. Biophys. Acta. 2010;1800:425–429. doi: 10.1016/j.bbagen.2009.11.015. [DOI] [PubMed] [Google Scholar]