Abstract
Background: Botulinum neurotoxins (BoNTs) comprise seven agreed-on serotypes, A through G. In 2014, a novel chimeric neurotoxin produced by clostridial strain IBCA10-7060 was reported as BoNT/H, with subsequent names of BoNT/FA or BoNT/HA based on sequence homology of the N-terminus to BoNT/F, the C-terminus to BoNT/A and neutralization studies. The purpose of this study was to define the immunologic identity of the novel BoNT. Methods: monoclonal antibodies (mAbs) to the novel BoNT/H N-terminus were generated by antibody repertoire cloning and yeast display after immunization with BoNT/H LC-HN or BoNT/F LC-HN. Results: 21 unique BoNT/H LC-HN mAbs were obtained; 15 from the BoNT/H LC-HN immunized library (KD 0.78 nM to 182 nM) and six from the BoNT/F-immunized libraries (KD 20.5 nM to 1490 nM). A total of 15 of 21 mAbs also bound catalytically inactive BoNT/H holotoxin. The mAbs bound nine non-overlapping epitopes on the BoNT/H LC-HN. None of the mAbs showed binding to BoNT serotypes A-G, nor any of the seven subtypes of BoNT/F, except for one mAb that weakly bound BoNT/F5. Conclusions: The results, combined with the chimeric structure and neutralization by anti-A, but not anti-F antitoxin indicate that immunologically the novel BoNT is BoNT/HA. This determination has significant implications for existing countermeasures and potential vulnerabilities.
Keywords: monoclonal antibodies, botulinum neurotoxin, botulism, Clostridium botulinum, botulinum neurotoxin HA, BoNT/HA
1. Introduction
Botulinum neurotoxins (BoNTs), the most poisonous substances known [1], consist of three functional domains [2]: a binding domain (HC), translocation domain (HN) and catalytic domain (LC). BoNTs exist as seven accepted serotypes (A–G), defined immunologically by the inability of polyclonal antibody that neutralizes one serotype to neutralize other serotypes [3]. Toxin serotypes A, B, E, and F can be further subdivided into subtypes or genetic variants (A1–A8, F1-9) based on sequence and immunologic differences [4]. Additional toxins with structural homology to BoNT have also been recently identified and include BoNT/X from a Clostridial species and eBoNT/J [5,6] from an Enterococcal species.
In 2014, a novel neurotoxin was reported that was produced by the bivalent Clostridium botulinum strain IBCA10-7060, which also produced BoNT/B2 [7,8]. The novel BoNT appeared to be chimeric with a HC most homologous to BoNT/A1 (~84%) and an HN and LC most homologous to BoNT/F5 (~64% and ~81% respectively) [8] (Figure S1). In the initial report, the novel neurotoxin was considered a new serotype, BoNT/H, based on failure of neutralization using the standard mouse bioassay [7] and also using a research antitoxin at antitoxin:toxin ratios as high as 595:1 [7]. Subsequent work demonstrated that the novel BoNT could be neutralized by a combination of anti-A and anti-B research antitoxin at ratios ranging from 20:1 to 200:1 [9,10]. Based on this neutralization and the mosaic structure of the novel toxin with its homology to parts of BoNT/A and BoNT/F5, these authors and others termed the novel toxin BoNT/FA [10,11]. However, the novel BoNT was not neutralized by anti-BoNT/F antitoxin [9,10] and was bound by only one of six anti-BoNT/F monoclonal antibodies (mAbs) and with an affinity more than 8000-fold lower than the affinity for BoNT/F1 (KD = 9.1 pM vs. ~75 nM) [12]. Based on these findings, the novel BoNT has also been termed BoNT/HA [13,14].
We sought to better define the immunologic nature of the novel BoNT by generating a panel of mAbs against the LC-HN portion of the novel toxin and determining their ability to bind other BoNT serotypes. The results indicate that immunologically, the novel BoNT is BoNT/HA, which has significant implications for existing countermeasures and potential vulnerabilities.
2. Results
Since the novel BoNT has been termed BoNT/H, BoNT/FA, BoNT/HA and the novel neurotoxin produced by strain IBCA10-7060, for clarity and brevity we will refer to the novel BoNT throughout the results section by the first name used to describe it, BoNT/H [7]. In the discussion and conclusion, we will use the name that is supported by the studies performed, BoNT/HA.
2.1. BoNT/H LC-HN Fragment Expression and Mouse Immunization
To focus the immune response on the portion of the novel BoNT not homologous to BoNT/A, the BoNT/H LC-HN gene encoding amino acids 1–859 was cloned into plasmid pET28b, expressed from Escherichia. coli and purified by IMAC (Figure S2A). The recombinant BoNT/H LC-HN was bound by the BoNT/H HN mAb 6F5.4 [12] and by anti-His tag IgG by ELISA (Figure S2B). The BoNT/H LC (amino acids 1–444) was produced similarly and was of the expected size by SDS-PAGE (Figure S2A). Mice were immunized with BoNT/H LC-HN and serum harvested at six weeks after the initial immunization. Immune serum bound recombinant BoNT/H LC-HN at dilutions greater than 1:325 as determined by ELISA (Figure S2C).
2.2. Isolation and Initial Characterization of mAbs from Mice Immunized with BoNT/H LC-HN
To generate antibodies to BoNT/H LC-HN, murine VH and VK genes were PCR amplified from cDNA prepared from the splenocytes of immunized mice and cloned into the yeast display vector pYD4 to create a single chain Fv (scFv) antibody gene repertoire as described [12]. The scFv gene repertoire in pYD4 was used to transform Saccharomyces cerevisiae EBY100 to create a yeast-displayed scFv library of 5 × 107 (Table 1). scFv display was induced, and the library sequentially sorted for three rounds after staining with BoNT/H LC-HN. After the final round of sorting, 96 individual colonies were analyzed for BoNT/H LC-HN binding. The scFv gene of each binding clone was sequenced resulting in 15 unique scFv binding BoNT/H LC-HN (Table 2). Equilibrium dissociation constants (KD) of each yeast-displayed scFv for BoNT/H LC-HN were measured by flow cytometry and ranged from 0.78 nM–182 nM with a median KD of 12.5 nM (Table 2).
Table 1.
Yeast-displayed scFv libraries used for monoclonal antibodies (mAb) isolation. The BoNT/F libraries are described in Reference [18].
| Library Name | Immunogens | Library Size |
|---|---|---|
| BoNT/F libraries | BoNT/F1HC and F1 holotoxin BoNT/F3 LC-HN BoNT/F5 LC-HN BoNT/F6 LC-HN BoNT/F7 LC-HN |
2 × 108 |
| BoNT/H library | BoNT/H LC-HN | 5 × 107 |
Table 2.
The characteristics of selected BoNT/H LC-HN mAbs.
| Domain Specificity | BoNT/F Binding1 | KD on Yeast (nM) | Epitope Group | ||
|---|---|---|---|---|---|
| H LC-HN | BoNT/Hi | ||||
| MAbs from BoNT/H Library | |||||
| 72A1 | H LC | No | 3.08 | 2.41 | II |
| 72A7 | H LC-HN | No | 0.78 | 1.79 | IX |
| 72B4 | H LC-HN | No | 12.5 | >200 | VI |
| 72B7 | H LC-HN | No | 25.1 | 32.85 | V/IX |
| 72B8 | H LC-HN | No | 14.2 | 52.4 | IX |
| 72B12 | H LC-HN | No | 29.4 | 19.5 | IX/IV |
| 72C4 | H LC-HN | No | 17.1 | 43.5 | VII |
| 72D10 | H LC-HN | No | 40.8 | No Binding | IX |
| 72E6 | H LC-HN | No | 0.99 | 0.88 | IX |
| 72G8 | H LC-HN | No | 5.00 | 2.4 | VIII |
| 72G9 | H LC | No | 6.62 | 53.9 | I/II |
| 73C4 | H LC-HN | No | 182 | >100 | V |
| 73D9 | H LC-HN | No | 30.0 | No Binding | VII/IX |
| 74F1 | H HN | No | 1.77 | 2.18 | II |
| 74G6 | H HN | No | 4.05 | 5.50 | III |
| MAbs from BoNT/F Libraries | |||||
| 71B8 | H LC-HN | No | 1490 | No Binding | VIII/IX |
| 71C7 | H LC-HN | No | 38.8 | No Binding | IV |
| 71D4 | H LC-HN | No | 184.8 | No Binding | III |
| 71E4 | H LC | F5 LC-HN | 70.65 (3984) | 68.8 | I |
| 6H12 | 2.89 (13.24) | 3.07 | |||
| 71F4 | H LC | No | 124.4 | No Binding | II |
| 71F4B3 | 34.6 | >200 | |||
| 71H6 | H LC-HN | No | 20.5 | No Binding | VII/IX |
1 No binding of BoNT/Hi means the scFv did not show detectable binding when tested with 200nM BoNT/Hi. 2 mAb 6H1 is an affinity improved derivative of mAb 71E4; 3 mAb 71F4B3 is an affinity improved derivative of mAb 71F4. 4 the KD values in parentheses are for the KD for binding to BoNT/F5 LC-HN.
To confirm that the epitopes bound by BoNT/H LC-HN mAbs exist in the holotoxin, we attempted to measure the KD of yeast displayed scFv for BoNT/H in crude IBCA10-7060 culture supernatants. However, the large amounts of BoNT/B in the supernatants cleaved the scFv from the yeast surface, even in the presence of zinc chelators and protease inhibitors, precluding KD measurement. We therefore purchased commercially available catalytically inactive BoNT/Hi holotoxin. To validate that the recombinant BoNT/Hi was properly folded, we measured the KD of two BoNT/A HC IgG (CR2 and RAZ1) and one BoNT/F mAb (6F5.4) that we have previously shown bound BoNT/H [12]. All three of these mAbs bind conformational epitopes as shown by alanine scanning and/or X-ray crystallography and do not bind linear epitopes [15,16,17]. As shown in Table S1, all three of these mAbs bound BoNT/Hi with a KD indistinguishable from their KD for BoNT/H.
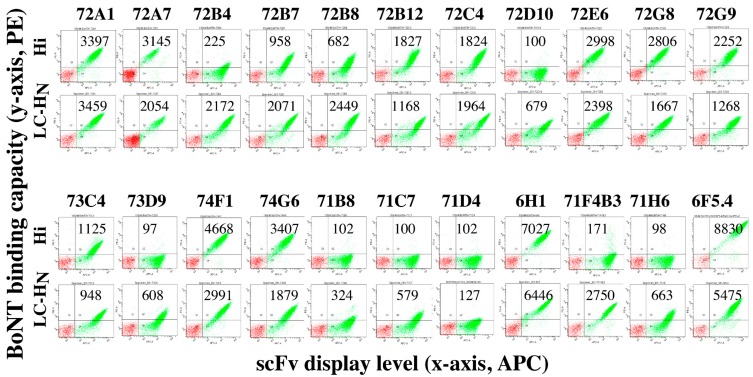
Thirteen of 15 scFv bound recombinant BoNT/Hi holotoxin, with the two non-binding scFv (72D10 and 73D9) having two of the three lowest affinities for BoNT/H LC-HN (Table 2 and Figure 1). This result indicates that the recombinant BoNT/H LC-HN represents the LC-HN structure as it exists in the holotoxin. Failure of scFvs to bind BoNT/Hi holotoxin could result from buried LC-HN epitopes due to packing of the HC against the HN in the holotoxin, or the scFv might bind epitopes on the BoNT/LC-HN that are not on the holotoxin. The scFv without BoNT/Hi binding also generally had lower affinity, and may not be detected with the current method.
Figure 1.
Dot-plots of the binding of single chain Fv (scFv) to botulinum neurotoxins (BoNT)/Hi. The ability of each yeast-displayed scFv to bind BoNT/Hi holotoxin was determined by flow cytometry. Binding of each scFv to BoNT/H LC-HN is shown as a positive control. Binding was detected using mAb 6F5.4 [12] or 6H1 (for 74F1 and 74G6) and is shown on the y-axis. The level of scFv display is shown on the x-axis. The green gated population are those yeast that display scFv on their surface. Mean fluorescent intensity (MFI) values are shown within each dot-plot. The negative control MFI (yeast stained with secondary antibody only) was 94.2 ± 15.7. A test MFI > two standard deviations from control (>125.6) was considered positive for binding BoNT/H LC-HN or BoNT/Hi.
2.3. Isolation and Initial Characterization of mAbs from Mice Immunized with BoNT/F LC-HN
Since the BoNT/H LC-HN is most homologous to the BoNT/F5 subtype, we sorted five yeast-displayed scFv libraries generated from mice immunized with recombinant BoNT/F domains (Table 1) [18] to determine if rare cross-reactive BoNT/H LC-HN mAbs could be isolated. After sorting on BoNT/H LC-HN, a total of six unique scFv were isolated with KD values of 20.5 nM–1490 nM (median of 127 nM, Table 2). Only one of six scFv bound recombinant BoNT/Hi holotoxin (Figure 1). The five non-binding scFv also generally had lower affinity for BoNT/H LC-HN than scFv that bound BoNT/Hi from the BoNT/H LC-HN library. One of the non-binding scFv had measurable binding to BoNT/Hi after its affinity was increased four-fold to 34.6 nM (Table 2).
2.4. The BoNT/H LC-HN scFv mAbs Bind Nine Non-Overlapping Epitopes
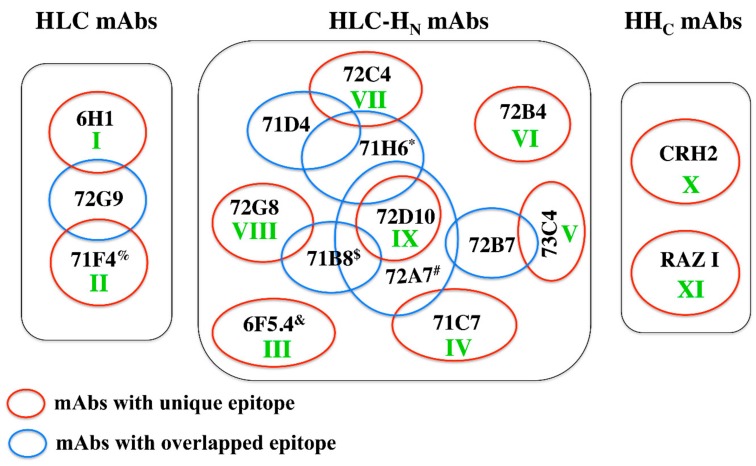
The 21 BoNT/H LC-HN scFv from the BoNT/H LC-HN and BoNT/F LC-HN immunized mice were clustered into non-overlapping epitope groups based on the ability of soluble scFv to bind BoNT/H LC-HN after capture by yeast-displayed scFv (Figure 2 and Table 2). The 15 scFv from the BoNT/H LC-HN library clustered into eight non-overlapping epitopes groups (groups II-IX). Two scFv (72A1 and 72G9) bound an overlapping epitope on the BoNT/H LC (Table 2 and Figure S3) while the remaining 13 bound the recombinant LC-HN but not the LC. The epitopes of scFv 74F1 and 74G6 were inferred to be on the BoNT/H HN since their epitopes overlapped with HN specific mAb 6F5.4 [12]. The remaining 11 scFv either bound epitopes spanning the LC-HN or were HN specific. Since we were unable to produce recombinant BoNT/H HN, we cannot distinguish between these two possibilities. Two of the six scFv (6H1 and 71F4) from the BoNT/F LC-HN libraries bound the BoNT/H LC (Table 2 and Figure S3). Overall, the epitopes of five of the scFv overlapped with one or more scFv from the BoNT/H LC-HN libraries, while one of the scFv (6H1) defined a new epitope (I) (Figure 2).
Figure 2.
Map of BoNT/H scFv domain specificity and epitope overlap. scFv were mapped to the BoNT/H LC or LC-HN based on their ability to bind each recombinant domain as determined by flow cytometry. Since recombinant HN was not produced, LC-HN binding scFv cannot be distinguished from HN binding scFv, except for scFv 74F1 and 74G6 which have epitopes that overlap with the HN binding scFv 6F5.4. The mAb clusters were based on ability of each scFv to bind simultaneously to BoNT/H LC, LC-HN or BoNT/A HC as determined by flow cytometry. The BoNT/H LC-HN scFv bound nine non-overlapping epitopes (I–IX), shown as red circles defined by a specific scFv. CRH2 and RAZ1 bind two unique epitopes on the BoNT/H HC. * shared epitope with scFv 73D9; # shared epitope with scFv 72E6 and scFv 72B12: $ shared epitope with scFv 72B8; & shared epitope with scFv 74F1 and scFv 74G6; % shared epitope with scFv 72A1.
2.5. mAbs Binding BoNT/H LC-HN do not Bind BoNT Serotypes A–G
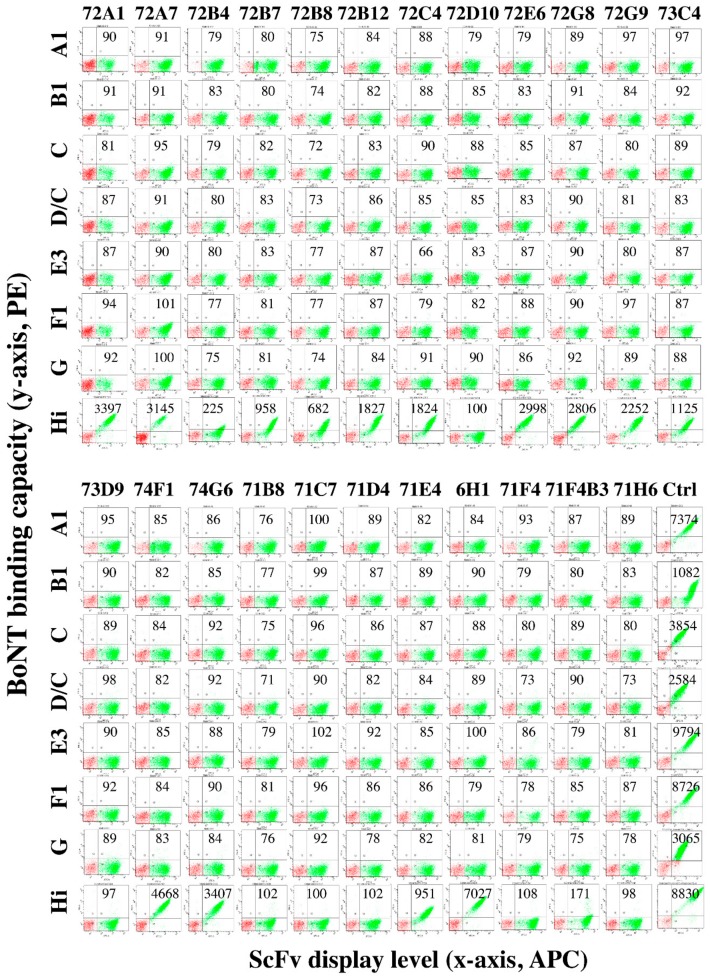
To determine the specificity of the 21 BoNT/H LC-HN scFv, each yeast-displayed scFv was stained with 100 nM of BoNT/A1, B1, C, D/C, E3, F1 and G holotoxin and binding detected by flow cytometry. None of the 21 BoNT/H LC-HN scFv bound any other BoNT serotypes, including BoNT/F1 (Figure 3), indicating that BoNT/H LC-HN is immunologically distinct from the seven other serotypes. This absence of binding to BoNT/F for scFv from mice immunized with BoNT/F LC-HN indicates that the scFv selected on BoNT/H LC-HN do not represent rare scFv binding both BoNT/H and BoNT/F but rather unique BoNT/H specificities arising most likely from non-cognate immunoglobulin VH-VL pairings created during library construction.
Figure 3.
Dot-plots of the specificity of BoNT/H LC-HN scFv for BoNT serotypes A, B, C, D/C, E, F, G and Hi. Yeast-displayed BoNT/H LC-HN scFv were incubated with 50 nM of the indicated BoNT holotoxin. BoNT binding was detected with IgG mAbs RAZ1/CR2 for BoNT/A1, 1B10.1/B6.1 for BoNT/B1, 4C2/4C4.2 for BoNT/C and D/C, 3E2/4E17.1 for BoNT/E3, hu6F11/hu6F13.4 for BoNT/F1, 7G1.1/7G3.1 for BoNT/G and 6F5.4 and 6H1 for BoNT/Hi followed by PE-conjugated goat-anti human IgG. Yeast-displayed 6F5.4 was used for the positive control for BoNT/A1, B1, E3, F1, Hi and yeast-displayed 4C2 or 7G1.1 was used for BoNT/C, D/C and BoNT/G. None of the scFv bound any BoNTs except BoNT/H. The level of BoNT binding is indicated on the Y-axis and the level of scFv display is shown on the X-axis. The gated populations shown in green are those yeasts displaying scFv on their surface. Mean fluorescent intensity (MFI) values are shown within each dot-plot. The negative control MFI (yeast stained with secondary antibody only) was 94.2 ± 15.7. A test MFI > two standard deviations from control (>125.6) was considered positive for binding BoNT.
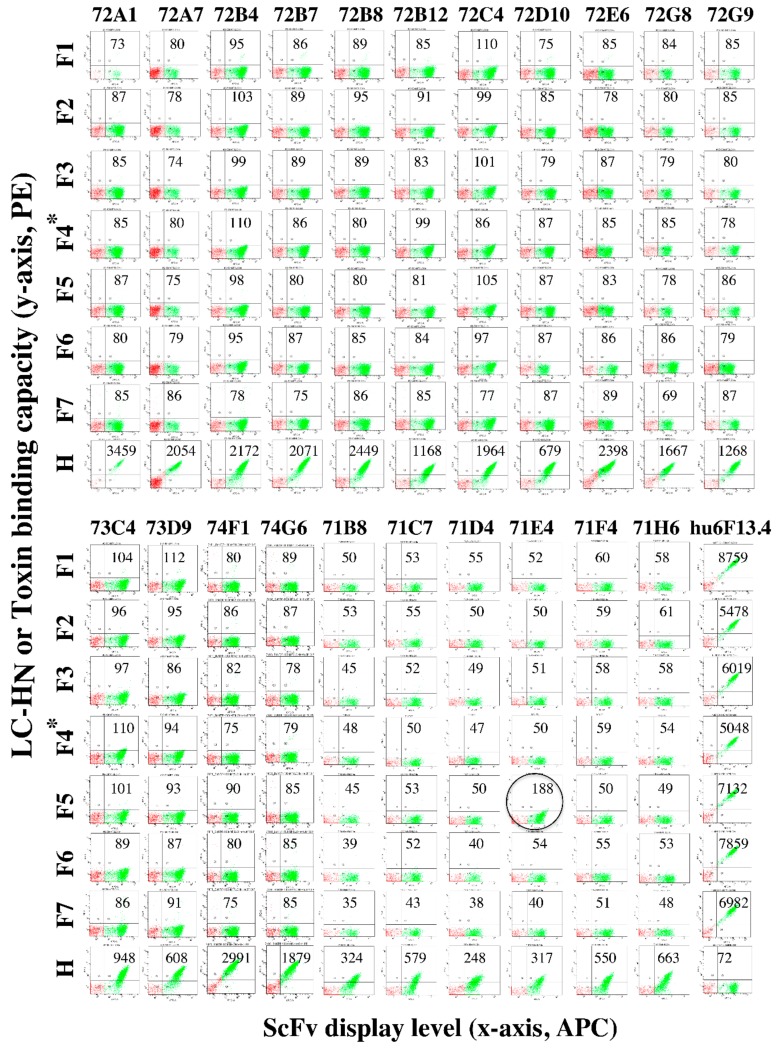
Since the BoNT/H LC-HN is most homologous with the BoNT/F serotype, we tested the ability of each mAb to bind each of seven BoNT/F subtypes. None of the mAbs derived from BoNT/H LC-HN immunization bound any BoNT/F subtype, including the light chain binders 72A1 and 72G9 (Figure 4). One of the six mAbs derived from BoNT/F LC-HN immunization (71E4) weakly bound BoNT/F5LC-HN (Figure 3) with a KD 5.6-fold lower than the KD for BoNT/H LC-HN (Table 2). When the affinity of 71E4 for BoNT/H LC-HN was increased 24.5-fold to obtain scFv 6H1, the affinity for BoNT/F5 LC-HN also increased 30.2-fold. However, neither the 71E4 scFv nor the higher affinity 6H1 scFv recognized other BoNT/F subtypes (Figure S4). The cross-reactivity of 71E4 and 6H1 for BoNT/H and only BoNT/F5 LC suggests that there exist common epitopes on the light chain of BoNT/H and BoNT/F5 that are not found on the other BoNT/F subtypes or other BoNT serotypes, consistent with an amino acid identity of 80.1% for BoNT/H LC and BoNT/F5 LC, and less than 50% between BoNT/H LC and other BoNT/F subtypes. However, the immunological similarity of BoNT/H and BoNT/F5 LC is relatively limited. scFv 72A1 and 72G9 also bound BoNT/H LC with high affinity (KD 3.08 nM and 6.62 nM, Table 2), but did not bind BoNT/F5LC-HN (Figure 4). Similarly, the scFv 71F4 binds BoNT/H LC (KD on yeast 128 nM) but not BoNT/F5LC-HN even after its affinity was improved to 34 nM (71F4B3) (Figure S4).
Figure 4.
Dot plots of the specificity of BoNT/H LC-HN scFv for BoNT/F subtypes. Yeast-displayed BoNT/H LC-HN scFv were incubated with 100 nM of the indicated BoNT/F LC-HN subtype, BoNT/F4 holotoxin (indicated with an * in the Figure) or BoNT/H LC-HN. Yeast-displayed hu6F13.4 was used for a control. BoNT/F subtype binding and BoNT/H LC-HN binding was detected with IgG 6F5.4 and 6H1 (for 74F1 and 74G6). Only scFv 71E4 (result circled) showed weak binding to BoNT/F5 LC-HN, while other scFv did not show any binding to BoNT/F subtypes. The level of BoNT binding is indicated on the y-axis and the level of scFv display is shown on the x-axis. The gated populations shown in green are those yeasts displaying scFv on their surface. Mean fluorescent intensity (MFI) values are shown within each dot-plot. The negative control MFI (yeast stained with secondary antibody only) was 94.2 ± 15.7. A test MFI > two standard deviations from control (>125.6) was considered positive for binding BoNT.
BoNT/H HN shares only 61.7% and 64.1% amino acid sequence identity with BoNT/F1 HN and BoNT/F5 HN respectively [11]; thus the immunological differences are likely greater than for the BoNT/F5 LC. While mAb 6F5.4 bound the HN domain of BoNT/H and all BoNT/F subtypes with high affinity [12], neither of the BoNT/H HN scFv that had overlapping epitopes with 6F5.4 (74F1 and 74G6) bound any BoNT/F subtype (Figure 4 and Figure S5). To determine whether BoNT/H and BoNT/F5 shared additional epitopes, the BoNT/H immune yeast-displayed library was sorted after incubation with BoNT/F5 LC-HN. Two scFv were identified, 77H2 and 78A2, that bound BoNT/F5 LC-HN with affinities of 80.9 nM and 110.4 nM respectively. However, neither of these scFv bound BoNT/H LC-HN (Figure S6).
2.6. Some mAbs can Bind both BoNT/A HC and BoNT/H
The BoNT HC comprises two functional domains HCN and HCC. The BoNT/A HCC and BoNT/H HCC have greater sequence identity (92.2%) than the BoNT/A HCN and BoNT/H HCN (73.1%) [7,12]. We previously evaluated four BoNT/A HC mAbs each binding non-overlapping epitopes (RAZ1, S25, CR2, and B4) for binding to BoNT/H. The two HCC binding mAbs (RAZ1 and S25) bound BoNT/H with affinities comparable to those for BoNT/A binding. One of the mAbs binding the BoNT/A HCN (B4) did not bind BoNT/H and the other, CR2, bound BoNT/H with a 400-fold lower affinity than BoNT/A [12]. To understand differences in the HCN epitope of CR2, we selected four yeast-displayed libraries of mutants of the CR2 parental antibody CR1 for higher affinity binding to BoNT/H, followed by additional rounds of random mutagenesis to further increase affinity. The resulting antibody, CRH2, had a 19-fold increased affinity for BoNT/H (KD 5.37 × 10−9 M vs. 2.9 × 10−10 M) but a 100-fold reduction in KD for BoNT/A1 (from 8.8 × 10−9 M to 8.9 × 10−7) demonstrating the challenges generating cross reactive antibodies to this BoNT epitope.
3. Discussion
A panel of 21 scFv mAbs binding the novel BoNT produced by C. botulinum strain IBCA10-7060 were isolated which bound nine non-overlapping epitopes spanning the HN and the LC domains. Despite this broad epitopic coverage, none of the 21 mAbs bound the other 7 BoNT serotypes except for one weakly binding the LC of BoNT/F5. We employed two selection strategies to enrich for potentially rare mAbs that bound both the novel BoNT and BoNT/F, the serotype whose subtype F5 has the closest LC-HN homology. In the first of these strategies, we used BoNT/F immune libraries and selected for BoNT/H LC-HN binding. Six mAbs binding the novel BoNT were obtained, but none bound any of the seven BoNT/F subtypes except the one mAb mentioned above that weakly bound to BoNT/F5 LC. In the second approach, the BoNT/H LC-HN immune library was selected for binding to the BoNT/F5 LC-HN. Two mAbs were obtained that bound BoNT/F5 LC-HN, but neither bound BoNT/H LC-HN. In both instances, the mAbs likely resulted from light chain shuffling which occurs during library construction and which creates new specificities that are not present in the original immune repertoire.
The results strongly indicate that the LC-HN of the novel BoNT is immunologically distinct from the other BoNT serotypes and thus should be termed BoNT/H LC-HN. This conclusion is supported by several other studies. First, only one of six mAbs (6F5.1) binding BoNT/F LC or HN domains bound the novel BoNT, and its novel toxin KD was 800-fold lower than the BoNT/F KD. This mAb, 6F5.1, binds an epitope at the tip of the HN that is shared by BoNT/A, B, E, F and the novel BoNT, and is the only mAb we have identified out of more than 100 mAbs studied that has this multi-serotype specificity [17]. Second, multiple studies have shown that BoNT/F polyclonal antisera alone does not neutralize the novel BoNT in mice [7,9]. This conclusion is consistent with the limited amino acid identity between the BoNT/H LC and HN and BoNT/F1 LC and HN (48.8% and 61.7% respectively, Figure S1), differences that are no different than the amino acid percentage identities between the seven BoNT serotypes (32.6–64.1%) [8]. Our use of a large panel of mAbs binding both the LC and HN provides confidence that both of these domains are immunologically distinct from the other BoNT serotypes. Such conclusions are less strongly supported using polyclonal antisera since there could be considerable epitopic bias from immunodominant epitopes. The HCC and HCN domains of the novel BoNT were bound by two of two and one of two anti-BoNT/A mAbs, respectively, a finding consistent with the amino acid percentage identities of each of these domains to BoNT/A1 (92.2% and 73.1% respectively) [12]. The novel BoNT is also neutralized in mice by anti-BoNT/A research polyclonal antitoxins, although 16-fold to 1080-fold less potently than BoNT/A1 [7,9]. Based on the above findings, immunologically the novel BoNT is BoNT/HA.
There is both precedent and value for the use of such chimeric nomenclature. For example, in 1924 it was found that anti-sera produced from “BoNT/C” from an avian species could neutralize BoNT/C from other avian strains and neutralize the Seddon type C strain causing botulism in cattle in Australia; however anti-sera from the Seddon strain did not neutralize avian BoNT/C [19]. This neutralization discrepancy was explained when DNA sequencing revealed the existence of chimeras between BoNT/C and BoNT/D that have been termed BoNT CD and BoNT DC. In BoNT CD, the N-terminal 2/3 is type C in sequence and the C-terminal 1/3 type D in sequence [20]. In BoNT DC the N-terminal 2/3 is type D in sequence and the C-terminal 1/3 is type C in sequence [21]. While BoNT CD and BoNT DC are not considered new serotypes, the use of the chimeric nomenclature both provides structural insights as well as helps explain otherwise inexplicable neutralization results. Such is the case with BoNT/HA, the nomenclature tells us that the N-terminus is unique and that neutralization by BoNT/A anti-sera will be less potent than anticipated (see ref [22] and conclusions below). The use of the chimeric nomenclature does beg the question how much of the structure needs to be immunologically unique (or different) before it is defined as a novel chimera. We would propose using some combination of precedent, the BoNT CD and DC studies where there is a domain level difference in structure, and whether there is an impact on potency of neutralization compared to the reference serotype. BoNT/HA meets both criteria.
The BoNT/HA gene is located at a unique site in the strain IBCA10-7060 chromosome that resulted from an insertion event facilitated by flanking IS 110 elements [8]. The chimeric nature of BoNT/HA with its sequence divergence from all known BoNTs except in the HCC region suggests that BoNT/HA is derived from an ancestral BoNT that has undergone multiple rounds of recombination and sequence mutation [8,23]. Of note, the higher homology of the BoNT/F5 LC to the BoNT/HA LC (80%) compared to the other BoNT/F subtypes (46.3–48.3%) suggests that BoNT/F5 may also represent one of these ancestral recombination events and would more accurately be termed BoNT/HF. This interpretation is consistent with the unique substrate cleavage site common to the BoNT/HA and BoNT/F5 LC that no other BoNTs have [24]. In addition, a Simplot showing that the first 1500 nucleotides encoding the BoNT/F5 LC gene have low similarity to the same nucleotides in the other six BoNT/F subtypes led to the conclusion that BoNT/F5 was in fact an unrecognized mosaic (“hybrid”) of the HC-HN of BoNT/F2 and an/H-like LC [8].
4. Conclusions
The above findings have implications for the use of existing BoNT countermeasures and the development of new countermeasures. 500-fold more equine botulinum antitoxin (BAT, serotypes A-G) [22] was required to neutralize BoNT/HA than to neutralize BoNT/A1 [9]. This is expected given that only 1/6 of BoNT/HA has high identity with any of the seven serotypes A-G used to generate BAT (the HCC of BoNT/HA with BoNT/A). Based on relative potencies, a one-vial dose of BAT in humans has been calculated to neutralize 50–54 mouse LD50s of BoNT/HA per mL of human serum [9,12]. Serum concentrations of BoNT in patients with foodborne botulism in one study were reported as high as 32 mouse LD50s/mL [25], with the highest foodborne botulism serum concentration reported being 1800 mouse LD50/mL of BoNT/A [26]. Serum concentrations of BoNT expected from bioterrorism are unknown. Comparing the estimated BoNT/HA neutralizing activity of BAT (<54 MLD50s/mL) with reported BoNT serum concentrations (up to 1800 LD50s/mL) indicates that for some patients a single vial of BAT might be inadequate. In addition, the ability of BoNT to persist in serum for as long as 25 days after exposure [26,27] together with the short serum half-life (8.6 h) of the anti-BoNT/A component of BAT [22] heightens the need to anticipate relapse and to consider early BAT re-administration where possible [28].
As a possible alternative to BAT, NTM-1631 [29], a combination of two human and one humanized BoNT/A mAbs (formerly XOMA 3AB), has been calculated to be at least ten times more potent than BAT for BoNT/HA using the mouse neutralization assay [12]. NTM-1631 has a serum half-life of >10 days and was well tolerated with no significant adverse events in a Phase 1 clinical trial [29]. Hence NTM-1631 could be developed as a treatment for botulism due to BoNT/HA.
Finally, if the ancestral BoNT that evolved into BoNT/HA still exists in nature and can cause botulism, it is likely that existing antitoxins would not neutralize it. The mAbs described here could be used for detection of the ancestral BoNT and could be developed into therapeutic antitoxins against both the ancestral BoNT and natural BoNT/HA or bioengineered derivatives.
5. Materials and Methods
5.1. Materials
Full length, genetically inactive BoNT/H with E227Q and Y366F mutation (BoNT/Hi) was purchased from Toxogen GmbH (Hannover, Germany). The pure toxins BoNT/A1, B1, C, D/C, E3, F1, and G were purchased from Metabiologics Inc. (Madison, WI, USA). mAbs 6H1 and 6F5.4 [12] were expressed from CHO cells as described [30]. All secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA, USA).
5.2. Protein Expression and Purification
The BoNT/H fragments BoNT/H LC (1–444) and HLC-HN (1–859) and soluble scFv were expressed and purified as described [18]. For ELISA, 100 μL of serially diluted BoNT/H LC-HN was coated on a plate, with F1 LC-HN as a control, followed by incubation with mAb 6F5.4 or anti-His tag IgG (1 μg/mL) with detection by using HRP-labeled goat anti-human or mouse IgG (0.1 μg/mL, Jackson ImmunoResearch) and 2,2’-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) as substrate.
5.3. Ethics Statement
The USDA Institutional Animal Care and Use Committee approved the animal care and use protocol to conduct the animal studies reported here under an IACUC-approved protocol (16-1) with an approval date of 7/27/2016 in compliance with the Animal Welfare Act, PHS Policy.
5.4. Mouse Immunization, Spleen Harvest, and Antiserum Titer
Mice were immunized with BoNT/H LC-HN three times. Briefly, 0.25–2.5 μg/mouse of BoNT/H LC-HN was intraperitoneally injected with adjuvant (Sigma, Burlington, MA, USA) at day one with boosting on days 21 and 42. Blood was drawn at the submandibular site from mice ten days after their final immunization and analyzed for binding to BoNT/H LC-HN by ELISA. Spleens from mice that had positive ELISA results were used to extract mRNA for scFv library construction.
5.5. scFv library Construction and FACS Sorting
The BoNT/H LC-HN scFv yeast display library was constructed as described [31]. BoNT/H specific binders were isolated using three rounds of FACS sorting in which 100 nM of BoNT/H LC-HN was used for yeast incubation, followed by Alexa-647-conjugated 6F5.4 IgG (6F5.4-647) for labeling. In parallel, yeast cells were labeled with BoNT/H LC-HN and co-stained with 6H1-647 and 6F5.4-488, from which 6F5.4 overlapping mAbs were isolated (6F5.4 negative but 6H1 positive colonies). In separate experiments, the BoNT/H LC-HN immunized library was also stained with 100 nM of BoNT/F5 LC-HN followed by incubation with mAb 6F5.4-647 to isolate BoNT/F5 specific mAbs. Individual colonies were picked from SD-CAA plates, cultured and scFv display induced for further characterization [31]. Libraries made from mice immunized with BoNT/F fragments [18] were sorted as described above (Table 1).
5.6. scFv binding Confirmation and Affinity Determination
Binding of individual yeast-displayed scFv was confirmed using BoNT/H LC-HN and full-length BoNT/Hi by using flow cytometry. Flow cytometry was used because it allows measurement of the scFv dissociation equilibrium constant (KD) without the need to subclone, express and purify the scFv as would be required for ELISA or surface plasmon resonance. The dissociation equilibrium constant (KD) of scFv was determined by using flow cytometer as described [18].
5.7. Epitope Overlap Determination
Epitope groups on BoNT/H surface were deduced from competition assays as described [18]. Briefly, yeast-displayed scFvs were incubated with 100 nM of BoNT/H LC-HN followed by incubation with soluble myc-tagged scFv. Anti-myc antibody was added for detection of soluble scFv binding and the MFI measured by flow cytometry. A positive signal indicated that both mAbs bound simultaneously.
5.8. MAb Affinity Maturation
Spiked oligonucleotide mutagenesis, light chain shuffling and error prone PCR-mediated random mutagenesis were used for affinity maturation of scFv 71E4, 71F4, and CR1. (See Supplementary Materials).
5.9. Measurement of Solution Phase Affinity at Equilibrium
For mAbs CR2, RAZ1 and 6F5.4, the solution phase affinity at equilibrium and binding kinetics were measured using flow fluorimetry in a KinExA (Sapidyne, Boise, ID, USA) as previously described [10,25] except that BoNT/Hi toxin was used. BoNT/Hi toxin solution was studied at a concentration estimated to be >10-fold above the value of the equilibrium dissociation binding constant [KD] of the mAb for the toxin to generate a concentration-controlled curve for greater accuracy in measuring BoNT/Hi concentrations. mAb-containing solutions were serially diluted in a constant concentration of culture filtrate, from >10-fold above to <0.1-fold below the estimated BoNT/Hi concentrations, to capture a complete binding curve. After equilibrium was achieved, samples were passed over a flow cell packed with Sepharose 4 Fast Flow beads (GE Healthcare, Marlborough, MA, USA) covalently coupled with the measuring mAb. An Alexa-647-labeled mAb binding a nonoverlapping BoNT epitope was then passed over the flow cell, producing a signal proportional to the free BoNT in each sample. An analysis curve yielding values for KD and the binding concentrations of the BoNTs was generated using KinExA Pro software (version 4.2.14, Sapidyne, Boise, ID, USA) and a 1:1 reverse-binding model.
6. Patents
The antibodies described here are the subject of patent application US20180208680A1, “Antibodies for Botulinum Neurotoxins” inventors James D. Marks, Yongfeng Fan, Jianlong Lou, Maria Consuelo Garcia Rodriguez, Shude Yan, Isin Nergiz Geren, Wenwu Zhai, Subhendu Chakraborti.
Acknowledgments
In this section you can acknowledge any support given which is not covered by the author contribution or funding sections. This may include administrative and technical support, or donations in kind (e.g., materials used for experiments).
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6651/12/1/9/s1, Table S1: mAbs 6F5.4, CR2 and RAZ1 bind BoNT/H and BoNT/Hi with comparable dissociation equilibrium constants (KD). Figure S1: amino acid identity of botulinum neurotoxin type H (BoNT/H) holotoxin and domains compared to BoNT/A1, BoNT/F1 and BoNT/F5. Figure S2: analysis of BoNT/H fragments and results of mouse immunization. Figure S3: dot plots of the specificity of BoNT/H LC binding scFv. Figure S4: affinity maturation of 71E4 and 71F4 did not extend their binding profile. Figure S5: dot plots of the binding of BoNT/H HN scFv to BoNT/F subtypes and BoNT/A. Figure S6: BoNT/F5 LC-HN scFv 77H2 and 78A5 bind the HN domain and do not bind BoNT/H LC-HN.
Author Contributions
Conceptualization, J.D.M. and S.S.A.; methodology, Y.F., J.L., J.R.B., L.W.C., C.T., F.C.; writing—original draft preparation, J.D.M.; writing—review and editing, J.D.M, S.S.A., Y.F.; visualization, Y.F.; supervision, J.D.M.; project administration, J.D.M.; funding acquisition, J.D.M., S.S.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded in part by a grant from the National Institute of Health (NIH), National Institute for Allergy and Infectious Diseases (NIAID), R01AI104579, PI, J.D.M.), the United States Department of Agriculture, Agricultural Research Service, National Program project NP108, CRIS 2030-42000-049-00D and in part by the California Department of Public Health Infant Botulism Treatment and Prevention Fund. The opinions, interpretations, and conclusions contained herein are those of the authors and are not necessarily endorsed by NIAID, the NIH, the USDA, or the California Department of Public Health.
Conflicts of Interest
Y.F., J.L., C.G-R. and J.D.M. are inventors of intellectual property in the field of botulinum antitoxins including intellectual property incorporated into NTM-1631. The other authors have no conflicts to disclose. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Key Contribution
The neurotoxin produced by clostridial strain IBCA10-7060 is immunologically identified in this report as BoNT/HA. This finding has implications for the use of existing BoNT countermeasures and the development of new countermeasures and suggests that recombinant antibodies that are in clinical development to treat botulism due to serotype A would have superior efficacy to equine heptavalent botulinum antitoxin.
References
- 1.Gill M.D. Bacterial toxins: A table of lethal amounts. Microbiol. Rev. 1982;46:86–94. doi: 10.1128/mr.46.1.86-94.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lacy D.B., Tepp W., Cohen A.C., DasGupta B.R., Stevens R.C. Crystal structure of botulinum neurotoxin type A and implications for toxicity. Nat. Struct. Mol. Biol. 1998;5:898. doi: 10.1038/2338. [DOI] [PubMed] [Google Scholar]
- 3.Lacy D.B., Stevens R.C. Sequence homology and structural analysis of the Clostridial neurotxins. J. Mol. Biol. 1999;291:1091–1104. doi: 10.1006/jmbi.1999.2945. [DOI] [PubMed] [Google Scholar]
- 4.Hill K.K., Smith T.J. Genetic diversity within Clostridium botulinum serotypes, botulinum neurotoxin gene clusters and toxin subtypes. Curr. Top. Microbiol. Immunol. 2013;364:1–20. doi: 10.1007/978-3-642-33570-9_1. [DOI] [PubMed] [Google Scholar]
- 5.Zhang S., Masuyer G., Zhang J., Shen Y., Lundin D., Henriksson L., Miyashita S.-I., Martínez-Carranza M., Dong M., Stenmark P. Identification and characterization of a novel botulinum neurotoxin. Nat. Commun. 2017;8:14130. doi: 10.1038/ncomms14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunt J., Carter A.T., Stringer S.C., Peck M.W. Identification of a novel botulinum neurotoxin gene cluster in Enterococcus. FEBS Lett. 2018;592:310–317. doi: 10.1002/1873-3468.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barash J.R., Arnon S.S. A novel strain of Clostridium botulinum that produces type B and type H botulinum toxins. J. Infect. Dis. 2014;209:183–191. doi: 10.1093/infdis/jit449. [DOI] [PubMed] [Google Scholar]
- 8.Dover N., Barash J.R., Hill K.K., Xie G., Arnon S.S. Molecular characterization of a novel botulinum neurotoxin type H gene. J. Infect. Dis. 2014;209:192–202. doi: 10.1093/infdis/jit450. [DOI] [PubMed] [Google Scholar]
- 9.Maslanka S.E., Luquez C., Dykes J.K., Tepp W.H., Pier C.L., Pellett S., Raphael B.H., Kalb S.R., Barr J.R., Rao A., et al. A novel botulinum neurotoxin, previously reported as serotype H, has a hybrid-like structure with regions of similarity to the structures of serotypes A and F and is neutralized with serotype A antitoxin. J. Infect. Dis. 2016;213:379–385. doi: 10.1093/infdis/jiv327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pellett S., Tepp W.H., Bradshaw M., Kalb S.R., Dykes J.K., Lin G., Nawrocki E.M., Pier C.L., Barr J.R., Maslanka S.E., et al. Purification and characterization of botulinum neurotoxin FA from a genetically modified Clostridium botulinum strain. mSphere. 2016;1:e00100–e00115. doi: 10.1128/mSphere.00100-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Escalona N., Thirunavukkarasu N., Singh A., Toro M., Brown E.W., Zink D., Rummel A., Sharma S.K. Draft genome sequence of bivalent Clostridium botulinum strain IBCA10-7060, encoding botulinum neurotoxin B and a new FA mosaic type. Genome Announc. 2014;2:e01275–14. doi: 10.1128/genomeA.01275-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan Y., Barash J.R., Lou J., Conrad F., Marks J.D., Arnon S.S. Immunological characterization and neutralizing ability of monoclonal antibodies directed against botulinum neurotoxin type H. J. Infect. Dis. 2016;213:1606–1614. doi: 10.1093/infdis/jiv770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao G., Zhang S., Mahrhold S., Lam K.-H., Stern D., Bagramyan K., Perry K., Kalkum M., Rummel A., Dong M. N-linked glycosylation of SV2 is required for binding and uptake of botulinum neurotoxin A. Nat. Struct. Mol. Biol. 2016;23:656. doi: 10.1038/nsmb.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao G., Lam K.-H., Perry K., Weisemann J., Rummel A., Jin R. Crystal structure of the receptor-binding domain of botulinum neurotoxin type HA, also known as type FA or H. Toxins. 2017;9:93. doi: 10.3390/toxins9030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy R., Forsyth C., LaPorte S., Geren I., Smith L., Marks J. Fine and domain-level epitope mapping of botulinum neurotoxin type A neutralizing antibodies by yeast surface display. J. Mol. Biol. 2007;365:196–210. doi: 10.1016/j.jmb.2006.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia C., Levy R., Arndt J., Forsyth C., Razai A., Lou J., Geren I., Stevens R., Marks J. Molecular evolution of antibody specificity and cross reactivity for type A botulinum neurotoxins. Nat. Biotech. 2007;25:107–116. doi: 10.1038/nbt1269. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Rodriguez C., Geren I.N., Lou J., Conrad F., Forsyth C., Wen W., Chakraborti S., Zao H., Manzanarez G., Smith T.J., et al. Neutralizing human monoclonal antibodies binding multiple serotypes of botulinum neurotoxin. Protein Eng. Des. Sel. Peds. 2011;24:321–331. doi: 10.1093/protein/gzq111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan Y., Garcia-Rodriguez C., Lou J., Wen W., Conrad F., Zhai W., Smith T.J., Smith L.A., Marks J.D. A three monoclonal antibody combination potently neutralizes multiple botulinum neurotoxin serotype F subtypes. PLoS ONE. 2017;12:e0174187. doi: 10.1371/journal.pone.0174187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfenninger W. Toxico Immunologic and Serologic Relationship of B. botulinus, Type C, and B. parabotulinus,” Seddon.” XXII. J. Infect. Dis. 1924;35:347–352. doi: 10.1093/infdis/35.4.347. [DOI] [Google Scholar]
- 20.Moriishi K., Koura M., Fujii N., Fujinaga Y., Inoue K., Syuto B., Oguma K. Molecular cloning of the gene encoding the mosaic neurotoxin, composed of parts of botulinum neurotoxin types C1 and D, and PCR detection of this gene from Clostridium botulinum type C organisms. Appl. Environ. Microbiol. 1996;62:662–667. doi: 10.1128/aem.62.2.662-667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moriishi K., Koura M., Abe N., Fujii N., Fujinaga Y., Inoue K., Ogumad K. Mosaic structures of neurotoxins produced from Clostridium botulinum types C and D organisms. Biochim. Biophys. Acta Gene Struct. Expr. 1996;1307:123–126. doi: 10.1016/0167-4781(96)00006-1. [DOI] [PubMed] [Google Scholar]
- 22.Botulism Antitoxin Heptavalent (A, B, C, D, E, F, G)—(Equine). Package Insert. Cangene Corp. [(accessed on 29 April 2019)]; Available online: https://www.fda.gov/vaccines-blood-biologics/approved-blood-products/bat-botulism-antitoxin-heptavalent-b-c-d-e-f-g-equine.
- 23.Hill K.K., Xie G., Foley B.T., Smith T.J. Genetic diversity within the botulinum neurotoxin-producing bacteria and their neurotoxins. Toxicon. 2015;107:2–8. doi: 10.1016/j.toxicon.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Lam K.-H., Sikorra S., Weisemann J., Maatsch H., Perry K., Rummel A., Binz T., Jin R. Structural and biochemical characterization of the protease domain of the mosaic botulinum neurotoxin type HA. Pathog. Dis. 2018;76:fty044. doi: 10.1093/femspd/fty044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatheway C.H., Snyder J.D., Seals J.E., Edell T.A., Lewis G.E., Jr. Antitoxin levels in botulism patients treated with trivalent equine botulism antitoxin to toxin types A, B, and E. J. Infect. Dis. 1984;150:407–412. doi: 10.1093/infdis/150.3.407. [DOI] [PubMed] [Google Scholar]
- 26.Sheth A.N., Wiersma P., Atrubin D., Dubey V., Zink D., Skinner G., Doerr F., Juliao P., Gonzalez G., Burnett C. International outbreak of severe botulism with prolonged toxemia caused by commercial carrot juice. Clin. Infect. Dis. 2008;47:1245–1251. doi: 10.1086/592574. [DOI] [PubMed] [Google Scholar]
- 27.Fagan R.P., McLaughlin J.B., Middaugh J.P. Persistence of botulinum toxin in patients’ serum: Alaska, 1959–2007. J. Infect. Dis. 2009;199:1029–1031. doi: 10.1086/597310. [DOI] [PubMed] [Google Scholar]
- 28.Fagan R.P., Neil K.P., Sasich R., Luquez C., Asaad H., Maslanka S., Khalil W. Initial recovery and rebound of type F intestinal colonization botulism after administration of investigational heptavalent botulinum antitoxin. Clin. Infect. Dis. 2011;53:e125–e128. doi: 10.1093/cid/cir550. [DOI] [PubMed] [Google Scholar]
- 29.Nayak S.U., Griffiss J.M., McKenzie R., Fuchs E.J., Jurao R.A., An A.T., Ahene A., Tomic M., Hendrix C.W., Zenilman J.M. Safety and pharmacokinetics of XOMA 3AB, a novel mixture of three monoclonal antibodies against botulinum toxin A. Antimicrob. Agents Chemother. 2014;58:5047–5053. doi: 10.1128/AAC.02830-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nowakowski A., Wang C., Powers D.B., Amersdorfer P., Smith T.J., Montgomery V.A., Sheridan R., Blake R., Smith L.A., Marks J.D. Potent neutralization of botulinum neurotoxin by recombinant oligoclonal antibody. Proc. Natl. Acad. Sci. USA. 2002;99:11346–11350. doi: 10.1073/pnas.172229899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan Y., Geren I.N., Dong J., Lou J., Wen W., Conrad F., Smith T.J., Smith L.A., Ho M., Pires-Alves M., et al. Monoclonal antibodies targeting the alpha-exosite of botulinum neurotoxin serotype/A inhibit catalytic activity. PLoS ONE. 2015;10:e0135306. doi: 10.1371/journal.pone.0135306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.