Abstract
Aspergillus niger produces a wide spectrum of extracellular polysaccharide hydrolases that hydrolyze cellulose into soluble glucose and cellodextrins. Transporters are essential for sugar uptake, yet it is not clear whether cellodextrin transporters exist in A. niger. Here, one cellulose inducible cellodextrin transporter CtA was identified in A. niger B2. It was found that CtA not only could transport cellobiose, but also cellotriose, cellotetraose, and cellopentaose. The yeast strain YPβG-CtA, expressing cellodextrin transporter CtA and an intracellular β-glucosidase, grew on cellobiose with the cell growth rate of 0.0830 ± 0.0113 h–1 under aerobic condition. Furthermore, the engineered yeast could produce 1.1 g/L ethanol anaerobically on cellobiose in 2 days. The identification of CtA provides evidence that the cellodextrin uptake is a complementary strategy of cellulose utilization in A. niger, and the CtA could be a strong transporter candidate for constructing engineered cellodextrin-utilizing microorganisms.
Keywords: Aspergillus niger, cellulose, cellodextrin transporter, cellobiose, cellodextrin
Introduction
The filamentous ascomycete fungus Aspergillus niger is ubiquitous in the environment and has been widely used in industry for the production of organic acids and extracellular enzymes (Okada, 1988). A. niger produces a wide spectrum of polysaccharide hydrolases, which enable it to grow on cellulosic biomass. The secreted cellulases hydrolyze cellulose into glucose and cellodextrins. The resulting sugars are then transported into the cells and get metabolized intracellularly. Characterization of the sugar transporters is therefore important for understanding the process of fungal plant biomass degradation.
Cellodextrin transporters from fungi commonly belong to the MFS (major facilitator superfamily) transporters, which are single polypeptide facilitating the movement of small soluble substrates in response to ion gradients (Yan, 2013). In the past years, several cellodextrin transporters have been identified in cellulolytic fungi. Two cellodextrin transporters (CDT-1 and CDT-2) were identified in cellulolytic fungus Neurospora crassa (Galazka et al., 2010), and three genes, CdtC, CdtD, and CdtG, encoding cellodextrin transporters have been reported in cellulolytic fungus Penicillium oxalicum (Li et al., 2013), and the transporter Stp1 and CltA involving cellobiose uptake were identified in Trichoderma reesei and Aspergillus nidulans, respectively (Zhang et al., 2013; dos Reis et al., 2016).
The genomic and transcriptomic analysis of A. niger showed that it contains 86 putative sugar transporters, which were classified as members of the Pfam family of “Sugar and other transporters” (PF00083) (Pel et al., 2007; de Vries et al., 2017; Peng et al., 2018). Until now, the high-affinity and low-affinity glucose transporters in A. niger have been identified and studied extensively (Torres et al., 1996; Vankuyk et al., 2004; Sloothaak et al., 2015), while the cellodextrin transporters in A. niger have not yet been identified. Recently, the abilities of transportation of 12 different sugars by 43 putative sugar transporters from A. niger were extensively analyzed by de Vries et al. (2017), yet these sugar transporters were not characterized for their cellodextrin transporter function. Several sugar transporters in A. niger were shown to be inducible by cellulosic biomass. Delmas et al. (2012) reported that 3 sugar transporter genes were up-regulated when A. niger grew on wheat straw and de Souza et al. (2011) found that 5 sugar transporter genes in A. niger were up-regulated on steam-exploded sugarcane bagasse, which indicate A. niger may have cellodextrin transporters.
The aim of this study was to identify and characterize the cellodextrin transporters in A. niger, to provide insight into the cellulose utilization mechanism of A. niger, and to provide potentially useful cellodextrin transporter for constructing efficient biofuel producer strains.
Materials and Methods
Strains, Media, and Culture Methods
Aspergillus niger B2 was isolated from rotting wheat straw in our laboratory (Yue et al., 2012), and it has been deposited in the China General Microbiological Culture Collection Center (CGMCC) with deposit number CGMCC No. 17078. Sugarcane bagasse was pretreated by steam explosion at 2.0 MPa for 150 s, and the steam-exploded sugarcane bagasse consists of 49 ± 0.3% cellulose, 23 ± 0.2% hemicellulose, 23 ± 0.2% lignin, and 5 ± 0.1% ash, which was determined by the NREL procedure: determination of structural carbohydrates and lignin in biomass (Sluiter et al., 2008). All the experiments were performed in technical and biological triplicates, and standard deviations of triplicates are represented by error bars. Cellobiose was purchased from Sigma-Aldrich (United States), cellotetraose was purchased from Elicityl (France), cellotriose and cellopentaose were purchased from TCI (Japan), and no glucose was detected in all the samples by HPLC (Supplementary Figure S1). All reagents were of analytical grade and were used without further purification, except where stated.
Aspergillus niger B2 was cultured on potato dextrose agar (PDA) solid medium 5 days for spores preparation. The modified Mandel’s medium for fungal spores germination and hyphal growth (growth media) contained glucose (20 g/L) and peptone (1 g/L) (Mandels and Weber, 1969). The modified Mandel’s media for inducing the transcription of cellodextrin transporters (induction media) contained steam-exploded sugarcane bagasse (5 g/L) and peptone (1 g/L).
Saccharomyces cerevisiae YPH499 strain was grown in synthetic media (SM) contained yeast nitrogen base without amino acids and ammonium sulfate (6.7 g/L), adenine 50 mg/L, histidine (20 mg/L), lysine (30 mg/L), tryptophan (20 mg/L), leucine (100 mg/L), uracil (20 mg/L), and glucose (20 g/L) or cellodextrins (5 g/L) as the sole carbon source at 30°C and 250 rpm. S. cerevisiae YPH499 harboring plasmid pRS425 was grown in the SM without leucine (SM-Leu), and S. cerevisiae YPH499 harboring plasmid pRS425 and plasmid pRS426 was grown in the SM without leucine and uracil (SM-Leu-Ura).
RNA Extraction and Expression Profiling by RT-qPCR
To characterize the expression of potential cellodextrin transporters (An12g09270, An14g01600, An13g03250, An03g05320, An08g09350, An16g06220, and An04g02790) upon the induction of cellulosic biomass, 50 mL Mandel’s growth medium was inoculated with fresh A. niger B2 spore suspension in Mandel’s salt solution (about 107 spores) and cultured at 200 rpm and 30°C for 48 h. Mycelia were harvested by vacuum filtration and washed with sterile Mandel’s salt solution three times, and then 1.5 g mycelia were transferred to 50 mL Mandel’s induction medium containing 0.5% of steam-exploded sugarcane bagasse, grown at 30°C and 200 rpm. Mycelia were harvested at 6 h, frozen in liquid nitrogen, stored in a freezer at −80°C, and then used for RNA isolation by using RNeasy Plant Mini Kit (Qiagen, Hilden, Germany, Cat. No. 74903) according to the manufacturer’s protocols.
The extracted mRNA samples were quantified on the NanoDrop® 1000 (Thermo Fisher Scientific). mRNA was then reversed transcribed to cDNA by using the ProtoScript® II Reverse Transcriptase (NEB), following the manufacturer’s menus. The primers used in RT-qPCR were listed in Supplementary Table S1. RT-qPCR reactions were performed using the Bio-Rad iQ5 Real-Time PCR System and the SYBR Green PCR Master Mix kit (Thermo Fisher Scientific, United States), following the manufacturer’s instructions. The reaction mixture containing 1.5 μL of cDNA (75 ng), 0.32 μL of primers mixture (100 μM forward primer and 100 μM reverse primer, 10 μM SYBR Green PCR Master Mix kit, and nuclease-free water up to 20 μL were dispensed into triplicate wells on a 384-well plate. The RT-qPCR protocol was initiated with 2 min activation at 60°C and a 5 min denaturation at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. The expression levels of 18s rRNA were used to normalize the data from the different samples, and the data analysis was performed using the relative quantitation/comparative CT (ΔΔCT) method.
Phylogenetic Analysis
Amino acid sequences of orthologs of CtA were obtained from NCBI database. Multiple sequence alignments were performed with ClustalX 2.0 (Larkin et al., 2007), and the phylogenetic trees by maximum likelihood was generated by Mega X with 1000 bootstrap replications using Jones-Taylor-Thornton (JTT) model, rates among sites were uniform, and gaps and missing data were completely deleted (Kumar et al., 2018).
Cloning of the Predicted Cellodextrin Transporter Genes Into S. cerevisiae
The predicted cellodextrin transporter genes, An12g09270 and An16g06220 (named as ctA and ctB), from A. niger B2 were amplified by using the cDNA as the template, and the primers were listed in Supplementary Table S2. The genes ctA and ctB were cloned into a pRS426 plasmid, creating plasmids pRS426-ctA and pRS426-ctB (Galazka et al., 2010). S. cerevisiae YPH499 (MATa ura3-52 lys2-801_amber ade2-101_ochre trp1-Δ63 his3-Δ200 leu2-Δ1) was used as the host for the cellodextrin transport assay (Sikorski and Hieter, 1989). The plasmid pRS425 harboring the N. crassa intracellular β-glucosidase gene (gh1-1) was transformed into the S. cerevisiae YPH499 competent cells by electroporation (1.5k V, 25 μF, 200 Ω, 5 ms), creating the strain YPβG (Galazka et al., 2010). The plasmid pRS426-ctA or pRS426-ctB was then transformed into YPβG in the same way mentioned above, creating the yeast strains YPβG-CtA and YPβG-CtB. The transformants were selected on SM plate containing glucose without leucine or leucine and uracil.
Fermentation and Cellodextrins Uptake Assay of YPβG-CtA and YPβG-CtB
In the fermentation experiments, the YPβG-CtA strain was first grown aerobically on SM-Leu-Ura medium (50 mL) containing 20 g/L glucose for 24 h. Cells were then harvested by centrifugation and were washed 3 times with sterile water 50 mL. Cell pellets were resuspended in SM-Leu-Ura medium (50 mL) containing cellobiose (20 g/L) and CuSO4 (0.2 mM) to a final OD600 of 0.5. The mixtures were transferred into serum bottles (100 mL), sealed and incubated at 30°C and 120 rpm. Samples were taken at various time intervals.
In cellodextrins uptake assay, flask (250 mL) containing SM-Leu-Ura medium (50 mL) with CuSO4 (0.2 mM) and cellobiose (5 g/L), cellotriose (2.5 g/L), cellotetraose (2.5 g/L), or cellopentaose (2.5 g/L) was inoculated with YPβG-CtA or YPβG-CtB to the final OD600 of 0.1. Flasks were incubated at 30°C and 250 rpm. The cell density was monitored by measuring the optical density at 600 nm. An aliquot of 1 mL culture was collected and analyzed by HPLC for measuring the residual cellodextrins.
Sample Analysis
Concentrations of cellodextrins (cellobiose, cellotriose, cellotetraose, or cellopentaose) and ethanol in the culture samples were analyzed using an UltiMate 3000 HPLC equipped with a refraction index detector and an Aminex HPX-87H (Bio-rad, Hercules, CA, United States). The eluent was H2SO4 (5 mM) and was eluted at a flow rate of 0.6 mL/min.
Results
Exploration of the Cellodextrin Transporters in A. niger
The analysis of transcriptome and genome sequencing data of A. niger CBS 513.88 showed that it had 86 putative sugar transporters (Pel et al., 2007; de Vries et al., 2017; Peng et al., 2018). The cellodextrin transporter gene candidates were selected based on their protein sequence identity to N. crassa CDT-1 or CDT-2 in the A. niger CBS 513.88 protein database (Galazka et al., 2010). Seven predicted sugar transporters (An12g09270, An14g01600, An13g03250, An03g05320, An08g09350, An16g06220, and An04g02790) having over 90% coverage and 30% identity to CDT-1 or CDT-2 were selected in this study (Supplementary Table S3). The selected seven putative sugar transporters were further analyzed in Transporter Classification Database (TCDB) (Saier et al., 2006), and the results showed that the gene at locus An12g09270 (36% identity to N. crassa CDT-1, e-value of 3e-113) and locus An14g01600 (38% identity to N. crassa CDT-2, e-value of 3e-112) were annotated as MFS lactose permease and MFS hexose transporter, respectively. The others having the best hit to CDT-2 were annotated as sugar transporters (Supplementary Table S3).
To shed more light on the transcription of these 7 predicted cellodextrin transporters’ gene in response to cellulosic biomass, a transcriptional analysis was performed by RT-qPCR for these transporter-coding genes in A. niger B2. The changes in transcriptional levels of those predicted cellodextrin transporter genes are shown in Table 1. The gene at locus An12g09270 was highly induced in the sugarcane bagasse medium, resulting in about a 12-fold up-regulation compared to the control. The gene at locus An16g06220 was also induced in the sugarcane bagasse medium, resulting in a two-fold up-regulation compared to the control. However, the other 5 tested genes were not induced by cellulosic biomass efficiently (less than a two-fold increase).
TABLE 1.
The relative transcription levels of predicted cellodextrin transporter genes in A. niger B2 on steam-exploded sugarcane bagasse 0.5%(w/v) compared to growth on glucose reference control.
| Gene | 2–ΔΔCt |
| An12g09270 | 12.6 ± 0.3 |
| An14g01600 | 0.35 ± 0.04 |
| An13g03250 | 0.18 ± 0.05 |
| An03g05320 | 0.14 ± 0.01 |
| An08g09350 | 0.57 ± 0.02 |
| An16g06220 | 1.68 ± 0.1 |
| An04g02790 | 0.16 ± 0.01 |
The here identified, putative lactose permease gene (lac12, An12g09270) and gene An16g06220 have identity with the N. crassa cellobiose transporter cdt-1 (36% identity, e-value of 3e-113) and cdt-2 (33% identity, e-value of 5e-91), respectively. These two genes were selected for further functional analysis, and here now were named ctA and ctB, respectively.
Investigation on the Ability of CtA and CtB to Transport Cellobiose
To evaluate the ability of CtA and CtB to transport cellobiose, both genes were amplified from A. niger B2 and were cloned into S. cerevisiae YPβG that harbored the N. crassa-glucosidase gene gh1-1 (NCU00130). The expression of a functional cellodextrin transporter will enable the YPβG strain to grow on the medium with cellobiose as the sole carbon source, which has been demonstrated in the previous work that we participated in Galazka et al. (2010). Herein, the strain YPβG-CtA and YPβG-CtB, harboring the predicted cellodextrin transporter gene ctA and ctB, respectively, were constructed.
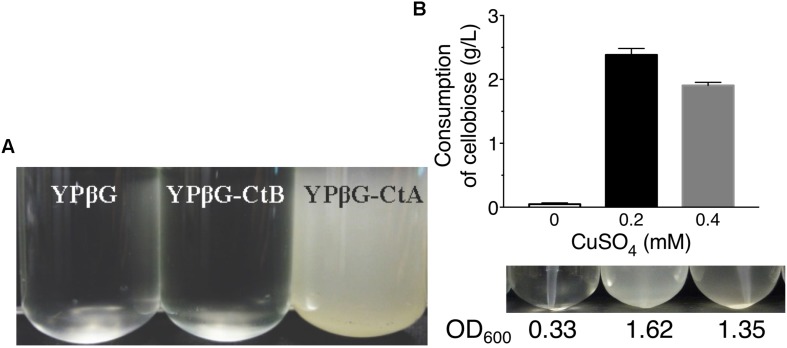
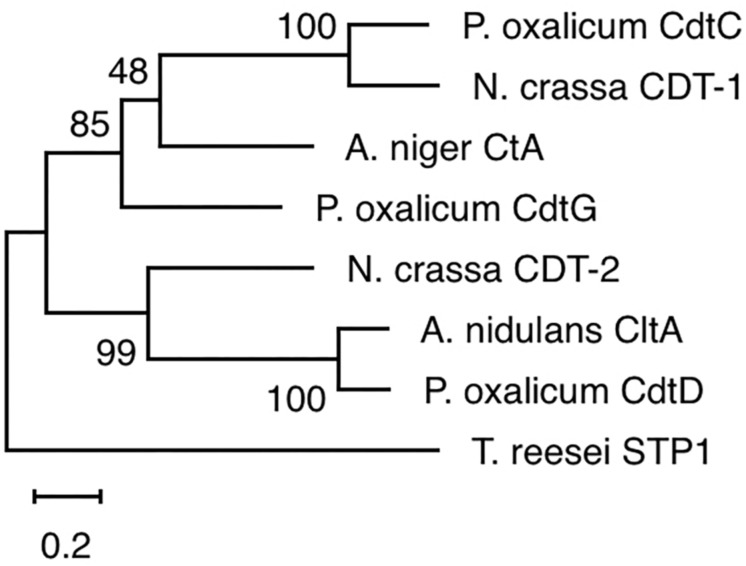
The yeast strains YPβG, YPβG-CtA and YPβG-CtB were inoculated into the SM-Leu or SM-Leu-Ura media with cellobiose as the sole carbon source to test the transportability of cellobiose (Figure 1A). The results showed that YPβG strain only harboring the β-glucosidase gene could not grow in the medium, while YPβG-CtA strain harboring the ctA gene and β-glucosidase gene could grow on cellobiose. Therefore, CtA’s function as a cellobiose transporter was confirmed. However, YPβG-CtB strain harboring the ctB gene and β-glucosidase gene could not grow in the medium, indicating that the CtB dose not have the ability to transport cellobiose into the cells. Herein, the sequence of ctA was deposited in GenBank under the accession number of MH648002. The cellodextrin transporter CtA in A. niger shares 36% identity with CDT-1 (Galazka et al., 2010), but only 29% identity with the cellobiose transporter CltA in A. nidulans (dos Reis et al., 2016). Phylogenetic analysis further revealed that CtA was closely related to the well characterized cellodextrin transporter CDT-1 from N. crassa and CdtC from P. oxalicum, but did not group with CltA from A. nidulans (Figure 2).
FIGURE 1.
The growth of yeast strains in the media with cellobiose as the sole carbon source. (A) Growths of yeast strain YPβG, YPβG-CtA, and YPβG-CtB in SM-Leu or SM-Leu-Ura media with cellobiose as the sole carbon source after 2 days. (B) The cellobiose utilization and cell growth of YPβG-CtA strain in the media containing a different concentration of copper.
FIGURE 2.
Phylogenetic analysis of CtA and other cellobiose/cellodextrin transporters. Sequence alignments were performed with ClustalX 2.0, and the maximum likelihood tree was generated with Mega X. Numbers on the tree branches represent the bootstrap support calculated per 1000 bootstrap replicates. CdtC, Penicillium oxalicum cellodextrin transporter (GenBank No. EPS25673.1); CDT-1, Neurospora crassa cellodextrin transporter (GenBank No. XP_963801.1); CdtG, Penicillium oxalicum cellodextrin transporter (GenBank No. EPS34431.1); CDT-2, Neurospora crassa cellodextrin transporter (GenBank No. XP_963873.1); CltA, Aspergillus nidulans cellobiose transporter (GenBank No. AN8347); CdtD, Penicillium oxalicum cellodextrin transporter (GenBank No. EPS25817.1); STP1, Trichoderma reesei cellodextrin transporter (GenBank No. ETS01554.1).
Since copper is the inducer for Cup1 promoter used in pRS426 plasmid expressing transporter genes, the copper concentration in the culture was investigated for efficient cellobiose utilization (Mascorro-Gallardo et al., 1996). As shown in Figure 1B, no obvious cell growth of YPβG-CtA and cellobiose consumption in the culture was observed when no copper was added in the media. When the copper concentration in the medium was 0.2 mM, about 50% cellobiose was consumed in 2 days, and the cell density in terms of OD600 was up to 1.6. However, when the copper concentration in the medium was increased to 0.4 mM, the strain utilized less cellobiose and achieved a lower cell density than those of 0.2 mM copper, indicating that 0.2 mM copper was optimal for CtA expression.
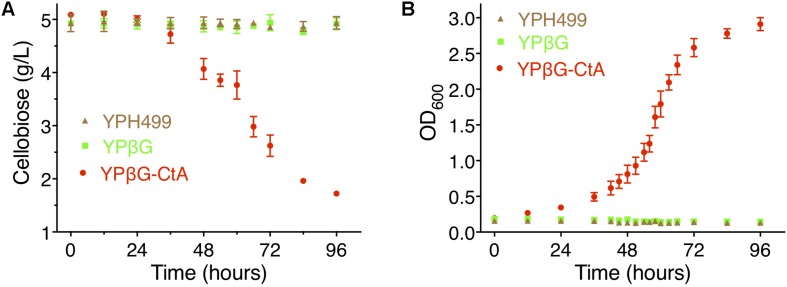
To further investigate the efficiency of CtA to transport cellobiose, the yeast strain YPH499, YPβG, and YPβG-CtA were cultured in the SM media, SM-Leu media or SM-Leu-Ura media with cellobiose as the sole carbon source, and the cell growth and concentration of cellobiose in the cultures were analyzed. It was shown that YPβG-CtA consumed about 3.5 g/L cellobiose in 4 days (Figure 3A). The cellobiose consumption rate during the exponential growth was 57.5 mg/L/h (Figure 3B). Cell density reached an OD600 of 3.0. However, no cellobiose consumption and cell growth with YPH499 and YPβG strains were observed.
FIGURE 3.
The cellobiose mediated growth of S. cerevisiae strain YPH499, YPβG, and YPβG-CtA. (A) The time course of cellobiose concentration of the YPH499, YPβG, and YPβG-CtA strains in the cultures with 0.5% cellobiose as the sole carbon source. (B) Growth curve of S. cerevisiae strains. The YPβG-CtA strain expressing transporter ctA and bgl consumed cellobiose, while the YPH499 and YPβG strains could not utilize cellobiose.
Investigation on the Ability of CtA and CtB to Transport Cellodextrins
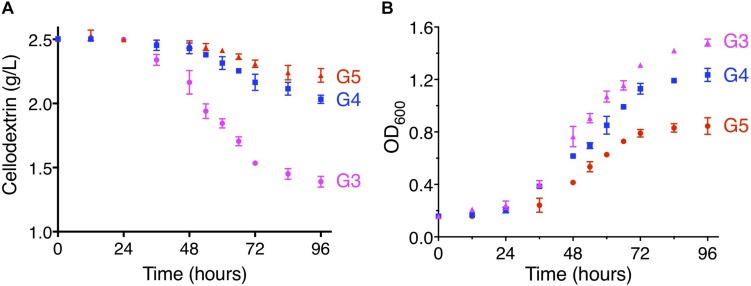
To further examine the ability to transport cellodextrins of CtA and CtB, the YPβG-CtA and YPβG-CtB were cultured in the SM-Leu-Ura media with cellotriose, cellotetraose, or cellopentaose as the sole carbon source. The YPβG-CtB strain could not grow in the medium containing cellotriose, cellotetraose, or cellopentaose and did not consume those cellodextrins, indicating that the CtB is not a cellodextrin transporter. The YPβG-CtA strain consumed 1.08 ± 0.10 g/L cellotriose, 0.5 ± 0.08 g/L cellotetraose, and 0.24 ± 0.09 g/L cellopentaose in 4 days (Figure 4). The OD600 reached 3.0, 1.5, 1.2, and 0.8 after 4 days culture when cellobiose, cellotriose, cellotetraose, and cellopentaose were used as the carbon source. The corresponding substrate consumption rates in the exponential growth phase are 57.5, 18.1, 8.3, and 4.1 mg/L/h (Figures 3, 4). Thus, CtA was able to transport all the tested G2-G5 cellodextrins into the cells. However, CtA had lower activity in the transportation of cellotriose, cellotetraose, and cellopentaose than that of cellobiose.
FIGURE 4.
The consumption of cellodextrins (A) and cell growth (B) of the YPβG-CtA strain. The yeast strain was cultured in the SM-Leu-Ura medium containing 0.25% (w/v) of cellotriose (G3), cellotetraose (G4), or cellopentaose (G5) as the sole carbon source at 30°C and 250 rpm.
Ethanol Production of YPβG-CtA on Cellobiose
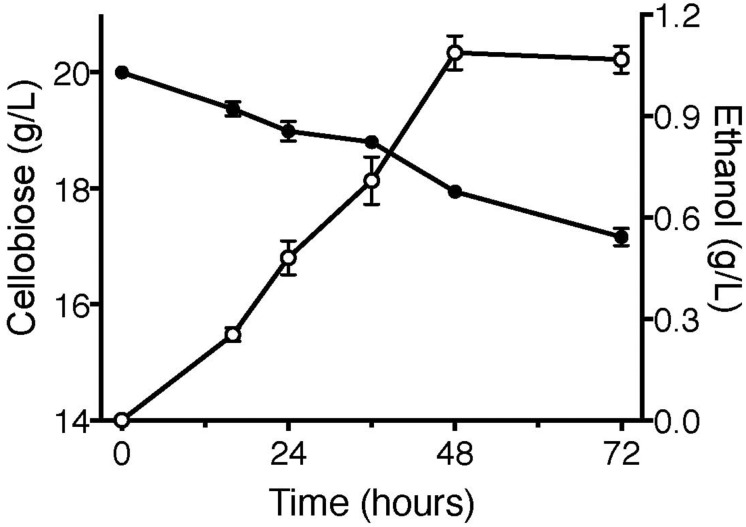
Cellodextrin transporters enable the non-cellobiose fermenting S. cerevisiae and E. coli to uptake cellodextrins, facilitating the engineered microorganisms to produce biofuels (Ha et al., 2011; Vinuselvi and Lee, 2011; Bae et al., 2014; Fan et al., 2016). To further confirm the function of CtA, the fermentation of YPβG-CtA on cellobiose (20 g/L) was conducted anaerobically, and the cellobiose consumption and ethanol production were analyzed (Figure 5). The YPβG-CtA utilized 2.2 g/L cellobiose in 2 days and yielded 1.1 g/L ethanol, which is 83.4% of the theoretical value. However, extending the fermentation time did not yield more ethanol with about 3 g/L cellobiose consumed after 3 days. The anaerobic fermentation of YPβG-CtA strain on cellobiose resulted in a slight lower ethanol production efficiency than the engineered strain harboring CDT-1 (Galazka et al., 2010). It is expected that the ethanol production efficiency of YPβG-CtA can be further improved by optimizing the fermentation conditions.
FIGURE 5.
The anaerobic fermentation of YPβG-CtA on cellobiose.
Discussion
In this study, two (An12g09270 and An16g06220) of the seven predicted sugar transporters genes were up-regulated when A. niger was grown on steam-exploded sugarcane bagasse, and the CtA (An12g09270) was confirmed to be a novel cellodextrin transporter. The phylogenetic analysis of the orthologs of CtA showed that it did not group with cellobiose transporter CltA from A. nidulans (Figure 2), which indicated CtA is a novel cellodextrin transporter in Aspergillus species. The further characterization of CtA showed that it was able to transport all the tested G2–G5 cellodextrins into the cells, while the transportation activities on the different substrates of CtA decreased slightly with increasing chain length (Figures 3, 4). The ability of CtA to transport G2–G5 cellodextrins, combined with the fact provided by de Vries et al. (2017) that none of the 12 tested sugars (including 8 monosaccharides, turanose, trehalose, maltose, and melitose) could be transported by An12g09270 (CtA), may suggest that CtA is a specific transporter for cellodextrin transportation in A. niger.
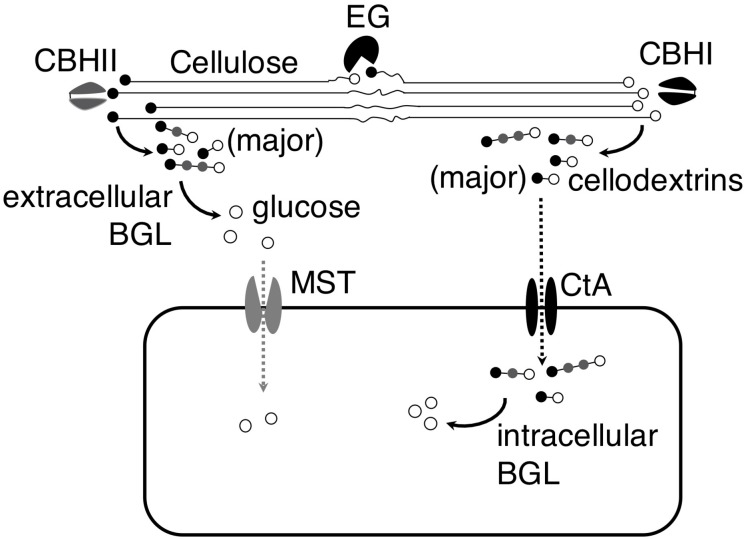
The identification of the cellodextrin transporter in A. niger led us to hypothesize that A. niger could import partial hydrolysis products, cellodextrins, and utilize them intracellularly, in addition to completely hydrolyzing cellodextrins to glucose and uptaking glucose. The analysis of A. niger CBS 513.88 genome showed that the fungus contains 5 putative BGLs without signal peptide (Supplementary Table S4; Pel et al., 2007; Petersen et al., 2011), and two (An03g03740 and An17g00520) of the 5 putative intracellular BGLs were showed as cellulosic inducible genes (de Souza et al., 2011), indicating their relevance to cellodextrins’ intracellular utilization. The existence of the putative intracellular BGLs in A. niger genome corroborated our hypothesis. The work provided a new understanding of the strategies of cellulose utilization in A. niger. When A. niger grows on cellulose, the fungus secrets cellulases to degrade cellulose into cellobiose and cellodextrins, and then cellobiose and cellodextrins are either subject to complete hydrolysis by extracellular BGLs into glucose followed by transportation into the cells by monosaccharide transporter (MST), or directly transported into the cells through cellodextrin transporter CtA followed by degradation by intracellular BGLs (Figure 6). Since A. niger secrets an enormous amount of BGLs, the complete hydrolysis strategy plays a major role in its cellulose utilization.
FIGURE 6.
Model of cellulose utilization in A. niger by both complete and partial hydrolysis strategies. In both strategies, cellulose is first transformed into cellobiose and cellodextrins by endoglucanase (EG) and exo-cellobiohydrolases (CBHI and CBHII). In the complete hydrolysis strategy, extracellular β-glucosidase (BGL) hydrolyzes the cellobiose and cellodextrins into glucose and then it is transferred into the cells by monosaccharide transporter (MST). However, the cellobiose and cellodextrins are transferred into the cells by cellodextrin transporter (CtA) and then they are hydrolyzed into glucose by intracellular BGL in the partial hydrolysis strategy.
The expression of ctA in the S. cerevisiae harboring a N. crassa β-glucosidase gene gh1-1 (YPβG-CtA) enabled the strain to use cellobiose efficiently and produced ethanol under the anaerobic fermentation. The YPβG-CtA strain had the cell growth rate of 0.0830 ± 0.0113 h–1 (Figure 3B), which was faster than the engineered yeast strain expressing the widely used N. crassa cdt-1 or cdt-2 and the same β-glucosidase gene gh1-1, which had the cell growth rate of 0.0341 ± 0.0010 h–1 and 0.0131 ± 0.0008 h–1, respectively (Galazka et al., 2010). It showed that CtA was a more efficient transporter, which enabled the engineered yeast strain to uptake cellobiose faster than N. crassa CDT-1 or CDT-2. Together with the ability to transport G2–G5 cellodextrins (Figure 4), CtA is an attractive choice of the cellodextrin transporter for engineering cellobiose or cellodextrin utilization microorganism.
Conclusion
In this study, the cellodextrin transporter CtA in A. niger was identified and characterized. Heterologously expressing the identified cellodextrin transporter CtA gene together with a β-glucosidase gene enabled the engineered S. cerevisiae strain to grow on cellobiose and cellodextrins up to chain length five. The engineered strain expressing CtA grew faster in cellobiose medium than the yeast strain harboring the widely used N. crassa cellobiose transporter CDT-1, which made it a strong transporter candidate for constructing engineered cellodextrin-utilizing microorganisms.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article (and its additional file). The sequence of CtA has been deposited in the GenBank under the accession number of MH648002.
Author Contributions
HC, CT, and HL designed the study. QZ, SC, and JZ carried out the experiments. HC, HL, QZ, and SC analyzed the data. HC, HL, and ZF wrote and edited the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (No. 31571775) and the Special Fund for Agro-Scientific Research in the Public Interest from the Ministry of Agriculture, China (No. 201503134) to Henan Agricultural University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00145/full#supplementary-material
References
- Bae Y. H., Kang K. H., Jin Y. S., Seo J. H. (2014). Molecular cloning and expression of fungal cellobiose transporters and beta-glucosidases conferring efficient cellobiose fermentation in Saccharomyces cerevisiae. J. Biotechnol. 169 34–41. 10.1016/j.jbiotec.2013.10.030 [DOI] [PubMed] [Google Scholar]
- de Souza W. R., de Gouvea P. F., Savoldi M., Malavazi I., Bernardes L. A. D., Goldman M. H. S., et al. (2011). Transcriptome analysis of Aspergillus niger grown on sugarcane bagasse. Biotechnol. Biofuels 4:40. 10.1186/1754-6834-4-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries R. P., Riley R., Wiebenga A., Aguilar-Osorio G., Amillis S., Uchima C. A., et al. (2017). Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 18:28. 10.1186/s13059-017-1151-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas S., Pullan S. T., Gaddipati S., Kokolski M., Malla S., Blythe M. J., et al. (2012). Uncovering the genome-wide transcriptional responses of the filamentous fungus Aspergillus niger to lignocellulose using RNA sequencing. Plos Genet. 8:e1002875. 10.1371/journal.pgen.1002875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis T. F., de Lima P. B. A., Parachin N. S., Mingossi F. B., Oliveira J. V. D., Ries L. N. A., et al. (2016). Identification and characterization of putative xylose and cellobiose transporters in Aspergillus nidulans. Biotechnol. Biofuels 9:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L. H., Zhang Z. J., Mei S., Lu Y. Y., Li M., Wang Z. Y., et al. (2016). Engineering yeast with bifunctional minicellulosome and cellodextrin pathway for co-utilization of cellulose-mixed sugars. Biotechnol. Biofuels 9:137. 10.1186/s13068-016-0554-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galazka J. M., Tian C. G., Beeson W. T., Martinez B., Glass N. L., Cate J. H. D. (2010). Cellodextrin transport in yeast for improved biofuel production. Science 330 84–86. 10.1126/science.1192838 [DOI] [PubMed] [Google Scholar]
- Ha S. J., Wei Q. S., Kim S. R., Galazka J. M., Cate J., Jin Y. S. (2011). Cofermentation of cellobiose and galactose by an engineered Saccharomyces cerevisiae strain. Appl. Environ. Microbiol. 77 5822–5825. 10.1128/AEM.05228-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Li J., Liu G. D., Chen M., Li Z. H., Qin Y. Q., Qu Y. B. (2013). Cellodextrin transporters play important roles in cellulase induction in the cellulolytic fungus Penicillium oxalicum. Appl. Microbiol. Biotechnol. 97 10479–10488. 10.1007/s00253-013-5301-3 [DOI] [PubMed] [Google Scholar]
- Mandels M., Weber J. (1969). “The production of cellulases,” in Cellulases and Their Applications, Vol. 95 eds Hajny G. J., Reese E. T., (Washington, DC: American Chemical Society; ), 391–414. 10.1021/ba-1969-0095.ch023 [DOI] [Google Scholar]
- Mascorro-Gallardo J. O., Covarrubias A. A., Gaxiola R. (1996). Construction of a CUP1 promoter-based vector to modulate gene expression in Saccharomyces cerevisiae. Gene 172 169–170. 10.1016/0378-1119(96)00059-5 [DOI] [PubMed] [Google Scholar]
- Okada G. (1988). “Cellulase of Aspergillus niger,” in Methods in Enzymology, Vol. 160 eds Abelson J., Simon M., Verdine G., Pyle A., (Cambridge, MA: Academic Press; ), 259–264. 10.1016/0076-6879(88)60128-5 [DOI] [Google Scholar]
- Pel H. J., de Winde J. H., Archer D. B., Dyer P. S., Hofmann G., Schaap P. J., et al. (2007). Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25 221–231. [DOI] [PubMed] [Google Scholar]
- Peng M., Aguilar-Pontes M. V., de Vries R. P., Mäkelä M. R. (2018). In silico analysis of putative sugar transporter genes in Aspergillus niger using phylogeny and comparative transcriptomics. Front. Microbiol. 9:1045. 10.3389/fmicb.2018.01045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Saier M. H., Tran C. V., Barabote R. D. (2006). TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 34 D181–D186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloothaak J., Odoni D. I., de Graaff L. H., Martins Dos Santos V. A. P., Schaap P. J., Tamayo-Ramos J. A. (2015). Aspergillus niger membrane-associated proteome analysis for the identification of glucose transporters. Biotechnol. Biofuels 8:150. 10.1186/s13068-015-0317-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluiter A., Hames B., Ruiz R., Scarlata C., Sluiter J., Templeton D., et al. (2008). Determination of Structural Carbohydrates and Lignin in Biomass. Golden, CO: National Renewable Energy Laboratory. [Google Scholar]
- Torres N. V., Riol-Cimas J. M., Wolschek M., Kubicek C. P. (1996). Glucose transport by Aspergillus niger: the low-affinity carrier is only formed during growth on high glucose concentrations. Appl. Microbiol. Biotechnol. 44 790–794. 10.1007/bf00178620 [DOI] [Google Scholar]
- Vankuyk P. A., Diderich J. A., MacCabe A. P., Hererro O., Ruijter G. J. G., Visser J. (2004). Aspergillus niger mstA encodes a high-affinity sugar/H+ symporter which is regulated in response to extracellular pH. Biochem. J. 379(Pt 2), 375–383. 10.1042/bj20030624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuselvi P., Lee S. K. (2011). Engineering Escherichia coli for efficient cellobiose utilization. Appl. Microbiol. Biotechnol. 92 125–132. 10.1007/s00253-011-3434-9 [DOI] [PubMed] [Google Scholar]
- Yan N. (2013). Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem. Sci. 38 151–159. 10.1016/j.tibs.2013.01.003 [DOI] [PubMed] [Google Scholar]
- Yue D., Liu X., Xu X., Wang M., Wang C., Chen H. (2012). Screening of high production strain of β-glucosidase and its enzyme characteristics. J. Henan Arg. Univ. 46 173–176. [Google Scholar]
- Zhang W. X., Kou Y. B., Xu J. T., Cao Y. L., Zhao G. L., Shao J., et al. (2013). Two major facilitator superfamily sugar transporters from Trichoderma reesei and their roles in induction of cellulase biosynthesis. J. Biol. Chem. 288 32861–32872. 10.1074/jbc.M113.505826 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article (and its additional file). The sequence of CtA has been deposited in the GenBank under the accession number of MH648002.