Osteomyelitis, an infection of the bone and bone marrow, imposes a heavy burden on public health care systems owing to its progressive bone destruction and sequestration. Human bone mesenchymal stem cells (hBMSCs) play a key role in the process of bone formation, and mounting evidence has confirmed that long noncoding RNAs (lncRNAs) are involved in hBMSC osteogenic differentiation.
KEYWORDS: osteomyelitis, FAM83H-AS1, osteogenic differentiation, miR-541-3p, WNT3A
ABSTRACT
Osteomyelitis, an infection of the bone and bone marrow, imposes a heavy burden on public health care systems owing to its progressive bone destruction and sequestration. Human bone mesenchymal stem cells (hBMSCs) play a key role in the process of bone formation, and mounting evidence has confirmed that long noncoding RNAs (lncRNAs) are involved in hBMSC osteogenic differentiation. Nevertheless, the exact function and molecular mechanism of lncRNAs in osteogenic differentiation during osteomyelitis development remain to be explored. In this study, hBMSCs were treated with staphylococcal protein A (SpA) during osteogenic differentiation induction to mimic osteomyelitis in vitro. The results of lncRNA microarray analysis revealed that FAM83H-AS1 presented the lowest expression among the significantly downregulated lncRNAs. Functionally, ectopic expression of FAM83H-AS1 contributed to osteogenic differentiation of SpA-induced hBMSCs. Additionally, our findings revealed that FAM83H-AS1 negatively regulated microRNA 541-3p (miR-541-3p), and WNT3A was validated as a target gene of miR-541-3p. Mechanically, FAM83H-AS1 elevated WNT3A expression by competitively binding with miR-541-3p. Lastly, it was demonstrated that FAM83H-AS1/miR-541-3p/WNT3A ameliorated SpA-mediated inhibition of the osteogenic differentiation of hBMSCs, which provided a novel therapeutic strategy for patients with osteomyelitis.
INTRODUCTION
Osteomyelitis is an infection of the bone and bone marrow resulting from bacteria, fungi, or mycobacteria that imposes a heavy burden on the survival ability of patients and on the public health care system (1). It is extensively acknowledged that osteomyelitis is generally segmented into hematogenous seeding secondary to direct inoculation and secondary to the spread of contiguous infection, according to the etiology (2, 3). The characteristics of osteomyelitis include progressive destruction of bone tissue and the formation of necrotic bone (4). Although great progress has been achieved in clinical interventions for osteomyelitis, it remains a huge challenge for orthopedists to cure it. Considering that osteoblasts serve as pivotal regulators in the physiological and pathological processes of bone formation and resorption (5), discerning the latent mechanism of osteoblast formation is necessary to improve the treatment of osteomyelitis.
Human bone mesenchymal stem cells (hBMSCs) are distinguished by their capability to self-renew, proliferate, and differentiate. A multitude of research has shown that hBMSCs play a critical role in the process of bone formation, because the osteogenic differentiation capacity of hBMSCs maintains the homeostasis of osteonecrosis and bone regeneration (6–8). Staphylococcal protein A (SpA) is a vital virulence factor of Staphylococcus aureus, which is the most ubiquitous microorganism found in osteomyelitis (9, 10). As SpA has been proven to impede the osteogenic differentiation capacity of hBMSCs, in order to elucidate the mechanism underlying osteomyelitis (11), we used SpA-treated hBMSCs to mimic osteomyelitis in vitro.
Long noncoding RNAs (lncRNAs) are a subset of non-protein-coding RNA transcripts with lengths of over 200 nucleotides (12). Mounting evidence has demonstrated that lncRNAs are involved in the development and evolution of numerous diseases by regulating the expression of different target genes through diverse mechanisms (13, 14). Many investigations have proven that lncRNAs play key roles in a wide range of biological processes, such as cell proliferation, metastasis, differentiation, and chemoresistance (15–17). Moreover, the function of lncRNAs in osteogenic differentiation of hBMSCs has been widely reported. For instance, the lncRNA MEG3 restrains the osteogenic differentiation of bone marrow mesenchymal stem cells from postmenopausal osteoporosis via modulation of microRNA 133a-3p (miR-133a-3p) (18). The lncRNA LINC00707 upregulates WNT2B to promote osteogenesis of hBMSCs by sponging miR-370-3p (19). The lncRNA NEAT1/miR-29b-3p/BMP1 pathway contributes to osteogenic differentiation in hBMSCs (20). Nevertheless, explorations into the exact role and regulatory mechanism of lncRNAs in osteomyelitis have come to a halt.
In the current study, our findings validated the notion that SpA repressed osteogenic differentiation of hBMSCs in a dose-dependent manner, and FAM83H-AS1 was discovered to be significantly downregulated in SpA-treated hBMSCs through lncRNA microarray assay. Further experiments revealed that FAM83H-AS1 ameliorated SpA-suppressed osteogenic differentiation of hBMSCs through targeting of the miR-541-3p/WNT3A pathway.
RESULTS
FAM83H-AS1 was downregulated in SpA-induced hBMSCs.
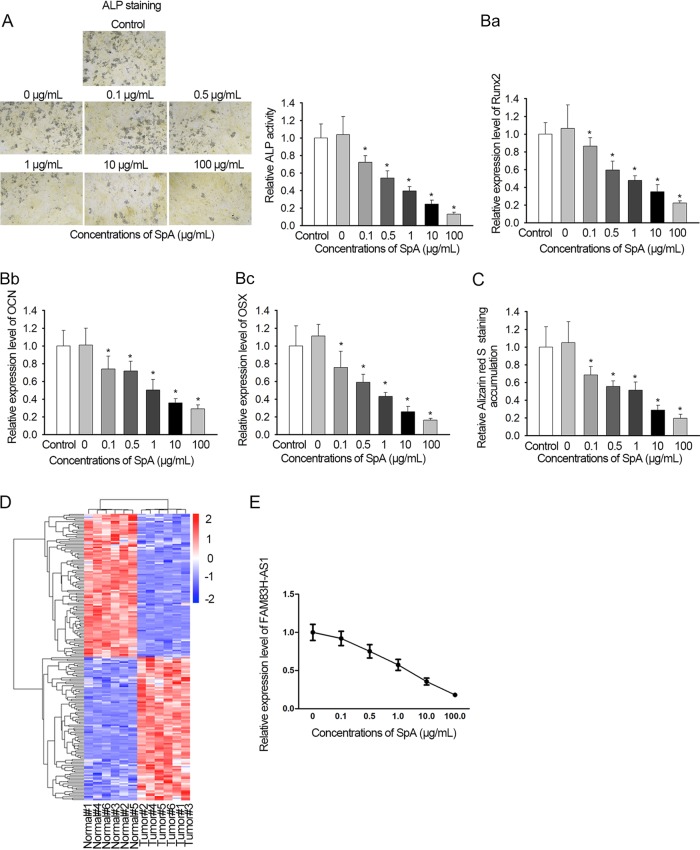
In order to validate the effect of SpA on osteogenic differentiation, hBMSCs were treated with different concentrations (0, 0.1, 0.5, 1, 10, or 100 μg/ml) of SpA during the induction of osteogenic differentiation. It was shown by ALP activity assay that SpA treatment led to suppression of ALP activity in a dose-dependent manner (Fig. 1A). The activity was lowered more than 80% at 100 μg/ml for 24 h. The expression levels of osteogenic markers (Runx2, OCN, and OSX) were also found to be gradually decreased in SpA-treated hBMSCs (Fig. 1B). Likewise, alizarin red S staining assays indicated that SpA-treated hBMSCs presented fewer mineralized nodules than untreated cells, and the degree of osteogenic differentiation inhibition rose with increasing SpA concentrations (Fig. 1C). To identify the expression profile of lncRNAs in SpA-treated hBMSCs, a lncRNA microarray assay was conducted by utilization of hBMSCs that had undergone 2 weeks’ treatment with osteogenic differentiation medium containing SpA or not. The lncRNA FAM83H-AS1 was found to exhibit the lowest expression among the significantly downregulated lncRNAs (Fig. 1D). Furthermore, the level of FAM83H-AS1 gradually became lower due to the augmentation of the SpA concentration (Fig. 1E). In a word, these results showed that SpA restrained hBMSC osteogenic differentiation, and FAM83H-AS1 was weakly expressed in hBMSCs after SpA treatment.
FIG 1.
FAM83H-AS1 was downregulated in SpA-induced hBMSCs. To stimulate osteomyelitis in vitro, hBMSCs were induced by SpA during osteogenic differentiation induction. (A and B) ALP activity detection (A) and qRT-PCR results of the levels of osteogenic markers (B) in SpA-treated hBMSCs. Magnification, ×100. (C) An alizarin red S staining assay was performed to assess the osteogenic differentiation ability of hBMSCs. Data were taken with a microscope at a magnification of ×100. (D) Microarray analysis was employed to expose differentially expressed lncRNAs in normal and induced hBMSCs. Osteogenic differentiation of hBMSCs was assessed. Scale numbers are log2 fold changes in lncRNA expression. (E) Effects of SpA treatment on FAM83H-AS1 expression in hBMSCs measured by qRT-PCR. *, P < 0.05. The error bars indicate SD.
Overexpression of FAM83H-AS1 ameliorated SpA-mediated inhibition of osteogenic differentiation of hBMSCs.
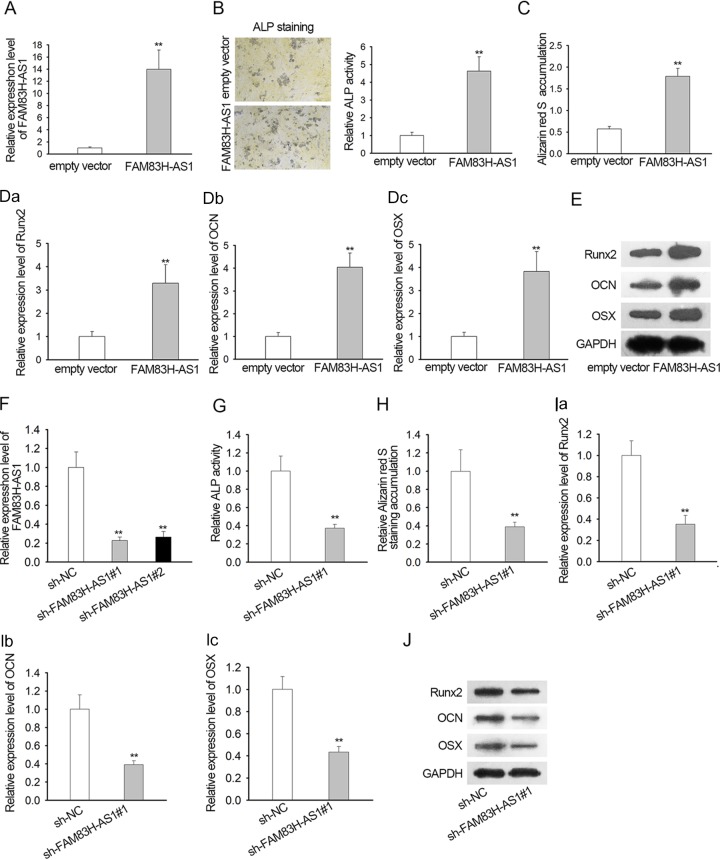
To explore the exact function of FAM83H-AS1 in SpA-regulated osteogenic differentiation in hBMSCs, FAM83H-AS1 was overexpressed in SpA-treated hBMSCs through transfection with the pcDNA3.1/FAM83H-AS1 vector (Fig. 2A). ALP activity detection demonstrated that upregulation of FAM83H-AS1 notably fortified the ALP activity of hBMSCs with SpA treatment (Fig. 2B). Meanwhile, the results of alizarin red S staining revealed that ectopic expression of FAM83H-AS1 contributed to the striking increase of calcium mineralization in SpA-treated hBMSCs (Fig. 2C). In agreement with the described findings, we observed that expression of the osteogenic markers Runx2, OCN, and OSX was enhanced in such hBMSCs due to FAM83H-AS1 overexpression (Fig. 2D). Western blotting also confirmed that the levels of Runx2, OCN, and OSX proteins showed the same trend described above when FAM83H-AS1 was upregulated in hBMSCs under SpA treatment (Fig. 2E). Moreover, we also conducted experiments by silencing FAM83H-AS1 in the indicated cells. The results showed that silencing of FAM83H-AS1 in hBMSCs decreased ALP activity and calcium mineralization (Fig. 2F to H). Meanwhile, the levels of Runx2, OCN, and OSX were greatly decreased after knockdown of FAM83H-AS1 (Fig. 2I and J). Altogether, forced expression of FAM83H-AS1 accelerated osteogenic differentiation in hBMSCs, along with SpA treatment.
FIG 2.
Overexpression of FAM83H-AS1 ameliorated SpA-mediated osteogenic differentiation inhibition in hBMSCs. (A) Effectiveness of transfection for FAM83H-AS1 detected by qRT-PCR analysis. (B) After transfection, ALP activity detection was conducted to examine the ALP activity of SpA-treated hBMSCs. Magnification, ×100. (C) The osteoblastic differentiation of hBMSCs under SpA treatment was also estimated by alizarin red S staining assay. Data were taken with a microscope at a magnification of ×100. (D) Expression of Runx2, OCN, and OSX tested by qRT-PCR. (E) Western blotting was performed to assess the protein levels of Runx2, OCN, and OSX. (F) Silencing of FAM83H-AS1 by specific shRNAs efficiently decreased the level of FAM83H-AS1 and was confirmed by qRT-PCR. (G) ALP activity in response to FAM83H-AS1 silence. (H) An alizarin red S staining assay was performed to detect the osteogenic differentiation ability of hBMSCs after FAM83H-AS1 knockdown. Data were taken with a microscope at a magnification of ×100. (I and J) mRNA and protein levels of osteogenic markers in response to silenced FAM83H-AS1. **, P < 0.01. The error bars indicate SD.
FAM83H-AS1 interacted with miR-541-3p.
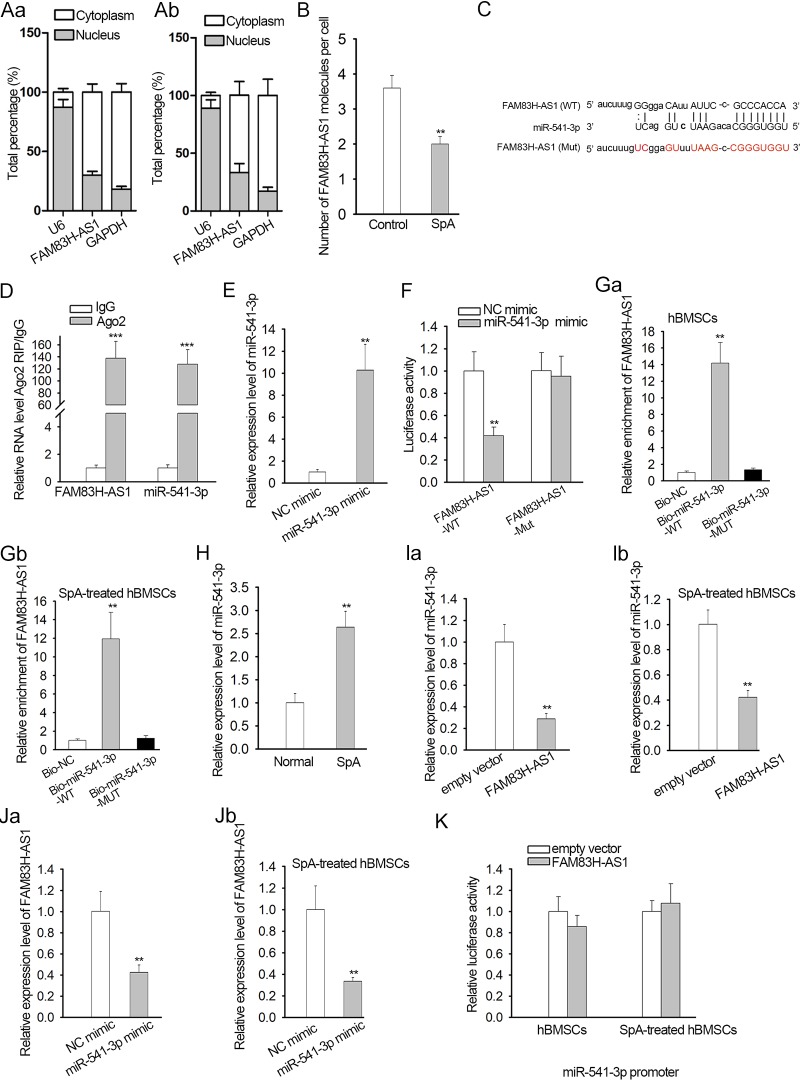
To gain a deeper understanding of the potential molecular mechanism governing FAM83H-AS1 in osteogenic differentiation, a subcellular fractionation assay was performed to identify the cellular localization of FAM83H-AS1 in control and SpA-treated hBMSCs. Cytoplasmic localization was identified for FAM83H-AS1 (Fig. 3A). The copy number of FAM83H-AS1 in control and SpA-treated hBMSCs was also determined and was shown to have dramatically declined under SpA treatment (Fig. 3B). Then, we implemented bioinformatics analysis using the DIANA database (http://diana.imis.athena-innovation.gr/DianaTools/index.php) and found that miR-541-3p possessed the predicted binding sites with FAM83H-AS1 (Fig. 3C). RNA immunoprecipitation (RIP) experiments showed that both FAM83H-AS1 and miR-541-3p were abundantly captured in compounds precipitated by Argonaute 2 (Ago2) antibody compared to IgG precipitates (Fig. 3D). Additionally, miR-541-3p was overexpressed in hBMSCs after transfection with an miR-541-3p mimic, and then it was disclosed that the luciferase activity of wild-type FAM83H-AS1 (FAM83H-AS1-WT) was lessened by upregulation of miR-541-3p, whereas no significant differences were noted in the luciferase activity of FAM83H-AS1-Mut (see Materials and Methods) (Fig. 3E and F). RNA pulldown assays further demonstrated the interaction between miR-541-3p and FAM83H-AS1 in both control hBMSCs and SpA-treated hBMSCs (Fig. 3G). We also examined the expression levels of miR-541-3p in cells treated or not treated with SpA. The level of miR-541-3p was obviously greatly enhanced in cells treated with SpA (Fig. 3H). Quantitative real-time (qRT)-PCR analysis suggested that overexpression of miR-541-3p resulted in the overt reduction of FAM83H-AS1 expression in both normal hBMSCs and SpA-treated hBMSCs, and overexpression of FAM83H-AS1 also reduced miR-541-3p expression (Fig. 3I and J). Since FAM83H-AS1 could regulate the expression level of miR-541-3p, we performed a luciferase reporter assay to prove the potential effect of FAM83H-AS1 on the activity of the miR-541-3p promoter. No significant effect of FAM83H-AS1 overexpression was found on the luciferase activity of the miR-541-3p promoter vector (Fig. 3K). Based on the above-mentioned findings, we concluded that FAM83H-AS1 worked as a sponge for miR-541-3p.
FIG 3.
FAM83H-AS1 interacted with miR-541-3p. (A) A subcellular fractionation assay was used to identify the localization of FAM83H-AS1 in control and SpA-treated hBMSCs. (B) Copy numbers of FAM83H-AS1 in control and SpA-treated hBMSCs. (C) Predicted binding sites of FAM83H-AS1 for miR-541-3p. Uppercase letters, binding bases; lowercase letters, bases that could not pair. Black letters, wild type; red letters, mutant. The lines indicate base pairing, while the dots mean that certain bases cannot pair. (D) A RIP experiment was conducted to evaluate the relationship between FAM83H-AS1 and miR-541-3p. (E) The overexpression efficiency of miR-541-3p was verified by qRT-PCR. (F) The interaction of FAM83H-AS1 with miR-541-3p was confirmed by luciferase reporter assay. (G) An RNA-RNA pulldown assay indicated the enrichment of FAM83H-AS1 in response to the biotin-labeled negative control (Bio-NC), biotin-labeled wild-type miR-541-3p (Bio-miR-541-3p-WT), or biotin-labeled mutant miR-541-3p (Bio-miR-541-3p-MUT). (H) The level of miR-541-3p in control or SpA-treated hBMSCs was analyzed by qRT-PCR. (I and J) The expression of FAM83H-AS1 and miR-541-3p in the indicated hBMSCs was assessed by qRT-PCR. (K) Luciferase reporter assays in the indicated hBMSCs were conducted to assess the promoter activity of miR-541-3p in response to the upregulation of FAM83H-AS1. **, P < 0.01; ***, P < 0.001. The error bars indicate SD.
FAM83H-AS1 regulated WNT3A expression by sponging miR-541-3p.
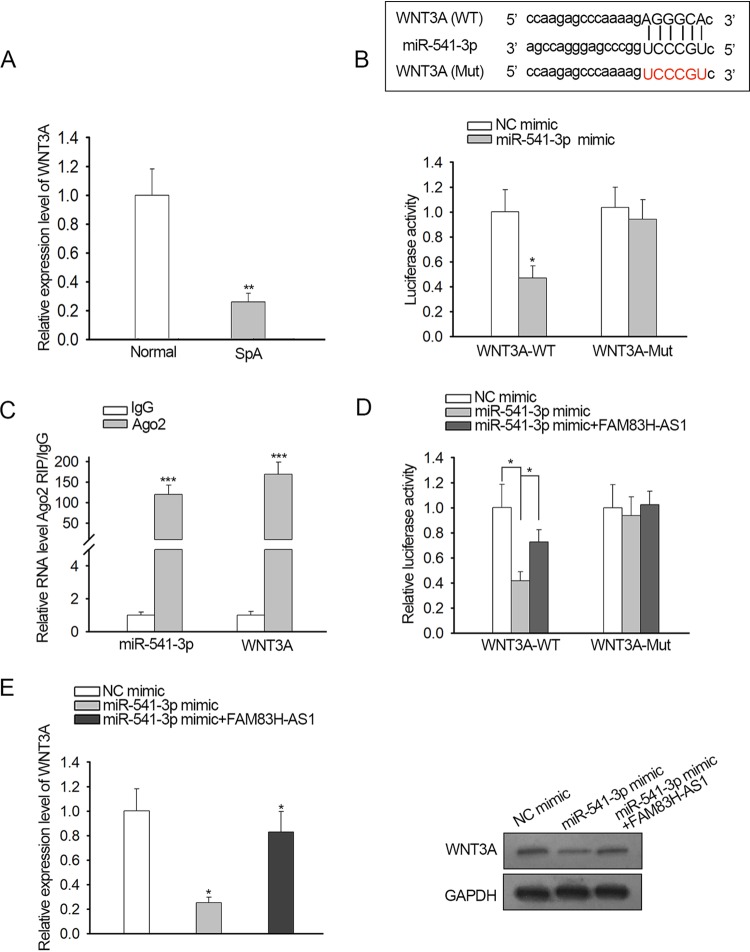
Previous research showed that WNT3A had a promoting function in hBMSC osteogenic differentiation, and we discovered that the expression of WNT3A was downregulated in SpA-treated hBMSCs (Fig. 4A). Browsing the starBase website (http://starbase.sysu.edu.cn/) revealed that WNT3A exhibited putative binding affinity with miR-541-3p (Fig. 4Ba). Therefore, we intended to probe whether the WNT3A gene was the downstream target gene of miR-541-3p. Luciferase reporter assay illustrated that the miR-541-3p mimic impaired only the luciferase activity of WNT3A-WT, manifesting the targeted relationship between miR-541-3p and WNT3A (Fig. 4Bb). RIP assay further validated the notion that miR-541-3p and WNT3A were enriched by the Ago antibody (Fig. 4C). In addition, our data showed that the decreased luciferase activity of WNT3A-WT induced by upregulation of miR-541-3p was reversed by ectopic expression of FAM83H-AS1 (Fig. 4D). Consistently, qRT-PCR and Western blot assays testified that miR-541-3p overexpression weakened the mRNA and protein expression of WNT3A and that upregulation of FAM83H-AS1 abolished the inhibitory effects of miR-541-3p (Fig. 4E). Overall, these findings provided strong evidence that FAM83H-AS1 positively modulated miR-541-3p-targeted WNT3A.
FIG 4.
FAM83H-AS1 regulated WNT3A expression by sponging miR-541-3p. (A) WNT3A expression in response to SpA treatment was detected by qRT-PCR. (B, top) miR-541-3p binding sites in the 3′ UTR of WNT3A. (Bottom) A luciferase reporter assay was carried out to certify the association between miR-541-3p and WNT3A in SpA-treated hBMSCs. (C) RIP assay further validated that the WNT3A gene was a target gene of miR-541-3p in SpA-treated hBMSCs. (D and E) The impacts of FAM83H-AS1 and miR-541-3p on WNT3A in SpA-treated hBMSCs were analyzed by luciferase reporter assay, qRT-PCR, and Western blotting. *, P < 0.05; **, P < 0.01; ***, P < 0.001. The error bars indicate SD.
FAM83H-AS1 improved SpA-repressed osteogenic differentiation of hBMSCs via the miR-541-3p/WNT3A pathway.
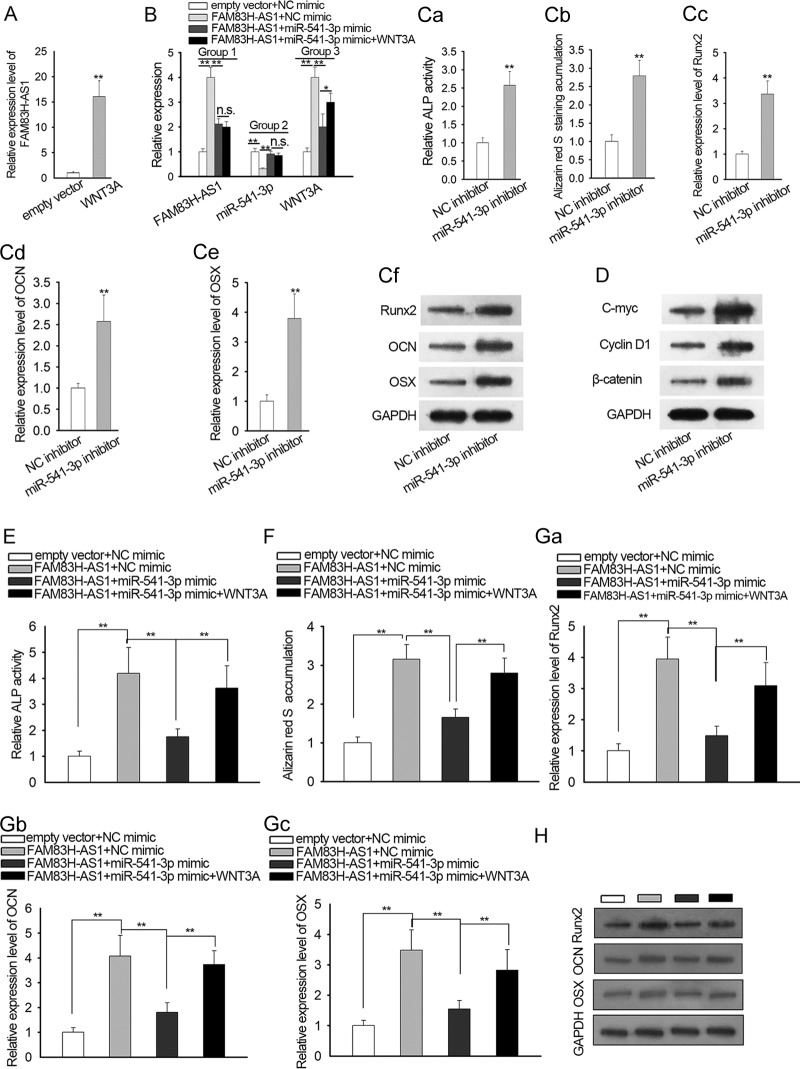
Rescue experiments were performed to explore whether FAM83H-AS1 played its role in SpA-mediated osteogenic differentiation inhibition of hBMSCs by regulation of the miR-541-3p/WNT3A axis. qRT-PCR analysis verified that WNT3A was highly expressed in SpA-treated hBMSCs after transfection (Fig. 5A). The expression levels of FAM83H-AS1, miR-541-3p, and WNT3A were separately assessed in transfected hBMSCs. The level of FAM83H-AS1 was upregulated in cells transfected with the FAM83H-AS1 expression vector, which was partially recovered after cotransfection with the miR-541-3p mimic. However, cotransfection with WNT3A had no significant effect on the level of FAM83H-AS1 (Fig. 5B, Group 1). On the other hand, downregulation of miR-541-3p induced by the upregulation of FAM83H-AS1 was recovered by the miR-541-3p mimic, while it was not affected by the overexpression of WNT3A (Fig. 5B, Group 2). As for WNT3A, its level was increased by upregulated FAM83H-AS1 and was rescued by supplementing with the miR-541-3p mimic. The reduction of WNT3A caused by the miR-541-3p mimic was further restored by overexpression of WNT3A (Fig. 5B, Group 3).
FIG 5.
FAM83H-AS1 improved SpA-repressed osteogenic differentiation of hBMSCs via the miR-541-3p/WNT3A pathway. (A) Overexpression of WNT3A in SpA-treated hBMSCs was certified by qRT-PCR analysis. (B) The expression levels of FAM83H-AS1, miR-541-3p, and WNT3A were separately examined in the indicated hBMSCs after transfection. (C) The effect of miR-541-3p inhibitor on osteogenic differentiation was evaluated via ALP staining (data were taken with a microscope at a magnification of ×100), alizarin red S staining (data were taken with a microscope at a magnification of ×100), and qRT-PCR and Western blot results for three osteogenic markers (Runx2, OCN, and OSX). (D) Western blot result for proteins associated with the Wnt/β-catenin signaling pathway in transfected SpA-treated hBMSCs. (E and F) The function of the FAM83H-AS1/miR-541-3p/WNT3A axis in osteogenic differentiation of SpA-treated hBMSCs was assessed by ALP activity detection and alizarin red S staining assay. Data were taken with a microscope at a magnification of ×100. (G and H) qRT-PCR assay and Western blotting were employed to examine the mRNA and protein expression of Runx2, OCN, and OSX in SpA-treated hBMSCs. *, P < 0.05; **, P < 0.01; n.s., not significant. The error bars indicate SD.
miR-542-3p inhibitor was used for subsequent experiments. Intriguingly, downregulation of miR-541-3p had the same effect on osteogenic differentiation as the overexpression of FAM83H-AS1 (Fig. 5C). We also evaluated whether inhibition of miR-541-3p had an effect on the levels of wnt/β-catenin pathway factors. The protein levels of the product of c-myc, cyclin D1, and β-catenin were enhanced after suppression of miR-541-3p expression (Fig. 5D). Data from ALP activity detection showed that the enhanced ALP activity caused by FAM83H-AS1 overexpression was diminished by upregulation of miR-541-3p and then recovered by forced expression of WNT3A (Fig. 5E). Similarly, alizarin red S staining revealed that the number of mineralized nodules, elevated by FAM83H-AS1 upregulation, was decreased by the miR-541-3p mimic, and the rebound of mineralization occurred with ectopic expression of WNT3A (Fig. 5F). Moreover, our results showed that the miR-541-3p mimic reduced the mRNA and protein levels of Runx2, OCN, and OSX promoted by FAM83H-AS1 overexpression and that upregulation of WNT3A abrogated the suppressive impacts of the miR-541-3p mimic on mRNA and protein expression of Runx2, OCN, and OSX in SpA-induced hBMSCs (Fig. 5G and H). To sum up, FAM83H-AS1/miR-541-3p/WNT3A facilitated osteogenic differentiation in SpA-treated hBMSCs.
DISCUSSION
Osteomyelitis is a bone disease arising from invading organisms, leading to progressive bone destruction and the formation of sequestra (21, 22). hBMSCs are of paramount significance in the process of bone formation, owing to their osteogenic differentiation ability. However, the molecular mechanisms underlying osteomyelitis in regard to hBMSC osteoblastic differentiation largely still need to be further investigated. SpA is considered a core virulence contributor of S. aureus, the most common microbe in osteomyelitis, and has been affirmed to trigger the osteoblastic differentiation inhibition of hBMSCs (11). As a result, in the present study, we adopted SpA treatment to simulate osteomyelitis in vitro.
Increasing evidence has demonstrated that lncRNAs are implicated in the occurrence and progression of various diseases, including osteoporosis and osteomyelitis (11, 23, 24). lncRNAs have been established as serving a crucial role in osteogenic differentiation. For example, the lncRNA KCNQ1OT1 promotes osteogenic differentiation by sponging miRNA-214 to regulate BMP2 expression (25). A novel lncRNA, PGC1β-OT1, suppresses miR-148a-3p to regulate adipocyte and osteoblast differentiation (26). The long noncoding RNA XIST inhibits the differentiation of BMSCs to accelerate osteoporosis (27). In order to reveal the function of lncRNAs in osteomyelitis, we carried out microarray analysis to detect the expression pattern of lncRNAs in osteomyelitis by using hBMSCs treated or not with SpA during the induction of osteogenic differentiation. Our findings showed that FAM83H-AS1 was the most differentially expressed lncRNA in SpA-treated hBMSCs and that the level of FAM83H-AS1 gradually dropped with an increase in the SpA concentration. Additionally, overexpression of FAM83H-AS1 promoted osteogenic differentiation of SpA-treated hBMSCs.
MicroRNAs (miRNAs) are a kind of highly conserved small noncoding RNA molecule 18 to 22 nucleotides in length (28). Increasing investigations have corroborated that miRNAs posttranscriptionally modulate target gene expression involving a wide spectrum of cellular activities by combining with the 3′ untranslated region (3′ UTR) of the target mRNA (29). The participation of miRNAs in osteogenic differentiation of hBMSCs has been confirmed by accumulating research (30–32). Although miR-541-3p has been certified to act as a tumor suppressor in prostate cancer and non-small cell lung cancer (33, 34), the specific function of miR-541-3p in hBMSC osteogenic differentiation has not yet been probed. In this study, we found that there were putative binding sites between FAM83H-AS1 and miR-541-3p. Functionally, it was shown that FAM83H-AS1 negatively regulated miR-541-3p. Moreover, our current study investigated whether the expression level of miR-541-3p could be negatively regulated by FAM83H-AS1 in hBMSCs. Through luciferase reporter assays, we determined that FAM83H-AS1 could not regulate the transcription of miR-541-3p. Therefore, we will try to investigate the mechanism by which miR-541-3p is regulated by FAM83H-AS1 in future studies.
WNT3A is an important member of the Wnt signaling pathway, participating in the regulation of embryonic development, cell proliferation, apoptosis, metastasis, and differentiation (35, 36). A growing body of studies has illustrated that WNT3A functions as a pivotal regulatory factor in the osteogenic differentiation capacity of hBMSCs (37, 38). Our findings suggested that weakly expressed WNT3A in SpA-induced hBMSCs was a downstream target of miR-541-3p. Furthermore, FAM83H-AS1 boosted WNT3A expression by competitively binding to miR-541-3p. Lastly, the results of rescue experiments revealed that FAM83H-AS1 ameliorated SpA-repressed osteogenic differentiation of hBMSCs through regulation of miR-541-3p/WNT3A. Lack of clinical samples and in vivo experimental data are two shortcomings of our current study. We decided to investigate the role of FAM83H-AS1 in clinical samples or animal studies in the future. However, our current study still provided comprehensive elucidation of the mechanism governing FAM83H-AS1 in SpA-repressed osteogenic differentiation and showed FAM83H-AS1 to be a promising therapeutic target for osteomyelitis. Finding a novel molecular pathway that affected osteogenic differentiation may also contribute to avoiding failed therapeutic outcomes.
MATERIALS AND METHODS
Cell culture and osteogenic differentiation.
hBMSCs acquired from Lonza (Walkersville, MD) were maintained in mesenchymal stem cell growth medium (MSCGM; BulletKit; Lonza) a moist atmosphere of 5% CO2 at 37°C. Until cell confluence at 80 to 90%, hBMSCs were grown in mesenchymal stem cell osteogenic differentiation medium (hMSC; BulletKit; Lonza). The osteogenic differentiation medium was refreshed every 3 days.
SpA treatment.
hBMSCs were treated with different concentrations of SpA (0, 0.1, 0.5, 1, 10, or 100 μg/ml) to construct a cell model of osteomyelitis. Briefly, hBMSCs were plated in 96-well plates and cultivated in osteogenic differentiation medium supplemented with different concentrations of SpA for 2 weeks. For lncRNA microarray analysis, hBMSCs were inoculated at a density of 2 × 106 cells/well in 6-well plates and incubated in osteogenic differentiation medium with or without 10 μg/ml SpA for 2 weeks.
lncRNA microarray analysis.
Total RNA from hBMSCs treated or not with SpA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA) as recommended by the manufacturer. The integrity of the RNA was determined with an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Afterward, reverse transcription was performed, and then double-stranded cDNA was synthesized into cRNA. The Cy3-dCTP-labeled cRNAs were hybridized onto a 4,000-by-180,000 human lncRNA microarray V4.0 (Agilent Technologies). Subsequently, the hybridized array mixtures were washed and scanned with a microarray scanner (Agilent Technologies). Feature Extraction software (Agilent Technologies) and GeneSpring software V13.0 (Agilent Technologies) were applied to complete the data processing. Differentially expressed lncRNAs were identified when the fold change was >2.0 and the P value was <0.05.
Cell transfection.
For overexpression of FAM83H-AS1 or WNT3A, the full-length of FAM83H-AS1 or WNT3A was ligated into pcDNA3.1 vectors procured from GenePhama Co. Ltd. (Shanghai, China), with the empty vector as a negative control. Short hairpin RNAs (shRNAs) targeting FAM83H-AS1 (sh-FAM83H-AS1 no. 1 and 2) were synthesized by GenePhama for silencing of FAM83H-AS1, and nontargeted shRNA (sh-NC) was used as a negative control. In addition, miR-541-3p mimic/inhibitor and negative control (NC) mimic/inhibitor were also purchased from GenePhama Co. Ltd. Cell transfection was carried out with Lipofectamine 3000 (Invitrogen) according to the vender’s recommendations. The shRNAs, MiR-541-3p mimic, or NC mimic was used at a final concentration of 20 nM. At 48 h after transfection, cells were collected for subsequent use.
qRT-PCR.
Total RNA extraction was implemented with TRIzol reagent (Invitrogen), and cDNA was generated using an iScript cDNA synthesis kit (Bio-Rad, Shanghai, China), in line with the manufacturer’s protocol. qRT-PCR was carried out with iQTM SYBR green supermix (Bio-Rad, China) on a 7500 HT fast real-time PCR system (Applied Biosystems, Foster City, CA). The relative gene expression was quantified using the 2−ΔΔCT method with U6 small nuclear RNA and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as internal normalization controls.
Western blotting.
RIPA buffer (Beyotime Biotechnology, Nantong, China) was applied to lyse hBMSCs, and the total protein concentration was detected with a bicinchoninic acid (BCA) protein assay kit (Beyotime Biotechnology). Total protein was loaded on SDS-10% PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA). Then, the membranes were sealed in 5% nonfat dry milk, followed by incubation with primary antibodies overnight at 4°C; probed with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature; and visualized with an ECL detection kit (Thermo Fisher Scientific, Rockford, IL). The primary antibodies against WNT3A, Runx2, OCN, OSX, and GAPDH were all obtained from Cell Signaling Technologies (Beverly, MA), and GAPDH served as the loading control. Goat anti-rabbit IgG (H+L) (horseradish peroxidase [HRP] preadsorbed) (ab7090), obtained from Abcam (United Kingdom), was used as a secondary antibody.
ALP activity detection.
An ALP activity assay was performed utilizing an ALP detection kit acquired from Nanjing Jiancheng Bioengineering Institute (Nanjing, China) according to the manufacturer’s directions. The total protein purity of each sample was verified with a BCA kit (Beyotime Biotechnology). Relative ALP activity was normalized to the total protein content.
Alizarin red S staining.
Cell mineralization post-osteogenic differentiation was estimated by alizarin red S staining assay. After different treatments, hBMSCs were seeded on 12-well plates, fixed with 4% paraformaldehyde for 20 min, and treated with 1% alizarin red S solution (Sigma) for 10 min. Thereafter, the cells were washed twice with phosphate-buffered saline (PBS) and photographed with an optical microscope (Olympus, Tokyo, Japan).
Luciferase reporter assay.
The segments of wild-type FAM83H-AS1 or WNT3A containing putative miR-541-3p binding sites were cloned into the luciferase reporter vector pmirGLO (Promega, Madison, WI) to synthesize FAM83H-AS1-WT or WNT3A-WT plasmids. To construct FAM83H-AS1-Mut or WNT3A-Mut vectors, the miR-541-3p binding sites were mutated. Then, cells were cotransfected with the indicated plasmids and the miR-541-3p mimic or NC mimic utilizing Lipofectamine 3000 (Invitrogen), in accordance with the product manuals. Forty-eight hours later, the luciferase activity of the collected cells was examined with the Dual-Luciferase reporter assay system (Promega).
RNA immunoprecipitation.
The RIP assay was implemented using the EZMagna RIP kit (Millipore) according to the supplier’s protocol. In short, RIP lysis buffer was adopted to harvest cell lysates of hBMSCs, and subsequently, cell extracts were incubated with magnetic beads and anti-Ago2 antibody or negative control anti-IgG antibody. After removing the proteins from the beads, precipitated RNA was purified and determined by qRT-PCR analysis.
Statistical analysis.
Experimental results are presented as means and standard deviations (SD) from three independent assays, and statistical analyses were conducted with SPSS 19.0 software (IBM, Chicago, IL). Comparisons between different groups were analyzed by Student's t test or one-way analysis of variance (ANOVA). A P value of <0.05 was identified as statistically significant.
ACKNOWLEDGMENTS
We acknowledge all support from the participants in this research.
We confirm that we have no conflicts of interest involved in this research.
REFERENCES
- 1.Fantoni M, Taccari F, Giovannenze F. 2019. Systemic antibiotic treatment of chronic osteomyelitis in adults. Eur Rev Med Pharmacol Sci 23:258–270. doi: 10.26355/eurrev_201904_17500. [DOI] [PubMed] [Google Scholar]
- 2.Spellberg B, Lipsky BA. 2012. Systemic antibiotic therapy for chronic osteomyelitis in adults. Clin Infect Dis 54:393–407. doi: 10.1093/cid/cir842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Graaf H, Sukhtankar P, Arch B, Ahmad N, Lees A, Bennett A, Spowart C, Hickey H, Jeanes A, Armon K, Riordan A, Herberg J, Hackett S, Gamble C, Shingadia D, Pallett A, Clarke SC, Henman P, Emonts M, Sharland M, Finn A, Pollard AJ, Powell C, Marsh P, Ballinger C, Williamson PR, Clarke NM, Faust SN. 2017. Duration of intravenous antibiotic therapy for children with acute osteomyelitis or septic arthritis: a feasibility study. Health Technol Assess 21:1–164. doi: 10.3310/hta21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maffulli N, Papalia R, Zampogna B, Torre G, Albo E, Denaro V. 2016. The management of osteomyelitis in the adult. Surgeon 14:345–360. doi: 10.1016/j.surge.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez CJ Jr, Ward CL, Romano DR, Hurtgen BJ, Hardy SK, Woodbury RL, Trevino AV, Rathbone CR, Wenke JC. 2013. Staphylococcus aureus biofilms decrease osteoblast viability, inhibits osteogenic differentiation, and increases bone resorption in vitro. BMC Musculoskelet Disord 14:187. doi: 10.1186/1471-2474-14-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JS, Lee JS, Roh HL, Kim CH, Jung JS, Suh KT. 2006. Alterations in the differentiation ability of mesenchymal stem cells in patients with nontraumatic osteonecrosis of the femoral head: comparative analysis according to the risk factor. J Orthop Res 24:604–609. doi: 10.1002/jor.20078. [DOI] [PubMed] [Google Scholar]
- 7.Glenn JD, Whartenby KA. 2014. Mesenchymal stem cells: emerging mechanisms of immunomodulation and therapy. World J Stem Cells 6:526–539. doi: 10.4252/wjsc.v6.i5.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei B, Wei W, Zhao B, Guo X, Liu S. 2017. Long non-coding RNA HOTAIR inhibits miR-17-5p to regulate osteogenic differentiation and proliferation in non-traumatic osteonecrosis of femoral head. PLoS One 12:e0169097. doi: 10.1371/journal.pone.0169097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Votintseva AA, Fung R, Miller RR, Knox K, Godwin H, Wyllie DH, Bowden R, Crook DW, Walker AS. 2014. Prevalence of Staphylococcus aureus protein A (Spa) mutants in the community and hospitals in Oxfordshire. BMC Microbiol 14:63. doi: 10.1186/1471-2180-14-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dayan GH, Mohamed N, Scully IL, Cooper D, Begier E, Eiden J, Jansen KU, Gurtman A, Anderson AS. 2016. Staphylococcus aureus: the current state of disease, pathophysiology and strategies for prevention. Expert Rev Vaccines 15:1373–1392. doi: 10.1080/14760584.2016.1179583. [DOI] [PubMed] [Google Scholar]
- 11.Cui Y, Lu S, Tan H, Li J, Zhu M, Xu Y. 2016. Silencing of long non-coding RNA NONHSAT009968 ameliorates the staphylococcal protein A-inhibited osteogenic differentiation in human bone mesenchymal stem cells. Cell Physiol Biochem 39:1347–1359. doi: 10.1159/000447839. [DOI] [PubMed] [Google Scholar]
- 12.Hung T, Chang HY. 2010. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol 7:582–585. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batista PJ, Chang HY. 2013. Long noncoding RNAs: cellular address codes in development and disease. Cell 152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinn JJ, Chang HY. 2016. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 15.Zhu L, Zhu Y, Han S, Chen M, Song P, Dai D, Xu W, Jiang T, Feng L, Shin VY, Wang X, Jin H. 2019. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis 10:383. doi: 10.1038/s41419-019-1585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Du M, Wang T, Chen W, Wu J, Li Q, Tian X, Qian L, Wang Y, Peng F, Fei Q, Chen J, He X, Yin L. 2019. Long non-coding RNA LINC01133 mediates nasopharyngeal carcinoma tumorigenesis by binding to YBX1. Am J Cancer Res 9:779–790. [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Yan J, Liu Q, Xie Z, Jiang H. 2019. LncRNA Rik-203 contributes to anesthesia neurotoxicity via microRNA-101a-3p and GSK-3β-mediated neural differentiation. Sci Rep 9:6822. doi: 10.1038/s41598-019-42991-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Li Y, Zhang Y, Ma L, Lin L, Meng J, Jiang L, Wang L, Zhou P, Zhang Y. 2017. LncRNA MEG3 inhibited osteogenic differentiation of bone marrow mesenchymal stem cells from postmenopausal osteoporosis by targeting miR-133a-3p. Biomed Pharmacother 89:1178–1186. doi: 10.1016/j.biopha.2017.02.090. [DOI] [PubMed] [Google Scholar]
- 19.Jia B, Wang Z, Sun X, Chen J, Zhao J, Qiu X. 2019. Long noncoding RNA LINC00707 sponges miR-370-3p to promote osteogenesis of human bone marrow-derived mesenchymal stem cells through upregulating WNT2B. Stem Cell Res Ther 10:67. doi: 10.1186/s13287-019-1161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Chen B, Li D, Zhou X, Chen Z. 2019. LncRNA NEAT1/miR-29b-3p/BMP1 axis promotes osteogenic differentiation in human bone marrow-derived mesenchymal stem cells. Pathol Res Pract 215:525–531. doi: 10.1016/j.prp.2018.12.034. [DOI] [PubMed] [Google Scholar]
- 21.Garzoni C, Kelley WL. 2009. Staphylococcus aureus: new evidence for intracellular persistence. Trends Microbiol 17:59–65. doi: 10.1016/j.tim.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Lew DP, Waldvogel FA. 2004. Osteomyelitis. Lancet 364:369–379. doi: 10.1016/S0140-6736(04)16727-5. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Zhu HM, Xu HD, Zhang B, Huang SM. 2018. CRNDE impacts the proliferation of osteoclast by estrogen deficiency in postmenopausal osteoporosis. Eur Rev Med Pharmacol Sci 22:5815–5821. doi: 10.26355/eurrev_201809_15907. [DOI] [PubMed] [Google Scholar]
- 24.Mei B, Wang Y, Ye W, Huang H, Zhou Q, Chen Y, Niu Y, Zhang M, Huang Q. 2019. LncRNA ZBTB40-IT1 modulated by osteoporosis GWAS risk SNPs suppresses osteogenesis. Hum Genet 138:151–166. doi: 10.1007/s00439-019-01969-y. [DOI] [PubMed] [Google Scholar]
- 25.Wang CG, Liao Z, Xiao H, Liu H, Hu YH, Liao QD, Zhong D. 2019. LncRNA KCNQ1OT1 promoted BMP2 expression to regulate osteogenic differentiation by sponging miRNA-214. Exp Mol Pathol 107:77–84. doi: 10.1016/j.yexmp.2019.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Yuan H, Xu X, Feng X, Zhu E, Zhou J, Wang G, Tian L, Wang B. 2019. A novel long noncoding RNA PGC1beta-OT1 regulates adipocyte and osteoblast differentiation through antagonizing miR-148a-3p. Cell Death Differ 26:2029–2045. doi: 10.1038/s41418-019-0296-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X, Yang L, Ge D, Wang W, Yin Z, Yan J, Cao X, Jiang C, Zheng S, Liang B. 2019. Long non-coding RNA XIST promotes osteoporosis through inhibiting bone marrow mesenchymal stem cell differentiation. Exp Ther Med 17:803–811. doi: 10.3892/etm.2018.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 29.Gomes BC, Rueff J, Rodrigues AS. 2016. MicroRNAs and cancer drug resistance. Methods Mol Biol 1395:137–162. doi: 10.1007/978-1-4939-3347-1_9. [DOI] [PubMed] [Google Scholar]
- 30.Li T, Li H, Wang Y, Li T, Fan J, Xiao K, Zhao RC, Weng X. 2016. microRNA-23a inhibits osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by targeting LRP5. Int J Biochem Cell Biol 72:55–62. doi: 10.1016/j.biocel.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Long H, Sun B, Cheng L, Zhao S, Zhu Y, Zhao R, Zhu J. 2017. miR-139-5p represses BMSC osteogenesis via targeting Wnt/beta-catenin signaling pathway. DNA Cell Biol 36:715–724. doi: 10.1089/dna.2017.3657. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Cai J, Cai XH, Chen L. 2013. miR-346 regulates osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by targeting the Wnt/β-catenin pathway. PLoS One 8:e72266. doi: 10.1371/journal.pone.0072266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long B, Li N, Xu XX, Li XX, Xu XJ, Liu JY, Wu ZH. 2018. Long noncoding RNA LOXL1-AS1 regulates prostate cancer cell proliferation and cell cycle progression through miR-541-3p and CCND1. Biochem Biophys Res Commun 505:561–568. doi: 10.1016/j.bbrc.2018.09.160. [DOI] [PubMed] [Google Scholar]
- 34.Lu YJ, Liu RY, Hu K, Wang Y. 2016. MiR-541-3p reverses cancer progression by directly targeting TGIF2 in non-small cell lung cancer. Tumour Biol 37:12685–12695. doi: 10.1007/s13277-016-5241-5. [DOI] [PubMed] [Google Scholar]
- 35.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. 2004. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet 5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 36.Yu JM, Kim JH, Song GS, Jung JS. 2006. Increase in proliferation and differentiation of neural progenitor cells isolated from postnatal and adult mice brain by Wnt-3a and Wnt-5a. Mol Cell Biochem 288:17–28. doi: 10.1007/s11010-005-9113-3. [DOI] [PubMed] [Google Scholar]
- 37.Liang T, Gao W, Zhu L, Ren J, Yao H, Wang K, Shi D. 2019. TIMP-1 inhibits proliferation and osteogenic differentiation of hBMSCs through Wnt/β-catenin signaling. Biosci Rep 39:BSR20181290. doi: 10.1042/BSR20181290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo Y, Zhang Y, Miao G, Zhang Y, Liu Y, Huang Y. 2019. Runx1 regulates osteogenic differentiation of BMSCs by inhibiting adipogenesis through Wnt/beta-catenin pathway. Arch Oral Biol 97:176–184. doi: 10.1016/j.archoralbio.2018.10.028. [DOI] [PubMed] [Google Scholar]