TRIF is an essential adaptor for Toll-like receptor 3/4 (TLR3/4) signaling to activate transcription factor interferon regulatory factor 3 (IRF-3). We examined the molecular mechanism of TLR3 signaling and found that TLR3 stimulation by double-stranded RNA (dsRNA) induces phosphorylation of TRIF at Ser210 and is required for IRF-3 recruitment. TANK-binding kinase 1 (TBK1) is known to be responsible for IRF-3 phosphorylation and activation.
KEYWORDS: innate immunity, TRIF, IRF-3, TBK1, IKKβ
ABSTRACT
TRIF is an essential adaptor for Toll-like receptor 3/4 (TLR3/4) signaling to activate transcription factor interferon regulatory factor 3 (IRF-3). We examined the molecular mechanism of TLR3 signaling and found that TLR3 stimulation by double-stranded RNA (dsRNA) induces phosphorylation of TRIF at Ser210 and is required for IRF-3 recruitment. TANK-binding kinase 1 (TBK1) is known to be responsible for IRF-3 phosphorylation and activation. We found that TBK1 is also responsible for phosphorylation of Ser210 in TRIF. Unexpectedly, we discovered that IκB kinase β (IKKβ) plays an essential role in TLR3/4 signaling using a pharmacological inhibitor and gene deletion. Of note, IKKβ is essential in TLR3/4 but not in retinoic acid-inducible gene I (RIG-I) signaling. Mechanistically, IKKβ transiently associates with and induces the phosphorylation of TBK1 upon TLR3 stimulation. These results suggest a phosphorylation cascade of IKKβ and TBK1, where priming phosphorylation of TBK1 by IKKβ is required to surpass the threshold to induce signaling, thereby activating IRF-3.
INTRODUCTION
Invading microbial pathogens, such as viruses and bacteria, are sensed by the innate immune system by recognizing pathogen-associated molecular patterns (PAMPs) through pattern recognition receptors (PRRs). PRRs play an essential role in initiating innate immune responses (1–3). Toll-like receptors (TLRs) are one of the PRRs that recognize bacterial and viral PAMPs. Ten members of the TLR family have been identified thus far. For example, TLR3 recognizes viral double-stranded RNA (dsRNA), including the synthetic dsRNA poly(I·C), and TLR4 detects the bacterial cell wall component lipopolysaccharide (LPS) to induce the intracellular signaling pathways to activate genes encoding type I interferons (IFNs) and inflammatory cytokines (4, 5). An adaptor protein, Toll-IL-1 receptor (TIR) domain-containing adaptor-inducing IFN-β (TRIF; also called TICAM-1), plays an important role in signaling through TLR3 and TLR4 (6, 7). TLR4 also recruits myeloid differentiation primary response 88 (MyD88) to induce TRIF-independent signaling. TRIF activates TANK-binding kinase 1 (TBK1). Artificial overexpression of either TRIF or TBK1 is sufficient to induce downstream signals; however, no sudden increase in these molecules is observed in physiological signaling. Phosphorylation of TBK1 at Ser172 in the activation loop is essential for its catalytic activity. However, activated TBK1 undergoes autotransphosphorylation to be phosphorylated at multiple sites and become further activated (8–10). TBK1 is a physiological kinase needed to activate interferon regulatory factor 3 (IRF-3) through specific phosphorylation at Ser386. Activated IRF-3 forms a dimer and translocates into the nucleus to induce transcription of target genes by forming a holocomplex with CREB-binding protein (CBP) or p300 (11–14). On the other hand, TRIF activates the canonical IκB kinase (IKK) complex consisting of IKKα, IKKβ, and the regulatory subunit NF-κB essential modulator (NEMO; also called IKKγ). The IKKα/IKKβ/NEMO complex induces the phosphorylation and degradation of IκBα, leading to the activation of NF-κB, which is responsible for the induction of inflammatory cytokines (15, 16). TRIF consists of 3 domains: the N-terminal domain, the TIR domain, and the receptor-interacting protein homotypic-interacting motif (RHIM) domain in the C-terminal region. The N-terminal domain plays an essential role in activating the IRF-3 and NF-κB promoters, whereas the TIR domain participates in the interaction with TLR3 (17). The RHIM domain is important for recruiting kinase receptor-interacting protein 1 (RIP1), which mediates activation of both NF-κB and cell death pathways (18–21).
As for the other PRRs, replicating viral RNA in the cytosol is sensed by retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), such as RIG-I and melanoma differentiation-associated protein 5 (MDA5) (22, 23). For example, RIG-I detects dsRNA produced by Sendai virus replication. RLR-dependent signaling is mediated by the adaptor protein mitochondrial antiviral signaling (MAVS) (also called IPS-1, Cardif, or VISA) (24–27). Moreover, dsDNA in the cytoplasm is sensed by cyclic GMP-AMP (cGAMP) synthase (cGAS), which catalyzes the production of cGAMP to activate the adaptor protein stimulator of interferon genes (STING) (28–30). In summary, activated TRIF, MAVS, and STING transduce TBK1–IRF-3 and IKK–IκBα–NF-κB cascades.
Although TRIF, MAVS, and STING signals activate IRF-3, the physiological relevance of these signaling pathways in host protection from infection or danger signals is unknown. Recently, STING and MAVS were reported to be phosphorylated at a conserved motif (pLXIS motif, where S is the phosphorylation site; p is a hydrophobic amino acid, and X is any amino acid) upon their respective signaling, and their phosphorylation promotes their interaction with IRF-3 (31). TBK1 is responsible for the phosphorylation of MAVS and STING. However, the precise role of TRIF in TLR3 signaling is unknown.
We demonstrated that TLR3 stimulation by dsRNA induces the phosphorylation of TRIF at Ser210, which is within the pLXIS-like motif, and is essential for the interaction between TRIF and IRF-3. TBK1 may be responsible for phosphorylation of Ser210. Furthermore, we discovered that IKKβ plays an essential role in the recruitment of IRF-3 to TRIF and subsequent IRF-3 activation by TBK1. Of note, IKKβ is essential in TLR3 but not in RIG-I signaling. Mechanistically, IKKβ is transiently associated with and induces phosphorylation of TBK1 upon TLR3 stimulation. These results suggest a phosphorylation cascade of IKKβ and TBK1, where priming phosphorylation of TBK1 by IKKβ is required to surpass the threshold to induce signaling, thereby activating IRF-3.
RESULTS
Activation of TLR3 results in the formation of the TRIF/IRF-3 complex in a phosphorylation-dependent manner.
A previous study reported that phosphorylation of IRF-3 at Ser386 is essential for IRF-3 dimerization and nuclear translocation (14). We mutated Ser386 to alanine (S386A) to make a phosphorylation-resistant mutant to prevent its nuclear translocation. To examine the interaction of TRIF and IRF-3 in response to TLR3 stimulation, we used 293T cells transiently expressing TLR3 and wild-type (WT) IRF-3 or IRF-3 S386A (Fig. 1A). TLR3 stimulation by poly(I·C) treatment transiently induced the interaction of endogenous TRIF and IRF-3 S386A, which culminated after 2 h, whereas association between WT IRF-3 and TRIF was undetectable. Similar results were obtained with the human monocyte cell line THP-1, which stably expresses IRF-3 S386A, as well as endogenous TLR3 and TLR4. TLR3 stimulation by poly(I·C) and TLR4 stimulation by lipopolysaccharide (LPS) induced transient formation of TRIF/IRF-3 S386A (Fig. 1B and C). TRIF/IRF-3 S386A complexes were also detected by pulldown of TRIF in poly(I·C)-treated cells (Fig. 1D). Moreover, transient association of TBK1 with TRIF was detected at 0.5 h after poly(I·C) stimulation, significantly earlier than TRIF/IRF-3 S386A association. The overexpression of the N-terminal region of TRIF (amino acids 1 to 540, TRIF-N540) resulted in the activation of IRF-3, without inducing cell death due to lack of the C-terminal region (20). The complex TRIF-N540/IRF-3 S385A/S386A was reported to form in 293T cells by transient expression and to dissociate upon phosphatase treatment (31). To examine if phosphorylation is involved in the complex formed by poly(I·C) stimulation, the immunoprecipitate was treated with lambda protein phosphatase (PP) (Fig. 1E). The TRIF/IRF-3 S386A complex dissociated upon PP treatment, suggesting that phosphorylation of either component is necessary. To identify the phosphorylated molecule required for association, an in vitro binding assay was performed. TRIF-N540 and WT IRF-3 were expressed in 293T and Escherichia coli cells, respectively. TRIF-N540 was purified by anti-Flag pulldown, and the precipitate was treated or not treated with PP and then washed. The precipitates were further incubated with purified recombinant IRF-3, washed, and then analyzed (Fig. 1F). PP treatment changed the gel mobility of TRIF-N540, suggesting its phosphorylation in 293T cells. IRF-3 associated with TRIF-N540, whereas PP-treated TRIF-N540 did not. Collectively, these results suggest that TLR3 stimulation induces TRIF phosphorylation, inducing its transient association with IRF-3, in which the phosphorylation of TRIF is essential for the association.
FIG 1.
TRIF and IRF-3 form a phosphorylation-dependent complex in response to TLR3/4 stimulation. (A) 293T cells were transfected with expression constructs for Flag-tagged TLR3, together with HA-tagged WT IRF-3 or S386A. Twenty-four hours later, cells were stimulated with 50 μg/ml of poly(I·C) for the indicated times. Cell lysates were subjected to immunoprecipitation (IP) by anti-HA, followed by immunoblotting (IB). Total lysates were analyzed similarly (Input). (B and C) THP-1 cells stably expressing HA-tagged IRF-3 S386A were cultured in medium containing 10 ng/ml of PMA for 72 h. Cells were stimulated with 50 μg/ml of poly(I·C) (B) or 1 μg/ml of LPS (C) for the indicated times. Cell lysates were prepared and subjected to immunoprecipitation with anti-HA, followed by immunoblotting as indicated. *, nonspecific band. Input: total lysates. (D) 293T cells were transfected with expression constructs for HA-tagged TLR3 and Flag-tagged IRF-3 S386A. Twenty-four hours later, cells were stimulated with 50 μg/ml of poly(I·C) for the indicated times. Cell lysates were subjected to immunoprecipitation with anti-TRIF, followed by immunoblotting with the indicated antibodies. (E) 293T cells were transfected with expression constructs for Flag-tagged TLR3 and HA-tagged IRF-3 S386A. Twenty-four hours later, cells were stimulated with 50 μg/ml of poly(I·C) for 2 h. Cell lysates were subjected to lambda protein phosphatase (PP) treatment, followed by anti-HA IP and immunoblotting. Input, total lysates. (F) 293T cells were transfected with empty vector or expression construct for Flag-tagged TRIF-N540 as indicated. Twenty-four hours later, Flag-TRIF-N540 proteins were affinity purified using anti-Flag beads, followed by lambda PP treatment as indicated. Lambda PP-treated TRIF-bound beads were washed and incubated with purified recombinant His6-tagged IRF-3 proteins for 4 h. The beads were washed, and bound proteins were analyzed by immunoblotting or staining with CBB.
Phosphorylation of TRIF at Ser210 is required for IRF-3 recruitment and subsequent IRF-3 activation.
Next, to identify the phosphorylated residue(s) of TRIF-N540, the 19 serine/threonine residues shown in Fig. 2A were replaced with alanine. The mutants were expressed in 293T cells, and activation of IRF-3, as judged by dimer formation, was examined. Overexpression of TRIF-N540 and the mutants resulted in IRF-3 dimerization, except for the mutant S210A, suggesting that phosphorylation of Ser210 is needed for IRF-3 recruitment and its activation. We next tried to obtain direct evidence for TRIF phosphorylation at Ser210 through liquid chromatography-tandem mass spectrometry (LC-MS/MS). Several phosphorylated serine residues, including Ser210, were identified from the purified TRIF-N540 expressed in 293T cells; however, it was difficult to discriminate phosphorylation at Ser210 from phosphorylation at Ser202, Ser205, and Ser212 because they were all predicted on one peptide at the same confidence level (Fig. 2B). We thus performed IFN-β promoter activation analysis for further confirmation. Overexpression of the wild type or the S202A or S205A mutant activated the reporter gene, but S210A exhibited reduced activity (Fig. 2C). Moreover, coimmunoprecipitation analysis revealed that S202A and S205A were associated with IRF-3 S386A but not with S210A (Fig. 2D). To examine if phosphorylation of TRIF Ser210 is responsible for poly(I·C)-induced IRF-3 activation, we generated TRIF knockout (KO) 293T cells by using the CRISPR-Cas9 system. We stably reconstituted TRIF KO cells with empty vector and WT Flag-TRIF-N540 or Flag-TRIF-N540 S210A. These cells transiently expressing TLR3 were stimulated with poly(I·C) and examined for IRF-3 dimerization (Fig. 2E). We found that IRF-3 dimer induction by TLR3 stimulation was abolished in TRIF KO 293T cells, which was complemented by WT Flag-TRIF-N540 but not Flag-TRIF-N540 S210A. We also observed that TBK1 phosphorylation was abrogated in TRIF KO cells, which was recovered by stable expression of WT Flag-TRIF-N540 or Flag-TRIF-N540 S210A. Combined, our results strongly suggest that phosphorylation of TRIF at Ser210 is required for IRF-3 recruitment and activation.
FIG 2.
Phosphorylation of Ser210 of TRIF is required for the interaction with IRF-3 and its subsequent activation. (A) 293T cells were transfected with expression constructs for Flag-tagged WT TRIF-N540 or the indicated mutants. Twenty-four hours later, cell lysates were subjected to native PAGE followed by immunoblotting. (B) Left, Flag-tagged TRIF-N540 was transiently expressed from 293T cells. Twenty-four hours later, cell lysates were subjected to anti-Flag IP and separated by SDS-PAGE. The gel was stained by CBB to visualize the Flag-TRIF-N540 band. Flag-TRIF-N540 was recovered from the gel and subjected to LC-MS/MS analysis. Right, representative MS/MS spectra of the phosphorylated TRIF peptide (STGSPAS*LAS*N*LEIS*QS*PTMPFLSLHR, residues 196 to 222). b fragment ions are shown in green and y fragment ions are shown in red. Asterisks indicate modification of amino acids in the peptide: Ser202 (phosphorylation), Ser205 (phosphorylation), Asp206 (deamidation, artifact), Ser210 (phosphorylation), and Ser212 (phosphorylation). The red line indicates the amino acid sequence conserved in MAVS and STING (pLXIS, where p is a hydrophobic amino acid and X is any amino acid). (C) 293T cells were transiently transfected with expression vectors for Flag-tagged WT TRIF-N540 or the indicated mutants and the reporter plasmids (p-125 luc and pRL-TK Renilla luc). Twenty-four hours later, cell lysates were subjected to dual-luciferase assay. Luciferase activity data are representative results from at least three independent experiments; means and standard deviations (SD) from triplicate experiments are shown. ***, P < 0.001; NS, not significant (by Student’s two-tailed t test compared with WT). (D) 293T cells were transfected with expression constructs for Flag-tagged WT TRIF-N540 or the indicated mutants and HA-tagged IRF-3 S386A. Twenty-four hours later, cell lysates were subjected to anti-HA immunoprecipitation, followed by immunoblotting for Flag or HA. (E) WT and TRIF KO 293T cells stably reconstituted with empty vector, WT Flag-TRIF-N540, or Flag-TRIF-N540 S210A were transfected with the expression construct for HA-tagged TLR3. Twenty-four hours later, cells were stimulated with 50 μg/ml of poly(I·C) for 2 h. Cell lysates were prepared and subjected to native PAGE, followed by immunoblotting as indicated.
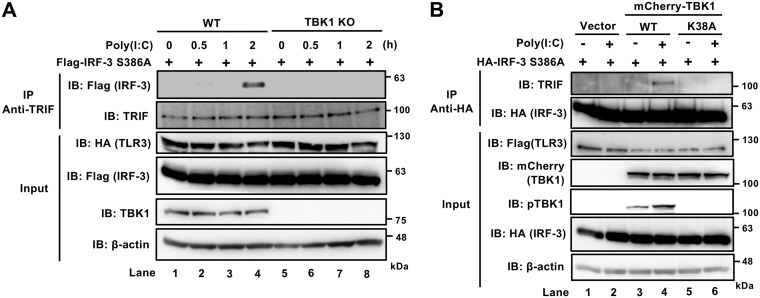
Kinase activity of TBK1 is required for the association of TRIF and IRF-3.
Next, we investigated which kinase is responsible for the phosphorylation of TRIF Ser210. A previous study found that TBK1 directly phosphorylates TRIF in cells (31). To confirm that TBK1 is responsible for TRIF phosphorylation and subsequent IRF-3 recruitment, we generated TBK1 KO 293T cells by using the CRISPR-Cas9 system. TLR3 and IRF-3 S386A were transiently expressed in WT and TBK1 KO 293T cells and stimulated with poly(I·C) treatment (Fig. 3A). Pulldown of endogenous TRIF demonstrated that IRF-3 S386A was recruited to TRIF in a TBK1-dependent manner. To explore the involvement of the kinase activity of TBK1, we stably complemented the TBK1 KO 293T cells with mCherry-TBK1 or mCherry-TBK1 K38A, a kinase-deficient mutant. Unlike transient overexpression, stable complementation by mCherry-TBK1 did not result in constitutive signaling (Hiroto Abe, unpublished observation). The cells were stimulated with poly(I·C) and examined for interaction between TRIF and IRF-3 S386A (Fig. 3B). TBK1 deficiency in inducing TRIF/IRF-3 interaction was efficiently complemented by mCherry-TBK1 but not by mCherry-TBK1 K38A. These results strongly suggest that upon TLR3 signaling, TBK1 activates TRIF through its kinase activity to recruit IRF-3.
FIG 3.
Kinase activity of TBK1 is required for the recruitment of IRF-3 to TRIF in response to TLR3 stimulation. (A) WT and TBK1 KO 293T cells were transfected with expression constructs for HA-tagged TLR3 and Flag-tagged IRF-3 S386A. Twenty-four hours later, cells were stimulated with 50 μg/ml of poly(I·C) for the indicated times. Cell lysates were subjected to immunoprecipitation with anti-HA, followed by immunoblotting for Flag and endogenous TRIF as indicated. Input, total lysates. (B) TBK1 KO 293T cells were stably reconstituted with empty vector or expression vector for mCherry-tagged WT TBK1 or TBK1 K38A and were transfected with expression constructs for Flag-tagged TLR3 and HA-IRF-3 S386A. Twenty-four hours later, cells were stimulated with 50 μg/ml of poly(I·C) for 2 h. Cell lysates were subjected to immunoprecipitation with anti-HA, followed by immunoblotting as indicated.
IKKβ is essential for IRF-3 activation via TLR3-TRIF-mediated but not RIG-I-mediated signaling.
During the course of this investigation, we tested pharmacological inhibition of TBK1 to block TLR3 signaling along with an inhibitor of IKKβ as a control (Fig. 4A). As expected, BX795, an inhibitor of TBK1, blocked the recruitment of IRF-3 S386A to TRIF. Unexpectedly, TPCA-1, an inhibitor of IKKβ, also inhibited the formation of the TRIF/IRF-3 S386A complex. Similar results were obtained with THP1 cells stimulated with LPS (Fig. 4B). To further corroborate the involvement of IKKβ in IRF-3-mediated gene induction, cells were treated with poly(I·C) (TLR3 stimulation) or infected with Sendai virus (RIG-I stimulation) and examined for IRF-3-responsive reporter gene expression. TLR3 signaling was inhibited by TPCA-1 in a dose-dependent manner (Fig. 4C); however, RIG-I signaling was unaffected (Fig. 4D) by the dose strongly inhibitory for TLR3 signaling. To exclude cross-reactivity of the inhibitors, we generated IKKα, IKKβ, and IKKα/β gene KO cells in addition to those of TBK1, IRF-3, and TRIF and examined them similarly (Fig. 4E and F). Stimulation with poly(I·C) treatment was dependent on TRIF, but Sendai virus-induced signaling was independent of TRIF, confirming that the stimuli were specific to TLR3 and RIG-I, respectively. These results demonstrated that TLR3 signaling requires IKKβ (Fig. 4E); however, RIG-I signaling was independent of IKKα and/or -β (Fig. 4F). NEMO, also known as IKKγ, was essential for TLR3 signaling but not for RLR signaling (Fig. 4E and F). As expected, the interaction between TRIF and IRF-3 S386A was abolished by IKKβ deficiency (Fig. 4G). IRF-3 dimer induction by TLR3 stimulation was dependent on IKKβ (Fig. 4H), but it was dispensable in RIG-I stimulation (Fig. 4I). To further confirm the involvement of kinase activity of IKKβ, the KO cells were complemented with WT IKKβ or kinase-deficient IKKβ K44A (Fig. 4J). IKKβ deficiency was complemented by WT IKKβ but not by the K44A mutant, suggesting that the kinase activity of IKKβ is essential for IRF-3 activation by TLR3 signaling.
FIG 4.
IKKβ plays an essential role in IRF-3 activation via TLR3/4 signaling but not RIG-I signaling. (A) 293T cells were transfected with expression constructs for Flag-tagged TLR3 and HA-tagged IRF-3 S386A. Twenty-four hours later, cells were pretreated with 20 μM TPCA-1 or 10 μM BX795 for 1 h. The cells were stimulated with 50 μg/ml of poly(I·C) for 2 h with or without inhibitors as indicated. Cell lysates were prepared and subjected to immunoprecipitation with anti-HA, followed by immunoblotting with anti-HA or anti-TRIF. (B) THP-1 cells stably expressing HA-tagged IRF-3 S386A were cultured in medium containing 10 ng/ml of PMA for 72 h. Cells were stimulated with 1 μg/ml of LPS for 2 h. Control solvent (DMSO), TPCA-1 (20 μM), or BX795 (10 μM) was added 1 h before stimulation and left until the end of the experiment. Cell lysates were prepared and subjected to immunoprecipitation with anti-HA, followed by immunoblotting as indicated. *, nonspecific band. Input, total lysates. (C) 293T cells were transfected with expression constructs for Flag-tagged TLR3 and reporter genes (p-55C1B luc and pRL-TK Renilla luc). Twenty-four hours later, cells were pretreated with TPCA-1 at the indicated dose for 1 h. Cells were stimulated with 50 μg/ml of poly(I·C) for 12 h in the presence of TPCA-1. (D) 293T cells were transfected with reporter genes (p-55C1B luc and pRL-TK Renilla luc). Twenty-four hours later, cells were pretreated with TPCA-1 at the indicated dose for 1 h. Cells were infected with Sendai virus (SeV) for 24 h in the presence of TPCA. Cell lysates were prepared and subjected to dual-luciferase assay. (E and F) WT 293T cells and IKKα, IKKβ, IKKα/β, NEMO, TBK1, IRF-3, and TRIF KO 293T cells were transfected with expression constructs for Flag-tagged TLR3 (E) and reporter genes (p-55C1B luc and pRL-TK Renilla luc). Twenty-four hours later, cells were stimulated with 50 μg/ml of poly(I·C) for 12 h (E) or infected with SeV for 24 h (F). Cell lysates were prepared and subjected to dual-luciferase assay. (G) WT 293T cells and IKKα, IKKβ, and IKKα/β KO 293T cells were transfected with expression constructs for HA-tagged TLR3 and Flag-tagged IRF-3 S386A. Twenty-four hours later, cells were stimulated with 50 μg/ml of poly(I·C) for 2 h. Cell lysates were subjected to immunoprecipitation with anti-HA, followed by immunoblotting. (H) WT and IKKβ KO 293T cells were transfected with expression constructs for Flag-tagged TLR3 and HA-IRF-3. Twenty-four hours later, cells were stimulated with 50 μg/ml of poly(I·C) for 2 h. Total cell lysates were subjected to native PAGE and SDS-PAGE, followed by immunoblotting. *, nonspecific band. (I) WT 293T cells and IKKα, IKKβ, or IKKα/β KO 293T cells were infected with SeV for 8 h. Total cell lysates were subjected to native PAGE and SDS-PAGE, followed by immunoblotting. (J) WT and IKKβ KO 293T cells were stably reconstituted for IKKβ or IKKβ K44A. The cells were transfected with expression constructs for Flag-tagged TLR3 and reporter genes (p-55C1B luc and pRL-TK Renilla luc). Twenty-four hours later, cells were stimulated with 50 μg/ml of poly(I·C) for 12 h. Cells were lysed and subjected to dual-luciferase assay. Luciferase activity data are representative results from at least three independent experiments; means and SD from triplicate experiments are shown. ***, P < 0.001; NS, not significant (by Student’s two-tailed t test compared with no treatment [C and D] or with WT [E and F] or by one-way ANOVA followed by Tukey’s test [J]).
IKKβ targets TBK1 to facilitate IRF-3 recruitment to TRIF and signaling.
Since the kinase activity of TBK1 is essential for the interaction of TRIF with IRF-3 (Fig. 2A), we examined the necessity of TBK1 kinase activity in phosphorylation of TRIF. We demonstrated that transient expression of WT mCherry-TBK1 but not its kinase-dead mutant (K38A) strongly enhanced phosphorylation of TRIF in 293T cells, as judged by the mobility shift in both normal and phosphate-affinity SDS-PAGE, which indicates TBK1-mediated TRIF phosphorylation (Fig. 5A) (32). In contrast, overexpression of IKKβ alone did not affect TRIF phosphorylation (Fig. 5A). We then hypothesized that IKKβ targets TBK1 to facilitate the signaling. IKKβ KO 293T cells complemented with WT IKKβ or the K44A mutant were examined for interaction with TBK1 upon TLR3 stimulation (Fig. 5B). Wild-type IKKβ, but not IKKβ K44A, interacted with TBK1 transiently, with a peak at 0.5 h after poly(I·C) stimulation when the TRIF/IRF-3 complex was still undetectable (Fig. 1D). Modest Ser172 phosphorylation of TBK1 was observed 1 h after stimulation in the absence of IKKβ, but robust phosphorylation was detected in cells complemented with WT IKKβ but not IKKβ K44A, strongly suggesting that IKKβ interacts with and phosphorylates TBK1 upon TLR3 stimulation. Artificial overexpression of wild-type TBK1 caused robust autophosphorylation, resulting in constitutive signaling. However, no such robust autophosphorylation was observed in the case of the kinase-dead K38A mutant. We then used TBK1 KO cells stably complemented with mCherry-TBK1 K38A (Fig. 3B) and examined its phosphorylation by exogenous IKKβ. Overexpression of wild-type IKKβ but not IKKβ K44A led to the phosphorylation of TBK1 (Fig. 5C). Next, the stable cells were stimulated with poly(I·C) and examined for TBK1 phosphorylation by an endogenous mechanism. Stimulation of TLR3 induced phosphorylation of TBK1 after 0.5 h (Fig. 5D); however, it was abrogated by IKKβ inhibition. In contrast, Sendai virus infection induced TBK1 phosphorylation independently of IKKβ (Fig. 5E).
FIG 5.
IKKβ targets TBK1 to facilitate IRF-3 recruitment to TRIF and signaling. (A) 293T cells were transfected with expression constructs for Flag-tagged TRIF-N540, together with mCherry-tagged TBK1 WT/K38A or IKKβ WT/K44A. Twenty-four hours later, cell lysates were prepared and subjected to Phos-tag SDS-PAGE and normal SDS-PAGE, followed by immunoblotting as indicated. (B) IKKβ KO 293T cells stably reconstituted with empty vector, IKKβ, or IKKβ K44A were transfected with the expression construct for Flag-tagged TLR3. Twenty-four hours later, cells were stimulated with 50 μg/ml of poly(I·C) for the indicated times. Cell lysates were subjected to immunoprecipitation with anti-TBK1, followed by immunoblotting. (C) TBK1 KO 293T cells stably reconstituted with mCherry-tagged TBK1 K38A were transfected with expression constructs for IKKβ or IKKβ K44A as indicated. Twenty-four hours later, cell lysates were prepared and subjected to immunoblotting. (D) TBK1 KO 293T cells stably reconstituted with mCherry-tagged TBK1 K38A were transfected with constructs for Flag-tagged TLR3. Twenty-four hours later, cells were stimulated with 50 μg/ml of poly(I·C) for the indicated times. Where indicated, TPCA-1 (20 μM) was added 1 h before and left during stimulation. Total cell lysates were subjected to immunoblotting. (E) WT, IKKβ KO, and TBK1 KO 293T cells were infected with Sendai virus (SeV) for the indicated times. Total cell lysates were subjected to immunoblotting. (F and G) 293T cells were transfected with expression constructs for Flag-tagged TLR3 and reporter genes (p-55A2 luc [F] or p-55C1B luc [G] and pRL-TK Renilla luc). Twenty-four hours later, cells were stimulated with 50 μg/ml of poly(I·C) together with 30 ng/ml of recombinant TNF-α for 12 h. Where indicated, TPCA-1 (20 μM) was added 1 h before and left during stimulation. Cells were lysed and subjected to dual-luciferase assay. Luciferase activity data are representative results from at least three independent experiments; means and SD from triplicate experiments are shown. *, P < 0.05; ***, P < 0.001; NS, not significant (by one-way ANOVA followed by Tukey’s test).
As the above results suggested an essential role of IKKβ in TLR3 signaling, we tested simultaneous stimulation by tumor necrosis factor alpha (TNF-α) and dsRNA. TNF-α strongly activates canonical IKK, resulting in the induction of NF-κB, but does not activate TBK1 or IRF-3. Activation of the NF-κB-driven reporter gene p-55A2 was not influenced by costimulation (Fig. 5F). However, activation of the IRF-responsive reporter p-55C1B was significantly altered by the costimulation (Fig. 5G). Furthermore, the observed induction of p-55A2 and p-55C1B was abolished by TPCA-1 (Fig. 5F and G). These results demonstrated cross talk by conventional NF-κB and TLR3 signaling.
DISCUSSION
In this study, we delineated the mechanism of TLR3 signaling. We propose a model for the sequential events after TLR3 stimulation (Fig. 6). When dsRNA is sensed by TLR3, adaptor molecules IKKβ, TBK1, and TRIF are recruited for signaling. These adaptors associate within 0.5 h after ligand stimulation and dissociate thereafter (see results for IKKβ and TBK1 in Fig. 5A and results for TBK1 and TRIF in Fig. 1B). The first cytoplasmic event needed for signal initiation is phosphorylation of Ser172 of TBK1 by IKKβ (Fig. 6, event 1). The absence of IKKβ or its inhibition by TPCA-1 markedly abrogated downstream events. However, we cannot exclude the possibility that IKKβ phosphorylates TBK1 at multiple sites, including Ser172, to initiate signaling. Of note, weak phosphorylation of Ser172 of TBK1 was observed even in the absence of IKKβ (Fig. 5B). This was likely due to autophosphorylation upon oligomerization of TBK1 upon TLR3 stimulation because kinase-dead TBK1 was not phosphorylated after poly(I·C) stimulation in the presence of TPCA-1 (Fig. 5D). This IKKβ-independent TBK1 phosphorylation is not sufficient to transmit signals, as evidenced by the absence of TRIF/IRF-3 interaction (Fig. 4A and G), IRF-3 dimerization (Fig. 4H), and reporter gene activation (Fig. 4J). We propose that phosphorylation of TBK1 by IKKβ primes TBK1 to facilitate robust autophosphorylation (Fig. 6, event 2). Next, the activated TBK1 phosphorylates TRIF (Fig. 6, event 3), presumably at multiple sites. Of several phosphorylated serine residues, Ser210 was identified as the essential site for signaling, as demonstrated by mutagenesis (Fig. 2). The surrounding residues fall into the consensus pLXIS (S is phosphorylated Ser), which was proposed for IRF-3 recruitment found in STING and MAVS (31). The next step takes place around 1 to 2 h after poly(I·C) treatment (Fig. 1D). The phosphorylated TRIF recruits IRF-3 by forming a complex (Fig. 6, event 4). This interaction is likely mediated by phosphorylated Ser210 and the phosphoacceptor pocket structure of IRF-3 (33) because mutagenesis of the pocket abrogated the binding (Abe, unpublished observation). TBK1 phosphorylates IRF-3 in the context of the TRIF/IRF-3 complex (Fig. 6, event 5). The most important serine residue of IRF-3 for activation is Ser386 (13). Upon phosphorylation, IRF-3 rapidly dissociates from TRIF (Fig. 6 event 6). Two phosphorylated IRF-3 molecules associate through phosphorylated Ser386 and the pocket structure to form a dimer (33) (Fig. 6, event 7) and then translocate to the nucleus for transcriptional activation (Fig. 6, event 8).
FIG 6.
Proposed model for TLR3 signaling to activate IRF-3-dependent genes. Once dsRNA binds to TLR3, signal is initiated. Although the precise nature of the signaling complex is not delineated, IKKβ and TBK1 are recruited to the signaling complex, where these kinases interact and the phosphorylation of TBK1 at Ser172 is catalyzed by IKKβ (1). The initial phosphorylation of TBK1 promotes robust autophosphorylation of TBK1 to increase its catalytic activity (2). TBK1 interacts with TRIF and phosphorylates Ser210 (3), and the phosphorylated TRIF dissociates from TBK1. The phosphorylated TRIF recruits IRF-3 through interaction between the phosphorylated Ser210 of TRIF and the phosphoacceptor pocket of IRF-3 (4). The conserved NLEIS sequence of TRIF is shown. TBK1 catalyzes the phosphorylation of Ser386 of IRF-3 (5), and the phosphorylated IRF-3 is released from TRIF (6). The phosphorylated IRF-3 forms a dimer (7) and translocates to the nucleus (8) to function in gene activation. Events 1 to 3 take place within 0.5 h of the initial trigger. Events 4 to 8 take place 2 h after signaling. Blue arrows indicate phosphorylation.
The above highlight the essential role of IKKβ for TBK1 priming in TLR3 signaling. Knockout analyses revealed that both IKKβ and NEMO, but not IKKα, are required (Fig. 4), suggesting that noncanonical IKK complexes containing IKKβ and NEMO are sufficient to prime TBK1. Further analyses are required to elucidate the nature of the responsible IKK complexes.
TPCA-1 inhibits LPS-induced TLR4 signaling (Fig. 4B), suggesting a similar mechanism for priming by IKKβ of TBK1 in the signaling in which TRIF acts as a signaling adaptor. In contrast, IKKβ is dispensable for RLR signaling (Fig. 4D, F, and I and 5E), in which several signaling adaptors, including MAVS, play important roles, suggesting that TBK1 priming by IKKβ is adaptor specific. The molecular mechanism underlying the absence of IKKβ involvement in RLR signaling is unknown, but IKKβ may be an additional safeguard to avoid excess innate immune responses. Alternatively, it may provide a cross talk or amplification mechanism for immune responses, as evidenced by cooperative IRF-3 activation by TNF-α and dsRNA (34) (Fig. 5G). In the case of RLR signaling, viral replication is ongoing within the cell, and it is needed to induce robust responses as a frontline defense mechanism.
MATERIALS AND METHODS
Reagents and antibodies.
Lipopolysaccharide (LPS) from E. coli O111:B4 and 4β-phorbol 12-myristate 13-acetate (PMA) were purchased from Sigma-Aldrich. TPCA-1 and BX795 were purchased from Tocris. Poly(I·C) was purchased from GE Healthcare. Recombinant human TNF-α was purchased from PeproTech. Rabbit polyclonal anti-IRF-3 and rabbit polyclonal anti-phospho IRF-3 (S386P) were generated previously (11, 13). Mouse monoclonal anti-Flag M2 (F1804) was purchased from Sigma-Aldrich. Mouse monoclonal anti-β-actin (sc-47778) and mouse monoclonal anti-IKKα (sc-7606) were purchased from Santa Cruz Biotechnology. Mouse monoclonal anti-HA.11 (901502) and mouse monoclonal anti-TICAM-1/TRIF (657102) were purchased from BioLegend. Rabbit monoclonal anti-TBK1 (ab40676) and rabbit polyclonal anti-mCherry (ab183628) were purchased from Abcam. Rabbit monoclonal anti-IKKβ (2370), rabbit monoclonal anti-phospho TBK1 (Ser172) (5483), and rabbit monoclonal anti-TRIF (4596) were purchased from Cell Signaling Technology.
Cell culture, transfection reagents, expression constructs, and lentiviral transduction.
293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM) (Nacalai Tesque) supplemented with 10% fetal bovine serum (Life Technologies) and penicillin-streptomycin (Nacalai Tesque) (100 U/ml and 100 μg/ml, respectively). THP-1 cells were cultured in RPMI 1640 medium (Nacalai Tesque) supplemented with 10% fetal bovine serum and penicillin-streptomycin. All cells were cultured in a humidified atmosphere with 5% CO2 at 37°C.
For transient expression in 293T cells, plasmids encoding each gene were transfected using polyethylenimine Max (Polysciences). cDNAs encoding hemagglutinin (HA)-tagged human IRF-3 and Flag-tagged human TLR3 were cloned into the pEF-BOS(+) vector, and cDNA encoding human TRIF was cloned into the pCMV-Flag vector. For lentiviral transduction, cDNA encoding HA-tagged human IRF-3, mCherry-tagged humanTBK1, human IKKβ, and Flag-tagged human TRIF were cloned into the pCSII-CMV-MCS-IRES2-Bsd vector. All mutant plasmids were generated by site-directed mutagenesis using the KOD-PLUS mutagenesis kit (Toyobo).
To generate cell lines stably expressing transgenes, lentiviral particles were produced by transfecting 293T cells with plasmids pCAG-HIVgp, pVSV-G-RSV-ReV, and pCSII-CMV-MCS-IRES2-Bsd containing the genes of interest. Plasmids were transfected into 293T cells in 10-cm dishes at a confluence of 50% to 70% using Lipofectamine 2000 (Life Technologies), media were changed at 8 h posttransfection, and viral supernatants were collected at 48 h posttransfection. Viral supernatants were centrifuged at 500 × g to remove cellular debris and then filtered (0.45 μm). Polybrene (Nacalai Tesque) was added to the filtered supernatants (10 μg/ml), and the viral supernatants were then used to infect cells. The infected cell lines were treated with blasticidin (10 μg/ml; InvivoGen) at 48 h after infection to select CSII-CMV-MCS-IRES2-Bsd containing cells.
Virus quantification and infection.
Sendai virus (Cantell strain) was inoculated into 9-day embryonated chicken eggs and incubated for 2 days at 37°C, followed by overnight incubation at 4°C. Allantoic fluid containing Sendai virus was collected from dead eggs. The virus titer was measured by the hemagglutination test using chicken red blood cells. Virus was added to cells at 3.2 × 102 hemagglutination units (HAU). After a 1-h incubation, the medium was replaced with fresh DMEM and incubated for the indicated time.
Purification of recombinant human IRF-3 proteins.
E. coli BL21(DE3) competent cells were transformed with the expression plasmid pET-GST-3C-human IRF-3-His6, kindly gifted by Fuyuhiko Inagaki (35). Protein expression was induced with 0.1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) overnight at 16°C with shaking (90 rpm). Glutathione-S-transferase- and His6-tagged IRF-3 proteins were purified using glutathione-Sepharose 4B (GE Healthcare), and His6-tagged IRF-3 proteins were obtained by cleavage with PreScission protease (GE Healthcare) in lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM dithiothreitol [DTT], and protease inhibitor cocktail [Roche]).
Generation of gene knockout 293T cells by CRISPR-Cas9.
All gene-specific knockout 293T cells were generated by transient transfection of the pSpCas9(BB)-2A-GFP (PX458) vector (Addgene no. 48138) (36) containing double-stranded oligonucleotides containing single guide RNA (sgRNA) sequences of each gene listed in Table 1 according to the manufacturer’s protocol for 24 h. Cells were subjected to sorting by green fluorescent protein (GFP) expression levels using the SH800 cell sorter (Sony), followed by limiting dilution in 96-well culture plates to obtain single clones. Knockout of each gene was examined by immunoblotting.
TABLE 1.
Target sequences of sgRNAs for CRISPR-Cas9 gene knockout
| Primer name | Sequence (5′→ 3′) |
|---|---|
| IKKα sgRNA forward | CACCGGTACCAAAAACAGAGAACGA |
| IKKα sgRNA reverse | AAACTCGTTCTCTGTTTTTGGTACC |
| IKKβ sgRNA forward | CACCGTCAGCCCCCGGAACCGAGAG |
| IKKβ sgRNA reverse | AAACCTCTCGGTTCCGGGGGCTGAC |
| TBK1 sgRNA forward | CACCGAAATATCATGCGTGTTATAG |
| TBK1 sgRNA reverse | AAACCTATAACACGCATGATATTTC |
| NEMO sgRNA forward | CACCGGCTGCACCTGCCTTCAGAACA |
| NEMO sgRNA reverse | AAACTGTTCTGAAGGCAGGTGCAGCC |
| TRIF sgRNA forward | CACCGATGAGGCCCGAAACCGGTGTGGG |
| TRIF sgRNA reverse | AAACCCCACACCGGTTTCGGGCCTCATC |
| IRF-3 sgRNA forward | CACCGTCCACCATTGGTGTCCGGAG |
| IRF-3 sgRNA reverse | AAACCTCCGGACACCAATGGTGGAC |
Immunoblotting and immunoprecipitation.
293T cells were washed with phosphate-buffered saline (PBS). Cells were lysed with lysis buffer (1% Nonidet P-40, 20 mM Tris-HCl [pH 7.5], and 150 mM NaCl supplemented with 0.1 mg/ml of leupeptin, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1 mM sodium orthovanadate). Cell extracts were centrifuged at 15,000 × g for 30 min at 4°C, and supernatants were collected as protein extracts, mixed with 2× SDS sample buffer (125 mM Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 0.01% bromophenol blue (BPB), and 10% 2-mercaptoethanol), and boiled for 4 min at 100°C for denaturation. For immunoprecipitation, protein extracts from 10-cm dishes were incubated with a mixture of 1 μg of antibody and protein G Mag Sepharose (GE Healthcare) overnight at 4°C. The immunocomplexes were washed three times in lysis buffer, followed by denaturation in 2× SDS sample buffer. Denatured proteins were resolved by SDS-PAGE. After proteins were transferred onto Immobilon-P polyvinylidene difluoride (PVDF) membranes (Millipore), the membranes were incubated in Tris-buffered saline with 0.1% Tween 20 (TBST) containing 5% skim milk for 30 min at room temperature for blocking, followed by incubation with a primary antibody diluted in the blocking buffer overnight at 4°C. After washing with TBST three times, the membranes were incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody in TBST for 1 h at room temperature. Proteins on the membranes were visualized with the a LAS-4000 instrument (Fujifilm) by chemiluminescence using Chemi-lumi One Super (Nacalai Tesque).
Native PAGE.
To detect IRF-3 dimerization, native PAGE was performed as described previously (13). Briefly, a 7.5% polyacrylamide gel was prerun with the running buffer (25 mM Tris-Cl [pH 8.4] and 192 mM glycine in the presence or absence of 0.2% deoxycholate in the cathode and anode buffers, respectively) at 40 mA for 30 min. Protein extracts were mixed with 5× native PAGE sample buffer (312.5 mM Tris-HCl [pH 6.8], 50% glycerol, and 0.01% BPB) and applied to the gel. The samples were electrophoresed at 26 mA for 50 min at room temperature. After being transferred onto membranes, proteins were detected by immunoblotting.
Phosphate-affinity SDS-PAGE.
For detecting TRIF phosphorylation, Phos-tag SDS-PAGE was performed (32). Briefly, cell extracts were separated by SDS-PAGE using a Phos-tag gel, which consists of a 5.0% polyacrylamide-SDS gel containing 50 μM Phos-tag acrylamide (Wako) and 100 μM MnCl2. After SDS-PAGE, the Phos-tag gel was washed with EDTA transfer buffer (25 mM Tris-HCl [pH 8.0], 192 mM glycine, 20% methanol, and 10 mM EDTA) twice for 10 min, following washing with EDTA-free transfer buffer (25 mM Tris-HCl [pH 8.0], 192 mM glycine, and 20% methanol) for 10 min. After being transferred onto membranes, proteins were detected by immunoblotting.
In vitro binding assay for TRIF-N540 and IRF-3 proteins.
TRIF proteins were purified from protein extracts of 1 × 107 293T cells transiently expressing Flag-TRIF-N540 by incubation with anti-Flag M2 affinity gel (A2220; Sigma-Aldrich) overnight for 4°C and then washed with lysis buffer (1% Nonidet P-40, 20 mM Tris-HCl [pH 7.5], and 150 mM NaCl supplemented with 0.1 mg/ml leupeptin, 1 mM PMSF, and 1 mM sodium orthovanadate) three times. Lambda protein phosphatase (Santa Cruz Biotechnology) at 2,000 units was added and incubated at 30°C for 1 h in a 50-μl reaction mixture, followed by washing three times. Five micrograms of recombinant His6-tagged IRF-3 proteins was added to 200 μl of binding buffer (0.1% Nonidet P-40, 20 mM Tris-HCl [pH 7.5], and 150 mM NaCl supplemented with 0.1 mg/ml leupeptin, 1 mM PMSF, and 1 mM sodium orthovanadate) mixed with the TRIF protein/anti-Flag M2 affinity gel complex, followed by incubation for 4 h at 4°C. Beads were washed five times with binding buffer, followed by denaturation in SDS.
Luciferase reporter assay.
293T cells were transfected with p-55C1B luc (IRF promoter) or p-55A2 luc (κB promoter) for firefly luciferase together with pRL-TK-Renilla luc (internal control) for Renilla luciferase in 48-well culture plates. Cells were lysed with passive lysis buffer (Promega), and firefly luciferase and Renilla luciferase activities were measured using a dual-luciferase assay kit (Promega).
Phosphomapping by LC-MS/MS.
For liquid chromatography-tandem mass spectrometry (LC-MS/MS), Coomassie brilliant blue (CBB)-stained bands were excised, destained, and digested in gel with trypsin using an in-gel tryptic digestion kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The protein digests were resuspended in 0.1% formic acid and separated using a Nano-LC-Ultra 2D-plus instrument equipped with cHiPLC Nanoflex (Eksigent) in trap-and-elute mode, with a trap column (200 μm by 0.5 mm [ChromXP C18-CL]; 3 μm by 120 Å [Eksigent]) and analytical column (75 μm by 15 cm [ChromXP C18-CL]; 3 μm by 120 Å [Eksigent]). The separation was carried out with a binary gradient as follows, where solvent A and B correspond to 0.1% formic acid–water and 0.1% formic acid–acetonitrile, respectively: 2 to 33.2% B in 125 min, 33.2% to 98% B in 2 min, 98% B for 5 min, 98% to 2% B in 0.1 min, and 2% B for 17.9 min. The eluates were infused on-line into a mass spectrometer (TripleTOF 5600+ system with a NanoSpray III source and heated interface [Sciex]) and ionized in electrospray ionization-positive mode. Data acquisition was carried out using an information-dependent acquisition method. The acquired data sets were analyzed by ProteinPilot software version 5.0.1 (Sciex) with the UniProtKB/Swiss-Prot database for humans (May 2018) appended with the amino acid sequences for Flag-TRIF-N540 and known common contaminants (Sciex). The identification was focused on biological modifications. Phosphorylation emphasis and gel-based identification were used as special factors. The reliability of peptide identifications was evaluated by the confidence values and the results of target-decoy-based false-discovery-rate analysis calculated and performed by ProteinPilot software (Sciex).
Statistical analysis.
Statistical analysis was performed using R3.6.0 software (The R Foundation for Statistical Computing). Student’s two-tailed t test was used to compare two groups (WT and other groups), and one-way analysis of variance (ANOVA) followed by Tukey’s test was used for multiple comparison. A P value of <0.05 was considered significant.
ACKNOWLEDGMENTS
We declare no conflicts of interest.
H. Abe performed the experiments and analyzed the data. J. Satoh performed the LC-MS/MS experiment, and S. Ito analyzed the LC-MS/MS data. Y. Shirasaka the designed CRISPR-Cas9 gene knockout assay. A. Kogure generated IRF-3 and TBK1 KO cells. H. Abe and S. Ito wrote the initial manuscript. H. Kato assisted in the study design. T. Fujita conceived the project, performed experiments, analyzed the data, and wrote the complete manuscript.
This study was supported by research grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Innovative Areas “Infection Competency” [24115004]), from the Japan Agency for Medical Research and Development (Research Program on Emerging and Re-emerging Infectious Diseases [19fk0108081h1001]), and from the Japan Society for the Promotion of Science (Core to Core Program, Grants-in-Aid for Scientific Research “B” [18H02344]; Fund for the Promotion of Joint International Research, Fostering Joint International Research [B] [18KK0232]).
REFERENCES
- 1.Janeway CA, Medzhitov R. 2002. Innate immune recognition. Annu Rev Immunol 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Yoneyama M, Fujita T. 2009. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev 227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 4.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 5.O'Neill LAJ, Bowie AG. 2007. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol 7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 7.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol 4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 9.Shu C, Sankaran B, Chaton CT, Herr AB, Mishra A, Peng J, Li P. 2013. Structural insights into the functions of TBK1 in innate antimicrobial immunity. Structure 21:1137–1148. doi: 10.1016/j.str.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma X, Helgason E, Phung QT, Quan CL, Iyer RS, Lee MW, Bowman KK, Starovasnik MA, Dueber EC. 2012. Molecular basis of Tank-binding kinase 1 activation by transautophosphorylation. Proc Natl Acad Sci U S A 109:9378–9383. doi: 10.1073/pnas.1121552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J 17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suhara W, Yoneyama M, Iwamura T, Yoshimura S, Tamura K, Namiki H, Aimoto S, Fujita T. 2000. Analyses of virus-induced homomeric and heteromeric protein associations between IRF-3 and coactivator CBP/p300. J Biochem 128:301–307. doi: 10.1093/oxfordjournals.jbchem.a022753. [DOI] [PubMed] [Google Scholar]
- 13.Mori M, Yoneyama M, Ito T, Takahashi K, Inagaki F, Fujita T. 2004. Identification of Ser-386 of interferon regulatory factor 3 as critical target for inducible phosphorylation that determines activation. J Biol Chem 279:9698–9702. doi: 10.1074/jbc.M310616200. [DOI] [PubMed] [Google Scholar]
- 14.Takahasi K, Horiuchi M, Fujii K, Nakamura S, Noda NN, Yoneyama M, Fujita T, Inagaki F. 2010. Ser386 phosphorylation of transcription factor IRF-3 induces dimerization and association with CBP/p300 without overall conformational change. Genes Cells 15:901–910. doi: 10.1111/j.1365-2443.2010.01427.x. [DOI] [PubMed] [Google Scholar]
- 15.Karin M, Cao Y, Greten FR, Li ZW. 2002. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer 2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 16.Li Q, Verma IM. 2002. NF-kappaB regulation in the immune system. Nat Rev Immunol 2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto M, Sato S, Mori K, Hoshino K, Takeuchi O, Takeda K, Akira S. 2002. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J Immunol 169:6668–6672. doi: 10.4049/jimmunol.169.12.6668. [DOI] [PubMed] [Google Scholar]
- 18.Cusson-Hermance N, Khurana S, Lee TH, Fitzgerald KA, Kelliher MA. 2005. Rip1 mediates the Trif-dependent toll-like receptor 3- and 4-induced NF-κB activation but does not contribute to interferon regulatory factor 3 activation. J Biol Chem 280:36560–36566. doi: 10.1074/jbc.M506831200. [DOI] [PubMed] [Google Scholar]
- 19.Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M, Tschopp J. 2004. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat Immunol 5:503–507. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- 20.Han KJ, Su X, Xu LG, Bin LH, Zhang J, Shu HB. 2004. Mechanisms of the TRIF-induced interferon-stimulated response element and NF-kappaB activation and apoptosis pathways. J Biol Chem 279:15652–15661. doi: 10.1074/jbc.M311629200. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser WJ, Offermann MK. 2005. Apoptosis induced by the toll-like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. J Immunol 174:4942–4952. doi: 10.4049/jimmunol.174.8.4942. [DOI] [PubMed] [Google Scholar]
- 22.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 23.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 24.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 25.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 26.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 27.Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 28.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishikawa H, Ma Z, Barber GN. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, Chen ZJ. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 32.Kinoshita E, Kinoshita-Kikuta E, Koike T. 2009. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat Protoc 4:1513–1521. doi: 10.1038/nprot.2009.154. [DOI] [PubMed] [Google Scholar]
- 33.Takahasi K, Suzuki NN, Horiuchi M, Mori M, Suhara W, Okabe Y, Fukuhara Y, Terasawa H, Akira S, Fujita T, Inagaki F. 2003. X-ray crystal structure of IRF-3 and its functional implications. Nat Struct Biol 10:922–927. doi: 10.1038/nsb1001. [DOI] [PubMed] [Google Scholar]
- 34.Clark K, Peggie M, Plater L, Sorcek RJ, Young ER, Madwed JB, Hough J, McIver EG, Cohen P. 2011. Novel cross-talk within the IKK family controls innate immunity. Biochem J 434:93–104. doi: 10.1042/BJ20101701. [DOI] [PubMed] [Google Scholar]
- 35.Horiuchi M, Takahasi K, Kobashigawa Y, Ochiai M, Inagaki F. 2012. A low-cost affinity purification system using β-1,3-glucan recognition protein and curdlan beads. Protein Eng Des Sel 25:405–413. doi: 10.1093/protein/gzs028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]